Abstract
Fourteen weeks after field release of luciferase gene-tagged Sinorhizobium meliloti L33 in field plots seeded with Medicago sativa, we found that the inoculant also occurred in bulk soil from noninoculated control plots. In rhizospheres of M. sativa plants, S. meliloti L33 could be detected in noninoculated plots 12 weeks after inoculation, indicating that growth in the rhizosphere preceded spread into bulk soil. To determine whether inoculation affected bacterial diversity, 1,119 bacteria were isolated from the rhizospheres of M. sativa and Chenopodium album, which was the dominant weed in the field plots. Amplified ribosomal DNA restriction analysis (ARDRA) revealed plant-specific fragment size frequencies. Dominant ARDRA groups were identified by 16S rRNA gene nucleotide sequencing. Database comparisons indicated that the rhizospheres contained members of the Proteobacteria (α, β, and γ subgroups), members of the Cytophaga-Flavobacterium group, and gram-positive bacteria with high G+C DNA contents. The levels of many groups were affected by the plant species and, in the case of M. sativa, by inoculation. The most abundant isolates were related to Variovorax sp., Arthrobacter ramosus, and Acinetobacter calcoaceticus. In the rhizosphere of M. sativa, inoculation reduced the numbers of cells of A. calcoaceticus and members of the genus Pseudomonas and increased the number of rhizobia. Cultivation-independent PCR–single-strand conformation polymorphism (SSCP) profiles of a 16S rRNA gene region confirmed the existence of plant-specific rhizosphere communities and the effect of the inoculant. All dominant ARDRA groups except Variovorax species could be detected. On the other hand, the SSCP profiles revealed products which could not be assigned to the dominant cultured isolates, indicating that the bacterial diversity was greater than the diversity suggested by cultivation.
Legume-nodulating, nitrogen-fixing soil bacteria belonging to the Rhizobium group, including members of the genera Rhizobium, Sinorhizobium, and Bradyrhizobium and other genera, have been used on a broad scale as inoculants to improve the nitrogen status of soils during crop rotation (33, 44). Recent developments based on our improving knowledge about plant-microorganism interactions and the availability of genetic engineering methods have yielded inoculants with potentially improved properties (9, 58). Before commercialization of such genetically modified organisms (GMO), their environmental safety should be demonstrated.
The performance and ecological effects of GMO can be evaluated in laboratory, greenhouse, or field release studies. Small-scale field releases have been found to be especially useful for studying GMO, since the complexity of environmental abiotic and biotic parameters cannot be simulated in contained systems without enormous expenditure (17, 55). In a number of recent field releases, the use of recombinant marker genes, which specifically tag bacterial cells, has allowed workers to specifically monitor the survival and dissemination of field-released GMO and to correlate ecological effects with the presence of these organisms (11, 12, 26, 27, 52, 62, 66, 67).
Overlapping nutritional requirements of inoculated bacteria and indigenous microorganisms might result in competition for niche colonization (2, 15, 23, 50). As a consequence, the survival of inoculated bacteria may be reduced or, on the other hand, indigenous microorganisms might be outcompeted (38, 48, 59). Such outcompetition can be regarded as an ecological risk if it affects microbiologically mediated functions (e.g., pathogen protection or nutrient cycling) (6, 7, 28, 29). Thus, the effect of inoculated bacteria on the resident microbial community in soil or rhizospheres could indicate both the potential of persistence and the risks associated with release of the organisms.
Determining the biodiversity of a microbial community is still a problem. Classical cultivation methods always favor the growth of some community members and do not detect other community members. Also, these methods are time-consuming and labor-intensive, which limits the number of samples which can be analyzed (30, 54). Recently developed genetic profiling techniques which are independent of cultivation have great potential for use in community analyses in the future. These techniques utilize DNA or RNA directly extracted from environmental samples and amplification of signature genes by PCR or reverse transcription-PCR with primers which bind to conserved gene regions and produce homologous gene fragments. Products can subsequently be analyzed to determine their nucleotide differences by electrophoretic techniques, such as denaturing gradient gel electrophoresis, terminal restriction fragment length polymorphism, or single-strand conformation polymorphism (SSCP) (31, 32, 39, 56). Due to their importance for phylogenetic classification (69) and the sequences available in databases (36), small-subunit RNAs or their encoding genes are the main target molecules used for community analyses (34, 43). Even though genetic profiling eliminates the bias of cultivation, it potentially has other biases which should be better characterized or eliminated, if possible, before this approach can replace cultivation-dependent techniques in risk assessment studies (65). The biases are related to the quality of the nucleic acids, primer selection, gene copy number, and the PCR process itself (21, 51). One approach to better understand the quality of genetic profiles is to compare the results obtained with this method with the results obtained by classical cultivation techniques (19).
Here, we report on results of a field release experiment performed with a chromosomally luciferase (luc) marker gene-tagged, Medicago sativa (alfalfa)-nodulating strain, Sinorhizobium meliloti L33 (10). At the beginning of the growing season, in April, replicate field plots that were seeded with M. sativa were inoculated with S. meliloti L33. Noninoculated plots that were also seeded with M. sativa were located between the inoculated plots. During the growing season, the flora in the plots comprised M. sativa and weeds, and Chenopodium album was the dominant weed. This close proximity allowed us to study the impact of S. meliloti L33 on the microbial communities in the rhizospheres of a target plant (M. sativa) and a non-target plant (C. album).
MATERIALS AND METHODS
Microorganisms and cultivation.
S. meliloti L33 (Smr) carrying a chromosomally inserted luciferase marker gene (10) was obtained from A. Pühler (Bielefeld, Germany). Other bacterial strains used as markers for SSCP genetic profiles were purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany). Unless indicated otherwise, all strains were cultivated at 28°C on Luria-Bertani agar, which contained (per liter) 10 g of tryptone (Difco Laboratories, Detroit, Mich.), 10 g of yeast extract (Difco), 5.0 g of NaCl, 1.0 g of glucose, and 15 g of agar (Oxoid, Unipath Ltd., Basingstoke, Hampshire, England) (pH 7.0).
S. meliloti L33 was cultivated on nutrient-poor agar (NPA) (8) amended with streptomycin (500 mg liter−1; Sigma Aldrich Chemie GmbH, Deisenhofen, Germany) and cycloheximide (150 mg liter−1; Sigma Aldrich Chemie GmbH).
Rhizosphere bacteria were isolated and subcultured on agar (1.0 g liter−1; concentration of agar [Oxoid], 15 g liter−1) containing Winogradski's mineral salts without inorganic nitrogen (49) and four amino acids, l-histidine, l-leucine, l-ornithine, and l-proline (all obtained from Sigma Aldrich Chemie GmbH). These four amino acids could all be utilized as sole carbon sources by S. meliloti L33, as indicated by characterization on BiologGN (Biolog Inc., Hayworth, Calif.). The final concentration of each amino acid in rhizophere amino acid agar (RAA) was 2.5 mM; the pH was adjusted to 7.0. Inoculated media were incubated at 28°C.
Field site and inoculation.
The field site (para brown earth, silty sand, even, no slope), which was located in Braunschweig, Germany, at our research center, consisted of 20 field plots, each 3 by 3 m, which were arranged in four rows of five plots with 3 m between plots. At this site, leguminous crops had not been cultivated for the previous 8 years and the titer of indigenous S. meliloti was low (37). Treatments were conducted in block randomized order (64). Five replicate plots were inoculated with S. meliloti L33, and five plots were not inoculated. Other treatments included inoculation with S. meliloti wild type and inoculation with S. meliloti L1 (RecA−).
The other treatments mentioned above were not relevant for this study. Inoculation with S. meliloti L33 was conducted in April 1995 by using cells grown to the late logarithmic phase. One day before inoculation each plot was sown by drilling M. sativa seeds according to agricultural practice. Bacterial cells, diluted in tap water, were sprayed onto field plots by using a device developed for quantitative, accurate pesticide application (Schachtner, Fahrzeug- und Gerätetechnik, Ludwigsburg, Germany) in order to obtain a titer of 106 S. meliloti L33 cells per g of soil in the Ap horizon (plough layer; depth, 0 to 25 cm).
Sampling, extraction, and isolation of soil and rhizosphere bacteria.
