Summary
The treatment of colorectal cancer (CRC) with FOLFOX shows some efficacy, but these tumors quickly develop resistance to this treatment. We have observed increased phosphorylation of AKT1/mTOR/4EBP1 and levels of p21 in FOLFOX-resistant CRC cells. We have identified a small molecule, NSC49L, that stimulates protein phosphatase 2A (PP2A) activity, downregulates the AKT1/mTOR/4EBP1-axis, and inhibits p21 translation. We have provided evidence that NSC49L- and TRAIL-mediated sensitization is synergistically induced in p21-knockdown CRC cells, which is reversed in p21-overexpressing cells. p21 binds with procaspase 3 and prevents the activation of caspase 3. We have shown that TRAIL induces apoptosis through the activation of caspase 3 by NSC49L-mediated downregulation of p21 translation, and thereby cleavage of procaspase 3 into caspase 3. NSC49L does not affect global protein synthesis. These studies provide a mechanistic understanding of NSC49L as a PP2A agonist, and how its combination with TRAIL sensitizes FOLFOX-resistant CRC cells.
Subject areas: Cell biology, Functional aspects of cell biology, Cancer
Graphical abstract

Highlights
-
•
A PP2A agonist has been identified that sensitizes FOLFOX-resistant CRC cells
-
•
It downregulates AKT1/mTOR/4EBP1 axis and p21 translation
-
•
Describes a link between p21 and procaspase 3 in TRAIL-resistance
-
•
Downregulation of p21 synergistically induces TRAIL-mediated apoptosis
Cell biology; Functional aspects of cell biology; Cancer
Introduction
Colorectal cancer (CRC) is the third most common cancer and the second leading cause of cancer death for men and women in the USA. The American Cancer Society has estimated about 151,030 new cases of CRC with an estimated 52,58 deaths in both sexes in the year 2022 (Siegel et al., 2022). Modern CRC therapy has achieved considerable progress with the use of aggressive surgical resection and chemotherapy. However, nearly 50% of patients with colonic tumors develop recurrent metastatic disease (Young et al., 2014). 5-Fluorouracil (5-FU) is the most integral systemic component of curative and palliative therapy for CRC, but the overall response rate in advanced CRC is only 10–15%. The current clinical practice utilizes the combination of 5-FU with folinic acid and oxaliplatin (FOLFOX) as first-line chemotherapy for metastatic CRC; however, CRC tumors quickly develop resistance to FOLFOX, which carries significant toxicity, cost, and patient morbidity (Yaffee et al., 2015). Despite the use of chemotherapy (oxaliplatin-based: FOLFOX or CAPOX, irinotecan-based: FOLFIRI) combined with bevacizumab (anti-VEGF), cetuximab (anti-EGFR), or panitumumab (for RAS wild-type tumors) to manage metastatic CRC tumors, the five-year survival rate remains low at 14% (Ikoma et al., 2017), which emphasizes the urgent need for better second-line chemotherapy for advanced-stage CRC.
At a molecular level, the activation of AKT1 signaling has been reported in 57% of CRCs as an early event during sporadic colon carcinogenesis, which can occur through gene amplification, mutation, or epigenetic manipulation (Roy et al., 2002). The increased levels of phosphorylated AKT1, mammalian target of rapamycin (mTOR), and eukaryotic translation initiation factor 4E-binding protein 1 (4EBP1) are linked with drug resistance and poor prognosis of patients with CRC (Li et al., 2018; Malinowsky et al., 2014; Melling et al., 2015). In the past few years, the development of several AKT1/mTOR/4EBP1-axis inhibitors has been described, but the success rate has been unsatisfactory (Xu et al., 2016). The discovery of perifosine (AKT1 inhibitor) in combination with capecitabine is showing some promise with CRC therapy (Richardson et al., 2012). The mTOR inhibitors have not been very successful for CRC therapy owing to the activation of upstream PI3K activity (Rozengurt et al., 2014). MAPK (Hoang et al., 2012) and EGFR (Wei et al., 2015) activation and 4EBP1 phosphorylation (T37/46P) prevention (Gerdes et al., 2013) are also reported with AKT1 and mTOR inhibitors. Furthermore, activation of the AKT1 pathway also enhances the function of eukaryotic translation initiation factor 4E (eIF4E), which selectively increases the translation of key mRNAs involved in tumor growth, angiogenesis, and cell survival (Graff et al., 2008). Besides the mentioned problems above, these therapeutic compounds are toxic to normal cells.
A possible mechanism to decrease drug resistance is via the dephosphorylation of AKT1 by PP2A, a tumor suppressor, belonging to the family of protein serine/threonine phosphatases (Ruvolo, 2016). PP2A is a heterotrimeric holoenzyme with three subunits: PP2A-C (catalytic), PP2A-B (regulatory), and PP2A-A (scaffolding). Each subunit further exists in multiple isoforms, such as PP2A-C exists as two isoforms—PP2A-CA and PP2A-CB (Janssens et al., 2008). Frequent inactivation of PP2A in CRC cell lines and patient tumor samples has been documented (Cristobal et al., 2014; Wang et al., 1998). In a recent clinical study, low expression of PP2A was linked with worse overall survival for patients with CRC and was suggested as an independent prognostic factor for CRC (Yong et al., 2018). In recent years, there has been a great focus toward the discovery of PP2A agonists for the downregulation of oncogenic phosphorylation of kinases, implicating PP2A as a useful target for therapeutic development (Mazhar et al., 2019; O'Connor et al., 2018; Sangodkar et al., 2017; Tohme et al., 2019; Westermarck, 2018). Different studies show the activation of PP2A by small molecules either indirectly removing the effect of the PP2A inhibitors or by directly interacting with PP2A holoenzyme complex and modulating its activity. The Suvar/Enhancer of zeste/Trithorax (SET) binds to PP2A and inhibits its activity. The SET inhibitors FTY720, OP449, and COG112 interact at the SET/PP2A interaction interface, release PP2A, and show a promising clinical implication (Agarwal et al., 2014; Cristobal et al., 2015; De Palma et al., 2019; Ishitsuka et al., 2014). In recent years, DT-061, a small molecule that directly activates PP2A (SMAP), has been shown to be binding and stimulating the PP2A activity by stabilizing the B56α-PP2A holoenzyme (Leonard et al., 2020). In this communication, we show the identification of a small molecule, NSC49L [NSC30049 ((1-(4-Chloro-2-butenyl)-1λ∼5∼,3,5,7-tetraazatricyclo [3.3.1.1–3,7∼]decane)] (Figure 3A), that interacts with the catalytic subunit C of the PP2A and stimulates its activity. Although a significant success has been achieved with new PP2A activators targeting different tumor types, studies with the FOLFOX-resistant CRC tumors are not yet reported. Furthermore, the in vivo toxicity of these PP2A activators remains a concern.
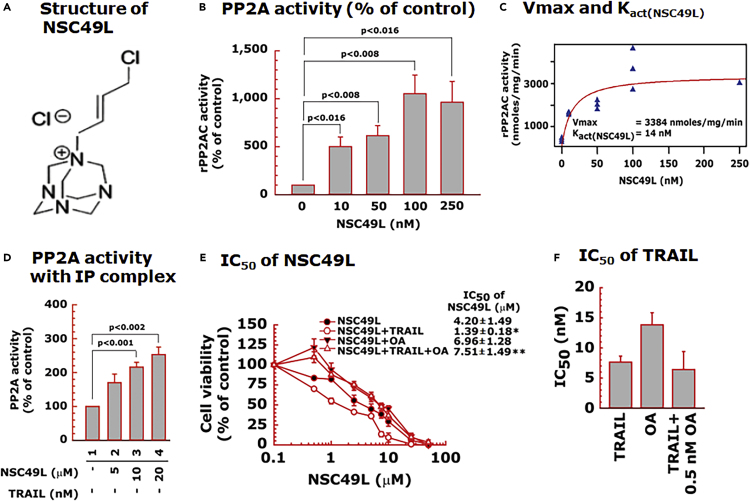
Figure 3.
NSC49L-mediated activation of PP2AC
(A) chemical structure of NSC49L.
(B) human rPP2AC activity determined with the malachite green assay.
(C) Kact(NSC49L) and Vmax values of NSC49L with PP2AC.
(D) PP2A activity using immune complexes isolated with anti-PP2AC antibody from control and NSC49L-treated FOLFOX-HT29 cells. Data are the mean ± SE of three experiments.
(E) Okadaic acid (OA), a PP2A inhibitor, blocks NSC49L and TRAIL-induced cytotoxicity to FOLFOX-HT29 cells. Cells were treated with different concentrations of NSC49L, TRAIL and OKA either alone are in combination as indicated in the figure. After 72 h treatment, cell viability was determined using the MTT-assay. ∗ = Significantly different as compared to NSC49L group, and ∗∗ = significantly different as compared to NSC49L+TRAIL group.
(F) effect of OA on the IC50 of the TRAIL. Data are the mean ± SE of four estimations. The p < 0.05 is considered significant.
Tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) is a member of a subset of the TNF receptor superfamily of protein ligands, which are physiological means of killing many cancer cells, including CRC cells, while sparing the normal cells (Ashkenazi et al., 1999). Thus, TRAIL has great promise as a tumor-selective anti-cancer agent. Unfortunately, the major limitation of the TRAIL therapy is the development of resistance in cancer cells, comprising CRC cells (Van der Jeught et al., 2018). TRAIL, a trimeric protein, binds with death receptor (DR) molecules and forms an assembly platform for the adaptor protein Fas-associated death domain (FADD) (Ralff and El-Deiry, 2018). Subsequently, FADD binds to procaspase 8, and together with DR and procaspase 8 forms the death-inducing signaling complex (DISC) that promotes the activation of caspase 8. The activated caspase 8 is released into the cytosol, where it activates the executioner caspase 3 which drives apoptotic breakdown of the cell (Carneiro and El-Deiry, 2020). Hence, the inactivation of one of these steps can contribute to TRAIL- or drug resistance via evasion of apoptosis. In previous studies, the interaction of p21 with procaspase 3 has been shown to block the cleavage of procaspase 3 into executioner caspase 3 which may prevent the executioner caspase 3 from inducing apoptosis (Suzuki et al., 2000). Increased levels of p21 have been identified as a drug-resistance factor in several cancer cells (Georgakilas et al., 2017), including 5-FU resistance to CRC cells (Maiuthed et al., 2018).
There are two types of drug-resistance mechanisms—intrinsic (primary) and acquired (secondary). The intrinsic drug resistance exists before the initiation of chemotherapy while the acquired resistance is induced during or after chemotherapy (Briffa et al., 2017; Gottesman, 2002). In recent studies, increased expression of p21 is considered an acquired resistant factor in cancer cells which functions by its interaction with and inactivation of procaspase 3 (Suzuki et al., 1998, 1999). The control of p21 expression in drug-resistant cancer cells is described at the level of translation through AKT/mTOR/4EBP1 axis (Beuvink et al., 2005; Tahmasebi et al., 2014). In the present study, we have identified p21 as an acquired FOLFOX resistance factor in CRC cells. We report that the increased FOLFOX-resistant CRC cells have increased levels of p21, which is independent of p53-mediated transcription. Here, we have discussed the role of the PP2A agonist, NSC49L that by inhibiting the AKT1/mTOR/4EBP1 pathway reduces p21 translation. The decreased level of p21 released procaspase 3 for TRAIL-dependent synergistic sensitization of FOLFOX-resistant CRC cells. Thus, downregulation of p21 can be a critical mechanism for abolishing drug resistance and inducing TRAIL-mediated apoptosis in FOLFOX-resistant CRC cells.
Results
PPP2CB was augmented in colorectal cancer tumors and closely associated with poor prognosis in patients with colorectal cancer
We used the Cancer Genome Atlas (TCGA) through the Genomic Data Commons Data Portal for determining the expression level of PPP2CB (catalytic subunit B) in CRC tumors compared to matched normal tissues. PPP2CB was significantly and steadily decreased during the progression from stage I to stage IV in CRC tumors compared to adjacent normal tissues (Figure 1A). We also analyzed PPP2CB levels in CRC cells spread in lymph nodes. There was a progressive significant decrease in the PPP2CB levels at different nodal stages (N0 to N2) of CRC development compared to normal tissues (Figure 1A). A similar decreased pattern of PPP2CB expression was also identified in mucinous and non-mucinous CRC tumors compared to normal tissues (Figure S1A). Then, we used the Clinical Proteomic Tumor Analysis Consortium (CPTAC) data portal and found a decreased PP2AC protein level in CRC tumors compared to normal tissues (Figures 1B and S1B). Furthermore, the low-level expression of PPP2CB was associated with poor prognosis in patients with CRC (Figures 1C and S1C). These results suggest a negative correlation of PPP2CB expression with CRC progression and poor prognosis of patients with CRC.
Figure 1.
PP2AC expression level is directly correlated with disease progression and the survival of patients with CRC
(A) Box plot depicting a summary of the TCGA data quantification of PPP2CB in normal tissue and CRC tumors (left), different stages (middle), and different nodal progressions (right).
(B) Box plot showing the CPTAC data quantitation of PP2AC protein levels in normal tissues and CRC tumors (left), and different stages of progression (right).
(C) Kaplan-Meier survival analysis of TCGA data was plotted for the CRC cases that were high and low in PPP2CB expression. The p-value is shown at the top of each graph. See also Figure S1.
PPP2CB expression is negatively linked with the expression of AKT1, mTOR, 4EBP1, and CDKN1A in FOLFOX-responsive and non-responsive colorectal cancer tumors
As AKT1 is a downstream target of PP2A and mTOR, 4EBP1, and p21 are on the same axis of regulation, we determined the correlation of their gene expression in FOLFOX-responsive and non-responsive CRC tumors. In this study, all publicly available databases were searched and three gene expression datasets of patients with CRC with FOLFOX therapy were combined, as described in an earlier study (Lin et al., 2018). We found that the PPP2CB gene expression level was significantly higher in FOLFOX-responsive versus non-responsive tumors (Figure 2). On the other hand, the gene expression level of AKT1, mTOR, 4EBP1, and CDKN1A was significantly lower in FOLFOX-responsive versus non-responsive tumors (Figure 2), suggesting a negative correlation of PPP2CB with AKT1, mTOR, 4EBP1and CDKN1A in FOLFOX-responsive versus non-responsive CRC tumors. As increased expression of AKT1 (Surov et al., 2018), mTOR (Wu et al., 2018), and 4EBP1 (Malinowsky et al., 2014) are linked with poor prognosis of patients with CRC and linked to increased levels of p21 with increased proliferative capacity of colon cancer cases (Palaiologos et al., 2019), we hypothesized that the upregulation of the PP2A activity and the downregulation of the AKT1, mTOR, 4EBP1 and p21 levels in CRC cells can be a suitable means of inducing the sensitization of the CRC tumors.
Figure 2.
PPP2CA/B expression is lower in FOLFOX-non-responsive (NR) versus responsive (R) CRC tumors, which negatively correlated with the expression of AKT1, mTOR, 4EBP1, and CDKN1A
Data were analyzed from the NCBI GEO accession number GSE72970. Number of samples in the NR and R groups was 61 and 63, respectively. The p value is shown at the top of each graph.
