Graphical abstract
Keywords: Genome-scale metabolic model, Cancer patient-specific metabolic model, Model extraction method, Model simulation method, Evaluation, Machine learning
Abbreviations: 1D CNN, one-dimensional convolutional neural network; E-Flux2, E-Flux method combined with minimization of L2norm; FBA, flux balance analysis; GEM, genome-scale metabolic model; GIMME, Gene Inactivity Moderated by Metabolism and Expression; GPR, gene-protein-reaction; LAD, least absolute deviation; MEM, model extraction method; MSM, model simulation method; pFBA, parsimonious flux balance analysis; SPOT, Simplified Pearson cOrrelation with Transcriptomic data; tINIT, task-driven Integrative Network Inference for Tissues; t-SNE, t-distributed stochastic neighbor embedding
Abstract
Genome-scale metabolic model (GEM) has been established as an important tool to study cellular metabolism at a systems level by predicting intracellular fluxes. With the advent of generic human GEMs, they have been increasingly applied to a range of diseases, often for the objective of predicting effective metabolic drug targets. Cancer is a representative disease where the use of GEMs has proved to be effective, partly due to the massive availability of patient-specific RNA-seq data. When using a human GEM, so-called context-specific GEM needs to be developed first by using cell-specific RNA-seq data. Biological validity of a context-specific GEM highly depends on both model extraction method (MEM) and model simulation method (MSM). However, while MEMs have been thoroughly examined, MSMs have not been systematically examined, especially, when studying cancer metabolism. In this study, the effects of pairwise combinations of three MEMs and five MSMs were evaluated by examining biological features of the resulting cancer patient-specific GEMs. For this, a total of 1,562 patient-specific GEMs were reconstructed, and subjected to machine learning-guided and biological evaluations to draw robust conclusions. Noteworthy observations were made from the evaluation, including the high performance of two MEMs, namely rank-based ‘task-driven Integrative Network Inference for Tissues’ (tINIT) or ‘Gene Inactivity Moderated by Metabolism and Expression’ (GIMME), paired with least absolute deviation (LAD) as a MSM, and relatively poorer performance of flux balance analysis (FBA) and parsimonious FBA (pFBA). Insights from this study can be considered as a reference when studying cancer metabolism using patient-specific GEMs.
1. Introduction
In studying cellular metabolism at a systems-level, a genome-scale metabolic model (GEM) has served as an important approach in the field of systems biology [[1], [2], [3]]. GEM is a stoichiometric computational model that contains information on entire metabolic genes and their corresponding proteins and reactions, and allows simulating genome-scale metabolic flux distributions under a specific genetic and environmental condition. With the availability of generic human GEMs [[4], [5], [6], [7]], GEMs started to be more increasingly applied to address various medical problems [[8], [9], [10], [11], [12], [13]]. A typical initial step of using a human GEM for a medical problem is to extract a context-specific GEM from a generic human GEM on the basis of omics data (often RNA-seq) from a target cell by using a model extraction method (MEM). The resulting context-specific GEM is subsequently simulated for various metabolic phenotypes (i.e., intracellular fluxes) via a model simulation method (MSM) [[14], [15], [16]]. Thus, successful application of a human GEM highly depends on both MEM and MSM.
At the moment, several MEMs and MSMs have been developed, which poses a technical challenge as to which combination would give the best prediction performance for a given human cell’s metabolism. MEMs have been comprehensively evaluated for studying cancer metabolism [[17], [18], [19], [20]], but, to the best of our knowledge, MSMs for human GEMs have not been systematically evaluated. For example, there was a recent study, arguing that metabolic fluxes can serve as fingerprints that can properly reflect distinct features of cancer types [21]. However, all the simulations were conducted using flux balance analysis (FBA), which motivates the use of other MSMs for possibly more accurate prediction of intracellular fluxes in cancer cells. Use of different combination of MEMs and MSMs is expected to heavily affect the simulation results from context-specific GEMs for various human cells.
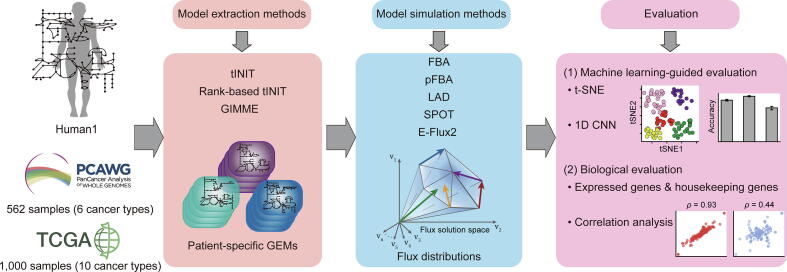
Thus, in this study, we systematically evaluated the effects of all pairwise combinations of three representative MEMs and five representative MSMs by examining biological features of the resulting cancer patient-specific GEMs (Fig. 1). Cancer was selected as a target disease because reprogrammed metabolism of cancer cells can be effectively addressed by using a human GEM; cancer cells show a wide spectrum of metabolic features that are specific to a tissue of origin [[22], [23], [24], [25]], and also share common metabolic features that are clearly distinct from normal cells [26]. To draw robust conclusions from the evaluation studies herein, a large volume of RNA-seq data and several different evaluation approaches, mainly machine learning, were considered; 562 samples from Pan-Cancer Analysis of Whole Genomes (PCAWG) [27] covering six cancer types, and 1,000 samples from The Cancer Genome Atlas (TCGA) covering 10 cancer types were considered in this study. Experimentally obtained 13C-metabolic flux data are also necessary for the rigorous evaluation of MEMs and MSMs [[28], [29]], but very few were available for cancers. To mitigate the limitation of this study to a certain extent, machine learning-guided and biological evaluations were newly devised, and applied to such a large volume of RNA-seq data. The evaluation studies revealed that ‘rank-based task-driven Integrative Network Inference for Tissues’ (rank-based tINIT) [5] or ‘Gene Inactivity Moderated by Metabolism and Expression’ (GIMME) [30], both MEMs, combined with least absolute deviation (LAD) as a MSM appeared to generate biologically most sound cancer patient-specific GEMs among all the pairwise combinations of MEMs and MSMs. This study also showed that FBA, still the most frequently used MSM, could not generate flux data that are sufficiently specific to a cancer type, and parsimonious FBA (pFBA) showed poorer performance than our initial expectation.
Fig. 1.
Workflow of the evaluation of model extraction and simulation methods using PCAWG and TCGA RNA-seq data.
2. Materials and methods
2.1. Data preparation
To reconstruct and simulate the patient-specific GEMs, a total of 1,562 personal RNA-seq data were obtained from the PCAWG Consortium [27] and TCGA (Supplementary Tables 1 and 2). The 562 PCAWG RNA-seq data represent six different cancer types, and the 1,000 TCGA RNA-seq data represent ten different cancer types. All these RNA-seq data correspond to primary tumors, and the samples from metastatic and recurrent tumors were not considered.
2.2. Generic human GEM
For a generic human GEM, Human1 version 1.6.0 was downloaded from a GitHub repository (https://github.com/SysBioChalmers/Human-GEM) [7]. Human1 is the most recent and comprehensive human GEM, which contains information on 13,082 reactions, 8,378 metabolites, and 3,625 genes.
2.3. Model extraction methods
All the MEMs process a generic human GEM and RNA-seq data as inputs, and generate a context-specific GEM by removing reactions associated with lowly expressed genes, and keeping reactions associated with highly expressed genes. In this process, reactions are given weight scores by subjecting gene expression values (Fragments per Kilobase of transcript per Million mapped reads, or FPKM in this study) to Boolean calculations of gene-protein-reaction (GPR) associations; reactions with negative weight scores are likely to be removed from a context-specific GEM (Supplementary Methods). MEMs can be categorized into three families, depending on the modeling logics [[17], [31]]: ‘integrative Metabolic Analysis Tool’ (iMAT)-like methods (e.g., iMAT, INIT, and tINIT), GIMME-like methods (e.g., GIMME, GIMMEp, and GIM3E), and ‘Model Building Algorithm’ (MBA)-like methods (e.g., MBA, mCADRE, FASTCORE and rFASTCORMICS). In this study, four MEMs were initially considered by selecting one representative method from each family (i.e., tINIT, GIMME and rFASTCORMICS), and additionally, rank-based tINIT, a modified version of tINIT [5]. All the MEMs were implemented in MATLAB R2018b (The Mathworks, Inc., Natick, MA). Using a computer with 2.30 GHz Intel Xeon CPU E5-2670 v3, tINIT and rank-based tINIT took around 90 min using 20 cores, and rFASTCORMICS required around 10 min using 10 cores. Finally, GIMME took around 35 s using a single core.