Soil samples were obtained from three replicate plots. Samples were obtained from each plot by inserting an auger five times to a depth of 25 cm randomly across the plot. The replicate soil samples from each plot were combined and sieved (mesh size, 2 mm). Soil samples (5 g each) were suspended in 10 ml of a sodium polyphosphate solution (0.1%, wt/vol) at 4°C with agitation at 45 rpm for 0.5 h by using an overhead shaker (KH, Guwina Hoffmann, Berlin, Germany). Samples were then immediately diluted in saline (0.85% [wt/vol] NaCl) and inoculated in triplicate onto the appropriate media.
Rhizosphere bacteria were isolated from root material of M. sativa and C. album plants which were carefully dug out from the field plots. All loosely adhering soil was removed, and the whole plants with roots were placed into separate plastic bags for transport to the laboratory. Roots were then cut from the plants and carefully dipped into sterile saline for 20 s in order to remove attached sand particles. A total of 10 g (wet weight) of root material from six to eight plants was then suspended in 40-ml portions of saline in 50-ml polypropylene tubes (Falcon Tubes; Becton Dickinson, N.J.). The rhizosphere bacteria were detached from the root material by shaking the material for 0.5 h at 45 rpm and 4°C with an overhead shaker (KH, Guwina Hoffmann). The resulting suspensions and 10-fold dilutions were inoculated in triplicate onto NPA and RAA and incubated as described above.
Colonies growing on RAA were counted after 3 days of incubation. Plates with less than 100 colonies were used to isolate pure bacterial cultures. This was done by transferring single colonies onto new RAA with sterile toothpicks and then incubating the preparations at 28°C for 2 days. Subculturing of isolates was repeated once before colonies were utilized for DNA extraction.
Monitoring of S. meliloti L33.
Colonies growing on NPA after 7 days of incubation at 28°C were transferred onto nylon membranes (Hybond-N; Amersham Pharmacia Biotech, Freiburg, Germany). The membranes were soaked with a luciferin solution (1 mM luciferin [Sigma Aldrich Chemie GmbH] in 100 mM sodium citrate, pH 5.0), and light emission was detected after overnight exposure by using Kodak T-MAT DG film (Integra Bioscience, Fernwald, Germany) (57).
DNA extraction.
Community DNA was obtained from the root washing suspensions which were used as inocula for cultivation of rhizosphere bacteria (see above). DNA was extracted as described previously (56), with the following modifications. DNA was recovered from agarose gels after electrophoresis and was purified with Elutip-d (Schleicher und Schüll, Dassel, Germany) by using the protocol recommended by the manufacturer. The eluted DNA was concentrated by ethanol precipitation and was resuspended in 10 mM Tris-HCl (pH 8.0) (53).
Crude DNA was extracted from pure cultures by using a boiling-lysis procedure. Cells from single colonies grown on agar plates were transferred with a toothpick to reaction tubes (Safe Lock; Eppendorf, Hamburg, Germany) filled with 50 μl of a 0.05 M NaOH–0.25% (wt/vol) sodium dodecyl sulfate solution. The tubes were heated at 95°C for 15 min, and then 450 μl of double-distilled water was added. Samples were mixed, centrifuged for 1 min at 8,000 × g to remove the cell debris, and then immediately frozen and stored at −20°C until analysis.
PCR.
Amplified ribosomal DNA restriction analysis (ARDRA) was used to differentiate cultivated isolates. Universal eubacterial primers 41f (Escherichia coli positions 22 to 41) and 1066r (E. coli positions 1066 to 1085) were used for PCR amplification. The reaction mixtures (total volume, 50 μl) contained 1× PCR buffer, 1.5 mM MgCl2 (Roche Diagnostics GmbH, Mannheim, Germany), 200 μM dATP, 200 μM dCTP, 200 μM dGTP, 200 μM dTTP, each primer at a concentration of 0.5 μM, and 1 U of Taq polymerase (Roche Diagnostics GmbH). The reaction mixtures were each overlaid with 1 drop of mineral oil, and 2 μl of crude bacterial lysate was pipetted through this layer as template DNA. Amplification reactions were performed with an OmniGene thermocycler (Hybaid, Teddington, United Kingdom) by using 96-well microtiter plates (Thermowell plates; Costar, Corington, N.Y.). An initial denaturation step consisting of 3 min at 94°C was followed by 35 cycles consisting of 25 s at 94°C, 40 s at 50°C, and 40 s at 72°C. For final primer extension the temperature was kept at 72°C for 5 min and finally decreased to 30°C.
For SSCP analysis, 16S rRNA gene sequences were amplified by using primer Com1 (5′ CAG CAG CCG CGG TAA TAC 3′), which was not phosphorylated at the 5′ end, and primer Com2-Ph (5′ CCG TCA ATT CCT TTG AGT TT 3′), which was phosphorylated at the 5′ end (56). These primers were used to amplify 16S rRNA genes from nucleotide 519 to nucleotide 926 (E. coli numbering), including two regions (regions V4 and V5) (42). Each PCR was performed by using a total volume of 100 μl in a micro test tube (Flat Cap Micro Tube; MWG-Biotech AG, Ebersberg, Germany). Each reaction mixture contained 1× PCR buffer, 2 mM MgCl2, 200 μM dATP, 200 μM dCTP, 200 μM dGTP, 200 μM dTTP, and primers Com1 and Com2-Ph (0.5 μM each). Each mixtures was overlaid with 40 μl of mineral oil. Approximately 1 to 2 ng of DNA obtained from bacterial consortia extracted from the rhizospheres or 4 μl of crude bacterial lysate was added to each PCR mixture. Thermocycling was conducted with a Primus 96 apparatus (MWG-Biotech) and was started with an initial denaturation step consisting of 5 min at 98°C. Samples were then kept at 80°C for 6 min, and 4 U of Taq DNA polymerase (Amersham Pharmacia Biotech) was added to each reaction mixture. A total of 35 cycles consisting of 60 s at 94°C, 60 s at 50°C, and 70 s at 72°C was followed by a final primer extension step consisting of 5 min at 72°C.
For detection of the luciferase gene, the PCR conditions used for SSCP analysis described above were modified as follows: the reaction volume and the amounts of reagents were each reduced by one-half, and primers lucP2 and lucP3 (10) were used. Taq polymerase was added directly to the master mixture, and 2 μl of crude bacterial lysate was used as the template.
The amplification products were analyzed by using 10 μl of the reaction mixture after agarose gel electrophoresis (1.3% [wt/vol] agarose gel containing 0.5 μg of ethidium bromide ml−1) (53).
Analyses of PCR products with restriction endonucleases.
For digestion of the amplified 16S ribosomal DNA fragments, the tetranucleotide-recognizing enzymes HhaI and HaeIII (New England Biolabs, Schwalbach/Taunus, Germany) were used. In a typical reaction, 5 μl of a PCR product was digested with 5 U of an enzyme in reaction buffers, as recommended by the manufacturer. The total reaction volume was 20 μl, and reaction mixtures were incubated at 37°C for 2 h. For fragment analysis, 6-μl portions of the digests were analyzed by gel electrophoresis.
The restriction fragments obtained from HhaI digests were separated on 5% (wt/vol) polyacrylamide (PAA) gels containing 7 M urea and 1× TBE (53). To separate the DNA fragments obtained after HaeIII digestion, nondenaturing PAA gels (10% T, 2% C) and 1× TAE (53) buffer was used. The buffer strips for the denaturing PAA gels contained 2× TBE, and the buffer strips for nondenaturing PAA gels contained 0.2 M Tris–Tricine [N-tris(hydroxymethyl)methylglycine; pH 8.2; Sigma Chemical Co., St. Louis, Mo.] buffer. The gels (thickness, 0.5 mm) were cast vertically on a carrier film (GelBond-PAG film; FMC Bioproducts, Rockland, Maine). Gel electrophoresis was performed horizontally by using a Multiphor II apparatus (Amersham Pharmacia Biotech) at 20°C, 400 V, and 50 mA. The gels were silver stained (3) and recorded in a digital format by using a ScanJet 4c printer (Hewlett-Packard, Böblingen, Germany) with a transparency adapter.
Preparation of single-stranded DNA and SSCP analyses.
