Summary
Clioquinol (CQ) is a hypoxic mimicker to activate hypoxia-inducible factor-1α (HIF-1α) by inhibiting HIF-1α specific asparaginyl hypoxylase (FIH-1). The structural similarity of the Jumonji C (JmjC) domain between FIH-1 and JmjC domain-containing histone lysine demethylases (JmjC-KDMs) led us to investigate whether CQ could inhibit the catalytic activities of JmjC-KDMs. Herein, we showed that CQ inhibits KDM4A/C, KDM5A/B, and KDM6B and affects H3K4me3, H3K9me3, and H3K27me3 marks, respectively. An integrative analysis of the histone methylome and transcriptome data revealed that CQ-mediated JmjC-KDM inhibition altered the transcription of target genes through differential combinations of KDMs and transcription factors. Notably, functional enrichment of target genes showed that CQ and hypoxia commonly affected the response to hypoxia, VEGF signaling, and glycolysis, whereas CQ uniquely altered apoptosis/autophagy and cytoskeleton/extracellular matrix organization. Our results suggest that CQ can be used as a JmjC-KDM inhibitor, HIF-α activator, and an alternative therapeutic agent in hypoxia-based diseases.
Subject areas: Biochemistry, Molecular biology, Transcriptomics
Graphical abstract

Highlights
-
•
Both hypoxia and clioquinol (CQ) inhibit histone lysine demethylases (KDMs)
-
•
CQ affects H3K4me3, H3K9me3, and H3K27me3 marks upon inhibition
-
•
CQ treatment-induced histone methylome changes affect target gene transcription
-
•
Histone methylome predicts TFs underlying transcription of CQ target genes
Biochemistry; Molecular biology; Transcriptomics
Introduction
The enzymes 2-oxoglutarate (2OG)-dependent dioxygenases (2OGXs) play important roles in the regulation of gene expression through their effects on transcription or histone/DNA demethylation. 2OGXs require O2, 2OG, Fe(II), and vitamin C for hydroxylation, releasing CO2, succinate, and Fe(III) in the process (Loenarz and Schofield, 2011). These 2OGXs include hypoxia inducible factor (HIF)-prolyl hydroxylases (PHDs), HIF-1α asparaginyl hydroxylase (factor inhibiting HIF-1, FIH-1), Jumonji C (JmjC) domain-containing histone lysine demethylases (JmjC-KDMs) (Islam et al., 2018), and ten-eleven translocation (TET) methylcytosine dioxygenases. PHD2 and FIH-1 modulate the transcriptional regulation of target genes under hypoxia through their interactions with HIF-1α, a major transcription factor (TF) activated by hypoxia. The hydroxylation of Pro402 and Pro564 of HIF-1α by PHD2 promotes the interaction between HIF-1α and the von Hippel-Lindau (VHL)-elongin C-B E3 ubiquitin-ligase complex, resulting in the degradation of HIF-1α (Kamura et al., 2000). The hydroxylation of HIF-1α at 803rd asparagine by FIH-1 prevents the binding of HIF-1α to CREB-binding protein (CBP)/p300 coactivators, thereby inhibiting the transactivation of HIF-1α (Lando et al., 2002). Moreover, JmjC-KDMs and TET enzymes modulate the transcription of their target genes by removing a methyl group from methylated lysines in histones and methylcytosine in DNA, respectively.
JmjC-KDMs have been identified as the largest family of 2OGXs, including more than 20 different members with distinctive and shared specificity for their substrates (Chang et al., 2019) as follows: KDM5A-D demethylates H3K4me1-3; KDM4A-D demethylates H3K9me1-3, H3K36me1-3, or H1.4K26me1-3; and KDM6A-C demethylates H3K27me1-3. JmjC-KDMs have been demonstrated to play important roles in the pathogenesis of various diseases. For example, the loss of KDM3A or KDM4B in mice results in obesity and hyperlipidemia (Tateishi et al., 2009), KDM3A promotes cardiac hypertrophy and fibrosis (Zhang et al., 2018), a deficiency of KDM3A in mice causes infertility via dysregulation of spermatogenesis (Okada et al., 2010), and the expression of KDM6B lacking the JmjC domain leads to delayed puberty and impaired fertility (Song et al., 2017). Moreover, the Cancer Genome Atlas (TCGA) project identified KDM5C and KDM6A as driver genes in various cancers, such as pancreatic, breast, lung, and prostate cancers (Bailey et al., 2018). These studies suggest that KDMs can serve as therapeutic targets for different diseases.
To suppress the activity of KDMs in pathological conditions, many inhibitors of 2OGXs have been introduced based on their structures or activities as evaluated using screening assays. These inhibitors can be categorized into two groups: those that compete with either 2OG or substrates of 2OGXs, and those that compete with both 2OG and its substrates (Islam et al., 2018). Growing evidence indicates that these inhibitors have poor cell permeability and present cellular toxicity and off-target effects. Thus, there is a need to discover improved inhibitors of JmjC-KDMs. We had found that clioquinol (CQ) inhibited the binding of 2OG to the JmjC domain of FIH-1 (Moon et al., 2010), thereby enhancing the activity of HIF-1α (Chen et al., 2007; Choi et al., 2006; Ding et al., 2005; Chen et al., 2007).
The structural similarity of the JmjC domain between FIH-1 and KDM4A (Chen et al., 2006b) prompted us to investigate whether CQ could inhibit the catalytic activities of KDM4A and other JmjC-KDMs. CQ has been identified as a membrane-permeable and hydrophobic metal chelator with favorable toxicity profiles (Cherny et al., 2001; Padmanabhan et al., 1990; Schimmer et al., 2012). Here, we investigated the CQ-mediated inhibition of KDM4A/C, KDM5A/B, and KDM6B by characterizing the effects of CQ on H3K4me3, H3K9me3, and H3K27me3, respectively, using chromatin immunoprecipitation sequencing (ChIP-seq). We further examined the effects of the CQ-mediated regulation of the methylation of these three histones on the transcription of target genes and cellular processes associated with these target genes by integrating ChIP-seq and gene expression data. We previously showed that hypoxia inhibited the activity of JmjC-KDMs in human adipocyte-derived stem cells (hADSCs) (Lee et al., 2017). We also used hADSCs in this study to examine whether CQ can have similar effects to hypoxia on histone methylation and the transcription of its target genes. Our results provide evidence that CQ is a novel inhibitor of JmjC-KDMs, including the molecular and functional characteristics of the CQ-mediated inhibition of JmjC-KDMs.
Results
CQ inhibits the activity of histone demethylases
To examine the effect of CQ on histone methylation, we treated human adipocyte-derived stem cells (hADSCs) with two different doses (25 and 50 μM) of CQ and a pan-inhibitor of histone demethylases — JIB-04 (0.1 μM) — under normoxic conditions (20% O2) and then measured the total amount of methylated histones in the CQ-treated hADSCs. CQ treatment increased total H3K4me3, H3K9me3, and H3K27me3 under normoxia, as well as under hypoxia (0.5% O2) and JIB-04 treatment (Figure 1A). Similar increases in the total amounts of H3K4me3, H3K9me3, and H3K27me3 were observed in human glioblastoma (U87) and cervical cancer (HeLa) cells (Figures S1A and S1E), suggesting that the CQ-induced increase in methylated histones is common in different cell types. Moreover, CQ also increased the protein levels of HIF-1α and the mRNA levels of its target genes, BCL2/adenovirus E1B 19kDa interacting protein 3 (BNIP3) and carbonic anhydrase IX (CA9), similar to hypoxia and JIB-04 treatment (Figures S1B and S1C).
Figure 1.
CQ inhibits the activity of histone demethylases
(A) Western blot analyses of histone extracts from hADSCs cultured under hypoxia (1.5%, 0.5% O2), treated with CQ (25 and 50 μM) or JIB-04 (0.1 μM) for 16 h using the indicated antibodies. H3 was detected as a loading control.
(B–F) Effects of CQ on the catalytic activities of histone demethylases. HeLa cells were transfected with the indicated KDMs. The transfected cells were exposed to hypoxia (0.5% O2) or CQ (50 μM) for 16 h, then immunostained using the indicated tag-antibodies (red) and indicated histone antibodies (green). The nuclei were stained with DAPI (blue). Arrows indicate cells transfected with the indicated histone demethylases. Using a pseudo-color 3D confocal fluorescence microscope, the fluorescence intensities of H3K4me3, H3K9me3, and H3K27me3 were estimated as shown in the contour maps (right columns) (Lee et al., 2013). The representative images of cells transfected with HA-tagged KDM5A among 20 cells (20% O2), 10 cells (0.5% O2), and six cells (CQ) (B); with Myc-tagged KDM5B among 16 cells (20% O2), 10 cells (0.5% O2), and 15 cells (CQ) (C); with HA-tagged KDM4A among 35 cells (20% O2), 17 cells (0.5% O2), and 16 cells (CQ) (D); with Myc-tagged KDM4C among 18 cells (20% O2), 15 cells (0.5% O2), and 21 cells (CQ) (E); with Myc-tagged KDM6B among 14 cells (20% O2), 29 cells (0.5% O2), and 13 cells (CQ) (F).
(G) The estimated activities of histone demethylases were visualized via immunofluorescence. For each histone demethylase, catalytic activity was defined as the difference in the estimated fluorescence intensity of its corresponding trimethylated histone between untransfected and transfected cells. The catalytic activity under hypoxia was normalized by that under normoxia. Data represent the average and standard deviations of the estimated activities of histone demethylases from 6 to 35 transfected cells. ∗∗∗ indicates p < 0.001 via one-way ANOVA with Tukey’s correction.
(H) Mass spectrometric analyses for measuring histone demethylase activity. Biotinylated histone H3K9me3 peptide (MW = 2,765 Da) was incubated with 5 μg of recombinant KDM4A protein (1–350 amino acids), vitamin C (2 μM), and 2-oxoglutarate (2-OG) (100 μM) in the absence or presence of CQ (50 μM) for 4 h then analyzed via MALDI-TOF. The detected mass of the major peptide is indicated above the associated peak.
