Abstract
Background & Aims
Inflammatory bowel disease (IBD) is a major intestinal disease. Excessive inflammation and increased endoplasmic reticulum (ER) stress are the key events in the development of IBD. Search of a genome-wide association study database identified a remarkable correlation between a TM9SF4 single-nucleotide polymorphism and IBD. Here, we aimed to resolve its underlying mechanism.
Methods
The role of TM9SF4 was determined with experimental mouse models of IBD. ER stress cascades, barrier functions, and macrophage polarization in colonic tissues and cells were assessed in vivo and in vitro. The expression of TM9SF4 was compared between inflamed regions of ulcerative colitis patients and normal colon samples.
Results
In mouse models of IBD, genetic knockout of the TM9SF4 gene aggravated the disease symptoms. In colonic epithelial cells, short hairpin RNA–mediated knockdown of TM9SF4 expression promoted inflammation and increased ER stress. In macrophages, TM9SF4 knockdown promoted M1 macrophage polarization but suppressed M2 macrophage polarization. Genetic knockout/knockdown of TM9SF4 also disrupted epithelial barrier function. Mechanistically, TM9SF4 deficiency may act through Ca2+ store depletion and cytosolic acidification to induce an ER stress increase. Furthermore, the expression level of TM9SF4 was found to be much lower in the inflamed colon regions of human ulcerative colitis patients than in normal colon samples.
Conclusions
Our study identified a novel IBD-associated protein, TM9SF4, the reduced expression of which can aggravate intestinal inflammation. Deficiency of TM9SF4 increases ER stress, promotes inflammation, and impairs the intestinal epithelial barrier to aggravate IBD.
Keywords: Inflammatory Bowel Disease, Inflammation, ER Stress, TM9SF4, Intestinal Homeostasis
Abbreviations used in this paper: ATP, adenosine triphosphate; BMDM, bone marrow-derived macrophage; cDNA, complementary DNA; CFSE, carboxyfluorescein succinimidyl ester; CHOP, C/EBP homologous protein; COX2, cyclooxygenase 2; DHE, dihydroethidium; DSS, dextran sulfate sodium; ER, endoplasmic reticulum; FACS, fluorescence-activated cell sorter; FBS, fetal bovine serum; FITC, fluorescein isothiocyanate; GRP78, glucose-regulated protein 78; HCEC, human colonic epithelial cell line; IBD, inflammatory bowel disease; IEC, intestinal epithelial cell; IFN, interferon; IL, interleukin; iNOS, inducible nitric oxide synthase; KEGG, Kyoto Encyclopedia of Genes and Genomes; KO, knockout; LPS, lipopolysaccharide; MerTK, –Mer tyrosine kinase; mRNA, messenger RNA; PBS, phosphate-buffered saline; PGN, peptidoglycan; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; ROS, reactive oxygen species; shRNA, short hairpin RNA; TER, transepithelial resistance; TM9SF, transmembrane 9 protein; TNBS, trinitrobenzene sulfonic acid; TNF, tumor necrosis; UC, ulcerative colitis; WT, wild-type; ZO-1, zonula occludens-1; 4-PBA, 4-phenylbutyric acid
Graphical abstract
Summary.
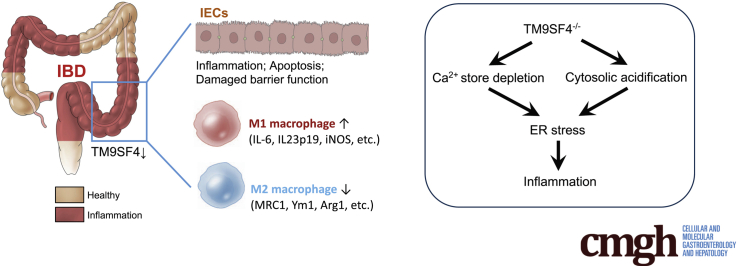
This study identified a novel inflammatory bowel disease–associated protein TM9SF4, which impacts inflammatory bowel disease progression via regulating endoplasmic reticulum stress and inflammation in both intestinal epithelial cells and macrophages.
Inflammatory bowel disease (IBD) is a common chronic inflammatory intestinal disorder of the gastrointestinal tract, which can be grouped into 2 principal types: ulcerative colitis (UC) and Crohn’s disease. Multiple factors, such as host genetic background, environmental and luminal factors, excessive inflammation, and inappropriate immune responses have been suggested to contribute to IBD pathogenesis.1,2 However, the detailed disease mechanisms of IBD are complicated and incompletely understood. Various animal models of IBD have been developed that provided indispensable insights into the pathogenesis of IBD. Dextran sulfate sodium (DSS)- and trinitrobenzene sulfonic acid (TNBS)-induced colitis are among the most widely used animal models of IBD.3, 4, 5 In terms of pathogenesis, the DSS model is more relevant to UC, whereas the TNBS model is more relevant to Crohn’s disease.3, 4, 5
Excessive colonic inflammation, both in intestinal epithelial cells (IECs) and colonic macrophages, underpins IBD.1,2,6 IECs constitute the surface barrier responsible for absorption of electrolytes and protection from gut microbiota.7 IECs are constantly exposed to various antigenic factors and bacterial toxins, such as lipopolysaccharides (LPS) from gram-negative bacteria and peptidoglycan (PGN) from gram-positive bacteria. In response, IECs release proinflammatory chemokines and cytokines, which are early signals to activate mucosal inflammatory responses.7 On the other hand, colonic macrophages originate mostly from circulating monocytes.8 After adhering and migrating to colonic sites, monocytes differentiate into 2 types of macrophages, namely, M1 and M2 macrophages.8 At the early stage of tissue injury or colonic infection, M1 macrophages secrete proinflammatory cytokines, such as interleukin (IL)1β, IL6, and tumor necrosis factor α (TNFα), to promote inflammation. However, at the late stage, M2 macrophages secrete anti-inflammatory mediators, such as arginase I and IL10, to antagonize inflammation and promote epithelial regeneration, consequently restoring barrier function.8
Endoplasmic reticulum (ER) stress also plays an important role in IBD.9 ER stress refers to the cellular condition characterized by accumulation of unfolded and misfolded proteins in the ER lumen. Excessive ER stress promotes inflammation and increases reactive oxygen species (ROS) production.10 Accumulation of misfolded proteins may also lead to formation of aggresome.11 ER stress can be restored by unfolded protein response, which encompasses 3 signaling branches, protein kinase RNA-like endoplasmic reticulum kinase–Activating transcription factor (ATF)4, ATF6, and Inositol-requiring enzyme 1–X-box-binding protein 1 (XBP1), under the regulation of glucose-regulated protein 78 (GRP78).9
Transmembrane 9 Superfamily isoform 4 (TM9SF4) proteins belong to the TM9SF family. TM9SF4 plays important roles in diverse cellular processes. Functionally, TM9SF4 homologs are involved in cell adhesion, phagocytosis, and innate immunity.12, 13, 14, 15, 16 TM9SF4 also can act on vacuolar H+-adenosine triphosphatase to regulate cytosolic pH,17 and regulate cell surface trafficking of glycine-rich transmembrane domains.18,19 We recently showed an important role of TM9SF4 in maintaining ER homeostasis. Short hairpin RNA (shRNA)-mediated TM9SF4 knockdown increased ER stress, probably via depleting ER Ca2+ stores, and increased the levels of ROS, consequently promoting cell death in chemoresistant breast cancer cells.20,21 In the present study, search of a genome-wide association study database identified a remarkable correlation between a single-nucleotide polymorphism (rs6142618) in the noncoding region of TM9SF4 and IBD, with a P value of 6 × 10-10 (genome-wide association study catalog European Molecular Biology Laboratory's European Bioinformatics Institute, EMBL-EBI).22,23 Based on the known association among TM9SF4, ER stress, and inflammation, we hypothesize that TM9SF4 may regulate ER stress and colonic inflammation to affect IBD. The role of TM9SF4 was mainly explored in a DSS-induced mouse model of IBD together with some supporting experiments from a TNBS-induced mouse model of IBD and pathologic samples from UC patients. Our results showed that TM9SF4 deficiency acts through multiple processes in colonic epithelial cells and macrophages to aggravate IBD.
Results
Genetic Knockout of TM9SF4 Aggravated DSS-Induced IBD
A DSS-induced mouse model of IBD was used.3,24 Wild-type and TM9SF4 knockout (KO) mice were fed with 2% DSS in drinking water for 7 days to induce colitis or with water as control. TM9SF4 was found to be expressed in the intestinal crypts (Figure 1A) and F4/80+ colonic macrophages, but not in Gr-1+ colonic neutrophils (Figure 1B). DSS treatment caused body weight loss, colon length decrease, diarrhea, and anal bleeding, all of which were more severe in KO mice than in wild-type (WT) mice (Figure 1C–G). In control mice fed with water for 7 days, there were no differences in body weight and colon length between WT and KO groups (Figure 1C–E). Quantitative reverse-transcription polymerase chain reaction (qRT-PCR) analysis of colon tissues showed higher messenger RNA (mRNA) expression of proinflammatory cytokines TNFα, IL1β, and IL6 in KO mice than in WT mice (Figure 1H).
Figure 1.
Knockout of TM9SF4 gene aggravated DSS-induced colitis in mice. (A) Representative TM9SF4 immunohistochemistry staining of colons from nontreated WT/KO mice showing distribution of TM9SF4. Brown, immunopositive signals; blue, nuclear counterstain. Red arrowheads indicate intestinal epithelial crypts, black arrows indicate infiltrating nonepithelial cells. Scale bar: 100 μm. (B) Colons from WT mice were stained with TM9SF4 antibody, macrophage marker F4/80 antibody, or neutrophil marker Gr-1 antibody. Expressions of TM9SF4 in infiltrating mononuclear cells are indicated as colocalization of TM9SF4 and specific cell markers. Scale bar: 50 μm. (C) Time courses of body weight loss in WT and KO mice treated with 2% DSS or water. Representative (D) images and (E) data summary of colon lengths in WT and KO mice. Time courses of (F) diarrhea and (G) fecal bleeding of WT and KO mice. (H) mRNA levels of TNFα, IL1β, and IL6 in colon tissues as assessed by qRT-PCR. (I) DSS-treated mice were gavaged with FITC–dextran (molecular weight, 40,000), followed by measurement of serum fluorescence intensity 4 hours later. Representative (J) images and (K) histopathologic scores of H&E-stained colon tissue sections from DSS-induced WT and KO mice. Scale bar: 200 μm. Means ± SEM. n = 6–8 mice per group in all experiments. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001.
To determine the role of TM9SF4 in intestinal integrity, fluorescein isothiocyanate (FITC)–dextran (molecular weight, 40,000 kilodaltons) were orally gavaged to DSS-treated mice. The serum level of FITC–dextran was found to be higher in KO mice than in WT mice, suggesting a more severely damaged intestinal epithelial barrier in KO mice (Figure 1I). Furthermore, histologic analysis of colon tissue slides indicated higher histopathologic scores in KO mice, which were scored based on depletion of goblet cells, epithelial erosion, and infiltration of proinflammatory cells into mucosa (Figure 1J and K). Together, these data suggested that genetic loss of TM9SF4 aggravated DSS-induced colitis in mice.