NSC49L activates PP2A. We have been investigating a small molecule, NSC49L, to understand the molecular mechanisms responsible for its anticancer activity. We examined whether it is a kinase inhibitor but the results of a panel of 366 kinases showed that it was not a kinase inhibitor (Narayan et al., 2019). However, from these experiments it appeared that NSC49L might be a phosphatase agonist, as it inhibited hydroxyurea- and 5-FU-induced phosphorylation of Chk1(S317P) and Chk1(S296) in HCT116 and HT29 cell lines (Narayan et al., 2017, 2019). In previous studies, the involvement of PP2A has been suggested in the ATR-dependent dephosphorylation of Chk1 at S317 and S296 residues (Leung-Pineda et al., 2006). Another link between NSC49L and Chk1 may be through the downregulation of mTORC1 activity by the downregulation of AKT1 activity, and in turn, the downregulation of the Chk1 phosphorylation (Zhou et al., 2017). The later possibility prompted us to examine whether NSC49L is a PP2A agonist that downregulates the AKT1/mTOR/4EBP1-axis in FOLFOX-resistant CRC cells.
First, we performed an in vitro PP2A activity assay using recombinant PP2A-Cα protein and threonine phosphopeptide (KRpTIRR) as substrate in a Malachite green assay (Lek et al., 2017). NSC49L stimulated PP2A-Cα activity in a dose-dependent manner with a Kact(NSC49L) of 14 nM and Vmax of 3384 nmoles/mg/min (Figures 3B and 3C). We also determined the effect of NSC49L treatment on the stimulation of PP2A-Cα in FOLFOX-HT29 cells. The immunocomplexes from the control and treated groups were isolated with anti-PP2A-C antibody and were used for PP2A-Cα activity (Lek et al., 2017). Results showed an increased PP2A activity in a dose-dependent manner (Figure 3D), suggesting that NSC49L acts as a PP2A agonist, which stimulates its activity at nanomolar concentrations.
Okadaic acid (OA) is a PP2A and PP1 inhibitor, but at a selective concentration of 2 nM or lower, it inhibits only PP2A activity (Lu et al., 2009). To determine whether the inhibition of PP2A activity by OA increases cell viability, we treated FOLFOX-HT29 cells with a fixed concentration of OA (0.5 nM) and TRAIL (1 nM) and with different concentrations of NSC49L. Then, we determined the IC50 of NSC49L by MTT-cell viability assays. Results showed that treatment with 0.5 nM of OA either alone or in combination with 1 nM of TRAIL increased the survival of FOLFOX-HT29 cells, as is evident from the increased IC50 of NSC49L (Figure 3E). The concentration of OA (0.5 nM) used in this experiment was much lower than its IC50 (∼15 nM). OA at the lower concentration did not affect the IC50 of the TRAIL (Figure 3F). These results suggest that NSC49L mediates its effect through the stimulation of the PP2A activity in FOLFOX-HT29 cells, which is further effective when treated in combination with TRAIL.
NSC49L and tumor necrosis factor (TNF)-related apoptosis-inducing ligand treatments sensitize FOLFOX-resistant HCT116 and HT29 cell lines
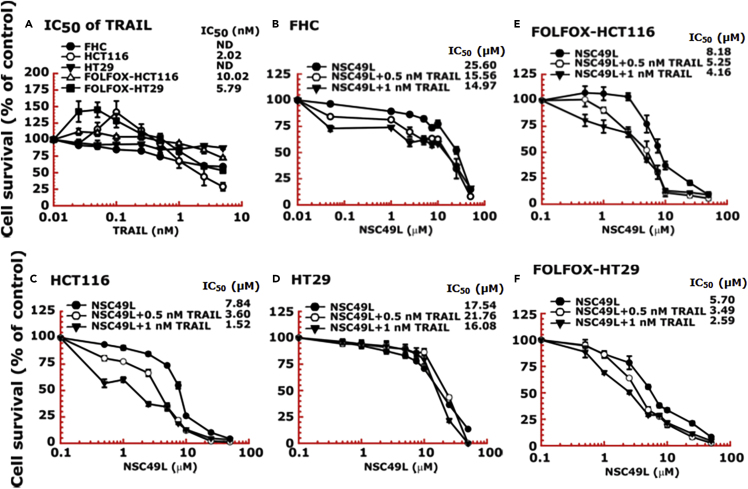
HCT116 and HT29 are TRAIL-sensitive and TRAIL-resistant CRC cell lines, respectively (Kim et al., 2015). NSC49L shows more toxicity to HCT116 cells than to HT29 cells, which were further sensitized after TRAIL treatment. The normal colonic epithelial cell line, FHC, was resistant to TRAIL (Figure 4A). To examine the effect of TRAIL on FOLFOX-resistant CRC cells, we used isogenic FOLFOX-resistant cells derived from HCT116 and HT29 cell lines (Narayan et al., 2017; Yu et al., 2009). TRAIL reduced the viability of both FOLFOX-HCT116 and FOLFOX-HT29 cell lines, while the TRAIL-resistant HT29-derived FOLFOX-HT29 cells were more sensitive than the TRAIL-sensitive HCT116-derived FOLFOX-HCT116 cells (Figure 4A). Then, we determined whether TRAIL in combination with NSC49L can enhance the cytotoxicity of FHC, HCT116, HT29, FOLFOX-HCT116, and FOLFOX-HT29 cell lines. The results showed little toxicity of NSC49L to FHC and HT29 cells below 10 μM, which was increased in combination with TRAIL, but to a much lower extent than to HCT116 cells (Figures 4B, C, and D). On the other hand, NSC49L sensitized both FOLFOX-HCT116 and FOLFOX-HT29 cell lines to TRAIL (Figures 4E and 4F). However, treatment with NSC49L in combination with TRAIL enhanced the FOLFOX sensitivity of FOLFOX-HCT116 and FOLFOX-HT29 cell lines. We observed a dose-dependent decrease in the IC50 (an indicator of increased sensitization or cytotoxicity) of NSC49L in the presence of TRAIL in both FOLFOX-resistant cell lines (Figures 4E and 4F).
Figure 4.
FOLFOX-HT29 and FOLFOX-HCT116 cell lines are much more sensitive than normal colonic epithelial cell lines (FHC) to NSC49L and TRAIL treatment
Cells were grown in 96-well plates and treated for 72 h with different concentrations of NSC49L and TRAIL either alone or in combination. Survival of cells was determined by the MTT-assay.
(A) IC50 of TRAIL.
(B–F) IC50 of NSC49L either alone or in the presence of TRAIL in different CRC cell lines. Data are the mean ± SE of four estimations.
NSC49L and tumor necrosis factor (TNF)-related apoptosis-inducing ligand treatments downregulate AKT1(S473P) and p21 levels in FOLFOX-HCT116 and FOLFOX-HT29 cell lines
Once we established that NSC49L stimulates PP2A activity, we examined its effect on PP2A expression levels. We performed RNASeq analysis of control and NSC49L/TRAIL-treated FOLFOX-HT29 cells. The results showed an increased expression of PPP2CA and PPP2CB, which are the catalytic subunits of PP2A (Figure 5A). However, the expression of other phosphatases was rather decreased as compared to treated groups. We also performed RNASeq analysis of TP53 and CDKN1A and other related genes. We found that the expression of TP53, TP63, and TP73 (also known as p53, p63, and p73, respectively) was absent in untreated FOLFOX-HT29 cells which were unchanged after the NSC49L or TRAIL treatments (Figure 5B). Whether the decreased expression level of p53, p63 and p73 in these cells after NSC49L/TRAIL treatment was regulated at the level of transcription or mRNA degradation is currently unknown. Furthermore, the expression of CDKN1A (p21Cip1/Kip1) and CDKN2A (p16INK4a) was increased but CDKN1B (p27Kip1) and CDKN1C (p57Kip2) were decreased (Figure 5B). Although the expression of p21 and p16 are linked with drug resistance (Kreis et al., 2019), we focused on p21 owing to its involvement in 5-FU resistance in CRC cells (Maiuthed et al., 2018).
Figure 5.
RNASeq and protein analysis of PP2A and downstream target proteins in CRC cells
(A and B) RNASeq analysis of FOLFOX-HT29 cells. Cells were treated with 20 μM of NSC49L and 1 nM of TRAIL either alone or in combination for 16 h. Total RNA was isolated by TRIzol reagent and processed for RNASeq analysis. Ward clustering algorithm with Euclidean distance measurement was used for heat map analysis.
(C) Correlation of PP2A expression level with AKT1(S473P) and p21 levels in CRC cell lines. To determine a correlation of PP2A with AKT1(S473P) and p21, we performed Western bots of these proteins using the whole-cell lysate of different CRC cell lines, including FOLFOX-resistant HCT116 and HT29 cells.
(D and E) Protein levels of AKT1, p53, p21, DR5, caspase 3, caspase 8, and PARP1 in HCT116, FOLFOX-CHT116, HT29, and FOLFOX-HT29 cell lines, respectively, treated with NSC49L and TRAIL either alone or in combination for 24 h. The blots are representative of three experiments. See also Figures S2 and S3.
We determined if there is any correlation between PP2AC, AKT1(S473P), and p21 levels in different CRC cell lines that are either resistant or sensitive to FOLFOX. The results showed a negative correlation between the PP2AC/AKT1(S473P) and PP2AC/p21 protein levels in most of the CRC cell lines, but a significant correlation was evident in FOLFOX-HCT116 and FOLFOX-HT29 cell lines (Figures 5C and S2). These results suggest that, at least, in FOLFOX-HCT116 and FOLFOX-HT29 cell lines, the reduced levels of PP2AC are clearly correlated with increased levels of AKT1(S473P) and p21.
Next, we determined whether the NSC49L and TRAIL-mediated sensitization of FOLFOX-HCT116 and FOLFOX-HT29 cell lines, as observed in Figure 4, are linked with increased PP2A activity and decreased AKT1(S473P) and p21 levels. We simultaneously compared these results with HCT116 and HT29 cell lines to compare the effects between the FOLFOX-HCT116 and FOLFOX-HT29 cell lines. In untreated HCT116 and HT29 cell lines we did not observe any increase in the level of AKT1(S473P) (Figures 5D and 5E, compare lanes 1-4, and 9–12, respectively); however, it was highly increased in FOLFOX-HCT116 and FOLFOX-HT29 cell lines, which was decreased after NSC49L and TRAIL treatments (Figures 5D and 5E, compare lanes 5–8, and 13–16, respectively). The expression of p53 was seen in both HCT116 and HT29 cell lines, which was completely absent in FOLFOX-HCT116 and FOLFOX-HT29 cell lines (Figures 5D and 5E). Whether the silencing of p53 in these cell lines is at the transcription or translation level is currently not known. As p21 is regulated by p53, we observed an increase in p21 in HCT116 cells that carry a wild-type TP53 gene, but not in HT29 cells that carry a mutant TP53 gene. Surprisingly, we observed a p53-independent increase in the level of p21 in FOLFOX-HCT116 and FOLFOX-HT29 cell lines, which was decreased after NSC49L and TRAIL treatments (Figures 5D and 5E, compare lanes 5–8 and 13–16, respectively). Thus, these results suggest that AKT(S473P) and p21 are associated with resistance of FOLFOX-HCT116 and FOLFOX-HT29 cell lines after 5-FU and oxaliplatin treatments. The level of cleaved caspase 8, caspase 3, and PARP1, indicators of apoptosis, were also more prominent when NSC49L and TRAIL were combined (Figures 5D and 5E, compare lanes 5–8 and 13–16, respectively). These results suggest that NSC49L-induced PP2A activity and the TRAIL-induced apoptotic pathway are linked with increased caspase 8, caspase 3, and PARP1 levels in FOLFOX-HCT116 and FOLFOX-HT29 cells. These results are supported by a TCGA analysis in which we found a positive correlation between PPP2CB expression and caspase8, caspase 3, and PARP1 expression (Figure S3).
As the activation of PP2A by NSC49L dephosphorylates AKT1, the effect of TRAIL on AKT1 dephosphorylation of these cells could be indirect. This observation is similar to TRAIL-induced sensitization of prostate cancer cells in response to the PIP3 phosphatase PTEN, suggesting that the activation of the AKT1 pathway may also be a common event in TRAIL-resistant cells (Xu et al., 2010). How TRAIL might induce its effect through the AKT1 pathway is not clearly understood. In these studies, we observed an increased effect of NSC49L on AKT1 dephosphorylation when combined with TRAIL. Hence, we expected that the increased expression of DR5, a TRAIL receptor, in FOLFOX-HCT116 and FOLFOX-HT29 cell lines might have been one of the reasons for the TRAIL/NSC49L-induced sensitization of these cells (Figures 5D and 5E, compare lanes 1 with 5 and 9 with 13, respectively). To understand if the low level of DR5 in HT29 cells might have been the reason for non-responsiveness to TRAIL (Figure 4A), we overexpressed tetracycline (tet)-inducible DR5 (Figure 6A) in these cells and determined the sensitivity in response to NSC49L and TRAIL treatments. The results showed that, indeed, the overexpression of DR5 increased TRAIL-mediated sensitization of HT29-tet-DR5+dox cells more than the HT29-tet-DR5 cells (Figure 6B). The NSC49L treatment also sensitized the HT29-tet-DR5+dox cells, suggesting a link between NSC49L and TRAIL actions (Figure 6C). However, the combination of TRAIL with NSC49L did not further affect the sensitization of these cells (Figure 6C). This may be owing to a saturating effect of NSC49L on the DR5 pathway that does not require TRAIL for further activation.
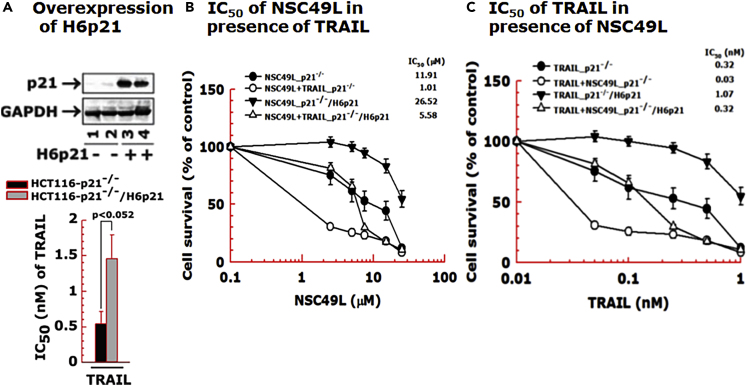
Figure 6.
Overexpression of DR5 induces the sensitization of HT29 cells to NSC49L and TRAIL
One set of the HT29-tet-DR5 cells was treated with DOX (1 μg/mL) to induce the expression of DR5. Then, control and DR5-induced cells were treated with different concentrations of NSC49L and TRAIL either alone or in combination for 72 h. The cell survival was determined by the MTT-assay.
(A) Doxycycline-induced DR5 expression was verified by immunoblot.
(B) IC50 of TRAIL.
(C) IC50 of NSC49L in the presence of 0, 0.5, and 1 nM of TRAIL, respectively. Data are the mean ± SE of four different estimations. The p-value is shown on the top of each graph.
