Abstract
Glioblastoma (GBM) is the most aggressive primary brain tumor in adults, which remains difficult to cure. The very high recurrence rate has been partly attributed to the presence of GBM stem-like cells (GSCs) within the tumors, which have been associated with elevated chemokine receptor 4 (CXCR4) expression. CXCR4 is frequently overexpressed in cancer tissues, including GBM, and usually correlates with a poor prognosis. We have created a CXCR4-retargeted oncolytic herpesvirus (oHSV) by insertion of an anti-human CXCR4 nanobody in glycoprotein D of an attenuated HSV-1 (ΔICP34.5, ΔICP6, and ΔICP47), thereby describing a proof of principle for the use of nanobodies to target oHSVs toward specific cellular entities. Moreover, this virus has been armed with a transgene expressing a soluble form of TRAIL to trigger apoptosis. In vitro, this oHSV infects U87MG CXCR4+ and patient-derived GSCs in a CXCR4-dependent manner and, when armed, triggers apoptosis. In a U87MG CXCR4+ orthotopic xenograft mouse model, this oHSV slows down tumor growth and significantly improves mice survival. Customizing oHSVs with diverse nanobodies for targeting multiple proteins appears as an interesting approach for tackling the heterogeneity of GBM, especially GSCs. Altogether, our study must be considered as a proof of principle and a first step toward personalized GBM virotherapies to complement current treatments.
Keywords: virotherapy, HSV, glioblastoma, glioblastoma stem-like cells, GBM, GSC, nanobody, oncolytic herpesvirus, virus retargeting, CXCR4
Graphical abstract

Sanchez Gil and colleagues show as a proof of principle that nanobodies can be used to retarget an oncolytic herpesvirus to a sub-population of GBM cells. An anti-CXCR4 nanobody-retargeted virus can infect U87MG and patient-derived GBM stem-like cells in a CXCR4-dependent manner and trigger apoptosis when armed with sTRAIL. In an orthotopic xenograft mouse model, this virus decreases tumor growth and increases survival.
Introduction
The chemokine receptor 4 (CXCR4), first described for its role in leukocyte trafficking or HIV infection,1 is a largely studied G-protein-coupled receptor that activates various signaling pathways upon binding of its unique ligand CXCL12, also known as stromal-cell-derived factor 1. CXCR4 overexpression has been reported in a wide range of tumors, including glioblastoma multiforme (GBM),2, 3, 4, 5 and increasing evidence has suggested its central role in cancer progression.6 Multiple preclinical or clinical studies have demonstrated that the disruption of CXCR4 downstream signaling via several approaches (CXCR4 short hairpin RNA [shRNA], CXCL12 mimetic peptide, anti-CXCR4 antibodies, or nanobodies) diminishes tumor growth and synergizes with chemo- or radiotherapy.7, 8, 9, 10, 11, 12, 13
GBM is the most frequent primary malignant brain tumor, classified by the World Health Organization as a grade 4 glioma.14 Despite standard therapies that associate surgical resection with radio- or chemotherapy, the prognosis remains dramatically poor, with a median survival of 16 months from diagnosis.15 GBM is indeed highly diffuse and tumor cells infiltrate healthy brain tissue, making the total resection of the tumor rather difficult or even impossible. GBM recurrences frequently develop within the margin of the resection cavity or at distant sites.16 In addition, GBM is characterized by a high degree of heterogeneity at the genetic, epigenetic, and transcriptomic levels. Many studies reported the presence of self-renewing, multipotent subsets of GBM cells endowed with high tumorigenic capacity, considered as GBM stem-like cells (GSCs).17, 18, 19 GSCs have been associated with the expression of specific markers, form tumorospheres in vitro upon limiting dilution, and are able to initiate a tumor when serially transplanted in mice brain. GSCs have long been considered as key actors in GBM relapse, and the mechanisms underlying GSC development, maintenance, and phenotypic plasticity yet remain intensively investigated.20 We previously have shown that, upon GBM xenotransplantation, CXCR4+ GSCs escape the tumor core and reach the subventricular zones (SVZs) based on a CXCR4/CXCL12-dependent signaling.21,22 GSCs hosted in the SVZ display an improved DNA double-strand break repair and hence are resistant to radiotherapy.22,23 These observations have been confirmed in GBM patients, in which GSCs can be found both in the tumor core, where the hypoxic environment constitutes an appropriate niche, and in the SVZ, reinforcing the role of these CXCR4+ cells in GBM recurrence.24,25 Importantly, a high expression of CXCR4 positively correlates with tumor size, tumor progression, recurrence, and ultimately with patient survival.3,5 Targeting GSCs and particularly CXCR4+ cells therefore provides an opportunity to reach tumor cells that escape current treatments.26
Over the last decade, virotherapy has emerged as a promising approach for cancer treatment.27 Oncolytic viruses (OVs) are currently at different stages of preclinical investigations, and numerous clinical trials are ongoing. In the context of GBM, virotherapy and oncolytic herpesviruses (oHSVs) in particular are currently being evaluated as an alternative or complementary therapeutic approach for patients resistant to traditional therapies.28 oHSV efficacy depends on the capacity of the virus to specifically infect cancer cells. However, it is estimated that about 20% of the GBM cells are not efficiently infected by oHSV, partly due to a low expression of CD111 (nectin-1, one of the HSV-1 natural receptors).29,30 A virus able to target cancer cells and GSCs in particular through its interaction with a membrane protein specifically expressed by these cells would thus allow to reach cells that have escaped standard therapeutic approaches. One strategy for oHSV retargeting is to replace the domain responsible for glycoprotein D (gD) interaction with its natural cellular receptors by a ligand able to interact with a protein of interest expressed by the target cells. Single-chain immunoglobulin (scFv) or ligands, such as cytokines or peptides, have been successfully introduced in gD to target cancer cells.31, 32, 33, 34, 35, 36 Nanobodies are a single heavy variable domain of camelid antibodies and constitute an interesting alternative to retarget an oHSV. They can be selected from a synthetic or immune library with a huge diversity and can recognize cryptic antigens with a high affinity. These nanobodies therefore open the possibility to develop a panel of tailored oHSVs for personalized therapy.
In this context, we have developed, as a proof of principle, an oncolytic HSV-1 specifically targeting CXCR4, thanks to the insertion in gD of an anti-human CXCR4 nanobody previously described for its capacity to efficiently recognize CXCR4 (WO2016156570Al). This virus (oHSV/Nb-gD) has been further armed with a transgene expressing the soluble form of TRAIL (oHSV/Nb-gD:sTRAIL), whose efficacy to trigger the extrinsic apoptosis pathway has been previously documented.37, 38, 39, 40 We demonstrated that the engineered virus infects U87MG CXCR4+ and patient-derived GSCs in a CXCR4-dependent manner and can replicate efficiently in these cells and lead to sTRAIL expression, thereby triggering apoptosis. When used in an in vivo orthotopic xenograft GBM model, oHSV/Nb-gD armed or not with sTRAIL had a clear impact on tumor progression and significantly improved mice survival. These results confirm nanobodies as appropriate tools for retargeting oHSVs toward specific cell subsets and constitute a proof of principle of an oHSV design strategy that could be considered for personalized treatment.
Results
Construction of a nanobody-retargeted and armed oncolytic herpesvirus
To specifically target GBM cells expressing CXCR4, we engineered an oHSV that was first detargeted from its natural receptors HVEM and nectin-1, prior to being retargeted to CXCR4 (Figure 1). These modifications were introduced within fQuick-1 (kind gift from Prof. E.A. Chiocca), a bacterial artificial chromosome (BAC) containing the HSV-1 genome (strain F; ΔICP34.5/ΔICP6/EGFP+). This backbone was further deleted from US12 coding for ICP47, this deletion being important to partly overcome the attenuation resulting from γ34.5 deletion.41 The detargeting and retargeting was achieved by replacing the residues 2–24 of gD within the HVEM-binding domain by an anti-human CXCR4 nanobody.42 In addition, the residue 38 of gD was mutated (Y38C) to impair gD interaction with nectin-1, another natural receptor.43 Moreover, two mutations (D285N and A549T) shown to improve the fusion capacity of glycoprotein B (gB) were introduced in UL27.44 Finally, the virus was armed with a transgene expressing sTRAIL45 under the control of a nestin promoter. After transfection of these constructs into Vero cells previously transduced with the human CXCR4, oHSVs were produced in the supernatant and further purified and titrated. In this publication, they are referred to as oHSV/gD (non-retargeted; non-armed), oHSV/Nb-gD (CXCR4 retargeted; non-armed), and oHSV/Nb-gD:sTRAIL (CXCR4 retargeted; sTRAIL armed).
Figure 1.