2.3.1. Task-driven integrative Network Inference for tissues (tINIT) and rank-based tINIT
Among the iMAT-like methods, tINIT is the most widely used method [32]. In the weight function of tINIT, a threshold for a gene expression level was set to be the median of all the metabolic gene expression levels for a single RNA-seq data. Rank-based tINIT is a slightly modified version that uses a rank-based weight function that minimizes the effects of outliers and sample variances by considering the rank of genes on the basis of their expression values [5]. tINIT was implemented through a GitHub repository (https://github.com/SysBioChalmers/Human-GEM), and two functions, getINITModel2 and scoreComplexModel, available for tINIT in this repository were modified to implement rank-based tINIT. A threshold for the gene rank was set to 0.25, which denotes metabolic genes ranked in the lowest 25 % of the expression levels; genes in the lowest 25 % are given negative weight scores from rank-based tINIT.
2.3.2. Gene Inactivity Moderated by metabolism and expression (GIMME)
GIMME minimizes the presence of reactions with lowly expressed genes, while keeping the growth rate beyond a certain value [30]. GIMME was implemented by using COBRA Toolbox v3.0 [33], and takes two parameters: a threshold for reaction weights, which was set to the median of all the reaction weights where the reaction weights were calculated from RNA-seq data by using the function mapExpressionToReactions in COBRA Toolbox; and a threshold for the allowed growth rate, which was set to 90 % of the maximum growth rate. Before running GIMME, specific constraints were first introduced to the generic Human1 model that reflect the composition of Ham’s medium (Supplementary Table 3); without the constraints, the reconstructed patient-specific GEMs did not show the growth under Ham’s medium.
2.3.3. rFASTCORMICS
The MBA-like methods use a predefined core set of reactions that should be retained and remove other remaining reactions as many as possible. Among the MBA-like methods, rFASTCORMICS [34] was selected in this study because it uses linear programming (LP) in contrast to other related methods (e.g., MBA and mCADRE) that use mixed-integer linear programming [35]. rFASTCORMICS directly uses gene expression values from RNA-seq data without using arbitrary thresholds, and discretizes the gene expression values to obtain core and non-core sets of reactions. rFASTCORMICS was implemented through a GitHub repository (https://github.com/sysbiolux/rFASTCORMICS). As with GIMME, constraints that reflect Ham’s medium were also provided to Human1 through the option optional_settings.medium because the resulting context-specific GEMs did not show the growth without these initial constraints.
2.4. Initial evaluation of the newly generated cancer patient-specific GEMs
For all the reconstructed patient-specific GEMs, their capability to grow in Ham’s medium was examined using FBA. Only the GEMs that showed the growth were considered for subsequent analyses. The patient-specific GEMs were also simulated to perform a total of 257 metabolic tasks, including 256 metabolic tasks previously defined by Uhlén et al. [36] using the checkTasks function of RAVEN Toolbox [37]; an additional task was additionally considered according to Robinson et al. [7] where the biosynthesis of vitamin C (ascorbate) should fail in the patient-specific GEMs. Finally, all the patient-specific GEMs were also evaluated using MEMOTE (metabolic model tests) [38].
2.5. Model simulation methods
The patient-specific GEMs were simulated using a total of five MSMs in this study: FBA [39], parsimonious FBA (pFBA) [40], least absolute deviation (LAD) [[41], [42], [43]], Simplified Pearson cOrrelation with Transcriptomic data (SPOT) [44], and E-Flux method combined with minimization of L2 norm (E-Flux2) [44]. These MSMs were run with constraints that reflect the Ham’s medium. Specifically, exchange reactions for main carbon sources in Ham’s medium were arbitrarily constrained with the maximum uptake rate of 10 mmol/gDCW/h, and exchange reactions for inorganic nutrients were set to have the maximum secretion and uptake rates of 1000 and −1000 mmol/gDCW/h, respectively. Uptakes of all the other nutrients were not allowed.
A threshold for a reaction flux to be considered as zero was set differently for each combination of the MEMs and the MSMs (Supplementary Table 4). A reason behind setting these threshold values is that the number of flux-carrying fluxes was significantly different across the MSMs; for example, 88 % of reactions in the patient-specific GEMs were predicted to carry fluxes according to SPOT and E-Flux2, which seems biologically unrealistic. Flux values at which the density of flux distributions rapidly decreases were set as the thresholds for each combination of MEM and MSM (Supplementary Figs. 1-6).
Using a computer with 2.30 GHz Intel Xeon CPU E5-2670 v3, implementation of the MSMs took a few seconds and up to around 1 min. All the MSMs were implemented by using Python 3.6 with Gurobi Optimizer 9.0.2 (Gurobi Optimization, LLC, Beaverton, OR) in the Linux Ubuntu environment. Reading, writing and manipulation of the COBRA-compliant MATLAB files were implemented using COBRApy 0.22.1 [45].
2.5.1. Flux balance analysis (FBA)
FBA is the most fundamental method to simulate a GEM that predicts intracellular flux values, based on the mass balance of metabolites, and the definition of an objective function and constraints that shape a cell’s metabolic objective [39] (Eq. S4 in Supplementary Methods). In this study, three different objective functions were considered, including biomass formation, energy (i.e., ATP) production, and reducing power (i.e., NADPH and NADH) production (Supplementary Table 5). The energy and reducing power productions were considered only for the TCGA samples.
2.5.2. Parsimonious flux balance analysis (pFBA)
pFBA is a variation of FBA that was coined to accurately predict intracellular flux values by minimizing a total sum of reaction fluxes [40]. The rationale behind pFBA is that cells attempt to efficiently (or minimally) use their energy to meet their metabolic objective. (Eq. S5 in Supplementary Methods) was relaxed to 99.999 % of an optimal solution (i.e., biomass formation, energy production, or reducing power production) in order to avoid infeasible solution.
2.5.3. Least absolute deviation (LAD)
As with pFBA, LAD also attempts to accurately predict intracellular flux values, often by using omics data (e.g., RNA-seq). In LAD, the Manhattan distance is minimized between a reference flux set (i.e., reaction weights) and a set of fluxes to be calculated [[42], [43]], and thus, theoretically, LAD is equivalent to linearized ‘minimization of metabolic adjustment’ (MOMA) [41], which attempts to accurately predict intracellular fluxes in genetically perturbed metabolism via gene knockout [46]. LAD can be a good alternative when a cellular objective is not clear, such as in normal human cells, as it does not require defining a cellular objective function (e.g., maximizing biomass formation rate) to predict fluxes. To predict more realistic fluxes in this study, the biomass formation rate was constrained to be greater than or equal to , which was set to 0.01 in this study (Eq. S6 in Supplementary Methods). In this study, so-called ‘the modified LAD’ was implemented as discussed in Supplementary Notes, which can generate reasonably accurate flux data in a short calculation time as a result of a slight mathematical modification of the original LAD. LAD hereafter refers to the modified LAD.
2.5.4. Simplified Pearson cOrrelation with Transcriptomic data (SPOT)
SPOT also attempts to predict flux values in accordance with omics data, but by maximizing cosine similarity between a reference flux set (i.e., reaction weights) and a set of fluxes to be calculated (Eq. S7 in Supplementary Methods).
2.5.5. E-Flux method combined with minimization of L2 norm (E-Flux2)
E-Flux2 also uses omics data (e.g., RNA-seq) to predict intracellular fluxes, but additionally modifies the upper and lower bounds of fluxes; greater and smaller values are given to the upper and lower bounds for reactions with highly expressed genes. was relaxed to 99.999 % of the optimal biomass formation rate for the modified constraints in order to avoid sub-optimal or infeasible solution (Eq. S8 in Supplementary Methods).