NSC49L and tumor necrosis factor (TNF)-related apoptosis-inducing ligand treatments downregulate AKT1/mTOR/4EBP1 signaling in FOLFOX-HT29 cells
In these experiments, we determined a link between AKT1, mTOR, and 4EBP1 in NSC49L- and TRAIL-treated FOLFOX-HT29 cells. The results showed that there was no effect of the NSC49L and TRAIL treatments on the total levels of AKT1, AKT2, and AKT3 in FOLFOX-HT29 cells; however, the AKT1(S473P) level was significantly reduced (Figure 7A). The AKT1(T308P) level was also reduced, although not as much (Figure 7A). These results suggest the importance of AKT1(S473P) in the induction of FOLFOX-resistance in HT29 cells. Furthermore, the phosphorylation of mTOR(S2448P) and its downstream target protein 4EBP1(T37/46P) was also decreased in NSC49L and TRAIL-treated FOLFOX-HT29 cells (Figure 7B). From these results, we conclude that NSC49L and TRAIL treatment affect the AKT1/mTOR/4EBP1/p21-axis in FOLFOX-resistant CRC cells.
Figure 7.
Western blot analysis of AKT/mTOR/4EBP1, apoptotic pathway proteins, and apoptosis of FOLFOX-HT29 cells treated with NSC49L and TRAIL either alone or in combination for 24 h
(A and B) Western blots demonstrating the regulation of the AKT/mTOR/4EBP1 pathway by NSC49L and TRAIL in FOLFOX-HT29 cells.
(C) Levels of apoptotic proteins. All of the blots are the representative of three experiments.
(D) Caspase 3/7 activity. FOLFOX-HT29 cells were treated with NSC49L and TRAIL either alone or in combination for 24 h, and the extent of apoptosis was determined by measuring the activation of caspase 3/7 using a flow cytometer. The experiment was repeated twice. Data are the mean ± SE of triplicate results. See also Figure S4.
NSC49L- and tumor necrosis factor (TNF)-related apoptosis-inducing ligand treatment increased the apoptosis of FOLFOX-HT29 cells
We examined the expression levels of pro-apoptotic (Bcl2 and BAD) and anti-apoptotic (BAX, Bim, and PUMA) proteins in FOLFOX-HT29 cells. The results showed a decrease in Bcl2 and BAD and an increase in Bax and Bim, but not PUMA, protein levels in FOLFOX-HT29 cells-treated with NSC49L and TRAIL (Figure 7C), which may be sufficient to drive these cells to cell death. To further confirm that NSC49L and TRAIL treatment induces apoptosis in FOLFOX-HT29 cells, we measured caspase 3/7 activation, which are executioner caspases that mediate apoptotic cell death (McIlwain et al., 2013). The results showed increased apoptosis after NSC49L treatment as compared to control, which was further increased when it was combined with TRAIL (Figure 7D). TRAIL alone also caused apoptosis of FOLFOX-HT29 cells, but less than NSC49L alone (Figure 7D). Similar results were also observed with FOLFOX-HCT116 cells (Figure S4). These results suggest that combining NSC49L and TRAIL decreases the levels of the pro-apoptotic proteins, increases the levels of the anti-apoptotic proteins, and induces apoptosis in FOLFOX-resistant cells.
NSC49L and tumor necrosis factor (TNF)-related apoptosis-inducing ligand synergistically enhance the sensitization of p21-knockout colorectal cancer cells
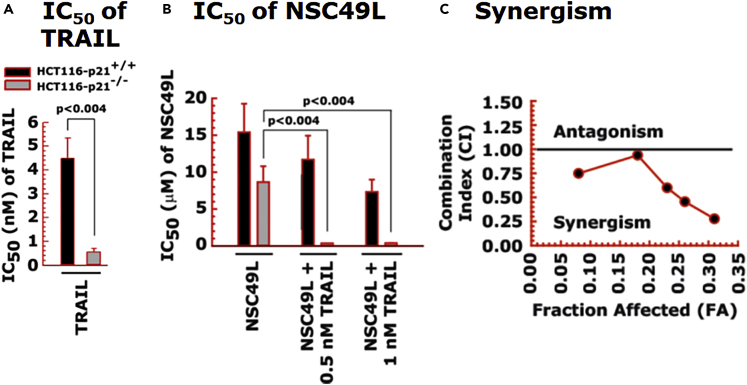
Next, we determined the importance of p21 in the sensitization of CRC cells in response to NSC49L and TRAIL treatments. For these experiments, we used isogenic wild-type and p21-knockout HCT116 cell lines. We also overexpressed H6p21 (6 his-tagged p21) to reverse the NSC49L- and TRAIL-mediated sensitization effect observed with p21-knockout cells. The HCT116-p21−/− cells were highly sensitive to TRAIL (IC50 of 0.54 ± 0.18 nM) (Figure 8A), and the sensitivity was decreased after the overexpression of H6p21 (IC50 of 1.46 ± 0.34 nM) (Figure 9A). Furthermore, the sensitization of HCT116-p21−/− cells drastically increased when the cells were treated together with NSC49L and TRAIL (Figure 8B). These results suggested that the combination of NSC49L and TRAIL has a profound effect on the sensitization of HCT116-p21−/− cells compared to HCT116-p21+/+ cells. To explore the possibility that the NSC49L and TRAIL treatments on HCT116-p21−/− cells showed a synergy, we determined the combination index (CI) by applying the Chou-Talalay method (Chou, 2010). The CI calculated by the Chou-Talalay approach provides a quantitative definition for synergism (CI < 1), additive effects (CI = 1), or antagonism (CI > 1). Based on this method, we performed MTT-cell viability assays with HCT116-p21−/− cells using increasing concentrations of NSC49L and TRAIL, where the increment of both concentrations was kept at a 1:1 ratio. The data analysis confirmed that the combined treatment with NSC49L and TRAIL has a synergistic effect on the HCT116-p21−/− cells (Figure 8C). This effect was reversed in the HCT116-p21−/−/H6p21 cells that overexpress p21. The HCT116-p21−/−/H6p21 cells showed more resistance to NSC49L and TRAIL than HCT116-p21−/− cells (Figure 9A). The IC50 of NSC49L in the presence of TRAIL became 5.5-fold higher in HCT116-p21−/−/H6p21 cells than the IC50 of HCT116-p21−/− cells (Figure 9B). Similarly, the IC50 of TRAIL in the presence of NSC49L became 10.67-fold higher in HCT116-p21−/−/H6p21 cells than the IC50 of HCT116-p21−/− cells (Figure 9C). These results support our hypothesis that the downregulation of p21 is critical for NSC49L- and TRAIL-mediated sensitization of CRC cells.
Figure 8.
Synergistic sensitization of p21-knockout cells in response to NSC49L and TRAIL treatment
HCT116-p21+/+ and HCT116-p21−/− cells were treated with different concentrations of NSC49L and TRAIL either alone or in combination for 72 h and the cell survival was determined by the MTT-assay.
(A and B) The IC50 of TRAIL and NSC49L cells, respectively. The IC50 is derived from the survival curves.
(C) The synergistic effect of NSC49L and TRAIL with HCT116-p21−/− cells. Data are the mean ± SE of four estimations. The p-value is shown on the top of each graph.
Figure 9.
Overexpression of H6p21 decreases the sensitization of p21-knockout cells in response to NSC49L and TRAIL treatment
The HCT116-p21−/− and HCT116-p21−/−/H6p21 cells were treated with different concentrations of NSC49L and TRAIL either alone or in combination for 72 h and the cell viability was determined by the MTT-assay.
(A) Western blot showing the overexpression of H6p21.
(B) The IC50 of NSC49L in combination with TRAIL.
(C) IC50 of TRAIL in combination with NSC49L. Different concentrations of NSC49L (0, 2.5, 5, 7.5, 15 and 25 μM) and TRAIL (0, 0.05, 0.1, 0.25, 0.5, and 1.0 nM) were used in the synergy experiments. Data are the mean ± SE of four estimations. The p-value is shown on the top of graph A.
NSC49L treatment increases CDKN1A mRNA levels and does not affect p21 protein degradation
The increased level of p21 in FOLFOX-HT29 cells is independent of p53 because the TP53 gene is mutated in these cells (Rodrigues et al., 1990). Although the mutant TP53 expression is higher in HT29 cells, it is drastically reduced in FOLFOX-HT29 cells, possibly owing to the transcriptional silencing of TP53 (Figure 10A, left). This supports the loss of the p53 protein level shown in Figure 5E. Now, the question arises whether the decreased level of p21 in FOLFOX-HT29 cells after NSC49L treatment is secondary to decreased transcription or increased protein degradation. First, we performed qRT-PCR to examine whether NSC49L-induced low levels of p21 protein are owing to decreased CDKN1A gene transcription. We treated HT29 and FOLFOX-HT29 cells with 20 μM of NSC49L for 16 h, then, determined CDKN1A mRNA levels by qRT-PCR. The results showed an increased level of CDKN1A mRNA in FOLFOX-HT29 cells that was not further altered after NSC49L treatment (Figure 10A, right). Similarly, we observed no effect of NSC49L treatment on CDKN1A levels in FOLFOX-HCT116 cells (Figure S5). These results indicate that NSC49L does not change CDKN1A gene expression in FOLFOX-HT29 and FOLFOX-HCT116 cells; however, the increased CDKN1A mRNA levels could be owing to acquired CDKN1A gene expression during the development of FOLFOX-induced resistance.
Figure 10.
Effect of NSC49L and TRAIL treatments on the TP53 and CDKN1A mRNA levels and protein kinetics in HT29 and FOLFOX-HT29 cells
(A) Cells were treated with NSC49L (20 μM) and TRAIL (1 nM) either alone or in combination for 16 h. Total RNA was isolated and used for qRT-PCR. Data are the mean ± SE of four estimations.
(B) Kinetics of p21 protein stability after CHX treatment. Cells were treated with NSC49L (20 μM) and TRAIL (1 nM) either alone or in combination for 24 h followed by the treatment with CHX (10 μg/mL) for different periods. The p21 protein level was determined by Western blot analysis. Blots shown are the representative of two experiments. See also Figure S5.
Next, we examined whether the decreased p21 protein level in FOLFOX-HT29 cells after NSC49L treatment was secondary to protein degradation. We treated HT29 and FOLFOX-HT29 cell lines with 20 μM NSC49L for 24 h. Then, we blocked nascent protein synthesis by treating the cells with 10 μg/mL of cycloheximide (CHX), which interferes with the translocation step in protein synthesis. As p21 protein has a short half-life (Beuvink et al., 2005), we followed its levels up to 4 h. The results showed no apparent difference in the kinetics of p21 protein degradation in FOLFOX-HT29 cells. Similar results were observed with HT29 cells as well (Figure 10B). These results suggest that the NSC49L treatment-mediated decrease of p21 protein levels in FOLFOX-HT29 cells is not owing to decreased mRNA levels or increased protein degradation, but it could be owing to a decrease in protein translation.
The rate of p21 protein synthesis is reduced by NSC49L treatment in FOLFOX-HT29 cells
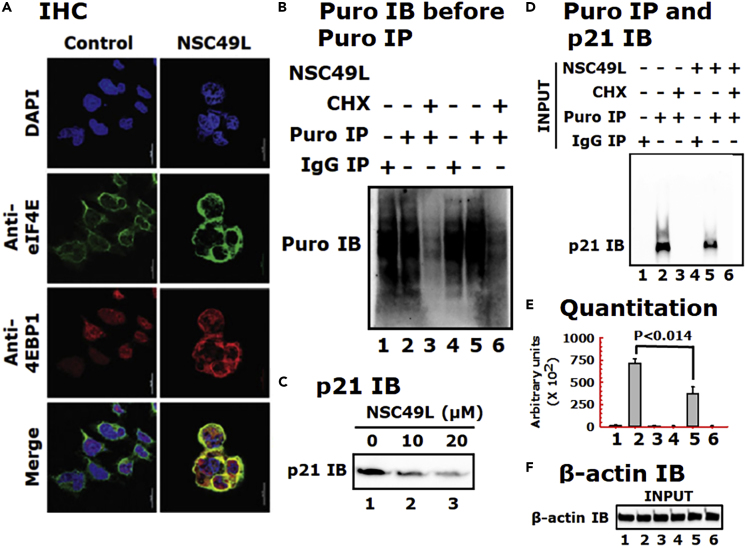
It is known that hypophosphorylated 4EBP1 binds to eIF4E and prevents it from interacting with eIF4G to promote ribosome recruitment to mRNAs. Hence, this can repress the initiation of mRNA translation. Once the phosphorylated 4EBP1(T37/46P) dissociates from eIF4E, it can then allow for the formation of the eIF4F complex and the initiation of cap-dependent translation (Lin et al., 1994). As discussed above, we observed that the increased level of 4EBP1(T37/46P) in FOLFOX-HT29 cells decreased after the treatment with NSC49L (Figure 7B). These results suggested that the decreased level of 4EBP1(T37/46P) may provide an opportunity to increase the interaction of hypophosphorylated 4EBP1 with eIF4E in these cells that may occur after treatment with NSC49L. To test this possibility, we performed immunohistochemistry (IHC) experiments with FOLFOX-HT29 cells. As untreated cells have higher levels of 4EBP1(T37/46P), we did not observe the colocalization of 4EBP1(T37/46P) and eIF4E. However, as the level of 4EBP1(T37/46P) decreased after NSC49L treatment (Figure 7B), we observed an increased colocalization of hypophosphorylated 4EBP1 with eIF4E (appearance of yellow color) (Figure 11A). In HT29 cells, we observed very poor colocalization of 4EBP1 with eIF4E after NSC49L treatment (Figure S6). This correlates with the low level of p21 in these cells owing to the presence of mutant p53, which could not regulate CDKN1A gene expression (Figure 5E). These results suggest that the NSC49L treatment decreases the levels of 4EBP1(T37/46P) and increases the colocalization of 4EBP1 with eIF4E, which may block eIF4F complex assembly, and hence decrease the translation of target protein(s), such as p21.
Figure 11.
NSC49L treatment increases the colocalization of eIF4E with unphosphorylated 4EBP1 and inhibits p21 protein synthesis in FOLFOX-HT29 cells
(A) IHC analysis. FOLFOX-HT29 cells were treated with 20 μM of NSC49L for 24 h. After treatment, the colocalization of eIF4E and 4EBP1 was determined by IHC. Yellow color in the merge column shows the colocalization of these two proteins. Images were captured with a Nikon A1RMPsi-STORM4.0 confocal microscope (Melville, NY) at 60× magnification.
(B) Global protein synthesis. FOLFOX HT29 cells were treated as above with 20 μM of NSC49L for 24 h. Cells were treated with 10 μg/mL of puromycin or puromycin plus 10 μg/mL of CHX for 15 min. Cells were washed and incubated in a fresh medium without drugs for an additional 2 h. Samples were blotted with anti-puromycin antibody.
(C) Validate the experimental conditions, a concentration-dependent effect on p21 expression was performed. Cells were mock treated (control) or treated with 10 and 20 μM of NSC49L for 24 h and p21 level was determined.
(D) Evaluate the effect of NSC49L on the regulation of p21 biosynthesis, we set up the experiment as in B. To determine the incorporation of puromycin into nascent p21, the pre-immunoprecipitated lysate was subjected to anti-puromycin immunoblot.
(E) Quantitative analysis of the blot shown in panel D, which is presented as the ImageJ arbitrary units. Data are Mean ± SE of three experiments. The p-value is shown on the top of the graph.
(F) β-actin was used as an internal control for equal loading. See also Figure S6.