The increase in methylated histones can be caused by the increased activity of methyltransferases or decreased activity of demethylases. To examine this, we measured the levels of H3K4me3, H3K9me3, and H3K27me3 after depleting methionine — a substrate of methyltransferases — in hADSCs and HeLa cells. The methylated histones were similarly increased after CQ treatment, regardless of the presence of methionine (Figures S1D and S1E) (Chen et al., 2006a), indicating that the increase of methylated histones was because of decreased demethylase activity. Next, we examined CQ-induced changes in the catalytic activities of JmjC-KDMs (KDM5A/B for H3K4me3, KDM4A/C for H3K9me3, and KDM6B for H3K27me3) using an immunofluorescence assay in cells overexpressing JmjC-KDMs. H3K4me3, H3K9me3, and H3K27me3 disappeared in JmjC-KDM-overexpressing cells under normoxia, despite the overexpression of JmjC-KDMs but were present under hypoxia and CQ treatment (Figures 1B–1F). Quantification of immunofluorescence intensity of the trimethylated histones confirmed that CQ significantly (p < 0.05) decreased the catalytic activity of JmjC-KDMs (Figure 1G).
To confirm this finding, we performed an in vitro demethylation assay using mass spectrometry. When recombinant KDM4A proteins were added to a solution containing vitamin C and 2OG, H3K9me3 peptides (m/z = 2,765, Figure 1H) were demethylated to H3K9me2 (m/z = 2,751, Figure 1H); however, they were not demethylated when CQ (50 μM) was added. Based on a known structure of the KDM4A-pyridine-2,4-dicarboxylic acid complex (PDB ID: 2VD7; Figure S1F, top), we performed molecular modeling of the KDM4A-CQ complex structure by matching the interactions of the rings in both CQ and pyridine-2, 4-dicarboxylic acid with Fe (Figure S1F, middle). In the modeled structure, CQ overlapped with the position of 2OG according to the KDM4A-2OG complex structure (PDB ID: 2Q8C), suggesting the competitive inhibition of KDM4A by CQ (Figure S1F, bottom). Furthermore, we performed molecular dynamics (MD) simulations of the KDMA4A-PDCA and KDM4A-CQ complex structures, which were obtained by matching the positions of the pyridine ring in both CQ and PDCA with Fe(II) (Figure S1G). During the 200-ns MD simulations, both complexes maintained a stable coordination structure, as shown in the representative snapshots and the distances between Fe(II) and the coordinating atoms, suggesting a competitive inhibition of KDM4A by CQ. We estimated the ratio (r = Kb(CQ)/Kb(PDCA)) of the binding constants from the MD simulation trajectory as follows:
where Kb(i) is the binding constant of substrate i (CQ or PDCA) and Nbound(unbound)i is the number of bound (unbound) states of substrate i during the MD simulation. We assumed that the substrate was bound to the enzyme if the distances between Fe(II) and both N and O atoms of the substrate were within 3 Å. From the MD simulation, we obtained r ≈ 1.1 (Kb(CQ) = 62.90 and Kb(PDCA) = 57.0), which implies that CQ can be a slightly better inhibitor than PDCA. Notably, analogs of CQ, such as broxyquinoline (BQ), hydroquinone (HQ), chloroacetoxyquinoline (CAQ), and iodoquinol (IQ), also inhibited the demethylation of H3K9me3, similar to CQ (Figure S1H). Overall, these data suggest that CQ inhibits the demethylation activities of JmjC-KDMs, increasing the methylated histones.
CQ affects the genomic distributions of methylated histones
To examine the genome-wide landscape of methylated histones affected by CQ, we performed ChIP-seq for H3K4me3, H3K9me3, and H3K27me3 in hADSCs under normoxia (N), hypoxia (H), or with CQ treatment (CQ). ChIP-seq yielded a total of 588.2 million reads for the three histone methylations in N, H, and CQ. These reads were aligned to the human genome (UCSC GRCh.38.91) using BOWTIE2, resulting in total mapped reads of 54.3 Giga base-pairs (16.5-fold coverage of the human genome) (Table S2). To investigate the changes in methylated histones by CQ, we first identified the peaks consistently detected in N, H, and CQ — called consensus peaks — and then examined the distribution of mapped reads across the genome or gene structure. Consistent with previous findings (Kooistra and Helin, 2012; Lee et al., 2017), the consensus peaks of H3K4me3 were enriched in gene bodies (Figure S2A) and near transcription start sites (TSS) (Figure 2A) under N, H, and CQ conditions. In contrast, the peaks of H3K9me3 and H3K27me3 were enriched in the intergenic regions (distal promoters) and gene bodies (Figures S2A and S2B). The gene body peaks of the three methylations were predominantly located in introns (Figure 2B).
Figure 2.
CQ affects histone trimethylation at the genomic level
(A) Distributions of H3K4me3 within the gene structures in hADSCs. The gene structure (−5 kb-TSS-TES-+5 kb) was parsed into 45 bins for each gene. The average RPKM read density profiles (y axis) of H3K4me3 for all the genes in individual bins (x axis) were plotted. Yellow, red, and blue lines represent profiles of the read densities under normoxia (N), hypoxia (H), and CQ (C), respectively.
(B) Numbers of consensus peaks of each histone methylation within the indicated regions. In this analysis, the gene body is further split into the coding sequence (CDS) and intron.
(C) Scatterplot showing the relationship of the log2-fold-changes of H3K4me3 levels in consensus peaks between CQ versus N (x axis) and H versus N (y axis). Red and green dots correspond to consensus peaks with increased and decreased H3K4me3 levels in both CQ and H compared to N, respectively.
(D) Distributions of the read counts in the neighboring regions of consensus peaks (−2 kb and +2 kb from the consensus peak centers). Red and blue bars indicate consensus peaks with increased and decreased H3K4me3 levels in both CQ and H compared to N, respectively. Color bar, gradient of the read counts at individual bases.
(E) Relationships of the DMGs identified for H3K4me3, H3K9me3, and H3K27me3. The numbers in parentheses denote the total numbers of DMGs identified for individual histone methylations.
(F) Relationship between the DMGs by CQ and hypoxia. The numbers in parentheses denote the total numbers of DMGs with alterations in at least one type of histone modification by CQ or hypoxia.
(G) GOBPs represented by the DMGs in CQ or hypoxia. x axis, -log10(p), where p is the enrichment p value obtained using DAVID software.
CQ and H strongly altered the read counts of H3K4me3, H3K9me3, and H3K27me3 peaks in many genes (−5 kb from the TSS and +5kb from the TES [transcription end site]; Figures 2C and S2C), as well as in their neighboring regions (Figures 2D and S2D), compared to N. We then identified 4,829 differentially methylated genes (DMGs) between N and CQ, which had altered peaks with average fold-changes larger than 1.5, including 1,147, 2,342, and 1939 genes with altered H3K4me3, H3K9me3, and H3K27me3 peaks, respectively (Figure 2E, top; Table S3). The number of DMGs by CQ (4,829 genes) (Figure 2E, top Venn diagram) was found comparable to that of the DMGs (4,921 genes) by hypoxia (Figure 2E, bottom Venn diagram). Among these DMGs, 51.2% (2,472 of 4,829 DMGs) was shared with those in hypoxia (Figure 2F and Table S4). Subsequently, we performed an enrichment analysis of gene ontology biological processes (GOBPs) for the DMGs under CQ or hypoxia (Figure 2G). CQ primarily affected cell adhesion/migration, cytoskeleton organization, angiogenesis, response to hypoxia, carbohydrate metabolism, and cell proliferation, which were consistently enriched in the DMGs by hypoxia (Figure 2G). In contrast, CQ predominantly affected apoptosis, whereas hypoxia affected fatty acid metabolism. Despite this difference, CQ and hypoxia generally have similar effects on the genomic landscapes of methylated histones and target genes.
CQ affects the expression of target genes and their associated cellular processes
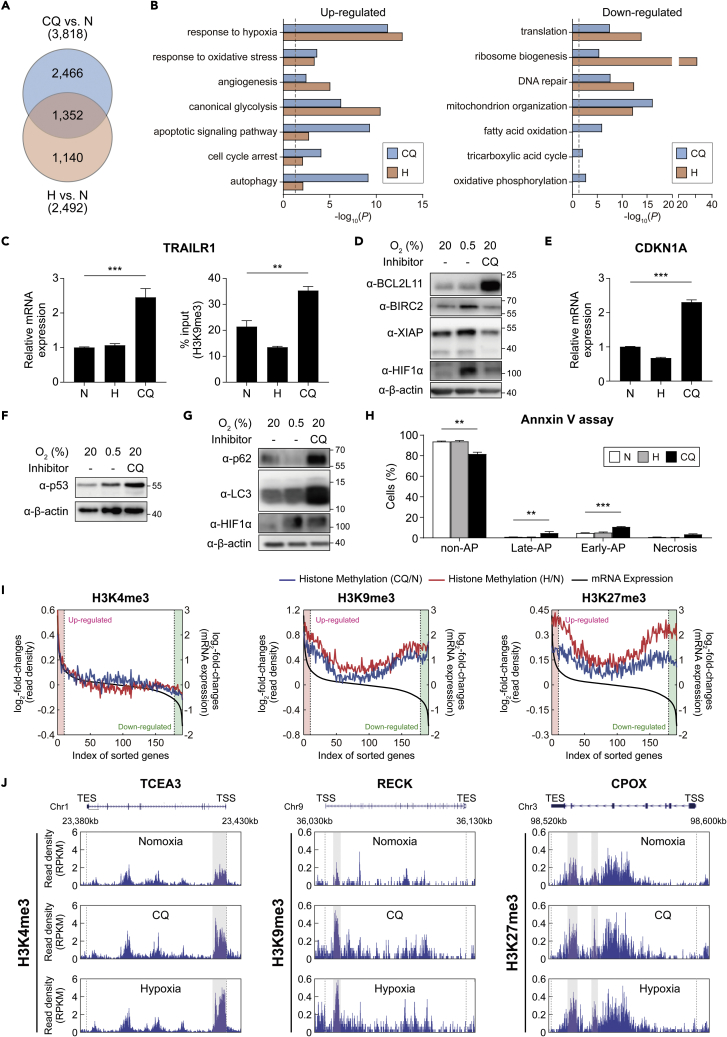
To examine the target genes of CQ, we performed gene expression profiling of hADSCs under N, H, and CQ conditions. H3K4me3 levels in CQ were positively correlated with mRNA expression levels, whereas H3K9me3 and H3K27me3 levels were negatively correlated (Figure S3A). We next identified 3,818 differentially expressed genes (DEGs; 1747 upregulated, 2071 downregulated) between CQ and N and 2,492 DEGs between H and N (Figures 3A and S3B; Table S5). Of the DEGs, 35.4% (1,352 of 3,818 DEGs) overlapped with those by hypoxia, consistent with a high overlap between the DMGs by CQ and hypoxia (Figure 2F).
