Abstract
The ubiquitin–proteasome system fulfills an essential role in regulating protein homeostasis by spatially and temporally controlling proteolysis in an ATP- and ubiquitin-dependent manner. However, the localization of proteasomes is highly variable under diverse cellular conditions. In yeast, newly synthesized proteasomes are primarily localized to the nucleus during cell proliferation. Yeast proteasomes are transported into the nucleus through the nuclear pore either as immature subcomplexes or as mature enzymes via adapter proteins Sts1 and Blm10, while in mammalian cells, postmitotic uptake of proteasomes into the nucleus is mediated by AKIRIN2, an adapter protein essentially required for nuclear protein degradation. Stressful growth conditions and the reversible halt of proliferation, that is quiescence, are associated with a decline in ATP and the reorganization of proteasome localization. Cellular stress leads to proteasome accumulation in membraneless granules either in the nucleus or in the cytoplasm. In quiescence, yeast proteasomes are sequestered in an ubiquitin-dependent manner into motile and reversible proteasome storage granules in the cytoplasm. In cancer cells, upon amino acid deprivation, heat shock, osmotic stress, oxidative stress, or the inhibition of either proteasome activity or nuclear export, reversible proteasome foci containing polyubiquitinated substrates are formed by liquid–liquid phase separation in the nucleus. In this review, we summarize recent literature revealing new links between nuclear transport, ubiquitin signaling, and the intracellular organization of proteasomes during cellular stress conditions.
Keywords: protein quality control, ubiquitin–proteasome system, nuclear transport, proteasome foci, proteasome storage granule, liquid–liquid phase separation, stress response
Abbreviations: CP, core particle; DUB, deubiquitinase; ER, endoplasmic reticulum; IDR, intrinsically disordered region; LLPS, liquid–liquid phase separation; NLS, nuclear localization sequence; PQC, protein quality compartment; PSG, proteasome storage granule; RP, regulatory particle
In 2004, the Nobel Prize in Chemistry was awarded to Aaron Ciechanover, Avram Hershko, and Irwin Rose for the discovery of the ubiquitin–proteasome system (UPS) as an essential pathway to maintain protein homeostasis in eukaryotic cells. The proteasome is the key protease complex of the UPS and is responsible for the selective degradation of short-lived proteins in an ATP- and ubiquitin-dependent manner (1). Well-known examples of proteasomal substrates are regulatory proteins, which control cell cycle progression and gene expression in the nucleus (2). Particularly under stress conditions, aberrant proteins accumulate as proteasomal substrates in the nucleus and cytoplasm. If not eliminated, proteasomal substrates tend to form soluble protein condensates or insoluble protein inclusion bodies. Temporary condensates dissolve upon stress relief, while protein inclusions are irreversible and can cause age-related neurodegenerative disorders (1, 3).
In this review, we will focus primarily on notable work in yeast, an excellent model organism to study mechanistic details of conserved pathways in the UPS. In addition, recent breakthroughs and implications for the mammalian system as well as differences between proteasome localizations in yeast and mammals will be discussed.
To contextualize our discussion on proteasome localization and movement, we introduce fundamental concepts of the UPS. Ubiquitin is a heat-stable 8.6 kDa protein, and its expression is highly induced upon stress (4). Repetitive conjugation of ubiquitin by isopeptide bonds through an ATP-consuming ubiquitin-activating E1 and E2 and E3 ligase cascade yields a proteasomal substrate with a polyubiquitin chain (5).
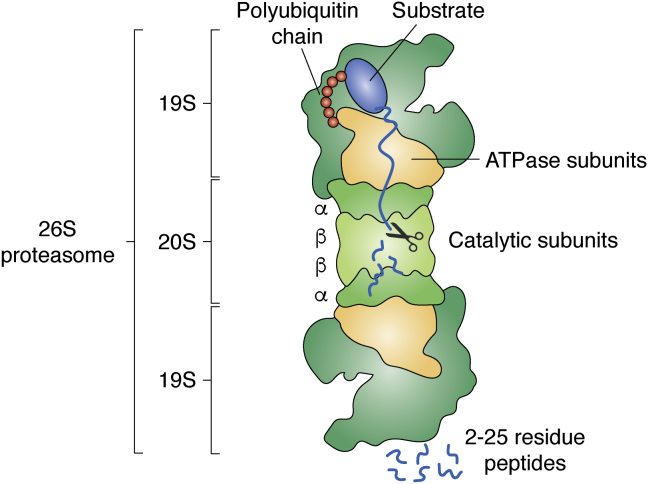
Proteasomes are multisubunit complexes that recognize polyubiquitinated substrates and are composed of the proteolytic core particle (CP) (20S CP) flanked by one or two regulatory particles (RPs) (19S RP) (Fig. 1) (6). The RP consists of the lid and base subcomplexes. Polyubiquitinated substrates are recognized by intrinsic ubiquitin receptors on the RP base (Rpn1, Rpn10, and Rpn13) and are often delivered by extrinsic ubiquitin shuttling proteins (Rad23, Dsk2, and Ddi1) (7). Before degradation, the isopeptide bond between the substrate and ubiquitin is cleaved by the RP lid subunit Rpn11 (8), releasing the polyubiquitin chain to be recycled to ubiquitin by deubiquitinases (DUBs). Substrate degradation is initiated by an intrinsically disordered region (IDR) of appropriate length and location within the protein (9). The polypeptide is unfolded by the ATPase ring of the RP base and translocated into the adjacent CP. The folding and size of the protein substrate determine whether its degradation is mediated by polyubiquitination, monoubiquitination, or neither (10).
Figure 1.
Structure and function of proteasome holoenzymes. The proteolytic 20S core particle (20S CP) is composed of 14 subunits arranged in four seven-membered subunit rings with α1–7, β1–7, β1–7, α1–7 configuration. The outer α rings contain the gates for substrate entry, while the inner β rings harbor the proteolytic active sites (11, 12). Proteins are degraded into peptides. The base of the 19S regulatory particle (19S RP) recognizes the polyubiquitinated substrate by intrinsic ubiquitin receptor subunits Rpn1, Rpn10, and Rpn13. The RP ATPase ring (Rpt1–6) opens the CP gate and translocates the unfolded polypeptide into the CP cavity, after the polyubiquitin chain has been cleaved off by the DUB Rpn11, a RP lid subunit. The dimensions of the CP are 15 nm in length and 11 nm in diameter; the holoenzyme with RP–CP–RP configuration is ∼45 nm in length (cartoon derived from PDB 4cr2 (115)).
