Abstract
SIN3/HDAC is a multi-protein complex that acts as a regulatory unit and functions as a co-repressor/co-activator and a general transcription factor. SIN3 acts as a scaffold in the complex, binding directly to HDAC1/2 and other proteins and plays crucial roles in regulating apoptosis, differentiation, cell proliferation, development, and cell cycle. However, its exact mechanism of action remains elusive. Using the Caenorhabditis elegans (C. elegans) model, we can surpass the challenges posed by the functional redundancy of SIN3 isoforms. In this regard, we have previously demonstrated the role of SIN-3 in uncoupling autophagy and longevity in C. elegans. In order to understand the mechanism of action of SIN3 in these processes, we carried out a comparative analysis of the SIN3 protein interactome from model organisms of different phyla. We identified conserved, expanded, and contracted gene classes. The C. elegans SIN-3 interactome -revealed the presence of well-known proteins, such as DAF-16, SIR-2.1, SGK-1, and AKT-1/2, involved in autophagy, apoptosis, and longevity. Overall, our analyses propose potential mechanisms by which SIN3 participates in multiple biological processes and their conservation across species and identifies candidate genes for further experimental analysis.
Subject terms: Regulatory networks, Systems analysis
Introduction
Cell death and aging are known to play a crucial role in development, health and disease. Biological processes such as cell death and aging are tightly controlled by gene expression, mediated by complex interactions between chromatin, epigenetic modifiers, and transcription regulatory proteins (TRPs). TRPs such as the SIN3 protein are associated with DNA-binding transcription factors in addition to different co-activator and co-repressor complexes. These protein–protein interactions function as writers, erasers, readers and modifiers of the chromatin design for the functioning of a cell1,2. SIN3 protein is a transcriptional regulator that functions as the central scaffold unit of the multi-protein SIN3/HDAC co-repressor complex3. This SIN3 core complex and its interaction partners play a significant role in several critical pathways such as autophagy, apoptosis and longevity.
A typical SIN3 protein harbours several paired amphipathic α-helix (PAH) domains, a prominent HDAC interacting domain (HID), and a highly conserved region (HCR)4. These evolutionarily conserved domains enable the protein to control transcription5,6. Though SIN3 has multiple roles in apoptosis, differentiation, cellular proliferation, development, cell cycle, cancer and aging7–10, the exact mechanism of regulation mediated by the protein is unknown.
The functional diversity of SIN3 can be facilitated by the diversity in its interactome. However, the mechanistic studies are impeded by the presence of multiple isoforms of SIN3 in mammals and their functional redundancy. The presence of SIN-3 and the absence of isoforms of SIN3 in C. elegans makes it a suitable model for understanding the functional diversity of SIN3.
In C. elegans, SIN-3 protein plays a vital role in male sensory organ development11, muscle integrity, motility, and longevity. It also controls several physiological parameters such as stress tolerance, protein homeostasis, muscle and mitochondrial functioning, cuticle integrity, accumulation of age-associated pigments, fecundity and fertility12. Remarkably, the protein modulates autophagy and lifespan in an unconventional way. Usually, an increase in autophagy leads to lifespan extension. However, reactive oxygen species (ROS) and intracellular oxidative stress in sin-3 deletion mutants are associated with uncoupling of autophagy and longevity such that increased autophagy leads to a shorter lifespan13. In order to understand the factors involved in SIN3 regulated autophagy and longevity, we carried out an analysis of the interactome of SIN3 in different model organisms from different phyla. Further analysis of gene ontology and the expansion/contraction of the gene classes led to the identification of conserved factors important for these functions that can serve as candidate genes for further analysis.
Methods
Software and databases
NCBI Protein database, Clustal Omega web server, MAFFT, MEGA-X v10, NCBI Conserved domain database, G-BLOCKS online web server v0.91b, GeneMANIA, DAVID Bioinformatics Resource v6.8, NCBI Conserved domain database, Ensembl genome, Orthofinder (version 2.3.12), RStudio v2021.09.0+351.pro6 (https://www.rstudio.com/)14, Microsoft Excel 2013 (https://www.microsoft.com/)15.
Retrieval of sequences and their analysis
The FASTA sequences of SIN3 homologs were downloaded from the NCBI Protein database16 and submitted to the Clustal Omega web server (http://www.ebi.ac.uk/Tools/msa/clustalo/) for proper sequence alignment and domain comparison17–19. The percent identity matrix was obtained for all the SIN3 homologs. Similarly, the percent identity matrix for SIN3 protein domains, such as PAH, SIN3A_C, and HDAC interaction domain, was also prepared for domain-specific comparison.
Phylogenetic analysis
The FASTA sequences of SIN3 homologs were downloaded and aligned in the MAFFT (L-INS-I method) online web server v720. L-INS-i is one of the most accurate multiple sequence alignments, particularly suitable to align 10–100 protein sequences. The amino acid alignment was curated in the G-BLOCKS online web server v0.91b21 with default parameters (except minimum block length set as 5 and allowed gap positions set as half). Phylogenetic tree was constructed through Maximum likelihood (ML) using MEGA-X v1022. Statistical significance was increased by bootstrapping with 1000 replicates23. The phylogenetic tree for SIN3 protein domains, PAH, SIN3A_C and HID, was obtained in the same manner.
Domain search
The FASTA sequences of SIN3 homologs were submitted to the NCBI Conserved domain database24 and analyzed for the different functional domains present in the SIN3 proteins.
Protein–protein interaction network
GeneMANIA (http://Genemania.org) was used to examine the various physical interactions of SIN3 homologs with a limitation on the maximum resultant genes, set to 10025. After selecting the specific organism under study, the official gene symbol of the protein of interest was submitted to the database. All default network attributes were de-selected, and only physical interactions were selected for generating the final SIN3 protein interactome.
Gene ontology analysis
DAVID (the information base for Annotation, Visualization, and Integrated Discovery) Bioinformatics Resource v6.8 was used for analyzing the physical protein interactors of C. elegans SIN-326–28. First, the gene list was uploaded to the database, and the saved list was then used as the input for the Functional Annotation tool. Finally, the protein list was separately used to extract and summarize the KEGG pathways29, InterPro domains30, and Gene ontology31 of the SIN-3 protein interactors using the different parameters.
Orthology
For evaluating the orthology of proteins of interest, the predicted proteomes of C. elegans (WBcel235), S. cerevisiae (R64-1-1), D. melanogaster (BDGP6.32), D. rerio (GRCz11), H. sapiens (GRCh38.p13), and M. musculus (GRCm39) from the Ensembl genome (ensembl.org) were used. Orthofinder (version 2.3.12) was run with default parameters to find orthogroups among the whole deduced proteomes of all six organisms listed above32. If there are multiple predicted proteins/transcripts for a gene, the primary transcript of the predicted protein was chosen for analysis.
Results and discussion
SIN3 is well-conserved across phyla due to the significant similarities in the functional domains of the protein. These domains and other protein motifs enable SIN3 to participate in multiple protein–protein interactions, hence regulating multiple pathways. The analysis we have carried out identifies candidate protein–protein interactions of SIN3 involved in the eukaryotic regulation of crucial biological processes like apoptosis, autophagy and longevity.
ceSIN3 shares sequence identity with homologs
One of the main approaches to understanding SIN3 function is the study of protein conservation determined via sequence homology between C. elegans SIN-3 protein (ceSIN3) and SIN3 isoforms present in other model organisms. Previous studies have reported that ceSIN3 has homology with human and mouse SIN311,13. Therefore, we carried out phylogenomic analyses of the SIN-3 protein of C. elegans and five other model organisms, namely, S. cerevisiae, D. melanogaster, D. rerio, H. sapiens, and M. musculus, to deduce the level of protein homology in the context of the functional domains. ceSIN3 is an orthologous member of the SIN3A phylogenetic family (Fig. 1A). Traditionally, C. elegans shares a close evolutionary relationship with Drosophila, followed by the yeast33. This is reflected in the phylogenetic tree of the SIN3 protein, which shows that SIN3 homologs of C. elegans, Drosophila, and yeast form a separate cluster within the SIN3A family, indicating high relatedness in the model organisms with lower complexity.