Knockout of TM9SF4 in Hematopoietic and Nonhematopoietic Cells Both Had a Detrimental Effect in DSS-Induced IBD In Vivo
To distinguish between the contribution of bone marrow–derived hematopoietic cells (later differentiated to monocytes, neutrophils, and so forth) and nonhematopoietic cells (including intestinal epithelial cells, endothelial cells, and so forth), we performed bone marrow transplantation experiments in the DSS model. Herein, bone marrow cells collected from WT or TM9SF4 KO mice were transferred to irradiated WT or KO recipient mice, yielding 4 experimental groups of chimeric mice, followed by DSS treatment at week 6 post-transplantation (Figure 2A). The results showed that compared with WT bone marrow cells, transplantation of KO bone marrow cells to WT mice (KO → WT vs WT → WT) caused more severe colitis symptoms, including colon shortening (Figure 2B and C), body weight loss (Figure 2D), diarrhea (Figure 2E), anal bleeding (Figure 2F), higher histopathologic scores (Figure 2G and H) and increased colonic levels of proinflammatory cytokines (Figure 2I). Furthermore, compared with KO bone marrow cells, transplantation of WT bone marrow cells to KO mice (WT → KO vs KO → KO) relieved the colitis symptoms (Figure 2B–I). We also examined colonic infiltration of bone marrow–derived (PKH67-labeled) circulating cells. KO of TM9SF4 in bone marrow cells promoted the infiltration of circulating cells into colonic lamina propria as indicated by fluorescence-activated cell sorter (FACS) analysis of PKH67+ cell infiltration to lamina propria (Figure 2J) and immunostaining of macrophage marker CD68 in colon tissues (Figure 2K). These data support the critical role of hematopoietic cell TM9SF4 in colitis.
Figure 2.
DSS-induced colitis in bone marrow–chimeric mice. (A) Schematic diagram of study design. Bone marrow cells of recipient WT/KO mice were depleted by radiation. Bone marrow cells from donor KO/WT mice were purified, stained, and injected into recipient WT/KO mice. Colitis was induced at 6 weeks post-transplantation. Representative (B) colon images, (C) colon length decrease, (D) body weight loss curve, (E) stool scores, (F) anal bleeding scores, and (G and H) H&E staining–based histopathologic examination in 2% DSS-treated bone marrow–chimeric mice. Scale bar: 100 μm. (I) mRNA levels of TNFα, IL1β, and IL6 in colon tissues as assessed by qRT-PCR. (J) Colon tissues in bone marrow–chimeras were digested for FACS analysis. Percentages of PKH67+CD45+ cells were determined by FACS analysis. (K) Representative images of CD68+ macrophages in colon sections. Brown, CD68+ signal; blue, nuclear counterstain. Scale bar: 100 μm. Means ± SEM. n = 6–8 mice per group in all experiments. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. BM cells, bone marrow cells.
On the other hand, TM9SF4 in nonhematopoietic cells also was important in colitis development, because when compared with WT recipient mice that received WT bone marrow cells, KO recipient counterparts developed more severe colitis symptoms (WT → KO vs WT → WT) (Figure 2B–I).
Knockdown of TM9SF4 Promoted Inflammation, Increased ER Stress, and Increased Cell Death in Colonic Epithelial Cells In Vitro
We examined the effect of shRNA-mediated TM9SF4 knockdown or overexpression on inflammatory response of colonic epithelial cells. LPS and PGN were used to trigger inflammation and release of proinflammatory cytokines in human colonic epithelial cells (HCECs)25 and HT-29 cells. Because the expression level of TM9SF4 was relatively high in HCECs but low in HT-29 cells (Figure 3A), we chose HCECs as the cell model for TM9SF4 knockdown although we used HT-29 as the cell model for TM9SF4 overexpression.
Figure 3.
Knockdown of TM9SF4 promoted inflammation, increased ER stress, and increased cell deaths in colon epithelial cells. (A) Expression levels of TM9SF4 in HCECs, Caco-2, and HT-29 cells were compared by Western blot (n = 3). HCECs with Scr-shRNA or TM9SF4-shRNAs were challenged with (B) 100 ng/mL LPS or (C) 1 μg/mL PGN for 24 hours. The mRNA levels of TNFα, IL1β, and IL6 were assessed by qRT-PCR (n = 3). (D) HCECs with Scr-shRNA or TM9SF4-shRNAs were challenged with LPS. The levels of TM9SF4, GRP78, ATF4, cleaved ATF6, spliced XBP1, CHOP, COX2, and cleaved caspase 3 were detected by Western blot. Representative from 3 experiments. (E) ROS measurement in HCECs by DHE fluorescence. Representative from 4 experiments. Scale bar: 100 μm. (F) Representative FACS analysis and data summary (right) of Annexin V and PI staining in LPS-challenged HCECs (n = 4). KEGG analysis of altered signaling pathways after TM9SF4 knockdown in (G) basal condition (vehicle) and (H) inflammatory condition (LPS) in HCECs. HCECs were transfected with scrambled-shRNA (SCR) or TM9SF4-shRNA 1 (KD) for RNA sequencing analysis. Shown are means ± SEM. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. ECM, extracellular matrix; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GnRH, gonadotropin releasing hormone; KD, knockdown clone; PI, propidium iodide; SCR, scrambled control; TGF, transforming growth factor; TRP, trasient receptor potential.
TM9SF4 knockdown in HCECs increased the LPS- and PGN-induced production of inflammatory cytokines, as indicated by mRNA levels of TNFα, IL1β, and IL6 (Figure 3B and C). TM9SF4 knockdown also increased the LPS-induced ER stress increase in HCECs, as indicated by ER stress markers GRP78, ATF4, cleaved ATF6, spliced XBP1, C/EBP homologous protein (CHOP), and cyclooxygenase 2 (COX2) (Figure 3D). Furthermore, TM9SF4 knockdown also increased ROS accumulation, as indicated by Dihydroethidium (DHE) staining (Figure 3E), and promoted apoptotic cell death, as indicated by Annexin V+ apoptotic cells (Figure 3F) and cleaved caspase 3 levels (Figure 3D). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis showed differential expression of several pathways related to TM9SF4 knockdown in HCECs in basal conditions (Figure 3G), as well as in LPS-stimulated conditions (Figure 3H). In basal conditions, knockdown of TM9SF4 altered the TNF signaling pathway (Figure 3G), suggesting that inflammation is an early process after TM9SF4 knockout. In the LPS-stimulated inflammatory condition, knockdown of TM9SF4 altered Ca2+ signaling–related pathways such as the phosphatidylinositol signaling system and Transient Receptor Potential (TRP) channels (Figure 3H), suggesting an involvement of Ca2+ signaling in the process.
4-phenylbutyric acid (4-PBA) is a small-molecule chaperone that can improve ER folding capacity and alleviate ER stress.26 4-PBA treatment reversed the TM9SF4 knockdown-induced production of inflammatory cytokines IL1β and IL6 (Figure 4A), reduced apoptotic cell death as indicated by cleaved caspase 3 level (Figure 4B), and diminished the TM9SF4 knockdown-induced ROS production (Figure 4C).
Figure 4.
4-PBA alleviated ER stress in vitro. 4-APB reversed the effect of TM9SF4 knockdown on (A) IL1β and IL6 production, (B) GRP78 increase, (B) caspase 3 activity increase, and (C) ROS production in HCECs (n = 3). Scale bar: 100 μm. Means ± SEM. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. SCR, scrambled control.
On the other hand, overexpression of exogenous TM9SF4 in HT-29 cells was found to reduce the LPS- and PGN-induced production of inflammatory cytokines (Figure 5A and B), and decreased the LPS-induced ER stress level in HT-29 cells (Figure 5C). TM9SF4 overexpression also decreased ROS accumulation (Figure 5D) and reduced apoptotic cell death (Figure 5E).
Figure 5.
Overexpression of TM9SF4 decreased inflammation, alleviated ER stress, and reduced cell deaths in colon epithelial cells. HT-29 cells were overexpressed with TM9SF4 or control plasmid, then challenged with (A) 100 ng/mL LPS or (B) 1 μg/mL PGN for 24 hours. The mRNA levels of TNFα, IL1β, and IL6 were assessed by qRT-PCR (n = 3). (C) HT-29 cells were overexpressed with TM9SF4 or control plasmid, then challenged with LPS. TM9SF4, GRP78, ATF4, cleaved ATF6, spliced XBP1, CHOP, COX2, and cleaved caspase-3 were detected by Western blot. Representative from 3 experiments. (D) Representative images of ROS measurement in LPS-treated HT-29 cells by DHE staining (n = 6). Scale bar: 100 μm. (E) Representative FACS analysis (left) and data summary (right) of Annexin V and PI staining in HT-29 cells (n = 5). Means ± SEM. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; pcDNA, plasmid cloning DNA; PI, propidium iodide.
Knockout of TM9SF4 Increased ER Stress and Increased Apoptosis in a DSS-Induced Colitis Model In Vivo
The role of TM9SF4 in ER stress and cell death was examined in vivo. Compared with WT mice, TM9SF4 KO mice showed a higher ER stress level in colon tissues as indicated by immunostaining of GRP78 and CHOP (Figure 6A), increased ROS production by DHE staining (Figure 6B), more apoptotic cells as indicated by terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling staining (Figure 6B), and immunostaining of cleaved caspase 3 (Figure 6A). Claudin-1 expression in the colonic epithelial layer also was reduced in TM9SF4 KO mice, an indication for impaired epithelial barriers (Figure 6B).
Figure 6.
Knockout of TM9SF4 gene increased ER stress and increased apoptotic cell death in DSS-induced colitis in mice. (A) Representative immunohistochemical staining of GRP78, CHOP, and cleaved caspase 3 in colon tissues of WT and KO mice after 2% DSS treatment. Scale bars: 100 μm. Brown, immunopositive signals; blue, nuclear counterstain. (B) Representative staining of ROS (DHE staining, red), terminal deoxynucleotidyl transferase–mediated deoxyuridine triphosphate nick-end labeling (TUNEL)-positive apoptotic cells (green), and claudin-1 (green) in colon tissues of WT and KO mice after 2% DSS treatment. Scale bars: 100 μm. 4-PBA reversed the TM9SF4 knockout-induced IBD disease aggravation in (C and D) colon lengths, (E) body weight, (F) diarrhea, and (G) anal bleeding (n = 7). (H) 4-PBA reversed the TM9SF4 knockout-induced colon tissue abnormality in H&E staining–based histopathologic examination, immunostaining-based expressional change of GRP78, CHOP, and claudin-1. Brown, immunopositive signals; blue, nuclear counterstain. Scale bar: 200 μm. Means ± SEM. n = 5–7 mice per group in all experiments. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. DAB, 3,3′-Diaminobenzidine.
Interestingly, 4-PBA treatment alleviated the DSS-induced colitis disease symptoms in both WT and TM9SF4 KO mice, including body weight loss, colon length decrease, diarrhea, and anal bleeding (Figure 6C–G). 4-PBA treatment also reduced the DSS-elicited histopathologic scores in KO colon tissue slides (Figure 6H), suppressed the DSS-elicited ER stress increases as indicated by immunostaining of GRP78 and CHOP, and restored Claudin-1 expression (Figure 6H).
TM9SF4 Regulated Colonic Infiltration of Macrophages and Phagocytosis
The role of TM9SF4 in macrophage infiltration and phagocytosis was examined. In the DSS-induced colitis model, immunostaining detected more infiltrated CD68+ macrophages in colonic tissue sections of TM9SF4 KO mice than those in WT mice (Figure 7A). FACS analysis indicated a higher percentage of colonic infiltration of CD45+CD11b+F4/80+ macrophages and a slightly reduced infiltration of CD4+ T cells in KO mice than in WT mice, but no difference in CD11C+MHCII+ dendritic cells, CD11b+Gr-1+ neutrophils, and CD8+ T cells between KO and WT mice (Figure 7B). These data indicated that TM9SF4 deficiency enhanced colonic infiltration of macrophages in colitis.
Figure 7.