2.6. Evaluation of model extraction and model simulation methods
2.6.1. t-SNE
Reaction contents and flux data from the patient-specific GEMs were visualized using t-distributed stochastic neighbor embedding (t-SNE) [47] to cluster the GEMs according to their cancer type. For MEMs, reaction content for each GEM was prepared as an input binary vector, indicating the presence and absence of a reaction as ‘1’ and ‘0’, respectively. For MSMs, flux values of reactions were standardized (the mean of ‘0’ and the standard deviation of ‘1’). When evaluating the MSMs using t-SNE, reactions absent in at least one patient-specific GEM were excluded from input flux data. For hyperparameters, ‘number of principal components’ and ‘perplexity’ were both set to be 20 after examining 10, 20, and 30 for both hyperparameters in pairwise combinations.
2.6.2. Convolutional neural network (CNN)
One-dimensional convolutional neural network (1D CNN) was used to classify the patient-specific GEMs to cancer types based on their reaction contents or flux data. 1D CNN uses the same types of input data (i.e., the binary vector of a reaction content and the standardized flux data) prepared for t-SNE, but additional preprocessing took place. For both types of input data, reactions in the patient-specific GEMs were first sorted according to their subsystem in order to capture meaningful features associated with each subsystem. For the flux data, they were first quantile-normalized [48] in order to remove any effects caused by large variations in the uptake rate of metabolites; quantile-normalized flux data were subsequently standardized. As with the input of flux data for t-SNE, reactions absent in at least one GEM were excluded from an input for 1D CNN.
The prepared datasets (Supplementary Table 6) were subjected to the stratified 10-fold cross-validation to develop 1D CNN. The dataset was first divided into 10 folds (i.e., outer loop) where the nine folds (i.e., inner loop) provided training and validation sets, and the remining single fold was used as a test set. This split was implemented by using the function StratifiedKFold from scikit-learn [49] to consider the relative sample size of each cancer type within each fold. An inner loop was further split into training and validation sets at a ratio of 8:1 by using the function StratifiedShuffleSplit from scikit-learn. This validation set was used to select the best model among the models generated at each epoch by using callbacks of Keras v2.3.0 [50]. The best model selected from an inner loop was further evaluated using a test set. This procedure was repeated ten times. It should be noted that the inner loops were not subjected to the cross-validation.
The input vector is directly fed into a 1D convolutional layer with L2 regularization and a rectified linear unit (ReLU) as an activation function. The convolutional layer used 32 filters, all having the same size of 1 × 30, which is equal to the average number of reactions in subsystems of Human1. Stride was set to 10. Output of the convolutional layer is subsequently passed to a max pooling layer, a fully connected layer with dropout rate of 0.5, and a softmax output layer. As a result, 1D CNN generates a probability distribution of six cancer types for input data from the PCAWG samples, and ten cancer types for input data from the TCGA samples (Supplementary Fig. 7). Performance of 1D CNN was also examined using a total of 27 different sets of hyperparameters: strides of 5, 10 and 20; 16, 32 and 64 filters; and kernel sizes of 15, 30 and 60. Each model was trained using Adam optimizer [51] with a batch size of 32 for 200 epochs with early stopping set at 20 epochs. 1D CNN models were developed using Python package Keras v2.3.0 [50] with TensorFlow backend [52] v1.14.0.
2.6.3. Preparation of gene expression data and housekeeping genes
‘Biological evaluation’ conducted in this study examines reactions in the patient-specific GEMs in comparison with 1) gene expression data from corresponding tissues of origin, and 2) housekeeping genes. Comparison with expressed genes from each tissue of origin is based on a previous finding that normal and cancer tissues still share tissue-specific metabolic features [[22], [23], [24], [25]]. For this, a set of expressed genes in a tissue (TPM 1 in each tissue of origin) was obtained from gene expression data available at Human Protein Atlas (HPA) [36]. The collected gene expression data cover the following ten tissues in accordance with cancer types considered in this study: brain, breast, colon, kidney, liver, lung, lymph node, ovary, pancreas, and urinary bladder. A full list of housekeeping genes was also obtained from HPA. Out of 8,839 housekeeping genes from HPA, 2,055 housekeeping genes appeared to be reflected in Human1, which correspond to 5,376 housekeeping reactions.
2.6.4. Spearman’s correlation coefficient
Spearman’s correlation coefficient (Spearman’s ρ) was computed using SciPy v1.5.4 [53] for the predicted flux data from pairwise combinations of the patient-specific GEMs. Reactions were removed from each model before computing Spearman’s ρ if their flux values were equal to zero or absent in both models. This correlation analysis is another part of the biological evaluation conducted in this study.
3. Results
3.1. Reconstruction of cancer patient-specific GEMs for 562 PCAWG samples using four different model extraction methods
Cancer patient-specific GEMs were reconstructed using 562 PCAWG RNA-seq data for six cancer types (Supplementary Tables 1 and 2), and the most recently developed generic human GEM ‘Human1′ as a template model [7]. In this study, tINIT [32], rank-based tINIT [5], GIMME [30] and rFASTCORMICS [34] were used to reconstruct a patient-specific GEM for each RNA-seq data. As a result, a total of 2,046 patient-specific GEMs were generated; 561 GEMs using tINIT, 562 GEMs using rank-based tINIT, 562 GEMs using GIMME, and 361 GEMs using rFASTCORMICS (Supplementary Table 2). Based on these reconstructions, rFASTCORMICS was no longer considered in this study because its reconstruction success rate (361 out of 562 RNA-seq data) was much lower than the other three methods (Supplementary Fig. 8). For both tINIT and rFASTCORMICS, failed reconstruction of the patient-specific GEMs was caused by infeasibility of the optimization problems.
The patient-specific GEMs were first evaluated for growth capability, metabolic tasks and MEMOTE [38] (Materials and methods). First, it was confirmed that all the patient-specific GEMs showed the growth under Ham’s medium. For the metabolic tasks, four patient-specific GEMs from rank-based tINIT and one GEM from GIMME were discarded as these five GEMs completed substantially fewer metabolic tasks than other GEMs (Supplementary Fig. 9); the remaining 1,680 patient-specific GEMs (Supplementary Table 1) built using tINIT, rank-based tINIT or GIMME successfully completed the averages of 206, 214 and 225 metabolic tasks, respectively. Also, according to MEMOTE, the 1,680 patient-specific GEMs showed high consistencies overall: on average, 99.47 % reactions mass-balanced, 100 % reactions charge-balanced and 100 % stoichiometric consistency.