The CP is composed of a barrel-shaped particle with two inner rings of β subunits harboring the proteolytic active sites, while the two outer rings of α subunits control the gates for substrate entry (11, 12). IDR loops can reach into the CP cavity rendering proteins susceptible for ATP- and ubiquitin-independent degradation (13). The CP is assembled from two inactive precursor complexes containing Ump1 in yeast/POMP in mammals (14, 15, 16). Ump1 is intrinsically disordered and becomes the first substrate during CP maturation (15, 17, 18). As the second most abundant protein complex in the cell, the proteasome is continuously synthesized during proliferation (19).
In yeast, prolonged stress such as nutrient limitation curbs ATP generation and leads to transient nonproliferation, named quiescence. Quiescence helps cells to overcome stress by reorganizing proteasome localization and possibly their structure and function (20, 21). Yeast proteasomes move from the nucleus into the cytoplasm with the transition from proliferation to quiescence, where they are sequestered into cytoplasmic storage granules to be protected from autophagic degradation (21, 22).
As a proteasome stress response, aberrant proteasome holoenzymes bound to Ecm29, a conserved 200 kDa protein, can be remodeled into a functional enzyme (23, 24). Upon oxidative stress Ecm29 disassembles the holoenzyme into CP and RP (25), possibly allowing the exchange of aberrant subcomplexes by functional subcomplexes with the reassembly of holoenzymes upon stress relief. In yeast, proteasome holoenzymes also appear to be unstable under nutrient limitation causing a decline in ATP (26, 27, 28). In general, sufficient amount of ATP is required to stabilize proteasome holoenzymes (29).
To understand the impact of proteasome movements on protein homeostasis, especially under stress conditions, which may cause temporary and local decreases in ATP, it will be important to know the nature and origin of proteasomal substrates. Soon after the discovery of the proteasome holoenzyme as an ATP- and ubiquitin-dependent protease, the finding that the CP exhibits an ATP- and ubiquitin-independent endoproteolytic activity against IDR-containing proteins (13) raised the possibility that the repertoire of proteasomal substrates might be much larger than initially assumed (13, 30, 31, 32).
Important in the context of the current review are recent mass spectrometry data showing that the repertoire of proteasomal substrates changes in response to different stress factors, for example, heat shock, oxidative, osmotic, UV stress, and proteasome inhibition (33).
Proteasome localizations without stress
Previous reports on proteasome localizations in mammalian cells were not always conclusive, as proteasome localizations varied from cell lines to growth conditions and were also dependent on antibodies used in indirect fluorescence microscopy (34). Compared with yeast proteasomes, the intracellular distribution of mammalian proteasomes was reported to be more complex with the majority of proteasomes localizing in the cytoplasm (35). In contrast to these studies, proteasomal antibodies such as MCP444 (anti-PSMB7 CP β7), which coimmunoprecipitate proteasome holoenzymes, yielded a major nuclear proteasome localization in HeLa, COS-7, U2OS, and KOLF cells, as shown by datasheets of different commercial providers (and our unpublished results). In parallel, a CP α3 subunit fusion with the GFP is suitable as proteasomal reporter in direct fluorescence microscopy and revealed a nuclear localization in HeLa and MelJuSo cells (36, 37). The predominant proteasome localization in the nucleus of mammalian cells was unexpected, as cytoplasmic localization had been the prevailing paradigm. Meanwhile, live cell–labeling techniques using tag-exchangeable Rpn11-Halo knock-in mice embryonic fibroblasts allowed the selective visualization of proteasome localization according to molecular age at the time after synthesis. Using this expression system, newly synthesized proteasomes were mainly detected in the nucleus, while 3-day-old proteasomes were mainly detected in the cytoplasm, suggesting that proteasomes move from the nucleus to the cytoplasm (38). Early observations linked the relocalization of mammalian proteasomes from the nucleus into the cytoplasm to high confluency (34). Particularly in cancer cell lines, which depend on glycolysis for ATP production (39), the shortage of growth factors in dense cell cultures may impact the synthesis of new proteasomes and the localization of proteasomes.
Recently, inducible CRISPR-Cas9 technologies allowed the replacement of proteasomal subunits PSMA5 CP α5, PSMB4 CP β4, and PSMD3 RP lid Rpn3 with fluorescent reporter proteins. All three reporter proteins consistently showed nuclear localization in the RKO colon cancer cell line (40). These technical advances based on controlled CRISPR-Cas9 screens will pave new ways to thoroughly analyze proteasome movements in living mammalian cells and to overcome limitations of classical biochemical fractionations. In yeast, the chromosomal replacement of proteasomal subunits by GFP-tagged versions via homologous recombination has been feasible for more than 2 decades and generated reliable reporters of proteasome holoenzymes (37, 41, 42, 43, 44, 45). All reporter subunits revealed a predominant nuclear localization of proteasomes in proliferating yeast.
Nuclear transport of proteasomes
In 1990, import-competent and import-incompetent CP conformations were proposed to have either accessible or masked nuclear localization sequences (NLSs) (46). According to studies in yeast, immature CP precursor complexes containing Ump1 represent an import-competent conformation, while mature CP is not imported on its own (41). RP base subunits have NLS that allow nuclear import of the RP base (47). Sts1 (in budding yeast/Cut8 in fission yeast) transiently confers an NLS to the RP lid (48) suggesting that the canonical NLS receptor importin/karyopherin αβ carries the RP lid, base, and CP independently into the nucleus (42). Intriguingly, Sts1 also facilitates nuclear import of holoenzymes, is extremely short-lived, and is degraded by the proteasome in the nucleus (49). Our studies suggest that growth conditions determine whether nuclear import of precursor complexes is favored over holoenzymes (50). In highly proliferating yeast cells, precursor complexes are abundant import cargoes, whereas with the transition to quiescence, the synthesis of precursor complexes is stalled. Upon the resumption of growth, in other words with the exit from quiescence, precursor complexes are unavailable. Then mature CP is the only nuclear import cargo and is piggybacked into the nucleus by either the RP or Blm10, an alternative regulatory protein serving as an adapter in yeast for nuclear import of the CP (28, 51).
Species-specific adapter proteins enable nuclear import of mature CP throughout the cell cycle in yeast having a closed mitosis as well as in postmitotic mammalian cells having an open mitosis. An ortholog of yeast Sts1/Cut8 does not exist in mammals. A recent study using a CRISPR screening array based on time-controlled Cas9 mutagenesis and live-cell time-lapse imaging revealed AKIRIN2, a protein without an ortholog in yeast, as nuclear import adapter for mature CP in three different cancer cell lines. AKIRIN2 confers an NLS to the mature CP and mediates binding to Ipo9, a member of the β karyopherin family. AKIRIN2 inhibits the CP and is degraded upon the arrival of the CP in the nucleus (40). The nuclear import mechanisms in yeast and mammalian cells have in common that the CP passes the nuclear pore complex either as inhibited or immature enzyme possibly to avoid the degradation of nuclear pore proteins that are rich in repetitive IDRs. How the proteasomal degradation of Sts1 and AKIRIN2 is blocked in the cytoplasm before nuclear transport and is then triggered once Sts1 and AKIRIN2 arrive in the nucleus remains elusive.