Figure 3.
CQ affects the expression of target genes and their associated cellular processes
(A) Comparison of the DEGs in hADSCs after CQ treatment and hypoxia. The numbers in the parentheses denote the total number of DEGs by CQ or hypoxia.
(B) GOBPs represented by the upregulated (left) and downregulated (right) genes by CQ or hypoxia. x axis, -log10(p), where p is the enrichment p value obtained using DAVID software. Dotted line, the cutoff value used to select the enriched GOBPs.
(C) Relative mRNA expression levels (left) and H3K9me3 levels (right) of the TRAILR1 gene in hADSCs, as measured using qRT-PCR and ChIP-PCR analyses, respectively, under N, H, or CQ. Data are shown as the mean ± SEM (n = 6 per condition).
(D) Representative western blot images showing the amounts of pro-apoptotic (BCL2L11) and anti-apoptotic (BIRC2 and XIAP) markers in hADSCs under N, H, or CQ (50 μM, 72 h).
(E and F) Relative mRNA expression levels of CDK1A. Western blot of p53 in hADSCs under N, H, or CQ. Data are shown as the mean ± SEM (n = 4 per condition). Statistical differences were assessed using a two-tailed paired Student’s t-test. ∗∗∗p < 0.001.
(G) Representative western blot images showing the amounts of an autophagy marker (p62/SQSTM1), LC3B, in hADSCs under N, H, and CQ (50 μM, 72 h).
(H) Percentages of non-apoptotic (AP), early-apoptotic, late-apoptotic, and necrotic hADSCs as measured via flow cytometry after annexin V/PI staining under normoxia (N), hypoxia (H) (0.5% O2, 48 h), or after treatment with CQ (CQ) (50 μM, 16 h). Data are presented as the mean ± SEM (n = 5 per condition).
(I) Correlation of log2-fold-changes in histone modifications with gene expression in individual histone methylations. The expressed genes were sorted using their log2-fold-changes and binned such that each bin includes 100 genes. For each type of histone methylation, the averaged log2-fold-changes of the RPKMs of the histone modification (y1-axis) and the averaged log2-fold-changes of gene expression levels (y2-axis) in individual bins (x axis) were plotted. Black lines represent the averaged log2-fold-changes of gene expression. Blue and red lines represent the averaged log2-fold-changes of histone methylation levels by CQ and hypoxia, respectively. Upregulated and downregulated genes (red and green backgrounds, respectively) were defined as those with fold-changes >1.5.
(J) The aligned reads of three representative genes (TCEA3, RECK, and CPOX) for the three histone methylations. The regions that include the differentially methylated peaks for H3K4me3, H3K9me3, and H3K27me3 were indicated by gray backgrounds. ∗∗p < 0.01 and ∗∗∗p < 0.001 by one-way ANOVA with Tukey’s correction (C and H).
To examine the cellular processes affected by CQ, we performed the GOBP enrichment analyses of the upregulated or downregulated genes. The upregulated genes were mainly associated with responses to hypoxia, angiogenesis, glycolysis, apoptosis/cell cycle arrest, and autophagy, similar to those upregulated by hypoxia (Figure 3B, left, Table S6). However, CQ more potently upregulated the genes associated with apoptosis, cell cycle arrest, and autophagy, which was consistent with the findings from the DMGs (Figure 2G). We also confirmed that CQ increased the mRNA and H3K9me3 levels of the TRAILR1 gene (Figure 3C) and protein levels of BCL2L11— a proapoptotic marker — compared to hypoxia. On the other hand, CQ decreased the protein levels of the antiapoptotic markers BIRC2 and XIAP compared to hypoxia (Figure 3D). Moreover, CQ treatment increased the levels of CDKN1A and p53, which induce cell cycle arrest and apoptosis, respectively (Figures 3E and 3F), and increased the expression of the autophagy markers p62 and LC3B (Figure 3G) to a greater extent than in hypoxia. Overall, these results indicated that CQ and hypoxia share many target genes; however, CQ induced the expression of genes involved in apoptosis, cell cycle arrest, and autophagy considerably greater than hypoxia. Consistent with these results, MTT assays showed that CQ (50 μM, 16 h) significantly reduced the viability of both hADSC and U87 cells by 59.12 and 69.64%, respectively (Figures S3C and S3D). Notably, FACS analyses showed that both CQ treatment (50 μM, 16 h) and hypoxia barely increased the number of apoptotic cells (Annexin V-positive/PI-negative cells) from 2.62% to 4.86 and 3.34%, respectively (Figures 3H, S3E, and S3F). These data suggest that CQ (50 μM) reduces cell viability not only by increasing apoptosis but also through other mechanisms, such as autophagy-related cell death, as previously reported (Mizutani et al., 2021). In contrast, the downregulated genes were associated with ribosome biogenesis/translation, DNA repair, and mitochondrion organization, similar to those downregulated by hypoxia (Figure 3B, right, Table S6). Notably, compared with hypoxia, CQ downregulated oxidative phosphorylation, the tricarboxylic acid cycle, and fatty acid oxidation (Table S6).
To explore the link between methylated histones and the mRNA expression of CQ target genes, we next analyzed the correlations between their histone methylation and mRNA expression changes (log2-fold-changes). For the upregulated genes (Figure 3I, red regions), there was a positive correlation (r = 0.99; p = 3.95 × 10−10) between the log2-fold-changes of H3K4me3 and mRNA expression levels (Figure 3I, left). In contrast, the downregulated genes (Figure 3I, green regions) showed increased levels (i.e., positive log2-fold-changes) of H3K9me3 (Figure 3I, middle) or H3K27me3 (Figure 3I, right). Unexpectedly, however, for the upregulated genes, the log2-fold-changes of H3K9me3 and mRNA levels showed a positive correlation (r = 0.69; p = 1.81 × 10−2), indicating the possibility that the upregulated genes could have increased H3K9me3 levels (Figure 3I, middle). The alteration patterns for the three trimethylations were consistent with those observed under hypoxic conditions. Moreover, an increased level of each trimethylation was observed in a representative gene in CQ compared to that in N (Figure 3J). Overall, these results suggest that CQ-induced alterations in H3K4me3, H3K9me3, and H3K27me3 are closely correlated with the changes in the expression of target genes, affecting cellular processes associated with the target genes.
Target genes show a differential association between histone methylation and mRNA expression
Next, we examined the relationship between CQ-induced alterations in histone methylation and mRNA expression at the individual gene level. A comparison of the DMGs and DEGs revealed that 789 DEGs (409 upregulated and 380 downregulated genes) had peaks with CQ-induced alterations of H3K4me3, H3K9me3, or H3K27me3 (Figure 4A). To systematically explore this relationship, we further categorized these 789 shared genes between the DEGs and DMGs into 29 groups based on their upregulation and downregulation patterns of histone methylation and mRNA expression by CQ (Figure S4A). Of the 29 groups, we focused on the top four major groups (G1-4 in Figure 4B), each of which included more than 5% (40 genes) of the 789 shared genes in both CQ and H for the subsequent comparison between CQ and H. Groups 1–4 included 63.6% of the 789 shared genes (Figure 4B; Table S7).
Figure 4.
Target genes show differential associations between histone methylation and mRNA expression in CQ-treated hADSCs
(A) Comparison of the DMGs and DEGs by CQ treatment. Total numbers of DEGs and DMGs, as well as the numbers of upregulated (U) and downregulated (D) genes are shown in parentheses.
(B) Four major groups (G1-4) of the shared genes between the DMGs and DEGs. The heatmap was generated after hierarchical clustering of the log2-fold-changes of gene expression and histone methylation levels (RPKMs) using the average linkage method and Euclidean distance as a dissimilarity measure. Colors in the heatmap represent upregulated (red) and downregulated (green) genes in CQ compared to normoxia. Color bar, gradient of the log2-fold changes in gene expression and histone methylation levels. The number of genes in each group is shown in parentheses.
(C) GOBPs represented by the genes in G1–4. Color bar, gradient of –log10(p), where p is the enrichment p value for individual GOBPs obtained using DAVID software.
(D) Relative methylated histone (left) and mRNA expression (right) levels of the indicated representative genes in G1-4 measured using ChIP-PCR and qRT-PCR analyses, respectively, under N and CQ. Data are shown as the mean ± SEM (n = 6 per condition, exceptionally for TMX4 gene n = 9 per condition of qRT-PCR, for ELOVL6 gene n = 3 per condition of H3K9me3 ChIP-PCR). ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001 using Students’ t-test.