PCR products were purified with Qiaquick columns by using a protocol recommended by the manufacturer (Qiagen, Hilden, Germany). Samples were eluted with 30 μl of Tris-HCl (pH 8.0). For digestion of the phosphorylated strand, 40 U of lambda exonuclease (New England Biolabs) was mixed with 28 μl of the eluted PCR product in an 80-μl (total volume) mixture containing 1× (final concentration) lambda exonuclease reaction buffer (New England Biolabs). The reaction mixtures were incubated at 37°C for 2 h, purified with Qiaquick columns (Qiagen), and eluted as previously described. For electrophoresis, 2.5-μl portions of denaturing loading buffer (95% [vol/vol] formamide, 10 mM NaOH, 0.25% [wt/vol] bromophenol blue, 0.25% xylene cyanol) were added to 7-μl portions of the single-stranded DNA solutions. Samples were incubated at 95°C for 2 min and immediately cooled on ice. After 5 min, samples were loaded onto the gel.
The samples were separated by electrophoresis in a 0.625× MDE gel (FMC Bioproducts) with 1× TBE buffer. The gels (length, 21 cm) were electrophoresed at 400 V and 10 mA for 18 h at 20°C with a Macrophor sequencing apparatus (Amersham Pharmacia Biotech). The gels were cast horizontally by using 0.4-mm spacers and a thermostatic plate as recommended by the manufacturer. Gels, which were fixed to the front glass plate by using Bind Silane (Amersham Pharmacia Biotech), were silver stained (3) and dried at room temperature. Interpretation of SSCP-based results in this study was based on evaluation of at least three independent replicate gel runs.
Analysis of ARDRA results.
The ARDRA patterns obtained from bacterial isolates with restriction endonuclease HhaI were analyzed by using WinCam 2.1 (Cybertech, Berlin, Germany). Each lane, which contained the fragment pattern of a single isolate, was calibrated with size standards and imported into a database. Pearson correlation, which incorporated both fragment size and amount, was used for a similarity analysis of the patterns. The resulting similarity matrix, which included all isolates, was converted into a dendrogram by using the unweighted pair group method with arithmetic averages. This procedure allowed us to identify ARDRA groups consisting of isolates that exhibited high levels of pattern similarity.
Sequencing of PCR products and sequence alignments.
A Thermo Sequenase kit (Amersham Pharmacia Biotech) was used for sequencing of PCR-amplified 16S rRNA genes. The primers used for the sequencing reaction were labeled with IR-800 (MWG Biotech AG). The reaction products were separated and analyzed by using a LI-COR Gene Read IR 4200 apparatus and software provided by the manufacturer. Sequences were edited and aligned by using DNasis 2.5 software (MWG Biotech AG) and were submitted to BLAST_N (1) for phylogenetic placement.
Nucleotide sequence accession numbers.
The nucleotide sequences of 16S rRNA gene regions that were sequenced in both directions have been deposited in the GenBank database under accession numbers AF214119 to AF214143.
RESULTS
Field inoculation and survival of S. meliloti L33 on inoculated and noninoculated field plots.
An average wind velocity of 1.6 m s−1 was measured while S. meliloti L33 cells were being inoculated (sprayed) onto the soil surfaces of the field plots. The inoculation procedure for each field plot lasted 0.5 h. Sedimentation plates placed around the field plots indicated that the average level of contamination of the surface soil was 1.25 cells per cm2 at a distance of 2 m. Thus, the level of contamination outside the inoculated areas was approximately 2 orders of magnitude below the threshold of detection by our cultivation-dependent technique for marker gene-tagged cells among soil bacteria (growth on NPA agar).
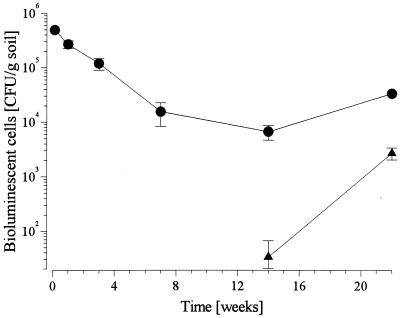
In bulk soil collected from inoculated field plots the concentration of S. meliloti L33 declined from 5 × 105 CFU g−1 2 days after inoculation to 104 CFU g−1 after 14 weeks (Fig. 1). After 22 weeks, at the end of the growing season in September, the concentration had increased to approximately 2 × 104 CFU g−1. Initially, no bioluminescent cells were found in noninoculated plots. After 14 weeks, however, several bioluminescent colonies, which were identified by PCR as strain L33, the inoculant, were detected. At this time, the titer was only slightly greater than the threshold of detection (2 × 10 CFU g of soil−1), but later, during the next two months, the titer increased by 2 orders of magnitude.
FIG. 1.
Survival of S. meliloti L33 in bulk soil after release in field plots seeded with alfalfa (M. sativa). The S. meliloti L33 titers in the Ap horizon (plough layer) of inoculated (●) and noninoculated (▴) plots are shown.
Detection of S. meliloti L33 in rhizospheres.
Rhizosphere microorganisms were isolated 12 weeks after field inoculation with S. meliloti L33 and seeding with M. sativa. In addition to M. sativa, the field plots were also colonized by several weeds, including C. album, the most abundant weed. In inoculated plots, the rhizospheres of M. sativa contained S. meliloti L33 at densities that were 2 orders of magnitude greater than the densities in C. album rhizospheres, indicating that enrichment due to the symbiotic partner plant occurred (Table 1). Surprisingly, although the inoculant was not detectable in bulk soil after 10 weeks, the rhizospheres of M. sativa plants grown in noninoculated plots also harbored cells of the inoculant. Depending on the plot, the densities ranged from 2 orders of magnitude less than the titer detected in inoculated plots (plot 6) to densities as high as the densities in the inoculated plots (plot 4) (Table 1). The numbers of the inoculant in the rhizospheres of C. album in noninoculated plots were almost 3 orders of magnitude less than the numbers in the rhizospheres of M. sativa. The numbers of bacterial cells detected on RAA medium were fourfold higher for M. sativa than for C. album. The total numbers of RAA-grown colonies were not affected significantly by inoculation.
Fragment frequency patterns of cultivated rhizosphere bacteria.
A total of 1,119 pure cultures were isolated on RAA from rhizospheres of M. sativa grown in inoculated plots (288 isolates), M. sativa grown in noninoculated plots (267 isolates), C. album grown in inoculated plots (288 isolates), and C. album grown in noninoculated plots (276 isolates). RAA contained four amino acids which could be utilized by S. meliloti L33 (see above). Growth on this agar, therefore, indicated that there was a metabolic capacity that was shared by the indigenous soil bacteria and the inoculant.
PCR amplification targeted the 16S rRNA gene region (size, approximately 1.060 bp). Amplified products were digested with HhaI, and the resulting DNA fragments were separated by electrophoresis. To do this, denaturing PAA gels were used to separate the products on the basis of size. For a small proportion of isolates (5.8%), PCR products were not obtained or not digested when this procedure was used. These isolates were omitted from the analysis.
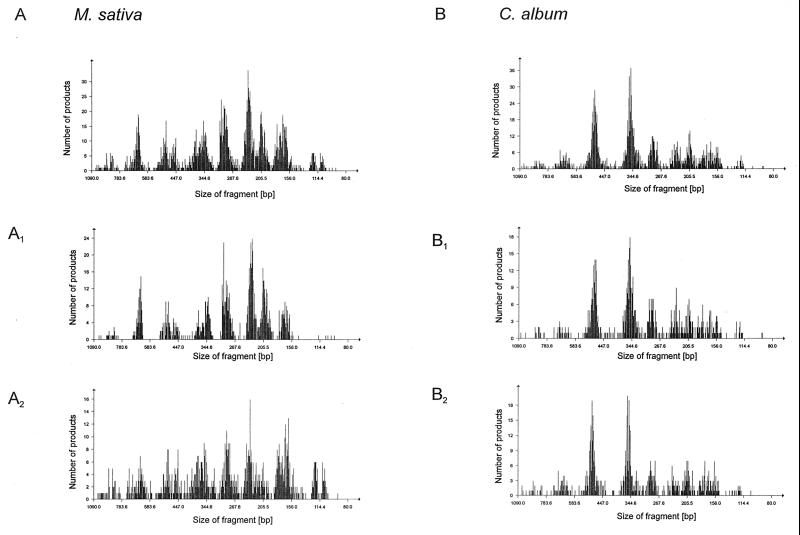
For the remaining isolates, the fragment patterns were calibrated on the basis of size and were imported into a database (WinCam) (see above). When all of the raw data fragment patterns (n = 1,054) in the database were combined to obtain an immediate graphical overview, the frequencies of fragment abundance indicated that there were common fragments. Fragment frequency patterns were distinguishable when isolates obtained from M. sativa and C. album rhizospheres were compared (Fig. 2A and B). This was a clear indication that there were plant-specific bacterial communities. On the other hand, there was also similarity between the two patterns, indicating that similar bacterial species (ARDRA types) occurred in the two rhizospheres, possibly at different levels. The fragment frequency patterns were further subdivided in order to detect whether S. meliloti L33 inoculation affected the diversity of isolates. In fact, the patterns obtained for noninoculated and inoculated M. sativa rhizospheres were different (Fig. 2A1 and A2), but the patterns obtained for C. album rhizospheres were very similar. We concluded from these results that inoculation had an effect on the structure of the rhizosphere bacteria from M. sativa but not on the structure of the rhizosphere bacteria from C. album.