In yeast, Blm10 is abundant in quiescence and facilitates nuclear import of the CP upon exit from quiescence (28). Mammalian PA200, the ortholog of Blm10, mediates nuclear trafficking of proteasomes into speckles in response to oxidative stress (52). Blm10/PA200 with a molecular mass of 240 kDa is primarily nuclear and capping the CP like a dome with few narrow openings. Blm10/PA200 has neither ubiquitin recognition motifs nor ATPase activity, suggesting an allosteric function on the activity of Blm10–CP–RP hybrids for specific substrates (53, 54, 55). The loss of Blm10/PA200 causes hypersensitivity toward stress and the decline of proteasome activity during aging (56, 57).
In addition to PA200, hexameric rings composed of either PA28α/β or PA28γ serve as regulators of nuclear proteasome activity in mammalian cells. PA28γ is present in nuclear speckles, accumulates in Cajal bodies following UV irradiation and causes the appearance of promyelocytic leukemia protein (PML)–nuclear bodies as reviewed by (58).
The question is why are proteasomes primarily nuclear in proliferating cells? Possibly, the majority of proteasomal substrates is nuclear during this cellular state. Their timely degradation by proteasomes in close proximity may expedite the progression through the cell cycle.
If the cell cycle progression is curbed, that is toward quiescence, proteasomes exit the nucleus. How proteasomes are exported from the nucleus into the cytoplasm is not understood. We searched for possible functions of the canonical exportin Crm1/Xpo1 in nuclear export of yeast proteasomes. Our data provide no evidence that Crm1/Xpo1 is required for bulk export of proteasomes under glucose starvation (our unpublished results). However, under nitrogen starvation nuclear export of proteasomes is blocked in xpo1-1 mutants (27). Since proteasomes are modified by ubiquitination during nitrogen starvation (59, 60), Crm1/Xpo1 could mediate nuclear export of ubiquitinated proteasomes for autophagic elimination.
In mammalian cells, polyubiquitinated substrates are exported from the nucleus to the cytoplasm by using Crm1/Xpo1 and UBIN, a protein with an ubiquitin-associated domain (61). In yeast, Rad23 harboring two ubiquitin-associated domains shuttles nuclear substrates to the cytoplasm for proteasomal degradation (62). On the opposite, certain cytoplasmic substrates are imported into the nucleus for proteasomal degradation (63, 64). The orchestration of nuclear transport of proteasomes and their substrates, especially under stress conditions, merit further investigation. Intriguingly, ubiquitin is required for the recovery of nuclear transport upon stress relief but the molecular mechanism is unknown (33).
Proteasome localization under stress
Early localization studies using indirect immunofluorescence microscopy of mammalian cells described the presence of microscopically visible nuclear foci, so called protein quality compartments (PQCs), which are stained by antibodies against ubiquitin and proteasomes. Whether proteasomes reside inside the PQCs or surround the PQCs was not resolved by light microscopy. At the very least, the presence of proteasomes suggested that PQCs function in protein degradation and confer resistance to cellular stress (2). Over the years evidence has been mounting that upon stress or in response to challenging growth conditions polyubiquitinated proteins are sequestered into bodies for spatial PQC (65, 66). PQC bodies have the capacity to dynamically exchange their components with the aqueous environment (67). The fluidity and plasticity of PQCs are due to sorting factors and chaperones (68) and is driven by ubiquitin (69). Few PQC structures have been visualized by electron microscopy and turned out to be enigmatic organelles not enclosed by lipid membranes. Membraneless organelles are proposed to be formed by liquid–liquid phase separation (LLPS), a mechanism allowing soluble proteins to condense in aqueous solution and still to be exchanged with their borderless environment (70, 71). Theoretically, the concept of LLPS is applicable to PQCs. In vitro, an increasing number of PQC components behave like LLPS drivers with multivalent interactions (72, 73, 74, 75, 76, 77).
Proteasome inhibition induces JUNQ in mammalian and yeast cells
One of the first PQC studied in more detail and containing polyubiquitinated substrates and proteasomes (Table 1) was the juxtanuclear compartment (JUNQ) (78). Formation of JUNQ was induced either by conditional proteasomal mutants in yeast or reversible proteasome inhibitors in HeLa cancer cells resulting in a temporary cell cycle arrest. Since proteasomes localize to JUNQ in an inactive state, there is no conclusive answer to how polyubiquitinated proteins are degraded within JUNQ. As soon as proteasomes were reactivated, the JUNQ rapidly dissolved (78). Possibly, the reversible cell cycle arrest under restrictive conditions was accompanied by a decline in ATP and GTP. Nucleotide shortage is disruptive for nuclear transport and may result in the accumulation of transport cargoes in reversible foci, in the case of JUNQ consisting of proteasomes and potential substrates, which are cleared under permissive growth conditions. In support of this interpretation are recent cryo-electron tomography data of nuclear pores. Nuclear pores dilate in cells with active metabolism and reversibly constrict upon energy depletion (79), suggesting that supramolecular assemblies might be trapped in the nucleus or on their way out of the nucleus under conditions of stress.
Table 1.
Reversible proteasome foci induced by different stress factors
| Name | Stress factor | Organism | Localization | Composition | Methods | Disassembly | Reference |
|---|---|---|---|---|---|---|---|
| JUNQ | proteasome inhibition | Y, M | NE | proteasome, poly-Ub proteins | FLIP, FRAP | proteasome activation | (78) |
| Foci | high glucose, hyperosmotic shock | M | N | proteasome, poly-Ub proteins, orphan ribosomal proteins, RAD23B | TEM, cryo-ET, FRAP, LLPS | isoosmosis, DUB | (76) |
| SIPAN | amino acid starvation | M | N | proteasome, poly-Ub proteins, RAD23B | Immuno-gold TEM, FRAP, LLPS | amino acids, DUB | (75) |
| p62 foci | heat, oxidation, inhibition of nuclear export by Crm1/Xpo1 | M | N | core: p62, poly-Ub proteins, orphan proteasomal subunits; shell: proteasome, E1, E2, E3 ubiquitination cascade |
FRAP, LLPS | (77) | |
| PSG | glucose starvation | Y | C | proteasome, ubiquitin | Immuno-gold EM | glucose, ATP | (20, 90) |
Abbreviations: C, cytoplasm; cryo-ET, cryo-electron tomography; FLIP, fluorescence loss in photobleaching; FRAP, fluorescence recovery after photobleaching; M, mammals; N, nucleus; NE, nuclear envelope; TEM, transmission electron microscopy; Y, yeast.