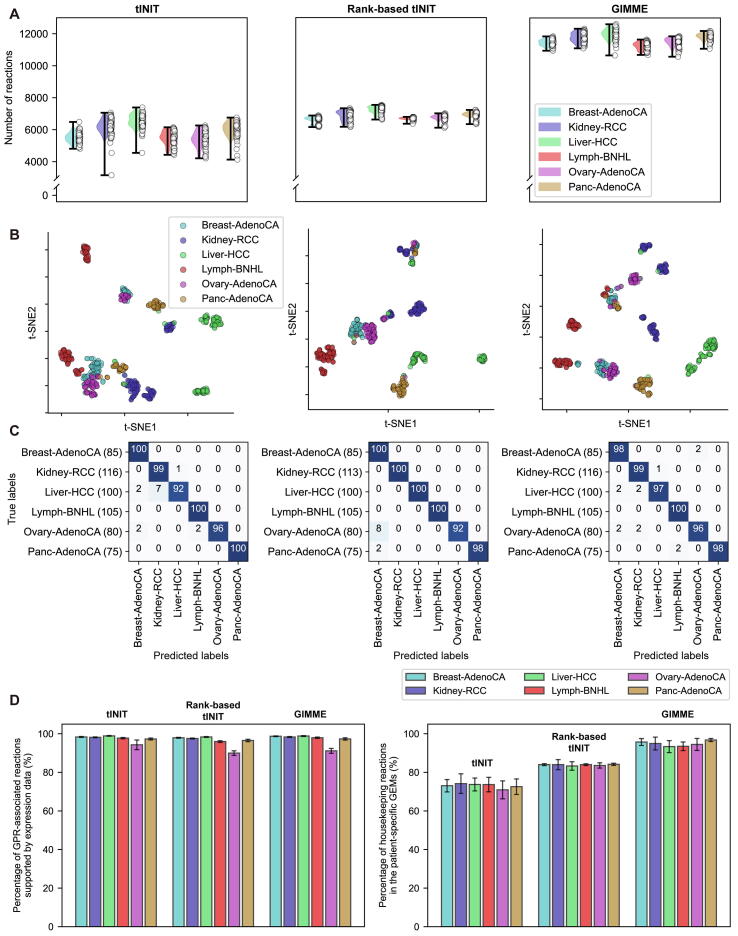
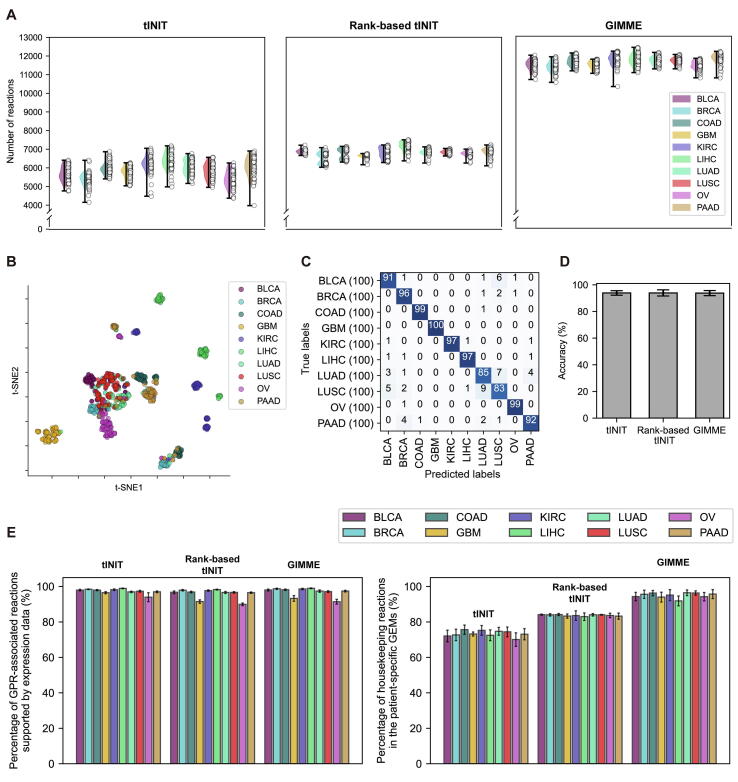
The average number of reactions in the patient-specific GEMs for each cancer type appeared to be heavily affected by the MEMs. GIMME overall generated the patient-specific GEMs with the greatest number of reactions, while the GEMs from rank-based tINIT showed the smallest variations in the number of reactions (Fig. 2A). However, the relative average number of reactions across the six cancer types appeared to be consistent throughout the three MEMs, for example liver cancer patient-specific GEMs having the greatest number of reactions (Liver-HCC in Fig. 2A) and blood cancer patient-specific GEMs with the lowest number of reactions (Lymph-BNHL in Fig. 2A).
Fig. 2.
Evaluation results of the three model extraction methods (MEMs). (A) Statistics of the cancer patient-specific genome-scale metabolic model (GEMs) built using the three MEMs and 562 PCAWG RNA-seq data. (B) t-SNE plots of the reaction contents of the patient-specific GEMs. (C) Classification of the patient-specific GEMs into six cancer types by using 1D CNN. Values in each confusion matrix correspond to the averaged percentage of cancer type predictions (i.e., true labels versus predicted labels); greater values in a diagonal indicate that the true and predicted labels are more consistent. The averaged percentage was obtained from 10 runs of 1D CNN using each fold of the 10 test datasets (Materials and methods). In (B) and (C), the evaluation results are presented using the patient-specific GEMs that were reconstructed using tINIT (left), rank-based tINIT (middle), and GIMME (right). (D) (left) Percentage of GPR-associated reactions supported by the expressed genes in each corresponding tissue of origin, among all the GPR-associated reactions in each patient-specific GEM. (right) Percentage of housekeeping reactions incorporated in each patient-specific GEM among all the known housekeeping reactions. Gene expression data and housekeeping genes were obtained from Human Protein Atlas (HPA).
3.2. Machine learning-guided and biological evaluation of the model extraction methods
The three MEMs (i.e., tINIT, rank-based tINIT and GIMME) were evaluated by using t-SNE and 1D CNN to examine to what extent these methods can generate biologically distinct patient-specific GEMs for each cancer type. To prepare input data for t-SNE and 1D CNN, reaction contents of the patient-specific GEMs were converted to binary values, indicating the presence and absence of a reaction as ‘1’ and ‘0’, respectively (Materials and methods). Implementation of t-SNE for three binary vectors, each corresponding to tINIT, rank-based tINIT and GIMME, showed that, overall, the patient-specific GEMs from all the MEMs could be clustered in accordance with cancer types (Fig. 2B). Also, 1D CNN models trained with the binary vectors from the three MEMs were able to classify the patient-specific GEMs to their corresponding cancer types at high accuracies: on average, 97.6 % for the patient-specific GEMs from tINIT, 98.5 % for rank-based tINIT, and 97.9 % for GIMME, respectively. The accuracy was calculated by dividing the sum of diagonal values in a confusion matrix (Fig. 2C) by the sum of entire values.
To more rigorously examine the MEMs in terms of biology, reactions in the patient-specific GEMs were examined in comparison with a set of expressed genes from corresponding tissues of origin as well as housekeeping genes; all the gene expression data were obtained from HPA [36]. For this biological evaluation, reactions that correspond to the expressed genes from tissues of origin and housekeeping genes were first identified through GPR associations (Materials and methods). Housekeeping genes are constitutively expressed in all the cell types in an organism [[36], [54]], and thus, their corresponding housekeeping reactions should be available in all the patient-specific GEMs. As expected, high percentages of reactions appeared to be present in the patient-specific GEMs, which are associated with the genes expressed in corresponding tissues, and the results did not seem to be affected by the MEMs (left graph in Fig. 2D). However, percentages of the housekeeping reactions in the patient-specific GEMs were affected by the MEMs (right graph in Fig. 2D); the patient-specific GEMs from GIMME included the greatest percentage of the housekeeping reactions (94.6 % on average), followed by rank-based tINIT (83.8 %) and tINIT (73.1 %). GIMME usually keeps a greater number of reactions in a context-specific GEM (Fig. 2A), which might have contributed to the inclusion of more housekeeping reactions than the other two methods.
Taken together, tINIT, rank-based tINIT and GIMME all appeared to have generated the patient-specific GEMs with acceptable qualities as a result of machine learning-guided and biological evaluations (i.e., classification of the patient-specific GEMs at an average accuracy of greater than 97 % by using 1D CNN; and high percentage of reactions supported by the expressed genes from tissues of origin and housekeeping genes).
