Summary
Compared with the conventional DNA probe immobilization on the planar surface, nanoparticles-based DNA probes enable more RNA molecules to be anchored to the sensor surface, thereby improving the detection sensitivity. In this work, we report phosphorodiamidate morpholino oligomers (PMO)-graphene quantum dots (GQDs)-functionalized reduced graphene oxide (RGO) field effect transistor (FET) biosensors for ultrasensitive detection of exosomal microRNAs. After the RGO FET sensor was fabricated, polylysine (PLL) film was deposited onto the RGO surface. GQDs-PMO hybrid was prepared and covalently bound to PLL surface, enabling detection of exosomal microRNAs (miRNAs). The method achieved a detection limit as low as 85 aM and high specificity. Furthermore, the FET sensor was able to detect exosomal miRNAs in plasma samples and distinguish breast cancer samples from healthy samples. Compared with other methods, we use GQDs to further improve the sensitivity of FET, making it a potential tool for early diagnosis of breast cancer.
Subject areas: Analytical chemistry, Sensor, Biotechnology, Nanotechnology
Graphical abstract

Highlights
-
•
Exosomal miRNAs are detected by GQDs-PMO-functionalized G-FET sensor
-
•
The sensor can specifically detect 85 aM exosomal miRNA
-
•
The results are in agreement with those of qRT-PCR
-
•
This method can also distinguish breast cancer patients from healthy people
Analytical chemistry; Sensor; Biotechnology; Nanotechnology
Introduction
In recent years, exosomes have become a research hotspot due to their important role in the occurrence, development, and metastasis of tumors (Jiang et al., 2019; Kalluri and LeBleu, 2020; Xia et al., 2017; Xu et al., 2018). More importantly, exosomes carry a variety of functional molecules with biological activity, such as nucleic acids, proteins, peptides and lipids, and so forth (Wang et al., 2017, 2020; Zhou et al., 2017). And exosomal miRNAs (Exo-miRs) are involved in the communication between cells (Tomasetti et al., 2017). Importantly, there is high correlation among Exo-miRs levels and disease development and malignant progression of tumors. Moreover, Exo-miRs secreted by donor cells could enter recipient cells and directly participate in regulating gene expression (Azmi et al., 2013; Li et al., 2016). In addition, Exo-miRs have an advantage as biomarkers over plasma-derived circulating miRNAs (Endzelins et al., 2017; Hannafon et al., 2016). First of all, miRNAs are protected by the phospholipid bilayer because they are inside the exosomes, preventing them from being degraded in circulation (Boriachek et al., 2018; Cheng et al., 2014; Guo et al., 2020). Secondly, due to the protected exosomes, Exo-miRs are still active even under repeated freeze-thaw cycles or unsuitable pH (Ren et al., 2016). Although Exo-miRs have gradually been used as biomarkers for the diagnosis of malignant tumors, it is still a challenge to detect them with high sensitivity (Shi et al., 2020; Zeng et al., 2018).
To improve the sensitivity of Exo-miRs detection, electrochemistry, Raman, fluorescence, and nanochannel-based sensors are used for the detection of Exo-miRs. Lee et al. showed a multifunctional magneto-plasmonic nanorod for detection of exosome and Exo-miRs (Lee et al., 2019). This method is simple and nondestructive, but its sensitivity limits its further application. Kang et al. used a DNA-functionalized Au octahedral array as a Raman sensing platform to detect Exo-miRs (Kang et al., 2021). This method requires fluorescent labeling, and the detection limit is not low enough. Miao et al. used dumbbell hybrid chain reaction to amplify the electrochemical signal for Exo-miRs analysis (Miao and Tang, 2020). This method can detect Exo-miRs with high sensitivity, but it needs amplification. In our group, Liao et al. demonstrated a phosphorodiamidate morpholino oligos (PMO)-functionalized nanochannel biosensor for detection of microRNAs in serum samples (Liao et al., 2017). In this system, PMO probe was modified on the channel surface, and miRNAs detection was realized through observing the change of surface charge density when PMO/miRNA hybridization occurred. Xiao et al. reported a PNA-functionalized nanochannel biosensor for pancreatic cancer exosomal miRNA10b detection (Xiao et al., 2021). In this work, PNA, as a capture probe, was anchored on the nanochannel surface. Although the PNA/exosomal miRNA hybridization took place, the membrane current change could be recorded, resulting in the surface charge density change of the nanochannel. Although the nanochannel biosensor can achieve a high sensitivity, the PET-film-based nanochannels are prepared by artificial etching, leading to uncontrollable uniformity of the sensor.
The field effect transistor (FET) biosensor, as a label-free detection tool, has been rapidly developed in recent years (Liang et al., 2020; Mao et al., 2017; Syedmoradi et al., 2019). The change in charge caused by the interaction of biomolecules on the surface of the sensing channel is converted into a readable electrical signal by the field-effect transistor. The detection platform has the characteristics of label-free detection, fast analysis speed, and high sensitivity. Graphene field effect transistor (G-FET) sensors have been used to detect a variety of biomolecules (Jin et al., 2019; Li et al., 2019b; Thakur et al., 2018; Xie et al., 2016; Yu et al., 2019). Usually, the probe molecules are immobilized on the graphene surface to detect the targets. To improve the detection sensitivity, nanoparticles-based DNA probes would be an option to allow more probe molecules to be modified on the surface and to enable high hybridization efficiency due to the three-dimensional structures compared with the planar surface.
Carbon dots (CDs) are one of the zero-dimensional members with promising applications in sensors, optronics, and electrochemical luminescence (Das et al., 2018). Ramadan et al. incorporated CDs on the graphene surface to fabricate the graphene FET sensor, after which specific antibody was immobilized on the CDs surface to capture exosome (Ramadan et al., 2021). Majd et al. prepared DNA-functionalized CDs and drop-casted the CDs on MoS2-FET chips to detect Hg2+ with high sensitivity (Majd and Ghasemi, 2020). In our group, gold nanoparticles (AuNPs) were deposited on the graphene surface to achieve various detections including SARS-COV-2 RNA (Li et al., 2021), exosomes (Wu et al., 2020), and miRNAs (Cai et al., 2015). In addition, Pt nanoparticles were also deposited on the graphene surface to detect BNP (Lei et al., 2017). Graphene quantum dots (GQDs) are a subset of carbon quantum dots (Xu et al., 2014). GQDs were first isolated and characterized during 2008–2010 (Sun et al., 2006; Xu et al., 2004). The size of GQDs is very small; generally, the particle size of GQDs is less than 10 nm and the thickness is less than 10 graphene layers (Li et al., 2019a). Compared with other noble metal nanoparticles, GQDs are inexpensive to manufacture, easy to obtain. More importantly, they have good electrical conductivity, making them more suitable for the functionalization of probe molecules for biosensing applications. For example, Lu et al. confined GQDs with different functionalities in nanopores to detect different ions, in which GQDs served as the recognition element and signal amplifier in nanopores (Lu et al., 2018). Chen et al. used antibody-functionalized nitrogen-doped GQDs electrochemiluminescence (ECL) biosensor for detection of Escherichia coli O 157:H7 (Chen et al., 2017). However, ultrasensitive detection of exosomal miRNAs by GQDs-PMO-functionalized graphene FET is desirable.