Then, to examine the effect of NSC49L on p21 protein synthesis, we used the puromycin-based surface sensing of translation (SUnSET) assay protocol for labeling nascent proteins (Schmidt et al., 2009). Puromycin functions as a structural analog of aminoacyl tRNAs and lead to the release of unfinished nascent polypeptide chains during the ribosomal elongation cycle and induce premature termination and subsequent drop-off of the ribosome from the mRNA. The relative rate of puromycylated nascent proteins can be detected by anti-puromycin antibody in control and drug-treated cells (Iwasaki and Ingolia, 2017). Here, we treated FOLFOX-HT29 cells with 20 μM of NSC49L for 24 h. Then, changed the medium and followed a 2 h chase with 10 μg/mL of puromycin, prepared whole-cell lysates, and performed immunoprecipitation (IP) and immunoblotting (IB). In the case of cycloheximide (CHX), after 24 h of NSC49L treatment, we treated the cells with 10 μg/mL of CHX for 1 h, followed by 2 h of chase with puromycin. In contrast to puromycin, CHX inhibits protein synthesis by binding exclusively to cytoplasmic (80S) ribosomes of eukaryotes (Stocklein and Piepersberg, 1980). Pretreatment with cycloheximide can limit puromycin incorporation by blocking peptidyl-tRNA transition from the A-site (Hobden and Cundliffe, 1978). First, we determined the effect of NSC49L on global protein synthesis in FOLFOX-HT29 cells. The results showed that global protein synthesis was unaffected by NSC49L treatment, while it was blocked by the CHX treatment (Figure 11B, compare lane 2 with 5 and 3 with 6, respectively). Treatment with NSC49L caused a dose-dependent decrease in p21 protein levels (Figure 11C), confirming the validity of the experimental conditions. Next, we determined the effect of NSC49L treatment on the rate of nascent p21 protein synthesis. We found a significantly decreased rate of nascent p21 synthesis in FOLFOX-HT29 cells after NSC49L treatment as compared to control (Figures 11D and 11E, compare lane 2 with 5). CHX treatment completely blocked p21 synthesis (Figures 11D and 11E, compare lane 3 with 6). Data in lanes 1 and 4 show the IgG control. The β-actin protein level indicates that the same amounts of whole-cell lysate were used in IP experiments (Figure 11F). These results thus suggest that NSC49L reduces the rate of p21 protein translation in FOLFOX-HT29 cells linked with the PP2A/AKT1/mTOR/4EBP1-axis.
The TRAIL pathway interacts with the NSC49L pathway at the level of procaspase 3 and p21. As discussed above, we consistently observed a profound effect of NSC49L when applied in combination with TRAIL on increased cytotoxicity, increased c-Caspase 8, c-Caspase 3, c-PARP1, and decreased p21 protein levels in FOLFOX-resistant CRCs. As TRAIL is not expected to affect PP2A activity, its coordination with NSC49L may occur at the p21 level. In previous studies, the interaction of p21 with procaspase 3, and thus the blockade of Fas-induced apoptosis has been shown (Suzuki et al., 1998, 1999). There is also evidence that caspase 3 cleaves p21, thereby, inducing apoptosis in various cell types (Jin et al., 2000). Based on these findings, we predicted that TRAIL might induce procaspase 3 cleavage into caspase 3 once free from p21 after downregulation through the NSC49L pathway. Then, the active caspase 3, through a feedback mechanism, may further cleave p21 to enhance the rate of apoptosis. If this is true, then the inhibition of caspase 3 activity may block NSC49L and TRAIL-induced sensitization of FOLFOX-HT29 cells and the cleavage of procaspase-3 and PARP1.
To test this possibility, we first examined the localization of p21 and procaspase-3 in response to NSC49L and TRAIL using immunofluorescence microscopy. We treated HT29 and FOLFOX-HT29 cells grown on coverslips with 20 μM of NSC49L and 1 nM of TRAIL either alone or in combination for 24 h. Cells were processed for IHC using anti-p21 and anti-procaspase antibodies with corresponding AlexaFluor-488 (green) and AlexaFluor-568 (red) secondary antibodies. Images were captured with a Leica DM IRBE microscope (Hawthorn, NY) at 20× magnification. As p21 levels are very low in HT29 cells, the colocalization of p21 and procaspase 3 was also poor; however, after NSC49L and TRAIL treatment it showed reduced levels and colocalization (Figure S7A). On the other hand, we observed a higher level of p21 and its colocalization with procaspase in FOLFOX-HT29 cells, which was drastically reduced after the NSC49L but not the TRAIL treatment (Figure S7B). The combination treatment of NSC49L and TRAIL even further reduced the levels and the colocalization of p21 and procaspase 3 (Figure S7B). These results suggest that p21 may interact with procaspase 3 and keep it in its inactive state in FOLFOX-HT29 cells. Procaspase 3 may become free and get cleaved to active caspase 3 after NSC49L-mediated p21 reduction in these cells.
Then, we examined whether the caspase 3 specific inhibitor, ZDEVD, will block NSC49- and TRAIL-induced sensitization of FOLFOX-HT29 cells. For these experiments, we pretreated FOLFOX-HT29 cells with 20 μM of ZDEVD for 3 h, followed by the treatment with NSC49L and TRAIL either alone or in combination for an additional 72 h. We performed MTT-cell viability assays and determined the IC50 of the drugs. Here, we have shown the sensitization effect of the drugs as IC50. Pretreatment with ZDEVD significantly increased the IC50 of TRAIL as compared to control, while it did not affect the IC50 of NSC49L (Figures 12A and 12B, respectively). However, pretreatment with ZDEVD significantly increased the IC50 of NSC49L when used in combination with TRAIL (Figure 12B). These results suggest that ZDEVD is acting through the TRAIL pathway, and the NSC49L pathway is coordinating with the TRAIL pathway. Next, we examined whether the coordination between the TRAIL and NSC49L pathways is at the procaspase 3 level. We used the same protocol as above, except that the experiments were terminated after 24 h. As we wanted to examine the effect of ZDEVD on TRAIL-induced effects, we treated FOLFOX-HT29 cells with 1 nM of TRAIL with or without 10 μM of ZDEVD and 20 μM of NSC49L. Western blot results showed no appreciable change in the level of DR5 after the combination treatment of TRAIL and NSC49L as compared to TRAIL alone treated cells (Figure 12C, compare lane 1 with 2). DR5 levels also remained unchanged after TRAIL and NSC49L treatment in the ZDEVD-pretreated cells (Figure 12C, compare lane 3 with 4). On the other hand, p21 levels were decreased in the TRAIL and NSC49L combination treatments as compared to TRAIL alone treated cells (Figure 12C, compare lane 1 with 2), which was restored in the ZDEVD-pretreated cells (Figure 12C, compare lane 3 with 4). Furthermore, the c-Caspase 8, c-Caspase 3, and c-PARP1 levels were increased, and the procaspase 3 level was decreased in (Figure 12C, compare lane 1 with 2). Interestingly, in the ZDEVD-pretreated cells, the combined effects of TRAIL and NSC49L on c-Caspase 8, c-Caspase 3 and c-PARP1, and procaspase 3 levels were reversed (Figure 12C, compare lane 3 with 4). Taken together, these results establish a relationship between p21 and caspase 3 in the regulation of NSC49L- and TRAIL-induced cytotoxic activities in FOLFOX-HT29 cells.
Figure 12.
Caspase 3 inhibitor ZDEVD reverses the cytotoxic effect of NSC49L and TRAIL in FOLFOX-HT29 cells
Cells were pretreated with 20 μM ZDEVD for 3 h followed by the treatment with different concentrations of NSC49L and TRAIL either alone or in combination for an additional 72 h. Cell viability was determined using the MTT-assay.
(A) The IC50 of TRAIL.
(B) The IC50 of NSC49L in a combination of TRAIL. Data are the mean ± SE of four estimations. The p-value is shown on the top of each graph.
(C) Western bots. For these experiments, after pretreatment with ZDEVD, the treatment with NSC49L and TRAIL either alone or in combination was carried out for 24 h. Blots are the representative of two different experiments. See also Figure S7.
Discussion
Access to TCGA data through the Genomic Data Commons Data Portal has provided an enormous opportunity to analyze important genes and pathways by which they can be implicated in developing precision medicine approaches for the diagnosis and treatment of cancer (Jensen et al., 2017). First, we found a decreased level of PPP2C in CRC tumors compared to normal tissues, and this pattern was linked with the progression of the tumor through different stages. Then, we analyzed CPTAC data and found a similar trend at the protein level. The low expression of PPP2CB was linked with a poor prognosis of patients with CRC. Second, we used a meta-analysis method to analyze three microarray datasets (accession number (GSE19860, GSE28702, and GSE72970)) of FOLFOX-responsive and non-responsive CRC tumors (Lin et al., 2018), and established a link between PPP2C and AKT1, mTOR, 4EBP1, and CDKN1A. We found a decreased level of PPP2CA/B in FOLFOX-non-responsive compared with responsive tumors. Furthermore, the expression levels of AKT1, mTOR, 4EBP1, and CDKN1A were increased in FOLFOX-non-responsive compared with responsive tumors. These results provided clear evidence that the upregulation of PP2A and downregulation of the AKT1, mTOR, 4EBP1, and p21 levels can be beneficial for the management of patients with CRC.
In the present study, we pursued the above concept and identified a novel PP2A agonist, NSC49L. We described the molecular mechanisms by which NSC49L downregulates AKT/mTOR/4EBP1 pathway-dependent p21 translation and induces TRAIL/caspase-3-dependent apoptosis of FOLFOX-resistant CRC cells. Decreased levels of PP2AC (Yong et al., 2018) and increased levels of phospho-AKT (Li et al., 2018), phospho-mTOR (Melling et al., 2015), phospho-4EBP1 (Malinowsky et al., 2014) and TRAIL (Zhang and Fang, 2005) have been implicated as drug-resistance factors for patients with metastatic CRC. We have identified p21 as an acquired resistance factor that is observed in HCT116 and HT29 cells during FOLFOX treatment. P21 levels were increased in both FOLFOX-HCT116 and FOLFOX-HT29 cell lines in a p53-independent manner. In fact, we observed a loss of p53 expression in both FOLFOX-HCT116 and FOLFOX-HT29 cell lines. This is the first report showing the upregulation of p21 and loss of p53 in FOLFOX-resistant CRC cells. The role of p21 is well-established in protecting CRC cells against a variety of stress stimuli, including exposure to radiation and chemotherapy, such as 5-FU (Bene and Chambers, 2009; Gorospe et al., 1996; Mahyar-Roemer and Roemer, 2001; Maiuthed et al., 2018; Sharma et al., 2005; Tian et al., 2000). Its upregulation correlates positively with tumor grade, invasiveness, and aggressiveness, and is considered a poor prognostic indicator (Abbas and Dutta, 2009). An anti-apoptotic and enhanced cell survival role of p21 is implicated by inhibiting initiator caspases directed by the TRAIL/DR4 pathway (Xu and El-Deiry, 2000). Considering the anti-apoptotic role of p21, it is, therefore, emerging as a therapeutic target for certain cancers (Weiss, 2003). Although in the current study we focused on p21 as a target of PP2A agonist-induced sensitization of FOLFOX-resistant CRC cells, the mechanism of the loss of p53 expression is still unclear.
By using inhibitors, a link between AKT1, mTOR, and 4EBP1 has been observed (Wang et al., 2020; Wang and Zhang, 2014); however, their efficacy in sensitizing FOLFOX-resistant CRC cells is not clear. Furthermore, the link between PP2A and the AKT1/mTOR/4EBP1-axis and with TRAIL is also obscure. As PP2A dephosphorylates AKT1 and induces the sensitization of cancer cells, it is desirable to develop a PP2A activator. In the past, several PP2A agonists have been developed that showed a significant therapeutic efficacy when used in combination with oncogenic kinase inhibitors, DNA damaging agents, and radiation (Mazhar et al., 2019). These agonists modulate the activity of PP2A by interacting with its scaffolding (PPP2R1A, PR65α) and regulatory (PPP2R2A, B55α) subunits, or by removing the antagonistic effects of the PP2A regulatory subunits (Mazhar et al., 2019; McClinch et al., 2018; Sangodkar et al., 2017). Recently, phenothiazine derivatives have been identified as PP2A agonists that regulate enzymes containing the PPP2R5E (B56ε) regulatory subunit (Morita et al., 2020). However, the targeting by agonists of PP2A’s catalytic (C) subunit is largely unknown. Furthermore, these agonists have not been tested against FOLFOX-resistant CRC cells and show a poor safety profile. On the other hand, our PP2A agonist, NSC49L, is specific to its catalytic subunit, has a safer profile, and sensitizes FOLFOX-resistant CRC cells in vitro (present data) and in vivo patient-derived xenograft models of CRC stem-like cell tumors (Narayan et al., 2017). NSC49L inhibits the AKT1/mTOR/4EBP-axis and interrupts p21 translation. The reduced level of p21 then links with the TRAIL pathway, thereby inducing caspase 3 activation and sensitization of FOLFOX-resistant CRC cells.
In previous studies, cap-dependent translational control has been implicated as a therapeutic target and biomarker for cancer (Vaklavas et al., 2017). Translational control of p21 has been reported for mTOR inhibitors (Beuvink et al., 2005), nutrient stress (Lehman et al., 2015), and UVB-irradiation (Collier et al., 2018). The first step in the cap-dependent translation initiation is the assembly of the trimolecular cap-binding complex, eIF4F, at the 5′UTR of the mRNA bound to the ribosome (Bhat et al., 2015). EIF4F consists of three subunits: eIF4E, the cap-binding protein; eIF4A, a bidirectional ATP-dependent RNA helicase; and eIF4G, a modular scaffolding protein that binds eIF4E, eIF4A, eIF3 and poly(A)-binding protein (PABP). A coordinated interaction between eIF4G, eIF4E, and PABP brings about mRNA circularization and promotes translation initiation (Kahvejian et al., 2005). The colocalization of eIF4E and eIF4G at the 5′-m7GpppN mRNA-cap is a critical step in translational control. The binding of 4EBP1 to the same region of eIF4E where eIF4G binds blocks translation (Bhat et al., 2015). Phosphorylated 4EBP1 dissociates from the complex and allows eIF4E to form an active translation initiation complex (Lin et al., 1994; Pause et al., 1994). The results of our studies supported this hypothesis. The data showed an increased colocalization of 4EBP1 and eIF4E with a decreased p21 translation in FOLFOX-resistant cells. As CDKN1A mRNA levels were increased in the FOLFOX-resistant cells, the possibility of CDKN1A sequestration into stress granules (SGs) is likely. SGs are cytoplasmic multimeric RNA bodies formed in response to anticancer drugs. The sequestered CDKN1A may become unavailable for protein translation in the FOLFOX-resistant cells after the NSC49L treatment. In recent years, the involvement of AKT/mTOR/4EBP1 has been described in the formation of SGs in response to anticancer drugs (Fournier et al., 2013). The formation of SGs is known to inhibit cap-dependent translation of p21 by the stabilization of CDKN1A mRNA in SGs (Gareau et al., 2011). The proteasome inhibitor, MG132, causes trapping of CDKN1A mRNA in SGs, resulting in the inhibition of p21 translation (Lian and Gallouzi, 2009). We have not examined the involvement of SGs in NSC49L-mediated regulation of p21 translation. Nonetheless, our results clearly suggest that NSC49L affects selective inhibition of p21 translation, instead of the global inhibition of protein translation, and thus can be a more therapeutically desirable agent than the ones affecting global protein synthesis. In other studies, rapamycin-dependent selective inhibition of mitogen-induced p21 translation (Gaben et al., 2004), as well as a global inhibition by mTOR inhibitor RAD001 (Beuvink et al., 2005) has been reported. The global inhibition of translation may preferentially affect cancer cells because the cancer cells have higher rates of translation than normal cells (Heys et al., 1991; Mills et al., 2008).