Schematic representation of oHSV/Nb-gD and oHSV/Nb-gD:sTRAIL genomes
Efficacy of the CXCR4 retargeting
To verify the detargeting efficacy, J1.1–2 hamster cells resistant to HSV due to the lack of HVEM or nectin-1 expression at the cell surface,46 as well as their modified version J/A and J/C expressing, respectively, human HVEM47 or nectin-148 (kind gift from Prof. G. Campadelli Fiume), were infected with oHSV/gD or oHSV/Nb-gD (MOI: 0.01, 0.1, and 1). Contrary to oHSV/gD, which led to numerous infectious foci in J/A and J/C, no foci were detected upon oHSV/Nb-gD infection, demonstrating that oHSV/Nb-gD was properly detargeted (Figure 2A). To evaluate the capacity of oHSV/Nb-gD to specifically infect CXCR4+ cells, glioblastoma U87MG cells that express CXCR4 at a very low level (Figures S1A and S1B) were transduced with a lentivirus expressing the human CXCR4. The ectopic expression of CXCR4 was confirmed by flow cytometry (Figures S1A and S1B). U87MG and U87MG CXCR4+ were then infected with oHSV/gD or oHSV/Nb-gD (MOI: 0.1), and the level of infection was evaluated by real-time GFP imaging and quantification with Incucyte S3 (Figures 2B, 2C, and S2). As expected, oHSV/gD efficiently replicated in both cell lines independently of CXCR4 expression. On the contrary, oHSV/Nb-gD infection remained very low in U87MG cells, with only very few cells infected, as reflected by a very weak EGFP expression and no statistical difference with the non-infected cells. This clearly contrasted with numerous foci and overtime increasing EGFP signal in oHSV/Nb-gD-infected U87MG CXCR4+ cells, confirming that oHSV/Nb-gD infection relies on the expression of CXCR4. Importantly, the efficacy of infection of oHSV/gD and oHSV/Nb-gD in U87MG CXCR4+ cells was similar. This was further confirmed by a growth curve of both oHSVs in U87MG-CXCR4+ cells. No statistical difference was observed (Figure S3).
Figure 2.
Efficacy of the oHSV detargeting and retargeting
(A) Detargeting was evaluated by infection of J1.1–2, J/A (J1.1 HVEM+), and J/C (J1.1 nectin-1+) cells infected for 72 h at different MOIs with the recombinant oHSVs expressing either wild-type (WT) gD (oHSV/gD) or gD modified by the insertion of an anti-hCXCR4 nanobody (oHSV/Nb-gD). Both viruses express EGFP under the control of pICP6, allowing the visualization of infected cells by epifluorescence microscopy. Scale bars represent 5 mm. (B and C) Retargeting was evaluated on U87MG (B) and U87MG CXCR4+ (C) cells. Cells were plated in 96-well plates, infected with oHSV/gD or oHSV/Nb-gD (MOI 0.1) and incubated in Incucyte S3 for real-time analyses during 72 h. EGFP expression and cell confluency were quantified every 6 h. Circles represent the ratio between the green and the phase area expressed as the mean ± SEM of four wells. Statistical significance was determined by ordinary two-way ANOVA with Bonferroni multiple comparisons of means with a single pooled variance (ns, non-significant; ∗ p < 0.05, ∗∗ p < 0.01, ∗∗∗∗ p < 0.0001). Images were taken every 6 h, and representative images taken at 72 hpi are shown. Scale bars represent 2 mm. Additional representative whole-well images taken at 24, 48, and 72 h are shown in Figure S2. See also growth curve of oHSV/gD and oHSV/Nb-gD in U87MG CXCR4+ cells in Figure S3
CXCR4-dependent infection of patient-derived GSCs by oHSV/Nb-gD
The efficacy of oHSV/gD and oHSV/Nb-gD was further evaluated on four different GSC cultures (T08, T013, T018, and T033) directly established from residual GBM tissue obtained from surgical resection (Department of Neurosurgery, CHU Liège, Belgium) and maintained as tumorospheres. In opposition to U87MG cells, GSCs express high levels of SOX2, POU3F2, and SALL2 (Figure S4). The percentage of CXCR4+ cells among the four different GSC cultures analyzed by flow cytometry was highly variable (Figures 3A and 3B). While less than 3% of T08 cells were positive for CXCR4, around 75% of T033 expressed this chemokine receptor, T013 and T018 being intermediate. As expected, the endogenous expression of CXCR4 was much lower than the ectopic expression by U87MG CXCR4+ cells (Figures 3A and 3B). To evaluate the efficacy of the retargeted oHSV and to compare it with the non-retargeted virus efficacy, primary GSCs were cultured as tumorospheres and infected with oHSV/gD or oHSV/Nb-gD (106 plaque-forming units [PFUs]/mL). Forty-eight hours post-infection, cells were dissociated and the percentage of EGFP-positive cells was analyzed by flow cytometry.
Figure 3.
Efficacy of the oHSV retargeting in patient-derived GSCs
(A) Patient-derived GSCs (T08, T013, T018, and T033), U87MG, or U87MG CXCR4+ cells were cultured as tumorospheres and further dissociated for flow cytometry quantification of the percentage of cells expressing CXCR4 (APC+) at the cell membrane. Bars represent the means ± SEM of four independent experiments. Statistical significance was determined by Kruskal-Wallis (primary cells, ∗∗p < 0.01) or Mann-Whitney (U87MG cells, ∗p < 0.05) test. (B) Overlayed histograms of a representative analysis allowing the comparison between endogenous and ectopic CXCR4 expression. Stemness features (expression of SOX2, POUF3, and SALL2) analyzed by qRT-PCR are depicted in Figure S4. (C) Tumorospheres cultured in 24-well plates were infected with oHSV/gD or oHSV/Nb-gD (106 PFUs/mL). Forty-eight hours post-infection, cells were dissociated and the EGFP fluorescence was analyzed by flow cytometry. Bars represent the means ± SEM of three independent experiments. Statistical significance was determined by ordinary two-way ANOVA with Bonferroni’s multiple comparisons of means (∗∗p < 0.01). Raw data (overlaid histograms) representative of one experiment are shown in Figure S5. (D) Tumorospheres cultures in 24-well plates and infected for 48 h by oHSV/Nb-gD (106 PFUs/mL) were either analyzed by epifluorescence for EGFP detection (left panels) or fixed for immunostaining of nestin (white) or CXCR4 (red) and GFP detection (green). Nuclei were labeled with DAPI (blue). Images were recorded with a NIKON A1R confocal microscope. Scale bars represent 100 μm. See also Figure S6 for real-time EGFP quantification and images of T033 tumorospheres infected with oHSV/gD or oHSV/Nb-gD at a higher titer (107 PFUs/mL).
Interestingly, the percentage of oHSV/Nb-gD-infected cells clearly reflected the level of CXCR4 expression (Figure 3C). T033 that expresses CXCR4 at a high level was the most infected (34.8% of EGFP cells on an average; 48 h post-infection [hpi]), while less than 2% of T08 cells that do not express CXCR4 or express it at a very low level were positive for EGFP. As expected, in most primary cells, oHSV/gD led to a higher percentage of infected cells compared with oHSV/Nb-gD (Figures 3C and S5). However, an Incucyte S5 overtime analysis of T033 cells infected with a high titer (107/mL) indicated that both the dynamics and the EGFP fluorescence were similar for both viruses (Figure S6). Finally, it is worth mentioning that, although all primary cell lines were infected by the non-retargeted virus, its efficacy greatly varied, with T013 being significantly less infected than the other cell lines.
In parallel, tumorospheres were infected with oHSV/Nb-gD (106 PFUs/mL) for immunostainings. Forty-eight hours post-infection, epifluorescence observation of oHSV-infected tumorospheres revealed that EGFP intensity was very low in T08 and much brighter in T033, confirming that the level of infection reflects the level of CXCR4 expression (Figure 3D, left panels). Tumorospheres were then fixed for immunostainings. Confocal microscopy of oHSV/Nb-gD-infected tumorospheres sections confirmed that only very few T08 cells were EGFP+, while more infected cells were observed in T013, T018, and T033 tumorospheres (Figure 3D). Although no clear co-localization between EGFP and CXCR4 was observed at the cellular level, infected cells were usually observed in the CXCR4+ area.
In vitro evaluation of the efficacy of the sTRAIL arming
oHSV/Nb-gD, shown to be efficiently retargeted and to specifically infect CXCR4+ cells, was further armed with the gene coding for sTRAIL under the control of the nestin promoter to trigger apoptosis upon viral infection. First, we showed that the armed and non-armed oHSVs replicated with the same efficacy in Vero CXCR4+ (data not shown) or U87MG CXCR4+ cells (Figure 4A), demonstrating that the arming does not impair oHSV replication. The efficacy of sTRAIL to trigger the apoptosis pathway was analyzed either by western blotting or using an annexin V/DAPI assay, while the viability was evaluated by measuring the cellular metabolism with resazurin. The expression of sTRAIL upon infection of U87MG CXCR4+ by oHSV/Nb-gD:sTRAIL led to the cleavage of PARP and caspase 3, while no cleavage was observed upon oHSV/Nb-gD infection (Figure 4B). The annexin V/DAPI assay further confirmed apoptosis in oHSV-infected U87MG CXCR4+ cells. sTRAIL-induced apoptosis was detectable 48 hpi and reached significance only 72 hpi, with an average of 36% of apoptotic cells upon oHSV/Nb-gD:sTRAIL infection compared with 12% upon oHSV/Nb-gD infection (Figure 4C). At 72 hpi, the percentage of apoptotic cells upon oHSV/Nb-gD:sTRAIL infection increased according to the MOI, which was not the case with the non-armed oHSV (Figure S7). Interestingly, the viability of the cells infected by oHSV/Nb-gD or oHDSV/Nb-gD:sTRAIL measured 24, 48, or 72 hpi was not statistically different (Figure 4C).
Figure 4.