Here, we report a GQDs-functionalized reduced graphene oxide (RGO) FET biosensor that can detect Exo-miRs in the blood of breast cancer patients with ultrasensitive sensitivity. As shown in Figure 1, the PMO-GQDs hybrid is obtained by incubating GODs with amine-modified PMO. After the RGO FET is fabricated, polylysine (PLL) is assembled onto the RGO surface, allowing the functionalized GQDs-PMO hybrid to be immobilized on the chip surface via amide formation. After that, Exo-miRs are specifically detected by hybridization of the immobilized PMO probes with miRNA. Upon binding of Exo-miRs with PMO on the chip surface, the net carrier density of the sensing channel changes due to contribution of negative charges from Exo-miRs, causing the Dirac point to shift to the left.
Figure 1.
Schematic principle of GQDs-PMO-functionalized PLL-RGO-FET (GPPR-FET) for detection of exosomal miRNA
Results
Characterization of the GQDs-PMO hybrid
The particle size of GQDs was characterized by transmission electron microscopy (TEM). As displayed in Figure 2A, GQDs were uniformly distributed in solution. Figure 2B shows the well-dispersed GQDs with the average size of about 4.4 nm.
Figure 2.
Characterization of GQDs-PMO
(A) TEM image of the GQDs. Scale bar, 20 nm.
(B) The size distribution of GQDs.
(C) Fluorescence spectra of GQDs and GQDs-PMO.
(D) Zeta potentials of GQDs and GQDs-PMO. Error bars represent the standard errors (n = 3).
To characterize the successful combination of GQDs and PMO, fluorescence and zeta potential were applied. As illustrated in the Figure 2C, after PMO (1 μM) was bound to GQDs (25 μg/mL), the fluorescence signal (365 nm) of GQDs-PMO was lower than that of GQDs, which was caused by the fluorescence quenching effect of PMO to GQDs. The results are consistent with those reported previously (Kermani et al., 2017; Wei et al., 2021), indicating that GQDs were successfully combined with PMO. Figure 2D was the zeta potential of GQDs and GQDs-PMO. The zeta potential of the GQDs was −21.22 mV. After PMO was combined with GQDs, the zeta potential (−10.37 mV) significantly reduced, also showing successful synthesis of GQDs-PMO hybrid.
Characterization of the GQDs-PMO functionalized PLL-RGO-FET
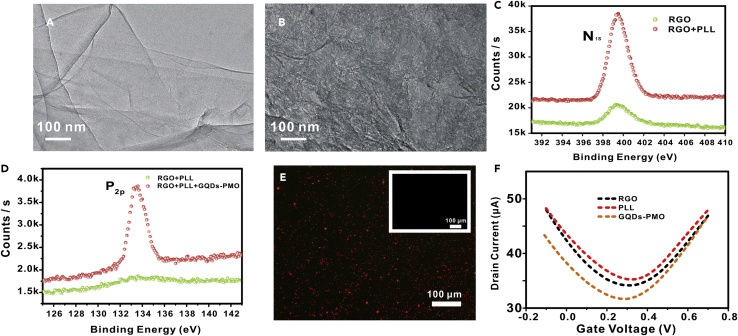
The constructed FET biosensor undergoes the modification of PLL and the immobilization of GQDs-PMO. Firstly, in order to prove that the PLL film was successfully deposited to the RGO surface, X-ray photoelectron spectroscopy (XPS), Raman spectrometer (Raman), and TEM were applied. As displayed in Figures 3A and 3B, compared with RGO, PLL-RGO hybrid reveals flat sheet with many wrinkled, folded structures. The phenomenon was consistent with that reported in the literatures (Li et al., 2018; Wang et al., 2013). As illustrated in the Figure 3C, the N1s (399.9 eV) peak had a significant increase, implying the successful assembly of PLL in the channel surface. Figure 3C shows that the small N1s of RGO may be the residue of hydrazine on the surface of graphene, because the chemical structure of hydrazine also contains N element (Wu et al., 2020). As shown in Figure S1, the Raman spectra show that the ID/IG ratio of RGO and PLL-RGO increased from 1.20 to 1.28, indicating that PLL film was modified on the RGO (Zhou et al., 2019).
Figure 3.
Fabrication and characterization of the GPPR-FET sensor
(A and B) TEM images of the RGO and RGO + PLL. Scale bar, 100 nm.
(C) X-ray photoelectron spectroscopic spectra of N1s for RGO and RGO + PLL.
(D) X-ray photoelectron spectroscopic spectra of P2p for RGO + PLL and RGO + PLL + GQDs-PMO.
(E) Fluorescence microscope images of GQDs-PMO (PMO labeled with Cy5). Inset images show the fluorescence of control. Scale bar, 100 μm.
(F) Id−Vg curves of the stepwise functionalization process of the FET chip including RGO assembly, PLL modification, and GQDs-PMO functionalization, respectively.
In order to prove that the GQDs-PMO was successfully modified on the GPPR-FET, XPS, fluorescence and AFM were conducted. As described in Figure 3D, after the modification of GQDs-PMO, P2p (134.4 eV) peak appeared; this is the contribution of GQDs-PMO hybrid, corroborating the assembly of GQDs-PMO. The GQDs-PMO (PMO labeled with Cy5) was dropped on the PLL-modified G-FET overnight at room temperature. Then, the chip was rinsed with deionized water and dried with nitrogen. The chip was observed by a fluorescence microscope. As shown in Figure 3E, a significant red fluorescent signal appeared on the chip. However, no fluorescent signal appeared in the control groups (without GQDs in the inset images), indicating the successful modification of GQDs-PMO. Atomic force microscope (AFM) was further used to characterize the PLL surface before and after deposition of GQDs-PMO. Compared with the PLL-RGO surface (Figure S2A), a large number of particles appeared after GQDs-PMO was deposited on the surface (Figure S2B). The results prove that GQDs-PMO has been successfully modified on the surface of the chip.
The change of the Id-Vg characteristic curves in Figure 3F also illustrates the PLL deposition and GQDs-PMO modification on the surface of the sensor channel. After PLL was deposited, the Dirac point shifted to the right. The reason for this phenomenon was that the RGO FET sensor is a p-type device (Zhang et al., 2016). The amino group of the PLL provides positive charges, causing the graphene to be p-doped and the Dirac point to be shifted to the right. When the GQDs-PMO was combined with the PLL-RGO chip, the contribution of the negatively charged GQDs-PMO caused the graphene to be n-doped and the Dirac point shifted to the left. Therefore, the change of Id-Vg characteristic curves shows that PLL deposition and GQDs-PMO functionalization have been successful.