Stress-induced nuclear foci recruit proteasomes in mammalian cells
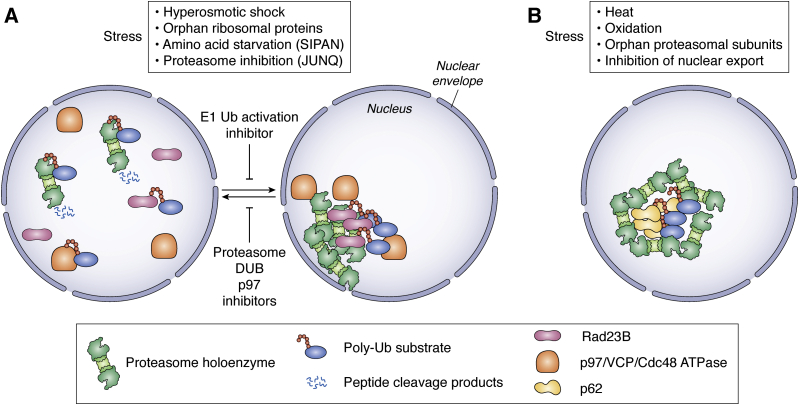
Hyperosmotic stress induced by glucose comparable with type II diabetes is linked with molecular crowding and causes the formation of transient proteasome foci in the nucleus of mammalian cells (76, 80). These proteasome foci behave like LLPS organelles (Fig. 2A). Their LLPS behavior is triggered by multivalent interactions involving the ubiquitin shuttling receptor RAD23B and polyubiquitin chains. The inhibition of the ubiquitin-activating enzyme E1 interfered with proteasome foci formation. Orphan or surplus ribosomal subunits, which represent the main source of defective ribosomal products (81), were detected as polyubiquitinated proteins within these proteasome foci (76). Upon stress relief, the foci dissolved and polyubiquitinated proteins were extracted by the ATPase p97/VCP (Cdc48 in yeast). Inhibitors of either p97/VCP, the proteasome, or proteasomal DUB Rpn11 interfered with the dissolution of these proteasome foci, suggesting that they serve as proteolytic centers for orphan ribosomal subunits (76). If polyubiquitinated substrates are simultaneously degraded with foci clearance (33), these foci could also be considered as storage compartments for UPS components during stress and ATP shortage. Notably, nucleoli where ribosomal subunits and rRNA are assembled into ribosomes do not contain proteasomes (81), showing that ribosome biogenesis and quality control of orphan ribosomal subunits are controlled by spatially distinct membraneless organelles (82).
Figure 2.
Intranuclear proteasome movements in mammalian cells in response to stress. The double line represents the nuclear envelope with nuclear pores. A, LLPS-driven nuclear proteasome foci are formed upon hyperosmotic and nucleolar stress resulting in the accumulation of nonincorporated ribosomal proteins (76). SIPAN in the nucleus and JUNQ at the ER/nuclear envelope are formed upon amino acid starvation and proteasome inhibition, respectively (75, 78). Rad23B with two ubiquitin-associated domains binds polyubiquitin chains of proteasomal substrates and mediates multivalent interactions for LLPS. ATPase p97/VCP/Cdc48 assists in foci clearance upon stress relief by removing polyubiquitinated proteins (76). B, intranuclear p62 foci are induced by inhibition of Crm1/Xpo1–mediated nuclear export, heat, and oxidative stress. Proteasomes are recruited to the periphery of p62 foci (77). ER, endoplasmic reticulum; LLPS, liquid–liquid phase separation.
In HeLa cells LLPS-driven nuclear condensates contain p62/sequestrome-1 and polyubiquitinated substrates such as orphan proteasomal subunits, which might represent a considerable fraction of defective ribosomal products under stress. These foci increase in numbers and size upon heat, oxidative stress, and the inhibition of Crm1/Xpo1 (77), which according to (61), is involved in nuclear export of polyubiquitinated proteins. Proteasomes, the E1, E2, and E3 ubiquitination cascade, and DUBs are recruited to the periphery of p62 foci. The coincidence of UPS components suggests a major function of p62 foci in nuclear protein homeostasis under basal and stress conditions and a new function of p62 unrelated to autophagy and lysosomal degradation (77) (Fig. 2B).
Amino acid deprivation also induces nuclear foci with proteasomes and polyubiquitinated proteins, named starvation-induced proteasome assemblies in the nucleus of mammalian cells (SIPAN), which behave like membraneless PQCs and quickly resolve upon amino acid supply (75). Comparable with proteasome foci induced by hyperosmotic stress, RAD23B is required for SIPAN formation. Again, DUB inhibition interfered with SIPAN clearance, providing further evidence that ubiquitin signaling drives SIPAN formation (75). Inhibition of the DUB UCH37 also resulted in the formation of nuclear proteasome foci, especially under osmotic and oxidative stress. Proteasome foci associated with active site mutation of UCH37 retained polyubiquitinated proteins and RAD23B. Again, this defect of substrate processing resulted in the immobility and irreversibility of proteasome foci (83). Here, it would be interesting to investigate whether nuclear foci either surrounded by or containing proteasomes share similar qualities or are even identical structures independently of the inducing stress factor, that is, osmotic stress or amino acid deprivation.
Cryo-electron tomography visualized proteasome microcompartments at the endoplasmic reticulum (ER)/nuclear envelope in association with the ATPase Cdc48/p97/VCP during the extraction of aberrant secretory proteins for ER-associated degradation (84), a process envisioned by multiple studies in the ER-associated degradation field (for review see (85)). Upon ER stress, misfolded secretory proteins accumulated in aggresomes, microtubule-dependent inclusion bodies close to the ER/nuclear envelope, and recruited proteasomes. Polyubiquitin chains released by the proteasomal DUB Rpn11/Poh1 facilitated aggresome clearance (86). Without DUBs, proteasomes overwhelmed by substrates remained trapped in densely packed aggresomes (81). Failures to eliminate aggresomes by autophagy are linked with age-related neurological disorders (34, 87).
In the nucleus, if aberrant proteins are not eliminated due to impaired proteasomal degradation, they can accumulate in PML-nuclear bodies, overflowing PQCs in response to inflammation induced proteotoxic stress (88). Again, proteasomes are recruited to PML bodies most likely for protein degradation. However, ATP shortage might become critical for the maintenance of the UPS. Under unfavorable conditions, PML bodies become toxic by immobilizing the UPS, thus threatening cell survival and leading to neurodegenerative diseases (81).