Efficacy of the oHSV arming
(A) The replication efficacy of the non-armed (oHSV/Nb-gD) and sTRAIL-armed (oHSV/Nb-gD:sTRAIL) oncolytic viruses was evaluated with a growth curve assay. U87MG CXCR4+ cells were infected at a MOI of 1, and supernatant was harvested 24, 48, and 72 h post-infection and used for titration as previously described.49 The number of foci was calculated based on Incucyte S3 imaging. Bars represent the means ± SEM (PFUs/mL) of three independent experiments. The lack of statistical difference is confirmed by unpaired t test analysis. (B) PARP and caspase 3 cleavage was evaluated by western blot analysis on total cell extracts from U87MG CXCR4+ cells infected for 18 h by oHSV/Nb-gD or oHSV/Nb-gD:sTRAIL (MOI: 0.5 or 1). gD and α-tubulin detection were used as infection or loading control, respectively. (C) Apoptosis was measured at different time points by flow cytometry using annexin V/DAPI labeling of U87MG CXCR4+ cells infected by oHSV/Nb-gD or oHSV/Nb-gD:sTRAIL (MOI: 5). The percentage of apoptotic cells corresponds to early (annexin V+/DAPI−) and late apoptotic (annexin V+/DAPI+) cells. Percentages of apoptotic cells upon infection at other MOI (1, 5, and 10) are shown in Figure S7. In parallel, cells were incubated with resazurin to evaluate the viability upon oHSV infection. Bars (percentage of apoptotic cells) and dots (percentage of viability) represent the means ± SEM of three independent experiments. Statistical significance was determined by ordinary two-way ANOVA with Bonferroni’s multiple comparisons of means (∗∗∗p < 0.001). (D and E) Patient-derived GSCs (T08, T013, T018, and T033) were cultured as tumorospheres in 24-well plates and infected with oHSV/Nb-gD or oHSV/Nb-gD:sTRAIL (106 PFUs/mL). gD and sTRAIL relative expression was analyzed 48 hpi by qRT-PCR as illustrated by a representative experiment. gD (D) and sTRAIL (E) mRNA level in oHSV/Nb-gD:sTRAIL-infected T08 are considered as the baseline (ND, not detected).
When used to infect patient-derived GSCs tumorospheres, oHSV/Nb-gD:sTRAIL led to the expression of gD and sTRAIL as measured by qRT-PCR, and this expression was significantly higher in T033 tumorospheres (Figures 4D and 4E).
Evaluation of the therapeutic efficacy of oHSV/Nb-gD and oHSV/Nb-gD:sTRAIL using an orthotopic xenograft GBM model
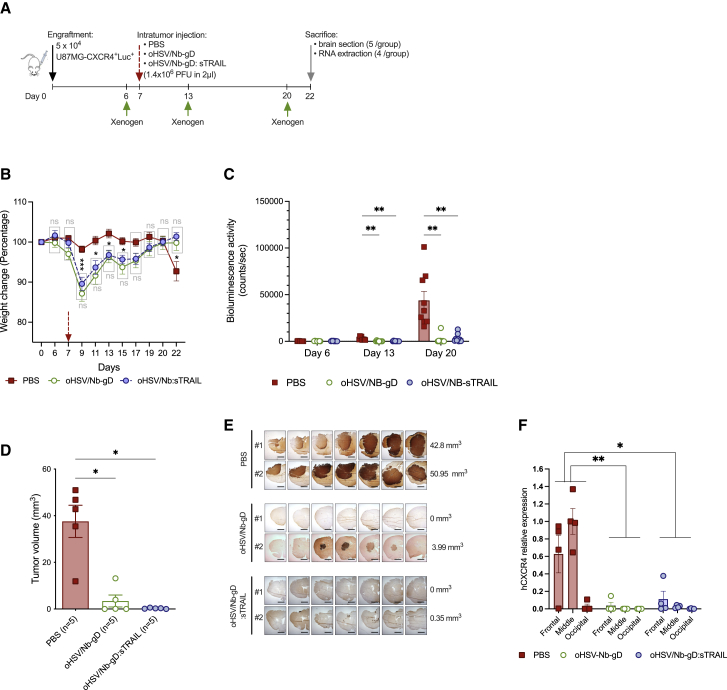
The capacity of oHSV/Nb-gD and oHSV/Nb-gD:sTRAIL to impact tumor growth was evaluated in vivo using an orthotopic xenograft GBM mouse model. A first experiment was set up with engraftment of 5 × 104 U87MG CXCR4+Luc+ into the right striatum under stereotactic control (Figure S8A). PBS or oHSVs (1.4 × 106 PFUs in 2 μL) were injected within the tumor on day 16. Weekly bioluminescence analysis revealed a very rapid tumor growth in all groups, even beyond oHSV intratumoral injection, although tumor growth appeared slightly reduced in oHSV/Nb-gD- or oHSV/Nb-gD:sTRAIL-treated mice compared with PBS-treated mice (Figure S8B). From day 19 on, PBS-treated mice health status rapidly evolved toward a critical point that justified sacrifice on day 24 (Figure S8C). Although not conclusive, these results paved the way for the design of another experiment, in which PBS or oHSVs (1.4 × 106 PFUs in 2 μL) were injected on day 7 after engraftment of 5 × 104 U87MG CXCR4+Luc+ GBM cells (Figure 5A). Body weight was monitored every 2nd day, and bioluminescence recording was performed weekly to evaluate the tumor size evolution. On day 22, mice were anesthetized and either perfused with saline solution only (for RNA extraction from brain tissue) or followed by paraformaldehyde to allow immunostaining analyses. Contrary to oHSV-treated mice which temporarily lost weight just after virus infection but showed a continuous weight gain until the end of the experiment, PBS-treated mice displayed a clear weight loss from day 20 on (Figure 5B). On day 6, the tumor size appeared homogeneous among groups, with no significant difference in the bioluminescent signal (Figures 5C and S9A). On day 13, bioluminescence in PBS-treated mice dramatically increased up to day 20, whereas the signal in oHSV-treated mice remained similar to day 6 or even decreased, becoming even undetectable in some mice (Figure 5C). All mice were sacrificed on day 22, and brains were harvested for either anti-human vimentin immunohistochemical staining and tumor size measurement (five mice/group) or RNA extraction and qRT-PCR analyses (four mice/group). The size of the tumor, calculated by measuring the area positive for human vimentin on serial sections and 3D volume reconstruction, clearly showed a significant impact of both oHSV/Nb-gD and oHSV/Nb-gD:sTRAIL treatment, even if no significant difference was observed between the two viruses (Figures 5D and 5E). For RNA extraction, right hemispheres, in which the cells were engrafted, were divided into three parts (frontal, middle, and occipital). Human CXCR4 expression, reflecting the presence of implanted human CXCR4+ GBM cells, was evaluated in each block individually and expressed as the relative expression to the level of expression in the middle part of PBS-treated mice brains (Figure 5F). Overall, human CXCR4 expression was significantly decreased in oHSV-treated mice compared with PBS-treated mice. In both oHSV-treated groups, differences in the level of expression of hCXCR4 were observed between the three blocks, with a higher abundance of human transcripts detected in samples corresponding to the frontal and middle samples, covering the initial site of engraftment. These results were confirmed by qRT-PCR for human nestin and TBP (data not shown) and corroborated bioluminescence analyses that showed some signal, although quite low in oHSV-treated mice (Figure S9A). At the end of the experiment (15 days after virus injection), we were unable to detect gD or sTRAIL neither by immunohistochemistry nor by qRT-PCR (data not shown).
Figure 5.
In vivo efficacy of oHSV/Nb-gD and oHSV/Nb-gD:sTRAIL
(A) Schematic representation of the experimental settings. Nude mice were engrafted with U87MG CXCR4+Luc+ cells and virus or PBS was injected in the tumor on day 7. Mice were sacrificed on day 22 (n = 9 in each group). (B) Mice were regularly weighed, and for each mouse, the weight change is expressed as a percentage to the weight on day 0, considered as equal to 100%. (C) Bioluminescence activity was recorded with Xenogen IVIS 50 on day 6, 13, and 20 after engraftment. See also Figure S9 for bioluminescence imaging. (B) and (C) represent the means ± SEM (n = 9 in each group). Statistical significance was determined by two-way ANOVA with Tukey’s multiple comparisons of means (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001). (D–F) On day 22, brain from five mice were sectioned for immunostaining of human vimentin and the measurement of the tumor volume by 3D reconstruction (D) Data represent the means ± SEM. Statistical significance was determined by Kruskal-Wallis test (∗p < 0.05). Representative pictures of serial sections of two mice/group as well as the estimated volume of the corresponding tumor are shown in (E). In parallel, brain from the four other mice were divided into three parts (frontal, middle, and occipital), which were frozen and treated independently for RNA extraction and qRT-PCR analysis of hCXCR4 expression (F). For each sample, PBS-treated mice (middle sample) are considered as the baseline. Bars represent the means ± SEM. Statistical significance was determined by two-way ANOVA with Tukey’s multiple comparisons of means with a single pooled variance (∗p < 0.05; ∗∗p < 0.01).