There is huge clinical potential for TRAIL in cancer therapy owing to its lack of toxicity to normal cells, but high toxicity to cancer cells (Deng and Shah, 2020). However, about 50% of cancer cells develop resistance to TRAIL (Walczak et al., 1999), thus limiting its therapeutic efficacy. Currently, multiple means, including the use of genotoxic agents, epigenetic modulators, synthetic small molecules, or immunotherapy either alone or in combination, have been implicated to circumvent TRAIL resistance and to induce apoptosis (Thorburn et al., 2008). TRAIL-mediated resistance can occur through two general mechanisms. First, through anti-apoptotic mechanisms, such as the reduced expression or epigenetic silencing of caspase 8, and increased expression of caspase inhibitors XIAP, cIAP2, and Mcl-1, or overexpression of anti-apoptotic protein Bcl2. Second, TRAIL resistance can result from decreased expression or cell surface localization of the TRAIL receptors DR4/DR5 or increased expression of DR4/DR5 inhibitors Fas-associated death domain-like interleukin-1β-converting enzyme-inhibitory protein (FLIP) and decoy receptor 1 and 2 (DcR1/DcR2) (Thorburn et al., 2008). In the present study, we have used Apo2L/TRAIL recombinant protein of 114–281 amino acids. Although its plasma half-life is very short, it selectively induces apoptosis and overcomes conventional drug resistance in preclinical in vitro and in vivo studies (Mitsiades et al., 2001). It has been already approved for use in clinical trials (Ashkenazi et al., 1999; Kelley and Ashkenazi, 2004; Kelley et al., 2001; Walczak et al., 1999). The Apo2L/TRAIL is untagged, less immunogenic in humans, and better at the induction of apoptosis (Ashkenazi et al., 1999). The Apo2/TRAIL also has the advantage of targeting both TRAIL receptors DR4 and DR5, rendering it less dependent on the expression of only one of them (von Karstedt et al., 2017). Although clinical trials with Apo2/TRAIL monotherapy did not produce significantly improved anticancer activity (Herbst et al., 2010; Lemke et al., 2014; Soria et al., 2011), we think that the combination strategy with NSC49L may be a better and feasible therapeutic strategy. Furthermore, we have observed that decreased expression of DR5 in CRC cells may have caused TRAIL resistance, which was reversed after DR5 overexpression.
In distinct cancer types, different mechanisms of TRAIL resistance may be operative. However, in FOLFOX-resistant CRC cells we show p21 as a major TRAIL resistance factor. This is consistent with our results showing a synergistic effect of NSC49L and TRAIL on the sensitization of CDKN1A (p21)-knockdown CRC cells that is reversed after p21-overexpression. TRAIL-induced DR5/caspase 8 signaling is active in FOLFOX-resistant cells but becomes trapped at the procaspase 3 step, as p21 bound procaspase 3 is not a suitable substrate for caspase 8. Then, the trapped procaspase 3 is not processed into caspase 3, resulting in the onset of apoptosis and induction of TRAIL resistance (Figure 12). After NSC49L treatment, PP2A activity was induced, which downregulated AKT1/mTOR/4EBP1 signaling and p21 translation in FOLFOX-resistant CRC cells. The reduced p21 level then relieved procaspase 3, which was then cleaved into caspase 3 through the TRAIL-induced DR5/caspase 8 pathway. Active caspase 3 then induced sensitization of FOLFOX-resistant CRC cells. Furthermore, active caspase 3 may through a feedback mechanism increase the cleavage of p21 to further potentiate the effect of NSC49L and TRAIL on the sensitization of these cells (Figure 12). A feedback mechanism of caspase 3-mediated cleavage of p21 is consistent with previous findings (Jin et al., 2000; Zhang et al., 1999). Thus, these preclinical studies provide a mechanistic explanation for how the novel PP2A agonist NSC49L can modulate the PP2A/AKT1/mTOR/4EBP1-axis and enhance the therapeutic efficacy of TRAIL by targeting the downregulation of p21 translation and caspase 3 activation in FOLFOX-resistant CRC cells.
Limitations of the study
Although PP2A activator, NSC49L, synergistically enhances TRAIL-mediated sensitization of FOLFOX-resistant CRC cells by the downregulation of the AKT1/mTOR/4EBP1 axis and p21 protein translation, its low plasma half-life remains a concern. To improve the affinity for the catalytic subunit of PP2A and to enhance the biological and pharmacological activities, including not only the half-life but cancer cell cytotoxicity, and therapeutic index, we plan on optimizing NSC49L structure by making appropriate single and combinational functional group alterations. We will develop NSC49L analogs with better bioavailability and having greater potency and safety profile that can affect both the intrinsic and acquired resistant CRCs. Other limitation of this study is understating the molecular mechanisms of NSC49L-mediated downregulation of p21 translation. In future studies, we will determine whether NSC49L through the AKT1/mTOR/4EBP1 axis affects the p21 translation at cap-dependent translation initiation complex assembly or the formation of stress granules (SGs). Nonetheless, our current studies provide a strong foundation for future investigations that will lead to the synthesis of a drug-like NSC49L analog with better therapeutic efficacy and will examine in detail the molecular mechanisms of p21 translational regulation in FOLFOX-resistant CRC cells. We will extend these studies with PP2A activator alone or in combination with TRAIL and other Food and Drug Administration (FDA) approved chemotherapeutic agents to both in vitro and in vivo models.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| 4EBP1 mAb (Rabbit) | Cell Signaling Technology | Cat# 9644; RRID:AB_2097841 |
| 4EBP1 (T37/46P) mAb (Rabbit) | Cell Signaling Technology | Cat# 2855; RRID:AB_560835 |
| AKT1 antibody (Rabbit) | Cell Signaling Technology | Cat# 9272; RRID:AB_329827 |
| AKT1 (S473P) mAb (Mouse) | Cell Signaling Technology | Cat# 4051; RRID:AB_331158 |
| AKT1 (T308P) antibody (Rabbit) | Cell Signaling Technology | Cat# 9275; RRID:AB_329828 |
| AKT2 antibody (Rabbit) | Cell Signaling Technology | Cat# 2962; RRID:AB_329872 |
| AKT3 antibody (Rabbit) | Cell Signaling Technology | Cat# 4059; RRID:AB_2225351 |
| Anti-p21 Antibody (F-5) (Mouse) | Santa Cruz Biotechnology | Cat# sc-6246; RRID:AB_628073 |
| Anti-puromycin antibody (Mouse) | Millipore | Cat# MABE343; RRID:AB_2566826 |
| BAD mAb (Rabbit) | Cell Signaling Technology | Cat# 9239; RRID:AB_2062127 |
| BAX mAb (Rabbit) | Cell Signaling Technology | Cat# 5023; RRID:AB_10557411 |
| Bcl2 mAb (Rabbit) | Cell Signaling Technology | Cat# 4223; RRID:AB_1903909 |
| β-Actin (13E5) mAb (Rabbit) | Cell Signaling Technology | Cat# 4970; RRID:AB_2223172 |
| Bim mAb (Rabbit) | Cell Signaling Technology | Cat# 2933; RRID:AB_1030947 |
| Caspase 3 (Rabbit) | Cell Signaling Technology | Cat# 9662; RRID:AB_331439 |
| Caspase 8 mAb (Rabbit) | Cell Signaling Technology | Cat# 4790; RRID:AB_10545768 |
| Cleaved caspase 3 (Rabbit) | Cell Signaling Technology | Cat# 9664; RRID:AB_2070042 |
| Cleaved caspase 8 mAb (Rabbit) | Cell Signaling Technology | Cat# 8592; RRID:AB_10891784 |
| Cleaved PARP1 mAb (Rabbit) | Cell Signaling Technology | Cat# 5625; RRID:AB_10699459 |
| DR5 mAb (Rabbit) | Cell Signaling Technology | Cat# 8074; RRID:AB_10950817 |
| GAPDH mAb (Rabbit) | Cell Signaling Technology | Cat# 8884; RRID:AB_11129865 |
| IgG (Rabbit) | Sigma-Aldrich | Cat# GENA934, RRID:AB_2722659 |
| IgG2a, kappa Isotype Control Antibody (Mouse) | Biolegend | Cat# 401502; RRID:AB_2800437 |
| mTOR (Rabbit) | Cell Signaling Technology | Cat# 2983; RRID:AB_2105622 |
| mTOR (S2448P) mAb (Rabbit) | Cell Signaling Technology | Cat# 5536; RRID:AB_10691552 |
| p53 (Mouse) | Santa Cruz Biotechnology | Cat# sc-126; RRID:AB_628082 |
| PARP1 mAb (Rabbit) | Cell Signaling Technology | Cat# 9532; RRID:AB_659884 |
| PP2AC mAb (Rabbit) | Cell Signaling Technology | Cat# 2259; RRID:AB_561239 |
| PUMA (Rabbit) | Cell Signaling Technology | Cat# 98672; RRID:N/A |
| Chemicals, peptides, and recombinant proteins | ||
| 10x Tris/glycine buffer | Bio-Rad | Cat# 1610734 |
| 10x Tris/glycine/SDS buffer | Bio-Rad | Cat# 1610732 |
| 5-FU (99% HPLC purified) | Sigma-Aldrich | Cat# F6627 |
| BCA protein assay kit | Thermo-Scientific | Cat# 23225 |
| Bolt LDS sample buffer (4X) | Life Technologies | Cat# B0007 |
| Cell Dissociation Buffer, enzyme-free | Thermo-Scientific | Cat#13151014 |
| Cycloheximide | Sigma-Aldrich | Cat# 01810 |
| Halt Protease and Phosphatase inhibitor cocktail | Thermo-Scientific | Cat# 78440 |
| Immobilon-P (PVDF) membrane | Merck Millipore | Cat# IPVH00010 |
| Mini-PROTEAN TGX precast gel 4–15% | Bio-Rad | Cat# 456–1085 |
| Muse Caspase-3/7 kitEMD | Millipore | Cat# MCH100108 |
| MTT (3-(4,5-dimethylthiazol-2yl-)-2,5-diphenyl tetrazolium bromide) | Fisher Scientific | Cat# M6494 |
| Oxaliplatin | TOCRIS Biosciences | Cat# 2623 |
| PBS with 0.5% Tween 20 pH 7.4 | Sigma-Aldrich | Cat# P3563 |
| Phosphate Buffer Saline | Sigma-Aldrich | Cat# P-5368 |
| Pierce RIPA buffer | Thermo-Scientific | Cat# 89901 |
| Protein A/G PLUS-Agarose | Santa Cruz Biotechnology | Cat# sc-2003 |
| Puromycin dihydrochloride | Santa Cruz Biotechnology | Cat# sc-108071A |
| rPP2A-Cα (recombinant) | Cayman Chemical | Cat# 10011237 |
| Threonine phosphopeptide (KRpTIRR) | MilliporeSigma | Cat# 12-219 |
| TRAIL/Apo2l | PreproTech, Inc. | Cat# 310-04 |
| TRIZOL reagent | Life Technologies | Cat# 15596 |
| Trypsin/EDTA (0.05%) | Thermo-Scientific | Cat# 25300054 |
| Deposited data | ||
| RNAseq data of FOLFOX-resistant HT29 cells | This paper: https://doi.org/10.5281/zenodo.6565046 | Zenoda data: https://zenodo.org/record/6565046#.YobdkKjMJPY |
| TCGA data were for CRC cells | This paper: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/studied-cancers/colorectal | Cancer Genome Atlas: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/studied-cancers/colorectal |
| Gene expression data for FOLFOX-resistant and non-resistant patient | This paper: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72970 | NCBI Gene expression Omnibus (GEO) under GSE72970 accession: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72970 |
| Experimental models: Cell lines | ||
| FHC | ATCC | N/A |
| FOLFOX-resistant HCT116 | Adhip P. Majumdar, John D. Dingell VA Medical Centre, Detroit, MI | N/A |
| FOLFOX-resistant HT29 | Adhip P. Majumdar, John D. Dingell VA Medical Centre, Detroit, MI | N/A |
| HCT116 | ATCC | N/A |
| HCT116-p21−/− cells | Bert Vogelstein, Johns Hopkins Medical School, Baltimore, MD | N/A |
| HCT116-p21−/−/H6p21 cells | Brian K. Law, University of Florida, Gainesville, FL | N/A |
| HT-29 | ATCC | N/A |
| LoVo | ATCC | N/A |
| LS174T | ATCC | N/A |
| RKO | ATCC | N/A |
| SW480 | ATCC | N/A |
| Oligonucleotides | ||
| actin B-F (GCTCCTCCTGAGCGCAAGTACTC | Sigma-Aldrich | N/A |
| actin B-R (GTGGACAGCGAGGCCAGGAT) | Sigma-Aldrich | N/A |
| p21-F- (CCGCGACTGTGATGCGCTAATG) | Sigma-Aldrich | N/A |
| p21-R (CTCGGTGACAAAGTCGAAGTTC) | Sigma-Aldrich | N/A |
| TP53-F (CACATGACGGAGGTTGTGAG) | Sigma-Aldrich | N/A |
| TP53-R (ACACGCAAATTTCCTTCCAC) | Sigma-Aldrich | N/A |
| Software and algorithms | ||
| CalcuSyn | Biosoft | http://www.biosoft.com/w/calcusyn.htm |
| GraphPad Prism v8.0 | Dotmatics | N/A |
| ImageJ | NIH Image | N/A |
| Sigma-Plot 9 | Systat Software, Inc. | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed and will be fulfilled by the lead contact, Prof. Satya Narayan (snarayan@ufl.edu) or Prof. Arun K. Sharma (asharm1@pennstatehealth.psu.edu).
Materials availability
FOLFOX-resistant HCT116 and HT29 cell lines were obtained from Dr. Adhip P. Majumdar (John D. Dingell VA Medical Center, Detroit, MI). Other CRC and normal cell lines were obtained from ATCC. ATCC source information is provided in the key resources table. HCT116-p21−/− cells were obtained from Dr. Bert Vogelstein (Johns Hopkins Medical School, Baltimore, MD). HCT116-p21−/−/H6p21 cells were created and characterized by Drs. Brian K. and Mary E. Law (University of Florida, Gainesville, FL) using previously described reagents and methods (Jahn et al., 2013; Law et al., 2006). HT29/tet-DR5 was constructed and characterized by Drs. Brian K. and Mary E. Law using the vectors and methods described previously (Wang et al., 2019). These cell lines can be requested from the above-mentioned investigators.