Figure 1.
SIN3 phylogeny based on alignments of the amino-acid sequences of Caenorhabditis elegans SIN-3 protein and its domains. Phylogenetic tree based on (A) SIN3 protein, (B) SIN3 HID domain, (C) SIN3 SIN3A_C domain, (D) SIN3 PAH domain, and (E) SIN3 PAH1 domain. The PAH1 domain was analysed separately due to its high sequence identity with PAH domain of C. elegans SIN-3 protein. The Phylogenetic tree was constructed using the Maximum Likelihood method of phylogenetic tree construction, with 1000 bootstrap replicates, using MEGA-X. The number mentioned at each node is the bootstrap percentage for a particular branch (Cele, Caenorhabditis elegans; Hsap, Homo sapiens; Mmus, Mus musculus; Drer, Danio rerio; Dmel, Drosophila melanogaster; Scer, Saccharomyces cerevisiae).
To determine the quantitative similarity, a percent identity (PID) analysis was carried out using ClustalW sequence alignment. ceSIN3 protein has a significant identity, ~ 21%, with all SIN3 isoforms present in different organisms that we analysed (Table 1, Fig. S1). It is generally believed that two sequences are homologous only if there is more than 30% identity at the protein sequence level34. However, the sequence length determines the possibility of correct alignment, and if proteins with large sequence length exhibit ≥ 20% identity, they are considered significant even in the twilight zone of evolutionary relatedness (20–30%)35. Therefore, the current study demonstrates that ceSIN3 protein shares homology with human and mice SIN3. It also shows significant similarity with the SIN3 isoforms of S. cerevisiae, D. melanogaster, and D. rerio.
Table 1.
Percentage sequence identity of C. elegans SIN-3 protein with the SIN3 isoforms in other model organisms (Hsap, Homo sapiens; Mmus, Mus musculus; Drer, Danio rerio; Dmel, Drosophila melanogaster; Scer, Saccharomyces cerevisiae).
| Scer Sin3 | Dmel Sin3A | Hsap SIN3A | Hsap SIN3B | Mmus SIN3A | Mmus SIN3B | Drer Sin3aa | Drer Sin3ab | Drer Sin3b | |
|---|---|---|---|---|---|---|---|---|---|
| C. elegans SIN-3 | 20.33 | 21.62 | 22.2 | 22.14 | 22 | 22.69 | 21.53 | 21.14 | 22.38 |
However, the multiple sequence alignment indicates that the sequence identity of ceSIN3 with the rest of the proteins is restricted to specific regions. While the PAH and HID domains contain the well-aligned regions of the protein, it was also seen that the C. elegans protein has large stretches of unaligned regions that seem to be only partially conserved in SIN3 proteins of yeast and Drosophila (Fig. S2). The close phylogenetic relationship among SIN3 homologs of these organisms could arise from the conservation of these specific regions. Interestingly, the nematode protein shares a closer relationship with SIN3A isoforms than SIN3B even though it shares the same sequence identity with both the isoforms (Fig. 1A). This might be because SIN3A and SIN3B are paralogs that evolved out of gene duplication. While SIN3A retained its similarity to the ancestral lineage, SIN3B underwent substantial diversification since its origin.
SIN3 homology for PAH and HID
SIN3 protein has multiple protein partners that interact through several structural and functional domains. In order to understand the nature of conservation in ceSIN3 protein, these unique domains were also analyzed for their sequence homology.
Earlier studies on SIN3 by Chaubal and Pile36 indicated the presence of PAH, HID and SIN3A_C domains in the isoforms of C. elegans, Drosophila, zebrafish and mouse. Our analysis supports this finding and further discloses that CeSIN3 protein is the only SIN3 homolog to harbor a single PAH domain. This domain has homology with all the three PAH domains of SIN3 isoforms. The CeSIN3 protein also contains one HDAC interaction domain and one SIN3A_C domain (Table 2).
Table 2.
Conservation of SIN-3 protein domains of C. elegans with those of other organisms expressed in percent similarity. Scer (Saccharomyces cerevisiae), Dmel (Drosophila melanogaster), Mmus (Mus musculus), Hsap (Homo sapiens), Drer (Danio rerio). Caenorhabditis elegans SIN-3 protein consists of a single PAH domain.
| Domain | Scer | Dmel | Mmus | Hsap | Drer | ||||
|---|---|---|---|---|---|---|---|---|---|
| Sin3 | Sin3A | SIN3A | SIN3B | SIN3A | SIN3B | Sin3aa | Sin3ab | Sin3b | |
| PAH1 | 55.56 | 48.89 | 53.33 | 51.11 | 53.33 | 51.11 | 53.33 | 53.33 | 54.55 |
| PAH2 | 31.11 | 33.33 | 31.11 | 31.11 | 31.11 | 31.11 | 29.55 | 33.33 | 31.11 |
| PAH3 | 27.91 | 26.67 | 28.89 | 28.89 | 28.89 | 28.89 | 28.89 | 28.89 | 28.89 |
| HID | 38.14 | 39.18 | 39.58 | 39.58 | 39.58 | 39.39 | 40.62 | 40.62 | 40.62 |
| SIN3A_C | 19.08 | 18.5 | 20.77 | 19.67 | 20.77 | 18.58 | 18.03 | 18.03 | 19.57 |
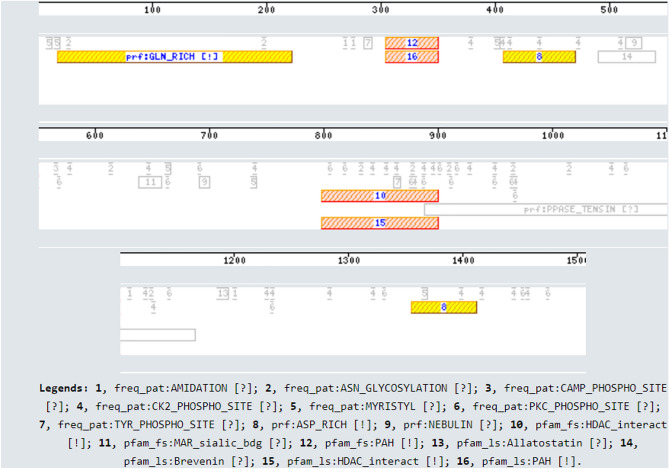
We found unique motifs, a large stretch of Glutamine rich repeats, and two interspersed regions of Aspartate rich repeats in the ceSIN3 protein (Fig. 3). These amino acid repeats seem to coincide with the unaligned regions of ceSIN3 (Table S2). These repeats are absent in other SIN3 homologs except yeast Sin3 protein, which contains a big stretch of Glutamine rich repeats. While excessive repeat expansion is pathogenic, Glutamine-rich repeats and Aspartate or Glutamate repeat with regular lengths help in gene regulation and protein–protein interaction37,38. Hence, ceSIN3 protein could participate in specific protein interactions involved in biological processes unique to the organism.
Figure 3.
Schematic representation of motifs in Caenorhabditis elegans SIN-3 protein. Motif Scan105 was used for finding all the known motifs that occur in the protein sequence of SIN-3 protein using HAMAP profiles [hamap], PROSITE patterns [pat], PROSITE patterns (frequent match producers) [freq_pat], Pfam HMMs (global models) [pfam_ls], Pfam HMMs (local models) [pfam_fs], PROSITE profiles [prf], More profiles [pre]. The legends at the bottom of the figure are provided for the key to the numbering of the protein domains/motifs. [!] represents a strong match (low likelihood of a false positive) while [?] represents a questionable or weak match (additional biological evidences are required for true/false negative status).
Proteins and transcription factors binding to the PAH domain in yeast or mammalian systems might also bind to C. elegans SIN-3 PAH (cePAH). The current study reveals that the PAH domain of ceSIN3 has ≥ 28% identity with all the three PAH domains of SIN3 isoforms. The cePAH domain is remarkably similar to the PAH1 (≥ 50%) domain of the other SIN3 homologs, especially the yeast Sin3 PAH1 (Table 2, Fig. S2). Surprisingly, despite the sequence homology of the cePAH domain, the protein domain is phylogenetically distant from all the three PAH domains, forming an out-group in the cluster containing PAH1 domains (Fig. 1D). But when the domain is studied exclusively in relation to PAH1 domains, it shows a close phylogenetic relationship with yeast Sin3 PAH1 domain (Fig. 1E).