G3 included genes with increased mRNA expression and H3K4me3 levels, which are consistent with the role of H3K4me3 as an active marker (Figure 4B). G1 included genes with decreased mRNA expression and increased H3K9me3 levels. Unlike the changes in the opposite direction, G2 and G4 included genes with changes in mRNA expression and H3K9me3 levels in the same direction. Next, we examined the localization of H3K9me3 marks in the gene structures of G1, G2, and G4. It was revealed that G1 showed the highest increase in H3K9me3 in the gene body but a relatively weak increase in the promoter region. In contrast, G4 showed decreased H3K9me3, predominantly in the promoter region, whereas G2 tended to show the most substantial increase in H3K9me3 in the promoter region and a comparable increase in the gene body (Figure S4B). These enrichments in the promoter or gene body of H3K9me3 may account for the difference in mRNA-H3K9me3 alteration patterns in G1, G2, and G4. Moreover, although many genes had multiple types of histone trimethylations under N (4,969 genes) or CQ (5,868 genes; Figure S4C), notably, the DEGs in G1–G4 showed changes in only one type of histone trimethylation, as reported in hypoxia (Lee et al., 2017).
Subsequently, we performed GOBP enrichment to examine whether the genes in G1-G4 were involved in different cellular processes (Figure 4C). Both upregulated G2 and G3 genes in CQ consistently represented a response to hypoxia. In contrast, G2 predominantly represented histone modification, whereas G3 predominantly represented VEGF signaling, autophagy/apoptosis, and glycolysis (Figure 4C). On the other hand, the downregulated G1 genes represented cell adhesion/migration, cytoskeleton organization, and extracellular matrix organization, whereas the downregulated G4 genes represented transcription, RNA processing, translation, and cell cycle. Of note, these processes were consistent with the upregulated and downregulated processes in CQ (Figure 3B), suggesting that G1-G4 encapsulates CQ-induced mRNA expression changes at the cellular process level. Notably, a comparison of the cellular processes represented by G1-G4 revealed that CQ and H consistently affected many of these processes, including extracellular matrix organization by G1, response to hypoxia by G2, and glycolysis by G3. CQ, but not H, uniquely regulated several processes (cytoskeleton organization by G1 and apoptosis by G3) (Figure S4D). We also confirmed CQ-induced alterations in mRNA expression and histone methylation for two representative genes in each group using qRT-PCR and ChIP-PCR analyses (Figure 4D). Overall, these data suggest that G1-4 represent four different CQ-induced alteration modes of histone methylation and mRNA expression, which define different target cellular processes.
Different TFs are associated with CQ-induced changes in mRNA expression
CQ regulates histone methylation through JmjC-KDMs without directly binding to DNA. KDMs are recruited to methylated histones for target genes by TFs, thereby regulating their mRNA expression (Benveniste et al., 2014). We then examined the TFs that could be involved in the recruitment of JmjC-KDMs to the differentially methylated histones for the genes in G1-4 as described previously (Lee et al., 2017). We identified a total of 62 TFs (45, 42, 46, and 31 TFs for G1-4, respectively) whose binding motifs were significantly (p < 0.01) enriched in the differentially methylated regions (peaks) for the genes in G1-4 (Figure 5A; Table S8). Of these, 45 TFs were shared in at least two groups between G1-4. In comparison, 17 TFs were uniquely identified in individual groups, suggesting that different sets of TFs could be associated with the recruitment of JmjC-KDMs to the differential trimethylation of histone three in G1-4.
Figure 5.
Different TFs are associated with CQ-induced changes in mRNA expression
(A) Relationships among the TFs whose binding motifs were significantly enriched in the methylated regions of the genes in G1-4 in hADSCs. Numbers in parentheses denote the numbers of TFs identified for G1-4.
(B) Binding motifs of the five representative TFs for the predicted target genes in G1-3. HIF1A and ARNT have a shared binding motif.
(C) GOBPs represented by the target genes of the five representative TFs in G1-3 (B). Color bar, gradient of –log10(p), where p is the enrichment p value for individual GOBPs obtained using DAVID software.
(D) Network model describing the interactions among the target genes of the five representative TFs. Node colors represent upregulation (red) and downregulation (green) in CQ, compared to normoxia. Green circles, red circles, and square nodes represent the target genes in G1-3, respectively. Large nodes denote the genes for which binding of the five representative TFs to their differentially methylated regions was experimentally confirmed. Arrows and inhibition symbols represent activation and inhibition information, respectively, as obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database. Solid and dotted lines denote direct and indirect activation/inhibition, respectively.
(E) Relative mRNA expression levels (left), methylated histone levels (middle), and TF-bound DNA amounts (right) of the indicated target genes in G1-3 as measured using qRT-PCR, histone ChIP-PCR, and TF ChIP-PCR analyses under N and CQ, respectively. Data are shown as the mean ± SEM (n = 6 per condition, exceptionally for TRIM39 gene n = 8 per condition of H3K9me3 ChIP-PCR, for MT3 gene n = 3 per condition of H3K4me3 ChIP-PCR).
(F) Effect of ARNT and HIF-1α knockdown. qRT-PCR analyses of ARNT and HIF-1α in hADSCs infected with lentiviruses encoding shRNAs against ARNT and HIF-1α (Table S1) (Lee et al., 2017).
(G) ChIP-PCR and qRT-PCR analyses of the indicated genes in shARNT- and shHIF-1α-hADSCs. (E–G) Data are shown as the mean ± SEM (n = 3 per condition). ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001 via Students’ t-test.
Of these enriched TFs, we focused on 60 TFs for G1-3 in the following analyses as there are fewer genes in G4 compared to G1-3. Among them, we selected the following representative TFs based on previously described criteria (Lee et al., 2017): RELA and RUNX2 for G1, JUN and STAT1 for G2, and ARNT/HIF1A and STAT1 for G3. The enriched binding motifs of these TFs are shown in Figure 5B. To understand the functional roles of these representative TFs, we performed GOBP enrichment analysis for their target genes in G1-3 (Figure 5C). We found that these target genes also represented most of the cellular processes in G1-3 (Figure 4C). To examine the collective associations of these TFs with these processes, we constructed a network model describing the interactions among their target genes (Figure 5D). The network model showed that the representative TFs primarily contributed to the downregulation of the actin cytoskeleton (TUBA1C, PARVA, CALD1, TLN2, WIPF1, MYH10, and KIF18A) together with its upstream extracellular matrix (ECM) (LUM, DDR2, SULF1, and COL8A1/12A1) and signaling (PDGFRC, FGF2, IRS1, ITGA11, and MYLK) molecules. They are also associated with the downregulation of cell adhesion (VCAN, CD99L2, CLDN11, and SDC2) and proliferation (GLI2/3, FIGNL1, NABP1, and EFEMP1) (Figure 5D, left). Moreover, the network model also showed that these TFs contributed to the upregulation of the HIF-1α pathway (BNIP3, MT3, VEGFB, APLN, LEP, PLIN2, and STC1), glucose metabolism (SLC2A3, PFKB4, ALDOC, and ENO2), autophagy (ATG14, RNF185, ATP6V0D1, and SLC17A9), apoptosis (ERO1A, MTFP1, NOL3, CDIP1, and BAG6), and proteolysis (UBE2O, SIAH2, UBR4/5, UBA6, TRIM39, UBQLN1, and NGLY1) (Figure 5D, right).
Next, we experimentally tested the validity of the following representative pairs of TF-target genes in the network model using ChIP-PCR experiments (Figure 5E): RELA-NABP1 and SULF1 (G1), JUN-TRIM39 and PLIN2 (G2), and ARNT-STC1 and MT3 (G3). To examine the effects of these predicted TFs on the expression and histone trimethylation of the selected target genes, we knocked down the mRNA expression of ARNT and its dimerization partner HIF-1α by 29.57 and 5.92% in hADSCs, respectively (Figure 5F). The knockdown of ARNT and HIF-1α significantly diminished the CQ-induced mRNA levels and H3K4me3 levels of the selected target genes, STC1 and MT3, in G3 (Figure 5G). These data suggest that representative TFs could be involved in the recruitment of KDMs to differentially methylated regions of the CQ target genes, contributing to the regulation of histone methylation and expression of target genes in G1–G3. Overall, these data suggest that different sets of TFs collectively contribute to the CQ-induced changes in key cellular processes (actin cytoskeleton, HIF-1α pathway, and glucose metabolism) associated with G1-3. In summary, our results demonstrate that CQ affects histone methylation (H3K4me3, H3K9me3, and H3K27me3) by inhibiting the activities of JmjC-KDMs, thereby contributing to the transcription of target genes and the cellular processes associated with them. The CQ-dependent regulation of histone methylation and target gene expression is characterized by multiple regulatory modes (i.e., four major modes represented by G1-4 in Figure 4B) involving different types of histone methylation and different sets of TFs involved in the recruitment of JmjC-KDMs. This further contributes to the differential regulation of the cellular processes associated with these individual regulatory modes.
Discussion
In this study, we investigated whether CQ could inhibit the catalytic activities of KDMs based on the binding of CQ to FIH-1 and the structural similarity of the JmjC domain in FIH-1 and KDMs (Choi et al., 2006). Our results revealed that CQ indeed inhibited the demethylation activities of KDM5A/B, KDM4A/C, and KDM6B for the three major histone methylation patterns, H3K4me3, H3K9me3, and H3K27me3, respectively (Figure 1). This CQ-mediated inhibition of KDMs increased the levels of all three histone methylation markers, as did the inhibition of KDMs by hypoxia. We previously developed an integrative analysis of methylated histones and mRNA expression data under hypoxia to understand the link between multidimensional histone methylation changes and the transcriptional regulation of target genes. Using this integrative analysis, we categorized the target genes of CQ into G1-4 based on the alteration patterns of methylated histones and mRNA expression. We revealed that the genes in G1-4 predominantly involved changes in a single type of histone trimethylation rather than complex combinatorial changes of multiple types of histone trimethylation, where the methylated histones corresponding to G1-4 were associated with different sets of TFs responsible for the transcriptional regulation of their target genes. This single-type histone methylation-based regulatory mechanism has been consistently observed under hypoxic conditions. Overall, our results suggest that KDM inhibitors, such as hypoxia and CQ, appear to regulate the mRNA expression of target genes by altering single types of histone trimethylations, although this regulatory mechanism should be investigated in other KDM inhibitors.