To verify whether, in vivo, oHSVs effectively replicate in tumor cells and sTRAIL is expressed, this experiment was repeated with the same settings, but mice were sacrificed 2 days after virus injection. Right hemispheres were divided into three parts (frontal, middle, and occipital), and total RNA was extracted from the brain tissue. gD and sTRAIL relative expression measured by qRT-PCR demonstrated the presence of gD transcripts in brains injected with oHSV/Nb-gD and oHSV/Nb-gD:sTRAIL, while sTRAIL transcripts were detected only in the oHSV/Nb-gD:sTRAIL group (Figures S9B and S9C).
Finally, a survival assay was set up with similar experimental settings (Figure 6A). U87MG CXCR4+Luc+ cells were injected under stereotactic control. All mice developed tumors (Figure S10A) and viral suspension, or PBS was injected within the tumor on day 7. Body weight was monitored every 2nd day, and mice were sacrificed when showing a significant weight loss or severe clinical signs. From day 19, all PBS-treated mice continuously lost weight, while oHSV-treated mice started to lose weight only on day 29, with the mice still alive 35 days after infection continuing to gain weight (Figure S10B). Again, tumor size appeared similar in all groups just before (day 5) virus injection (Figures 6B and S10A). However, one week after the intratumoral injection (day 13), bioluminescence signal in oHSV-treated mice was significantly reduced compared with the PBS group. In these oHSV-injected tumors, bioluminescence was very low and even undetectable in four of six and three of five mice in oHSV/Nb-gD and oHSV/Nb-gD:sTRAIL, respectively (Figures 6B and S10A). However, no significant difference was observed between oHSV/Nb-gD and oHSV/Nb-gD:sTRAIL-treated mice (Figure 6B). Importantly, while all PBS-treated mice died between day 21 and 27, the oHSV-treated mice death was significantly delayed, with the first deaths observed on day 31 (Figure 6C). At day 61, one of six oHSV/Nb-gD- and two of five oHSV/Nb-gD:sTRAIL-treated mice were still alive.
Figure 6.
Survival assay upon oHSV/Nb-gD or oHSV/Nb-gD:sTRAIL treatment
(A) Schematic representation of the survival assay experimental settings. (B) Bioluminescence activity of nude mice engrafted with 5 × 104 U87MG CXCR4 Luc+ cells was recorded with Xenogen IVIS 50 on day 5 (2 days before treatment) and 13 (6 days after treatment). Bars represent the means ± SEM. Statistical significance was determined by two-way ANOVA with Tukey’s multiple comparisons of means. (∗∗p < 0.01). See also Figure S10B for bioluminescence imaging. (C) Probability of survival of mice treated with PBS (n = 7), oHSV/Nb-gD (n = 6), or oHSV/Nb-gD:sTRAIL (n = 5). The red arrow indicates the day of treatment (day 7). Statistical significance was determined by log rank (Mantel-Cox) test (∗∗∗∗p < 0.0001). See also Figure S10A for weight follow-up.
Taken together, all these results show that oHSV/Nb-gD and oHSV/Nb-gD:sTRAIL are suited for intratumoral injection in GBM orthotopic models and exert a potent oncolytic activity in vivo.
Discussion
GBM remains the most aggressive form of adult brain cancer, associated with a dismal prognosis. Therapeutic failure and high recurrence rate endorse the need for novel, alternative, or add-on approaches to improve the standard-of-care therapy. GBM exhibits a wide cellular diversity, with malignant cells being highly heterogeneous in terms of molecular profile, phenotype, tumorigenic potential, and resistance to treatment. Such heterogeneity is largely accountable for tumor recurrence.
GSCs display stemness features, appear more resistant to radio- and chemotherapies, and are endowed with increased tumorigenicity.50 Targeting GSCs thus appears as an opportunity for new therapeutic approaches. A wide variety of therapeutic strategies aiming to target GSCs have been evaluated in preclinical models and are being clinically translated.26 However, considering the biological complexity and phenotypic plasticity of those cells, the main hurdle is to target GSCs without impairing normal tissue. In the perspective of eradicating peculiar GBM cell entities, such as GSCs, highly specific and targeted strategies should be considered.
Oncolytic virotherapy has been proposed as a promising avenue for GBM therapy, and herpesviruses offer numerous opportunities for tailored design and targeting strategies. oHSVs are the first viruses approved by the U.S. Food and Drug Administration (FDA) for virotherapy. Their mechanism of cell entry is well documented51 and can be modified to restrict oHSV entry into cells that specifically express a receptor of interest at their surface. oHSV retargeting requires the replacement of the viral glycoprotein domain important for their interaction with either the heparan sulfate or the natural receptors by a ligand specific for a protein of interest. Single-chain antibodies (scFv), cytokines, or specific ligands have been described for their efficacy to retarget oHSV.31, 32, 33, 34, 35 In our study, we describe oHSV retargeting using a nanobody. Nanobodies correspond to the single heavy variable domain of camelid antibodies. They can be quite easily obtained by screening either immune or artificial libraries characterized by a huge sequence diversity and thereby constitute an interesting tool for oHSV customization and specific targeting.
In this study, GBM has been chosen as a model to evaluate the nanobody-based oHSV retargeting. As a proof of principle, we considered to genetically engineer an oHSV whose gD is modified by the insertion of a nanobody able to recognize hCXCR4, a chemokine receptor expressed on several GBM cell subtypes, including GSCs. CXCR4 has been associated with cancer cell proliferation, tumorigenesis, and migration, and its expression correlates with a poor prognosis.52 In addition, we have previously shown CXCR4+ cells as able to move away from the tumor core and specifically invade the subventricular zones,21 and targeting of CXCR4 therefore appears as an encouraging approach. The CXCR4-retargeted oHSV described in this paper (namely oHSV/Nb-gD) has been engineered from an attenuated backbone (ΔICP34.5, ΔICP6, and ΔICP47), whose safety in GBM treatment has been largely documented.41 Other oHSVs retargeted to the epidermal growth factor receptor (EGFR), the human receptor tyrosine-protein kinase erbB-2 (hHER2), the interleukin-13 receptor, the epithelial cell adhesion molecule (EpCAM), or the urokinase plasminogen activator receptor; all described to be overexpressed in cancer tissues have been constructed and characterized.31, 32, 33, 34, 35, 36 Contrarily to the oHSVs described in this paper, all these retargeted viruses were engineered in a non-attenuated HSV background, inducing a higher level of viral replication. However, their safety only relies on the tight control of their entry into cancer cells and consequently requires an absence or a very low expression of the target of interest on healthy cells. Similarly, the CXCR4-retargeted oHSVs entry depends on the capacity of the virus to specifically interact with a receptor, but its attenuated character limits its replication in non-cancer cells, improving its safety. We show that the CXCR4-retargeted virus (oHSV/Nb-gD) can specifically infect in a CXCR4-dependent manner, not only U87MG CXCR4+ but also patient-derived GSCs, despite a much lower CXCR4 endogenous expression. In vitro, when armed with a secreted form of TRAIL (oHSV/Nb-gD:sTRAIL), this virus is able to trigger apoptosis. The replication of these oncolytic viruses in cells transduced with CXCR4 is not impaired by the retargeting or the arming. Importantly, when inoculated at high titers (107 PFUs/mL) on primary GBM cells expressing a high level of endogenous CXCR4 (T033), both the retargeted and the non-retargeted viruses show the same kinetics and the same efficacy of infection.
When used in vivo in an orthotopic xenograft model of GBM, in which U87MG CXCR4+ cells were engrafted, both sTRAIL-armed and non-armed oHSVs were able to limit the tumor progression and to significantly improve mice survival. Even though sTRAIL triggers apoptosis in vitro, its impact in the xenograft model seems to be limited. Contrarily to the sTRAIL-armed oHSV previously described in the literature and whose expression is driven by the HSV immediate-early promoter IE4/5,37,40 sTRAIL expression in oHSV/Nb-gD:sTRAIL is driven by the nestin promoter. Although nestin is overexpressed in most GBM tumors,26 it might not be activated at the same level in all GBM cells and hence be too restrictive for an optimal expression of sTRAIL. Moreover, in vitro, the percentage of apoptotic cells as measured by flow cytometry does not reflect the strong impact of oHSV infection on U87MG viability (Figure 4C). The oncolysis mediated by the virus itself may hide the sTRAIL-induced apoptosis when high MOI are used.37,40 The efficacy of the arming should be further evaluated in vivo in the xenograft model after engraftment of patient-derived GSCs. If needed, a stronger promoter should be considered to drive sTRAIL expression. U87MG CXCR4+ cells engrafted in the xenograft model have a very rapid growth kinetics. Such a rapid growth can hamper the total elimination of the tumor after a single virus injection and could explain the regrowth observed in some mice. In this context, it would be worth evaluating the impact of repeated injections or of continuous delivery of the virus thanks to a mini-osmotic pump system.53 In addition, the role of the tumor microenvironment and especially of the innate immune response should not be underestimated. oHSV virotherapy has been shown to rapidly activate natural killer (NK) cells that diminish the virotherapy efficacy,54 while adenovirus virotherapy has been shown to induce a phenotypic shift of macrophages from pro-tumoral M2-like toward the anti-tumoral and pro-inflammatory M1-like phenotype.55 A deeper characterization of the tumor microenvironment upon virotherapy will provide important information that might help to improve the treatment.