TM9SF4 regulated colonic infiltration of macrophages and phagocytosis. (A) Immunohistochemical staining (left) and summary (right) of CD68+ cells in colons of WT and KO mice after 2% DSS treatment (n = 6). Brown, CD68+-positive signals; blue, nuclear counterstain. Scale bar: 100 μm. (B) Representative FACS analysis showing percentages of F4/80+CD11b+ macrophages, CD11c+MHCII+ dendritic cells, CD11b+GR-1+ neutrophils, CD4+ T cells, and CD8+ T cells in total CD45+ cells from colons of WT and KO mice after 2% DSS treatment (n = 6). (C) RNA sequencing analysis of colonic F4/80+CD11b+ macrophages from DSS-treated WT and KO mice. KEGG analysis of altered signaling pathways after TM9SF4 knockdown in colonic F4/80+CD11b+ macrophages from (D) water-treated and (E) DSS-treated mice. Mouse peritoneal macrophages were treated with CFSE-labeled apoptotic cells. Percentage of F4/80+ macrophages with green CFSE fluorescence was assessed by (F) FACS (n = 8) and (G) confocal microscopy analysis (n = 6). Scale bar: 100 μm. (H) The mRNA levels of Mfge8, Cd36, and Gas6 of WT/KO peritoneal macrophages were assessed by qRT-PCR (n = 6). (I) Raw264.7 cells were transfected with TM9SF4 plasmid or control plasmid pcDNA6, followed by incubation with CFSE-labeled apoptotic Jukart cells. The percentage of phagocytosis was determined by CFSE+ cells in FACS analysis (n = 8). Means ± SEM. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. BF, bright field; ECM, extracellular matrix; MAPK, mitogen-activated protein kinase; NF-kappa B, nuclear factor-kappa B; pcDNA, plasmid cloning DNA; PI3K-Akt, phosphatidylinositol 3-kinase-protein kinase B; RIG-I, retinoic acid-inducible gene I.
RNA sequencing then was used to compare the global gene expression between WT and KO colonic macrophages. The results showed that, compared with WT counterparts, KO colonic macrophages showed high levels of proinflammatory genes, including CCR7, IL12b, CCL5, IL1b, and TNF (Figure 7C), but low levels of anti-inflammatory genes, such as CCL20, Vsig4, and IL17f. The KEGG pathway enrichment analysis showed differential expression of several pathways related to TM9SF4 knockdown in macrophages in basal condition (Figure 7D) and DSS-induced inflammatory condition (Figure 7E). In basal conditions, knockdown of TM9SF4 altered several inflammatory pathways including TNF- and IL17-signaling pathways, suggesting that inflammation is an early event after TM9SF4 KO (Figure 7D). In DSS-induced inflammatory conditions, TM9SF4 knockdown altered the expression of several pathways related to macrophage function, such as cytokine–cytokine–receptor interaction, the nucleotide-binding and oligomerization domain (NOD)-like receptor signaling pathway, and phagosomes (Figure 7E). Phagocytotic ability was analyzed in vitro using carboxyfluorescein succinimidyl ester (CFSE)-labeled apoptotic Jurkat cells (Figure 7F–G). The results showed that KO of TM9SF4 markedly decreased the phagocytotic ability of primary mouse peritoneal macrophages, as indicated by CFSE+F4/80+ populations in FACS analysis (Figure 7F) and by CFSE fluorescence visualization under confocal microscopy (Figure 7G). Furthermore, the expression of several phagocytic genes, including Mfge8, CD36, and Gas6, was lower in KO macrophages than in WT macrophages (Figure 7H). In contrast, exogenous overexpression of TM9SF4 in Raw264.7 macrophages promoted the phagocytotic ability of the macrophages as measured by CFSE-labeled apoptotic Jurkat cells (Figure 7I).
Knockout/Knockdown of TM9SF4 Increased Macrophage and Promoted M1 Macrophage Polarization, but Inhibited M2 Macrophage
The role of TM9SF4 in macrophage polarization was investigated. qRT-PCR analysis showed that in CD11b+ myeloid cells isolated from the colons of DSS-treated mice, knockout of TM9SF4 increased M1 macrophages, as indicated by M1 macrophage markers IL23p19, IL6, and inducible nitric oxide synthase (iNOS) (Figure 8A), but reduced M2 macrophages, as indicated by M2 macrophage markers Mrc1, Ym1, and Arg1 (Figure 8B). Immunostaining confirmed that TM9SF4 KO increased the CD80+ M1 macrophages (Figure 8C), but decreased CD206+ M2 macrophages in colon tissue sections (Figure 8D).
Figure 8.
KO of TM9SF4 promoted M1 macrophage polarization but inhibited M2 macrophage polarization. (A and B) Colonic CD11b+ cells from 2% DSS-challenged WT or KO mice were purified by magnetic-activated cell sorting. The mRNA levels of (A) M1 macrophages markers IL23p19, IL6, iNOS, and (B) M2 macrophages markers MRC1, Ym1, Arg1 were assessed by qRT-PCR (n = 4). Representative images and data summary for immunohistochemical staining of (C) CD80 and (D) CD206 in the colon tissues of DSS-treated WT/KO mice (n = 7). Brown, CD80/CD206-positive signals; blue, nuclear counterstain. Scale bars: 100 μm. (E) Expression levels of TM9SF4 in BMDMs and Raw264.7 macrophages were compared by Western blot (n = 3). BMDMs were treated with LPS/IFNγ or vehicle, followed by (F) qRT-PCR–based mRNA level analysis of IL23p19, IL6, IL12a, and iNOS (n = 4–6) or by (H) FACS analysis of CD11c+ M1 BMDMs (n = 4). (G and I) BMDMs were treated with IL4/IL13 or vehicle, followed by (G) qRT-PCR–based mRNA level analysis of Mrc1, Ym1, Il10, and Arg1 (n = 4–6) or by (I) FACS analysis of CD206+ M2 BMDMs (n = 4). (J) BMDMs were challenged with LPS/IFNγ. The levels of GRP78, ATF4, cleaved ATF6, spliced XBP1, CHOP, IL1β, and iNOS were detected by Western blot (n = 3). (K) Enhanced aggresome formation in BMDMs from KO mice by flow cytometry analysis (n = 5). Means ± SEM. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MFI, mean fluorescent intensity.
Next, bone marrow–derived macrophages (BMDMs) and Raw264.7 macrophages were treated with LPS and interferon γ (IFNγ) to stimulate M1 macrophage polarization, or treated with IL4 and IL13 to induce M2 macrophage polarization. Because the expression level of TM9SF4 was relatively high in BMDMs but low in Raw264.7 (Figure 8E), we chose BMDMs as the cell model for TM9SF4 gene KO while using Raw264.7 as the cell model for TM9SF4 overexpression.
In BMDM experiments, KO of TM9SF4 increased the LPS/IFNγ-induced M1 polarization, as indicated by qRT-PCR analysis of M1 markers IL23p19, IL6, IL12a, and iNOS (Figure 8F), but decreased the IL4/IL13-induced M2 polarization of BMDMs, as indicated by M2 markers MRC1, Ym1, IL10, and Arg-1 (Figure 8G). In agreement with the qRT-PCR results, FACS analysis also indicated that TM9SF4 KO increased CD11c+ M1 macrophages (Figure 8H), but decreased CD206+ M2 macrophages (Figure 8I).
TM9SF4 knockdown also was found to enhance the LPS/IFNγ-induced ER stress increase in BMDMs, as indicated by ER stress markers GRP78, ATF4, cleaved ATF6, spliced XBP1, CHOP, and COX2, and proinflammatory markers IL1β and iNOS (Figure 8J). Furthermore, TM9SF4 knockdown increased aggresome formation, an indication for accumulation of misfolded proteins (Figure 8K).
On the other hand, overexpression of exogenous TM9SF4 in Raw264.7 cells was found to decrease the LPS/IFNγ-induced M1 polarization (Figure 9A), but increase the IL4/IL13-induced M2 polarization (Figure 9B).
Figure 9.
Overexpression of TM9SF4 decreased M1 macrophage polarization but increased M2 macrophage polarization. (A) Raw264.7 cells overexpressing TM9SF4 or control plasmids were treated with LPS/IFNγ to induce M1 polarization. The mRNA levels of IL23p19, IL6, TNFα, and iNOS were assessed by qRT-PCR (n = 4–6). (B) Raw264.7 cells overexpressing TM9SF4 or control plasmids were treated with IL4/IL13 to induce M2 polarization. The mRNA levels of MRC1, Ym1, IL10, and Arg1 were determined by qRT-PCR (n = 3–6). Means ± SEM. ∗∗P < .01, ∗∗∗P < .001. pcDNA, plasmid cloning DNA.
KO/Knockdown of TM9SF4 Reduced ER Ca2+ Content
We explored whether TM9SF4 can modulate ER Ca2+ content in colonic epithelial cells and macrophages as a means of regulating ER stress, as we reported previously in other cell types.20 HCECs, HT-29 cells, or BMDMs were bathed in Ca2+-free saline and stimulated with adenosine triphosphate (ATP) or ionomycin to induce Ca2+ release from ER Ca2+ stores. Indeed, TM9SF4 knockdown/KO reduced the magnitude of cytosolic Ca2+ response to ATP and ionomycin in HCECs (Figure 10A and B) and BMDMs (Figure 10E and F), indicating that TM9SF4 knockdown reduced the ER Ca2+ content in these cells. Conversely, overexpression of TM9SF4 increased the ER Ca2+ content in response to ATP or ionomycin stimulation in HT-29 cells (Figure 10C and D).
Figure 10.
Knockdown of TM9SF4 depleted ER calcium content in colonic epithelial cells. (A and B) HCECs with Scr-shRNA or TM9SF4-shRNAs were bathed in Ca2+-free physiological saline. Shown are time courses and a data summary of cytosolic calcium change in response to (A) 10 μmol ATP or (B) 5 μmol ionomycin (n = 4–5). Change in cytosolic calcium concentration was monitored with the Fluo-4 AM indicator. (C and D) HT-29 cells were overexpressed with TM9SF4 or pcDNA plasmids as control. Shown are time courses and a data summary of cytosolic calcium change in response to (C) 10 μmol ATP or (D) 5 μmol ionomycin (n = 4). (E and F) BMDMs with Scr-shRNA or TM9SF4-shRNAs were bathed in Ca2+-free physiological saline. Shown are time courses and a data summary of cytosolic calcium change in response to (E) 10 μmol ATP or (F) 5 μmol ionomycin (n = 4). Means ± SEM. ∗P < .05, ∗∗P < .01. pcDNA, plasmid cloning DNA; SCR, scrambled control.
KO/Knockdown of TM9SF4 Caused Cytosolic Acidification and Decreased Cell Surface Targeting of Two Phagocytosis-Related Proteins
We examined whether TM9SF4 can modulate cytosolic pH in colonic epithelial cells and macrophages. Cytosolic pH was measured with pHrodo Red Indicator (Invitrogen, Waltham, MA) by flow cytometry. The results showed that TM9SF4 knockdown/KO induced cytoplasmic acidification in HCECs (Figure 11A) and BMDMs (Figure 11B), as indicated by increased pHrodo intensity.
Figure 11.
TM9SF4 regulated cytosolic pH value and cell surface expression of 2 phagocytosis-related proteins. Cytosolic pH values of (A) HCECs and (B) BMDMs were measured with pHrodo Red Indicator by flow cytometry. Shown are representative flow cytometer experiments and a data summary. (C) Whole-cell lysates of WT or KO BMDMs were collected, and cell surface proteins were isolated using surface biotinylation methods. The total and surface expressions of CD36 and MerTK were measured. ATP1A1 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were the loading control for cell membrane proteins and total cellular proteins, respectively (n = 3). Means ± SEM. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. max, maximum; SCR, scrambled control.