3.3. Evaluation of the model simulation methods using machine learning
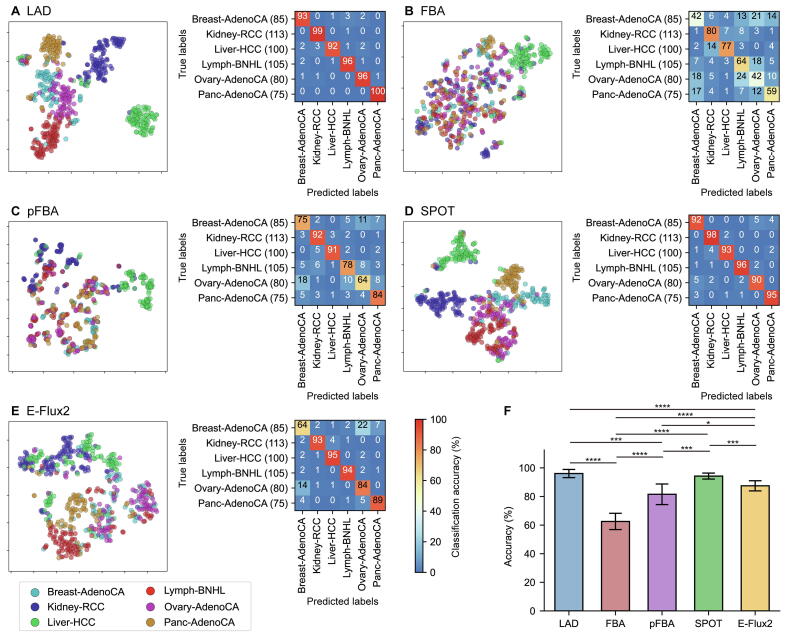
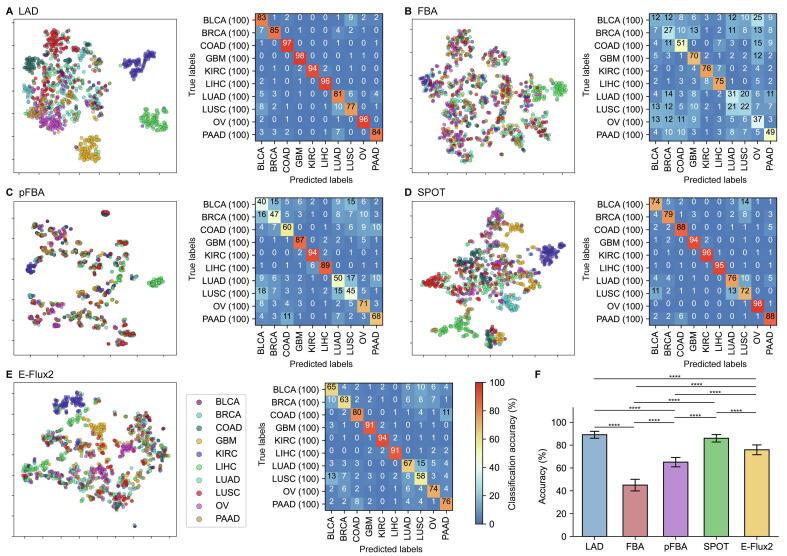
Next, five MSMs (i.e., FBA [39], pFBA [40], LAD [[41], [42], [43]], SPOT [44], and E-Flux2 [44]) were examined by using the patient-specific GEMs constructed above (Supplementary Tables 1 and 2). Here, the five MSMs were implemented in combination with each of the three MEMs, and their prediction results were evaluated by using machine learning. For machine learning-guided evaluation, a total of 8,398 flux data were used as input for t-SNE and 1D CNN; 8,398 flux data were prepared by simulating the 1,680 patient-specific GEMs using each of the five MSMs; two flux data from a combination of tINIT and E-Flux2 were not obtained due to numerical difficulties. First, the patient-specific GEMs were clustered by t-SNE [47] on the basis of their flux data from each MSM (left plots in Fig. 3A-E, and left plots in Supplementary Figs. 10A-E and 11A-E). Overall, LAD and SPOT generated more distinct clusters than the other three MSMs (i.e., FBA, pFBA, and E-Flux2) regardless of the MEMs used.
Fig. 3.
Evaluation results of the five model simulation methods combined with rank-based tINIT for the PCAWG samples. (A-E) (left) t-SNE plots, and (right) confusion matrices showing the classification results of 1D CNN for flux data from the 558 patient-specific GEMs. Values in each confusion matrix correspond to the averaged percentage of cancer type predictions (i.e., true labels versus predicted labels); greater values in a diagonal indicate that the true and predicted labels are more consistent. The averaged percentage was obtained from 10 runs of 1D CNN using each fold of the 10 test datasets (Materials and methods). Flux data of the 558 patient-specific GEMs were generated using (A) LAD, (B) FBA, (C) pFBA, (D) SPOT, and (E) E-Flux2. (A-E) Figures for quantitative analysis of the t-SNE plots are available at https://doi.org/10.6084/m9.figshare.19810927.v1. (F) The mean classification accuracies and standard deviations from 10 runs of 1D CNN. The accuracy was calculated by dividing the sum of diagonal values in a confusion matrix (A-E) by the sum of entire values. P values were calculated by using two-sided Welch’s t-test (*P < 0.05, ***P < 0.001, and ****P < 0.0001).
1D CNN was also used to evaluate the MSMs, which classified the patient-specific GEMs into six cancer types (confusion matrices in Fig. 3A-E, and confusion matrices in Supplementary Figs. 10A-E and 11A-E). In contrast to the MEM evaluation using 1D CNN, 1D CNN models were trained using 15 different flux data generated from pairwise combinations of the three MEMs and the five MSMs (Materials and methods). As a result, with the GEMs from rank-based tINIT, LAD, SPOT and E-Flux2 showed relatively high classification accuracies, on average, 97.3 %, 95.3 % and 87.8 %, respectively, while FBA and pFBA showed lower classification accuracies (Fig. 3F). These results are somewhat consistent with the clustering results from t-SNE where LAD, SPOT, and E-Flux2 generated more distinct clusters. If tINIT and GIMME were additionally considered, a combination of rank-based tINIT and LAD (average accuracy of 97.3 %) as well as a combination of GIMME and LAD (average accuracy of 98.0 %) generated the best prediction results (Fig. 3F, and Supplementary Figs. 10F and 11F). It was interesting to note that pFBA was initially thought to outperform other MSMs according to a previous study [28], but showed moderate prediction performance; pFBA showed better predictions when combined with tINIT and GIMME than rank-based tINIT (Fig. 3F, and Supplementary Figs. 10F and 11F).
3.4. Biological evaluation of the five model simulation methods
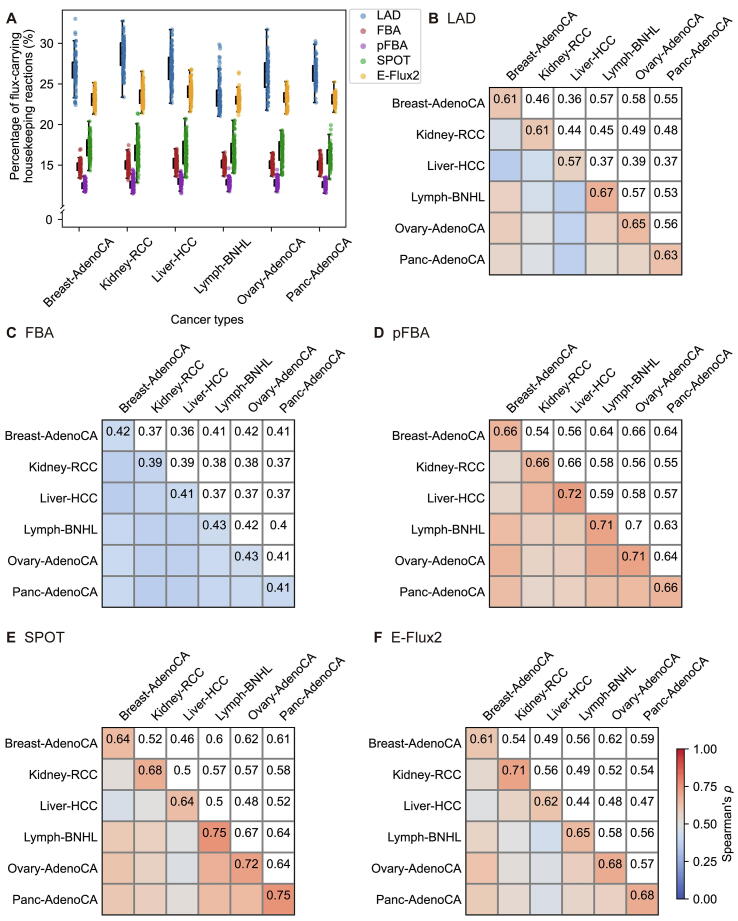
As with the MEMs, the patient-specific GEMs were also evaluated with respect to the set of expressed genes from corresponding tissues of origin as well as the housekeeping genes, both from HPA [36]. First, most of the patient-specific GEMs from all pairwise combinations of the MEMs and the MSMs had greater than 90 % of their flux-carrying reactions that were associated with the expressed genes from corresponding tissues of origin (Supplementary Fig. 12). For the housekeeping reactions, which should carry fluxes to perform essential functions, LAD showed the best results (i.e., the greatest percentage of flux-carrying reactions among all the housekeeping reactions in the patient-specific GEMs) with rank-based tINIT and GIMME across the six cancer types, and E-Flux2 additionally performed good when combined with tINIT (Fig. 4A, and Supplementary Figs. 13A and 14A). In contrast, FBA and pFBA overall showed poor results for the patient-specific GEMs from the three MEMs. It should be noted that, among all the known housekeeping reactions, more than 70 % of them were included in the patient-specific GEMs (right graph in Fig. 2D), but only <40 % of them carried fluxes (Fig. 4A, and Supplementary Figs. 13A and 14A). This high discrepancy may be largely due to the limited prediction capacity of the currently available MSMs; reaction fluxes are highly dependent on objective functions and constraints that do not necessarily consider housekeeping reactions.
Fig. 4.