Proteasome storage granules in the cytoplasm of quiescent yeast cells
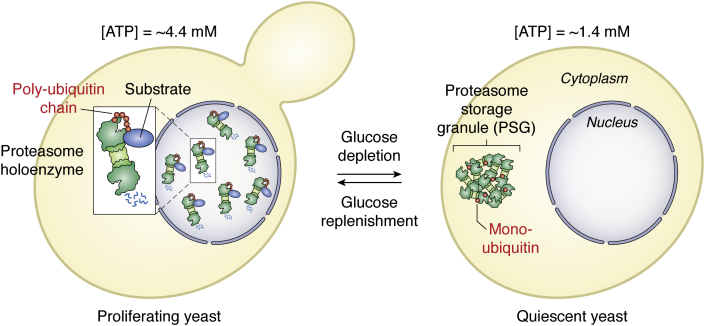
In yeast, membraneless organelles, named proteasome storage granules (PSGs), exist in the cytoplasm of quiescent cells (21). Metabolomics analysis revealed that ATP levels decline from 4.4 mM during proliferation to 1.4 mM with glucose depletion in quiescence (21). Nuclear proteasomes respond to metabolic cues. In quiescence, protein synthesis is stalled. As a consequence not only less short-lived proteins regulating cell cycle progression but also less defective ribosomal products will accrue as potential proteasomal substrates. Further, proteasomes migrate to the nuclear envelope and accumulate in PSGs, which pinch off the nuclear pore and move as stable entities through the cytoplasm (21) (Fig. 3). Notably, ubiquitin availability is important for PSG formation. PSG formation depends on free ubiquitin, while nuclear proteasome foci depend on polyubiquitin.
Figure 3.
Proteasome movement between proliferation and quiescence in yeast. In proliferating cells with high ATP, proteasome holoenzymes are mainly localized in the nucleus and engaged in the degradation of polyubiquitinated proteins. With glucose depletion toward quiescence, the ATP level declines. Proteasomes then exit the nucleus through nuclear pores and are sequestered into PSGs, which move as membraneless organelles through the cytoplasm (21). PSGs are mainly composed of proteasomal components and monoubiquitin (90). Since proteasome holoenzymes are unstable at low ATP, CP and RP are dissociated in lysates of quiescent yeast cells (26). With the addition of glucose, cells receive the signal to resume growth. The ATP level increases and PSGs clear within a few min. Proteasomes are rapidly relocated to the nucleus where ATP- and ubiquitin-dependent proteolysis promotes cell cycle progression. CP, core particle; PSG, proteasome storage granule; RP, regulatory particle.
Under normal growth conditions, de novo synthesized ubiquitin is a side product of ribosome biogenesis (5). Under acute stress, ubiquitin abundance relies on UBI4, a gene expressing multiple ubiquitin molecules behind a promoter with responsiveness to heat, oxidation, and heavy metals (4). Consequently, yeast ubi4Δ mutants are hypersensitive toward stress and have a shortened life span (89). However, during prolonged stress and quiescence, global protein translation is stalled and ubiquitin is not synthesized de novo (90); the ubiquitin pool must be replenished by DUBs hydrolyzing polyubiquitin chains. Therefore, PSGs were not formed in ubi4Δ mutants, proteasomal DUB mutants of UBP6 (90) or RPN11 (91) and the ubiquitin shuffling receptor mutant rad23Δ (75).
To buffer ubiquitin fluctuations, proteasomes alter their composition, for example, by incorporating Ubp6 to enable polyubiquitin chain remodeling (92, 93). Ubp6 (human USP14) shortens polyubiquitin chains (94) and inhibits proteasome activity (93) possibly to avoid futile degradation of essential proteins during stress.
Interestingly, the CP and RP seem to be separately sequestered into PSGs, the CP by Blm10 (28) and the RP by Spg5 (95). Since proteasome holoenzymes are unstable in quiescence, PSGs hardly serve as proteolytic centers for polyubiquitinated substrates. UPS downregulation including proteasome disassembly was discussed as a mechanism to cope with stress and to maintain cell viability during quiescence (26). However, ATP- and ubiquitin-independent proteolysis through the CP could conform to the metabolic state of quiescence (96, 97).
The mechanism underlying PSG formation has not yet been investigated. Is the concept of LLPS based on multivalent interactions between low complexity domains or IDRs applicable to PSGs (67, 71, 98)? No protein with low complexity domains or rich in IDRs was identified as a significant PSG constituent (90). PSGs are mainly composed of proteasomes with folded subunits and ubiquitin. Extrinsic ubiquitin receptors Rad23, Dsk2, and Ddi1 have IDRs (99) but were not identified within PSGs.
So far, the function of PSGs is the storage of proteasomes during quiescence and their protection from autophagic elimination (22, 100). Under carbon starvation AMP-dependent kinase (AMPK), a key regulator of energy homeostasis, triggers microautophagy of aberrant proteasomes, thereby saving intact proteasomes within PSGs. If AMPK is inhibited, both aberrant and functional proteasomes are sequestered into the irreversible protein deposit compartment, which is eliminated by lysosomal degradation (101). PSGs transiently interact with irreversible protein deposit compartments (102), possibly allowing the sorting of intact from aberrant proteasomes.
The common signal for proteasome sequestration into PSGs is the drop in ATP levels (21). With ATP regeneration, proteasomes are released from PSGs and immediately imported into the nucleus to resume cell cycle progression (21, 28). ATP is an amphiphilic molecule with properties of a biological hydrotrope (103). Millimolar concentrations of ATP prevent protein aggregation not only by the amphiphilic structure of ATP but also by fueling protein folding through chaperones. If proliferating yeast cells are shifted to anaerobic conditions by uncoupling the mitochondrial oxidative phosphorylation, the pH changes from ∼7.2 to 6.7, less ATP is gained compared with aerobic conditions (20, 104) resulting in premature PSG formation (105).
Multiple condensates like stress granules containing polyubiquitinated proteins coexist with PSGs (90). Possibly, upon PSG clearance, proteasomes clear polyubiquitinated proteins from other PQCs and maintain the PQC reversibility and plasticity. How the organization of PSGs and related PQCs is synchronized could be tested by high-throughput screens in the yeast mutant collection by using colocalization studies with differently labeled fluorescent PQC reporter proteins.
An important question is whether PSGs are formed in mammalian cells. Quiescence is poorly studied in mammalian cells, since quiescence exists only in a subset of adult stem cells including neural stem cells (106). Interestingly, dendrites of primary neuronal cells revealed motile proteasome foci upon excitotoxicity (107). These foci resemble CP clusters at the plasma membrane of dendrite spines, which were proposed to serve as proteolytic hubs for ATP- and ubiquitin-independent degradation to regulate synaptic communication (108, 109, 110). Excitatory stimulation may result in a local drop of ATP, which forces the reorganization of ATP-consuming machineries such as proteasomes.
In general, the accumulation of undegradable proteasomal substrates in neuronal cells bears the risk of recruiting proteasomes from the protoplasm. This depletion of proteasomes leads to disturbed protein homeostasis associated with neurodegenerative diseases (111, 112). Taken together, it is essential to understand intracellular movements of proteasomes and polyubiquitinated proteins, their equilibrium between free diffusion, and condensation into liquid organelles in response to stress, before becoming immotile as irreversible and toxic protein inclusions.