Synthesis of NSC49L was according to the previously published methods (Warmus et al., 1993). Compounds can be generated according to the described procedure or will be shared upon request by contacting Dr. Arun K. Sharma (Department of Pharmacology, Penn State University College of Medicine, Hershey, PA). Oxaliplatin and 5-FU were purchased from TOCRIS Biosciences and Sigma-Aldrich, respectively. TRAIL/Apo2L and PP2A-Cα were purchased from PreproTech, Inc. (Cranbury, NJ) and Cayman Chemical (Ann Arbor, MI), respectively.
Methods details
Experimental model and subject details
In these studies, human colorectal cancer (CRC) and normal colonic epithelial cell lines have been used a model system. The CRC cell lines HCT-116, LoVo, SW480, LS174T, RKO, HT-29, and the normal colonic epithelial cell line FHC were obtained from the American Type Culture Collection (ATCC, Rockville, MD). Cells were maintained as recommended by ATCC either in McCoy’s 5a or Dulbecco’s modified Eagle medium (DMEM; 4.5 g/L D-glucose) supplemented with 10% FBS and 1% antibiotic/antimycotic in tissue culture flasks in a humidified incubator at 37 °C in an atmosphere of 95% air and 5% carbon dioxide. The medium was changed two times a week and cells were passaged using 0.05% trypsin/EDTA. The normal colonic epithelial cell line FHC was maintained in ATCC-formulated DMEM:F12 Medium Catalog #30–2006, containing 25 mM HEPES, 10 ng/mL cholera toxin, 0.005 mg/mL insulin, 0.005 mg/mL transferrin, 100 ng/mL hydrocortisone and fetal bovine serum 10% (v/v). The FOLFOX-resistant HCT-116 and HT29 cell lines were obtained from Dr. Adhip P. Majumdar (John D. Dingell VA Medical Center, Detroit, MI) (Yu et al., 2009), as described in (Narayan et al., 2017).
Cell viability assay
The viability of cells was determined by MTT (3-(4,5-dimethylthiazol-2yl-)-2,5-diphenyl tetrazolium bromide) assay. In principle, the viable cell number is directly proportional to the purple formazan color of the reduced MTT dye, which can be quantitatively measured by spectrophotometry. Briefly, 1,500 cells were plated in quadruplets in 96-well flat-bottom tissue culture plates. After treatment with compounds for certain periods as described in respective figure legends, 10 μL of MTT reagent was added to each well and incubated at 37 °C for 4 h to allow the formation of purple color crystals of formazan. In total, 100 μL of detergent solution was added to each well, and the reaction mixture was incubated in the dark for 2–4 h at room temperature. The developed color density was then measured spectrophotometrically at 570 nm using the POLARstar Omega micro-plate reader (BMG Labtech, Inc., Cary, NC).
Apoptosis assay
The apoptosis assay using Muse Caspase-3/7 AAD assay kit was performed following manufacturer’s protocol. Briefly, cells were seeded in 6-well plate at a density of 1 × 105 cells/well. Next day, cells were treated with 20 μM NSC49L and 1 nM TRAIL either alone or in combination for 24 h. Then, cells were harvested using enzyme-free Cell Dissociation Buffer. The cells were resuspended in 1× Assay buffer BA. A 50 μL cell suspension and 5 μL Muse Caspase-3/7 reagent working solution was added in each tube. Samples were mixed thoroughly and gently by pipetting up and down, followed by incubation for 30 min at 37 °C with 5% CO2. After incubation, 150 μL of Muse Caspase 7-AAD working solution was added to each tube and mixed thoroughly. Samples were incubated for 5 min in dark at room temperature, and then analyzed in Muse Cell Analyzer. Events in each of the four quadrants were as follows. Lower-left: Viable cells, not undergoing detectable apoptosis [caspase-3/7 (−) and dead cell marker (−)]; Lower-right: cells in early stages of apoptosis [caspase-3/7 (+) and dead cell marker (−)]; Upper-right: cells in late stages of apoptosis or dead by apoptotic mechanism [caspase-3/7 (+) and dead cell marker (+)], and Upper-left: cells that have died via necrosis but not through apoptotic pathway [caspase-3/7 (−) and dead cell marker (+)].
PP2A assay
We determined PP2A activity by using recombinant human PP2A-Cα protein and threonine phosphopeptide (KRpTIRR) substrate following the Malachite green protocol as previously described (Lek et al., 2017). The exact protocol we followed for the PP2A assay is given at: https://www.jove.com/pdf/55361/jove-protocol-55361-recombinant-synucleins-stimulate-protein-phosphatase-2a-catalytic. We used 0.0007 U/μL of human rPP2Ac, 0, 1, 10, 50, 100, 500 and 1,000 nM NSC49L, and 2 mM threonine phosphopeptide (KRpTIRR) substrate in this assay. PO4 standards (0, 150, 300, 600, 1,200, and 2,400 pmol) were run at the same time. Reaction was started by the addition of the Malachite green solution, incubated for 10 min at room temperature, and the change in color was read at 630 nm using POLARstar Omega micro-plate reader (BMG Labtech, Inc., Cary, NC).
We also determined PP2A activity in the immunocomplexes isolated from the FOLFOX-HT29 cells using anti-PP2A-C antibody as described (Lek et al., 2017). Cells were treated with NSC49L (0, 5, 10 and 20 μM) for 24 h. Whole lysates were prepared and immunocomplexes were isolated on Protein A Sepharose CL-4B beads. A 250 mg beads were diluted in 4 mL cold PBS containing 0.1% NaN3 (w/v), vortexed briefly, and incubated at room temperature in agitation (rotation) for 1 h. Then, the beads were spined down at 5,000 rpm for 1 min, washed 3-times with Co-IP buffer. Resuspend the beads in twice-volume of the Co-IP buffer. A 250 mg beads made 1.5 mL of the bed volume. Resuspend the pellet into 1.5 mL of Co-IP buffer with 1:1 ratio. Precleared the whole cell lysate by adding 100 μL of beads slurry (50 μL packed beads) per 100 μL of the whole cell lysate (containing 100 μg of protein) and incubated at 4 °C for 10–30 min on a rocker or orbital shaker. Beads were removed by centrifugation at 14,000 × g at 4 °C for 10 min. The supernatant was transferred to a fresh centrifuge tube and the pellet was discarded. To the 100 μg of precleared whole cell lysate, 2 μg Anti-PP2A, C subunit (52F8) rabbit monoclonal antibody was added and incubated at 4 °C for 1–2 h or overnight on a rotator. Immunocomplexes were captured by adding 50 μL Protein A sepharose CL-4B beads slurry (25 μL packed beads), and gently rocking on a rocker for 1 h or overnight at 4 °C. The volume was brought to 500 μL with of ice-cold lysis buffer, and then centrifuged at 2,500 × g for 30 s at 4 °C. Carefully removed the supernatant and washed the beads 5-times with 500 μL of pNPP assay buffer. Finally, resuspended the beads into a pNPP buffer and determined the PP2A activity as described above.
qRT-PCR
The mRNA levels of CDKN1A (p21) and TP53 (p53) genes were determined by qRT-PCR. The total RNA was isolated from control and treated cells by using TRIZOL reagent. The nucleotide sequences used for the forward and reverse primers are described in the key resources table. The qRT-PCR was run on a real-time PCR machine (ABT 7500 fast) with standard cycle protocol. The relative mRNA abundance was determined using the differential cycle threshold (ΔCt) between the Ct values of cDNA from experiment and control cells.
RNAseq analysis
For RNA-seq data analysis, we used the RNAseq data analysis pipeline reported previously. Briefly, Briefly, FASTQ files were aligned to Genome Reference Consortium Human Build 38 (GRCh38) using HISAT2; the transcripts assembling was performed using StringTie with RefSeq as transcripts ID; and the normalized counts (by FPKM) was called using Ballgown. The differential expression analysis was performed using R package limma; and the gene enrichment analysis was performed using R heatmap package.
Western blot analysis
The levels of various proteins were determined by western blot analysis using 5-bromo-4-chloro-3-indolyl phosphate (BCIP)/nitro blue tetrazolium (NBT) substrate. Thirty micrograms of the whole cell lysate were separated by SDS-PAGE and transferred onto the PVDF membrane. After the transfer, the membrane was Incubated in the blocking solution (0.2 g/L NaN3, 50 g/L BSA and 2 mL/L Tween 20) for 60 min. Then, incubated with primary antibody (1:1,000, v/v in the blocking solution) at 4 °C for overnight. Washed the membrane 3-times, each for 10 min, in a washing buffer ((Tris-base 6.05 g/L, CaCl2 (Dihydrate) 0.222 g/L, NaCl 4.75 g/L and Tween 20 500 μL/L, pH 8)). Incubated the membrane in a secondary antibody (1:5,000, v/v in the blocking solution) for 60 min. Washed the membrane 3-times, each for 10 min, in a washing buffer as described above. Layered the membrane with alkaline phosphatase substrate ((33 μL BCIP (5 mg/mL in 100% dimethylformamide) and 66 μL NBT (10 mg/mL in 70% dimethylformamide)) for 5–15 min. Once the desired signal appeared, the membrane washed 3-times with water. After membrane dries, the signal was scanned and recorded. In some cases, we used chemiluminescence method of western blotting as well.
SUnSET assays
Following 24 h treatment with 20 μM of NSC49L, FOLFOX-HT29 cells were subjected to SUnSET assay (Schmidt et al., 2009), whereby cells were incubated with puromycin that was incorporated into the newly translated proteins. Briefly, after the onset of the treatment period, the cells were treated with vehicle control or with puromycin (10 μg/mL) or with puromycin and cycloheximide (CHX; 10 μg/mL) for 15 min (pulse). Further, the medium was removed, and the cells were washed thrice with fresh medium and incubated in fresh growth medium for an additional 2 h chase. The proteins were extracted, and 500 μg of cell lysate was used for immunoprecipitations (IP). The cell lysate was incubated with 5 μg of mouse control IgG2a (401,501), K isotype, or 5 μg of mouse anti-puromycin antibody (MABE343) for 2 h at 4 °C. Resuspended Protein A/G PLUS-Agarose (20 μL) was added to the cell lysate and mixed at 4 °C overnight. Immunocomplexes were collected by centrifugation at 2,500 rpm for 5 min at 4 °C. Pellets were washed 4-times with 1 mL PBS. After the final wash, the supernatant was discarded, and pellet was resuspended in 40 μL of 2× electrophoresis sample buffer. Proteins were then resolved by SDS-PAGE and analyzed by immunoblotting using an anti-p21 (F-5) antibody. The pre-immunoprecipitated lysate was used to assess the loading control, puromycin incorporation and p21 biosynthesis using β-actin antibody (1:1,000, v/v), anti-puromycin antibody (1:2,500, v/v) and p21 antibody (1:200, v/v), respectively.
Small molecule synthesis
NSC49L was synthesized following the procedure of Warmus et al. (Narayan et al., 2019; Warmus et al., 1993). The final product was purified, crystallized, and characterized based on NMR and mass spectral data. The purity was determined to be at the level of ≥99%. Briefly, a mixture of 1,3,5,7-tetraazaadamantane (1.0 mmol) and 1 4-dichloro-2-butene (1.0 mmol) in CH2Cl2 (10 mL) was refluxed 6 h. After completion of the reaction as indicated by TLC, reaction mixture was cooled to room temperature, filtered, and the residue washed with excess methylene dichloride. The crude solid thus obtained was recrystallized in an EtOH/CH2Cl2 mixture and was dried under vacuum (1 mm) to afford NSC49L. Yield 92% (0.243 g); white solid; mp 199–201 °C; 1H NMR (500 MHz, D2O) δ [ppm]: 6.31–6.27 (m,1H), 5.97–5.93 (m,1H), 5.08 (s, 6H), 4.71 (s, 3H), 4.54 (d, J = 11.0 Hz, 3H), 4.20 (d, J = 5.0 Hz, 2H), 3.57 (d, J = 6.0 Hz, 2H); 13C NMR (125 MHz, DMSO-d6) δ [ppm]: 140.6, 117.1, 78.1, 70.13, 57.8, 43.0; HRMS (ESI, M+) calcd. for C10H18ClN4 229.1219, found 229.1218.
Quantification and statistical analysis
Statistical analyses for synergies between NSC49L and TRAIL treatments and p21 status were performed using CalcuSyn software (http://www.biosoft.com/w/calcusyn.htm). The combination index (CI) was calculated by applying Chou-Talalay method and was used for synergy quantification (Chou, 2010). Student’s t test was used for comparisons in both in vitro and in vivo experiments. All experiments were repeated at least three times and results were expressed as mean ± SE. One-way analysis of variance (ANOVA) was calculated with Sigma-Plot 9. A one-tailed t-test was used to compare any significant differences between control and treated groups. The criterion for statistical significance was p < 0.05. For western blotting data, band intensities were measured using ImageJ and normalized with GAPDH. Graph Pad Prism version 9.1.0 was also used for data visualization and p value calculation.
Acknowledgments
This work was supported in part by the Department of Anatomy and Cell Biology and by Royalty Funds (#00126956), University of Florida, Gainesville, FL to S.N. A.K.S thanks to the Department of Pharmacology, Penn State College of Medicine, and Penn State Cancer Institute (PSCI) for financial support. A.K.S. is also funded by NIH/NCI grant R21 CA234681 and CDMRP Lung Cancer Research Program (LCRP) Award Number W81XWH-19-1–0231. The authors thank the Organic Synthesis Shared Resource of the Penn State Cancer Institute for the synthesis of NSC49L. B.L. was supported in part by grants from the Florida Breast Cancer Foundation, the Ocala Royal Dames for Cancer Research, and the Office of the Assistant Secretary of Defense for Health Affairs through the Breast Cancer Research Program under Award No. W81XWH-15-1–0199, and NIH/NCI grant CA252400.
Author contributions
S.N. developed the concept, performed experiments, and wrote the article; A.K.S., synthesis of the NSC49L, was consistently involved during the development of the study; N.K. and A.R. performed experiments; I.M. and T.J.G., performed bioinformatic analysis; and B.K.L. and M.E.L., provided reagents and made intellectual contributions. All authors read the article and provided their feedback.
Declaration of interests
The authors declare no competing interests.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104518.
Supplemental information
Data and code availability
-
•
TCGA data were for CRC’s are available at The Cancer Genome Atlas (TCGA) data repository (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/studied-cancers/colorectal).
-
•
Gene expression data for FOLFOX-resistant and non-resistant patient are available at the NCBI Gene expression Omnibus (GEO) under GSE72970 accession (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72970).