FIG. 2.
DNA fragment frequencies for ARDRA products (raw data) obtained from cultivated bacteria isolated from the rhizospheres of M. sativa (A) and C. album (B) collected from inoculated and noninoculated field plots 12 weeks after field release of S. meliloti L33. (A and B) All isolates cultivated from the rhizospheres. (A1 and B1) Isolates obtained from noninoculated field plots. (A2 and B2) Isolates obtained from inoculated field plots.
Diversity of cultivated rhizosphere bacteria.
Similarity analyses of ARDRA patterns (n = 1,054) obtained with HhaI yielded 39 groups, which included 1 to 289 isolates. Ten of these groups consisted of more than 11 isolates (>1% of the total isolates). To increase the resolution of the groups, each of the large groups was further characterized by ARDRA by using an additional enzyme (HaeIII). When this was done, the number of groups increased from 10 to 25, not including the subgroups with less than four members (0.5% of the remaining isolates; n = 859). One randomly chosen isolate belonging to each of these groups was characterized by sequencing the 16S rRNA gene PCR product. Isolates belonging to α, β, and γ subgroups of the Proteobacteria, as well as members of the Flavobacterium-Cytophaga group and gram-positive bacteria with high G+C DNA contents (Table 2), were identified.
The largest groups were related to the genera Variovorax and Arthrobacter and the species Acinetobacter calcoaceticus (24.7, 14.4, and 12.7% of all isolates, respectively). Some groups exhibited clear plant specificity. Members related to Agrobacterium rubi, Variovorax paradoxus, and the genus Burkholderia dominated the rhizosphere of C. album but were rarely found in M. sativa rhizospheres. M. sativa-specific groups were also detected; these groups included bacteria related to S. meliloti, A. calcoaceticus, and Arthrobacter ramosus.
In order to decide whether the inoculation process modified the rhizospheres, we compared the numbers of bacterial isolates belonging to each ARDRA group obtained from inoculated and noninoculated plots. A high correlation value for group sizes (r = 0.949) was obtained for isolates from C. album rhizospheres, but a low correlation value (r = 0.270) was obtained for isolates from M. sativa (Table 2). This difference in correlation factors clearly indicated that M. sativa rhizospheres, but not C. album rhizospheres, were affected by inoculation. As a consequence of inoculation, S. meliloti was detected in the cultivated population obtained from rhizospheres of M. sativa. PCR detection of the luc marker gene confirmed that these isolates were the inoculants themselves (data not shown). Members of the genus Rhizobium were also detected in M. sativa rhizospheres from inoculated plots, but almost no Rhizobium cells were detected in rhizospheres of plants grown in noninoculated plots. In contrast, A. calcoaceticus, the most dominant species, was almost eliminated from the cultivated bacteria as a consequence of inoculation. Other groups negatively affected by inoculation were related to the genus Pseudomonas.
Cultivation-independent genetic profiles for bacterial communities in the rhizospheres.
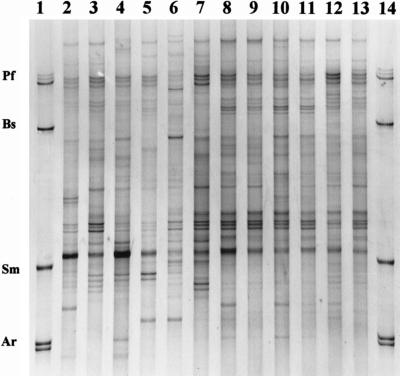
Total DNA was directly extracted from the rhizosphere samples which were used for cultivation of bacteria (see above). The heterogeneity of PCR products amplified from community DNA with eubacterial primers spanning the V4 and V5 regions of 16S rRNA genes was analyzed by the SSCP method. The SSCP patterns of the rhizosphere samples consisted of 20 to 32 distinguishable bands of different intensities. Most of the bands were produced by all samples. A typical SSCP gel is shown in Fig. 3. The M. sativa rhizosphere samples (Fig. 3, lanes 2 to 7) exhibited greater variation than the C. album rhizosphere samples (Fig. 3, lanes 8 to 13). For C. album, the variation in the patterns could not be correlated with the treatments (inoculation versus control), but M. sativa-derived patterns were affected. The patterns for rhizospheres from noninoculated plots were very similar and contained one dominant product. In contrast, the patterns of replicates of inoculated M. sativa rhizospheres were less similar. As suggested by comparison with the species standard (lanes 1 and 14), S. meliloti products were detected in the samples collected from inoculated M. sativa (more dominant in lanes 6 and 7 than in lane 5) but not in M. sativa rhizospheres collected from noninoculated plots.
FIG. 3.
Cultivation-independent PCR-SSCP profiles of bacterial communities isolated from rhizospheres of M. sativa (lanes 2 to 7) and C. album (lanes 8 to 13). Results from three replicate field plots are shown for each treatment. The results for plants from noninoculated field plots are shown in lanes 2 to 4 and 8 to 10. The results for plants from S. meliloti-inoculated plots are shown in lanes 5 to 7 and 11 to 13. The results for SSCP species standards, consisting of single-stranded DNA products obtained from PCR-amplified 16S rRNA gene regions (including regions V4 and V5) (see Materials and Methods) of selected bacterial species (Pf, Pseudomonas fluorescens; Bs, Bacillus subtilis; Sm, Sinorhizobium meliloti; Ar, Agrobacterium radiobacter), are shown in lanes 1 and 14.
Comparison of SSCP products of cultivated and noncultivated isolates.
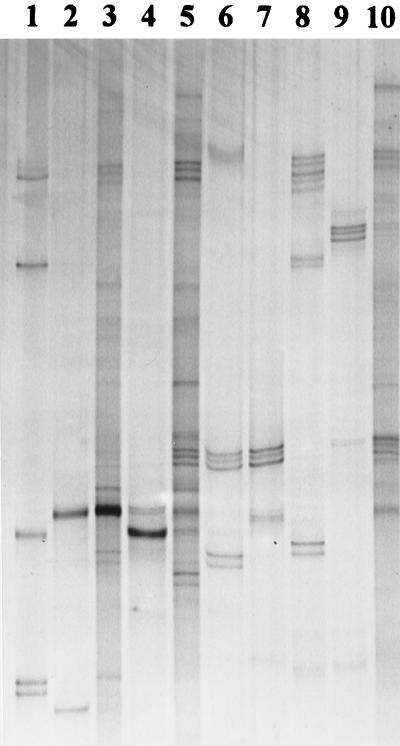
Both ARDRA combined with DNA sequencing of 16S rRNA genes and SSCP community analysis indicated that the bacterial diversity in rhizospheres was qualitatively and quantitatively affected by the plant species and, probably, in the case of M. sativa, by inoculation with S. meliloti L33. In order to link the two methods, SSCP products were generated from bacterial isolates belonging to the dominant ARDRA groups. Based on replicate electrophoretic runs, these products were compared to SSCP products found in community patterns. Analyses of the electrophoretic gels indicated that 6 of 25 ARDRA groups could be detected in the community profiles. Besides S. meliloti, SSCP products could be assigned to quantitatively important ARDRA groups, including A. calcoaceticus, Pseudomonas sp., Pseudomonas putida, Arthrobacter sp., and A. ramosus (Fig. 4). The ARDRA group which included the largest number of individual isolates was related to the genus Variovorax. This group and other ARDRA groups containing fewer isolates were not detected in the community profiles, as shown for some examples in Fig. 5. On the other hand, several products found in community profiles could not be attributed to the selected, cultivated isolates.