Future directions
While many studies on proteasome dynamics have thus far relied on targeting UPS components to infer functions based on educated guess, the availability of unbiased reporter-based genetic screens initially in yeast and nowadays in mammalian cells using the CRISPR-Cas system as a more integrative method will elucidate unpredictable mechanisms of proteasome and ubiquitin movements between the nucleus and cytoplasm (40, 113). Stress consumes ATP and influences nucleocytoplasmic transport, UPS dynamics by impaired ubiquitin fluctuations, and nucleotide shortages. Interconvertible posttranslational modifications in response to nucleotide changes may influence intracellular movements of UPS components. Knowing how intracellular movements of the UPS are regulated will provide the basis for timely and spatially controlled interference, which is required to overcome metabolic challenges and to protect cells from burden in disturbed protein homeostasis. Upon stress, proteasomes and polyubiquitinated proteins accumulate in PQCs presumably driven by LLPS. If proteasomes and polyubiquitinated proteins colocalize within the PQC, we may wonder what are the advantages of protein degradation in membraneless organelles? Countless in vitro experiments in the past have taught us that proteasomal proteolysis is efficient in free diffusion of enzymes and substrates.
Single particle cryo-electron microscopy and in situ cryo-electron tomography are emerging technologies in structural biology, which allow to reveal large scale changes within proteasome configurations and conformations in the protoplasm and membraneless organelles. Faced with the limitations of conventional biochemical methods to resolve supramolecular structures, cryo-electron tomography will provide insight into unprecedented concepts of proteasome organization underlying the degradation of dysfunctional proteins under various forms of stress (114).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Daniela Rotin for critical reading of the manuscript.
Author contributions
F. W. and C. E. conceptualization; R. W. K., F. W., and C. E. writing–original draft; F. W. and C. E. visualization; F. W. and C. E. supervision; O. P. E. funding acquisition.
Funding and additional information
This work was supported in part by grants from the Natural Sciences and Engineering Research Council RGPIN/5974-2019 (to C. E.), RGPIN/2017-06862 (to O. P. E.), Mitacs IT17088 (to C. E.), and the Canadian Institute for Advanced Research. O. P. E. was supported as Anne and Max Tanenbaum Chair in Neuroscience.
Edited by George DeMartino
References
- 1.Hershko A., Ciechanover A., Varshavsky A. Basic medical research award. The ubiquitin system. Nat. Med. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 2.von Mikecz A. The nuclear ubiquitin-proteasome system. J. Cell Sci. 2006;119:1977–1984. doi: 10.1242/jcs.03008. [DOI] [PubMed] [Google Scholar]
- 3.Goldberg A.L. Protein degradation and protection against misfolded or damaged proteins. Nature. 2003;426:895–899. doi: 10.1038/nature02263. [DOI] [PubMed] [Google Scholar]
- 4.Finley D., Ozkaynak E., Varshavsky A. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell. 1987;48:1035–1046. doi: 10.1016/0092-8674(87)90711-2. [DOI] [PubMed] [Google Scholar]
- 5.Finley D., Ulrich H.D., Sommer T., Kaiser P. The ubiquitin-proteasome system of Saccharomyces cerevisiae. Genetics. 2012;192:319–360. doi: 10.1534/genetics.112.140467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baumeister W., Walz J., Zuhl F., Seemuller E. The proteasome: paradigm of a self-compartmentalizing protease. Cell. 1998;92:367–380. doi: 10.1016/s0092-8674(00)80929-0. [DOI] [PubMed] [Google Scholar]
- 7.Shi Y., Chen X., Elsasser S., Stocks B.B., Tian G., Lee B.H., et al. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science. 2016;351 doi: 10.1126/science.aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Worden E.J., Dong K.C., Martin A. An AAA motor-driven mechanical switch in Rpn11 controls deubiquitination at the 26S proteasome. Mol. Cell. 2017;67:799–811.e8. doi: 10.1016/j.molcel.2017.07.023. [DOI] [PubMed] [Google Scholar]
- 9.Davis C., Spaller B.L., Matouschek A. Mechanisms of substrate recognition by the 26S proteasome. Curr. Opin. Struct. Biol. 2021;67:161–169. doi: 10.1016/j.sbi.2020.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shabek N., Herman-Bachinsky Y., Buchsbaum S., Lewinson O., Haj-Yahya M., Hejjaoui M., et al. The size of the proteasomal substrate determines whether its degradation will be mediated by mono- or polyubiquitylation. Mol. Cell. 2012;48:87–97. doi: 10.1016/j.molcel.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 11.Groll M., Bajorek M., Kohler A., Moroder L., Rubin D.M., Huber R., et al. A gated channel into the proteasome core particle. Nat. Struct. Mol. Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 12.Orlowski M., Wilk S. Catalytic activities of the 20 S proteasome, a multicatalytic proteinase complex. Arch. Biochem. Biophys. 2000;383:1–16. doi: 10.1006/abbi.2000.2036. [DOI] [PubMed] [Google Scholar]
- 13.Liu C.W., Corboy M.J., DeMartino G.N., Thomas P.J. Endoproteolytic activity of the proteasome. Science. 2003;299:408–411. doi: 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen P., Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86:961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- 15.Ramos P.C., Hockendorff J., Johnson E.S., Varshavsky A., Dohmen R.J. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell. 1998;92:489–499. doi: 10.1016/s0092-8674(00)80942-3. [DOI] [PubMed] [Google Scholar]
- 16.Murata S., Yashiroda H., Tanaka K. Molecular mechanisms of proteasome assembly. Nat. Rev. Mol. Cell Biol. 2009;10:104–115. doi: 10.1038/nrm2630. [DOI] [PubMed] [Google Scholar]
- 17.Uekusa Y., Okawa K., Yagi-Utsumi M., Serve O., Nakagawa Y., Mizushima T., et al. Backbone (1)H, (1)(3)C and (1)(5)N assignments of yeast Ump1, an intrinsically disordered protein that functions as a proteasome assembly chaperone. Biomol. NMR Assign. 2014;8:383–386. doi: 10.1007/s12104-013-9523-1. [DOI] [PubMed] [Google Scholar]