Optimization
In order to obtain the best performance of GPPR-FET, we explored the optimization regarding concentration ratio of GQDs to PMO, concentrations of the PLL, hybridization time, and temperature.
The ratio of GQDs to PMO governs the density of GQDs-PMO nanocomposites assembled on the chip surface and further the hybridization efficiency. Too low concentration of PMO decreases the efficiency of hybridization, whereas too high concentration of PMO results in less remaining carboxylic groups of GQDs-PMO (Figure S3A). In this work, we changed the ratio of PMO to GQDs by adjusting the following parameters (the PMO concentration from 0.5 to 2.5 μM, and the concentration of the GQDs were 500 μg/mL, 1:1 mixed). When the PMO concentration was 1 μM, we observed maximal signal (-ΔVCNP) in the presence of the target miRNA21 (1 nM). Then, we tested the effect of different concentrations of polylysine (PLL) film on the detection and found that 0.5 mg/mL was the optimal concentration (Figure S3B). We also found that with the increase of hybridization time, the signal change (-ΔVCNP) gradually increased and reached equilibrium at 1 h (Figure S3C). At the same time, we also found that the detection signal reached its maximum value when the temperature was 37°C (Figure S3D).
Sensitivity
We examined the sensitivity of the GPPR-FET sensor by detecting the target miRNA21 at different concentrations from 100 aM to 1 nM. We dropped 10 μL of miRNA21 with different concentrations from low to high concentration onto the FET sensor surface. The incubation conditions were 37°C for 1 h. Figure 4A shows the Dirac point change (-ΔVCNP) of the GPPR-FET chip at different conditions. We could observe that the Dirac point shifted to the left as the concentration of miRNA21 increased. In detail, the average values of -ΔVCNP (11, 23, 37, 51, 65, 78, 88, and 104 mV, respectively) gradually increased. This phenomenon is due to the increase of the number of miRNA21 captured on the surface of the sensing channel of the GPPR-FET sensor. Thus, the negative charges of miRNA21 are accumulated on the surface, causing continuous n-doping of graphene. The results are in good agreement with those reported before (Cai et al., 2015; Li et al., 2021). In Figure 4B, there is a good linear relationship between the target miRNA21 concentration and -ΔVCNP, and the correlation coefficient (R2) is 0.99. The regression equation is -ΔVCNP = 13.23 lgC+222.64 (C is the concentration of miRNA21). The dotted line here represents 3-fold of the signal-to-noise ratio (10 mV). According to the regression equation and 3-fold of the signal-to-noise ratio, the detection limit was calculated to be 85 aM.-
Figure 4.
Sensitivity and specificity
(A) Transfer curves of the GPPR-FET sensor to different concentrations of miRNA21.
(B) Shift of VCNP of the GPPR-FET sensor at a series of miRNA21 concentrations. The dashed line represents the signal/noise ratio of 3. Error bars represent the standard errors (n = 3).
(C) Transfer curves of the sensor toward the random sequences (miRNA10b), one-base mismatched sequences (OM), three-base mismatched sequences (TM), and complementary sequence. The concentration of the target miRNA21 is 100 pM and that of other miRNAs are 1 nM.
(D) The VCNP values of the GPPR-FET sensor at detection of blank and four sequences.
Error bars represent the standard errors (n = 3).
We also compared the sensing performance between the GPPR-FET sensor and the PMO-functionalized FET sensor (PMO was immobilized on the sensing surface without the GQDs) (Figure S4). At the same concentration of PMO (1 μM), a range of various concentrations of target miRNA21, from 1 fM to 1 nM, were dropped to the PMO-functionalized FET sensor. Figure S4 shows the good linear relationship between miRNA21 concentrations and -ΔVCNP. The regression equation is -ΔVCNP = 6.67 lgC+110, where C is the concentration of miRNA21, and the correlation coefficient R2 is 0.991. Based on a signal that is 3-fold higher than the baseline, the LOD was calculated to be 2.04 fM, which was about 2-fold magnitude higher than that of the GPPR-FET sensor. The result shows that the GPPR-FET sensor has a higher sensitivity than the PMO-functionalized FET sensor.
In order to better show the sensitivity of the sensor, we compared detection range and LOD of the GPPR-FET sensor with those of sensors (Caputo et al., 2019; Cogal et al., 2021; Gao et al., 2020; Huang et al., 2021; Kim et al., 2021; Li et al., 2021; Wang et al., 2021; Wu et al., 2021). As shown in Table S2, it can be seen that the method established in this article is superior to other sensors in terms of detection range and LOD. We believe that the high sensitivity of GPPR-FET comes from the following aspects: (1) neutrally charged PMO probes have higher hybridization efficiency with miRNA (Zhang et al., 2010). (2) GQDs enable more PMO to be immobilized on the chip surface than the planar substrate.
Specificity
In order to test the specificity of the GPPR-FET sensor, we successively investigated the response of the GPPR-FET sensor to one-base mismatched (OM), three-base mismatched (TM), and random miRNA sequences (miRNA10b) and target miRNA21, respectively. In Figures 4C and 4D, the Dirac point shift (-ΔVCNP) generated by OM was much smaller than that obtained by miRNA21. As seen in Figures 4C and 4D, the Dirac point shift induced by TM and miRNA10b was much smaller than that obtained by miRNA21, which clearly demonstrates the specificity of our GPPR-FET sensor. In short, the GPPR-FET biosensor can effectively distinguish miRNA21 from OM, TM, and random miRNA sequences (miRNA10b), showing good specificity for detecting exosomal miRNA21.
Stability
To prove the stability of PMO-GQDs RGO FET, we dropped 1×PBS on the surface of the GPPR-FET sensor and measured Id-Vg after incubation for 2 h, 4 h, 6 h, and 12 h (Figure S5). We recorded the rate of change of its Dirac point of the Id-Vg curve after 2 h, 4 h, 6 h, and 12 h. We found that even after incubation after 12 h, the sensor’s Dirac point still maintained above 95%. Meanwhile, we put the GPPR-FET sensor in a vacuum oven and kept it for 7 days and tested the change of GPPR-FET every day. As shown in the Figure S6, the current signal did not change much at the Dirac point after 7 days, maintaining more than 88% of the original value. The results indicate that the device has good stability.