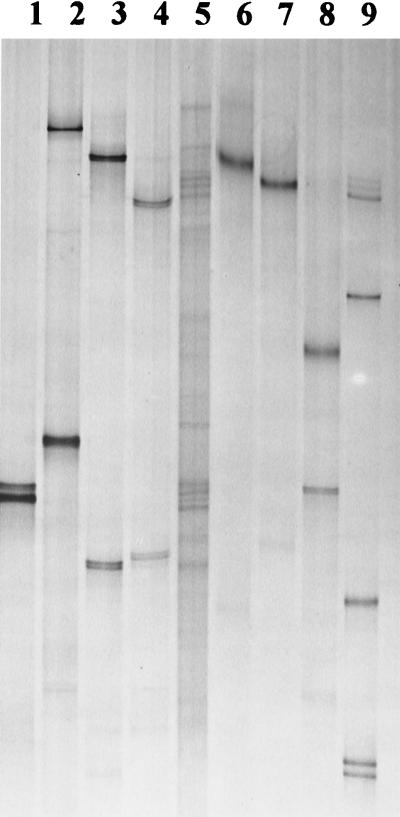
FIG. 4.
Matching of SSCP products obtained from cultivated isolates (lanes 2, 4, and 6 to 9) with rhizosphere bacterial communities extracted from M. sativa roots collected from noninoculated field plots (lane 3), inoculated field plots (lane 5), and rhizospheres of C. album (noninoculated plots) (lane 10). Pure-culture products were obtained from isolates related to A. calcoaceticus (ARDRA group G) (lane 2), S. meliloti (ARDRA group CD) (lane 4), A. ramosus (ARDRA group JA) (lane 6), Arthrobacter sp. (ARDRA group JB) (lane 7), Pseudomonas sp. (ARDRA group FA) (lane 8), and P. putida (ARDRA group FB) (lane 9). For SSCP species standards (lane 1), see Fig. 3; for ARDRA groups, see Table 2.
FIG. 5.
SSCP products of cultivated bacterial isolates (lanes 1 to 4 and 6 to 9) which could not be found in community profiles. A community profile obtained from C. album rhizospheres (lane 5) collected from S. meliloti L33-inoculated field plots is shown as an example. Pure-culture isolates were related to A. rubi (ARDRA group AA) (lane 1), Phyllobacterium myrsinacearum (ARDRA group AB) (lane 2), Burkholderia glathei (ARDRA group EB) (lane 3), Burkholderia sp. (ARDRA group EC) (lane 4), Variovorax sp. (ARDRA group DA) (lane 6), V. paradoxus (ARDRA group DB) (lane 7), and V. paradoxus (ARDRA group DC) (lane 8). Lane 9 contained SSCP species standards (for composition see Fig. 3). ARDRA groups are listed in Table 2.
DISCUSSION
The diversity and numbers of microorganisms in rhizospheres are, to a large extent, determined by the composition and concentration of root exudates excreted by plants (35). These exudates mainly serve as nutrient sources for microorganisms (14). Additionally, roots can also excrete substrates which specifically interact with bacteria; these substrates include flavones excreted by M. sativa, which attract S. meliloti cells (4, 16, 25, 45, 47). In our study, we wanted to determine whether the composition of the bacterial types in rhizospheres was affected by inoculation with S. meliloti. Transiently occurring changes in microbial community structure in response to inoculation have been detected in studies performed with nonsymbiotic rhizosphere bacteria (5, 13, 40, 41). To understand the importance of the symbiotic relationship, we included analyses of bacterial populations in the rhizospheres of a host plant, M. sativa, and a nonhost plant, the weed C. album.
Surprisingly, M. sativa plants collected 12 weeks after inoculation from noninoculated control plots also harbored populations of marker gene-tagged S. meliloti cells in their rhizospheres. The S. meliloti titers were as high as 4 to 64% of the titers found in rhizospheres of M. sativa plants grown in inoculated field plots. After 14 weeks, spread of the inoculant to noninoculated control plots was also detected with bulk soil. The numbers of S. meliloti L33 cells, however, were only slightly greater than the threshold of detection. During further plant development, the bulk soil titer of the inoculant increased, indicating that S. meliloti L33 was capable of surviving and growing in the noninoculated plots. Sedimentation plates, which were utilized during the inoculation procedure to detect aerial spread, indicated that small amounts of S. meliloti L33 escaped. Escape of bacterial cells during inoculation was also reported in a field release study performed with a marker gene-tagged Pseudomonas fluorescens strain (12). Consistent with our results, the inoculant was also recovered later in plant-associated niches in the vicinity of the inoculated area. On the basis of the development of the titer of S. meliloti L33 in bulk soil, we concluded that growth of the escaped cells in the rhizospheres of M. sativa plants in noninoculated plots preceded the spread of the inoculant into bulk soil.
Despite the existence of S. meliloti L33 populations in the rhizospheres of M. sativa and C. album plants in the noninoculated control plots, these samples were still valid controls for community analysis because the previous histories of plant exposure to the inoculant were different; in inoculated plots, the M. sativa roots were immediately exposed to high densities of inoculant cells (105 to 106 cells g of soil−1) after germination, whereas in noninoculated plots the levels of S. meliloti L33 were below the threshold of detection (<10 cells g−1). Thus, the factors acting on the selection of bacterial communities in rhizospheres in the inoculated and noninoculated plots were, in fact, different, and community structure, as detected 12 weeks after inoculation, was likely to be affected by this history.
Regarding the detection of luciferase-tagged S. meliloti cells, there was a difference between the numbers obtained by cultivation on selective agar (NPA) and the numbers obtained by cultivation on nonselective agar (RAA) (Table 1). On nonselective agar, bioluminescent cells were detected in rhizospheres of M. sativa collected from inoculated plots but not in rhizospheres collected from noninoculated plots. Based on the fact that 8% of the isolates obtained from M. sativa rhizospheres from inoculated plots were identified as the inoculant, we calculated that the total concentration of S. meliloti L33 (8% of 6.6 × 107 CFU g−1) was 5.3 × 106 CFU g−1 and, thus, 17-fold greater than the concentrations obtained after selective cultivation. The selective agar, NPA, contained low concentrations of nutrients and the compounds Congo red and pentachloronitrobenzene, which inhibit the growth of many bacteria and are probably also stressful for S. meliloti (8). It has been shown that immediate cultivation of environmental samples on selective growth agar can result in underrepresentation of the targeted populations by orders of magnitude (61). However, in this study it was not clear why the differences between the S. meliloti L33 populations in the rhizospheres of M. sativa plants from noninoculated and inoculated field plots were greater on nonselective agar than on selective agar. The results obtained after cultivation on nonselective agar are supported by the cultivation-independent community profiles. Products which comigrated with the S. meliloti pure-culture product were detected only in rhizospheres of M. sativa plants collected from inoculated plots. The lack of competing indigenous S. meliloti probably explains the capacity of the inoculant to spread and grow in the rhizospheres of M. sativa plants and bulk soil in the noninoculated field plots.
The ARDRA results for cultivated bacteria indicated that the plant species and, in the case of M. sativa but not in the case of C. album, the inoculant affected the bacterial community structure in the rhizospheres. Our description of community structure was based on ARDRA patterns obtained with two restriction endonucleases. In this way we identified groups which differentiated isolates belonging to the same genus (e.g., the genus Phyllobacterium, Rhizobium, Variovorax, or Arthrobacter) and, in the case of V. paradoxus, isolates belonging to the same species. On the other hand, we cannot rule out the possibility that the large ARDRA groups (groups DA, JA, and G, which were later determined to be related to Variovorax sp., A. ramosus, and A. calcoaceticus, respectively) consisted of more than one species. However, for the purposes of this ecological study, the resolution achieved with two restriction enzymes was appropriate for detecting microbial community changes. ARDRA typing was also found to be useful in other studies of the community structure of cultivated soil bacteria (20, 68).