An important issue that must be carefully studied when targeting tumor cells is the fact that healthy cells might express the protein of interest and thus be infected by the oncolytic virus. Although in our study oHSVs are attenuated, this issue must be taken into consideration. CXCR4 is mainly expressed in the bone marrow or lymphoid tissues and poorly expressed in the brain (https://www.proteinatlas.org/ENSG00000121966-CXCR4). Taking into consideration that the oHSV is injected within the tumor, CXCR4 expression on non-tumoral cells in the vicinity of the tumor must, however, be considered. Based on publicly available patient-derived transcriptomic data, CXCR4 is expressed in malignant cells, in endothelial cells within the tumor, and on tumor-associated macrophages (TAMs) and tumor-infiltrating lymphocytes (TILs).56 The capacity of the CXCR4-retargeted virus to infect and potentially destroy these cells, especially endothelial cells and M2-like macrophages, would certainly be of interest; still, the benefit and risk balance has to be assessed very carefully. Unfortunately, the anti-hCXCR4 nanobody used in this study does not recognize the murine CXCR4, which limits the questions that could be addressed in the human GBM xenograft model. We are currently screening a nanobody library to identify nanobodies that recognize both the human and murine CXCR4 receptor. Such nanobodies would allow not only to address important issues, such as the undesired targeting of healthy cells, but also to evaluate the importance of the immune response and particularly of the adaptive immune response, this latest requiring a syngeneic GBM murine model.
Altogether, the results described in this proof of principle study show that the retargeting of oHSVs by the insertion of a nanobody appears highly encouraging and constitutes an interesting approach for the targeting of GBM cell subsets, e.g., GSCs, expressing specific proteins of interest. Our data support the idea that a set of nanobodies specific for diverse GSCs markers may be used to customize oHSVs that could be exploited as an add-on to complement the current standard-of-care therapeutic approaches.
Material and methods
Cell lines
Vero cells (ATCC; no. CCL-81) and human glioblastoma U87MG (ATCC; no. HTB-14) cells were maintained in Dulbecco’s modified Eagle medium (DMEM) (Biowest, VWR Inernational, Leuven, Belgium) supplemented with 10% fetal bovine serum (FBS). J1.1–2 cells are HSV-1-resistant baby hamster kidney cells lacking both HVEM and nectin-1, two natural HSV-1 receptors. J/A and J/C cells are J1.1 transduced with HVEM and nectin-1, respectively (kind gift of Prof. G. Campadelli-Fiume [University of Bologna, Italy]). They were cultured with DMEM supplemented with 5% of FBS. J/A and J/C cells were treated with 400 μg/mL of G418 (Invivogen, Belgium). Vero CXCR4+ and U87MG CXCR4+ obtained by transduction of a lentivirus (Viral Vector platform, University of Liège) were treated with 20 ng/mL and 10 ng/mL of blasticidin (Invivogen, Belgium), respectively. Primary GBM primary cultures (T08, T013, T018, and T033) were established from freshly resected human GBM tissue obtained from GBM patients. They were cultured as tumorospheres in stem cell medium (DMEM/F-12 with GlutaMAX [Gibco, Fisher Scientific, Belgium] supplemented with B27 [1/50] without vitamin A [Gibco, Fisher Scientific, Belgium], 1% penicillin-streptomycin [Biowest, VWR Inernational, Leuven, Belgium], 1 μg/mL of heparin [no. 7692.1; Carl Roth, Belgium], human EGF [20 ng/mL; BioLegend, Amsterdam, The Netherlands], and βFGF [20 ng/mL; BioLegend, Amsterdam, The Netherlands]).
Construction of recombinant oHSVs
Recombinant viruses were engineered in fHsvQuik-1 BAC containing an attenuated strain F HSV-1 (Δγ34.5, ΔUL39, GFP+; kind gift from A. Chiocca from the University of Pittsburgh, USA). Recombinants were obtained by the two-step Red recombination technique “en passant.”57 ICP47 deletion was done as described by Todo et al.58 The detargeting of gD from its natural receptors was performed according to Uchida et al.43 For retargeting, we inserted a patented sequence coding for a nanobody against human CXCR4 receptor (CXCR4-NB; WO2016156570Al) in the gD coding sequence. The “arming” sequence containing a soluble form of TRAIL (sTRAIL)45 under the nestin promoter was inserted before the ICP6 promoter as shown in Figure 1. A double mutation (D285N and A549T) was inserted within gB to compensate the loss of infectivity generally observed upon gD retargeting.44 CXCR4+ Vero cells were plated in 6-well plate at 40% confluence and transfected with 3 μg of BAC using JETPEI (Polyplus, Illkirch, France). Viral replication was detected 48 h after transfection by the visualization of fluorescent foci. Virus stocks were produced and concentrated as previously described.59 Briefly, cells were infected at low MOI (0.005) and cultured for 4 to 5 days at 33°C. The day before the experiment, cells were treated with 0.45 M of NaCl and 100 μg/mL of dextran sulfate. Supernatant was collected and centrifuged at 2,200 g for 10 min at 4°C and then filtered with 0.8-μm filter to discard cell debris. Then, viral particles were ultracentrifugated at 47.850g at 4°C using Beckman SW27 rotor. Centrifugated virus was resuspended in PBS with 10% glycerol, aliquoted, and stored at −80°C. Plaque assay in Vero CXCR4+ was used to titrate the virus and determine the amount of PFUs/mL.49
Viral growth assay
U87MG CXCR4+ or Vero CXCR4+ cells were seeded in a 12-well plate and infected with oHSV/gD, oHSV/Nb-gD, or oHSV/Nb-gD:sTRAIL at a MOI of 1 for 24, 48, or 72 h. Supernatant was then harvested, and titer (PFUs/mL) was determined by plaque assay as previously described.49 The number of foci was calculated based on Incucyte S3 imaging.
Entry assay
J1.1–2, J/A, and J/C cells were seeded in a 24-well plate the day before infection. Cells were infected with a MOI of 1, 0.1, and 0.01. After 48 h, cells were fixed with 4% paraformaldehyde and washed with PBS. Images were collected with the Incucyte S3 (Sartorius).
qRT-PCR
Total RNA was isolated using the RNA isolation Nucleospin kit (Macherey-Nagel) according to the manufacturer’s protocol. Five hundred nanograms of RNA was reverse transcribed using RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific) with random primers (for gD or sTRAIL transcripts detection) or oligo-dT primers (for stemness markers transcripts detection). TBP or 18S were used as controls. qRT-PCR reaction samples were prepared as follows: 4 μL of the diluted cDNA (2.5 ng in total for gD and sTRAIL or 10 ng in total for stemness markers) were mixed with 5 μL of SYBR green (TAKYON, Eurogentec, Liege, Belgium) and 0.2μM of each primer in a final volume of 10 μL. Primers used for transcripts detection are described in Table 1. Quantitative real-time PCR was done using the Roche LightCycler 480 (3 min at 95°C of activation; 45 cycles: denaturation 95°C, 3 s; hybridization and elongation 60°C, 25 s).
Table 1.
Primers used for qRT-PCR
| Forward | Reverse | |
|---|---|---|
| HSV-1 gD | 5ʹ-GCCCCGCTGGAACTACTATG-3ʹ | 5ʹ-TTATCTTCACGAGCCGCAGG-3ʹ |
| sTRAIL | 5ʹ-CATCGAGAACGAGATCGCCC-3ʹ | 5ʹ-TGTGTTGCTTCTTCCTCTGGT-3ʹ |
| SOX2 | 5ʹ-AGTCTCCAAGCGACGAAAAA-3ʹ | 5ʹ-TTTCACGTTTGCAACTGTCC-3ʹ |
| POU3F2 | 5ʹ-CTGACGATCTCCACGCAGTA-3ʹ | 5ʹ-GGCAGAAAGCTGTCCAAGTC-3ʹ |
| SALL2 | 5ʹ-ACTCCTCTGGGGTGACCTTT-3ʹ | 5ʹ-GGAGTGGTAGTGGAGGTGGA-3ʹ |
| 18S | 5ʹ-AACTTTCGATGGTAGTCGCCG-3ʹ | 5ʹ-CCTTGGATGTGGTAGCCGTTT-3ʹ |
| hTBP | 5ʹ-ACAGCCTGCCACCTTACG-3ʹ | 5ʹ-TGCCATAAGGCATCATTGGACTA-3ʹ |
Flow cytometry
For CXCR4 detection by flow cytometry, cells were plated in 6-well plate 2 days before analysis or cultured as tumorospheres. Tumorospheres and cells cultured as monolayers were washed with PBS and dissociated by incubating the cells for 10 min at 37°C with Accutase (Biowest, Nuaillé, France). Dissociated cells were centrifugated at 350g for 5 min at 4°C and washed with flow cytometry buffer (PBS with BSA 1%, EDTA 1 mM, and ADE 0.1%). Five microliters of antigen-presenting cell (APC)-conjugated anti-CXCR4 antibody (BioLegend, Amsterdam, the Netherlands) were added to 1 × 105 cells in 100 μL of flow cytometry buffer (dilution 1/20) and kept at 4°C for 1 h in the dark. Cells were washed by adding 1 mL of flow buffer and centrifugated at 400g for 4 min at 4°C. After a second wash, cells were resuspended in 200 μL of flow buffer and directly analyzed with the FACS CANTO II (BD Biosciences). Data were analyzed with FlowJo software.