TM9SF4 is reported to regulate cell surface trafficking of glycine-rich transmembrane domains.18,19 Therefore, we also determined the effect of TM9SF4 knockdown on cell surface expression of 2 glycine-rich proteins, CD36 and Mer tyrosine kinase (MerTK), in BMDMs. CD36 contains 5 glycine residues in 1 transmembrane domain and 3 glycine residues in another transmembrane domain, whereas MerTK contains 3 glycine residues in 1 transmembrane domain. CD36 and MerTK both participate in phagocytosis in macrophages.27,28 KO of TM9SF4 reduced the total cellular expression level of CD36 and MerTK, as well as their cell surface expression (Figure 11C). Interestingly, a reduction in cell surface expression of CD36 and MerTK was more than the reduction in total cellular expression (Figure 11C), suggesting that TM9SF4 knockdown decreases the cell surface targeting of these 2 proteins.
TM9SF4 Knockdown Disrupted Intestinal Epithelial Barrier
We explored whether TM9SF4 in epithelial cells could regulate intestinal epithelial barrier function. Caco-2 and HT-29 epithelial cell monolayers were established, followed by measurement of transepithelial electrical resistance. Because TM9SF4 expression was relatively high in Caco-2 but low in HT-29 cells (Figure 3A), Caco-2 was used as the cell model for shRNA-mediated TM9SF4 knockdown while HT-29 was used as the cell model for TM9SF4 overexpression. The results showed that TM9SF4 knockdown in Caco-2 cells decreased the transepithelial resistance of monolayer (Figure 12A) and reduced the expression of tight junction proteins Claudin-1 and zonula occludens-1 (ZO-1) at both mRNA (Figure 12B) and protein levels (Figure 12C). In contrast, overexpression of TM9SF4 in HT-29 cells increased the transepithelial resistance of epithelial monolayer (Figure 12D) and increased the expression of tight junction proteins Claudin-1 and ZO-1 at mRNA (Figure 12E) and protein levels (Figure 12F).
Figure 12.
TM9SF4 knockdown disrupted the intestinal epithelial barrier. (A–C) Caco-2 cells were stably transfected with Scr-shRNA or TM9SF4-shRNAs. Relative transepithelial electrical resistance was measured (A) (n = 5). The levels of claudin-1 and ZO-1 were detected by (B) qRT-PCR and (C) Western blot (n = 3). (D–F) HT-29 cells were overexpressed with TM9SF4 or vector pcDNA6. (D) Relative transepithelial electrical resistance was measured (n = 5). The levels of claudin-1 and ZO-1 were detected by (E) qRT-PCR (n = 3) and (F) Western blot (n = 3). (G–J) Phorbol 12-myristate 13-acetate (PMA)-primed THP-1 cells were treated with LPS, IL1β, and IFNγ, and co-cultured with Caco-2 cells. Shown are the schematic diagram of (G) co-culture study, (H) relative transepithelial resistance (n = 5), (I) expressional level of claudin-1 and ZO-1 by qRT-PCR (n = 3), and (J) Western blot (n = 3). (K) Data summary of serum FITC–dextran concentration in bone marrow–chimeras (n = 6). (L) Immunofluorescence staining of claudin-1 (green) in colon tissue sections from 2% DSS-treated bone marrow–chimeras (n = 6). Blue, Hoechst nuclear stain. Scale bars: 100 μm. (M and N) HCECs were transfected with Scr-shRNA or TM9SF4-shRNA1. Supernatants were collected from IL4/IL13-treated PMA-primed THP-1 cells that were stably expressed with Scr-shRNA or with TM9SF4-shRNA1. The supernatants were added into HCECs. (M) Cell migration was measured by wound healing assays (n = 8–15 per group). (N) Cell proliferation was quantified by Cell Counting Kit-8 (CCK8) from day 0 to day 4 after THP-1 conditional medium-treatment (n = 4). Means ± SEM. ∗∗P < .01, ∗∗∗P < .001. pcDNA, plasmid cloning DNA; SCR, scrambled control.
We next examined whether TM9SF4 in macrophages also could regulate epithelial barrier function. Herein, we established a macrophage–epithelial cell co-culture system using Caco-2 epithelial cells and LPS/IL1β/IFNγ-treated phorbol 12-myristate 13-acetate–primed THP-1 macrophages (Figure 12G). Co-culture with THP-1 cells decreased the transepithelial resistance of Caco-2 monolayer (Figure 12H), which was aggravated by TM9SF4 knockdown in THP-1 cells (Figure 12H). Co-culture with THP-1 cells also decreased the expression of tight junction proteins Claudin-1 and ZO-1 in Caco-2 cells (Figure 12I and J), which was exacerbated by TM9SF4 knockdown in THP-1 cells (Figure 12I and J). These data suggest that TM9SF4 expressed in macrophages could regulate epithelial barrier function in vitro.
To confirm whether macrophages could regulate epithelial barriers in vivo, we used bone marrow transplantation experiments. After induction of colitis, transplantation of KO bone marrow cells to WT mice damaged the intestinal permeability (KO → WT vs WT → WT), as indicated by serum FITC–dextran assay (Figure 12K) and Claudin-1 immunostaining (Figure 12L). In contrast, transplantation of WT bone marrow cells to KO mice (WT → KO vs KO → KO) showed a redemptive effect by decreasing intestinal permeability (Figure 12K and L).
TM9SF4 Knockdown Impaired the Migration and Proliferation of Colonic Epithelial Cells
Migration and proliferation of colonic epithelial cells are correlated positively with epithelial layer regeneration.29 Thus, we also examined the role of TM9SF4 in migration and proliferation of colonic epithelial cells. TM9SF4 knockdown in HCECs reduced epithelial cell migration (Figure 12M) and proliferation (Figure 12N). In addition, HCECs were incubated with conditional media derived from THP-1 cells with or without TM9SF4 knockdown. The results showed that the conditional media from THP-1 cells could rescue the epithelial cell migration (Figure 12M) and proliferation (Figure 12N). However, the conditional media from TM9SF4 gene silenced THP-1 cells only had a limited rescue effect (Figure 12M and N).
Genetic KO of TM9SF4 Aggravated TNBS-Induced IBD
A TNBS-induced model of IBD also was used to verify the role of TM9SF4 in IBD. TNBS treatment caused body weight loss, colon length decrease, and diarrhea, all of which were more severe in KO mice than in WT mice (Figure 13A–D). In control mice fed with vehicle for 4 days, there were no differences in colon length and body weight between WT and KO groups (Figure 13A–C). qRT-PCR analysis of colon tissues showed higher mRNA expression of proinflammatory cytokines TNFα, IL1β, and IL6 in KO mice than in WT mice (Figure 13E). Histologic analysis of colon tissue slides indicated higher histopathologic scores in KO mice (Figure 13F). TM9SF4 KO mice also showed a higher ER stress level in colon tissues as indicated by immunostaining of GRP78 and CHOP (Figure 13G), more cell death as indicated by immunostaining of cleaved caspase 3 (Figure 13G), and more macrophage infiltration in colon tissue (Figure 13G). Together, these data suggested that genetic loss of TM9SF4 aggravated TNBS-induced colitis in mice.
Figure 13.
Knockout of TM9SF4 exacerbated TNBS-induced colitis in mice. (A) Time courses of body weight loss in WT and KO mice injected intrarectally with TNBS or vehicle (50% ethanol in 100 μL). Representative (B) images and (C) data summary of colon lengths in WT and KO mice. (D) Time courses of diarrhea of WT and KO mice. (E) mRNA levels of TNFα, IL1β, and IL6 in colon tissues as assessed by qRT-PCR. (F) Representative images (top) and histopathologic scores (bottom) of H&E-stained colon tissue sections from TNBS-induced WT and KO mice. Scale bars: 100 μm. (G) Expressions of GRP78, CHOP, cleaved caspase 3, and CD68 in colons of TNBS-treated WT or KO mice as detected by immunohistochemistry. Shown are representative images (left) and data summary (right). Brown, immunopositive signals; blue, nuclear counterstain. Scale bars: 100 μm. Means ± SEM. n = 5–6 mice per group in all experiments. ∗P < .05, ∗∗P < .01, and ∗∗∗P < .001. DAB, 3,3′-Diaminobenzidine.
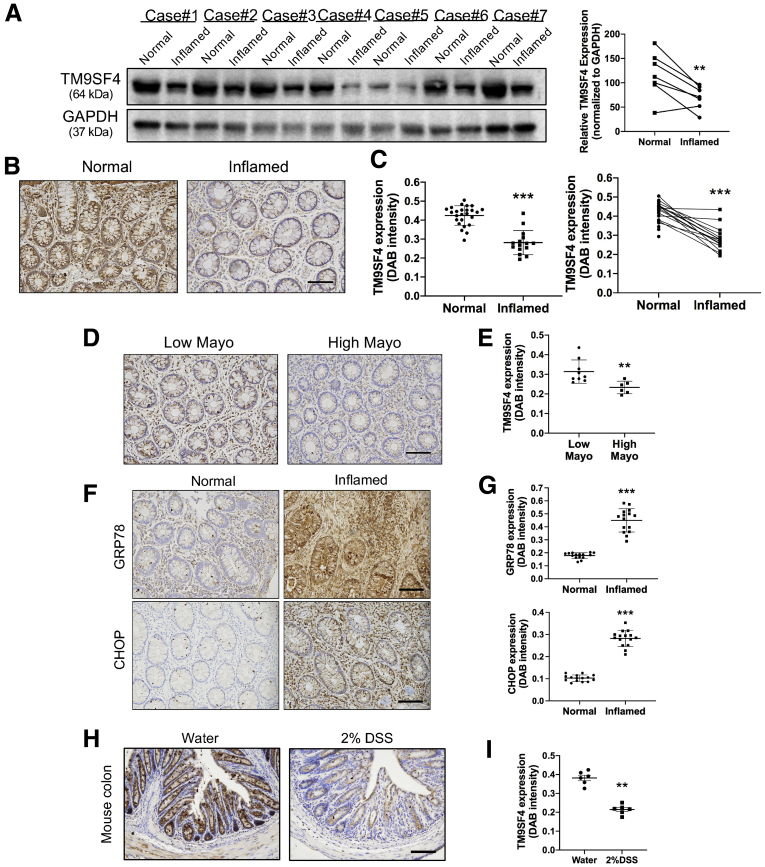
TM9SF4 Expression Was Down-Regulated in Colonic Samples From Patients With UC and Mice With DSS-Induced Colitis
To explore the clinical significance of TM9SF4 in intestinal inflammation, we recruited 25 patients who were diagnosed with UC. The disease severity of UC was assessed with the Mayo score (range, 0 to 3). Among 25 patients, 15 patients showed inflammation in the colon. Interestingly, Western blot (Figure 14A) and tissue section immunostaining (Figure 14B and C) showed abundant expression of TM9SF4 proteins in normal colon tissues. However, the expression of TM9SF4 decreased markedly in the inflamed colonic regions of UC patients (Figure 14A–C). Comparison also was made in inflamed regions of colon between patients with low Mayo scores (Mayo score, 0–1) and high Mayo scores (Mayo score, 2–3). The results showed that the high Mayo score group expressed relative lower TM9SF4 than the low Mayo score group (Figure 14D and E).
Figure 14.
TM9SF4 expression was down-regulated in human UC and mouse experimental colitis. (A) Tissue biopsy specimens from normal and inflamed regions of colons from patients with UC were collected. Levels of TM9SF4 from case 1 to case 7 were determined by Western blot. Representative (B) images and (C) data summary of TM9SF4 immunostaining in normal (n = 25) and inflamed (n = 15) regions of colons from UC patients. Scale bar: 100 μm. Representative (D) images and (E) data summary of TM9SF4 staining in colons from UC patients with low Mayo scores (n = 9) and high Mayo scores (n = 6). Scale bar: 100 μm. Representative immunostaining (F) images and (G) data summary of GRP78 and CHOP in normal (n = 15) and inflamed (n = 15) regions of colons from UC patients. Scale bars: 100 μm. Representative TM9SF4 immunohistochemistry staining (H) images and (I) data summary of colons from WT mice with or without 2% DSS treatment. Scale bar: 100 μm. Means ± SEM. ∗∗P < .01, ∗∗∗P < .001. DAB, 3,3′-Diaminobenzidine; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Furthermore, we found that ER stress level in inflamed colonic regions of UC patients was high as indicated by immunostaining of GRP78 and CHOP (Figure 14F and G), but was extremely low in noninflamed colonic regions (Figure 14F and G).