Inoculation with S. meliloti L33 affected the composition of the bacterial community in the rhizosphere of M. sativa by reducing the number of members of the γ subgroup of the Proteobacteria and increasing the number of members of the α subgroup of the Proteobacteria. This shift can be interpreted as replacement of more general bacteria (A. calcoaceticus, Pseudomonas sp.) by specialists (rhizobia). Pseudomonas spp. were also displaced in rhizosphere communities in other studies (5, 13). It was suggested that this effect is related to the potential for fast growth, which allows the organisms to respond quickly to changing conditions in the rhizosphere (13). The enrichment of other rhizobia as a consequence of S. meliloti inoculation in this study may have been a result of increased production of root exudates by the nodulated plants. It is known that rhizobia can be attracted by roots of nonsymbiotic plants (18, 24).
The single-stranded PCR–SSCP approach (56) was used to characterize the bacterial community independent of cultivation. The profiles obtained confirmed that the plant species and, in the case of M. sativa, the inoculant affected the structural diversity in the rhizospheres. Our results corroborate those of a recent study in which Dunbar et al. found that the microbial communities of soils exhibited similar relationships with both cultivation and 16S rRNA gene cloning (19). This linking of two methods, which was applied in the study of Dunbar et al., was also possible in our study, since the same target gene (16S rRNA gene) was used for characterization. The comparison of products of cultivated isolates and community profiles as determined by SSCP analysis indicated that the most dominant groups identified by cultivation were also detected in the community profiles. However, there was one exception; the most dominant group, which represented 24% of the total cultivated isolates and was related to the genus Variovorax, was not detected. DNA extraction and cell lysis efficiencies (63, 70), preferential PCR amplification (51, 60), or rRNA gene copy numbers (21, 22) might have affected this lack of detection.
The fact that the Variovorax group was not detected in SSCP profiles revealed limitations of the SSCP technique which probably also occur with other PCR-dependent profiling techniques. On the other hand, our results encourage the use of genetic profiles, since, independent of cultivation, plant-specific rhizosphere communities and an inoculation effect were detected. Additionally, SSCP profiles revealed the presence of organisms which were not among the dominant cultivated bacteria. In a recent study we utilized different primers to monitor eubacteria, actinomycetes, and fungi during a composting process in parallel and, thus, increased the resolution of the diversity analysis (46). A similar strategy should be useful if genetic profiles are used in future studies on ecological effects and potential risks of inocula in agricultural biotechnology.
TABLE 1.
Sizes of populations of cultivated bacteria from rhizospheres of plants grown in field plots and collected 12 weeks after field release of S. meliloti L33
| Plot | No. of bacteria in M. sativa rhizospheres (CFU g [wet wt] of roots−1)
|
No. of bacteria in C. album rhizospheres (CFU g [wet wt] of roots−1)
|
||
|---|---|---|---|---|
| S. meliloti L33a | Total bacteriab | S. meliloti L33a | Total bacteriab | |
| Plots inoculated with S. meliloti L33 | ||||
| 1 | 3.3 × 105 | 6.4 × 107 | 4.2 × 103 | 1.5 × 107 |
| 2 | 2.4 × 105 | 6.4 × 107 | 2.1 × 103 | 1.8 × 107 |
| 3 | 3.5 × 105 | 6.9 × 107 | 1.9 × 103 | 2.0 × 107 |
| Avg | 3.1 × 105 | 6.6 × 107 | 2.7 × 103 | 1.8 × 107 |
| Noninoculated plots | ||||
| 4 | 2.1 × 105 | 7.4 × 107 | 2.2 × 102 | 2.1 × 107 |
| 5 | 5.6 × 104 | 5.6 × 107 | 3.6 × 101 | 1.7 × 107 |
| 6 | 1.3 × 103 | 9.4 × 107 | NDc | 1.3 × 107 |
| Avg | 8.9 × 104 | 7.5 × 107 | 1.0 × 102 | 1.7 × 107 |
Number of luciferase-positive colonies grown on NPA.
Number of colonies grown on RAA.
ND, not detected.
TABLE 2.
Diversity of cultivated bacteria extracted from rhizospheres of M. sativa and C. album plants collected from S. meliloti L33-inoculated and noninoculated field plots and characterized by using ARDRA and the nucleotide sequence of PCR-amplified 16S rRNA genes
| ARDRA groupa | No. of isolatesb
|
Phylogenetic assignmentc | Closest relative | % Similarity to closest relative | Representative strain (GenBank accession no.) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Total | M. sativa, inoculated | M. sativa, noninoculated | C. album, inoculated | C. album, noninoculated | |||||
| AA | 25 | 2 | 1 | 9 | 13 | α-Proteobacteria | Agrobacterium rubi | 99.5 | FSC127 (AF214119) |
| AB | 19 | 9 | 0 | 5 | 5 | α-Proteobacteria | Phyllobacterium myrsinacearum | 98.0 | FSC650 (AF214120) |
| B | 9 | 6 | 0 | 2 | 1 | α-Proteobacteria | Phyllobacterium rubiacearum | 98.9 | FSC635 (AF214121) |
| CA | 9 | 5 | 1 | 1 | 2 | α-Proteobacteria | Rhizobium leguminosarum | 99.5 | FSC115 (AF214122) |
| CB | 25 | 10 | 0 | 9 | 6 | α-Proteobacteria | Rhizobium sp. (partial) strain 21321 | 99.1 | FSC117 (AF214123) |
| CC | 17 | 6 | 0 | 9 | 2 | α-Proteobacteria | Rhizobium sp. strain USDA 1920 | 100 | FSC177 (AF214124) |
| CD | 25 | 25 | 0 | 0 | 0 | α-Proteobacteria | Sinorhizobium meliloti | 100 | FSC302 (AF214125) |
| CE | 4 | 2 | 1 | 1 | 0 | α-Proteobacteria | Ensifer adhaerens | 99.3 | FSC375 (AF214126) |
| DA | 212 | 25 | 13 | 85 | 89 | β-Proteobacteria | Variovorax sp. strain WFF052 | 99.3 | FSC781 (AF214127) |
| DB | 46 | 8 | 5 | 17 | 16 | β-Proteobacteria | Variovorax paradoxus | 99.1 | FSC933 (AF214128) |
| DC | 14 | 0 | 1 | 8 | 5 | β-Proteobacteria | Variovorax paradoxus | 99.6 | FSC917 (AF214129) |
| EA | 5 | 0 | 0 | 1 | 4 | β-Proteobacteria | Burkholderia sp. isolate N3P2 | 98.1 | FSC1134 (AF214130) |
| EB | 16 | 2 | 2 | 7 | 5 | β-Proteobacteria | Burkholderia glathei | 99.9 | FSC1120 (AF214131) |
| EC | 15 | 2 | 0 | 10 | 3 | β-Proteobacteria | Burkholderia sp. strain LB400 | 99.0 | FSC1145 (AF214132) |
| ED | 7 | 0 | 0 | 5 | 2 | γ-Proteobacteria | Erwinia amylovora | 98.9 | FSC1186 (AF214133) |
| FA | 61 | 9 | 32 | 1 | 19 | γ-Proteobacteria | Pseudomonas sp. strain IC038 | 99.0 | FSC408 (AF214134) |
| FB | 50 | 6 | 31 | 3 | 10 | γ-Proteobacteria | Pseudomonas putida | 99.3 | FSC413 (AF214135) |
| FC | 11 | 0 | 8 | 0 | 3 | γ-Proteobacteria | Pseudomonas sp. strain S2 | 100 | FSC410 (AF214136) |
| FD | 6 | 4 | 2 | 0 | 0 | γ-Proteobacteria | Pseudomonas sp. strain PsF | 99.2 | FSC472 (AF214137) |
| G | 109 | 8 | 101 | 0 | 0 | γ-Proteobacteria | Acinetobacter calcoaceticus | 100 | FSC001 (AF214138) |
| H | 9 | 7 | 0 | 1 | 1 | γ-Proteobacteria | Stenotrophomonas maltophilia | 99.8 | FSC630 (AF214139) |
| IA | 17 | 4 | 1 | 10 | 2 | F-C group | Flavobacterium johnsonae | 97.4 | FSC841 (AF214140) |
| IB | 15 | 7 | 0 | 6 | 2 | F-C group | Flexibacter columnaris | 98.1 | FSC231 (AF214141) |
| JA | 124 | 63 | 35 | 18 | 8 | GP bacteria | Arthrobacter ramosus | 99.7 | FSC1047 (AF214142) |
| JB | 9 | 7 | 1 | 0 | 1 | GP bacteria | Arthrobacter oxidans | 99.2 | FSC285 (AF214143) |
| Total | 859 | 217 | 235 | 208 | 199 | ||||
The on-line letter indicates the group based on fragment length polymorphism obtained after digestion of the PCR-amplified 16S rRNA gene product with HhaI. The subscript letter indicates the subgroup based on results obtained with restriction endonuclease HaeIII.