Similarly, C. elegans HDAC interaction domain shows 38.14–40.62% sequence identity with all SIN3 homologs HDAC domains, principally the SIN3 isoforms in zebrafish (Table 2). However, it shares the closest phylogenetic relationship with the yeast Sin3 protein (Fig. 1B). In addition, C. elegans HDAC interaction domain has almost perfect sequence alignment with its homologs (Fig. S2). Due to this alignment, a good percentage of identity exists between the HDAC interaction domain of ceSIN3 protein and homologs. Unlike HID and PAH domains, the percent identity matrix and poor sequence alignment of the evolutionarily well-conserved SIN3A_C domain hint at a low sequence identity between the nematode protein and the other SIN3 homologs (Table 2, Fig. S2). This also accounts for the phylogenetic relationship between SIN3A_C domains of the worm and yeast (Fig. 1C). Overall, the conservation in CeSIN3 protein seems to be localized in the PAH and HID domains.
Orthologs in C. elegans genome
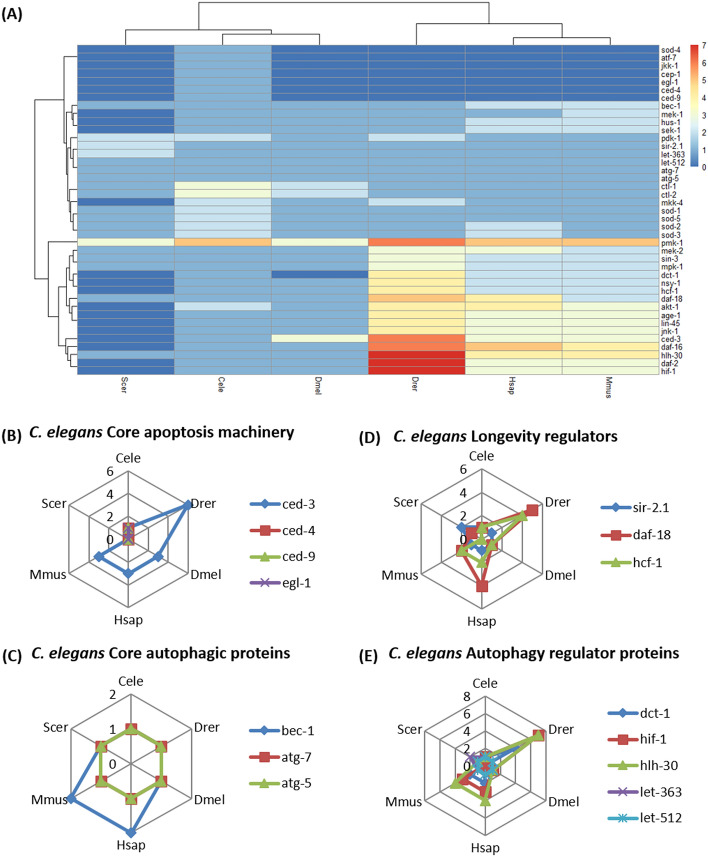
The gain or loss of gene function or its regulation contributes to the adaptation and survival of organisms. As the study focuses on understanding SIN3 mediated regulation of apoptosis, autophagy and longevity, the orthology of the major nematode proteins associated with these crucial biological processes was examined. The data derived from 18,002 comparative analyses of proteins using OrthoFinder, that are over-represented or under-represented in the proteome of C. elegans, Yeast, Drosophila, Zebrafish, Human, and Mouse was used to construct orthogroups (Fig. 2). Of the 41 proteins investigated, we identified 34 different orthogroups having conservation, expansion, and contraction and 7 single copy orthologue sequences (Fig. 2, Fig. S3).
Figure 2.
Abundance of proteins involved in apoptosis, autophagy and longevity in C. elegans. (A) Heatmap depicting the orthology of 41 proteins derived from 18,002 comparative analyses of gene classes using OrthoFinder, in the proteome of Caenorhabditis elegans (Cele), Danio rerio (Drer), Drosophila melanogaster (Dmel), Homo sapiens (Hsap), Mus musculus (Mmus), and Saccharomyces cerevisiae (Scer). Color scale key is depicted at the side. The spider plots further indicate the number of proteins in the different species; C. elegans specific pathways like core apoptosis machinery (B), longevity regulators (C), core autophagic proteins (D) and autophagic regulator proteins) (E) are indicated at the top of each spider plot.
Protein orthogroups involved in signaling pathways are well conserved in higher eukaryotes. While the number of proteins attributed to the different orthogroups is similar for the invertebrates, C. elegans and Drosophila, proteins involved in important pathways such as the ERK-MAPK pathway (LIN-45, MEK-2, MPK-1), p38-MAPK pathway (NSY-1, PMK-1), and insulin signaling pathway (DAF-2, DAF-16, AGE-1, AKT-1) are conserved across phyla and expanded in proteomes of three or more species. These pathway proteins are crucial for cellular processes preceding apoptosis and longevity, and are expanded in the eukaryotic system. The ERK-MAPK pathway is the primary signaling network involved in regulating cell division, growth, development, apoptosis39 and tumor formation, and the p38-MAPK pathway works in response to stress to regulate cellular functions such as differentiation, apoptosis, and senescence40. The insulin/IGF signaling (IIS) pathway is involved in stress resistance and longevity along with nutrient regulation and growth41. Interestingly, orthogroups of signaling proteins (MKK-4, PDK-1, AKT-1 AND PMK-1) are also expanded in the nematode proteome, revealing the need for increased signaling proteins even in lower eukaryotes.
Orthogroups of nematode proteins that show poor conservation in other model organisms are mainly associated with the main apoptotic pathway. While the main cell death protein CED-3 (caspase 2/3/6 homolog) protein is expanded in higher eukaryotes, rest of the apoptosis proteins (CED-4, CED-9, EGL-1, CEP-1) are contracted in the species included in the study. This is likely because of the functional redundancy of the proteins associated with essential functions. For example, CEP-1, a functional p53 homolog, has very low sequence identity with p53 isoforms of different model organisms42.
Most of the orthogroups do not have any yeast protein as the unicellular organism lacks homologs for most eukaryotic proteins involved in apoptosis, autophagy, and longevity. Yeast does not have distinct cell death pathways similar to mammalian ones43; even the MAPK cascade components are only conserved in function44 but not in terms of sequence identity. Certain zebrafish proteins are over-represented due to genome duplication, which precedes the teleost evolution. As a result, many mammalian genes have more than one ortholog in the zebrafish genome45,46.
Protein–protein interactions in the SIN3 network
Protein–protein interactions are crucial for the basic functionality of cells (40). The detailed study of the protein interactome holds the key to identifying biological pathways and predicting protein functions47–50. Therefore, identifying the interaction repertoire of transcriptional regulator SIN-3 will give a better understanding of the regulation of various biological processes in C. elegans51.
The SIN3 interactome is well studied in yeast, human, and mouse52,53. Therefore, the protein–protein interactions of these eukaryotic systems can be extrapolated to ceSIN3 ortholog with computational methods54. GeneMANIA was used to obtain protein–protein interactions of SIN3 in different eukaryotic systems. The database extracted 100 physical interactions, represented in the form of a network image (Fig. S4), of the SIN3 protein for each species included in the study. In the nematode SIN-3 interactome, we found several protein interactions conserved across evolution (Fig. 3).
21 proteins out of the 100 C. elegans SIN-3 interactors were found to be conserved in one or more SIN3 interactomes of other model organisms (Table 3). While the number of conserved protein interactions seems low, this could be due to the limitations of studying only 100 protein–protein interactions within the database used to generate the protein interactome and ambiguity in the available data. These interactors are involved in essential biological processes like transcriptional regulation, signal transduction, development and lifespan regulation55. Notably, ceSIN3 protein also participates in many unique protein interactions (Table S1). Some of these interactions might be mediated by Glutamine-rich and Aspartate repeats present outside the protein's functional domains (PAH and HID) and via similar repeats occurring in SIN-3 interacting nematode proteins.