- 18.Sa-Moura B., Simoes A.M., Fraga J., Fernandes H., Abreu I.A., Botelho H.M., et al. Biochemical and biophysical characterization of recombinant yeast proteasome maturation factor ump1. Comput. Struct. Biotechnol. J. 2013;7 doi: 10.5936/csbj.201304006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marguerat S., Schmidt A., Codlin S., Chen W., Aebersold R., Bahler J. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–683. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laporte D., Lebaudy A., Sahin A., Pinson B., Ceschin J., Daignan-Fornier B., et al. Metabolic status rather than cell cycle signals control quiescence entry and exit. J. Cell Biol. 2011;192:949–957. doi: 10.1083/jcb.201009028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laporte D., Salin B., Daignan-Fornier B., Sagot I. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J. Cell Biol. 2008;181:737–745. doi: 10.1083/jcb.200711154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marshall R.S., Vierstra R.D. Proteasome storage granules protect proteasomes from autophagic degradation upon carbon starvation. Elife. 2018;7 doi: 10.7554/eLife.34532. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Lehmann A., Niewienda A., Jechow K., Janek K., Enenkel C. Ecm29 fulfils quality control functions in proteasome assembly. Mol. Cell. 2010;38:879–888. doi: 10.1016/j.molcel.2010.06.016. [DOI] [PubMed] [Google Scholar]
- 24.Park S., Kim W., Tian G., Gygi S.P., Finley D. Structural defects in the regulatory particle-core particle interface of the proteasome induce a novel proteasome stress response. J. Biol. Chem. 2011;286:36652–36666. doi: 10.1074/jbc.M111.285924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X., Chemmama I.E., Yu C., Huszagh A., Xu Y., Viner R., et al. The proteasome-interacting Ecm29 protein disassembles the 26S proteasome in response to oxidative stress. J. Biol. Chem. 2017;292:16310–16320. doi: 10.1074/jbc.M117.803619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bajorek M., Finley D., Glickman M.H. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr. Biol. 2003;13:1140–1144. doi: 10.1016/s0960-9822(03)00417-2. [DOI] [PubMed] [Google Scholar]
- 27.Nemec A.A., Howell L.A., Peterson A.K., Murray M.A., Tomko R.J., Jr. Autophagic clearance of proteasomes in yeast requires the conserved sorting nexin Snx4. J. Biol. Chem. 2017;292:21466–21480. doi: 10.1074/jbc.M117.817999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weberruss M.H., Savulescu A.F., Jando J., Bissinger T., Harel A., Glickman M.H., et al. Blm10 facilitates nuclear import of proteasome core particles. EMBO J. 2013;32:2697–2707. doi: 10.1038/emboj.2013.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu C.W., Li X., Thompson D., Wooding K., Chang T.L., Tang Z., et al. ATP binding and ATP hydrolysis play distinct roles in the function of 26S proteasome. Mol. Cell. 2006;24:39–50. doi: 10.1016/j.molcel.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Erales J., Coffino P. Ubiquitin-independent proteasomal degradation. Biochim. Biophys. Acta. 2014;1843:216–221. doi: 10.1016/j.bbamcr.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsvetkov P., Reuven N., Shaul Y. The nanny model for IDPs. Nat. Chem. Biol. 2009;5:778–781. doi: 10.1038/nchembio.233. [DOI] [PubMed] [Google Scholar]
- 32.Ben-Nissan G., Sharon M. Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules. 2014;4:862–884. doi: 10.3390/biom4030862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maxwell B.A., Gwon Y., Mishra A., Peng J., Nakamura H., Zhang K., et al. Ubiquitination is essential for recovery of cellular activities after heat shock. Science. 2021;372 doi: 10.1126/science.abc3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wojcik C., DeMartino G.N. Intracellular localization of proteasomes. Int. J. Biochem. Cell Biol. 2003;35:579–589. doi: 10.1016/s1357-2725(02)00380-1. [DOI] [PubMed] [Google Scholar]
- 35.Brooks P., Fuertes G., Murray R.Z., Bose S., Knecht E., Rechsteiner M.C., et al. Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem. J. 2000;346 Pt 1:155–161. [PMC free article] [PubMed] [Google Scholar]
- 36.Salomons F.A., Acs K., Dantuma N.P. Illuminating the ubiquitin/proteasome system. Exp. Cell Res. 2010;316:1289–1295. doi: 10.1016/j.yexcr.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Enenkel C. Proteasome dynamics. Biochim. Biophys. Acta. 2014;1843:39–46. doi: 10.1016/j.bbamcr.2013.03.023. [DOI] [PubMed] [Google Scholar]
- 38.Tomita T., Hirayama S., Sakurai Y., Ohte Y., Yoshihara H., Saeki Y., et al. Specific modification of aged proteasomes revealed by tag-exchangeable knock-in mice. Mol. Cell. Biol. 2019;39 doi: 10.1128/MCB.00426-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 40.de Almeida M., Hinterndorfer M., Brunner H., Grishkovskaya I., Singh K., Schleiffer A., et al. AKIRIN2 controls the nuclear import of proteasomes in vertebrates. Nature. 2021;599:491–496. doi: 10.1038/s41586-021-04035-8. [DOI] [PubMed] [Google Scholar]
- 41.Lehmann A., Janek K., Braun B., Kloetzel P.M., Enenkel C. 20 S proteasomes are imported as precursor complexes into the nucleus of yeast. J. Mol. Biol. 2002;317:401–413. doi: 10.1006/jmbi.2002.5443. [DOI] [PubMed] [Google Scholar]
- 42.Isono E., Nishihara K., Saeki Y., Yashiroda H., Kamata N., Ge L., et al. The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol. Biol. Cell. 2007;18:569–580. doi: 10.1091/mbc.E06-07-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Russell S.J., Steger K.A., Johnston S.A. Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J. Biol. Chem. 1999;274:21943–21952. doi: 10.1074/jbc.274.31.21943. [DOI] [PubMed] [Google Scholar]
- 44.Wilkinson C.R., Wallace M., Morphew M., Perry P., Allshire R., Javerzat J.P., et al. Localization of the 26S proteasome during mitosis and meiosis in fission yeast. EMBO J. 1998;17:6465–6476. doi: 10.1093/emboj/17.22.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Enenkel C., Lehmann A., Kloetzel P.M. Subcellular distribution of proteasomes implicates a major location of protein degradation in the nuclear envelope-ER network in yeast. EMBO J. 1998;17:6144–6154. doi: 10.1093/emboj/17.21.6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tanaka K., Yoshimura T., Tamura T., Fujiwara T., Kumatori A., Ichihara A. Possible mechanism of nuclear translocation of proteasomes. FEBS Lett. 1990;271:41–46. doi: 10.1016/0014-5793(90)80367-r. [DOI] [PubMed] [Google Scholar]
- 47.Wendler P., Lehmann A., Janek K., Baumgart S., Enenkel C. The bipartite nuclear localization sequence of Rpn2 is required for nuclear import of proteasomal base complexes via karyopherin alphabeta and proteasome functions. J. Biol. Chem. 2004;279:37751–37762. doi: 10.1074/jbc.M403551200. [DOI] [PubMed] [Google Scholar]