The expression of TM9SF4 in colonic tissues of DSS-induced IBD mice also was determined. The results showed that TM9SF4 expression was lower in colonic tissue of DSS-induced colitic mice than those of normal mice (Figure 14H and I).
We also determined whether inflammation could regulate the expression of TM9SF4 proteins. Under LPS/IFNγ-induced inflammation, TM9SF4 expression was reduced in colonic epithelial cells (HCECs) and macrophages (BMDMs) (Figure 15A and B). In contrast, under IL4/IL13-induced anti-inflammatory conditions, TM9SF4 expression was increased in macrophages Raw264.7 (Figure 15C).
Figure 15.
The effect of LPS/IFNγ or IL4/IL13 treatment on TM9SF4 expression. (A) LPS-induced decrease of TM9SF4 expression in HCECs as assessed by Western blot. (B) LPS/IFNγ-induced decrease of TM9SF4 expression in BMDMs as assessed by Western blot. (C) IL4/IL13-induced increase of TM9SF4 expression in Raw264.7 cells as assessed by Western blot. Means ± SEM. n = 3. ∗P < .05. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Discussion
Advances in colitis genetics have identified several IBD-associated genes, including CARD15, SLC22A4, SLC22A5, and MDR1.3,30 These genes play functional roles in immunoregulation, maintenance of mucosal barrier integrity, and microbial clearance.30 In the present study, search of a GWAS database found 1 single-nucleotide polymorphism (rs6142618) of TM9SF4 that is highly associated with IBD with a P value of 6 × 1010 (https://www.ebi.ac.uk/gwas/variants/rs6142618).22,23 This prompted us to explore the role of TM9SF4 in colitis. In a DSS-induced mouse model of IBD, we found that KO of TM9SF4 aggravated the disease symptoms of colitis, including severely disrupted crypts in colon tissue sections, aggravated body weight reduction, shortened colon length, worsen anal bleeding score, and leakier intestinal epithelial barrier. Similar results were obtained in a TNBS-induced model of IBD. As such, we provided the experimental evidence for TM9SF4 as a novel IBD-associated gene, the deficiency of which contributes to colitis pathogenesis. Bone marrow reconstitution experiments further confirmed that TM9SF4 deficiency in both bone marrow–derived hematopoietic cells and nonhematopoietic cells (intestinal epithelial cells, endothelial cells, and so forth) contribute to disease progression.
Deficiency of TM9SF4 in IECs may promote colitis progression. In colitis, IECs release proinflammatory cytokines to promote inflammation.31 In the present study, TM9SF4 knockdown was found to enhance the LPS- or PGN-induced inflammation in colonic epithelial cells HT-29 and HCECs. Interestingly, previous studies have shown an association between ER stress/unfolded protein response and inflammation in IECs.32 We have reported that TM9SF4 knockdown may increase ER stress in other cell types.20 Therefore, we next asked the question of whether TM9SF4 may act through ER stress to regulate inflammation in IECs. Indeed, TM9SF4 knockdown boosted ER stress and increased ROS level in colonic cells, whereas overexpression of TM9SF4 had the opposite effect (Figure 3). Importantly, in the presence of 4-PBA, which is a chemical chaperone that can improve ER folding capacity and alleviate ER stress,26 TM9SF4 knockdown could no longer promote inflammation and only barely increase ROS. These data strongly support that TM9SF4 acts through ER stress and ROS production to regulate inflammation in colonic epithelial cells.
Deficiency of TM9SF4 in macrophages also may contribute to colitis. An important function of macrophages is to phagocytose invading pathogens and apoptotic cells (efferocytosis).33,34 Previous studies have reported that TM9SF4 is critical for phagocytosis in Drosophila hemocytes and mammalian cancer cells.13,14,16 In agreement, we also found that TM9SF4 knockdown markedly reduced the phagocytosis of peritoneal macrophages, supporting the critical importance of TM9SF4 in macrophage phagocytosis.
The role of TM9SF4 in macrophages may not be limited to phagocytosis. At the early stage of tissue injury or colonic infection, monocytes are mostly differentiated to proinflammatory M1 macrophages to neutralize invading pathogens, whereas at the late stage, M2 macrophages dominate to resolve inflammation and initiate repair.35 It is well documented that defects in macrophage polarization to M1 or M2 types may contribute to colitis pathogenesis.35,36 We thus explored whether TM9SF4 could affect macrophage polarization to M1 or M2 types. The results showed that TM9SF4 gene KO increased M1 macrophages but decreased M2 macrophages in colonic tissue in vivo. Furthermore, in isolated BMDMs, TM9SF4 gene KO enhanced the LPS/IFNγ-induced M1 macrophage polarization but inhibited the IL4/IL13-induced M2 macrophage polarization. These data indicate a crucial role of TM9SF4 in macrophage polarization.
Disruption of the intestinal epithelial barrier is a hallmark of colitis, which is characterized by induction of epithelial cell apoptosis, altered tight junction protein expression, and increased intestinal permeability.2,3,37 Intestinal inflammation, excessive ER stress, and increased macrophage polarization to M1 rather than M2 types are all documented to promote intestinal barrier dysfunction.2,3,35,38 Therefore, we explored the role of TM9SF4 in intestinal barrier function. The results showed that knockdown of TM9SF4 in either epithelial cells or macrophages each decreased the transepithelial resistance of epithelial monolayers, reduced the expression of tight junction proteins Claudin-1 and ZO-1, and reduced epithelial cell migration and proliferation. Together, these data support that deficiency of TM9SF4 in epithelial cells and macrophages disrupted epithelial barrier function.
Intriguingly, we found that the expression level of TM9SF4 in inflamed colon regions of human UC patients was much lower compared with that of normal colon samples. In addition, the patients with more severe disease (high Mayo score) had lower expression of TM9SF4 compared with those with mild disease (low Mayo score). Similarly, TM9SF4 expression was lower in colonic tissue of DSS-induced IBD mice than those of normal mice. We also found that treatment with LPS/IFNγ, which stimulates inflammation, suppressed the expression of TM9SF4 in colonic epithelial cells (HCECs) and macrophages (BMDMs). Based on these lines of evidence, we hypothesize an existence of a vicious cycle consisting of inflammation → reduced TM9SF4 expression → more severe inflammation. On the other hand, we also found that treatment of BMDMs with anti-inflammatory cytokines IL4 and IL13 greatly enhanced the expression of TM9SF4. Because TM9SF4 has anti-inflammatory actions, we further hypothesized that stimulating the signaling axis of IL4/IL13–TM9SF4 could be a means to reduce inflammation and alleviate IBD symptoms. Further experiments are needed to verify these hypotheses.
Because deficiency of TM9SF4 elicited multiple pathophysiological responses that can aggravate IBD progression. An intriguing next question is: what is the immediate downstream event after TM9SF4 that eventually triggers all other dysregulations? We speculate that ER stress increase could be the key event. It is well documented that an increase in ER stress can promote inflammation,32 enhance M1 macrophage polarization,39,40 and disrupt the epithelial barrier.41 As to the mechanisms of how TM9SF4 deficiency can increase ER stress, we propose 2 mechanisms. First, TM9SF4 deficiency may deplete ER Ca2+ content, causing an ER stress increase; and, second, TM9SF4 deficiency may cause cytoplasmic acidification, subsequently leading to an ER stress increase. Previous studies have established that Ca2+ store depletion20,42 and cytosolic acidification43 can increase ER stress. Our results also clearly showed that TM9SF4 deficiency could deplete intracellular Ca2+ stores and cause cytoplasmic acidification. Therefore, a likely overall scenario is that TM9SF4 deficiency acts through Ca2+ store depletion and cytoplasmic acidification to increase ER stress, subsequently triggering other dysregulation, leading to IBD aggravation. To its support, we also found that ER stress alleviation by 4-PBA greatly reduced the IBD symptoms (Figure 4, Figure 6). However, an additional mechanism also may exist. We found that TM9SF4 deficiency may reduce the cell surface targeting of 2 phagocytosis-related proteins, CD36 and MerTK, in macrophages, which also is expected to affect inflammation.
In summary, TM9SF4 is a key regulatory protein in the maintenance of colonic homeostasis. Deficiency of TM9SF4 in colonic epithelial cells and macrophages increases ER stress, promotes inflammation, and disrupts the intestinal epithelial barrier, consequently aggravating IBD symptoms.
Materials and Methods
Human Tissues
Human tissue experiments were approved by the Joint Chinese University of Hong Kong–New Territories East Cluster Clinical Research Ethics Committee (2019.671). All patients with UC were recruited from the Department of Medicine and Therapeutics, Prince of Wales Hospital, from June 2020 through April 2021. Biopsy samples were obtained from the actively inflamed (n = 15) and/or noninflamed (n = 25) normal regions of colons from patients undergoing a colonoscopy with the patient’s consent. The diagnoses of UC were based on clinical, radiologic, endoscopic, and histologic criteria. The disease severity was assessed with the Mayo Endoscopic subscore according to standard criteria during endoscopy (Table 1).
Table 1.
Clinical Characteristics of Patients With Ulcerative Colitis
| Colonic UC | |
|---|---|
| Patients, n | 25 |
| Age, y | 50.02 ± 14.25 |
| Sex | |
| Male | 10 |
| Female | 15 |
| Disease duration, mo | 124.82 ± 110.15 |
| Current therapy | |
| No treatment | 3 |
| Topical treatment | 5 |
| Mesalamine | 17 |
| Prednisolone | 1 |
| Immunosuppressants | 3 |
| Biologics | 4 |
| Disease extent | |
| E1 | 8 |
| E2 | 7 |
| E3 | 10 |
Mice
Tm9sf4tm1a(KOMP)Wtsi mice were purchased from KOMP Repository, The Jackson Laboratory (Davis, CA). Tm9sf4tm1a(KOMP)Wtsi were created on a C57BL/6N background and were heterozygous for TM9SF4 KO. TM9SF4-/- (KO) mice were generated by crossing heterozygous mice as described elsewhere.21 Standard tail DNA genotyping was used to identify TM9SF4 KO mice. All animal experiments were approved by the Animal Experimentation Ethics Committee, The Chinese University of Hong Kong (20-138-MIS), and performed in compliance with the Guide for the Care and Use of Laboratory Animals published by the US National Institute of Health (publication no. 85-23, revised 1996).
The bone marrow chimeras were generated with age- and sex-matched WT and TM9SF4 KO mice. Briefly, recipients were injected intraperitoneal with clodronate liposomes (Liposoma BV, Amsterdam, Netherlands) and irradiated with 8 Gy (Gammacell 3000; MDS Nordion, Ottawa, Ontario). Bone marrow cells (2 × 106 cells for each recipient) from WT or KO donor mice were harvested and stained with PKH67 dyes (Sigma, Burlington, MA), and then injected intravenously into recipient mice. DSS-induced colitis was induced 6 weeks after bone marrow transplantation.
Mouse Models of Colitis
Male WT and TM9SF4 KO littermates aged 8–10 weeks were used for induction of colitis. For DSS-induced colitis, mice received 2% DSS (molecular weight, 36,000–50,000 kilodaltons; MP Biomedicals, Irvine, CA) in drinking water for 7 days. Body weights and colitis index were monitored daily. In some cases, 4-PBA (MedChemExpress, Monmouth Junction, NJ) dissolved in corn oil was given orally to mice at a dose of 500 mg/kg body weight twice daily during induction of colitis.