Table 3.
Caenorhabditis elegans SIN-3 protein interactors conserved in SIN3 interactome of other model organisms (Cele, Caenorhabditis elegans Hsap, Homo sapiens; Mmus, Mus musculus; Drer, Danio rerio; Dmel, Drosophila melanogaster; Scer, Saccharomyces cerevisiae).
| S. no. | Cele | Drer | Dmel | Hsap | Mmus | Scer |
|---|---|---|---|---|---|---|
| 1 | AKT-1, AKT-2, SGK-1 | Sch9 | ||||
| 2 | CEH-27 | Nkx2.2a | NKX2-2 | |||
| 3 | CEH-28 | NKX3-2 | ||||
| 4 | CEH-62 | Bcd | ||||
| 5 | DCP-66 | Gatad2ab | Simj | GATAD2A | ||
| 6 | FOS-1 | Sko1 | ||||
| 7 | HCF-1 | Hcfc1b | Hcf | HCFC1 | ||
| 8 | HLH-3 | Tal1 | TAL1 | |||
| 9 | JMJD-3.2 | Lid | KDM5A | KDM5B | Cyc8 | |
| 10 | KLF-2 | Sp3a | KLF9, KLF16 | SP3, KLF1 | Swi5 | |
| 11 | MED-2 | GATA4 | ||||
| 12 | MXL-3 | Mntb, Mxd1, Mxi1 | MNT, MXD1, MXI1, MAX, MXD3 | MNT, MXD1, MXD4, MXI1, MAX | ||
| 13 | NHR-23 | NR1D2 | THRAB | |||
| 14 | SIN-3 | Sin3ab, Sin3b, Sin3aa | Sin3A | SIN3A, SIN3B | SIN3A, SIN3B | Sin3 |
| 15 | SOX-3 | Sox2 | SOX2 | SOX2 | ||
| 16 | TAB-1 | NANOG | ||||
| 17 | TAG-97 | ETV6 | ||||
| 18 | UNC-130 | Fkh2, Fkh1 | ||||
| 19 | ZTF-14 | Gli1 |
Missing data is a major problem in large-scale profiling experiments, and their adverse effect on the downstream analysis is beyond the capacity of simple computational methods56. To find reliable protein–protein interactions of ceSIN3, interactions with experimental evidence should be analyzed along with the GeneMANIA dataset57. Therefore, ceSIN3 protein–protein interactions were manually curated from published data and represented in a network constructed using Cytoscape (Fig. S5).
Orthology of the nematode proteins in the SIN-3 interactome, obtained via GeneMANIA and literature based evidence method, was checked in other model organisms included in the analysis using the Orthofinder tool. It was found that many of the SIN-3 interactors found in C. elegans were not conserved in other organisms (Fig. S6). This contraction in protein orthogroups could be due to the lower sequence identity of nematode proteins with higher eukaryotic proteins. Further, SIN3 might be involved in multiple transient interactions to be able to regulate several pathways at a time. This poses a challenge in the in-vitro characterization of SIN3 protein–protein interactions. Some C. elegans proteins involved in the SIN-3 interaction network are quite expanded in the nematode proteome. These proteins (HDA-1, NHR-67, MAB-3, ODD-2, AKT-1, AKT-2, EGL-38, ELT-7, LIN-15A, FKH-6, FOZI-1 etc) are involved in important processes like sex differentiation, multicellular development, cell differentiation and cell signaling pathways, thereby enabling SIN-3 mediated regulation via protein–protein interaction.
It has been well-established that both SIN3 isoforms, SIN3A and SIN3B, form distinct protein complexes. Phylogenetic analysis has revealed that ceSIN3 shares a closer relationship with SIN3A isoforms. Therefore, it was imperative to evaluate the nematode protein for conservation of SIN3A/SIN3B specific protein interactions in model organisms which harbor multiple isoforms of the SIN3 protein. While it was observed that very few SIN3A/SIN3B specific interactions are conserved in ceSIN3, no particular preference for SIN3A specific interactions is seen in humans, mice, or zebrafish. This might be due to the similarity in the protein sequence identity of the ceSIN3 protein with SIN3A and SIN3B isoforms.
Context-specific protein annotation
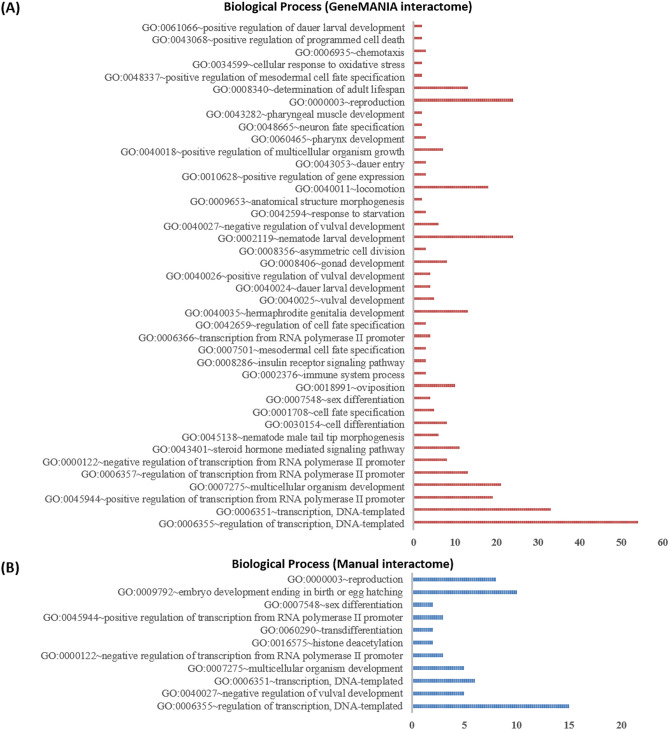
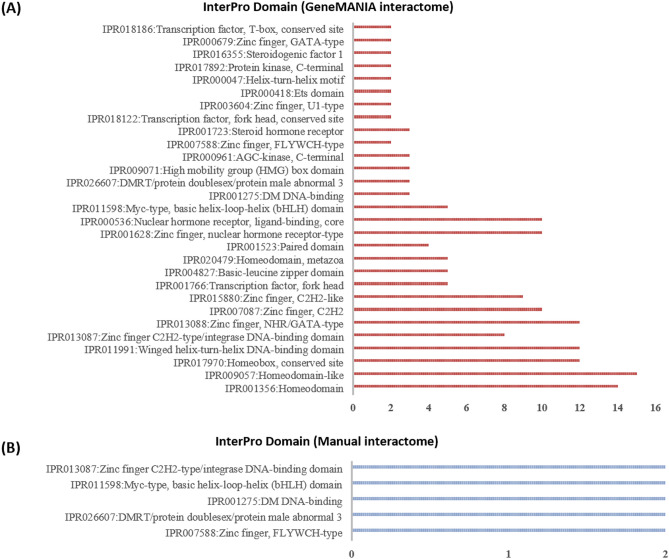
A protein can have multiple functions depending on the tissue/cellular context in which the protein is present. Some of the functions assigned to a protein are more relevant in certain conditions, such as programmed cell death, determination of adult lifespan and cellular response to oxidative stress. In this study, we investigated protein–protein functional associations by using GO terms, InterPro domains and KEGG pathways29,58 to identify the participation of SIN-3 protein interactors in apoptosis, autophagy and longevity. GO ‘biological process’ classification of ceSIN3 protein interactome reveals that 53% interactors are associated with the regulation of transcription, the majority being transcription factors (Fig. 4). Furthermore, when annotated with InterPro domains, most of the interactor proteins indicated the presence of DNA binding domains, homeobox domain, zinc finger domain and winged helix-turn-helix DNA-binding domain (Fig. 5) that assist in transcriptional regulation59,60. Previous studies also supported this by establishing SIN3 interaction with many transcriptional factors3,9.