-
•
RNAseq data are deposited at Zenodo Data Repository (https://zenodo.org/record/6565046#.YobdkKjMJPY) and data are publicly available. Link to this data is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abbas T., Dutta A. p21 in cancer: intricate networks and multiple activities. Nat. Rev. Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal A., MacKenzie R.J., Pippa R., Eide C.A., Oddo J., Tyner J.W., Sears R., Vitek M.P., Odero M.D., Christensen D.J., Druker B.J. Antagonism of SET using OP449 enhances the efficacy of tyrosine kinase inhibitors and overcomes drug resistance in myeloid leukemia. Clin. Cancer Res. 2014;20:2092–2103. doi: 10.1158/1078-0432.ccr-13-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkenazi A., Pai R.C., Fong S., Leung S., Lawrence D.A., Marsters S.A., Blackie C., Chang L., McMurtrey A.E., Hebert A., et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J. Clin. Invest. 1999;104:155–162. doi: 10.1172/jci6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bene A., Chambers T.C. p21 functions in a post-mitotic block checkpoint in the apoptotic response to vinblastine. Biochem. Biophys. Res. Commun. 2009;380:211–217. doi: 10.1016/j.bbrc.2009.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuvink I., Boulay A., Fumagalli S., Zilbermann F., Ruetz S., O'Reilly T., Natt F., Hall J., Lane H.A., Thomas G. The mTOR inhibitor RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell. 2005;120:747–759. doi: 10.1016/j.cell.2004.12.040. [DOI] [PubMed] [Google Scholar]
- Bhat M., Robichaud N., Hulea L., Sonenberg N., Pelletier J., Topisirovic I. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 2015;14:261–278. doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- Briffa R., Langdon S.P., Grech G., Harrison D.J. Acquired and intrinsic resistance to colorectal cancer treatment. IntechOpen Chapter. 2017;14:24. [Google Scholar]
- Carneiro B.A., El-Deiry W.S. Targeting apoptosis in cancer therapy. Nat. Rev. Clin. Oncol. 2020;17:395–417. doi: 10.1038/s41571-020-0341-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou T.C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.can-09-1947. [DOI] [PubMed] [Google Scholar]
- Collier A.E., Spandau D.F., Wek R.C. Translational control of a human CDKN1A mRNA splice variant regulates the fate of UVB-irradiated human keratinocytes. Mol. Biol. Cell. 2018;29:29–41. doi: 10.1091/mbc.e17-06-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristobal I., Manso R., Rincón R., Caramés C., Senin C., Borrero A., Martínez-Useros J., Rodriguez M., Zazo S., Aguilera O., et al. PP2A inhibition is a common event in colorectal cancer and its restoration using FTY720 shows promising therapeutic potential. Mol. Cancer Therapeut. 2014;13:938–947. doi: 10.1158/1535-7163.mct-13-0150. [DOI] [PubMed] [Google Scholar]
- Cristobal I., Rincon R., Manso R., Carames C., Zazo S., Madoz-Gurpide J., Rojo F., Garcia-Foncillas J. Deregulation of the PP2A inhibitor SET shows promising therapeutic implications and determines poor clinical outcome in patients with metastatic colorectal cancer. Clin. Cancer Res. 2015;21:347–356. doi: 10.1158/1078-0432.ccr-14-0724. [DOI] [PubMed] [Google Scholar]
- De Palma R.M., Parnham S.R., Li Y., Oaks J.J., Peterson Y.K., Szulc Z.M., Roth B.M., Xing Y., Ogretmen B. The NMR-based characterization of the FTY720-SET complex reveals an alternative mechanism for the attenuation of the inhibitory SET-PP2A interaction. Faseb. J. 2019;33:7647–7666. doi: 10.1096/fj.201802264r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng D., Shah K. TRAIL of hope meeting resistance in cancer. Trends in cancer. 2020;6:989–1001. doi: 10.1016/j.trecan.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier M.J., Coudert L., Mellaoui S., Adjibade P., Gareau C., Côté M.F., Sonenberg N., Gaudreault R.C., Mazroui R. Inactivation of the mTORC1-eukaryotic translation initiation factor 4E pathway alters stress granule formation. Mol. Cell Biol. 2013;33:2285–2301. doi: 10.1128/mcb.01517-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaben A.M., Saucier C., Bedin M., Barbu V., Mester J. Rapamycin inhibits cdk4 activation, p 21(WAF1/CIP1) expression and G1-phase progression in transformed mouse fibroblasts. Int. J. Cancer. 2004;108:200–206. doi: 10.1002/ijc.11521. [DOI] [PubMed] [Google Scholar]
- Gareau C., Fournier M.J., Filion C., Coudert L., Martel D., Labelle Y., Mazroui R. p21(WAF1/CIP1) upregulation through the stress granule-associated protein CUGBP1 confers resistance to bortezomib-mediated apoptosis. PLoS One. 2011;6:e20254. doi: 10.1371/journal.pone.0020254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgakilas A.G., Martin O.A., Bonner W.M. p21: a two-faced genome guardian. Trends Mol. Med. 2017;23:310–319. doi: 10.1016/j.molmed.2017.02.001. [DOI] [PubMed] [Google Scholar]
- Gerdes M.J., Sevinsky C.J., Sood A., Adak S., Bello M.O., Bordwell A., Can A., Corwin A., Dinn S., Filkins R.J., et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl. Acad. Sci. U S A. 2013;110:11982–11987. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorospe M., Wang X., Guyton K.Z., Holbrook N.J. Protective role of p21(Waf1/Cip1) against prostaglandin A2-mediated apoptosis of human colorectal carcinoma cells. Mol. Cell Biol. 1996;16:6654–6660. doi: 10.1128/mcb.16.12.6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M.M. Mechanisms of cancer drug resistance. Annu. Rev. Med. 2002;53:615–627. doi: 10.1146/annurev.med.53.082901.103929. [DOI] [PubMed] [Google Scholar]
- Graff J.R., Konicek B.W., Carter J.H., Marcusson E.G. Targeting the eukaryotic translation initiation factor 4E for cancer therapy. Cancer Res. 2008;68:631–634. doi: 10.1158/0008-5472.can-07-5635. [DOI] [PubMed] [Google Scholar]
- Herbst R.S., Eckhardt S.G., Kurzrock R., Ebbinghaus S., O'Dwyer P.J., Gordon M.S., Novotny W., Goldwasser M.A., Tohnya T.M., Lum B.L., et al. Phase I dose-escalation study of recombinant human Apo2L/TRAIL, a dual proapoptotic receptor agonist, in patients with advanced cancer. J. Clin. Oncol. 2010;28:2839–2846. doi: 10.1200/jco.2009.25.1991. [DOI] [PubMed] [Google Scholar]
- Heys S.D., Park K.G., McNurlan M.A., Calder A.G., Buchan V., Blessing K., Eremin O., Garlick P.J. Measurement of tumour protein synthesis in vivo in human colorectal and breast cancer and its variability in separate biopsies from the same tumour. Clin. Sci. (Lond.) 1991;80:587–593. doi: 10.1042/cs0800587. [DOI] [PubMed] [Google Scholar]
- Hoang B., Benavides A., Shi Y., Yang Y., Frost P., Gera J., Lichtenstein A. The PP242 mammalian target of rapamycin (mTOR) inhibitor activates extracellular signal-regulated kinase (ERK) in multiple myeloma cells via a target of rapamycin complex 1 (TORC1)/eukaryotic translation initiation factor 4E (eIF-4E)/RAF pathway and activation is a mechanism of resistance. J. Biol. Chem. 2012;287:21796–21805. doi: 10.1074/jbc.m111.304626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobden A.N., Cundliffe E. The mode of action of alpha sarcin and a novel assay of the puromycin reaction. Biochem. J. 1978;170:57–61. doi: 10.1042/bj1700057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikoma N., Raghav K., Chang G. An update on randomized clinical trials in metastatic colorectal carcinoma. Surg. Oncol. Clin. 2017;26:667–687. doi: 10.1016/j.soc.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitsuka A., Fujine E., Mizutani Y., Tawada C., Kanoh H., Banno Y., Seishima M. FTY720 and cisplatin synergistically induce the death of cisplatin-resistant melanoma cells through the downregulation of the PI3K pathway and the decrease in epidermal growth factor receptor expression. Int. J. Mol. Med. 2014;34:1169–1174. doi: 10.3892/ijmm.2014.1882. [DOI] [PubMed] [Google Scholar]
- Iwasaki S., Ingolia N.T. The growing toolbox for protein synthesis studies. Trends Biochem. Sci. 2017;42:612–624. doi: 10.1016/j.tibs.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn S.C., Corsino P.E., Davis B.J., Law M.E., Nørgaard P., Law B.K. Constitutive Cdk2 activity promotes aneuploidy while altering the spindle assembly and tetraploidy checkpoints. J. Cell Sci. 2013;126:1207–1217. doi: 10.1242/jcs.117382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V., Longin S., Goris J. PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail) Trends Biochem. Sci. 2008;33:113–121. doi: 10.1016/j.tibs.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Jensen M.A., Ferretti V., Grossman R.L., Staudt L.M. The NCI Genomic Data Commons as an engine for precision medicine. Blood. 2017;130:453–459. doi: 10.1182/blood-2017-03-735654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y.H., Yoo K.J., Lee Y.H., Lee S.K. Caspase 3-mediated cleavage of p21 associated with the cyclin A-cyclin-dependent kinase 2 complex is a prerequisite for apoptosis in SK-HEP-1 cells. J. Biol. Chem. 2000;275:30256–30263. doi: 10.1074/jbc.m001902200. [DOI] [PubMed] [Google Scholar]
- Kahvejian A., Svitkin Y.V., Sukarieh R., M'Boutchou M.N., Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes & development. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley S.K., Ashkenazi A. Targeting death receptors in cancer with Apo2L/TRAIL. Curr. Opin. Pharmacol. 2004;4:333–339. doi: 10.1016/j.coph.2004.02.006. [DOI] [PubMed] [Google Scholar]
- Kelley S.K., Harris L.A., Xie D., Deforge L., Totpal K., Bussiere J., Fox J.A. Preclinical studies to predict the disposition of Apo2L/tumor necrosis factor-related apoptosis-inducing ligand in humans: characterization of in vivo efficacy, pharmacokinetics, and safety. J. Pharmacol. Exp. Therapeut. 2001;299:31–38. [PubMed] [Google Scholar]
- Kim S.L., Liu Y.C., Park Y.R., Seo S.Y., Kim S.H., Kim I.H., Lee S.O., Lee S.T., Kim D.G., Kim S.W. Parthenolide enhances sensitivity of colorectal cancer cells to TRAIL by inducing death receptor 5 and promotes TRAIL-induced apoptosis. Int. J. Oncol. 2015;46:1121–1130. doi: 10.3892/ijo.2014.2795. [DOI] [PubMed] [Google Scholar]
- Kreis N.N., Louwen F., Yuan J. The multifaceted p21 (Cip1/Waf1/CDKN1A) in cell differentiation, migration and cancer therapy. Cancers. 2019;11:1220. doi: 10.3390/cancers11091220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M., Forrester E., Chytil A., Corsino P., Green G., Davis B., Rowe T., Law B. Rapamycin disrupts cyclin/cyclin-dependent kinase/p21/proliferating cell nuclear antigen complexes and cyclin D1 reverses rapamycin action by stabilizing these complexes. Cancer Res. 2006;66:1070–1080. doi: 10.1158/0008-5472.can-05-1672. [DOI] [PubMed] [Google Scholar]
- Lehman S.L., Cerniglia G.J., Johannes G.J., Ye J., Ryeom S., Koumenis C. Translational upregulation of an individual p21Cip1 transcript variant by GCN2 regulates cell proliferation and survival under nutrient stress. PLoS Genet. 2015;11:e1005212. doi: 10.1371/journal.pgen.1005212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek S., Vargas-Medrano J., Villanueva E., Marcus B., Godfrey W., Perez R.G. Recombinant alpha- beta- and gamma-synucleins stimulate protein phosphatase 2A catalytic subunit activity in cell free assays. JoVE. 2017;13:55361. doi: 10.3791/55361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemke J., von Karstedt S., Zinngrebe J., Walczak H. Getting TRAIL back on track for cancer therapy. Cell Death Differ. 2014;21:1350–1364. doi: 10.1038/cdd.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard D., Huang W., Izadmehr S., O'Connor C.M., Wiredja D.D., Wang Z., Zaware N., Chen Y., Schlatzer D.M., Kiselar J., et al. Selective PP2A enhancement through biased heterotrimer stabilization. Cell. 2020;181:688–701.e16. doi: 10.1016/j.cell.2020.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung-Pineda V., Ryan C.E., Piwnica-Worms H. Phosphorylation of Chk1 by ATR is antagonized by a Chk1-regulated protein phosphatase 2A circuit. Mol. Cell Biol. 2006;26:7529–7538. doi: 10.1128/mcb.00447-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wang J., Li J.G., Wu Z., Ma P. Expression of Ki-67 and pAKT in colorectal cancer tissues and their clinical significance. Int. J. Clin. Exp. Med. 2018;11:8. [Google Scholar]
- Lian X.J., Gallouzi I.E. Oxidative stress increases the number of stress granules in senescent cells and triggers a rapid decrease in p21 translation. J. Biol. Chem. 2009;284:8877–8887. doi: 10.1074/jbc.m806372200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H., Qiu X., Zhang B., Zhang J. Identification of the predictive genes for the response of colorectal cancer patients to FOLFOX therapy. OncoTargets Ther. 2018;11:5943–5955. doi: 10.2147/ott.s167656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T.A., Kong X., Haystead T.A.J., Pause A., Belsham G., Sonenberg N., Lawrence J.C., Jr. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 1994;266:653–656. doi: 10.1126/science.7939721. [DOI] [PubMed] [Google Scholar]
- Lu J., Kovach J.S., Johnson F., Chiang J., Hodes R., Lonser R., Zhuang Z. Inhibition of serine/threonine phosphatase PP2A enhances cancer chemotherapy by blocking DNA damage induced defense mechanisms. Proc. Natl. Acad. Sci. U S A. 2009;106:11697–11702. doi: 10.1073/pnas.0905930106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahyar-Roemer M., Roemer K. p21 Waf1/Cip1 can protect human colon carcinoma cells against p53-dependent and p53-independent apoptosis induced by natural chemopreventive and therapeutic agents. Oncogene. 2001;20:3387–3398. doi: 10.1038/sj.onc.1204440. [DOI] [PubMed] [Google Scholar]
- Maiuthed A., Ninsontia C., Erlenbach-Wuensch K., Ndreshkjana B., Muenzner J.K., Caliskan A., Ahmed P H., Husayn A.P., Chaotham C., Hartmann A., et al. Cytoplasmic p21 mediates 5-fluorouracil resistance by inhibiting pro-apoptotic Chk2. Cancers. 2018;10:373. doi: 10.3390/cancers10100373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinowsky K., Nitsche U., Janssen K.P., Bader F.G., Bader F.G., Spath C., Drecoll E., Keller G., Keller G., Hofler H., et al. Activation of the PI3K/AKT pathway correlates with prognosis in stage II colon cancer. BJC (Br. J. Cancer) 2014;110:2081–2089. doi: 10.1038/bjc.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazhar S., Taylor S.E., Sangodkar J., Narla G. Targeting PP2A in cancer: combination therapies. Biochim. Biophys. Acta Mol. Cell Res. 2019;1866:51–63. doi: 10.1016/j.bbamcr.2018.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClinch K., Avelar R.A., Callejas D., Izadmehr S., Wiredja D., Perl A., Sangodkar J., Kastrinsky D.B., Schlatzer D., Cooper M., et al. Small-molecule activators of protein phosphatase 2A for the treatment of castration-resistant prostate cancer. Cancer Res. 2018;78:2065–2080. doi: 10.1158/0008-5472.can-17-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain D.R., Berger T., Mak T.W. Caspase functions in cell death and disease. Cold Spring Harbor Perspect. Biol. 2013;5:a008656. doi: 10.1101/cshperspect.a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melling N., Simon R., Izbicki J.R., Terracciano L.M., Bokemeyer C., Sauter G., Marx A.H. Expression of phospho-mTOR kinase is abundant in colorectal cancer and associated with left-sided tumor localization. Int. J. Clin. Exp. Pathol. 2015;8:7009–7015. [PMC free article] [PubMed] [Google Scholar]