For TNBS-induced colitis, mice were anesthetized and received an intrarectal injection of 2.5 mg TNBS in 100 μL 50% ethanol, or 100 μL 50% ethanol alone as control. After infusion, mice were hold in a vertical position for 3 minutes. Body weights and colitis index were monitored daily. On day 4, the mice were killed and colons were excised.
For both DSS- and TNBS- colitis, the severity of colitis was interpreted or scored as follows: body weight loss, colon length on the end day of treatment, stool consistency (score, 0–3: 0, normal; 1, slightly soft stools; 2, loose stools; 3, watery stools), anal bleeding (score, 0–3: 0, normal; 1, minimal bleeding; 2, slight bleeding; 3, gross bleeding). For histologic analysis, colons were harvested on the end day of treatment for paraffin section preparation and stained with H&E. Histopathologic scores were determined as follows: score, 0–4: 0, normal tissues; 1, mild inflammation in the mucosa with some infiltrating mononuclear cells; 2, increased level of inflammation in the mucosa with more infiltrating cells, damaged crypt glands and epithelium, mucin depletion from goblet cells; 3, extensive infiltrating cells in the mucosa and submucosa area, crypt abscesses present with increased mucin depletion and epithelial cell disruption; and 4, massive infiltrating cells in the tissue, complete loss of crypts.
Cell Culture, Lentiviral Infection, and Plasmid Transfection
HEK293T, HT-29, Caco-2, THP-1, and Raw 264.7 cell lines were purchased from the American Type Culture Collection and were maintained under 95% O2 and 5% CO2 in a humidified incubator at 37°C according to instructions. HCECs were a gift from Professor Jerry W. Shay (University of Texas Southwestern Medical Center, Dallas, TX) and were cultured in the conditions as previously reported.25
Lentiviral-based shRNAs were used to create TM9SF4 stable knockdown cell lines under the selection of 2 μg/mL puromycin (Invitrogen, Waltham, MA). TM9SF4–shRNA1 sequence was 5’-GCGGATCACAGAAGACTACTA-3’; TM9SF4–shRNA2 sequence was 5’-CGGTGGTACATGAAC CGATTT-3’. Scrambled-shRNA was used as control.
For TM9SF4 overexpression, TM9SF4 complementary DNA (cDNA) was subcloned in pcDNA6/V5-His A vector (Addgene, Watertown, MA) by EcoRI and XbaI. Plasmids were transfected into cells with Lipofectamine 3000 (Invitrogen, MA) according to the manufacturer’s instructions.
RNA Sequencing Analysis and qRT-PCR
Total RNA from colon tissues or cells was extracted with NucleoSpin RNA Plus (Macherey-Nagel, Allentown, PA). RNA sequencing was performed with the BGISEQ-500 platform by BGI (Wuhan, China). The Gene Ontology and KEGG databases were used to extrapolate differentially expressed pathways in a knowledge base–driven pathway analysis approach. Heatmaps were drawn using GraphPad Prism 9 software (San Diego, CA).
For qRT-PCR analysis, cDNA was synthesized with the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Bedford, MA). Real-time PCR analysis was performed on ABI QuantStudio 7 (QS7) Flex Real Time PCR System (Bedford, MA), using SYBR select mater mix (Applied Biosystems) and following the standard protocol. The gene expression levels were normalized with the housekeeping gene GAPDH and relative expression levels were measured using comparative CT (ΔΔCt) analysis (Table 2).
Table 2.
Primers for qRT-PCR
| Gene | Species | Sequence |
|---|---|---|
| Il1b | Mouse | AAAAAAGCCTCGTGCTGTCG |
| GTCGTTGCTTGGTTCTCCTTG | ||
| Il6 | Mouse | TCCATCCAGTTGCCTTCTTG |
| TTCCACGATTTCCCAGAGAAC | ||
| Tnfa | Mouse | AAGCCTGTAGCCCACGTCGTA |
| AGGTACAACCCATCGGCTGG | ||
| Il1b | Human | AATCTGTACCTGTCCTGCGTGTT |
| TGGGTAATTTTTGGGATCTACACTCT | ||
| Il6 | Human | AGCCCTGAGAAAGGAGACATGTA |
| AGGCAAGTCTCCTCATTGAATCC | ||
| Tnfa | Human | TCTCGAACCCCGAGTGACAA |
| TATCTCTCAGCTCCACGCCA | ||
| Gapdh | Human | CCACCCATGGCAAATTCC |
| TGGGATTTCCATTGATGACAAG | ||
| Gapdh | Mouse | TGTGTCCGTCGTGGATCTGA |
| CCTGCTTCACCACCTTCTTGAT | ||
| Mfge8 | Mouse | GGACATCTTCACCGAATACATCTGC |
| TGATACCCGCATCTTCCGCAG | ||
| Cd36 | Mouse | TCGGAACTGTGGGCTCATTG |
| CCTCGGGGTCCTGAGTTATATTTTC | ||
| Gas6 | Mouse | TCTTCTCACACTGTGCTGTTGCG |
| GGTCAGGCAAGTTCTGAACACAT | ||
| Il23p19 | Mouse | CATGCTAGCCTGGAACGCACAT |
| ACTGGCTGTTGTCCTTGAGTCC | ||
| Il11 | Mouse | CTGACGGAGATCACAGTCTGGA |
| CAGCTTGTACCAGAAGCAAGGG | ||
| Mrc1 | Mouse | ACAACAGACAGGAGGACTGCGT |
| AACCCATGCCGTTTCCAGCCTT | ||
| Ym1 | Mouse | CAGGGTAATGAGTGGGTTGG |
| AAGTAGATGTCAGAGGGAAATGTC | ||
| Il12a | Mouse | CACAAGAACGAGAGTTGCCTGGCTACTA |
| TAAGGGTCTGCTTCTCCCACAGGAGGTT | ||
| Il10 | Mouse | TGGCCCAGAAATCAAGGAGC |
| CAGCAGACTCAATACACACT | ||
| Tm9sf4 | Mouse | CTGGAGTCGCGCCAATCAAT |
| GGCAGAAGGGCAATGAGTAGT | ||
| Tm9sf4 | Human | GATTGGTTGCCGTGGTCTTTA |
| TTCTACGGGATCGTTCTGGTG | ||
| Zo1 | Human | CAACATACAGTGACGCTTCACA |
| CACTATTGACGTTTCCCCACTC | ||
| Cldn1 | Human | CCTCCTGGGAGTGATAGCAAT |
| GGCAACTAAAATAGCCAGACCT | ||
| Inos | Mouse | AGGGACAAGCCTACCCCTC |
| CTCATCTCCCGTCAGTTGGT | ||
| Arg1 | Mouse | CCCTGGGGAACACTACATTTTG |
| GCCAATTCCTAGTCTGTCCACTT |
Immunoblotting
Cells were lysed in radioimmunoprecipitation assay lysis buffer supplemented with 1 mmol/L phenylmethylsulfonyl fluoride. Cell plasma membrane was labeled with sulfo-NHS-SS-biotin, and plasma membrane proteins were isolated with Pierce Cell Surface Protein Biotinylation and Isolation Kit (ThermoFisher, Waltham, MA). Cell lysates were separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA). The membranes were immunoblotted with primary antibodies at 4°C overnight and incubated with secondary antibodies at room temperature for 1 hour. The blots were visualized with ECL substrates (GE Healthcare, Chicago, IL) using the ChemiDoc XRS + system (Bio-Rad). The following antibodies were used: TM9SF4 (25595-1-AP; Proteintech, Rosemont, IL), GRP78 (11587-1-AP; Proteintech), ATF4 (10835-1-AP; Proteintech), ATF6 (24169-1-AP; Proteintech), XBP1 (25997-1-AP; Proteintech), CHOP (15204-1-AP; Proteintech), COX2 (12282; Cell Signaling Technology, Danvers, MA), cleaved caspase 3 (9664; Cell Signaling Technology), claudin-1 (13050-1-AP; Proteintech), ZO-1 (8193; Cell Signaling Technology), glyceraldehyde-3-phosphate dehydrogenase (97166; Cell Signaling Technology), tubulin (10068-1-AP; Proteintech), β-actin (66009-1-Ig; Proteintech), MerTK (DF7344; Affinity Biosciences, Melbourne, Australia), and CD36 (18836-1-AP; Proteintech).
Immunostaining
For immunohistochemistry staining of human and mouse colon sections, paraffin sections were heated for antigen retrieval in citrate solutions, blocked with 5% bovine serum albumin for 30 minutes, incubated with primary antibodies at 4°C overnight, and horseradish peroxidase–conjugated secondary antibodies at 37°C for 1 hour. Immunosignals were reacted with 3,3’-diaminobenzidine and sections were finally counterstained with hematoxylin. Pictures were taken under the microscope.
For immunofluorescence staining, tissue cryosections were washed in phosphate-buffered saline (PBS) 3 times to remove optimal cutting temperature (OCT) gels, blocked with 5% bovine serum albumin supplemented with 0.01% Triton-X-100 (Sigma, Burlington, MA) for 2 hours, followed by staining with primary antibodies at 4°C overnight, and fluorescence-conjugated secondary antibodies at room temperature for 2 hours. Images were acquired under a confocal microscope. The following antibodies were used: TM9SF4 (25595-1-AP; Proteintech), GRP78 (11587-1-AP; Proteintech), CHOP (15204-1-AP; Proteintech), cleaved caspase 3 (9664; Cell Signaling Technology), claudin-1 (13050-1-AP; Proteintech), CD68 (28058-1-AP; Proteintech), CD206 (60143-1-Ig; Proteintech), and CD80 (66406-1-Ig; Proteintech).
Measurement of ER Ca2+ Release
Cells were loaded with Ca2+-sensitive fluorescence dye 10 μmol/L Fluo-4 AM (Invitrogen) and 0.02% Pluronic F-127 at 37°C for 30 minutes in the dark. To measure the Ca2+ release from ER Ca2+ stores, the cells were incubated in 0Ca2+ (calcium-free) physiological saline solutions and challenged with 10 μmol ATP or 5 μmol ionomycin. 0Ca2+-PSS contained in 140 mmol/L NaCl, 5 mmol/L KCl, 1 mmol/L MgCl2, 10 mmol/L glucose, 0.2 mmol/L ethylene glycol-bis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid, and 5 mmol/L HEPES, pH 7.4. Fluorescence intensity was recorded by an Olympus FV1000 confocal microscope (Tokyo, Japan) and the signals relative to the starting signal (F1/F0) were calculated to quantify the cytosolic Ca2+ change.
ROS Measurement
The cell-permeable fluorescent dye DHE (Beyotime Biotechnology, Shanghai, China) was used to measure ROS levels in vitro and in vivo. Briefly, OCT-embedded freshly isolated colon tissues were cut into cryosections. Tissue sections or live cells were washed with PBS and stained with DHE dye according to the manufacturer’s instructions. DHE fluorescent images were taken under a confocal microscope.
Intestinal Permeability Assay
The DSS-challenged mice were deprived of food and water for 3 hours before experiments. FITC–dextran (molecular weight, 40,000kDa) was gavaged into mice at a dose of 0.6 mg/g body weight in 100 μL PBS. Four hours later, mice were killed. Serum FITC concentration was measured with a standard curve using a SpectraMax platereader (Silicon Valley, CA).
In Vitro Transepithelial Resistance Measurement
Transepithelial resistance (TER) of Caco-2 cells or HT-29 cells was measured as previously described.3 Monolayers were cultured on Transwell–collagen membranes (culture area, 0.2 cm2) for 2 weeks to reach confluence. To measure the TER, the monolayers were mounted in an Ussing chamber and perfused at 37°C with normal bicarbonate–buffered Krebs–Henseleit solution, which contained 117 mmol/L NaCl, 25 mmol/L NaHCO3, 4.7 mmol/L KCl, 1.2 mmol/L MgCl2, 1.2 mmol/L KH2PO4, 2.5 mmol/L CaCl2, and 11 mmol/L D-glucose, pH 7.4, with continuous bubbling of 95% O2 and 5% CO2. The TER was measured using a voltage clamp amplifier (MC6; Physiologic Instruments, Reno, NV). A voltage pulse of 2 mV was applied periodically, and the average change in current was used to calculate the TER according to Ohm's law.