Biological evaluation results of the five model simulation methods combined with rank-based tINIT for the PCAWG samples. (A) Percentage of the housekeeping reactions that carry fluxes among all the housekeeping reactions in the patient-specific GEMs. (B-F) Spearman’s correlation coefficients (Spearman’s ρ) between two flux data generated from all pairwise combinations of the patient-specific GEMs. Values in each cell represent the average of Spearman’s ρ. Flux data of the 558 patient-specific GEMs were generated using (B) LAD (P < 0.0001; mean difference of 0.14), (C) FBA (P < 0.001; mean difference of 0.028), (D) pFBA (P < 0.0001; mean difference of 0.081), (E) SPOT (P < 0.0001; mean difference of 0.13), and (F) E-Flux2 (P < 0.0001; mean difference of 0.13). P value for each matrix (B-F) was calculated by using one-sided Welch’s t-test, which indicates that the diagonal values are significantly greater than the off-diagonal values. The mean difference for each matrix (B-F) refers to the difference between the mean of the diagonal values and the mean of the off-diagonal values.
Finally, Spearman’s ρ were calculated between two flux data generated from all pairwise combinations of the patient-specific GEMs built with rank-based tINIT. Ideally, the patient-specific GEMs from two different cancer types should have lower Spearman’s ρ, whereas the GEMs from the same cancer type should have higher values. As expected, use of all the MSMs resulted in lower Spearman’s ρ for the patient-specific GEMs from two different cancer types than the GEMs from the same cancer type (Fig. 4B-F); however, this trend was less clear for FBA according to ‘the mean difference’ (defined in Fig. 4B-F). FBA, in contrast to the other four MSMs, showed relatively similar Spearman’s ρ across all pairwise combinations of the patient-specific GEMs regardless of cancer types (Fig. 4C). Combinations of MSMs with tINIT or GIMME also showed similar results as rank-based tINIT (Supplementary Figs. 13B-F and 14B-F).
3.5. Evaluation of the model extraction and simulation methods using TCGA samples
To confirm whether the observations made using the PCAWG samples are robust, 1,000 RNA-seq data across ten cancer types from TCGA were additionally considered in this study. First, a total of 2,997 cancer patient-specific GEMs were reconstructed using the three MEMs (i.e., tINIT, rank-based tINIT, and GIMME); three out of 3,000 patient-specific GEMs could not be developed using tINIT (Supplementary Table 2). As expected, GIMME-generated GEMs had much greater numbers of reactions, and rank-based tINIT generated the GEMs with the smallest variations in the number of reactions (Fig. 5A). Except for one patient-specific GEM from GIMME (Supplementary Figs. 15 and 16), all the other patient-specific GEMs built using tINIT, rank-based tINIT and GIMME successfully completed a high number of metabolic tasks: i.e., averages of 208, 213, and 225 metabolic tasks, respectively. Also, according to MEMOTE, the 2,996 patient-specific GEMs showed high consistencies: on average, 99.48 % reactions mass-balanced, 100 % reactions charge-balanced and 100 % stoichiometric consistency. As with the PCAWG samples, all the three MEMs generated distinct clusters overall, depending on a cancer type, as a result of implementing t-SNE (Fig. 5B and Supplementary Fig. 17). Also, all the patient-specific GEMs were well classified into their corresponding cancer types via 1D CNN (Fig. 5C,D and Supplementary Fig. 17). As to the presence of reactions that correspond to the expressed genes in corresponding tissues of origin as well as housekeeping genes, patterns observed with the PCAWG samples (Fig. 2D) were consistently observed from the TCGA samples (Fig. 5E).
Fig. 5.
Evaluation results of the three model extraction methods (MEMs) using the TCGA samples. (A) Statistics of the cancer patient-specific GEMs built using the three MEMs and 1,000 TCGA RNA-seq data. (B) t-SNE plots of the reaction contents of the patient-specific GEMs. (C) Classification of the patient-specific GEMs into ten cancer types by using 1D CNN. Values in the confusion matrix correspond to the averaged percentage of cancer type predictions (i.e., true labels versus predicted labels); greater values in a diagonal indicate that the true and predicted labels are more consistent. The averaged percentage was obtained from 10 runs of 1D CNN using each fold of the 10 test datasets (Materials and methods). In (B) and (C), the data are presented only for rank-based tINIT, and those from tINIT and GIMME are available in Supplementary Fig. 17. (D) The mean classification accuracies and standard deviations from 10 runs of 1D CNN for the patient-specific GEMs built with tINIT, rank-based tINIT, and GIMME. The accuracy was calculated by dividing the sum of diagonal values in a confusion matrix (C) by the sum of entire values. (E) (left) Percentage of GPR-associated reactions supported by the expressed genes in each corresponding tissue of origin, among all the GPR-associated reactions in each patient-specific GEM. (right) Percentage of housekeeping reactions incorporated in each patient-specific GEM among all the known housekeeping reactions. Gene expression data and housekeeping genes were obtained from Human Protein Atlas (HPA).
Subsequently, flux data were generated from all the 2,992 patient-specific GEMs by using the five MSMs (i.e., LAD, FBA, pFBA, SPOT, and E-Flux2) for further analysis using machine learning (Fig. 6, and Supplementary Figs. 18 and 19); four flux data from a combination of E-Flux2 and tINIT were additionally discarded at this stage because they could not generate optimal solutions (Supplementary Table 2). When t-SNE was applied to these flux data, LAD and SPOT generated more distinct clusters with respect to cancer types than FBA and pFBA (left plots in Fig. 6A-E, and left plots in Supplementary Fig. 19A-E). Clusters from the five MSMs were overall slightly less distinct when tINIT was used (left plots in Supplementary Figs. 18A-E). Using 1D CNN for all the three MEMs, LAD and SPOT consistently showed the best classification performances, while FBA showed the worst performance, and pFBA and E-Flux2 showed moderate performances (confusion matrices in Fig. 6A-E, and confusion matrices in Supplementary Figs. 18A-E and 19A-E). In particular, LAD combined with rank-based tINIT (average accuracy of 89.1 %; Fig. 6F) or GIMME (average accuracy of 89.9 %; Supplementary Fig. 19F) generated the best prediction results. Despite the use of rank-based tINIT, FBA still generated the worst prediction results (average accuracy of 45.0 %; Fig. 6F). To examine whether FBA and pFBA would show improved prediction results by using different objective functions, maximizing energy (i.e., ATP) production and maximizing reducing power (i.e., NADPH and NADH) production were additionally considered for the TCGA samples. However, FBA or pFBA with these new objective functions did not outperform FBA or pFBA with maximization of biomass formation according to 1D CNN (Supplementary Fig. 20).
Fig. 6.
Evaluation results of the five model simulation methods combined with rank-based tINIT for the TCGA samples. (A-E) (left) t-SNE plots, and (right) confusion matrices showing the classification results of 1D CNN for flux data from the 1,000 patient-specific GEMs. Values in each confusion matrix correspond to the averaged percentage of cancer type predictions (i.e., true labels versus predicted labels); greater values in a diagonal indicate that the true and predicted labels are more consistent. The averaged percentage was obtained from 10 runs of 1D CNN using each fold of the 10 test datasets (Materials and methods). Flux data of the 1,000 patient-specific GEMs were generated using (A) LAD, (B) FBA, (C) pFBA, (D) SPOT, and (E) E-Flux2. (A-E) Figures for quantitative analysis of the t-SNE plots are available at https://doi.org/10.6084/m9.figshare.19810927.v1. (F) The mean classification accuracies and standard deviations from 10 runs of 1D CNN. The accuracy was calculated by dividing the sum of diagonal values in a confusion matrix (A-E) by the sum of entire values. P values were calculated by using two-sided Welch’s t-test (****P < 0.0001). (A and C) An outlier was removed from each t-SNE plot for better presentation of data points.
Finally, the flux data from the patient-specific GEMs built using the TCGA RNA-seq data were subjected to the biological evaluation. First, the flux data generated from LAD and E-Flux2 showed greater percentages of flux-carrying housekeeping reactions than the other three MSMs (Supplementary Fig. 21), which was consistent with the overall patterns observed with the PCAWG samples (Fig. 4A, and Supplementary Figs. 13A and 14A). Next, all the MSMs showed lower Spearman’s ρ for the patient-specific GEMs from two different cancer types than the GEMs from the same cancer type, regardless of the MEMs used (Supplementary Figs. 22–24). In particular, Spearman’s ρ differences as a function of cancer types were clearer for LAD and SPOT (Supplementary Figs. 22F, 23F, and 24F). Taken together, the prediction outcomes with the TCGA samples were found to be consistent with the observations made with the PCAWG samples where LAD often generated more ‘cancer type-specific’ flux data, and FBA did not.