Annexin/DAPI assay
For annexin V/DAPI apoptosis assay, 92,000 cells were seeded in a 12-well plate and infected with a MOI of 1, 5, or 10 for 72 h. Cells were collected and resuspended in 140 μL of 1X Binding Buffer (ref. 556,454; BD Pharmingen). Ten microliters of DAPI (Invitrogen; 1:100) and 5 μL of annexin V-PE (ref. AB 2869071; BD Biosciences) were added, and cells were incubated for 15 min at room temperature (RT) in the dark. Finally, 200 μL of 1X Binding Buffer was added and samples were directly analyzed with the FACS FORTESSA (BD Biosciences). Data were analyzed with FlowJo software.
Viability assay
U87MG and U87MG CXCR4+ cells were plated in a 12-well plate and infected with the different viruses at a MOI of 5. Measure of viability was done at 24, 48, and 72 hpi by evaluating the metabolic activity using a resazurin assay. At each time point, media were removed and replaced by 500 μL of resazurin (20% [v/v] in DMEM-10% FBS), and cells were further incubated for 4 h at 37°C. Metabolized media were transferred into a 96-well flat-bottom black plate and read (λ ex = 535 nm; λ em = 595 nm) using the multi-mode microplate reader (FilterMax F5). Results are expressed as a percentage of the control.
Real-time measure of the GFP fluorescence
U87MG and U87MG CXCR4+ cells were plated in a 24-well flat bottom plate (46,000 cells/well). After 24 h of monolayer culture, cells were infected with oHSV/gD or oHSV/Nb-gD (MOI: 0.1) and incubated in the Incucyte S3 for real-time analyses of the mean EGFP fluorescence intensity with the whole well module (magnification 4×).
Patient-derived GSCs were seeded in 96-well round bottom plate (100,000 cells/well) in stem cell medium. Twenty-four hours after seeding, tumorospheres were infected with oHSV/gD or oHSV/Nb-gD (104 PFUs/well) and incubated in the Incucyte S5 for a real-time analysis of the mean EGFP fluorescence intensity with the organoid module (magnification 4×).
Immunofluorescence staining on tumorospheres
Tumorospheres were infected with 106 PFUs/mL. Forty-eight hours post-infection, cells were washed and fixed with 4% paraformaldehyde for 20 min and incubated overnight with 20% PBS-sucrose before being embedded with colored OCT (Neg-50). Spheroids were cut into 5-μm-thick cryosections (Microm HM 560, Thermo Scientific) and placed onto SuperFrost slides (Thermo Scientific). Sections were permeabilized with 0.3% Triton X-100 PBS solution for 10 min, and unspecific binding sites were blocked with 5% BSA for 30 min. Tumorospheres sections were incubated overnight at 4°C with primary antibodies diluted in 5% BSA (rabbit anti-CXCR4 [ref. AB124824; Abcam; 1:200]; mouse anti-nestin [ref. sc-23927; Santa Cruz; 1:250]). After two washes, slides were incubated for 1 h at RT in the dark with secondary antibodies (goat anti-mouse Alexa Fluor 633 and goat anti-rabbit Alexa Fluor 568; 1:500). Nuclei were stained by incubation with Hoechst for 10 min at 1:50,000. Finally, Mowiol (Sigma) was added, and sections were covered by a coverslip. Images were recorded with Nikon A1R confocal microscope. Figures were composed and examined with ImageJ software.
Western blot assay
Cells were lysed with radioimmunoprecipitation assay (RIPA)-modified buffer (50 mM of Tris-HCl, 150 mM of NaCl, 1 mM of EDTA, 1% NP40, and 0.25% of DOC). Eighty micrograms of proteins were loaded on a 6% (for PARP and gD detection) or 12% (for caspase 3 and α-tubulin detection) SDS-acrylamide gel. After electrophoresis, proteins were transferred on a polyvinylidene fluoride (PVDF) membrane (GE Healthcare) according to standard protocols. Mouse anti-gD was used to determine viral infection level (ref. sc-21719; Santa Cruz; 1:1,000), and rabbit anti-PARP (ref. 9532; Cell Signaling; 1:1,000) and mouse anti-caspase 3 (CC3) (ref. ALX-804-305; Enzo, Life Sciences, Brussels, Belgium; 1:1,000) were used to detect the activation of the apoptotic pathway. Mouse anti-α-tubulin (ref. T6199; Sigma, 1:2,000) was used as loading control. Horseradish peroxidase (HRP)-conjugated anti-rabbit-immunoglobulin G (IgG) (ref. 7074; Cell Signaling) and HRP-conjugated anti-mouse-IgG (ref. 7076; Cell Signaling) were used as secondary antibodies. Signals were revealed using enhanced chemiluminescence (ECL) and imaged with LAS4000 charge-coupled device (CCD) camera (GE Healthcare).
In vivo experiments
Adult 6 weeks female immunodeficient Crl:NU-Foxn1nu mice (Charles River Laboratories, Brussels, Belgium) were used for xenograft experiments. The athymic nude mice were housed in sterilized, filter-topped cages at the Animal Facility at the University of Liège, and all experiments were performed as previously approved by the Animal Ethical Committee of the University of Liège, in accordance with the Declaration of Helsinki and following the guidelines of the Belgium Ministry of Agriculture in agreement with European Commission Laboratory Animal Care and Use Regulation. Intrastriatal grafts were performed following the previously described procedures.60 Briefly, 50,000 U87MG CXCR4+Luc+ cells resuspended in 2 μL of PBS were injected into the right striatum of mice previously anesthetized with an intraperitoneal injection of a Rompun (Sedativum 2%; Bayer, Brussels, Belgium) and Ketalar (ketamine 50 mg/mL, Pfizer, Brussels, Belgium) solution (V/V) prepared just before injection. Injection was performed according to stereotactic coordinates (0.5 mm anterior and 2.5 mm lateral from the bregma and at a depth of 3 mm), allowing a precise and reproducible injection site. Later, oncolytic viruses resuspended in 2 μL of PBS were injected, under similar anesthesia, within the tumor using the same stereotactic coordinates. Mice health status was evaluated daily, and mice were weighed regularly.
Bioluminescence activity
Immunodeficient nude mice bearing intracranial U87MG CXCR4+Luc+ xenografts were injected intraperitoneally with beetle luciferin potassium salt (ref. E1605; Promega; 150 mg/kg). Under anesthesia using 2.5% isoflurane, mice were imaged with a camera-based bioluminescence imaging system (Xenogen IVIS 50; exposure time 1 min, 15 min after intraperitoneal injection). Regions of interest were defined manually, and images were processed using Living Image and IgorPro Software (v.2.60.1). Raw data were expressed as total counts/s or total counts/min.
Brain tissue processing and tumor volume measurement
Mice were euthanized with intraperitoneal (i.p.) injection of Euthasol Vet (140 mg/kg) and intracardiac perfusion of ice-cold saline solution, followed by paraformaldehyde 4% in PBS (for histology). Brains were extracted, placed in sucrose 30% for tissue cryopreservation, and sectioned into 14-μm-thick serial sections using a cryostat. Tumor volume analysis was performed by immunohistochemistry for human vimentin detection (mouse anti-human vimentin; MAB3400; Merck; 1:200) with PolyviewPlus HRP-DAB kit (Enzo Life Sciences, Brussels, Belgium). Tumor was delineated based on anti-vimentin positivity. Ten to twelve serial brain sections were analyzed using the Mercator software (ExploraNova, La Rochelle, France). 3D reconstitution and extrapolation of tumor volume were performed using Map3D software.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 9. Data are displayed as mean ± SEM. Depending on the experiments, paired t test, Kruskal-Wallis, or two-way ANOVAs were performed as indicated in the figure legends. Statistical significance of survival assay was analyzed by log rank (Mantel-Cox) test.
Acknowledgments
J.S.G. and P.D. are, respectively, a PhD student and a PhD funded by the TELEVIE-FNRS; M.D. is a Research Fellow of the FNRS-Belgium; and A.L. is Clinical Post-doctoral Researcher of the FNRS-Belgium. This work was supported by grants from the National Fund for Scientific Research (FNRS, Télévie); the Special Funds of the University of Liège; the Leon Fredericq Foundation, Liège, Belgium; and the Neurological Foundation of New Zealand. The authors would like to thank Prof. A. Chiocca (University of Pittsburgh, USA) for fHsvQuik-1 BAC, Prof. G. Campadelli-Fiume (University of Bologna, Italy) for J1.1-2, J/A and J/C cells, A. Deward (GIGA, Université de Liège) for the graphical abstract and all the members of the GIGA Viral Vector, Imaging and Flow Cytometry, and Genomics platforms and animal facilities for valuable technical support.
Author contributions
Conception of the project, C.S.-D.; funding acquisition, B.R. and C.S.-D.; design of the experiments, J.S.G., V.N., A.L., N.C., M.L., and C.S.-D.; experiments, J.S.G., D.I., M.D., and P.D.; technical assistance, C.L., T.A., and B.B.; writing, C.S.-D. and J.S.G.; reviewing, P.D., V.N., B.R., A.L., M.L., and N.C.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omto.2022.06.002.
Supplemental information
Data and availability
All raw data are available upon request.