Isolation of Murine Colonic Lamina Propria Mononuclear Cells and Magnetic Activated Cell Sorting
Colons were isolated, opened longitudinally, and cut into approximately 0.5-cm pieces. The pieces were incubated with Hank’s balanced salt solution buffer supplemented with 5% fetal bovine serum (FBS), 10 mmol/L HEPES, and 5 mmol/L EDTA at 37ºC in a horizontal shaker to remove intestinal epithelium. The remaining tissues were washed and digested with Hank’s balanced salt solution containing 1 mg/mL collagenase D and 0.1 mg/mL DNAse for 20 minutes at 37ºC. The suspensions were filtered through 70-μm nylon mesh and centrifuged for pellets. After centrifugation, the supernatant was discarded and the pellets were resuspended in 30% Percoll (GE Healthcare), overlaid on 80% Percoll, and centrifuged at 2000 rpm for 20 minutes at 22ºC. Lamina propria mononuclear cells from the white interface were collected and washed twice for FACS analysis.
Lamina propria mononuclear cells were incubated with CD11b microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer’s instructions. The CD11b+ cells were collected for Western blot and qRT-PCR analysis.
Flow Cytometry
IEC cell lines or digested cell suspensions were washed with FACS buffer (PBS containing 5% FBS and 2 mmol/L EDTA) and centrifuged. Cell pellets were incubated with fragment crystallizable (Fc) receptor blocker (CD16/32; BioLegend, San Diego, CA) at 4ºC for 15 minutes and stained with antibodies or the matching isotype control at 4ºC for 25 minutes in the dark. For staining of intracellular markers, the cells were incubated with antibodies against surface markers, fixed, and permeated with Perm/Wash Buffer (BD Biosciences, Franklin Lakes, NJ) before intracellular staining. The following antibodies were used: CD45 (Brilliant Violet 711, 30-F11; BioLegend), CD11b (allophycocyanin, M1/70; BioLegend), F4/80 (phycoerythrin, BM8; BioLegend), CD11c (PE, N418; BioLegend), CD206 (Alexa Fluor 488, C068C2; BioLegend), MHCII (FITC, M5/114.15.2; BioLegend) CD4 (FITC, GK1.5; BioLegend), CD8 (PE/Cy5, SK1; BioLegend), and Gr-1 (PE/Cy7, RB6-8C5; BioLegend). Analysis or cell sorting was performed with the Aria Fusion Cell sorter and Cell Analyzer (BD Biosciences). The acquired data were analyzed by FlowJo software.
Primary Culture of Peritoneal Macrophages and BMDMs
To generate peritoneal macrophages, 10- to 12-week-old mice were injected intraperitoneally with 1 mL 3.5% Brewer’s thioglycollate (Sigma-Aldrich). After 3 days, peritoneal macrophages were isolated from peritonitis exudates by peritoneal lavage with 5 mL ice-cold RPMI 1640 medium (Gibco, Waltham, MA). The peritoneal macrophages were allowed to rest overnight in RPMI 1640 supplemented with 10% FBS at 37ºC in a 5% CO2 incubator before the start of experiments.
To generate BMDMs, tibias and femurs from 6- to 8-week-old mice were harvested. Bone marrows were flushed and passed through 70-μm mesh, resuspended in Dulbecco’s modified Eagle medium (Gibco), and overlaid on Ficoll-Paque Plus (GE Healthcare). The mixtures were centrifuged at 1800 rpm for 20 minutes at 22ºC. BMDMs from the interface of the red Dulbecco’s modified Eagle medium layer and the colorless clear layer were collected and cultured in high-glucose Dulbecco’s modified Eagle medium supplemented with 10% FBS, 2 mmol/L L-glutamine (Gibco), 0.5% 2-mercaptoethanol (Gibco), and 10 ng/mL recombinant murine macrophage colony stimulating factor (Peprotech, Cranbury, NJ) for 7 days before experiments.
Phagocytosis Assay
In vitro phagocytosis was measured with mouse peritoneal macrophages. Jurkat cells were treated with staurosporine (Sigma) to induce apoptosis and stained with CFSE dye (Invitrogen). Phagocytosis was induced by incubating peritoneal macrophages with CFSE-labeled apoptotic cells at a ratio of 1:5 in RPMI1640 for 1 hour at 37ºC. After incubation, adherent macrophages were detached and stained with PE-conjugated F4/80 for flow cytometry or confocal microscopy analysis.
Intracellular pH Measurement
Cells were digested and stained with pHrodo AM Ester (Invitrogen) for 30 minutes. The cytosolic pH values were evaluated using a LSRFortessa Cell Analyzer (BD Biosciences) with excitation/emission of 560/585 nm.
Wound Healing Assay
Two-well culture-insert (ibidi, Gräfelfing, Germany) was used for the wound healing assay. Briefly, HCECs were pre-incubated with conditional medium from IL4/IL13-induced THP-1 macrophages or control medium for 72 hours. Cells (2 × 105) were added into each well of the insert. Inserts were removed after appropriate cell attachment. The cells were fixed and stained with crystal violet at 0, 12, and 24 hours. The fields were selected randomly under an IX83 Inverted Microscope (Olympus) and wound healing was assessed by Fiji ImageJ software (National Institutes of Health, Bethesda, MD).
Cell Viability Assay
Cell growth was detected by a commercial cell counting kit-8 (CCK-8; Beyotime). Cells were seeded at a density of 2 × 103 per well in the 96-well plates and cultured at 37°C. Cell counts at different indicated times were determined with CCK-8 under a microplate reader following the manufacturer’s instructions.
Co-culture of THP-1 and Caco-2 Cells
Differentiation of THP-1 macrophages was induced with 100 ng/mL phorbol 12-myristate 13-acetate (Sigma) for 48 hours at 37°C. Co-culture of THP-1 and Caco-2 cells was conducted using Transwell chambers with a 0.4-μm Pore Polycarbonate Membrane (Corning, Corning, NY). THP-1 cells with a mixture of IL1β, IL18, and LPS were added to the upper chamber, while Caco-2 cells at 80% confluence were grown on the basal side. After 72 hours, Caco-2 cells were used for TER measurement or gene expression analysis.
Statistical Analysis
Data from at least 3 independent experiments were calculated with GraphPad Prism 9. All data are presented as the means ± SEM and Student t test, 1-way analysis of variance or 2-way analysis of variance was used for comparison between groups.
Acknowledgment
The authors wish to thank Professor Jerry W. Shay for kindly gifting HCECs.
CRediT Authorship Contributions
Mingxu Xie, PhD (Data curation: Lead; Formal analysis: Lead; Investigation: Lead; Methodology: Lead; Project administration: Lead; Writing – original draft: Equal)
Joyce Wing Yan Mak, MBBS, MRCP (UK), FHKCP, FHKAM (Medicine) (Conceptualization: Lead; Data curation: Lead; Funding acquisition: Lead; Investigation: Equal; Project administration: Equal; Validation: Equal; Writing – review & editing: Equal)
Hongyan Yu, PhD (Investigation: Equal; Resources: Equal)
Cherry Tsz Yan Cheng, BSc (Investigation: Equal)
Heyson Chi Hey Chan, MBBS (Investigation: Equal)
Ting Ting Chan, MBBS (Investigation: Equal)
Louis Ho Shing Lau, MBBS (Investigation: Equal)
Marc Ting Long Wong, MBBS (Investigation: Equal)
Wing-Hung Ko, PhD (Funding acquisition: Lead; Methodology: Equal; Resources: Equal)
Liwen Jiang, PhD (Funding acquisition: Lead; Methodology: Equal; Resources: Equal)
Xiaoqiang Yao, PhD (Conceptualization: Lead; Data curation: Equal; Funding acquisition: Lead; Methodology: Equal; Project administration: Lead; Supervision: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by the Hong Kong Research Grant Committee (AoE/M-05/12, 14100619, and RIF/R4005-18F), Hong Kong Health and Medical Research Fund (06170176), and The National Natural Science Foundation of China (82102973).
Contributor Information
Mingxu Xie, Email: xmx0606@126.com.
Joyce Wing Yan Mak, Email: wingyanmak@cuhk.edu.hk.
Xiaoqiang Yao, Email: yao2068@cuhk.edu.hk.