Our results revealed that most of the target genes and their associated cellular processes were shared between CQ and hypoxia. However, several cellular processes are affected by either CQ or hypoxia. Representative cellular processes associated with hypoxia, such as responses to hypoxia and glycolysis, were shared. We have previously demonstrated that CQ, similar to hypoxia, stabilizes and activates HIF-1α (Choi et al., 2006). Therefore, the CQ-mediated inhibition of KDMs and the activation of HIF-1α are expected to result in a large overlap of cellular processes shared between CQ and hypoxia. However, unlike hypoxia, CQ uniquely enhanced apoptosis and autophagy and decreased oxidative phosphorylation, tricarboxylic acid cycle, and fatty acid oxidation. Consistent with our findings from FACS analyses (Figures 3H, S3E, and S3F) and MTT assays (Figures S3C and S3D), Mizutani et al. (2021) also showed that CQ (50 μM, 24 h) induced cell death in astrocyte-derived KT-5 cells in an apoptosis-independent manner. They found that although CQ increased the expression of LC3-II and p62 autophagy-related proteins, it inhibited lysosomal function, resulting in an incomplete execution of autophagy. In addition to impairing autophagy degradation, CQ increased ROS and decreased ATP levels, leading to cell death.
Our results showed that CQ treatment induced alterations in the expression of target genes and cellular processes associated with diseases, for which CQ could be used for therapeutic purposes (Perez et al., 2019). Many genes (128 genes) encoding transition metal ion-binding proteins, including KDMs, were found to be significantly upregulated by CQ, suggesting the possibility that CQ can modulate metal ion-dependent cellular processes. CQ, as an ion chelator or ionophore, has been previously shown to effectively reduce Cu(II) and Zn(II), which are used for amyloid-beta plaque formation (Cherny et al., 2001). Accordingly, CQ effectively dissolved β-amyloid deposits in mouse and human brains with Alzheimer’s disease (Helmuth, 2000) and prevented the deterioration of their cognitive abilities with no adverse effects (Ritchie et al., 2003). Similarly, CQ and PBT2 (5,7-dichloro-2-[(dimethylamino)methyl]-8-hydroxyquinoline) — a CQ derivative — also have protective effects against Parkinson’s disease (Billings et al., 2016) and Huntington’s disease (Huntington Study Group Reach, 2015), respectively, via ion chelating or ionophore properties. Moreover, ATP-binding cassette transporter subfamily C (ABCC) members have been reported to be targets of CQ (Perez et al., 2019). Notably, ABCC4 and ABCC5 were significantly downregulated by CQ, suggesting that CQ may inhibit the efflux of molecules transported by these ABCCs, such as nucleotides and cAMP (Copsel et al., 2011), and reduce the efflux of therapeutic agents, thereby improving the reversal of multidrug resistance (Robey et al., 2018).
During the early 1900s, CQ was considered a safe drug and was clinically used as an antimicrobial, with therapeutic doses varying from 250 mg to 3.5 g per day (Woodward and Rahman, 1969; Richards, 1971; Tsubaki et al., 1971). In the 1970s, subacute myelo-optic neuropathy (SMON) was reported as an adverse effect of CQ in Japan but not elsewhere (Kono, 1971). Notably, recent studies have revealed that single-nucleotide polymorphisms in ABCCs (in particular, ABCC4 and ABCC11) might be responsible for SMON in the Japanese population (Perez et al., 2019). In a pilot phase 2 clinical trial in patients with Alzheimer’s disease, the total daily doses of 250, 500, and 750 mg of CQ ranged from 13 to 25 μmol/L (4–8 μg/mL) as measured from basal plasma levels (Ritchie et al., 2003). In a phase 1 clinical trial in patients with hematologic malignancies, the maximum tolerated dose of CQ was 2,400 mg/day (estimated to be 80 μmol/L in plasma). Administration of 3,200 mg CQ/day was dose-limiting, showing neurotoxicity and abdominal pain (Schimmer et al., 2012). The regulation of the CQ dose in other applications (such as cancer and neurological diseases) should be carefully monitored to avoid potential adverse effects. This study provides a large amount of data on histone methylation and putative TFs for the expression of CQ target genes in comparison with hypoxic target genes, which can be used to predict the potential beneficial and adverse effects of CQ as a new therapy for malignancies (Chen et al., 2007; Schimmer et al., 2012) and neurodegenerative diseases (Ritchie et al., 2003).
Limitations of the study
CQ has a relatively broad spectrum of target molecules, including FIH-1, ABCC, and proteasomes. This study identified novel targets of CQ, the JmjC-KDM family, which has more than 20 different isoforms with distinct and shared specificities for methylated histones. Owing to the diverse targets of CQ, undesirable off-target effects of CQ have been a concern. Nonetheless, CQ has been evaluated for therapeutic use in treating many diseases, such as Alzheimer’s disease and various cancers, owing to its favorable toxicity profiles and lipid solubility.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-Histone H3 | Abcam | Cat# ab1791; RRID: AB_302613 |
| Rabbit polyclonal anti-H3K9me3 | Abcam | Cat# ab8898; RRID: AB_306848 |
| Rabbit monoclonal anti-H3K4me3 | Cell signaling Technology | Cat# 9751S; RRID: AB_2616028 |
| Rabbit polyclonal anti-H3K27me3 | Millipore | Cat# 07-449; RRID: AB_310624 |
| Rabbit monoclonal BCL2L11(Bim) | Cell Signaling Technology | Cat# 2933; RRID: AB_1030947 |
| Rabbit monoclonal BIRC2 (cIAP1) | Cell Signaling Technology | Cat# 7065; RRID: AB_10890862 |
| Rabbit monoclonal XIAP | Cell Signaling Technology | Cat# 2045; RRID: AB_2214866 |
| Mouse monoclonal anti-HIF-1α (Figure 3D) | BD biosciences | Cat# 610959; RRID: AB_398272 |
| Mouse monoclonal anti-β-actin (Figure 3D) | Sigma-Aldrich | Cat# A5441; RRID: AB_476744 |
| Mouse monoclonal p53 (clone DO-1) | Santa Cruz Biotechnology | Cat# sc-126; RRID: AB_628082 |
| mouse monoclonal mono/polyclonal p62 (clone: D-3) | Santa Cruz Biotechnology | sc-28359; RRID: AB_628279 |
| Rabbit polyclonal anti-LC3B | Cell Signaling Technology | Cat# 2775; RRID: AB_915950 |
| Bacterial and virus strains | ||
| E. coli BL21(DE3) | Thermo Fisher | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Clioquinol (CQ) | Sigma-Aldrich | 24880; CAS: 130-26-7 IUPAC Name: 5-chloro-7-iodoquinolin-8-ol |
| JIB-04 | Sigma-Aldrich | SML0808; CAS: 199596-05-9, IUPAC Name: 5-chloro-N-[(E)-[phenyl(pyridin-2-yl)methylidene]amino]pyridin-2-amine |
| Broxyquinoline (BQ) | Sigma-Aldrich | D41600; CAS: 521-74-4, IUPAC Name: 5,7-dibromoquinolin-8-ol |
| Chloroacetoxyquinoline (CAQ) | MicroSource Discovery Systems | The NINDS custom collection library, CAS: 10173-02-1, IUPAC Name: (5-chloroquinolin-8-yl) acetate |
| Hydroxyquinol (HQ) | Sigma-Aldrich | H6878, CAS: 148-24-3, IUPAC Name: benzene-1,2,4-triol |
| Iodoquinol (IQ) | Sigma-Aldrich | D123609; CAS: 83-73-8, IUPAC Name: 5,7-diiodoquinolin-8-ol |
| 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenylte trazolium bromide (MTT) | Sigma-Aldrich | M2128; CAS: 298-93-1, IUPAC Name: 2-(3,5-diphenyltetrazol-2-ium-2-yl)-4,5-dimethyl-1,3-thiazole; bromide |
| Trimethyl-histone H3K9 peptide | Upstate | #12-568 |
| Critical commercial assays | ||
| SYBR Green PCR master mix | Applied Biosystems | 4367659 |
| FITC Annexin V Apoptosis Detection Kit I | BD bioscience | 556547 |
| Deposited data | ||
| Raw and processed microarray data | This paper | GEO: GSE69495 |
| Raw and processed ChIP-seq data | This paper | GEO: GSE69493 |
| Experimental models: Cell lines | ||
| Human: Human adipose-derived stem cells | Invitrogen | R7788115 |
| Human: HeLa cells | ATCC | CCL-2 |
| Human: U87 cells | Korean Cell Line Bank | Human primary glioblastoma cell line (U87 MG) |
| Oligonucleotides | ||
| Primers for RT-PCR, see Table S1 | This paper | N/A |
| Primers for ChIP-qPCR, see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| pET-32a-KDM4A(1–350aa) | (Whetstine et al., 2006) | HGNC:22978 |
| pcDNA-HA tagged-KDM4A | (Whetstine et al., 2006) | HGNC:22978 |
| pcDNA-Myc tagged-KDM4C | Addgene ID: 24214 | HGNC:17071 |
| pcDNA-HA tagged-KDM5A | (Lee et al., 2013) | HGNC:9886 |
| pcDNA-Myc tagged-KDM5B | (Xiang et al., 2007b) | HGNC:18039 |
| pcDNA-Myc tagged-KDM6B | (Xiang et al., 2007a) | HGNC:29012 |
| Software and algorithms | ||
| ImageJ | (Schneider et al., 2012) | https://imagej.nih.gov/ij/ |
| BD FACSDivaTM | BD bioscience | https://www.bdbiosciences.com/ko-kr/products/software/instrument-software/bd-facsdiva-software#Overview |
| Genome Studio | Illumina | https://support.illumina.com/array/array_software/genomestudio/downloads.html |
| Cutadapt | (Martin, 2011) | https://cutadapt.readthedocs.io/en/stable/ |
| BOWTIE2 | (Langmead and Salzberg, 2012) | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| Bedtools | (Quinlan and Hall, 2010) | https://bedtools.readthedocs.io/en/latest/ |
| MACS2 | (Zhang et al., 2008) | https://pypi.org/project/MACS2/ |
| DiffBind | (Ross-Innes et al., 2012) | https://bioconductor.org/packages/release/bioc/html/DiffBind.html |
| Other | ||
| https://github.com/SehyunChae/Clioquinol | In this paper | https://doi.org/10.5281/zenodo.6580328 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Hyunsung Park at hspark@uos.ac.kr.