The Pearson correlation coefficient for inoculated and noninoculated M. sativa was 0.270. The Pearson correlation coefficient for inoculated and noninoculated C. album was 0.949.
α-Proteobacteria, α subgroup of the Proteobacteria; β-Proteobacteria, β subgroup of the Proteobacteria; γ-Proteobacteria, γ subgroup of the Proteobacteria; F-C group, Flavobacterium-Cytophaga group; GP bacteria, gram-positive bacteria with high G+C DNA contents.
ACKNOWLEDGMENT
This work was supported by grant 0311203 from the German Ministry for Education and Research (BMBF).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anyango B, Wilson K, Giller K. Competition in Kenyan soils between Rhizobium leguminosarum biovar phaseoli strain Kim5 and R. tropici strain CIAT899 using the gusA marker gene. Plant Soil. 1998;204:69–78. [Google Scholar]
- 3.Bassam B J, Caetano-Anolles G, Greshoff P M. Fast and sensitive staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196:80–83. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- 4.Bauer W D, Caetano-Anolles G. Chemotaxis, induced gene-expression and competitiveness in the rhizosphere. Plant Soil. 1990;129:45–52. [Google Scholar]
- 5.Bolton H J, Frederickson J K, Thomas J M, Li S W, Workmann D J, Bentjen S A, Smith J L. Field calibration of soil-core microcosms: ecosystem structural and functional comparisons. Microb Ecol. 1991;21:175–189. doi: 10.1007/BF02539152. [DOI] [PubMed] [Google Scholar]
- 6.Brimecombe M J, De Leij F A A M, Lynch J M. Effect of genetically modified Pseudomonas fluorescens strains on the uptake of nitrogen by pea from N-15 enriched organic residues. Lett Appl Microbiol. 1998;26:155–160. [Google Scholar]
- 7.Brimecombe M J, De Leij F A A M, Lynch J M. Effect of introduced Pseudomonas fluorescens strains on the uptake of nitrogen by wheat from N-15-enriched organic residues. World J Microbiol Biotechnol. 1999;15:417–423. [Google Scholar]
- 8.Bromfield E S P, Wheatcroft R, Barran L R. Medium for direct isolation of Rhizobium meliloti from soils. Soil Biol Biochem. 1994;26:423–428. [Google Scholar]
- 9.Caetano-Anollés G. Molecular dissection and improvement of the nodule symbiosis in legumes. Field Crop Res. 1997;53:47–68. [Google Scholar]
- 10.Dammann-Kalinowski T, Niemann S, Keller M, Selbitschka W, Tebbe C C, Pühler A. Characterization of two bioluminescent Rhizobium meliloti strains constructed for field releases. Appl Microbiol Biotechnol. 1996;45:509–512. doi: 10.1007/BF00578463. [DOI] [PubMed] [Google Scholar]
- 11.Dane F, Shaw J J. Survival and persistence of bioluminescent Xanthomonas campestris pv. campestris on host and non-host plants in the field environment. J Appl Bacteriol. 1996;80:73–80. [Google Scholar]
- 12.De Leij F A A M, Sutton E J, Whipps J M, Fenlon J S, Lynch J M. Field release of a genetically modified Pseudomonas fluorescens on wheat—establishment, survival and dissemination. Bio/Technology. 1995;13:1488–1492. [Google Scholar]
- 13.De Leij F A A M, Sutton E J, Whipps J M, Fenlon J S, Lynch J M. Impact of field release of genetically modified Pseudomonas fluorescens on indigenous microbial populations of wheat. Appl Environ Microbiol. 1995;61:3443–3453. doi: 10.1128/aem.61.9.3443-3453.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Troch P, Vanderleyden J. Surface properties and motility of Rhizobium and Azospirillum in relation to plant root attachment. Microb Ecol. 1996;32:149–169. doi: 10.1007/BF00185885. [DOI] [PubMed] [Google Scholar]
- 15.De Weger L A, von der Bij A J, Dekkers L C, Simons M, Wijffelman C A, Lugtenberg B J J. Colonization of rhizosphere of crop plants by plant-beneficial pseudomonads. FEMS Microbiol Ecol. 1995;17:221–228. [Google Scholar]
- 16.Dharmatilake A J, Bauer W D. Chemotaxis of Rhizobium meliloti towards nodulation gene-inducing compounds from alfalfa roots. Appl Environ Microbiol. 1992;58:1153–1158. doi: 10.1128/aem.58.4.1153-1158.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dighton J, Jones H E, Robinson C H, Beckett J. The role of abiotic factors, cultivation practices and soil fauna in the dispersal of genetically-modified microorganisms in soils. Appl Soil Ecol. 1997;5:109–131. [Google Scholar]
- 18.Dowling D N, Broughton W J. Competition for nodulation of legumes. Annu Rev Microbiol. 1986;40:131–157. doi: 10.1146/annurev.mi.40.100186.001023. [DOI] [PubMed] [Google Scholar]
- 19.Dunbar J, Takala S, Barns S M, Davis J A, Kuske C R. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl Environ Microbiol. 1999;65:1662–1669. doi: 10.1128/aem.65.4.1662-1669.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dunbar J, White S, Forney L. Genetic diversity through the looking-glass—effect of enrichment bias. Appl Environ Microbiol. 1997;63:1326–1331. doi: 10.1128/aem.63.4.1326-1331.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrelly V, Rainey F A, Stackebrandt E. Effect of genome size and rrn gene copy number on PCR amplification of 16S ribosomal RNA genes from a mixture of bacterial species. Appl Environ Microbiol. 1995;61:2798–2801. doi: 10.1128/aem.61.7.2798-2801.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fogel G B, Collins C R, Li J, Brunk C F. Prokaryotic genome size and SSU rDNA copy number: estimation of microbial relative abundance from a mixed population. Microb Ecol. 1999;38:93–113. doi: 10.1007/s002489900162. [DOI] [PubMed] [Google Scholar]
- 23.Fukui R, Schroth M N, Hendson M, Hancock J G. Interaction between strains of pseudomonads in sugar-beet spermospheres and their relationship to pericarp colonization by Pythium ultimum in soil. Phytopathology. 1994;84:1322–1330. [Google Scholar]
- 24.Gaworzewska E T, Carlile M J. Positive chemotaxis of Rhiziobium leguminosarum and other bacteria towards root exudates from legumes and other plants. J Gen Microbiol. 1982;128:1179–1188. [Google Scholar]
- 25.Gyorgypal Z, Kondorosi E, Kondorosi A. Diverse signal sensitivity of nodD protein homologs from narrow and broad host range rhizobia. Mol Plant-Microbe Interact. 1991;4:356–364. [Google Scholar]
- 26.Hattemer-Frey H A, Brandt E J, Travis C C. Small-scale field test of the genetically engineered lacZY marker. Reg Toxicol Pharmacol. 1990;11:253–261. doi: 10.1016/0273-2300(90)90025-7. [DOI] [PubMed] [Google Scholar]
- 27.Hirsch P R. Population dynamics of indigenous and genetically modified rhizobia in the field. New Phytol. 1996;133:159–171. [Google Scholar]
- 28.Jones R A, Broder M W, Stotzky G. Effects of genetically engineered microorganisms on nitrogen transformations and nitrogen-transforming microbial populations in soil. Appl Environ Microbiol. 1991;57:3212–3219. doi: 10.1128/aem.57.11.3212-3219.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennedy A C. Bacterial diversity in agroecosystems. Agric Ecosyst Environ. 1999;74:65–76. [Google Scholar]
- 30.Kluepfel D A. The behavior and tracking of bacteria in the rhizosphere. Annu Rev Phytopathol. 1993;31:441–472. [Google Scholar]
- 31.Lee D-H, Zo Y-G, Kim S-J. Nonradioactive method to study genetic profiles of natural bacterial communities by PCR-single-strand conformation polymorphism. Appl Environ Microbiol. 1996;62:3112–3120. doi: 10.1128/aem.62.9.3112-3120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu W T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowendorf H S. Factors affecting survival of Rhizobium in soil. In: Alexander M, editor; Alexander M, editor. Advances in microbial ecology. Vol. 4. New York, N.Y: Plenum Press; 1980. pp. 87–124. [Google Scholar]
- 34.Ludwig W, Schleifer K-H. Phylogeny of Bacteria beyond the 16S rRNA standard. ASM News. 1999;65:752–757. [Google Scholar]