References
- 1.Kobayashi T., Siegmund B., Le Berre C., Wei S.C., Ferrante M., Shen B., Bernstein C.N., Danese S., Peyrin-Biroulet L., Hibi T. Ulcerative colitis. Nat Rev Dis Primers. 2020;6:74. doi: 10.1038/s41572-020-0205-x. [DOI] [PubMed] [Google Scholar]
- 2.Roda G., Chien Ng S., Kotze P.G., Argollo M., Panaccione R., Spinelli A., Kaser A., Peyrin-Biroulet L., Danese S. Crohn’s disease. Nat Rev Dis Primers. 2020;6:22. doi: 10.1038/s41572-020-0156-2. [DOI] [PubMed] [Google Scholar]
- 3.Eichele D.D., Kharbanda K.K. Dextran sodium sulfate colitis murine model: an indispensable tool for advancing our understanding of inflammatory bowel diseases pathogenesis. World J Gastroenterol. 2017;23:6016. doi: 10.3748/wjg.v23.i33.6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melgar S., Karlsson A., Michaëlsson E. Acute colitis induced by dextran sulfate sodium progresses to chronicity in C57BL/6 but not in BALB/c mice: correlation between symptoms and inflammation. Am J Physiol Gastrointest Liver Physiol. 2005;288:G1328–G1338. doi: 10.1152/ajpgi.00467.2004. [DOI] [PubMed] [Google Scholar]
- 5.Antoniou E., Margonis G.A., Angelou A., Pikouli A., Argiri P., Karavokyros I., Papalois A., Pikoulis E. The TNBS-induced colitis animal model: an overview. Ann Med Surg (Lond) 2016;11:9–15. doi: 10.1016/j.amsu.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones G.-R., Bain C.C., Fenton T.M., Kelly A., Brown S.L., Ivens A.C., Travis M.A., Cook P.C., MacDonald A.S. Dynamics of colon monocyte and macrophage activation during colitis. Front Immunol. 2018;9:2764. doi: 10.3389/fimmu.2018.02764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turner J.R. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 8.Isidro R.A., Appleyard C.B. Colonic macrophage polarization in homeostasis, inflammation, and cancer. Am J Physiol Gastrointest Liver Physiol. 2016;311:G59–G73. doi: 10.1152/ajpgi.00123.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao S.S. Epithelial ER stress in Crohn's disease and ulcerative colitis. Inflamm Bowel Dis. 2016;22:984–993. doi: 10.1097/MIB.0000000000000660. [DOI] [PubMed] [Google Scholar]
- 10.Dandekar A., Mendez R., Zhang K. Cross talk between ER stress, oxidative stress, and inflammation in health and disease. Methods Mol Biol. 2015;1292:205–214. doi: 10.1007/978-1-4939-2522-3_15. [DOI] [PubMed] [Google Scholar]
- 11.Park J., Park Y., Ryu I., Choi M.H., Lee H.J., Oh N., Kim K., Kim K.M., Choe J., Lee C., Baik J.H., Kim Y.K. Misfolded polypeptides are selectively recognized and transported toward aggresomes by a CED complex. Nat Commun. 2017;8:15730. doi: 10.1038/ncomms15730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benghezal M., Cornillon S., Gebbie L., Alibaud L., Brückert F., Letourneur F., Cosson P. Synergistic control of cellular adhesion by transmembrane 9 proteins. Mol Biol Cell. 2003;14:2890–2899. doi: 10.1091/mbc.E02-11-0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bergeret E., Perrin J., Williams M., Grunwald D., Engel E., Thevenon D., Taillebourg E., Bruckert F., Cosson P., Fauvarque M.O. TM9SF4 is required for Drosophila cellular immunity via cell adhesion and phagocytosis. J Cell Sci. 2008;121:3325–3334. doi: 10.1242/jcs.030163. [DOI] [PubMed] [Google Scholar]
- 14.Fais S., Fauvarque M.O. TM9 and cannibalism: how to learn more about cancer by studying amoebae and invertebrates. Trends Mol Med. 2012;18:4–5. doi: 10.1016/j.molmed.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 15.Paolillo R., Spinello I., Quaranta M.T., Pasquini L., Pelosi E., Lo Coco F., Testa U., Labbaye C. Human TM9SF4 is a new gene down-regulated by hypoxia and involved in cell adhesion of leukemic cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0126968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lozupone F., Perdicchio M., Brambilla D., Borghi M., Meschini S., Barca S., Marino M.L., Logozzi M., Federici C., Iessi E., de Milito A., Fais S. The human homologue of Dictyostelium discoideum phg1A is expressed by human metastatic melanoma cells. EMBO Rep. 2009;10:1348–1354. doi: 10.1038/embor.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lozupone F., Borghi M., Marzoli F., Azzarito T., Matarrese P., Iessi E., Venturi G., Meschini S., Canitano A., Bona R., Cara A., Fais S. TM9SF4 is a novel V-ATPase-interacting protein that modulates tumor pH alterations associated with drug resistance and invasiveness of colon cancer cells. Oncogene. 2015;34:5163–5174. doi: 10.1038/onc.2014.437. [DOI] [PubMed] [Google Scholar]
- 18.Perrin J., Le Coadic M., Vernay A., Dias M., Gopaldass N., Ouertatani-Sakouhi H., Cosson P. TM9 family proteins control surface targeting of glycine-rich transmembrane domains. J Cell Sci. 2015;128:2269–2277. doi: 10.1242/jcs.164848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vernay A., Lamrabet O., Perrin J., Cosson P. TM9SF4 levels determine sorting of transmembrane domains in the early secretory pathway. J Cell Sci. 2018;131:jcs220830. doi: 10.1242/jcs.220830. [DOI] [PubMed] [Google Scholar]
- 20.Zhu Y., Xie M., Meng Z., Leung L.K., Chan F.L., Hu X., Chi K., Liu C., Yao X. Knockdown of TM9SF4 boosts ER stress to trigger cell death of chemoresistant breast cancer cells. Oncogene. 2019;38:5778–5791. doi: 10.1038/s41388-019-0846-y. [DOI] [PubMed] [Google Scholar]
- 21.Sun L., Meng Z., Zhu Y., Lu J., Li Z., Zhao Q., Huang Y., Jiang L., Yao X. TM9SF4 is a novel factor promoting autophagic flux under amino acid starvation. Cell Death Differ. 2018;25:368–379. doi: 10.1038/cdd.2017.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A., Essers J., Mitrovic M., Ning K., Cleynen I., Theatre E., Spain S.L., Raychaudhuri S., Goyette P., Wei Z., Abraham C., Achkar J.P., Ahmad T., Amininejad L., Ananthakrishnan A.N., Andersen V., Andrews J.M., Baidoo L., Balschun T., Bampton P.A., Bitton A., Boucher G., Brand S., Büning C., Cohain A., Cichon S., D'Amato M., De Jong D., Devaney K.L., Dubinsky M., Edwards C., Ellinghaus D., Ferguson L.R., Franchimont D., Fransen K., Gearry R., Georges M., Gieger C., Glas J., Haritunians T., Hart A., Hawkey C., Hedl M., Hu X., Karlsen T.H., Kupcinskas L., Kugathasan S., Latiano A., Laukens D., Lawrance I.C., Lees C.W., Louis E., Mahy G., Mansfield J., Morgan A.R., Mowat C., Newman W., Palmieri O., Ponsioen C.Y., Potocnik U., Prescott N.J., Regueiro M., Rotter J.I., Russell R.K., Sanderson J.D., Sans M., Satsangi J., Schreiber S., Simms L.A., Sventoraityte J., Targan S.R., Taylor K.D., Tremelling M., Verspaget H.W., De Vos M., Wijmenga C., Wilson D.C., Winkelmann J., Xavier R.J., Zeissig S., Zhang B., Zhang C.K., Zhao H., International IBD Genetics Consortium (IIBDGC) Silverberg M.S., Annese V., Hakonarson H., Brant S.R., Radford-Smith G., Mathew C.G., Rioux J.D., Schadt E.E., Daly M.J., Franke A., Parkes M., Vermeire S., Barrett J.C., Cho J.H. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Lange K.M., Moutsianas L., Lee J.C., Lamb C.A., Luo Y., Kennedy N.A., Jostins L., Rice D.L., Gutierrez-Achury J., Ji S.G., Heap G., Nimmo E.R., Edwards C., Henderson P., Mowat C., Sanderson J., Satsangi J., Simmons A., Wilson D.C., Tremelling M., Hart A., Mathew C.G., Newman W.G., Parkes M., Lees C.W., Uhlig H., Hawkey C., Prescott N.J., Ahmad T., Mansfield J.C., Anderson C.A., Barrett J.C. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mizoguchi A. Animal models of inflammatory bowel disease. Progress Mol Biol Transl Sci. 2012;105:263–320. doi: 10.1016/B978-0-12-394596-9.00009-3. [DOI] [PubMed] [Google Scholar]
- 25.Roig A.I., Eskiocak U., Hight S.K., Kim S.B., Delgado O., Souza R.F., Spechler S.J., Wright W.E., Shay J.W. Immortalized epithelial cells derived from human colon biopsies express stem cell markers and differentiate in vitro. Gastroenterology. 2010;138:1012–1021.e1–5. doi: 10.1053/j.gastro.2009.11.052. [DOI] [PubMed] [Google Scholar]
- 26.Welch W.J., Brown C.R. Influence of molecular and chemical chaperones on protein folding. Cell Stress Chaperones. 1996;1:109. doi: 10.1379/1466-1268(1996)001<0109:iomacc>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Greenberg M.E., Sun M., Zhang R., Febbraio M., Silverstein R., Hazen S.L. Oxidized phosphatidylserine-CD36 interactions play an essential role in macrophage-dependent phagocytosis of apoptotic cells. J Exp Med. 2006;203:2613–2625. doi: 10.1084/jem.20060370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemke G. Biology of the TAM receptors. Cold Spring Harb Perspect Biol. 2013;5:a009076. doi: 10.1101/cshperspect.a009076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taniguchi K., Wu L.W., Grivennikov S.I., de Jong P.R., Lian I., Yu F.X., Wang K., Ho S.B., Boland B.S., Chang J.T., Sandborn W.J., Hardiman G., Raz E., Maehara Y., Yoshimura A., Zucman-Rossi J., Guan K.L., Karin M. A gp130–Src–YAP module links inflammation to epithelial regeneration. Nature. 2015;519:57–62. doi: 10.1038/nature14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sartor R.B. Mechanisms of disease: pathogenesis of Crohn's disease and ulcerative colitis. Nat Clin Pract Gastroenterol Hepatol. 2006;3:390–407. doi: 10.1038/ncpgasthep0528. [DOI] [PubMed] [Google Scholar]
- 31.Blumberg R., Li L., Nusrat A., Parkos C.A., Rubin D.C., Carrington J.L. Recent insights into the integration of the intestinal epithelium within the mucosal environment in health and disease. Mucosal Immunol. 2008;1:330–334. doi: 10.1038/mi.2008.29. [DOI] [PubMed] [Google Scholar]
- 32.Kaser A., Blumberg R.S. Endoplasmic reticulum stress and intestinal inflammation. Mucosal Immunol. 2010;3:11–16. doi: 10.1038/mi.2009.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Z., Li Q., Wang X., Jiang X., Zhao D., Lin X., He F., Tang L. C-type lectin receptor LSECtin-mediated apoptotic cell clearance by macrophages directs intestinal repair in experimental colitis. Proc Natl Acad Sci U S A. 2018;115:11054–11059. doi: 10.1073/pnas.1804094115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo B., Gan W., Liu Z., Shen Z., Wang J., Shi R., Liu Y., Liu Y., Jiang M., Zhang Z., Wu Y. Erythropoeitin signaling in macrophages promotes dying cell clearance and immune tolerance. Immunity. 2016;44:287–302. doi: 10.1016/j.immuni.2016.01.002. [DOI] [PubMed] [Google Scholar]
- 35.Na Y.R., Stakenborg M., Seok S.H., Matteoli G. Macrophages in intestinal inflammation and resolution: a potential therapeutic target in IBD. Nat Rev Gastroenterol Hepatol. 2019;16:531–543. doi: 10.1038/s41575-019-0172-4. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Y., Li X., Luo Z., Ma L., Zhu S., Wang Z., Wen J., Cheng S., Gu W., Lian Q., Zhao X., Fan W., Ling Z., Ye J., Zheng S., Li D., Wang H., Liu J., Sun B. ECM1 is an essential factor for the determination of M1 macrophage polarization in IBD in response to LPS stimulation. Proc Natl Acad Sci U S A. 2020;117:3083–3092. doi: 10.1073/pnas.1912774117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee Y., Sugihara K., Gillilland M.G., 3rd, Jon S., Kamada N., Moon J.J. Hyaluronic acid–bilirubin nanomedicine for targeted modulation of dysregulated intestinal barrier, microbiome and immune responses in colitis. Nat Mater. 2020;19:118–126. doi: 10.1038/s41563-019-0462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lissner D., Schumann M., Batra A., Kredel L.I., Kühl A.A., Erben U., May C., Schulzke J.D., Siegmund B. Monocyte and M1 macrophage-induced barrier defect contributes to chronic intestinal inflammation in IBD. Inflamm Bowel Dis. 2015;21:1297–1305. doi: 10.1097/MIB.0000000000000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang F., Liu Y., Ren H., Zhou G., Yuan X., Shi X. ER-stress regulates macrophage polarization through pancreatic EIF-2alpha kinase. Cell Immunol. 2019;336:40–47. doi: 10.1016/j.cellimm.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 40.Zhou Y., Dong B., Kim K.H., Choi S., Sun Z., Wu N., Wu Y., Scott J., Moore D.D. Vitamin D receptor activation in liver macrophages protects against hepatic endoplasmic reticulum stress in mice. Hepatology. 2020;71:1453–1466. doi: 10.1002/hep.30887. [DOI] [PubMed] [Google Scholar]
- 41.Akiyama T., Oishi K., Wullaert A. Bifidobacteria prevent tunicamycin-induced endoplasmic reticulum stress and subsequent barrier disruption in human intestinal epithelial Caco-2 monolayers. PloS One. 2016;11 doi: 10.1371/journal.pone.0162448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mekahli D., Bultynck G., Parys J.B., De Smedt H., Missiaen L. Endoplasmic-reticulum calcium depletion and disease. Cold Spring Harb Perspect Biol. 2011;3:a004317. doi: 10.1101/cshperspect.a004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Melnik S., Dvornikov D., Müller-Decker K., Depner S., Stannek P., Meister M., Warth A., Thomas M., Muley T., Risch A., Plass C., Klingmüller U., Niehrs C., Glinka A. Cancer cell specific inhibition of Wnt/β-catenin signaling by forced intracellular acidification. Cell Discov. 2018;4:37. doi: 10.1038/s41421-018-0033-2. [DOI] [PMC free article] [PubMed] [Google Scholar]