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Cell culture
Human adipose-derived stem cells (hADSCs) were purchased from Invitrogen (Carlsbad, CA, USA) and isolated from human adipose tissue collected during liposuction procedures and cultured in MesenPRO RSTM Basal Medium (Invitrogen, Carlsbad, CA, USA) with MesenPRO RSTM Growth supplement and L-glutamine according to manufacturer’s instruction. Human HeLa cervical epithelium cells (CCL-2) were purchased from the American Type Culture Collection (Manassas, VA, USA) and cultured in DMEM supplemented with 10% fetal bovine serum (Life Technologies, Grand Island, NY, USA). Human primary glioblastoma cell line (U87 MG) were obtained from Korean Cell Line Bank. Cells were cultured in Eagle’s minimum essential medium (10-009, Corning, Glendale, AZ, USA) supplemented with 10% fetal bovine serum (FBS), 100 IU/mL penicillin, and 100 μg/mL streptomycin. The cells were exposed to hypoxic conditions (<0.5% O2) in an anaerobic incubator (Model 1029; Forma Scientific, Waltham, MA, USA) or 1.5% O2 using an In Vivo 200 Hypoxia Workstation (Ruskin Technology, Leeds, UK). The oxygen level in the anaerobic incubator was verified to be <0.5% using a Fyrite Analyzer (Bacharach, New Kensington, PA, USA).
Method details
Quantitative real-time revere transcription-polymerase chain reaction (qRT-PCR) and ChIP analysis
qRT-PCR and ChIP-PCR were performed using a SYBR Green PCR master mix (Applied Biosystems, USA) and a QuantStudio™ 3 Real-Time PCR System (Applied Biosystems). ChIP was performed, with the following modifications (Park and Park, 2010). After cross-linking, the nuclei were isolated, treated with MNase (300 gel units, 15 min; NEB, Ipswich, MA, USA), and sonicated to shear the chromatin. After centrifugation at 12,000 rpm for 10 min, the DNA concentration in the chromatin supernatant was measured to ensure that an equal amount of chromatin was used. For each immunoprecipitation, 1–1.5 mg of soluble chromatin was diluted in ChIP dilution buffer at a ratio of 1:10. Diluted lysates were immunoprecipitated with 10–20 μg of the indicated antibodies at 4°C overnight. The antibody-chromatin complexes were recovered via incubation with 200 μL of protein G agarose (50% slurry) and then washed. The immunocomplexes were then eluted with 300 μL of elution buffer, and cross-linking was reversed. Immunoprecipitated DNA was purified by phenol/chloroform extraction and ethanol precipitation using a QIAquick PCR purification kit (QIAGEN, Chatsworth, CA, USA). The primer sets used for qRT-PCR and ChIP-PCR are listed in Table S1.
FACS of apoptotic cells
hADSCs were incubated under hypoxia (0.5% O2, 48 h) or treated with clioquinol (50 μM, 48 h), then trypsinized and resuspended in 100 μL annexin V-binding buffer containing 5 μL of annexin V-FITC. After 15 min, 5 μL of propidium iodide (PI) was added to the cells, which were further incubated for 15 min in the dark at room temperature, according to the manufacturer’s instructions (556547; BD PharmingenTM, Franklin Lakes, NJ). PI-positive and annexin V-positive cells were counted on a FACSCantoII system and analyzed using BD FACSDivaTM software (BD Biosciences, Piscataway, NJ).
Molecular dynamics simulations
In the molecular dynamics (MD) simulation study, we used OpenMM (Eastman et al., 2017) with the CHARMM force field (Vanommeslaeghe et al., 2010). The crystal structure of KDM4A-pyridine-2,4-dicarboxylic acid (PDBID: 2VD7) was used, and Ni(II) was replaced with Fe(II). The protonation states of KDM4A under the corresponding experimental conditions (pH = 7.3) were set using PROPKA 3.1 (Sondergaard et al., 2011); (Olsson et al., 2011). The location of CQ inside the active site of KDM4A was initially set by fitting the pyridine ring and the N and O positions of pyridine-2,4-dicarboxylic acid and CQ. NaCl salts (0.15 M; physiological conditions) were then randomly added to the simulation box 9.3 × 9.3 × 9.3 nm3. Finally, the empty space was filled with TIP3P water molecules. The initial KDM4A-CQ complex was energy-minimized for 5,000 steps and then annealed from 0 K to 310.15 K at 1 bar for 30 ps. Equilibrium MD simulations of the resulting complex were run for 200 ns with a time step of 2 fs. The thermostat and barostat used in the MD simulation were the Langevin dynamics and the Monte Carlo method, respectively. The periodic boundary condition was applied in all directions, and both the Lennard-Jones and the real part of the electrostatic interaction used a cutoff radius of 1.2 Å. In addition, the particle mesh Ewald (PME) method was used for the reciprocal part of the electrostatic interaction.
MTT assay
MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) was purchased from Sigma-Aldrich (St. Louis, MO). Briefly, 50 μL of the MTT reagent (5 mg/mL) was added to the culture medium and incubated at 37°C for 3 h. DMSO was added to dissolve the formazan aggregates. The absorbance of the resulting solution was then measured at 550 nm using a microplate reader. The control was set to 100% viability for each assay, and all other treatments were corrected against this value. Cells were seeded onto each well of a 24-well plate at a density of 4×104 cells/well.
shRNA
The following lentiviral constructs were used in this study. For shRNA studies, shRNA against ARNT and HIF-1α cloned into the lentiviral vector pLKO.1 TRC cloning vector (Addgene, Watertown, MA). hADSCs were infected with lentiviral constructs encoding hHIF1 alpha shRNAs (5′-CCAGTTATGATTGTGAAGTTA-3′), hARNT shRNAs (5′-GCCTACACTCTCCAACACAAT-3′), or scramble shRNAs (5′- CCTAAGGTTAA-GTCGCCCTCG-3′) by standard procedures.
Gene expression profiling experiments
Gene expression profiles of hADSCs were generated using an Illumina HumanHT-12 v4 Expression BeadChip (Illumina, San Diego, CA), which includes 47,323 probes corresponding to 30,500 annotated genes. According to Illumina’s standard protocol, biotinylated cRNA was prepared from total RNA using the Illumina Total Prep RNA Amplification Kit (Ambion, Austin, TX, USA). Following fragmentation, the cRNA was hybridized to the Illumina HumanHT-12 Expression BeadChip. The arrays were scanned using the Illumina bead array reader confocal scanner. After the microarray experiments, the log2-intensities of all probes and their annotations were acquired using Illumina Genome Studio v2009.2 (Gene Expression Module v1.5.4). The raw data were deposited in the Gene Expression Omnibus (GEO) database with accession ID: GSE176557.
Identification of differentially expressed genes (DEGs)
The log2-intensities of the probes from the arrays were normalized using the quantile normalization method (Bolstad et al., 2003). To identify the DEGs, a previously reported statistical hypothesis test was performed (Lee et al., 2017) Briefly, for each gene, a T-statistic value was calculated using Student’s t-test, and the log2-median-ratio was calculated. The empirical distributions of the T-statistics and log2-median-ratios for the null hypothesis (i.e., the genes were not differentially expressed) were estimated by performing all possible combinations of random permutations of the samples. Using the estimated empirical distributions, the adjusted p-values for the t-test and log2-median-ratio test for each gene were computed and then combined with Stouffer’s method (Hwang et al., 2005). Finally, the DEGs were identified as genes with combined p-values < 0.05 and fold-changes ≥1.5.
Library construction and ChIP-seq data analyses
ChIP-seq was performed on the sequencing library prepared according to Illumina’s standard protocols (Lee et al., 2017). Chromatin immunoprecipitated DNA (ChIP DNA) was processed using Klenow polymerase and T4 polynucleotide kinase to create 3-dA. ChIP DNA was then ligated with Illumina adaptors and amplified using Illumina primers for 18 cycles. Following the isolation of 100–500 bp of ChIP DNA from the agarose gel, an Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA) was used to confirm the size, purity, and concentration of DNA, and library quantitation was performed using the HiSeq™ 2000 platform to produce 101-bp paired-end samples. From the sequencing data, adapter sequences (TruSeq universal and indexed adapters) were removed using cutadapt software (version 2.7) (Martin, 2011)(ref), and the remaining reads were aligned to the human genome (GRCh38) using BOWTIE2 software (version 2.2.6) with default parameters (Langmead and Salzberg, 2012). After alignment, the number of reads mapped to gene features (GTF file of GRCh38.89) were counted using CoverageBed from Bedtools (Quinlan and Hall, 2010), and the reads per kilobase of target per million mapped reads (RPKM) were computed for the gene features. The total read counts obtained for individual samples and the alignment results are summarized in Table S2. Raw ChIP-seq data were deposited in the GEO database (accession ID: GSE176379).
Locational distribution analysis of histone methylations
For each gene, we first divided the promoter region (–5kb ∼ transcription start site, TSS), gene body (TSS ∼ transcription end site, TES), and downstream region (TES ∼ +5kb) into 10, 25, and 10 bins, respectively. In each sample, we obtained the read counts in individual bins using CoverageBed from Bedtools (Quinlan and Hall, 2010) and computed the read densities as RPKMs by dividing the read counts in the bins using the following equation: (bin size/1,000) × (total number of mapped reads in the sample/106). For each histone methylation (H3K4me3, H3K9me3, or H3K27me3), we obtained the average read density profile over the bins by computing the average read densities of all the genes in the individual bins.