References
- 1.Feng Y., Broder C.C., Kennedy P.E., Berger E.A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 2.Alimohammadi M., Rahimi A., Faramarzi F., Alizadeh-Navaei R., Rafiei A. Overexpression of chemokine receptor CXCR4 predicts lymph node metastatic risk in patients with melanoma: a systematic review and meta-analysis. Cytokine. 2021;148:155691. doi: 10.1016/j.cyto.2021.155691. [DOI] [PubMed] [Google Scholar]
- 3.Salmaggi A., Gelati M., Pollo B., Marras C., Silvani A., Balestrini M.R., Eoli M., Fariselli L., Broggi G., Boiardi A. CXCL12 expression is predictive of a shorter time to tumor progression in low-grade glioma: a single-institution study in 50 patients. J. Neurooncol. 2005;74:287–293. doi: 10.1007/s11060-004-7327-y. [DOI] [PubMed] [Google Scholar]
- 4.Katsumoto K., Kume S. The role of CXCL12-CXCR4 signaling pathway in pancreatic development. Theranostics. 2013;3:11–17. doi: 10.7150/thno.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gagliardi F., Narayanan A., Reni M., Franzin A., Mazza E., Boari N., Bailo M., Zordan P., Mortini P. The role of CXCR4 in highly malignant human gliomas biology: current knowledge and future directions. Glia. 2014;62:1015–1023. doi: 10.1002/glia.22669. [DOI] [PubMed] [Google Scholar]
- 6.Santagata S., Ieranò C., Trotta A.M., Capiluongo A., Auletta F., Guardascione G., Scala S. CXCR4 and CXCR7 signaling pathways: a focus on the cross-talk between cancer cells and tumor microenvironment. Front. Oncol. 2021;11:591386. doi: 10.3389/fonc.2021.591386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gagner J.P., Sarfraz Y., Ortenzi V., Alotaibi F.M., Chiriboga L.A., Tayyib A.T., Douglas G.J., Chevalier E., Romagnoli B., Tuffin G., et al. Multifaceted C-X-C chemokine receptor 4 (CXCR4) inhibition interferes with anti–vascular endothelial growth factor therapy–induced glioma dissemination. Am. J. Pathol. 2017;187:2080–2094. doi: 10.1016/j.ajpath.2017.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gatti M., Pattarozzi A., Bajetto A., Würth R., Daga A., Fiaschi P., Zona G., Florio T., Barbieri F. Inhibition of CXCL12/CXCR4 autocrine/paracrine loop reduces viability of human glioblastoma stem-like cells affecting self-renewal activity. Toxicology. 2013;314:209–220. doi: 10.1016/j.tox.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Mercurio L., Ajmone-Cat M.A., Cecchetti S., Ricci A., Bozzuto G., Molinari A., Manni I., Pollo B., Scala S., Carpinelli G., Minghetti L. Targeting CXCR4 by a selective peptide antagonist modulates tumor microenvironment and microglia reactivity in a human glioblastoma model. J. Exp. Clin. Cancer Res. 2016;35:55. doi: 10.1186/s13046-016-0326-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gil M., Seshadri M., Komorowski M.P., Abrams S.I., Kozbor D. Targeting CXCL12/CXCR4 signaling with oncolytic virotherapy disrupts tumor vasculature and inhibits breast cancer metastases. Proc. Natl. Acad. Sci. U S A. 2013;110:E1291–E1300. doi: 10.1073/pnas.1220580110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scala S. Molecular pathways: targeting the CXCR4-CXCL12 Axis-Untapped potential in the tumor microenvironment. Clin. Cancer Res. 2015;21:4278–4285. doi: 10.1158/1078-0432.ccr-14-0914. [DOI] [PubMed] [Google Scholar]
- 12.Kioi M., Vogel H., Schultz G., Hoffman R.M., Harsh G.R., Brown J.M. Inhibition of vasculogenesis, but not angiogenesis, prevents the recurrence of glioblastoma after irradiation in mice. J. Clin. Invest. 2010;120:694–705. doi: 10.1172/jci40283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C.C., Lai J.H., Hueng D.Y., Ma H.I., Chung Y.C., Sun Y.Y., Tsai Y.J., Wu W.B., Chen C.L. Disrupting the CXCL12/CXCR4 axis disturbs the characteristics of glioblastoma stem-like cells of rat RG2 glioblastoma. Cancer Cell Int. 2013;13:85. doi: 10.1186/1475-2867-13-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostrom Q.T., Gittleman H., Truitt G., Boscia A., Kruchko C., Barnholtz-Sloan J.S. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2011-2015. Neuro. Oncol. 2018;20:1–86. doi: 10.1093/neuonc/noy131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stupp R., Taillibert S., Kanner A., Read W., Steinberg D.M., Lhermitte B., Toms S., Idbaih A., Ahluwalia M.S., Fink K., et al. Effect of tumor-treating fields plus maintenance temozolomide vs maintenance temozolomide alone on survival in patients with glioblastoma: a randomized clinical trial. JAMA. 2017;318:2306. doi: 10.1001/jama.2017.18718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lara-Velazquez M., Al-Kharboosh R., Jeanneret S., Vazquez-Ramos C., Mahato D., Tavanaiepour D., Rahmathulla G., Quinones-Hinojosa A. Advances in brain tumor surgery for glioblastoma in adults. Brain Sci. 2017;7:166. doi: 10.3390/brainsci7120166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 18.Galli R., Binda E., Orfanelli U., Cipelletti B., Gritti A., De Vitis S., Fiocco R., Foroni C., Dimeco F., Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.can-04-1364. [DOI] [PubMed] [Google Scholar]
- 19.Chen R., Nishimura M.C., Bumbaca S.M., Kharbanda S., Forrest W.F., Kasman I.M., Greve J.M., Soriano R.H., Gilmour L.L., Rivers C.S., et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17:362–375. doi: 10.1016/j.ccr.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 20.Singh S.K., Clarke I.D., Terasaki M., Bonn V.E., Hawkins C., Squire J., Dirks P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 21.Goffart N., Kroonen J., Di Valentin E., Dedobbeleer M., Denne A., Martinive P., Rogister B. Adult mouse subventricular zones stimulate glioblastoma stem cells specific invasion through CXCL12/CXCR4 signaling. Neuro. Oncol. 2015;17:81–94. doi: 10.1093/neuonc/nou144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goffart N., Lombard A., Lallemand F., Kroonen J., Nassen J., Di Valentin E., Berendsen S., Dedobbeleer M., Willems E., Robe P., et al. CXCL12 mediates glioblastoma resistance to radiotherapy in the subventricular zone. Neuro. Oncol. 2017;19:66–77. doi: 10.1093/neuonc/now136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dedobbeleer M., Willems E., Lambert J., Lombard A., Digregorio M., Lumapat P.N., Di Valentin E., Freeman S., Goffart N., Scholtes F., Rogister B. MKP1 phosphatase is recruited by CXCL12 in glioblastoma cells and plays a role in DNA strand breaks repair. Carcinogenesis. 2020;41:417–429. doi: 10.1093/carcin/bgz151. [DOI] [PubMed] [Google Scholar]
- 24.Cheray M., Bégaud G., Deluche E., Nivet A., Battu S., Lalloué F., et al. In: Glioblastoma. De Vleeschouwer S., editor. Chapter 4. Codon Publications; 2017. Cancer stem-like cells in glioblastoma; pp. 59–71. [PubMed] [Google Scholar]
- 25.Lombard A., Digregorio M., Delcamp C., Rogister B., Piette C., Coppieters N. The subventricular zone, a hideout for adult and pediatric high-grade glioma stem cells. Front. Oncol. 2020;10:614930. doi: 10.3389/fonc.2020.614930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang X., Zuo C., Fang P., Liu G., Qiu Y., Huang Y., Tang R. Targeting glioblastoma stem cells: a review on biomarkers, signal pathways and targeted therapy. Front. Oncol. 2021;11:701291. doi: 10.3389/fonc.2021.701291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fountzilas C., Patel S., Mahalingam D. Review: oncolytic virotherapy, updates and future directions. Oncotarget. 2017;8:102617–102639. doi: 10.18632/oncotarget.18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyauchi J.T., Tsirka S.E. Advances in immunotherapeutic research for glioma therapy. J. Neurol. 2018;265:741–756. doi: 10.1007/s00415-017-8695-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman G.K., Langford C.P., Coleman J.M., Cassady K.A., Parker J.N., Markert J.M., Yancey Gillespie G. Engineered herpes simplex viruses efficiently infect and kill CD133+ human glioma xenograft cells that express CD111. J. Neuro. Oncol. 2009;95:199–209. doi: 10.1007/s11060-009-9926-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rueger M.A., Winkeler A., Miletic H., Kaestle C., Richter R., Schneider G., Hilker R., Heneka M.T., Ernestus R.I., Hampl J.A., et al. Variability in infectivity of primary cell cultures of human brain tumors with HSV-1 amplicon vectors. Gene Ther. 2005;12:588–596. doi: 10.1038/sj.gt.3302462. [DOI] [PubMed] [Google Scholar]
- 31.Menotti L., Nicoletti G., Gatta V., Croci S., Landuzzi L., De Giovanni C., Nanni P., Lollini P.-L., Campadelli-Fiume G. Inhibition of human tumor growth in mice by an oncolytic herpes simplex virus designed to target solely HER-2-positive cells. Proc. Natl. Acad. Sci. U S A. 2009;106:9039–9044. doi: 10.1073/pnas.0812268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamiyama H., Zhou G., Roizman B. Herpes simplex virus 1 recombinant virions exhibiting the amino terminal fragment of urokinase-type plasminogen activator can enter cells via the cognate receptor. Gene Ther. 2006;13:621–629. doi: 10.1038/sj.gt.3302685. [DOI] [PubMed] [Google Scholar]