- 35.Lynch J M. The rhizosphere. New York, N.Y: Wiley; 1990. [Google Scholar]
- 36.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik P, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP (Ribosomal Database Project) Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miethling, R., G. Wieland, H. Backhaus, and C. C. Tebbe. Variation of microbial rhizosphere communities in response to crop species, soil origin and inoculation with Sinorhizobium meliloti L33. Microb. Ecol., in press. [DOI] [PubMed]
- 38.Moenne-Loccoz Y, McHugh B, Stephens P M, McConnell F I, Glennon J D, Dowling D N, O'Gara F. Rhizosphere competence of fluorescent Pseudomonas sp. B24 genetically modified to utilise additional ferric siderophores. FEMS Microbiol Ecol. 1996;19:215–225. [Google Scholar]
- 39.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nacamulli C, Bevivino A, Dalmastri C, Tabacchioni S, Chiarini L. Perturbation of maize rhizosphere microflora following seed bacterization with Burkholderia cepacia MCI 7. FEMS Microbiol Ecol. 1997;23:183–193. [Google Scholar]
- 41.Natsch A, Keel C, Hebecker N, Laasik E, Défago G. Influence of biocontrol strain Pseudomonas fluorescens CHA0 and its antibiotic overproducing derivative on the diversity of resident root colonizing pseudomonads. FEMS Microbiol Ecol. 1997;23:341–352. [Google Scholar]
- 42.Neefs J-M, Van der Peer Y, De Rejk P, Chapelle S, De Wachter R. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 1993;21:3025–3049. doi: 10.1093/nar/21.13.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Donnell A G, Goodfellow M, Hawksworth D L. Theoretical and practical aspects of the quantification of biodiversity among microorganisms. Phil Trans R Soc London Ser B Biol Sci. 1994;345:65–73. doi: 10.1098/rstb.1994.0087. [DOI] [PubMed] [Google Scholar]
- 44.Paau A S. Improvement of Rhizobium inoculants. Appl Environ Microbiol. 1989;55:862–865. doi: 10.1128/aem.55.4.862-865.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peters N K, Frost J W, Long S R. A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science. 1986;233:977–980. doi: 10.1126/science.3738520. [DOI] [PubMed] [Google Scholar]
- 46.Peters S, Koschinsky S, Schwieger F, Tebbe C C. Succession of microbial communities during hot composting as detected by PCR-single-strand-conformation polymorphism-based genetic profiles of small-subunit rRNA genes. Appl Environ Microbiol. 2000;66:930–936. doi: 10.1128/aem.66.3.930-936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phillips D A, Joseph C M, Hirsch P R. Occurrence of flavonoids and nucleosides in agricultural soils. Appl Environ Microbiol. 1997;63:4573–4577. doi: 10.1128/aem.63.11.4573-4577.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips D A, Streit W R. Modifying rhizosphere microbial communities to enhance nutrient availability in cropping systems. Field Crop Res. 1998;56:217–221. [Google Scholar]
- 49.Pochon J, Tardieux P. Techniques d'analyse en microbiologie du sol. Edition de la Tourelle, St. Mandé (Seine) 1962. France. [Google Scholar]
- 50.Rao J R, Fenton M, Jarvis B D W. Symbiotic plasmid transfer in Rhizobium leguminosarum biovar trifolii and competition between the inoculant strain Icmp2163 and transconjugant soil bacteria. Soil Biol Biochem. 1994;26:339–351. [Google Scholar]
- 51.Reysenbach A-L, Giver L J, Wickham G S, Pace N R. Differential amplification of rRNA genes by polymerase chain reaction. Appl Environ Microbiol. 1992;58:3417–3418. doi: 10.1128/aem.58.10.3417-3418.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ryder M. Key issues in the deliberate release of bacteria. FEMS Microbiol Ecol. 1994;15:139–145. [Google Scholar]
- 53.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 54.Sato K, Jiang J-Y. Gram-negative bacterial flora on the root surface of wheat (Triticum aestivum) grown under different soil conditions. Biol Fertil Soils. 1996;23:273–281. [Google Scholar]
- 55.Schwieger F, Dammann-Kalinowski T, Dresing U, Selbitschka W, Munch J C, Pühler A, Keller M, Tebbe C C. Field lysimeter investigation with luciferase-gene (luc)-tagged Sinorhizobium meliloti strains to evaluate the ecological significance of soil inoculation and a recA-mutation. Soil Biol Biochem. 2000;32:859–868. [Google Scholar]
- 56.Schwieger F, Tebbe C C. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl Environ Microbiol. 1998;64:4870–4876. doi: 10.1128/aem.64.12.4870-4876.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Selbitschka W, Pühler A, Simon R. The construction of recA deficient containment Rhizobium meliloti and R. leguminosarum strains marked with gusA or luc cassettes for use in risk assessment studies. Mol Ecol. 1992;1:9–19. [Google Scholar]
- 58.Sitrit Y, Barak Z, Kapulnik Y, Oppenheim A B, Chet I. Expression of Serratia marcescens chitinase gene in Rhizobium meliloti during symbiosis on alfalfa roots. Mol Plant-Microbe Interact. 1993;6:293–298. [Google Scholar]
- 59.Streeter J G. Failure of inoculant rhizobia to overcome the dominance of indigenous strains for nodule formation. Can J Microbiol. 1994;40:513–522. [Google Scholar]
- 60.Suzuki M T, Giovannoni S J. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl Environ Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tebbe C C, Ogunseitan O A, Rochelle P A, Tsai Y-L, Olson B H. Varied responses in gene expression of culturable heterotrophic bacteria isolated from the environment. Appl Microbiol Biotechnol. 1992;37:818–824. [Google Scholar]
- 62.Tebbe C C, Schwieger F, Munch J C, Pühler A, Keller M. Field release of genetically engineered, bioluminescent Sinorhizobium meliloti strains. In: El Bassam N, Behl R K, Prochnow B, editors; El Bassam N, Behl R K, Prochnow B, editors. Sustainable agriculture for food, energy and industry. London, United Kingdom: James & James; 1998. pp. 450–452. [Google Scholar]
- 63.Tebbe C C, Vahjen W. Interference of humic acids and DNA extracted directly from soil in detection and transformation of recombinant DNA from bacteria and a yeast. Appl Environ Microbiol. 1993;59:2657–2665. doi: 10.1128/aem.59.8.2657-2665.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.United States Environmental Protection Agency. Monitoring small-scale field tests of microorganisms. Publication EPA 700 R-92-008. Washington, D.C.: Environmental Protection Agency; 1992. [Google Scholar]
- 65.van Elsas J D, Duarte G F, Rosado A S, Smalla K. Microbiological and molecular biological methods for monitoring microbial inoculants and their effects in the soil environment. J Microbiol Methods. 1998;32:133–154. [Google Scholar]
- 66.Van Overbeek L, van Veen J A, van Elsas J D. Induced reporter gene activity, enhanced stress resistance, and competitive ability of a genetically modified Pseudomonas fluorescens strain released into a field plot planted with wheat. Appl Environ Microbiol. 1997;63:1965–1973. doi: 10.1128/aem.63.5.1965-1973.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Villadas P J, Burgos P, Jording D, Selbitschka W, Pühler A, Toro N. Comparative analysis of the genetic structure of a Rhizobium meliloti field population before and after environmental release of the highly competitive R. meliloti strain GR4. FEMS Microbiol Ecol. 1996;21:37–45. [Google Scholar]
- 68.Wenderoth D F, Reber H H. Correlation between structural diversity and catabolic versatility of metal-affected prototrophic bacteria in soil. Soil Biol Biochem. 1999;31:345–352. [Google Scholar]
- 69.Woese C R, Kandler R O, Wheelis M L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eukarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou J Z, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]