Figure 4.
Biological processes associated with protein interactome of C. elegans SIN-3 protein, obtained from (A) GeneMANIA database and (B) Direct Evidence method using the NIH LHRI DAVID 6.8 service. In order to find reliable protein–protein interactions, experimental evidence was analyzed along with the GeneMANIA dataset via literature based evidence method. The y-axis represents the GO biological processes, while the x-axis represents the number of proteins associated with a particular biological process.
Figure 5.
InterPro domain analysis of protein interactome of C. elegans SIN-3 protein, obtained from (A) GeneMANIA database and (B) direct evidence method using the NIH LHRI DAVID 6.8 service. The y-axis represents the InterPro domains, while the x-axis represents the number of proteins associated with a particular domain.
GO term analysis revealed that some ceSIN3 protein interactors are involved in the positive regulation of programmed cell death, cellular response to oxidative stress and determination of adult lifespan in C. elegans (Table 4). HLH-3 and CES-2 are involved in the positive regulation of programmed cell death, while DAF-16 and BAR-1 are involved in the cellular response to oxidative stress. Further, 12% of SIN-3 interactors are involved in the determination of adult lifespan. Among these SIN-3 interactors, BAR-1, DAF-12, JUN-1, and PHA-4 undergo a significant transcriptional up regulation in case of sin-3 deletion (q value < 0.05, log2 Fold Change > 1)61. The rest of the proteins are also modulated upon sin-3 deletion but at a lower statistical significance (q value > 0.05).
Table 4.
C. elegans SIN-3 protein interactors involved in apoptosis, autophagy and longevity.
| S. no. | Name of protein | Function | References |
|---|---|---|---|
| 1 | HLH-3 | Induction of programmed cell death, regulation of axon extension involved in axon guidance; regulation of oviposition | 62,63 |
| 2 | CES-2 | Promotes programmed cell death and autophagy | 63–65 |
| 3 | DAF-16 | Supports resistance to oxidative stress and longevity, apoptosis, defense response to other organism; regulation of dauer larval development; regulation of primary metabolic process | 66–72 |
| 4 | BAR-1 | Life-span regulation, multicellular organism development; reproductive behavior; signal transduction | 73,74 |
| 5 | AKT-1 | Induction of DNA damage-dependent apoptosis, inhibition of apoptosis, cellular protein modification process; determination of adult lifespan; signal transduction | 75–78 |
| 6 | AKT-2 | Induction of DNA damage-dependent apoptosis, inhibition of apoptosis, cellular protein modification process; determination of adult lifespan; insulin receptor signaling pathway | 75–77 |
| 7 | SIR-2.1 | Chromosome organization, determination of adult lifespan, induction of p53/cep-1 independent apoptosis, and autophagy | 79–82 |
| 8 | SGK-1 | Determination of adult lifespan, mesendoderm development; peptidyl-serine phosphorylation; regulation of protein localization to basolateral plasma membrane | 83–86 |
| 9 | RLE-1 | Determination of adult lifespan, response to heat; and ubiquitin-dependent protein catabolic process | 87,88 |
| 10 | FOS-1 | Cell invasion, lifespan regulation | 89,90 |
| 11 | SET-1 | Embryo development, histone methylation, apoptosis lifespan regulation | 91–94 |
| 12 | PHA-4 | Multicellular organism development; determination of adult lifespan; positive regulation of cellular metabolic process; response to caloric restriction | 68,95,96 |
| 13 | HCF-1 | Dauer exit; determination of adult lifespan | 97 |
| 14 | DAF-12 | Determination of adult lifespan; heat acclimation; positive regulation of dauer larval development | 98–101 |
| 15 | JUN-1 | Determination of adult lifespan; response to starvation | 102 |
Notable KEGG pathways corresponding to cell death and longevity were associated with some of these ceSIN3 interactors. The FOXO (controls starvation-induced autophagy along with Foxk1/2 and SIN3 proteins), MAPK and Jak-STAT (Fig. 6) pathways involve DAF-16, SIR-2.1, AKT-1/2, SGK-1, MXL-3 from the SIN-3 interactome.
Figure 6.
KEGG pathway analysis of protein interactome of C. elegans SIN-3 protein, obtained from (A) GeneMANIA database and (B) direct evidence method using the NIH LHRI DAVID 6.8 service. The y-axis represents the KEGG Pathways, while the x-axis represents the number of proteins associated with a particular KEGG Pathway.
PAH domain interaction motif in nematode protein interactors
Motifs are used as scaffolds by proteins and transcription factors interacting with SIN3 PAH domains. LxxLL (x indicates any amino acid) motif is an acidic leucine (L)–rich protein binding motif used by proteins to interact with the PAH1 domain of SIN3 protein103. SIN3 interactors like SAP25 and MAD1 interact with the PAH1 domain of SIN3 protein via this unique sequence104. Such motifs are likely to be conserved in the case of ceSIN3 PAH domain protein interactions. This unique motif was checked for its presence in the SIN-3 interactor protein sequences. It was found that AKT-2, AKT-1, SIR-2.1, CES-2, and BAR-1, which are associated with apoptosis, autophagy, and longevity, harbor one or more LxxLL-type motifs (Table 5). Since the ceSIN3 PAH domain is primarily similar to the PAH1 domain of other SIN3 homologs, C. elegans proteins might interact with the SIN-3 protein via the LxxLL motif. In addition, it was seen that six nematode proteins (DCP-66, FOZI-1, TAF-12, GEI-13, SMA-9, CEH-36) interacting with ceSIN3 protein contain a stretch of Glutamine-rich repeats or Aspartate repeats (Table S3).
Table 5.
Caenorhabditis elegans SIN-3 protein interactors containing LxxLL motifs. LxxLL motif is an acidic leucine (L)–rich protein binding motif used by proteins to interact with the PAH1 domain of SIN3 protein. The protein sequences were obtained from NCBI protein and Uniprot databases, and the motifs were scanned using the ScanProsite106 tool.
| Gene name | Accession no. | No. of LxxLL motifs | Name of the LXXLL motif |
|---|---|---|---|
| sin-3 | A5JYW9.1 | 2 | LgkLL, LmtLL |
| sir-2.1 | NP_001255484.1 | 1 | LsdLL |
| ceh-62 | Q09602.1 | 1 | LrlLL |
| f57c9.4 | NP_491459.1 | 1 | LpnLL |
| mab-3 | CAB16489.1 | 1 | LasLL |
| ztf-2 | CAA95789.1 | 1 | LasLL |
| wbgene00017431 | CCD69466.1 | 1 | LryLL |
| ets-5 | CCJ09391.1 | 1 | LleLL |
| nhr-43 | CDR32746.1 | 1 | LenLL |
| akt-1 | NP_001023646.1 | 2 | LenLL, LtgLL |
| ces-2 | CAB05032.1 | 1 | LrfLL |
| imb-3 | CCD67936.1 | 2 | LetLL, LvcLL |
| fkh-7 | CCD67108.1 | 1 | LskLL |
| nhr-67 | Q9XVV3.1 | 1 | LnfLL |
| lin-15a | Q27365.1 | 2 | LvaLL, LneLL |
| t04h1.2 | CAB01578.1 | 1 | LrtLL |
| nurf-1 | CAA0059157.1 | 4 | LvvLL, LyqLL, LleLL, LelLL |
| bar-1 | AAC17424.1 | 6 | LrdLL, LlmLL, LanLL, LksLL, LhkLL, LslLL |
| eyg-1 | NP_496116.1 | 1 | LslLL |
| akt-2 | CAA20936.1 | 2 | LenLL, LsgLL |
| gei-13 | CAA80171.2 | 2 | LaaLL, LnaLL |
| ztf-11 | CAB05737.2 | 1 | LmsLL |
Effect of mutation in protein interactors
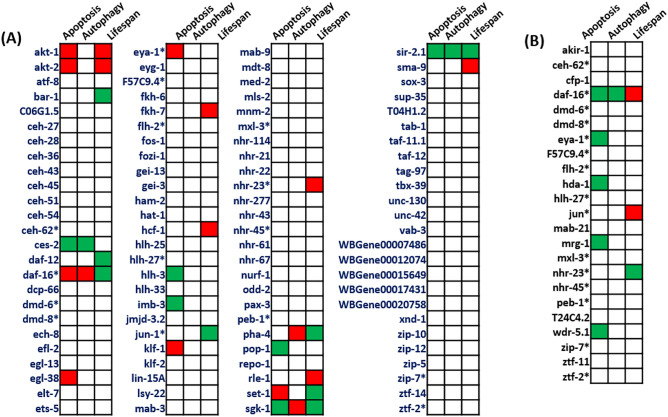
Consequential mutant phenotypes provide evidence-based annotations for a majority of proteins. Such an approach helps to investigate the presence of SIN3 mutant-like phenotypes. Therefore, the mutant phenotypes associated with apoptosis, autophagy, and longevity were curated for all the proteins in the ceSIN3 interactome (Fig. 7). We used extensive text mining to identify such phenotypes. We further wanted to check whether phenotypes associated with mutants of the interactor proteins were phenocopies known for the sin-3 mutants. Mutant phenotypes of multiple proteins show sin-3 mutant-like phenotypes with respect to apoptosis, autophagy, and longevity. Remarkably, four mutants, pha-4 (ortholog of human FOXA1 and FOXA2), daf-16 (ortholog of human FOXO1; FOXO3; and FOXO4), sgk-1 (ortholog of human SGK2), and sir-2.1 mutant (ortholog of human SIRT1), show elevated autophagy, programmed cell death and shortened lifespan like that of sin-3 mutants. Other mutants such as akt-2, akt-1, ces-2, and bar-1 also show phenotypes related to apoptosis and longevity. These interactor proteins seem to have a well-established connection with the apoptosis-autophagy-longevity axis (Konwar et. al., unpublished results)13.