4. Discussion
Use of MEMs and MSMs has been considered critical for the successful development and simulation of context-specific GEMs. Therefore, in this study, all pairwise combinations of three MEMs (i.e., tINIT, rank-based tINIT, and GIMME) and five MSMs (i.e., FBA, pFBA, LAD, SPOT, and E-Flux2) were evaluated using machine learning (i.e., t-SNE and 1D CNN) and a total of 1,562 RNA-seq data from PCAWG and TCGA. Besides machine learning, the biological evaluation was also conducted for the patient-specific GEMs from pairwise combinations of the MEMs and the MSMs, which involved the use of gene expression data, housekeeping gene list, and correlation analysis. These evaluations revealed to what extent the resulting patient-specific GEMs would be biologically valid. Noteworthy findings from these evaluations include the high performance of rank-based tINIT or GIMME paired with LAD, and relatively poorer performance of FBA and pFBA regardless of the MEMs used.
These evaluation studies also provided insights on the MSMs applied to cancer patient-specific GEMs. First, it was our surprise to learn that FBA and pFBA did not perform well for predicting cancer type-specific fluxes despite their wide usage in various human metabolism studies [[11], [12], [55], [56]]; relative poor performance of FBA and pFBA became clearer as all the three MEMs generated highly cancer type-specific GEMs (Fig. 2B,C). These findings suggest that FBA and pFBA may not be suitable for predicting fluxes in cancer metabolism. In case of FBA, introduction of more sophisticated constraints, such as availability of tumor microenvironment-specific nutrients, enzyme kinetics [[7], [57]], or thermodynamic irreversibility [58], may help improve the prediction performance; however, use of such complex constraints for a large network model such as Human1 is another challenge. Second, generation of both RNA-seq and 13C-metabolic flux data will certainly help advancing MSMs for cancer metabolism studies. To more precisely evaluate MSMs, 13C-metabolic flux data are ideal as they clearly reveal metabolic phenotypes of a cell [[59], [60], [61]]. However, 13C-metabolic flux data could not be used in this study because there are very few studies on cancers that generated both RNA-seq data and 13C-metabolic flux data. Greater availability of RNA-seq data coupled with 13C-metabolic flux data will allow simulating patient-specific GEMs in various manners, which can be validated using corresponding 13C-metabolic flux data. Also, our conclusions derived in this study will be able to serve as a reference for integrative analysis of RNA-seq and 13C-metabolic flux data to be generated from cancer samples. Finally, it remains to be seen whether our findings from this study can also be applied to other diseases, for example diabetes. The findings from rigorous evaluations in this study will serve as a useful reference when simulating cancer metabolism using a GEM.
CRediT authorship contribution statement
Sang Mi Lee: Conceptualization, Methodology, Software, Visualization, Formal analysis, Writing – original draft, Writing – review & editing. GaRyoung Lee: Conceptualization, Methodology, Software, Visualization, Formal analysis, Writing – original draft, Writing – review & editing. Hyun Uk Kim: Conceptualization, Writing – review & editing, Supervision, Project administration, Funding acquisition.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.06.027.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.O'Brien E.J., Monk J.M., Palsson B.O. Using genome-scale models to predict biological capabilities. Cell. 2015;161(5):971–987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yizhak K., Chaneton B., Gottlieb E., Ruppin E. Modeling cancer metabolism on a genome scale. Mol Syst Biol. 2015;11(6):817. doi: 10.15252/msb.20145307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu C., Kim G.B., Kim W.J., Kim H.U., Lee S.Y. Current status and applications of genome-scale metabolic models. Genome Biol. 2019;20(1):121. doi: 10.1186/s13059-019-1730-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duarte N.C., Becker S.A., Jamshidi N., Thiele I., Mo M.L., et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proc Natl Acad Sci U S A. 2007;104(6):1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryu J.Y., Kim H.U., Lee S.Y. Framework and resource for more than 11,000 gene-transcript-protein-reaction associations in human metabolism. Proc Natl Acad Sci U S A. 2017;114(45):E9740–E9749. doi: 10.1073/pnas.1713050114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunk E., Sahoo S., Zielinski D.C., Altunkaya A., Drager A., et al. Recon3D enables a three-dimensional view of gene variation in human metabolism. Nat Biotechnol. 2018;36(3):272–281. doi: 10.1038/nbt.4072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson J.L., Kocabas P., Wang H., Cholley P.E., Cook D., et al. An atlas of human metabolism. Sci Signal. 2020;13(624) doi: 10.1126/scisignal.aaz1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frezza C., Zheng L., Folger O., Rajagopalan K.N., MacKenzie E.D., et al. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature. 2011;477(7363):225–228. doi: 10.1038/nature10363. [DOI] [PubMed] [Google Scholar]
- 9.Mardinoglu A., Agren R., Kampf C., Asplund A., Uhlen M., et al. Genome-scale metabolic modelling of hepatocytes reveals serine deficiency in patients with non-alcoholic fatty liver disease. Nat Commun. 2014;5:3083. doi: 10.1038/ncomms4083. [DOI] [PubMed] [Google Scholar]
- 10.Rohlenova K., Goveia J., Garcia-Caballero M., Subramanian A., Kalucka J., et al. Single-cell RNA sequencing maps endothelial metabolic plasticity in pathological angiogenesis. Cell Metab. 2020;31(4):862–77 e14. doi: 10.1016/j.cmet.2020.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Lewis J.E., Forshaw T.E., Boothman D.A., Furdui C.M., Kemp M.L. Personalized genome-scale metabolic models identify targets of redox metabolism in radiation-resistant tumors. Cell Syst. 2021;12(1):68–81 e11. doi: 10.1016/j.cels.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lewis J.E., Kemp M.L. Integration of machine learning and genome-scale metabolic modeling identifies multi-omics biomarkers for radiation resistance. Nat Commun. 2021;12(1):2700. doi: 10.1038/s41467-021-22989-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S.M., Kim H.U. Development of computational models using omics data for the identification of effective cancer metabolic biomarkers. Mol Omics. 2021;17(6):881–893. doi: 10.1039/d1mo00337b. [DOI] [PubMed] [Google Scholar]
- 14.Jerby L., Ruppin E. Predicting drug targets and biomarkers of cancer via genome-scale metabolic modeling. Clin Cancer Res. 2012;18(20):5572–5584. doi: 10.1158/1078-0432.CCR-12-1856. [DOI] [PubMed] [Google Scholar]
- 15.Ryu J.Y., Kim H.U., Lee S.Y. Reconstruction of genome-scale human metabolic models using omics data. Integr Biol (Camb) 2015;7(8):859–868. doi: 10.1039/c5ib00002e. [DOI] [PubMed] [Google Scholar]
- 16.Nilsson A., Nielsen J. Genome scale metabolic modeling of cancer. Metab Eng. 2017;43(Pt B):103–112. doi: 10.1016/j.ymben.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 17.Opdam S., Richelle A., Kellman B., Li S., Zielinski D.C., et al. A systematic evaluation of methods for tailoring genome-scale metabolic models. Cell Syst. 2017;4(3):318–29 e6. doi: 10.1016/j.cels.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richelle A., Chiang A.W.T., Kuo C.C., Lewis N.E. Increasing consensus of context-specific metabolic models by integrating data-inferred cell functions. PLoS Comput Biol. 2019;15(4):e1006867. doi: 10.1371/journal.pcbi.1006867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jamialahmadi O., Hashemi-Najafabadi S., Motamedian E., Romeo S., Bagheri F. A benchmark-driven approach to reconstruct metabolic networks for studying cancer metabolism. PLoS Comput Biol. 2019;15(4):e1006936. doi: 10.1371/journal.pcbi.1006936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joshi C.J., Schinn S.M., Richelle A., Shamie I., O'Rourke E.J., et al. StanDep: Capturing transcriptomic variability improves context-specific metabolic models. PLoS Comput Biol. 2020;16(5):e1007764. doi: 10.1371/journal.pcbi.1007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jalili M., Scharm M., Wolkenhauer O., Damaghi M., Salehzadeh-Yazdi A. Exploring the metabolic heterogeneity of cancers: A benchmark study of context-specific models. J Pers Med. 2021;11(6) doi: 10.3390/jpm11060496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yuneva M.O., Fan T.W., Allen T.D., Higashi R.M., Ferraris D.V., et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab. 2012;15(2):157–170. doi: 10.1016/j.cmet.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu J., Locasale J.W., Bielas J.H., O'Sullivan J., Sheahan K., et al. Heterogeneity of tumor-induced gene expression changes in the human metabolic network. Nat Biotechnol. 2013;31(6):522–529. doi: 10.1038/nbt.2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayers J.R., Torrence M.E., Danai L.V., Papagiannakopoulos T., Davidson S.M., et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science. 2016;353(6304):1161–1165. doi: 10.1126/science.aaf5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jun S., Mahesula S., Mathews T.P., Martin-Sandoval M.S., Zhao Z., et al. The requirement for pyruvate dehydrogenase in leukemogenesis depends on cell lineage. Cell Metab. 2021;33(9):1777–92 e8. doi: 10.1016/j.cmet.2021.07.016. [DOI] [PubMed] [Google Scholar]