Exo-miRNA detection
In order to detect exosomal miRNA, we cultured MCF-7 and MCF-10A cancer cells and collected the exosomes of MCF-7 and MCF-10A, respectively. Then, we used the kit to extract exosomal total RNA. In order to prove the successful acquisition of exosomes, we used western blot, NTA, and TEM for characterization. Western blot of CD63, EpCAM, and TSG101 shows the successful isolation and extraction of exosomes (Figure 5A). And the TEM image shows that the exosomes had a clear phospholipid bilayer structure (Figure 5B). The NTA analysis in Figure 5C shows that the exosomes had a particle size between 50 and 150 nm, mainly around 125 nm. Then, we further extracted total RNA from exosomes. The extracted total RNA was applied to the GPPR-FET sensor. We dropped the total RNA of exosomes derived from MCF-10A onto the GPPR-FET sensor, and the mean value of -ΔVCNP was 54.6 mV, which was lower than the total RNA extracted from exosomes derived from MCF-7 (78 mV) (Figure 5D). The results show that the sensor can effectively distinguish breast cancer from a normal control group. We further used qRT-PCR to detect the expression level of miRNA21 in exosomes derived from MCF-7 and MCF-10A. And we compared the concentration of exosomal miRNA21 detected by our sensor with the result of qRT-PCR and found that our sensor could achieve a consistent result with gold standard (Figure 5D).
Figure 5.
Characterization of exosome
(A) Western blotting of CD63, EpCAM, and TSG101 in exosomes derived from MCF-7 and MCF-10A cell lines.
(B) TEM image of exosomes. Scale bar, 100 nm.
(C) Characterize the particle size distribution of exosomes by NTA. Error bars represent the standard errors (n = 3).
(D) Using qRT-PCR and GPPR-FET to detect miRNA21 in exosomes derived from MCF-7 and MCF-10A cell lines.
Error bars represent the standard errors (n = 3).
Clinical sample assays
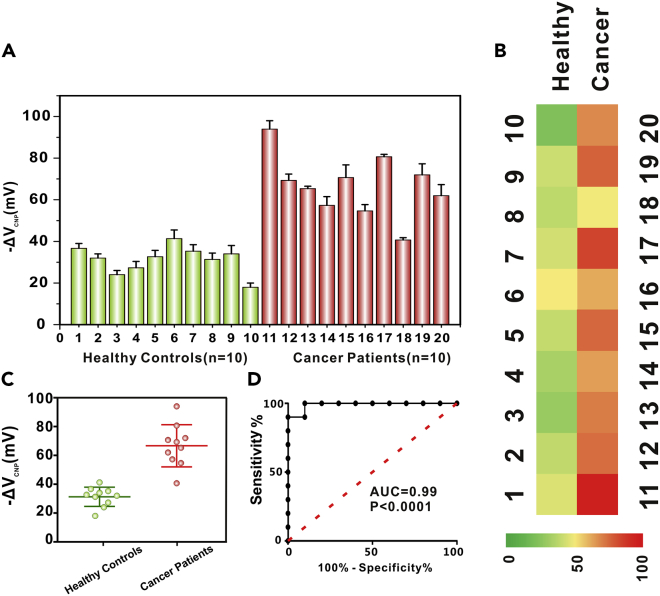
In order to verify the application potential of our GPPR-FET biosensor in clinical samples, we collected plasma samples from 10 breast cancer patients and 10 normal patients. We used the GPPR-FET sensor to detect miRNA21 from the exosomal RNA of human plasma samples. The method of extracting exosomal miRNA from plasma samples was described in the supporting information. It can be seen from Figure 6A that the signal change of breast cancer patients was higher than that of healthy people, proving that the concentration of miRNA21 in the plasma exosomes carried by breast cancer patients was higher than that of healthy people. The heatmap analysis of Figure 6B shows that the GPPR-FET sensor can distinguish normal samples from cancer samples. The scatterplot analysis in Figure 6C shows that the levels of exosomal miRNA21 in normal samples and cancer samples were different. The performance of our GPPR-FET sensor was further evaluated by ROC (receiver operating characteristic) curve analysis in Figure 6D, which further demonstrates the high accuracy (AUC = 0.99, p < 0.0001). These results reflect that our sensor can distinguish breast cancer patients from healthy people by detecting Exo-miR21 level.
Figure 6.
Clinical sample analysis
(A) Using GPPR-FET sensor to detect the level of exosomal miRNA21 in plasma of 10 normal people and 10 breast cancer patients. Error bars represent the standard errors (n = 3).
(B) Analysis of heatmap of 10 normal people and 10 breast cancer patients.
(C) Significant expression difference of Exo-miR21 levels between cancer patients and healthy controls (p < 0.0001).
(D) ROC analysis of 10 normal persons and 10 breast cancer patients detected by the GPPR-FET sensor.
AUC: area under the curve. Error bars represent the standard errors (n = 3).
Discussion
We have designed a GPPR-FET biosensor that enables ultrasensitive detection of exosomal miRNA21 in clinical samples. This GPPR-FET biosensor holds several unique merits: (1) compared with the direct probe modifications in previous reports, GQDs make more PMO probes attached to the sensing surface. (2) GQDs provide a three-dimensional structure to enable high hybridization efficiency, ensuring GPPR-FET sensor to have lower sensitivity. (3) Our sensor can distinguish the level of miRNA21 in the blood of breast cancer patients and normal people, showing clinical application potential.
Limitations of the study
This study proves that the exosomal miRNA21 level of breast cancer patients is higher than that of healthy people, but the accuracy of the approach needs to be verified in the real applications due to the interference of other substances in blood.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| graphene oxide (GO, 99.99995%) | XFNANO | CAS:7440-44-0 |
| poly-L-lysine solution | Macklin | CAS:25,988-63-0 |
| bovine serum | Sigma-Aldrich | CAS:9048-46-8 |
| 1-ethyl-3-(3-dimethyllaminopropyl) carbodiimide | Sigma-Aldrich | CAS:25,952-53-8 |
| N-hydroxysulfosuccinimide | Sigma-Aldrich | CAS:6066-82-6 |
| Software and algorithms | ||
| Origin | OriginLab Corporation | https://www.originlab.com/ |
Resource availability
Lead contact
Further information and requests for resources and materials should be directed to and will be fulfilled by the lead contact, Prof. Guo-Jun Zhang (zhanggj@hbtcm.edu.cn).
Materials availability
This study did not generate new unique reagents.
Method details
Materials and chemicals
XFNANO (Nanjing, China) provided graphene oxide (GO, 99.99995%). Concentrated H2SO4, 35 wt % H2O2 was offered by Generay Biotech Co. Ltd. (Shanghai, China). Hydrazine (98%) was provided from Aladdin Co. Ltd. (Shanghai, China). Carboxylated GQDs used in this work was ordered from XFNANO (Nanjing, China). Macklin (Shanghai, China) provided poly-L-lysine solution (PLL). Sigma-Aldrich (Shanghai, China) provided 1-ethyl-3-(3-dimethyllaminopropyl) carbodiimide (EDC, 98%), N-hydroxysulfosuccinimide (sulfo-NHS, 98.5%), bovine serum (BSA), RNaseZap and 1-Pyrenebutanoic acid succinimidyl ester (PASE). PMO and PMO-Cy5 used in this work were offered by Sangon Biotech (Shanghai, China). Takara Biotechnology (Dalian, China) provided all miRNAs. All the sequences are listed in Table S1. Antibodies were obtained by Abcam (Cambridge, USA). The exoRNeasy Serum/Plasma Midi Kit was offered by QIAGEN (Germany). All aqueous solutions were derived from ultrapure water (18.2 MΩ, Millipore Crop, Besford, MA). All surfaces were deRNase treated with RNaseZAP solution before use. Blood samples of breast cancer patients and healthy people were provided by Hubei Provincial Hospital of Traditional Chinese Medicine (Wuhan, China) and approved by the hospital Ethics Committee.