Figure 7.
Analysis of SIN-3 protein interactome of mutants of C. elegans obtained from (A) GeneMANIA database and (B) direct evidence method using WormBase107 phenotype option. Green indicates an increase, while red indicates a decrease in apoptosis/autophagy/ longevity of C. elegans mutants.
Conclusion
Our study identifies novel protein interactors of the SIN3 protein. The protein homology and phylogenetic relationship between the ceSIN3 and other eukaryotic SIN3 isoforms provides a basis for the genetic and functional correlation of the C. elegans protein with the other homologs. Protein–protein interactions of ceSIN3 protein revealed many interacting proteins associated with the regulation of programmed cell death, autophagy, and adult life span. Some of these interacting proteins, such as SIR-2.1, AKT-1/2, BAR-1, and CES-2, contain the SIN3 PAH domain interaction (LXXLL) motif, while others, like SMA-9, FOZI-1, and CEH-36, harbor Glutamine rich repeats known to be involved in protein–protein interactions. A few of these protein interactors also display mutant phenotypes similar to that of sin-3 mutants, indicating their potential in regulating apoptosis, autophagy, and longevity. However, experimental evidence will provide better insight. It is important to check the protein interactions of the ceSIN3 protein by performing a protein immunoprecipitation (IP) assay followed by Mass Spectroscopic (MS) analysis. This will confirm SIN3 protein interactions, which can be genetically studied for their role in apoptosis, autophagy, and longevity.
Supplementary Information
Acknowledgements
The authors thank the University of Delhi for providing the infrastructure. The support from Department of Biotechnology (DBT), Govt. of India for Bioinformatics Facility to D.S. (BT/PR40153/BTIS/137/8/2021(BIF) at Dr. B.R. Ambedkar Center for Biomedical Research is highly acknowledged. JRF Fellowship to C.K. from the University of Delhi and SRF fellowship to S.K. from ICMR is gratefully acknowledged.
Author contributions
C.K., J.M. and D.S. conceived and designed the study. C.K. performed the experiments. C.K., J.M., S.K., V.B. and D.S. analyzed the data. C.K. wrote the manuscript. All authors read, revised and approved the manuscript. V.M. and D.S. provided overall supervision throughout the study.
Data availability
All relevant data are within the manuscript and its Supporting Information file.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-13864-0.
References
- 1.Kuo M-H, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. BioEssays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 2.Wray GA, et al. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 3.Adams GE, Chandru A, Cowley SM. Co-repressor, co-activator and general transcription factor: The many faces of the Sin3 histone deacetylase (HDAC) complex. Biochem. J. 2018;475:3921–3932. doi: 10.1042/BCJ20170314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van Ingen H, Baltussen MAH, Aelen J, Vuister GW. Role of structural and dynamical plasticity in Sin3: The free PAH2 domain is a folded module in mSin3B. J. Mol. Biol. 2006;358:485–497. doi: 10.1016/j.jmb.2006.01.100. [DOI] [PubMed] [Google Scholar]
- 5.Pile LA, Spellman PT, Katzenberger RJ, Wassarman DA. The SIN3 deacetylase complex represses genes encoding mitochondrial proteins: Implications for the regulation of energy metabolism. J. Biol. Chem. 2003;278:37840–37848. doi: 10.1074/jbc.M305996200. [DOI] [PubMed] [Google Scholar]
- 6.Silverstein RA, Ekwall K. Sin3: A flexible regulator of global gene expression and genome stability. Curr. Genet. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- 7.Bansal N, David G, Farias E, Waxman S. Emerging roles of epigenetic regulator Sin3 in cancer. Adv. Cancer Res. 2016;130:113–135. doi: 10.1016/bs.acr.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 8.Barnes VL, Laity KA, Pilecki M, Pile LA. Systematic analysis of SIN3 histone modifying complex components during development. Sci. Rep. 2018;8:17048. doi: 10.1038/s41598-018-35093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kadamb R, Mittal S, Bansal N, Batra H, Saluja D. Sin3: Insight into its transcription regulatory functions. Eur. J. Cell Biol. 2013;92:237–246. doi: 10.1016/j.ejcb.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 10.Saha N, Liu M, Gajan A, Pile LA. Genome-wide studies reveal novel and distinct biological pathways regulated by SIN3 isoforms. BMC Genom. 2016;17:111. doi: 10.1186/s12864-016-2428-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choy SW, Wong YM, Ho SH, Chow KLC. Elegans SIN-3 and its associated HDAC corepressor complex act as mediators of male sensory ray development. Biochem. Biophys. Res. Commun. 2007;358:802–807. doi: 10.1016/j.bbrc.2007.04.194. [DOI] [PubMed] [Google Scholar]
- 12.Pandey R, Sharma M, Saluja D. SIN-3 as a key determinant of lifespan and its sex dependent differential role on healthspan in Caenorhabditis elegans. Aging (Albany NY) 2018;10:3910–3937. doi: 10.18632/aging.101682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharma M, Pandey R, Saluja D. ROS is the major player in regulating altered autophagy and lifespan in sin-3 mutants of C. elegans. Autophagy. 2018;14:1239–1255. doi: 10.1080/15548627.2018.1474312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.RStudio Team . RStudio: Integrated Development for R. RStudio PBC; 2020. [Google Scholar]
- 15.Microsoft Corporation . Microsoft Excel. One Microsoft Way; 2013. [Google Scholar]
- 16.Geer LY, et al. The NCBI BioSystems database. Nucl. Acids Res. 2010;38:D492–D496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higgins DG, Sharp PM. CLUSTAL: A package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 18.Sievers F, Higgins DG. Clustal Omega. Curr. Protoc. Bioinform. 2014;48:3.13.1–3.13.16. doi: 10.1002/0471250953.bi0313s48. [DOI] [PubMed] [Google Scholar]
- 19.Thompson, J. D., Gibson, T. J. & Higgins, D. G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform.00, 2.3.1–2.3.22 (2003). [DOI] [PubMed]
- 20.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 22.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemoine F, et al. Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature. 2018;556:452–456. doi: 10.1038/s41586-018-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchler-Bauer A, et al. CDD: NCBI’s conserved domain database. Nucl. Acids Res. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warde-Farley D, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucl. Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang DW, et al. DAVID bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucl. Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucl. Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan F, et al. Analysis of protein–protein functional associations by using gene ontology and KEGG pathway. Biomed. Res. Int. 2019;2019:e4963289. doi: 10.1155/2019/4963289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter S, et al. InterPro: The integrative protein signature database. Nucl. Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gene Ontology Consortium The gene ontology (GO) database and informatics resource. Nucl. Acids Res. 2004;32:D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emms DM, Kelly S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaidel-Bar R. Evolution of complexity in the integrin adhesome. J. Cell Biol. 2009;186:317–321. doi: 10.1083/jcb.200811067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson WR. An introduction to sequence similarity (“Homology”) searching. Curr. Protoc. Bioinform. 2013 doi: 10.1002/0471250953.bi0301s42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson JD, Plewniak F, Poch O. A comprehensive comparison of multiple sequence alignment programs. Nucl. Acids Res. 1999;27:2682–2690. doi: 10.1093/nar/27.13.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaubal A, Pile LA. Same agent, different messages: Insight into transcriptional regulation by SIN3 isoforms. Epigenet. Chromatin. 2018;11:17. doi: 10.1186/s13072-018-0188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chou C-C, Wang AH-J. Structural D/E-rich repeats play multiple roles especially in gene regulation through DNA/RNA mimicry. Mol. BioSyst. 2015;11:2144–2151. doi: 10.1039/C5MB00206K. [DOI] [PubMed] [Google Scholar]
