Figure 3.
Evaluation of CYP3A4 by reporter gene assay in organoid-derived IECs
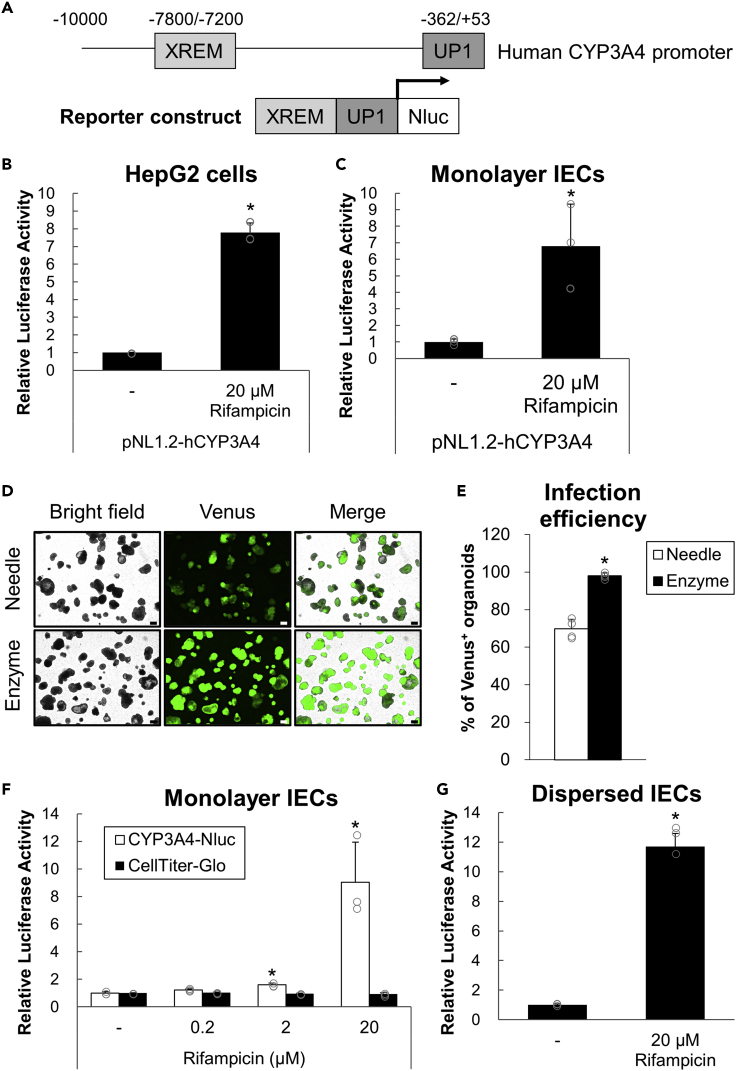
(A) Part of a reporter gene construct (pNL1.2-hCYP3A4) with human PXR promoter combined with two regions (XREM and UP1) containing functional PXR response elements.
(B and C) HepG2 cells (B) or hiPSO-derived monolayer IECs (C) were transiently transfected with pNL1.2-hCYP3A4 with pSV40-β-Gal. After 24 h, cells were treated with 0 or 20 μM rifampicin for 24 h. Luciferase activities were measured and normalized to β-galactosidase activities. Assays were performed in n = 3 independent biological replicates (mean ± S.D.). Statistical significance was determined by Student’s t test. ∗p < 0.05. (D, E) After being disrupted by physical breaking with a 26-gauge needle or dispersed by enzymatic digestion with TrypLE Express solution followed by vigorous pipetting, hiPSO-derived intestinal organoids were seeded on collagen I-coated plates. Cells were cultured for four days (physical breaking, “Needle”) or one day (enzymatic digestion, “Enzyme”), infected with undiluted CSII-EF-MCS-IRES2-Venus lentiviruses, and embedded in Matrigel to regenerate organoids.
(D) bright-field or fluorescent images of organoids after 9 days of infection were taken. Scale bar, 200 μm.
(E) The proportions of Venus-positive organoids per microscopic bright field after 9 days of infection were calculated. Assays were performed in n = 4 independent images (mean ± S.D.). Statistical significance was determined by Student’s t test. ∗p < 0.05 (versus Needle).
(F and G) Monolayer (F) or dispersed (G) IECs developed from hiPSOs stably expressing human CYP3A4-Nluc were treated with 0-20 μM rifampicin for 24 h, and luciferase activities were determined. Assays were performed in n = 3 independent biological replicates (mean ± S.D.). Statistical significance was determined by one-way analysis of variance with the Bonferroni test (F) or by Student’s t test (G). ∗p < 0.05 (versus no treatment group).
See also Figure S4.