Preparation of GQDs-PMO hybrid
For the preparation of GQDs-PMO hybrid, GQDs were interacted with PMO in the presence of EDC and NHS. In general, a mixture of EDC (50 μL, 50 mg/mL) and NHS (50 μL, 12.5 mg/mL) were added to a GQDs solution (100 μL, 1 mg/mL) for 30 min. Then, the activated GQDs (200 μL, 500 μg/mL) were reacted with the PMO (200 μL, 1 μM) for 1 h, forming GQDs-PMO hybrid.
Fabrication of RGO-PLL FET sensors
The FET chip was fabricated and the RGO-FET chip was also prepared according to the procedures produced in our laboratory. To make GQDs-PMO hybrid modified on the sensing surface better, 0.5 μL of 0.5 mg/mL PLL solution was drop-casted on the chip at 80 °C for 2 h. Next, the device was thoroughly rinsed by DI water and dried with nitrogen.
Fabrication of the GQDs-PMO hybrid functionalized RGO FET sensor
10 μL of GQDs-PMO were dropped on the RGO-PLL chip. Then, the chip was stored at room temperature for overnight. Via reaction of carboxyl groups on GQDs-PMO with amino groups on PLL, GQDs-PMO was successfully assembled on the chip surface. Then, the chip was rinsed with ultrapure water, and dried with nitrogen. In order to bind additional non-specific sites, a 10 μL of 1 mg/mL BSA solution was dropped on the device for 1 h. And finally the chip was washed and dried with deionized water and nitrogen flow.
Detection of exosomal miRNA
Firstly, we used ultracentrifugation to extract exosomes. Then, the exoRNeasy serum/plasma Midi kit (Qiagen) was used to further extract exosomal total RNA. The total RNA samples were extracted from different cellular exosomes, and they were divided into two aliquots separately and measured by qRT-PCR and the established sensor, respectively. For miRNA21 detection, the total RNAs (10 μL) were dropped on the GQDs-PMO hybrid functionalized PLL-RGO-FET at 37 °C for 1 h. Then the chip was rinsed with nuclease-free water, and dried with nitrogen.
Measurement
The semiconductor characterization system (Keithley 4200-SCS) and a probe station (EverBeing BD-6) were used to conduct the electrical measurement of the chip, and record the Id-Vg curves. Throughout the experiment, the drain current was measured in 0.01×PBS as a liquid-gate and a bias of 0.1 V. Moreover, the value of -ΔVCNP was used as the signal (VCNP is the gate voltage of Dirac point).
Exosome purification
MCF-7 was cultured with DMEM medium, which contained 10% (v/v) FBS, 1% (v/v) penicillin, and 100 μg/mL streptomycin. MCF-10A was cultured with MCF-10A cell culture medium (procell), and maintained under a humidified atmosphere of 5% CO2 at 37 °C. The cells were collected after being cultured for 48 h in a serum-free medium. Subsequently, exosomes were purified according to the steps of standard overspeed centrifugation. The centrifugation method was as follows: the sample was first centrifuged at 300 g for 15 min in order to remove impurities such as cell fragments, and then the supernatant was centrifuged at 2000 g for 20 min to remove the cell debris. The filtrate was collected, filtered with a 0.22 μm filter membrane (Millipore), and centrifuged at 1000 g for 30 min. Finally, the supernatant solution was centrifuged at 110000 g for 70 min to precipitate exosomes. In order to obtain higher purity exosomes, the exosomes were re-centrifuged after re-suspension (11000 g, 70 min). Finally, the exosome pellet was re-suspended in 400 μL of PBS and stored at −80°C for use.
TEM characterization of exosomes
Firstly, 15 μL of the exosome suspension was loaded in the 400 mesh carbon-coated copper grid and fixed with 2% glutaraldehyde. The grid was stained with 2% phosphotungstic acid for 10 min and then washed twice with PBS. After drying at room temperature, the grid was observed under a transmission electron microscope (TEM) (JEM-2100, Japan) at 80 kV.
NTA analysis
Nanoparticle tracking analysis (NTA) can characterize exosomes suspended in liquids. 20 μL portion of exosomes was diluted 100-fold with PBS and then filtered with a 0.22 μm filter membrane. When the sample was irradiated by the laser in NTA, scattered light signals caused by the Brownian motion of nanoparticles were collected by an optical microscope and CMOS camera. The concentration and particle size distribution of exosomes were measured by NTA (Zeta view Ltd., Germany) under optimal parameter settings.
Western Blot
Exosomes were lysed in a buffer containing protease inhibitors, and protein quantification was performed with a BCA Protein Assay Kit (Beyotime Biotechnology Co. Ltd., Shanghai, China). Proteins were denatured and separated by gel electrophoresis, transferred to a polyvinylidene fluoride membrane (PVDF) after cooling to room temperature, and then blocked with 5% (w/v) bovine albumin in TBST for 2 h. Blots were further immune-blotted with primary antibodies against CD63 (Abcam, 1:5000 solution), EpCAM (Abcam, 1:1000 solution), TSG-101 (Abcam, 1:5000 solution) overnight at 4 °C. Then blots were incubated with shaking with HRP-conjugated secondary antibody (Abcam, 1:5000 solution) for 2 h at room temperature. Finally, GeneGnome XQR chemiluminescence imaging system (Synoptics Ltd, England) was used to visualize protein bands.
RNA extraction
The cell derived exosomes were obtained by the overspeed centrifugation. The plasma exosomes were isolated according to the recommended protocol of the exoRNeasy Serum/Plasma Midi Kit (Qiagen). Subsequently, exosomal total RNA was further extracted by the exoRNeasy Serum/Plasma Midi Kit (Qiagen). The extracted total RNA samples from different cellular exosomes were divided into two aliquots separately measured by qRT-PCR and the established GPPR-FET sensor.
qRT-PCR protocol
Total RNA was extracted from exosomes using ExoRNeasy Serum/Plasma Midi Kit (QIAGEN). About 2 μg total RNA was converted into cDNA (RT product) with the Super Script III reverse transcriptase (Invitrogen) according to the manufacturers’ instructions, yielding 20 μL of cDNA solutions. The reverse transcription program was 42°C for 60 min and 95°C for 3 min. The RT products were individually diluted 10-fold to be used as templates for the subsequent RT-PCR analysis. Amplification reaction occurred with the participation of 10 μL of 2×miRNA Premix (SYBR&ROX) and 2 μL of RT products using 0.4 μL of designed forward primer and 0.4 μL of reverse primer, in a total volume of 20 μL. PCR amplification were conducted at 95°C for 15 min, followed by 40 cycles at 94°C for 20 s and 60°C for 34 s, and melting curve was then recorded.