Peak enrichment
The enriched peak regions were determined using the MACS2 algorithm with the following parameters: -f BAMPE -g hs -p 1e-3 --keep-dup = 1 (Zhang et al., 2008). Consensus peaks detected consistently in normoxia, hypoxia, and CQ were identified using DiffBind (version 2.14.0) (Ross-Innes et al., 2012) with the following parameters: minOverlap = 1 and score = DBA_SCORE_READS.
Identification of differentially methylated genes (DMGs)
For each gene, RPKMs for the consensus peaks within the promoter + gene body region (-5kb ∼ TES) were calculated for H3K4me3, H3K9me3, and H3K27me3 in normoxia, hypoxia, and CQ, respectively. For each consensus peak of the gene, the mean log2-fold-changes of the RPKMs were computed for the three histone methylations. Finally, the DMGs were identified as the genes for which the absolute mean log2-fold-changes of H3K4me3, H3K9me3, or H3K27me3 for all consensus peaks in the gene was >0.58 (1.5-fold increase).
Correlation analysis of gene expression and histone methylation data
Correlation analysis between gene expression and histone methylation was performed as previously reported (Lee et al., 2017). Briefly, the annotated genes were sorted in a descending manner according to their expression levels or log2-fold-changes and then binned such that each bin included 100 genes. The mean levels or log2-fold-changes of histone methylation (H3K4me3, H3K9me3, or H3K27me3) were computed in individual bins. A scatter plot of gene expression versus histone methylation data (mean levels or log2-fold-changes) in individual bins was generated. The Pearson’s correlation coefficient between gene expression and histone methylation data in the scatter plot was computed. p-values were computed using a t-test for the correlation coefficients.
Functional enrichment analysis
Enrichment analysis of gene ontology biological processes (GOBPs) was performed for a list of genes (e.g., selected DEGs) using DAVID software (Huang et al., 2009). GOBPs with p-values < 0.1 (default cutoff in DAVID) were selected as the processes enriched by the list of genes analyzed. A network model was constructed for selected DEGs using the interactions between these DEGs in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (Kanehisa et al., 2012). The constructed network model was visualized using Cytoscape software (Cline et al., 2007), and the nodes were arranged based on the locations and relationships of the corresponding genes in the KEGG pathway database.
TF binding site analysis
To identify TFs with binding motifs enriched in differentially methylated peak regions in a list of genes, we collected 161 TF matrices from the JASPAR CORE database (Mathelier et al., 2014). Sequences for differentially methylated peaks (input sequences) were scanned using MotifLocator software (Thijs et al., 2002); (Lee et al., 2017). The 2nd order custom background model was created using the CreateBackgroundModel software in MotifSuite, using all peak sequences. To assess the statistical significance of the MotifLocator scores, we randomized the input sequence and applied MotifLocator 1000 times to the randomized sequence using the parameters –b (custom background model) and –t −1. The resulting scores from the random experiments were used to estimate the empirical distribution of the MotifLocator scores for each TF. Based on the empirical distribution, we computed the false discovery rates (FDRs) for the observed scores using Storey’s method (Storey and Tibshirani, 2003). The binding sites of each TF were selected as those with an FDR<0.001, and target genes of the TF were identified as those with >1 binding site. Next, we identified TFs with a significant number of target genes in a list of genes. We first randomly shuffled the input sequence 10,000 times, counted the number of targets for each TF in individual random experiments, and estimated an empirical distribution for the number of targets using a Gaussian kernel density estimation. Using empirical distribution, we computed the FDR for the observed number of targets of each TF in a list of genes using Storey’s method (Storey and Tibshirani, 2003). Finally, we selected TF candidates with an FDR <0.01.
Evaluation of in vitro histone demethylase activity and mass spectrometric analyses
Recombinant His-tagged truncated KDM4A (1–350 amino acids) proteins were produced in E. coli BL21(DE3) transformed with pET-32a-KDM4A(1–350aa) and isolated Cusing Ni-NTA agarose (QIAGEN) and then purified. The purified KDM4A proteins were incubated with 2 μM of trimethyl-histone H3K9 peptide (ARTKQTAR(me3K)STGGKAPRKQLA-GCK-biotin, upstate #12-568, 2,765.3 Da) in demethylase reaction buffer (20 mM Tris-HCl [pH 7.3], 150 mM NaCl, 100 μM 2OG, and 2 μM ascorbic acid) for 4 hours at 37°C. After the demethylase reaction, excess salts were removed with ZipTipC18 (Millipore). The peptide was eluted from the tip with cyano-4-hydroxycinnamic acid in acetonitrile/water containing 0.1% trifluoroacetic acid (50:50, v/v) followed by extensive washing with 0.1% trifluoroacetic acid in water. The eluted peptide solution was transferred to a MALDI sample plate and MALDI-TOF measurements were performed with a Voyager analyzer (Applied Biosystems).
Quantification and statistical analysis
Statistical analyses were performed using Graphpad Prism software. For comparisons between two groups, we used a two-tailed Student’s t-test. We used one-way ANOVA with Tukey’s correction as a post-hoc test for comparisons among multiple groups. The sample sizes, statistical tests, and parameters are listed in the corresponding figure legends. Data are shown as the mean ± SEM. The criterion of significance was set at p < 0.05 in all cases.
Acknowledgments
This research was supported by grants from the National Research Foundation of Korea (NRF-2012M3A9B6055343, NRF-2018R1A4A1025985, NRF-2019R1A2B5B01069366, NRF-2019R1A6A1A10073079, and NRF-2019M3A9D5A01102794) and the Basic Research Program (22-BR-01-02) through the Korea Brain Research Institute, funded by the Ministry of Science and ICT.
Author contributions
Conceptualization, H.P., D.H., Y.M., and S.C.; Experiments, Y.M., S.Y., E.G.Y., and J.C.; Data Analysis, D.H., J.C., J.H., R.C., and S.C.; Investigation, H.P., S.C., and D.H.; Writing – Original Draft, H.P. and D.H.; Writing – Review & Editing, H.P., D.H., Y.M., S.C., and S.Y.; Supervision, H.P. and D.H.
Declaration of interests
The authors declare no competing interests.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104517.
Contributor Information
Daehee Hwang, Email: daehee@snu.ac.kr.
Hyunsung Park, Email: hspark@uos.ac.kr.
Supplemental information
Data and code availability
Raw and processed microarray data have been deposited at GEO and are publicly available at the date of publication. Accession numbers are listed in the key resources table [GSE69495]. Raw and processed ChIP-seq data have been deposited at GEO and are publicly available at the date of publication. Accession numbers are listed in the key resources table [GSE69493] . All source codes have been deposited at Github and are publicly available at https://github.com/SehyunChae/Clioquinol as of the date of publication. A DOI is listed in the key resources table. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Bailey M.H., Tokheim C., Porta-Pardo E., Sengupta S., Bertrand D., Weerasinghe A., Colaprico A., Wendl M.C., Kim J., Reardon B., et al. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;174:1034–1035. doi: 10.1016/j.cell.2018.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benveniste D., Sonntag H.J., Sanguinetti G., Sproul D. Transcription factor binding predicts histone modifications in human cell lines. Proc. Natl. Acad. Sci. U S A. 2014;111:13367–13372. doi: 10.1073/pnas.1412081111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings J.L., Hare D.J., Nurjono M., Volitakis I., Cherny R.A., Bush A.I., Adlard P.A., Finkelstein D.I. Effects of neonatal iron feeding and chronic clioquinol administration on the Parkinsonian human A53T transgenic mouse. ACS Chem. Neurosci. 2016;7:360–366. doi: 10.1021/acschemneuro.5b00305. [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Chang S., Yim S., Park H. The cancer driver genes IDH1/2, JARID1C/KDM5C, and UTX/KDM6A: crosstalk between histone demethylation and hypoxic reprogramming in cancer metabolism. Exp. Mol. Med. 2019;51:1–17. doi: 10.1038/s12276-019-0230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D., Cui Q.C., Yang H., Barrea R.A., Sarkar F.H., Sheng S., Yan B., Reddy G.P.V., Dou Q.P. Clioquinol, a therapeutic agent for Alzheimer's disease, has proteasome-inhibitory, androgen receptor-suppressing, apoptosis-inducing, and antitumor activities in human prostate cancer cells and xenografts. Cancer Res. 2007;67:1636–1644. doi: 10.1158/0008-5472.can-06-3546. [DOI] [PubMed] [Google Scholar]
- Chen H., Yan Y., Davidson T.L., Shinkai Y., Costa M. Hypoxic stress induces dimethylated histone H3 lysine 9 through histone methyltransferase G9a in mammalian cells. Cancer Res. 2006;66:9009–9016. doi: 10.1158/0008-5472.can-06-0101. [DOI] [PubMed] [Google Scholar]
- Chen Z., Zang J., Whetstine J., Hong X., Davrazou F., Kutateladze T.G., Simpson M., Mao Q., Pan C.H., Dai S., et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- Cherny R.A., Atwood C.S., Xilinas M.E., Gray D.N., Jones W.D., Mclean C.A., Barnham K.J., Volitakis I., Fraser F.W., Kim Y.S., et al. Treatment with a copper-zinc chelator markedly and rapidly inhibits beta-amyloid accumulation in Alzheimer's disease transgenic mice. Neuron. 2001;30:665–676. doi: 10.1016/s0896-6273(01)00317-8. [DOI] [PubMed] [Google Scholar]