- 38.Gemayel R, et al. Variable glutamine-rich repeats modulate transcription factor activity. Mol. Cell. 2015;59:615–627. doi: 10.1016/j.molcel.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guo Y-J, et al. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 2020;19:1997–2007. doi: 10.3892/etm.2020.8454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asih PR, et al. Functions of p38 MAP kinases in the central nervous system. Front. Mol. Neurosci. 2020;13:172. doi: 10.3389/fnmol.2020.570586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan H, Finkel T. Key proteins and pathways that regulate lifespan. J. Biol. Chem. 2017;292:6452–6460. doi: 10.1074/jbc.R116.771915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koonin EV, Galperin MY. Evolutionary Concept in Genetics and Genomics. Sequence–Evolution–Function: Computational Approaches in Comparative Genomics. Kluwer Academic; 2003. [PubMed] [Google Scholar]
- 43.Váchová L, Palková Z. Caspases in yeast apoptosis-like death: Facts and artefacts. FEMS Yeast Res. 2007;7:12–21. doi: 10.1111/j.1567-1364.2006.00137.x. [DOI] [PubMed] [Google Scholar]
- 44.Banuett F. Signalling in the yeasts: An informational cascade with links to the filamentous fungi. Microbiol. Mol. Biol. Rev. 1998;62:249–274. doi: 10.1128/MMBR.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu J, Peatman E, Tang H, Lewis J, Liu Z. Profiling of gene duplication patterns of sequenced teleost genomes: Evidence for rapid lineage-specific genome expansion mediated by recent tandem duplications. BMC Genom. 2012;13:246. doi: 10.1186/1471-2164-13-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Postlethwait JH, et al. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 2000;10:1890–1902. doi: 10.1101/gr.164800. [DOI] [PubMed] [Google Scholar]
- 47.Ding Z, Kihara D. Computational identification of protein–protein interactions in model plant proteomes. Sci. Rep. 2019;9:8740. doi: 10.1038/s41598-019-45072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hawkins T, Chitale M, Kihara D. New paradigm in protein function prediction for large scale omics analysis. Mol. BioSyst. 2008;4:223–231. doi: 10.1039/b718229e. [DOI] [PubMed] [Google Scholar]
- 49.Hawkins T, Kihara D. Function prediction of uncharacterized proteins. J. Bioinform. Comput. Biol. 2007;05:1–30. doi: 10.1142/S0219720007002503. [DOI] [PubMed] [Google Scholar]
- 50.Kuzmanov U, Emili A. Protein–protein interaction networks: Probing disease mechanisms using model systems. Genome Med. 2013;5:37. doi: 10.1186/gm441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Khan IK, Kihara D. Genome-scale prediction of moonlighting proteins using diverse protein association information. Bioinformatics. 2016;32:2281–2288. doi: 10.1093/bioinformatics/btw166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams MK, et al. Differential complex formation via paralogs in the human Sin3 protein interaction network. Mol. Cell. Proteom. 2020;19:1468–1484. doi: 10.1074/mcp.RA120.002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hasan T, Saluja D. Structural allostery and protein–protein interactions of Sin3. In: Singh LR, Dar TA, Ahmad P, editors. Proteostasis and Chaperone Surveillance. Springer; 2015. pp. 3–24. [Google Scholar]
- 54.Liu L, et al. Combining sequence and network information to enhance protein–protein interaction prediction. BMC Bioinform. 2020;21:537. doi: 10.1186/s12859-020-03896-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Plewczyński D, Ginalski K. The interactome: Predicting the protein–protein interactions in cells. Cell. Mol. Biol. Lett. 2008;14:1–22. doi: 10.2478/s11658-008-0024-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liebeskind BJ, Aldrich RW, Marcotte EM. Ancestral reconstruction of protein interaction networks. PLoS Comput. Biol. 2019;15:e1007396. doi: 10.1371/journal.pcbi.1007396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: A real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9:S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen CD, Gardiner KJ, Cios KJ. Protein annotation from protein interaction networks and gene ontology. J. Biomed. Inform. 2011;44:824–829. doi: 10.1016/j.jbi.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: Survey and summary. Nucl. Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teichmann M, Dumay-Odelot H, Fribourg S. Structural and functional aspects of winged-helix domains at the core of transcription initiation complexes. Transcription. 2012;3:2–7. doi: 10.4161/trns.3.1.18917. [DOI] [PubMed] [Google Scholar]
- 61.Müthel, S., et al. The conserved histone chaperone LIN‐53 is required for normal lifespan and maintenance of muscle integrity in Caenorhabditis elegans. Aging cell.18, e13012 (2019). [DOI] [PMC free article] [PubMed]
- 62.Doonan R, Hatzold J, Raut S, Conradt B, Alfonso A. HLH-3 is a C. elegans Achaete/Scute protein required for differentiation of the hermaphrodite-specific motor neurons. Mech. Dev. 2008;125:883–893. doi: 10.1016/j.mod.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 63.Thellmann M, Hatzold J, Conradt B. The snail-like CES-1 protein of C. elegans can block the expression of the BH3-only cell-death activator gene egl-1 by antagonizing the function of bHLH proteins. Development. 2003;130:4057–4071. doi: 10.1242/dev.00597. [DOI] [PubMed] [Google Scholar]
- 64.Erdélyi P, et al. Shared developmental roles and transcriptional control of autophagy and apoptosis in Caenorhabditis elegans. J. Cell Sci. 2011;124:1510–1518. doi: 10.1242/jcs.080192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hatzold J, Conradt B. Control of apoptosis by asymmetric cell division. PLoS Biol. 2008;6:e84. doi: 10.1371/journal.pbio.0060084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hesp K, Smant G, Kammenga JE. Caenorhabditis elegans DAF-16/FOXO transcription factor and its mammalian homologs associate with age-related disease. Exp. Gerontol. 2015;72:1–7. doi: 10.1016/j.exger.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 67.Kondo M, et al. The p38 signal transduction pathway participates in the oxidative stress-mediated translocation of DAF-16 to Caenorhabditis elegans nuclei. Mech. Ageing Dev. 2005;126:642–647. doi: 10.1016/j.mad.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 68.Lapierre LR, Gelino S, Meléndez A, Hansen M. Autophagy and lipid metabolism coordinately modulate life span in germline-less C. elegans. Curr. Biol. 2011;21:1507–1514. doi: 10.1016/j.cub.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee RYN, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr. Biol. 2001;11:1950–1957. doi: 10.1016/S0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- 70.Sun, X., Chen, W.-D. & Wang, Y.-D. DAF-16/FOXO transcription factor in aging and longevity. Front. Pharmacol.8, 548 (2017). [DOI] [PMC free article] [PubMed]