Quantification and statistical analysis
Origin software was used to compile and analyze data.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 21974035).
Author contributions
Kun Li: Methodology, Writing—Original Draft. Jiyuan Tu: Investigation, Sample treatment. Yulin Zhang: Investigation. Dan Jin: Investigation. Tingxian Li: Investigation. Jiahao Li: Investigation. Wei Ni: Sample Treatment. Meng-Meng Xiao: Investigation. Zhi-Yong Zhang: Supervision, Writing—Review and Editing. Guo-Jun Zhang: Supervision, Writing—Review and Editing, Project Administration, Funding Acquisition.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104522.
Contributor Information
Zhi-Yong Zhang, Email: zyzhang@pku.edu.cn.
Guo-Jun Zhang, Email: zhanggj@hbtcm.edu.cn.
Supplemental information
Data and code availability
Data reported in this paper will be shared by the lead contact upon request. This study did not generate/analyze code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Azmi A.S., Bao B., Sarkar F.H. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32:623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boriachek K., Islam M.N., Möller A., Salomon C., Nguyen N.T., Hossain M.S.A., Yamauchi Y., Shiddiky M.J.A. Biological functions and current advances in isolation and detection strategies for exosome nanovesicles. Small. 2018;14:1702153. doi: 10.1002/smll.201702153. [DOI] [PubMed] [Google Scholar]
- Cai B., Huang L., Zhang H., Sun Z., Zhang Z., Zhang G.J. Gold nanoparticles-decorated graphene field-effect transistor biosensor for femtomolar MicroRNA detection. Biosens. Bioelectron. 2015;74:329–334. doi: 10.1016/j.bios.2015.06.068. [DOI] [PubMed] [Google Scholar]
- Caputo T.M., Battista E., Netti P.A., Causa F. Supramolecular microgels with molecular beacons at the interface for ultrasensitive amplification-free, and SNP-selective miRNA fluorescence detection. ACS Appl. Mater. Interfaces. 2019;11:17147–17156. doi: 10.1021/acsami.8b22635. [DOI] [PubMed] [Google Scholar]
- Chen S., Chen X., Zhang L., Gao J., Ma Q. Electrochemiluminescence detection of Escherichia coli O157:H7 based on a novel polydopamine surface imprinted polymer biosensor. ACS Appl. Mater. Interfaces. 2017;9:5430–5436. doi: 10.1021/acsami.6b12455. [DOI] [PubMed] [Google Scholar]
- Cheng L., Sharples R.A., Scicluna B.J., Hill A.F. Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles. 2014;3:23743. doi: 10.3402/jev.v3.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celik Cogal G., Das P.K., Yurdabak Karaca G., Bhethanabotla V.R., Uygun Oksuz A. Fluorescence detection of miRNA-21 using Au/Pt bimetallic tubular micromotors driven by chemical and surface acoustic wave forces. ACS Appl. Bio. Mater. 2021;4:7932–7941. doi: 10.1021/acsabm.1c00854. [DOI] [PubMed] [Google Scholar]
- Das R., Bandyopadhyay R., Pramanik P. Carbon quantum dots from natural resource: a review. Mater. Today Chem. 2018;8:96–109. doi: 10.1016/j.mtchem.2018.03.003. [DOI] [Google Scholar]
- Endzeliņš E., Berger A., Melne V., Bajo-Santos C., Soboļevska K., Abols A., Rodriguez M., Santare D., Rudņickiha A., Rudnickiha A., et al. Detection of circulating miRNAs: comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer. 2017;17:730. doi: 10.1186/s12885-017-3737-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Gao Y., Han Y., Pang J., Wang C., Wang Y., Liu H., Zhang Y., Han L. Ultrasensitive label-free MiRNA sensing based on a flexible graphene field-effect transistor without functionalization. ACS Appl. Electron. Mater. 2020;2:1090–1098. doi: 10.1021/acsaelm.0c00095. [DOI] [Google Scholar]
- Guo Q., Yu Y., Zhang H., Cai C., Shen Q. Electrochemical sensing of exosomal MicroRNA based on hybridization chain reaction signal amplification with reduced false-positive signals. Anal. Chem. 2020;92:5302–5310. doi: 10.1021/acs.analchem.9b05849. [DOI] [PubMed] [Google Scholar]
- Hannafon B.N., Trigoso Y.D., Calloway C.L., Zhao Y.D., Lum D.H., Welm A.L., Zhao Z.J., Blick K.E., Dooley W.C., Ding W.Q. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. 2016;18:90. doi: 10.1186/s13058-016-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C.H., Huang W.T., Huang T.T., Ciou S.H., Kuo C.F., Hsieh A.H., Hsiao Y.S., Lee Y.J. Dual-gate enhancement of the sensitivity of miRNA detection of a solution-gated field-effect transistor featuring a graphene oxide/graphene layered structure. ACS Appl. Electron. Mater. 2021;3:4300–4307. doi: 10.1021/acsaelm.1c00439. [DOI] [Google Scholar]
- Jiang Q., Liu Y., Wang L., Adkins G.B., Zhong W. Rapid enrichment and detection of extracellular vesicles enabled by CuS-enclosed microgels. Anal. Chem. 2019;91:15951–15958. doi: 10.1021/acs.analchem.9b04485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Zhang H., Li Y.T., Xiao M.M., Zhang Z.L., Pang D.W., Wong G., Zhang Z.Y., Zhang G.J. A field effect transistor modified with reduced graphene oxide for immunodetection of Ebola virus. Mikrochim. Acta. 2019;186:223. doi: 10.1007/s00604-019-3256-5. [DOI] [PubMed] [Google Scholar]
- Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T., Zhu J., Luo X., Jia W., Wu P., Cai C. Controlled self-assembly of a close-packed gold octahedra array for SERS sensing exosomal MicroRNAs. Anal. Chem. 2021;93:2519–2526. doi: 10.1021/acs.analchem.0c04561. [DOI] [PubMed] [Google Scholar]
- Kermani H.A., Hosseini M., Dadmehr M., Hosseinkhani S., Ganjali M.R. DNA methyltransferase activity detection based on graphene quantum dots using fluorescence and fluorescence anisotropy. Sens. Actuat. B Chem. 2017;241:217–223. doi: 10.1016/j.snb.2016.10.078. [DOI] [Google Scholar]