- Choi S.M., Choi K.O., Park Y.K., Cho H., Yang E.G., Park H. Clioquinol, a Cu(II)/Zn(II) chelator, inhibits both ubiquitination and asparagine hydroxylation of hypoxia-inducible factor-1α, leading to expression of vascular endothelial Growth factor and erythropoietin in normoxic cells. J. Biol. Chem. 2006;281:34056–34063. doi: 10.1074/jbc.m603913200. [DOI] [PubMed] [Google Scholar]
- Cline M.S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B., et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copsel S., Garcia C., Diez F., Vermeulem M., Baldi A., Bianciotti L.G., Russel F.G., Shayo C., DaviO C. Multidrug resistance protein 4 (MRP4/ABCC4) regulates cAMP cellular levels and controls human leukemia cell proliferation and differentiation. J. Biol. Chem. 2011;286:6979–6988. doi: 10.1074/jbc.m110.166868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding W.Q., Liu B., Vaught J.L., Yamauchi H., Lind S.E. Anticancer activity of the antibiotic clioquinol. Cancer Res. 2005;65:3389–3395. doi: 10.1158/0008-5472.can-04-3577. [DOI] [PubMed] [Google Scholar]
- Eastman P., Swails J., Chodera J.D., Mcgibbon R.T., Zhao Y., Beauchamp K.A., Wang L.P., Simmonett A.C., Harrigan M.P., Stern C.D., et al. OpenMM 7: rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 2017;13:e1005659. doi: 10.1371/journal.pcbi.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmuth L. Neuroscience. An antibiotic to treat Alzheimer's? Science. 2000;290:1273–1274. doi: 10.1126/science.290.5495.1273a. [DOI] [PubMed] [Google Scholar]
- Huang Da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huntington Study Group Reach H.D.I. Safety, tolerability, and efficacy of PBT2 in Huntington's disease: a phase 2, randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2015;14:39–47. doi: 10.1016/S1474-4422(14)70262-5. [DOI] [PubMed] [Google Scholar]
- Hwang D., Rust A.G., Ramsey S., Smith J.J., Leslie D.M., Weston A.D., De Atauri P., Aitchison J.D., Hood L., Siegel A.F., Bolouri H. A data integration methodology for systems biology. Proc. Natl. Acad. Sci. U S A. 2005;102:17296–17301. doi: 10.1073/pnas.0508647102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.S., Leissing T.M., Chowdhury R., Hopkinson R.J., Schofield C.J. 2-Oxoglutarate-dependent oxygenases. Annu. Rev. Biochem. 2018;87:585–620. doi: 10.1146/annurev-biochem-061516-044724. [DOI] [PubMed] [Google Scholar]
- Kamura T., Sato S., Iwai K., Czyzyk-Krzeska M., Conaway R.C., Conaway J.W. Activation of HIF1α ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc. Natl. Acad. Sci. U S A. 2000;97:10430–10435. doi: 10.1073/pnas.190332597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kono R. Subacute myelo-optico-neuropathy, a new neurological disease prevailing in Japan. Jpn. J. Med. Sci. Biol. 1971;24:195–216. doi: 10.7883/yoken1952.24.195. [DOI] [PubMed] [Google Scholar]
- Kooistra S.M., Helin K. Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 2012;13:297–311. doi: 10.1038/nrm3327. [DOI] [PubMed] [Google Scholar]
- Lando D., Peet D.J., Gorman J.J., Whelan D.A., Whitelaw M.L., Bruick R.K. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.Y., Yang E.G., Park H. Hypoxia enhances the expression of prostate-specific antigen by modifying the quantity and catalytic activity of Jumonji C domain-containing histone demethylases. Carcinogenesis. 2013;34:2706–2715. doi: 10.1093/carcin/bgt256. [DOI] [PubMed] [Google Scholar]
- Lee S., Lee J., Chae S., Moon Y., Lee H.Y., Park B., Yang E.G., Hwang D., Park H. Multi-dimensional histone methylations for coordinated regulation of gene expression under hypoxia. Nucleic Acids Res. 2017;45:11643–11657. doi: 10.1093/nar/gkx747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loenarz C., Schofield C.J. Physiological and biochemical aspects of hydroxylations and demethylations catalyzed by human 2-oxoglutarate oxygenases. Trends Biochem. Sci. 2011;36:7–18. doi: 10.1016/j.tibs.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011;17:10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- Mathelier A., Zhao X., Zhang A.W., Parcy F., Worsley-Hunt R., Arenillas D.J., Buchman S., Chen C.Y., Chou A., Ienasescu H., et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani Y., Maeda T., Murate K., Ito S., Watanabe H., Mutoh T. Clioquinol kills astrocyte-derived KT-5 cells by the impairment of the autophagy-lysosome pathway. Arch. Toxicol. 2021;95:631–640. doi: 10.1007/s00204-020-02943-8. [DOI] [PubMed] [Google Scholar]
- Moon H., Han S., Park H., Choe J. Crystal structures of human FIH-1 in complex with quinol family inhibitors. Mol. Cell. 2010;29:471–474. doi: 10.1007/s10059-010-0058-3. [DOI] [PubMed] [Google Scholar]
- Okada Y., Tateishi K., Zhang Y. Histone demethylase JHDM2A is involved in male infertility and obesity. J. Androl. 2010;31:75–78. doi: 10.2164/jandrol.109.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson M.H., Sondergaard C.R., Rostkowski M., Jensen J.H. PROPKA3: consistent treatment of internal and surface residues in empirical pKa predictions. J. Chem. Theor. Comput. 2011;7:525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- Padmanabhan G., Becue I., Smith J.B. In: Analytical Profiles of Drug Substances. Florey K., Al-Badr A., Forcier G., Brittain H., Grady L.T., editors. Academic Press; 1990. Clioquinol. [Google Scholar]
- Park Y.K., Park H. Prevention of CCAAT/enhancer-binding protein beta DNA binding by hypoxia during adipogenesis. J. Biol. Chem. 2010;285:3289–3299. doi: 10.1074/jbc.m109.059212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez D.R., Sklar L.A., Chigaev A. Clioquinol: to harm or heal. Pharmacol. Ther. 2019;199:155–163. doi: 10.1016/j.pharmthera.2019.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards D.A. Prophylactic value of clioquinol against travellers' diarrhœa. Lancet. 1971;297:44–45. doi: 10.1016/s0140-6736(71)80056-9. [DOI] [PubMed] [Google Scholar]
- Ritchie C.W., Bush A.I., Mackinnon A., Macfarlane S., Mastwyk M., Macgregor L., Kiers L., Cherny R., Li Q.X., Tammer A., et al. Metal-protein attenuation with iodochlorhydroxyquin (clioquinol) targeting Abeta amyloid deposition and toxicity in Alzheimer disease: a pilot phase 2 clinical trial. Arch. Neurol. 2003;60:1685. doi: 10.1001/archneur.60.12.1685. [DOI] [PubMed] [Google Scholar]
- Robey R.W., Pluchino K.M., Hall M.D., Fojo A.T., Bates S.E., Gottesman M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer. 2018;18:452–464. doi: 10.1038/s41568-018-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R., et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmer A.D., Jitkova Y., Gronda M., Wang Z., Brandwein J., Chen C., Gupta V., Schuh A., Yee K., Chen J., et al. A phase I study of the metal ionophore clioquinol in patients with advanced hematologic malignancies. Clin. Lymphoma Myeloma Leuk. 2012;12:330–336. doi: 10.1016/j.clml.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondergaard C.R., Olsson M.H., Rostkowski M., Jensen J.H. Improved treatment of ligands and coupling effects in empirical calculation and rationalization of pKa values. J. Chem. Theor. Comput. 2011;7:2284–2295. doi: 10.1021/ct200133y. [DOI] [PubMed] [Google Scholar]
- Song A., Jiang S., Wang Q., Zou J., Lin Z., Gao X. JMJD3 is crucial for the female AVPV RIP-cre neuron-controlled Kisspeptin-estrogen feedback loop and reproductive function. Endocrinology. 2017;158:1798–1811. doi: 10.1210/en.2016-1750. [DOI] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateishi K., Okada Y., Kallin E.M., Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–761. doi: 10.1038/nature07777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thijs G., Moreau Y., De Smet F., Mathys J., Lescot M., Rombauts S., Rouze P., De Moor B., Marchal K. INCLUSive: integrated clustering, upstream sequence retrieval and motif sampling. Bioinformatics. 2002;18:331–332. doi: 10.1093/bioinformatics/18.2.331. [DOI] [PubMed] [Google Scholar]
- Tsubaki T., Honma Y., Hoshi M. Neurological syndrome associated with clioquinol. Lancet. 1971;297:696–697. doi: 10.1016/s0140-6736(71)92699-7. [DOI] [PubMed] [Google Scholar]
- Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., Mackerell A.D., Jr. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whetstine J.R., Nottke A., Lan F., Huarte M., Smolikov S., Chen Z., Spooner E., Li E., Zhang G., Colaiacovo M., Shi Y. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Woodward W.E., Rahman A.S. Trial of clioquinol in cholera. Lancet. 1969;294:270. doi: 10.1016/s0140-6736(69)90037-3. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Zhu Z., Han G., Lin H., Xu L., Chen C.D. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007;17:850–857. doi: 10.1038/cr.2007.83. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Zhu Z., Han G., Ye X., Xu B., Peng Z., Ma Y., Yu Y., Lin H., Chen A.P., Chen C.D. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc. Natl. Acad. Sci. U S A. 2007;104:19226–19231. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.J., Tran T.A.T., Wang M., Ranek M.J., Kokkonen-Simon K.M., Gao J., Luo X., Tan W., Kyrychenko V., Liao L., et al. Histone lysine dimethyl-demethylase KDM3A controls pathological cardiac hypertrophy and fibrosis. Nat. Commun. 2018;9:5230. doi: 10.1038/s41467-018-07173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., Liu X.S. Model-based analysis of ChIP-seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw and processed microarray data have been deposited at GEO and are publicly available at the date of publication. Accession numbers are listed in the key resources table [GSE69495]. Raw and processed ChIP-seq data have been deposited at GEO and are publicly available at the date of publication. Accession numbers are listed in the key resources table [GSE69493] . All source codes have been deposited at Github and are publicly available at https://github.com/SehyunChae/Clioquinol as of the date of publication. A DOI is listed in the key resources table. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.