- 26.Vander Heiden M.G., DeBerardinis R.J. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168(4):657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Consortium ITP-CAoWG Pan-cancer analysis of whole genomes. Nature. 2020;578(7793):82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machado D., Herrgard M. Systematic evaluation of methods for integration of transcriptomic data into constraint-based models of metabolism. PLoS Comput Biol. 2014;10(4):e1003580. doi: 10.1371/journal.pcbi.1003580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bhadra-Lobo S., Kim M.K., Lun D.S. Assessment of transcriptomic constraint-based methods for central carbon flux inference. PLoS ONE. 2020;15(9):e0238689. doi: 10.1371/journal.pone.0238689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becker S.A., Palsson B.O. Context-specific metabolic networks are consistent with experiments. PLoS Comput Biol. 2008;4(5):e1000082. doi: 10.1371/journal.pcbi.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robaina Estevez S., Nikoloski Z. Generalized framework for context-specific metabolic model extraction methods. Front Plant Sci. 2014;5:491. doi: 10.3389/fpls.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agren R., Mardinoglu A., Asplund A., Kampf C., Uhlen M., et al. Identification of anticancer drugs for hepatocellular carcinoma through personalized genome-scale metabolic modeling. Mol Syst Biol. 2014;10:721. doi: 10.1002/msb.145122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heirendt L., Arreckx S., Pfau T., Mendoza S.N., Richelle A., et al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox vol 3.0. Nat Protoc. 2019;14(3):639–702. doi: 10.1038/s41596-018-0098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pacheco M.P., Bintener T., Ternes D., Kulms D., Haan S., et al. Identifying and targeting cancer-specific metabolism with network-based drug target prediction. EBioMedicine. 2019;43:98–106. doi: 10.1016/j.ebiom.2019.04.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vlassis N., Pacheco M.P., Sauter T. Fast reconstruction of compact context-specific metabolic network models. PLoS Comput Biol. 2014;10(1):e1003424. doi: 10.1371/journal.pcbi.1003424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347(6220):1260419. [DOI] [PubMed]
- 37.Wang H, Marcisauskas S, Sanchez BJ, Domenzain I, Hermansson D et al. RAVEN 2.0: A versatile toolbox for metabolic network reconstruction and a case study on Streptomyces coelicolor. PLoS Comput Biol 2018;14(10):e1006541. [DOI] [PMC free article] [PubMed]
- 38.Lieven C., Beber M.E., Olivier B.G., Bergmann F.T., Ataman M., et al. MEMOTE for standardized genome-scale metabolic model testing. Nat Biotechnol. 2020;38(3):272–276. doi: 10.1038/s41587-020-0446-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orth J.D., Thiele I., Palsson B.O. What is flux balance analysis? Nat Biotechnol. 2010;28(3):245–248. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis NE, Hixson KK, Conrad TM, Lerman JA, Charusanti P et al. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol Syst Biol 2010;6:390. [DOI] [PMC free article] [PubMed]
- 41.Becker S.A., Feist A.M., Mo M.L., Hannum G., Palsson B.O., et al. Quantitative prediction of cellular metabolism with constraint-based models: The COBRA Toolbox. Nat Protoc. 2007;2(3):727–738. doi: 10.1038/nprot.2007.99. [DOI] [PubMed] [Google Scholar]
- 42.Kim H.U., Kim T.Y., Lee S.Y. Framework for network modularization and Bayesian network analysis to investigate the perturbed metabolic network. BMC Syst Biol. 2011;5(Suppl 2):S14. doi: 10.1186/1752-0509-5-S2-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee D., Smallbone K., Dunn W.B., Murabito E., Winder C.L., et al. Improving metabolic flux predictions using absolute gene expression data. BMC Syst Biol. 2012;6:73. doi: 10.1186/1752-0509-6-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim M.K., Lane A., Kelley J.J., Lun D.S. E-Flux2 and SPOT: Validated methods for inferring intracellular metabolic flux distributions from transcriptomic data. PLoS ONE. 2016;11(6):e0157101. doi: 10.1371/journal.pone.0157101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ebrahim A., Lerman J.A., Palsson B.O., Hyduke D.R. COBRApy: COnstraints-Based Reconstruction and Analysis for Python. BMC Syst Biol. 2013;7:74. doi: 10.1186/1752-0509-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Segre D., Vitkup D., Church G.M. Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci U S A. 2002;99(23):15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lvd M., Hinton G.E. Visualizing data using t-SNE. J Machine Learn Res. 2008;9:2579–2605. [Google Scholar]
- 48.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 49.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., et al. Scikit-learn: Machine learning in Python. J Machine Learn Res. 2011;12:2825–2830. [Google Scholar]
- 50.Chollet F Keras. 2015. https://keras.io. Accessed 2022 March 24.
- 51.Kingma DP, Ba J. Adam: A method for stochastic optimization (2014). arXiv:1412.6980.
- 52.Abadi M., Barham P., Chen J.M., Chen Z.F., Davis A., et al. Proceedings of Osdi'16: 12th Usenix Symposium on Operating Systems Design and Implementation. 2016. TensorFlow: A system for large-scale machine learning; pp. 265–283. [Google Scholar]
- 53.Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., et al. Scipy 1.0: Fundamental algorithms for scientific computing in Python. Nat Methods. 2020;17(3):261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eisenberg E., Levanon E.Y. Human housekeeping genes, revisited. Trends Genet. 2013;29(10):569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 55.Maoz B.M., Herland A., FitzGerald E.A., Grevesse T., Vidoudez C., et al. A linked organ-on-chip model of the human neurovascular unit reveals the metabolic coupling of endothelial and neuronal cells. Nat Biotechnol. 2018;36(9):865–874. doi: 10.1038/nbt.4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Puniya B.L., Amin R., Lichter B., Moore R., Ciurej A., et al. Integrative computational approach identifies drug targets in CD4(+) T-cell-mediated immune disorders. NPJ Syst Biol Appl. 2021;7(1):4. doi: 10.1038/s41540-020-00165-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nilsson A., Bjornson E., Flockhart M., Larsen F.J., Nielsen J. Complex i is bypassed during high intensity exercise. Nat Commun. 2019;10(1):5072. doi: 10.1038/s41467-019-12934-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Masid M., Ataman M., Hatzimanikatis V. Analysis of human metabolism by reducing the complexity of the genome-scale models using redHUMAN. Nat Commun. 2020;11(1):2821. doi: 10.1038/s41467-020-16549-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jeong S.M., Xiao C., Finley L.W., Lahusen T., Souza A.L., et al. SIRT4 has tumor-suppressive activity and regulates the cellular metabolic response to DNA damage by inhibiting mitochondrial glutamine metabolism. Cancer Cell. 2013;23(4):450–463. doi: 10.1016/j.ccr.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen W.L., Wang Y.Y., Zhao A., Xia L., Xie G., et al. Enhanced fructose utilization mediated by SLC2A5 is a unique metabolic feature of acute myeloid leukemia with therapeutic potential. Cancer Cell. 2016;30(5):779–791. doi: 10.1016/j.ccell.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ye S., Xu P., Huang M., Chen X., Zeng S., et al. The heterocyclic compound Tempol inhibits the growth of cancer cells by interfering with glutamine metabolism. Cell Death Dis. 2020;11(5):312. doi: 10.1038/s41419-020-2499-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.