- Kim S., Park S., Cho Y.S., Kim Y., Tae J.H., No T.I., Shim J.S., Jeong Y., Kang S.H., Lee K.H. Electrical cartridge sensor enables reliable and direct identification of MicroRNAs in urine of patients. ACS Sens. 2021;6:833–841. doi: 10.1021/acssensors.0c01870. [DOI] [PubMed] [Google Scholar]
- Lee J.H., Choi J.H., Chueng S.T.D., Pongkulapa T., Yang L., Cho H.Y., Choi J.W., Lee K.B. Nondestructive characterization of stem cell neurogenesis by a magneto-plasmonic nanomaterial-based exosomal miRNA detection. ACS Nano. 2019;13:8793–8803. doi: 10.1021/acsnano.9b01875. [DOI] [PubMed] [Google Scholar]
- Lei Y.M., Xiao M.M., Li Y.T., Xu L., Zhang H., Zhang Z.Y., Zhang G.J. Detection of heart failure-related biomarker in whole blood with graphene field effect transistor biosensor. Biosens. Bioelectron. 2017;91:1–7. doi: 10.1016/j.bios.2016.12.018. [DOI] [PubMed] [Google Scholar]
- Li J., Wu D., Yu Y., Li T., Li K., Xiao M.M., Li Y., Zhang Z.Y., Zhang G.J. Rapid and unamplified identification of COVID-19 with morpholino-modified graphene field-effect transistor nanosensor. Biosens. Bioelectron. 2021;183:113206. doi: 10.1016/j.bios.2021.113206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li C., Wang S., Wang Z., Jiang J., Wang W., Li X., Chen J., Liu K., Li C., Zhu G. Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR-21 to normoxic cells to elicit a prometastatic phenotype. Cancer Res. 2016;76:1770–1780. doi: 10.1158/0008-5472.can-15-1625. [DOI] [PubMed] [Google Scholar]
- Li M., Chen T., Gooding J.J., Liu J. Review of carbon and graphene quantum dots for sensing. ACS Sens. 2019;4:1732–1748. doi: 10.1021/acssensors.9b00514. [DOI] [PubMed] [Google Scholar]
- Li X., Zhang Y., Ma Z., Fu M., An Q. The fabrication of rGO/(PLL/PASP)3 @DOX nanorods with pH-switch for photothermal therapy and chemotherapy. Chem. Eur J. 2018;24:13830–13838. doi: 10.1002/chem.201801884. [DOI] [PubMed] [Google Scholar]
- Li Y.T., Jin X., Tang L., Lv W.L., Xiao M.M., Zhang Z.Y., Gao C., Zhang G.J. Receptor-mediated field effect transistor biosensor for real-time monitoring of glutamate release from primary hippocampal neurons. Anal. Chem. 2019;91:8229–8236. doi: 10.1021/acs.analchem.9b00832. [DOI] [PubMed] [Google Scholar]
- Liang Y., Xiao M., Wu D., Lin Y., Liu L., He J., Zhang G.J., Peng L.M., Zhang Z. Wafer-scale uniform carbon nanotube transistors for ultrasensitive and label-free detection of disease biomarkers. ACS Nano. 2020;14:8866–8874. doi: 10.1021/acsnano.0c03523. [DOI] [PubMed] [Google Scholar]
- Liao T.B., Li X.R., Tong Q., Zou K., Zhang H., Tang L.N., Sun Z.Y., Zhang G.J. Ultrasensitive detection of MicroRNAs with morpholino-functionalized nanochannel biosensor. Anal. Chem. 2017;89:5511–5518. doi: 10.1021/acs.analchem.7b00487. [DOI] [PubMed] [Google Scholar]
- Lu L., Zhou L., Chen J., Yan F., Liu J., Dong X., Xi F., Chen P. Nanochannel-confined graphene quantum dots for ultrasensitive electrochemical analysis of complex samples. ACS Nano. 2018;12:12673–12681. doi: 10.1021/acsnano.8b07564. [DOI] [PubMed] [Google Scholar]
- Mansouri Majd S., Ghasemi F., Salimi A., Sham T.K. Transport properties of a molybdenum disulfide and carbon dot nanohybrid transistor and its applications as a Hg2+ aptasensor. ACS Appl. Electron Mater. 2020;2:635–645. doi: 10.1021/acsaelm.9b00632. [DOI] [Google Scholar]
- Mao S., Chang J., Pu H., Lu G., He Q., Zhang H., Chen J. Two-dimensional nanomaterial-based field-effect transistors for chemical and biological sensing. Chem. Soc. Rev. 2017;46:6872–6904. doi: 10.1039/c6cs00827e. [DOI] [PubMed] [Google Scholar]
- Miao P., Tang Y. Dumbbell hybridization chain reaction based electrochemical biosensor for ultrasensitive detection of exosomal miRNA. Anal. Chem. 2020;92:12026–12032. doi: 10.1021/acs.analchem.0c02654. [DOI] [PubMed] [Google Scholar]
- Ramadan S., Lobo R., Zhang Y.Z., Xu L.Z., Shaforost O., Kwong Hong Tsang D., Tsang D.K.H., Feng J.Y., Yin T.Y., Qiao M., et al. Carbon-dot-enhanced graphene field-effect transistors for ultrasensitive detection of exosomes. ACS Appl. Mater. Interfaces. 2021;13:7854–7864. doi: 10.1021/acsami.0c18293. [DOI] [PubMed] [Google Scholar]
- Ren J., He W., Zheng L., Duan H. From structures to functions: insights into exosomes as promising drug delivery vehicles. Biomater Sci. 2016;4:910–921. doi: 10.1039/c5bm00583c. [DOI] [PubMed] [Google Scholar]
- Shi Y., Wang Z., Zhu X., Chen L., Ma Y., Wang J., Yang X., Liu Z. Exosomal miR-1246 in serum as a potential biomarker for early diagnosis of gastric cancer. Int. J. Clin. Oncol. 2020;25:89–99. doi: 10.1007/s10147-019-01532-9. [DOI] [PubMed] [Google Scholar]
- Sun Y.P., Zhou B., Lin Y., Wang W., Fernando K.A.S., Pathak P., Meziani M.J., Harruff B.A., Wang X., Wang H., et al. Quantum-sized carbon dots for bright and colorful photoluminescence. J. Am. Chem. Soc. 2006;128:7756–7757. doi: 10.1021/ja062677d. [DOI] [PubMed] [Google Scholar]
- Syedmoradi L., Ahmadi A., Norton M.L., Omidfar K. A review on nanomaterial-based field effect transistor technology for biomarker detection. Mikrochim. Acta. 2019;186:739. doi: 10.1007/s00604-019-3850-6. [DOI] [PubMed] [Google Scholar]
- Thakur B., Zhou G., Chang J., Pu H., Jin B., Sui X., Yuan X., Yang C.H., Magruder M., Chen J. Rapid detection of single E. coli bacteria using a graphene-based field-effect transistor device. Biosens. Bioelectron. 2018;110:16–22. doi: 10.1016/j.bios.2018.03.014. [DOI] [PubMed] [Google Scholar]