- 33.Zhou G., Roizman B. Construction and properties of a herpes simplex virus 1 designed to enter cells solely via the IL-13α2 receptor. Proc. Natl. Acad. Sci. U S A. 2006;103:5508–5513. doi: 10.1073/pnas.0601258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grandi P., Fernandez J., Szentirmai O., Carter R., Gianni D., Sena-Esteves M., Breakefield X.O. Targeting HSV-1 virions for specific binding to epidermal growth factor receptor-vIII-bearing tumor cells. Cancer Gene Ther. 2010;17:655–663. doi: 10.1038/cgt.2010.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shibata T., Uchida H., Shiroyama T., Okubo Y., Suzuki T., Ikeda H., Yamaguchi M., Miyagawa Y., Fukuhara T., Cohen J.B., et al. Development of an oncolytic HSV vector fully retargeted specifically to cellular EpCAM for virus entry and cell-to-cell spread. Gene Ther. 2016;23:479–488. doi: 10.1038/gt.2016.17. [DOI] [PubMed] [Google Scholar]
- 36.Alessandrini F., Menotti L., Avitabile E., Appolloni I., Ceresa D., Marubbi D., Campadelli-Fiume G., Malatesta P. Eradication of glioblastoma by immuno-virotherapy with a retargeted oncolytic HSV in a preclinical model. Oncogene. 2019;38:4467–4479. doi: 10.1038/s41388-019-0737-2. [DOI] [PubMed] [Google Scholar]
- 37.Jahan N., Lee J.M., Shah K., Wakimoto H. Therapeutic targeting of chemoresistant and recurrent glioblastoma stem cells with a proapoptotic variant of oncolytic herpes simplex virus. Int. J. Cancer. 2017;141:1671–1681. doi: 10.1002/ijc.30811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim M.-H., Billiar T.R., Seol D.-W. The secretable form of trimeric TRAIL, a potent inducer of apoptosis. Biochem. Biophys. Res. Commun. 2004;321:930–935. doi: 10.1016/j.bbrc.2004.07.046. [DOI] [PubMed] [Google Scholar]
- 39.Kock N., Kasmieh R., Weissledery R., Shah K. Tumor therapy mediated by lentiviral expression of shBcl-2 and S-TRAIL. Neoplasia. 2007;9:435–442. doi: 10.1593/neo.07223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tamura K., Wakimoto H., Agarwal A.S., Rabkin S.D., Bhere D., Martuza R.L., Kuroda T., Kasmieh R., Shah K. Multimechanistic tumor targeted oncolytic virus overcomes resistance in brain tumors. Mol. Ther. 2013;21:68–77. doi: 10.1038/mt.2012.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wakimoto H., Kesari S., Farrell C.J., Curry W.T., Zaupa C., Aghi M., Kuroda T., Stemmer-Rachamimov A., Shah K., Liu T.C., et al. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res. 2009;69:3472–3481. doi: 10.1158/0008-5472.can-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.WO2016156570A1- Bispecific CXCR4-CD4 polypeptides with potent anti-HIV activity (https://patents.google.com/patent/WO2016156570A1/en?oq=WO2016156570) Google Patents 2016.
- 43.Uchida H., Marzulli M., Nakano K., Goins W.F., Chan J., Hong C.-S., Mazzacurati L., Yoo J.Y., Haseley A., Nakashima H., et al. Effective treatment of an orthotopic xenograft model of human glioblastoma using an EGFR-retargeted oncolytic herpes simplex virus. Mol. Ther. 2013;21:561–569. doi: 10.1038/mt.2012.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uchida H., Chan J., Goins W.F., Grandi P., Kumagai I., Cohen J.B., Glorioso J.C. A double mutation in glycoprotein gB compensates for ineffective gD-dependent initiation of herpes simplex virus type 1 infection. J. Virol. 2010;84:12200–12209. doi: 10.1128/jvi.01633-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah K., Tung C.H., Yang K., Weissleder R., Breakefield X.O. Inducible release of TRAIL fusion proteins from a proapoptotic form for tumor therapy. Cancer Res. 2004;64:3236–3242. doi: 10.1158/0008-5472.can-03-3516. [DOI] [PubMed] [Google Scholar]
- 46.Cocchi F., Menotti L., Mirandola P., Lopez M., Campadelli-Fiume G. The ectodomain of a novel member of the Immunoglobulin subfamily related to the poliovirus receptor has the attributes of a bona fide receptor for herpes simplex virus types 1 and 2 in human cells. J. Virol. 1998;72:9992–10002. doi: 10.1128/jvi.72.12.9992-10002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uchida H., Shah W.A., Ozuer A., Frampton A.R., Goins W.F., Grandi P., Cohen J.B., Glorioso J.C., Glorioso J.C. Generation of herpesvirus entry mediator (HVEM)-Restricted herpes simplex virus type 1 mutant viruses: resistance of HVEM-expressing cells and identification of mutations that rescue nectin-1 recognition. J. Virol. 2009;83:2951–2961. doi: 10.1128/jvi.01449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frampton A.R., Stolz D.B., Uchida H., Goins W.F., Cohen J.B., Glorioso J.C. Equine herpesvirus 1 enters cells by two different pathways, and infection requires the activation of the cellular kinase ROCK1. J. Virol. 2007;81:10879–10889. doi: 10.1128/jvi.00504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marconi P., Manservigi R. Herpes simplex virus growth, preparation, and assay. Methods Mol. Biol. 2014;1144:19–29. doi: 10.1007/978-1-4939-0428-0_2. [DOI] [PubMed] [Google Scholar]
- 50.Lathia J.D., Mack S.C., Mulkearns-Hubert E.E., Valentim C.L.L., Rich J.N. Cancer stem cells in glioblastoma. Genes Dev. 2015;29:1203–1217. doi: 10.1101/gad.261982.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Connolly S.A., Jardetzky T.S., Longnecker R. The structural basis of herpesvirus entry. Nat. Rev. Microbiol. 2021;19:110–121. doi: 10.1038/s41579-020-00448-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bian X.w., Yang S.x., Chen J.h., Ping Y.f., Zhou X.d., Wang Q.l., Jiang X.f., Gong W., Xiao H.l., Du L.l., et al. Preferantial expression of chemokine receptor CXCR4 by highly malignant human gliomas and its association with poor patient survival. Neurosurgery. 2007;61:570–579. doi: 10.1227/01.neu.0000290905.53685.a2. [DOI] [PubMed] [Google Scholar]
- 53.Tang C., Guo W. Implantation of a mini-osmotic pump plus stereotactical injection of retrovirus to study newborn neuron development in adult mouse hippocampus. STAR Protoc. 2021;2:100374. doi: 10.1016/j.xpro.2021.100374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alvarez-Breckenridge C.A., Yu J., Price R., Wojton J., Pradarelli J., Mao H., Wei M., Wang Y., He S., Hardcastle J., et al. NK cells impede glioblastoma virotherapy through NKp30 and NKp46 natural cytotoxicity receptors. Nat. Med. 2012;18:1827–1834. doi: 10.1038/nm.3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Den Bossche W.B.L., Kleijn A., Teunissen C.E., Voerman J.S.A., Teodosio C., Noske D.P., Van Dongen J.J.M., Dirven C.M.F., Lamfers M.L.M. Oncolytic virotherapy in glioblastoma patients induces a tumor macrophage phenotypic shift leading to an altered glioblastoma microenvironment. Neuro. Oncol. 2018;20:1494–1504. doi: 10.1093/neuonc/noy082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Isci D., D’uonnolo G., Wantz M., Rogister B., Lombard A., Chevigné A., Szpakowska M., Neirinckx V. Patient-oriented perspective on chemokine receptor expression and function in glioma. Cancers. 2021;14:130. doi: 10.3390/cancers14010130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tischer B.K., Kaufer B.B., Sommer M., Wussow F., Arvin A.M., Osterrieder N. A self-excisable infectious bacterial artificial chromosome clone of varicella-zoster virus allows analysis of the essential tegument protein encoded by ORF9. J. Virol. 2007;81:13200–13208. doi: 10.1128/jvi.01148-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Todo T., Martuza R.L., Rabkin S.D., Johnson P.A. Oncolytic herpes simplex virus vector with enhanced MHC class I presentation and tumor cell killing. Proc. Natl. Acad. Sci. U S A. 2001;98:6396–6401. doi: 10.1073/pnas.101136398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goins W.F., Huang S., Hall B., Marzulli M., Cohen J.B., Glorioso J.C. In: Herpes Simplex Virus: Methods and Protocols, Methods Mol. Biol. Diefenbach R.J., Fraefel C., editors. Springer Science; 2020. Engineering HSV-1 vectors for gene therapy; p. 2060. Chapter 4. [DOI] [PubMed] [Google Scholar]
- 60.Kroonen J., Nassen J., Boulanger Y.G., Provenzano F., Capraro V., Bours V., Martin D., Deprez M., Robe P., Rogister B. Human glioblastoma-initiating cells invade specifically the subventricular zones and olfactory bulbs of mice after striatal injection. Int. J. Cancer. 2011;129:574–585. doi: 10.1002/ijc.25709. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data are available upon request.