- 71.van der Bent ML, et al. Loss-of-function of β-catenin bar-1 slows development and activates the Wnt pathway in Caenorhabditis elegans. Sci. Rep. 2014;4:4926. doi: 10.1038/srep04926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yanase S, Yasuda K, Ishii N. Interaction between the ins/IGF-1 and p38 MAPK signaling pathways in molecular compensation of sod genes and modulation related to intracellular ROS levels in C. elegans. Biochem. Biophys. Rep. 2020;23:100796. doi: 10.1016/j.bbrep.2020.100796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Byrne AB, et al. A global analysis of genetic interactions in Caenorhabditis elegans. J. Biol. 2007;6:8. doi: 10.1186/jbiol58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Essers MAG, et al. Functional interaction between ß-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- 75.Chiang W-C, Ching T-T, Lee HC, Mousigian C, Hsu A-L. HSF-1 regulators DDL-1/2 link insulin-like signaling to heat-shock responses and modulation of longevity. Cell. 2012;148:322–334. doi: 10.1016/j.cell.2011.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gatsi R, et al. Prohibitin-mediated lifespan and mitochondrial stress implicate SGK-1, insulin/IGF and mTORC2 in C. elegans. PLoS ONE. 2014;9:e107671. doi: 10.1371/journal.pone.0107671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Perrin AJ, et al. Noncanonical control of C. elegans germline apoptosis by the insulin/IGF-1 and Ras/MAPK signaling pathways. Cell Death Differ. 2013;20:97–107. doi: 10.1038/cdd.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Quevedo C, Kaplan DR, Derry WB. AKT-1 regulates DNA-damage-induced germline apoptosis in C. elegans. Curr. Biol. 2007;17:286–292. doi: 10.1016/j.cub.2006.12.038. [DOI] [PubMed] [Google Scholar]
- 79.Greiss S, Schumacher B, Grandien K, Rothblatt J, Gartner A. Transcriptional profiling in C. elegans suggests DNA damage dependent apoptosis as an ancient function of the p53 family. BMC Genom. 2008;9:334. doi: 10.1186/1471-2164-9-334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hashimoto Y, Ookuma S, Nishida E. Lifespan extension by suppression of autophagy genes in Caenorhabditis elegans. Genes Cells. 2009;14:717–726. doi: 10.1111/j.1365-2443.2009.01306.x. [DOI] [PubMed] [Google Scholar]
- 81.Morselli E, et al. Caloric restriction and resveratrol promote longevity through the Sirtuin-1-dependent induction of autophagy. Cell Death Dis. 2010;1:e10–e10. doi: 10.1038/cddis.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Morselli E, et al. The life span-prolonging effect of Sirtuin-1 is mediated by autophagy. Autophagy. 2010;6:186–188. doi: 10.4161/auto.6.1.10817. [DOI] [PubMed] [Google Scholar]
- 83.Aspernig H, et al. Mitochondrial perturbations couple mTORC2 to autophagy in C. elegans. Cell Rep. 2019;29:1399.e5–1409.e5. doi: 10.1016/j.celrep.2019.09.072. [DOI] [PubMed] [Google Scholar]
- 84.Jones KT, Greer ER, Pearce D, Ashrafi K. Rictor/TORC2 regulates Caenorhabditis elegans fat storage, body size, and development through sgk-1. PLoS Biol. 2009;7:e1000060. doi: 10.1371/journal.pbio.1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhou B, et al. Mitochondrial permeability uncouples elevated autophagy and lifespan extension. Cell. 2019;177:299–314.e16. doi: 10.1016/j.cell.2019.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhu M, et al. Serum- and glucocorticoid-inducible kinase-1 (SGK-1) plays a role in membrane trafficking in Caenorhabditis elegans. PLoS ONE. 2015;10:e0130778. doi: 10.1371/journal.pone.0130778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Antebi A. Genetics of aging in Caenorhabditis elegans. PLoS Genet. 2007;3:e129. doi: 10.1371/journal.pgen.0030129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li W, Gao B, Lee S-M, Bennett K, Fang D. RLE-1, an E3 ubiquitin ligase, regulates C. elegans aging by catalyzing DAF-16 polyubiquitination. Dev. Cell. 2007;12:235–246. doi: 10.1016/j.devcel.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 89.Sherwood DR, Butler JA, Kramer JM, Sternberg PW. FOS-1 promotes basement-membrane removal during anchor-cell invasion in C. elegans. Cell. 2005;121:951–962. doi: 10.1016/j.cell.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 90.Zhang Z, Liu L, Twumasi-Boateng K, Block DHS, Shapira M. FOS-1 functions as a transcriptional activator downstream of the C. elegans JNK homolog KGB-1. Cell. Signal. 2017;30:1–8. doi: 10.1016/j.cellsig.2016.11.010. [DOI] [PubMed] [Google Scholar]
- 91.Andersen EC, Horvitz HR. Two C. elegans histone methyltransferases repress lin-3EGF transcription to inhibit vulval development. Development. 2007;134:2991–2999. doi: 10.1242/dev.009373. [DOI] [PubMed] [Google Scholar]
- 92.Checchi PM, Engebrecht J. Caenorhabditis elegans histone methyltransferase MET-2 shields the male X chromosome from checkpoint machinery and mediates meiotic sex chromosome inactivation. PLoS Genet. 2011;7:e1002267. doi: 10.1371/journal.pgen.1002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kramer M, et al. Developmental dynamics of X-chromosome dosage compensation by the DCC and H4K20me1 in C. elegans. PLoS Genet. 2015;11:e1005698. doi: 10.1371/journal.pgen.1005698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ni Z, Ebata A, Alipanahiramandi E, Lee SS. Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging Cell. 2012;11:315–325. doi: 10.1111/j.1474-9726.2011.00785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Greer EL, Brunet A. Different dietary restriction regimens extend lifespan by both independent and overlapping genetic pathways in C. elegans. Aging Cell. 2009;8:113–127. doi: 10.1111/j.1474-9726.2009.00459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.O’Rourke EJ, Kuballa P, Xavier R, Ruvkun G. ω − 6 Polyunsaturated fatty acids extend life span through the activation of autophagy. Genes Dev. 2013;27:429–440. doi: 10.1101/gad.205294.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li J, et al. Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator. PLoS Biol. 2008;6:e233. doi: 10.1371/journal.pbio.0060233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ewald CY, Marfil V, Li C. Alzheimer-related protein APL-1 modulates lifespan through heterochronic gene regulation in Caenorhabditis elegans. Aging Cell. 2016;15:1051–1062. doi: 10.1111/acel.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Karp X, Ambros V. The developmental timing regulator hbl-1 modulates the Dauer formation decision in Caenorhabditis elegans. Genetics. 2011;187:345–353. doi: 10.1534/genetics.110.123992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Larsen PL, Albert PS, Riddle DL. Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics. 1995;139:1567–1583. doi: 10.1093/genetics/139.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li S, et al. Specific regulation of thermosensitive lipid droplet fusion by a nuclear hormone receptor pathway. PNAS. 2017;114:8841–8846. doi: 10.1073/pnas.1704277114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Khan MH, et al. TAF-4 is required for the life extension of isp-1, clk-1 and tpk-1 Mit mutants. Aging (Albany NY) 2013;5:741–758. doi: 10.18632/aging.100604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Le Guezennec X, Vermeulen M, Stunnenberg HG. Molecular characterization of Sin3 PAH-domain interactor specificity and identification of PAH partners. Nucl. Acids Res. 2006;34:3929–3937. doi: 10.1093/nar/gkl537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zanier K, et al. Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins. Science. 2013;339:694–698. doi: 10.1126/science.1229934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pagni, M. et al. MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic acids research. 35, W433-W437 (2007). [DOI] [PMC free article] [PubMed]
- 106.Hulo, N. et al. The PROSITE database. Nucleic acids research. 34, D227-D230 (2006). [DOI] [PMC free article] [PubMed]
- 107.Davis, P. et al. WormBase in 2022—data, processes, and tools for analyzing Caenorhabditis elegans. Genetics.220, iyac003 (2022). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information file.