- Tomasetti M., Lee W., Santarelli L., Neuzil J. Exosome-derived microRNAs in cancer metabolism: possible implications in cancer diagnostics and therapy. Exp. Mol. Med. 2017;49:e285. doi: 10.1038/emm.2016.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Wang C., Jin D., Yu Y., Yang F., Zhang Y., Yao Q., Zhang G.J. AuNP-amplified surface acoustic wave sensor for the quantification of exosomes. ACS Sens. 2020;5:362–369. doi: 10.1021/acssensors.9b01869. [DOI] [PubMed] [Google Scholar]
- Wang J., Zhao Y., Ma F.X., Wang K., Wang F.B., Xia X.H. Synthesis of a hydrophilic poly-l-lysine/graphene hybrid through multiple non-covalent interactions for biosensors. J. Mater. Chem. B. 2013;1:1406. doi: 10.1039/c2tb00454b. [DOI] [PubMed] [Google Scholar]
- Wang Q., Liu Y.Q., Yan J.X., Liu Y.Q., Gao C.M., Ge S.G., Yu J.H. 3D DNA walker-assisted CRISPR/Cas12a trans-cleavage for ultrasensitive electrochemiluminescence detection of miRNA-141. Anal. Chem. 2021;93:13373–13381. doi: 10.1021/acs.analchem.1c03183. [DOI] [PubMed] [Google Scholar]
- Wang S., Zhang L., Wan S., Cansiz S., Cui C., Liu Y., Cai R., Hong C., Teng I.T., Shi M., et al. Aptasensor with expanded nucleotide using DNA nanotetrahedra for electrochemical detection of cancerous exosomes. ACS Nano. 2017;11:3943–3949. doi: 10.1021/acsnano.7b00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q.Y., Zhang P.Y., Liu T., Pu H.B., Sun D.W. A fluorescence biosensor based on single-stranded DNA and carbon quantum dots for acrylamide detection. Food Chem. 2021;356:129668. doi: 10.1016/j.foodchem.2021.129668. [DOI] [PubMed] [Google Scholar]
- Wu C.J., Huang S.Q., Wang Y.Y., Chai Y.Q., Yuan R., Yang X. DNA structure-stabilized liquid-liquid self-assembled ordered Au nanoparticle interface for sensitive detection of MiRNA 155. Anal. Chem. 2021;93:11019–11024. doi: 10.1021/acs.analchem.1c02336. [DOI] [PubMed] [Google Scholar]
- Wu D., Yu Y., Jin D., Xiao M.M., Zhang Z.Y., Zhang G.J. Dual-aptamer modified graphene field-effect transistor nanosensor for label-free and specific detection of hepatocellular carcinoma-derived microvesicles. Anal. Chem. 2020;92:4006–4015. doi: 10.1021/acs.analchem.9b05531. [DOI] [PubMed] [Google Scholar]
- Xia Y., Liu M., Wang L., Yan A., He W., Chen M., Lan J., Xu J., Guan L., Chen J. A visible and colorimetric aptasensor based on DNA-capped single-walled carbon nanotubes for detection of exosomes. Biosens. Bioelectron. 2017;92:8–15. doi: 10.1016/j.bios.2017.01.063. [DOI] [PubMed] [Google Scholar]
- Xiao P.P., Wan Q.Q., Liao T.B., Tu J.Y., Sun Z.Y., Zhang G.J. Peptide nucleic acid-functionalized nanochannel biosensor for the highly sensitive detection of tumor exosomal MicroRNA. Anal. Chem. 2021;93:10966–10973. doi: 10.1021/acs.analchem.1c01898. [DOI] [PubMed] [Google Scholar]
- Xie H., Li Y.T., Lei Y.M., Liu Y.L., Xiao M.M., Gao C., Pang D.W., Huang W.H., Zhang Z.Y., Zhang G.J. Real-time monitoring of nitric oxide at single-cell level with porphyrin-functionalized graphene field-effect transistor biosensor. Anal. Chem. 2016;88:11115–11122. doi: 10.1021/acs.analchem.6b03208. [DOI] [PubMed] [Google Scholar]
- Xu R., Rai A., Chen M., Suwakulsiri W., Greening D.W., Simpson R.J. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018;15:617–638. doi: 10.1038/s41571-018-0036-9. [DOI] [PubMed] [Google Scholar]
- Xu X.Y., Ray R., Gu Y.L., Ploehn H.J., Gearheart L., Raker K., Scrivens W.A. Electrophoretic analysis and purification of fluorescent single-walled carbon nanotube fragments. J. Am. Chem. Soc. 2004;126:12736–12737. doi: 10.1021/ja040082h. [DOI] [PubMed] [Google Scholar]
- Xu Y., Liu J., Gao C., Wang E. Applications of carbon quantum dots in electrochemiluminescence: a mini review. Electrochem. Commun. 2014;48:151–154. doi: 10.1016/j.elecom.2014.08.032. [DOI] [Google Scholar]
- Yu Y., Li Y.T., Jin D., Yang F., Wu D., Xiao M.M., Zhang H., Zhang Z.Y., Zhang G.J. Electrical and label-free quantification of exosomes with a reduced graphene oxide field effect transistor biosensor. Anal. Chem. 2019;91:10679–10686. doi: 10.1021/acs.analchem.9b01950. [DOI] [PubMed] [Google Scholar]
- Zeng Z., Li Y., Pan Y., Lan X., Song F., Sun J., Zhou K., Liu X., Ren X., Wang F., et al. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat. Commun. 2018;9:5395. doi: 10.1038/s41467-018-07810-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Xu J.Q., Li Y.T., Huang L., Pang D.W., Ning Y., Huang W.H., Zhang Z., Zhang G.J. Photocatalysis-induced renewable field-effect transistor for protein detection. Anal. Chem. 2016;88:4048–4054. doi: 10.1021/acs.analchem.6b00374. [DOI] [PubMed] [Google Scholar]
- Zhang G.J., Luo Z.H., Huang M.J., Tay G.K.I., Tay G.K., Lim E.J.A., Lim E.J. Morpholino-functionalized silicon nanowire biosensor for sequence-specific label-free detection of DNA. Biosens. Bioelectron. 2010;25:2447–2453. doi: 10.1016/j.bios.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Zhou L., Lv T., Zhang Q., Zhu Q., Zhan P., Zhu S., Zhang J., Song Y. The biology, function and clinical implications of exosomes in lung cancer. Cancer Lett. 2017;407:84–92. doi: 10.1016/j.canlet.2017.08.003. [DOI] [PubMed] [Google Scholar]
- Zhou X., Huang H., Zhu R., Sheng X., Xie D., Mei Y. Facile modification of graphene oxide with Lysine for improving anti-corrosion performances of water-borne epoxy coatings. Prog. Org. Coat. 2019;136:105200. doi: 10.1016/j.porgcoat.2019.06.046. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data reported in this paper will be shared by the lead contact upon request. This study did not generate/analyze code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.