- Mills J.R., Hippo Y., Robert F., Chen S.M.H., Malina A., Lin C.J., Trojahn U., Wendel H.G., Charest A., Bronson R.T., et al. mTORC1 promotes survival through translational control of Mcl-1. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10853–10858. doi: 10.1073/pnas.0804821105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsiades C.S., Treon S.P., Mitsiades N., Shima Y., Richardson P., Schlossman R., Hideshima T., Anderson K.C. TRAIL/Apo2L ligand selectively induces apoptosis and overcomes drug resistance in multiple myeloma: therapeutic applications. Blood. 2001;98:795–804. doi: 10.1182/blood.v98.3.795. [DOI] [PubMed] [Google Scholar]
- Morita K., He S., Nowak R.P., Wang J., Zimmerman M.W., Fu C., Durbin A.D., Martel M.W., Prutsch N., Gray N.S., et al. Allosteric activators of protein phosphatase 2A display broad antitumor activity mediated by dephosphorylation of MYBL2. Cell. 2020;181:702–715.e20. doi: 10.1016/j.cell.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Narayan S., Jaiswal A.S., Sharma R., Nawab A., Duckworth L.V., Law B.K., Zajac-Kaye M., George T.J., Sharma J., Sharma A.K., Hromas R.A. NSC30049 inhibits Chk1 pathway in 5-FU-resistant CRC bulk and stem cell populations. Oncotarget. 2017;8:57246–57264. doi: 10.18632/oncotarget.19778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayan S., Ramisetti S., Jaiswal A.S., Law B.K., Singh-Pillay A., Singh P., Amin S., Sharma A.K. ASR352, A potent anticancer agent: synthesis, preliminary SAR, and biological activities against colorectal cancer bulk, 5-fluorouracil/oxaliplatin resistant and stem cells. Eur. J. Med. Chem. 2019;161:456–467. doi: 10.1016/j.ejmech.2018.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor C.M., Perl A., Leonard D., Sangodkar J., Narla G. Therapeutic targeting of PP2A. Int. J. Biochem. Cell Biol. 2018;96:182–193. doi: 10.1016/j.biocel.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaiologos P., Chrysikos D., Theocharis S., Kouraklis G. The prognostic value of G1 cyclins, p21 and Rb protein in patients with colon cancer. Anticancer Res. 2019;39:6291–6297. doi: 10.21873/anticanres.13839. [DOI] [PubMed] [Google Scholar]
- Pause A., Belsham G.J., Gingras A.C., Donzé O., Lin T.A., Lawrence J.C., Jr., Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5'-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- Ralff M.D., El-Deiry W.S. TRAIL pathway targeting therapeutics. Expert review of precision medicine and drug development. 2018;3:197–204. doi: 10.1080/23808993.2018.1476062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson P.G., Eng C., Kolesar J., Hideshima T., Anderson K.C. Perifosine , an oral, anti-cancer agent and inhibitor of the Akt pathway: mechanistic actions, pharmacodynamics, pharmacokinetics, and clinical activity. Expet Opin. Drug Metabol. Toxicol. 2012;8:623–633. doi: 10.1517/17425255.2012.681376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues N.R., Rowan A., Smith M.E., Kerr I.B., Bodmer W.F., Gannon J.V., Lane D.P., RowAn A. p53 mutations in colorectal cancer. Proc. Natl. Acad. Sci. U. S. A. 1990;87:7555–7559. doi: 10.1073/pnas.87.19.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy H.K., Olusola B.F., Clemens D.L., Karolski W.J., Ratashak A., Lynch H.T., Smyrk T.C. AKT proto-oncogene overexpression is an early event during sporadic colon carcinogenesis. Carcinogenesis. 2002;23:201–205. doi: 10.1093/carcin/23.1.201. [DOI] [PubMed] [Google Scholar]
- Rozengurt E., Soares H.P., Sinnet-Smith J. Suppression of feedback loops mediated by PI3K/mTOR induces multiple overactivation of compensatory pathways: an unintended consequence leading to drug resistance. Mol. Cancer Therapeut. 2014;13:2477–2488. doi: 10.1158/1535-7163.mct-14-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvolo P.P. The broken "Off" switch in cancer signaling: PP2A as a regulator of tumorigenesis, drug resistance, and immune surveillance. BBA Clin. 2016;6:87–99. doi: 10.1016/j.bbacli.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangodkar J., Perl A., Tohme R., Kiselar J., Kastrinsky D.B., Zaware N., Izadmehr S., Mazhar S., Wiredja D.D., O'Connor C.M., et al. Activation of tumor suppressor protein PP2A inhibits KRAS-driven tumor growth. J. Clin. Invest. 2017;127:2081–2090. doi: 10.1172/jci89548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E.K., Clavarino G., Ceppi M., Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- Sharma R.R., Ravikumar T.S., Raimo D., Yang W.L. Induction of p21WAF1 expression protects HT29 colon cancer cells from apoptosis induced by cryoinjury. Ann. Surg Oncol. 2005;12:743–752. doi: 10.1245/aso.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA A Cancer J. Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- Soria J.C., Márk Z., Zatloukal P., Szima B., Albert I., Juhász E., Pujol J.L., Kozielski J., Baker N., Smethurst D., et al. Randomized phase II study of dulanermin in combination with paclitaxel, carboplatin, and bevacizumab in advanced non-small-cell lung cancer. J. Clin. Oncol. 2011;29:4442–4451. doi: 10.1200/jco.2011.37.2623. [DOI] [PubMed] [Google Scholar]
- Stöcklein W., Piepersberg W. Binding of cycloheximide to ribosomes from wild-type and mutant strains of Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 1980;18:863–867. doi: 10.1128/aac.18.6.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surov A., Clauser P., Chang Y.W., Li L., Martincich L., Partridge S.C., Kim J.Y., Meyer H.J., Wienke A. Can diffusion-weighted imaging predict tumor grade and expression of Ki-67 in breast cancer? A multicenter analysis. Breast Cancer Res. 2018;20:58. doi: 10.1186/s13058-018-0991-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Ito T., Kawano H., Hayashida M., Hayasaki Y., Tsutomi Y., Akahane K., Nakano T., Miura M., Shiraki K. Survivin initiates procaspase 3/p21 complex formation as a result of interaction with Cdk4 to resist Fas-mediated cell death. Oncogene. 2000;19:1346–1353. doi: 10.1038/sj.onc.1203429. [DOI] [PubMed] [Google Scholar]
- Suzuki A., Tsutomi Y., Akahane K., Araki T., Miura M. Resistance to Fas-mediated apoptosis: activation of caspase 3 is regulated by cell cycle regulator p21WAF1 and IAP gene family ILP. Oncogene. 1998;17:931–939. doi: 10.1038/sj.onc.1202021. [DOI] [PubMed] [Google Scholar]
- Suzuki A., Tsutomi Y., Miura M., Akahane K. Caspase 3 inactivation to suppress Fas-mediated apoptosis: identification of binding domain with p21 and ILP and inactivation machinery by p21. Oncogene. 1999;18:1239–1244. doi: 10.1038/sj.onc.1202409. [DOI] [PubMed] [Google Scholar]
- Tahmasebi S., Alain T., Rajasekhar V.K., Zhang J.P., Prager-Khoutorsky M., Khoutorsky A., Dogan Y., Gkogkas C.G., Petroulakis E., Sylvestre A., et al. Multifaceted regulation of somatic cell reprogramming by mRNA translational control. Cell Stem Cell. 2014;14:606–616. doi: 10.1016/j.stem.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Thorburn A., Behbakht K., Ford H., Thorburn A. TRAIL receptor-targeted therapeutics: resistance mechanisms and strategies to avoid them. Drug Resist. Updates. 2008;11:17–24. doi: 10.1016/j.drup.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., Wittmack E.K., Jorgensen T.J. p21WAF1/CIP1 antisense therapy radiosensitizes human colon cancer by converting growth arrest to apoptosis. Cancer Res. 2000;60:679–684. [PubMed] [Google Scholar]
- Tohmé R., Izadmehr S., Gandhe S., Tabaro G., Vallabhaneni S., Thomas A., Vasireddi N., Dhawan N.S., Ma'ayan A., Sharma N., et al. Direct activation of PP2A for the treatment of tyrosine kinase inhibitor-resistant lung adenocarcinoma. JCI Insight. 2019;4:125693. doi: 10.1172/jci.insight.125693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaklavas C., Blume S.W., Grizzle W.E. Translational dysregulation in cancer: molecular insights and potential clinical applications in biomarker development. Front. Oncol. 2017;7:158. doi: 10.3389/fonc.2017.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Jeught K., Xu H.C., Li Y.J., Lu X.B., Ji G. Drug resistance and new therapies in colorectal cancer. World J. Gastroenterol. : WJG. 2018;24:3834–3848. doi: 10.3748/wjg.v24.i34.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Karstedt S., Montinaro A., Walczak H. Exploring the TRAILs less travelled: TRAIL in cancer biology and therapy. Nat. Rev. Cancer. 2017;17:352–366. doi: 10.1038/nrc.2017.28. [DOI] [PubMed] [Google Scholar]
- Walczak H., Miller R.E., Ariail K., Gliniak B., Griffith T.S., Kubin M., Chin W., Jones J., Woodward A., Le T., et al. Tumoricidal activity of tumor necrosis factor-related apoptosis-inducing ligand in vivo. Nat. Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- Wang H., Liu Y., Ding J., Huang Y., Liu J., Liu N., Ao Y., Hong Y., Wang L., Zhang L., et al. Targeting mTOR suppressed colon cancer growth through 4EBP1/eIF4E/PUMA pathway. Cancer Gene Ther. 2020;27:448–460. doi: 10.1038/s41417-019-0117-7. [DOI] [PubMed] [Google Scholar]
- Wang M., Law M.E., Davis B.J., Yaaghubi E., Ghilardi A.F., Ferreira R.B., Chiang C.W., Guryanova O.A., Kopinke D., Heldermon C.D., et al. Disulfide bond-disrupting agents activate the tumor necrosis family-related apoptosis-inducing ligand/death receptor 5 pathway. Cell Death Dis. 2019;5:153. doi: 10.1038/s41420-019-0228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.S., Esplin E.D., Li J.L., Huang L., Gazdar A., Minna J., Evans G.A. Alterations of the PPP2R1B gene in human lung and colon cancer. Science. 1998;282:284–287. doi: 10.1126/science.282.5387.284. [DOI] [PubMed] [Google Scholar]
- Wang X.W., Zhang Y.J. Targeting mTOR network in colorectal cancer therapy. World J. Gastroenterol. : WJG. 2014;20:4178. doi: 10.3748/wjg.v20.i15.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmus J.S., Dilley G.J., Meyers A.I. A modified procedure for the preparation of 2, 5-dihydropyrrole (3-pyrroline) J. Org. Chem. 1993;58:270–271. doi: 10.1021/jo00053a053. [DOI] [Google Scholar]
- Wei F., Zhang Y., Geng L., Zhang P., Wang G., Liu Y. mTOR inhibition induces EGFR feedback activation in association with its resistance to human pancreatic cancer. Int. J. Mol. Sci. 2015;16:3267–3282. doi: 10.3390/ijms16023267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss R.H. p21Waf1/Cip1 as a therapeutic target in breast and other cancers. Cancer Cell. 2003;4:425–429. doi: 10.1016/s1535-6108(03)00308-8. [DOI] [PubMed] [Google Scholar]
- Westermarck J. Targeted therapies don't work for a reason; the neglected tumor suppressor phosphatase PP2A strikes back. FEBS J. 2018;285:4139–4145. doi: 10.1111/febs.14617. [DOI] [PubMed] [Google Scholar]
- Wu N., Du Z., Zhu Y., Song Y., Pang L., Chen Z. The expression and prognostic impact of the PI3K/AKT/mTOR signaling pathway in advanced esophageal squamous cell carcinoma. Technol. Cancer Res. Treat. 2018;17 doi: 10.1177/1533033818758772. 153303381875877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Wang P., Yang H., Zhou J., Li Y., Li X., Xue W., Yu C., Tian Y., Zhu F. Comparison of FDA approved kinase targets to clinical trial ones: insights from their system profiles and drug-target interaction networks. BioMed Res. Int. 2016;2016:1–9. doi: 10.1155/2016/2509385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Zhou J.Y., Wei W.Z., Wu G.S. Activation of the Akt survival pathway contributes to TRAIL resistance in cancer cells. PLoS One. 2010;5:e10226. doi: 10.1371/journal.pone.0010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S.Q., El-Deiry W.S. p21(WAF1/CIP1) inhibits initiator caspase cleavage by TRAIL death receptor DR4. Biochem. Biophys. Res. Commun. 2000;269:179–190. doi: 10.1006/bbrc.2000.2247. [DOI] [PubMed] [Google Scholar]
- Yaffee P., Osipov A., Tan C., Tuli R., Hendifar A. Review of systemic therapies for locally advanced and metastatic rectal cancer. J. Gastrointest. Oncol. 2015;6:185–200. doi: 10.3978/j.issn.2078-6891.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong L., YuFeng Z., Guang B. Association between PPP2CA expression and colorectal cancer prognosis tumor marker prognostic study. Int. J. Surg. 2018;59:80–89. doi: 10.1016/j.ijsu.2018.09.020. [DOI] [PubMed] [Google Scholar]
- Young P.E., Womeldorph C.M., Johnson E.K., Maykel J.A., Brucher B., Stojadinovic A., Avital I., Nissan A., Steele S.R. Early detection of colorectal cancer recurrence in patients undergoing surgery with curative intent: current status and challenges. J. Cancer. 2014;5:262–271. doi: 10.7150/jca.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Kanwar S.S., Patel B.B., Nautiyal J., Sarkar F.H., Majumdar A.P. Elimination of colon cancer stem-like cells by the combination of curcumin and FOLFOX. Translational oncology. 2009;2:321–328. doi: 10.1593/tlo.09193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Fang B. Mechanisms of resistance to TRAIL-induced apoptosis in cancer. Cancer Gene Ther. 2005;12:228–237. doi: 10.1038/sj.cgt.7700792. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Fujita N., Tsuruo T. Caspase-mediated cleavage of p21Waf1/Cip1 converts cancer cells from growth arrest to undergoing apoptosis. Oncogene. 1999;18:1131–1138. doi: 10.1038/sj.onc.1202426. [DOI] [PubMed] [Google Scholar]
- Zhou X., Liu W., Hu X., Dorrance A., Garzon R., Houghton P.J., Shen C. Regulation of CHK1 by mTOR contributes to the evasion of DNA damage barrier of cancer cells. Sci. Rep. 2017;7:1535. doi: 10.1038/s41598-017-01729-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
TCGA data were for CRC’s are available at The Cancer Genome Atlas (TCGA) data repository (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga/studied-cancers/colorectal).
-
•
Gene expression data for FOLFOX-resistant and non-resistant patient are available at the NCBI Gene expression Omnibus (GEO) under GSE72970 accession (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE72970).
-
•
RNAseq data are deposited at Zenodo Data Repository (https://zenodo.org/record/6565046#.YobdkKjMJPY) and data are publicly available. Link to this data is listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.