- 48.Chen L., Romero L., Chuang S.M., Tournier V., Joshi K.K., Lee J.A., et al. Sts1 plays a key role in targeting proteasomes to the nucleus. J. Biol. Chem. 2011;286:3104–3118. doi: 10.1074/jbc.M110.135863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Budenholzer L., Breckel C., Hickey C.M., Hochstrasser M. The Sts1 nuclear import adapter uses a non-canonical bipartite nuclear localization signal and is directly degraded by the proteasome. J. Cell Sci. 2020;133 doi: 10.1242/jcs.236158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wendler P., Enenkel C. Nuclear transport of yeast proteasomes. Front. Mol. Biosci. 2019;6:34. doi: 10.3389/fmolb.2019.00034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pack C.G., Yukii H., Toh-e A., Kudo T., Tsuchiya H., Kaiho A., et al. Quantitative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome. Nat. Commun. 2014;5:3396. doi: 10.1038/ncomms4396. [DOI] [PubMed] [Google Scholar]
- 52.Ustrell V., Hoffman L., Pratt G., Rechsteiner M. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 2002;21:3516–3525. doi: 10.1093/emboj/cdf333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sadre-Bazzaz K., Whitby F.G., Robinson H., Formosa T., Hill C.P. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol. Cell. 2010;37:728–735. doi: 10.1016/j.molcel.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toste Rego A., da Fonseca P.C.A. Characterization of fully recombinant human 20S and 20S-PA200 proteasome complexes. Mol. Cell. 2019;76:138–147.e5. doi: 10.1016/j.molcel.2019.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guan H., Wang Y., Yu T., Huang Y., Li M., Saeed A., et al. Cryo-EM structures of the human PA200 and PA200-20S complex reveal regulation of proteasome gate opening and two PA200 apertures. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Doherty K.M., Pride L.D., Lukose J., Snydsman B.E., Charles R., Pramanik A., et al. Loss of a 20S proteasome activator in Saccharomyces cerevisiae downregulates genes important for genomic integrity, increases DNA damage, and selectively sensitizes cells to agents with diverse mechanisms of action. G3 (Bethesda) 2012;2:943–959. doi: 10.1534/g3.112.003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen L.B., Ma S., Jiang T.X., Qiu X.B. Transcriptional upregulation of proteasome activator Blm10 antagonizes cellular aging. Biochem. Biophys. Res. Commun. 2020;532:211–218. doi: 10.1016/j.bbrc.2020.07.003. [DOI] [PubMed] [Google Scholar]
- 58.Cascio P. PA28gamma: new insights on an ancient proteasome activator. Biomolecules. 2021;11:228. doi: 10.3390/biom11020228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marshall R.S., Vierstra R.D. Dynamic regulation of the 26S proteasome: from synthesis to degradation. Front. Mol. Biosci. 2019;6:40. doi: 10.3389/fmolb.2019.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cohen-Kaplan V., Livneh I., Avni N., Cohen-Rosenzweig C., Ciechanover A. The ubiquitin-proteasome system and autophagy: coordinated and independent activities. Int. J. Biochem. Cell Biol. 2016;79:403–418. doi: 10.1016/j.biocel.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 61.Hirayama S., Sugihara M., Morito D., Iemura S.I., Natsume T., Murata S., et al. Nuclear export of ubiquitinated proteins via the UBIN-POST system. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E4199–E4208. doi: 10.1073/pnas.1711017115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okeke E., Chen L., Madura K. The cellular location of Rad23, a polyubiquitin chain-binding protein, plays a key role in its interaction with substrates of the proteasome. J. Mol. Biol. 2020;432:2388–2404. doi: 10.1016/j.jmb.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 63.Park S.H., Kukushkin Y., Gupta R., Chen T., Konagai A., Hipp M.S., et al. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 2013;154:134–145. doi: 10.1016/j.cell.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 64.Prasad R., Kawaguchi S., Ng D.T. A nucleus-based quality control mechanism for cytosolic proteins. Mol. Biol. Cell. 2010;21:2117–2127. doi: 10.1091/mbc.E10-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schneider K.L., Nystrom T., Widlund P.O. Studying spatial protein quality control, proteopathies, and aging using different model misfolding proteins in S. cerevisiae. Front. Mol. Neurosci. 2018;11:249. doi: 10.3389/fnmol.2018.00249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ciechanover A., Kwon Y.T. Protein quality control by molecular chaperones in neurodegeneration. Front. Neurosci. 2017;11:185. doi: 10.3389/fnins.2017.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alberti S., Hyman A.A. Biomolecular condensates at the nexus of cellular stress, protein aggregation disease and ageing. Nat. Rev. Mol. Cell Biol. 2021;22:196–213. doi: 10.1038/s41580-020-00326-6. [DOI] [PubMed] [Google Scholar]
- 68.Alberti S. Molecular mechanisms of spatial protein quality control. Prion. 2012;6:437–442. doi: 10.4161/pri.22470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Franzmann T., Alberti S. Ubiquitin protein helps cells to recover from stress. Nature. 2021;597:183–184. doi: 10.1038/d41586-021-02197-z. [DOI] [PubMed] [Google Scholar]
- 70.Shin Y., Brangwynne C.P. Liquid phase condensation in cell physiology and disease. Science. 2017;357 doi: 10.1126/science.aaf4382. [DOI] [PubMed] [Google Scholar]
- 71.Forman-Kay J.D., Kriwacki R.W., Seydoux G. Phase separation in biology and disease. J. Mol. Biol. 2018;430:4603–4606. doi: 10.1016/j.jmb.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun D., Wu R., Zheng J., Li P., Yu L. Polyubiquitin chain-induced p62 phase separation drives autophagic cargo segregation. Cell Res. 2018;28:405–415. doi: 10.1038/s41422-018-0017-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zaffagnini G., Savova A., Danieli A., Romanov J., Tremel S., Ebner M., et al. p62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 2018;37 doi: 10.15252/embj.201798308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dao T.P., Kolaitis R.M., Kim H.J., O'Donovan K., Martyniak B., Colicino E., et al. Ubiquitin modulates liquid-liquid phase separation of UBQLN2 via disruption of multivalent interactions. Mol. Cell. 2018;69:965–978.e6. doi: 10.1016/j.molcel.2018.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Uriarte M., Sen Nkwe N., Tremblay R., Ahmed O., Messmer C., Mashtalir N., et al. Starvation-induced proteasome assemblies in the nucleus link amino acid supply to apoptosis. Nat. Commun. 2021;12:6984. doi: 10.1038/s41467-021-27306-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yasuda S., Tsuchiya H., Kaiho A., Guo Q., Ikeuchi K., Endo A., et al. Stress- and ubiquitylation-dependent phase separation of the proteasome. Nature. 2020;578:296–300. doi: 10.1038/s41586-020-1982-9. [DOI] [PubMed] [Google Scholar]
- 77.Fu A., Cohen-Kaplan V., Avni N., Livneh I., Ciechanover A. p62-containing, proteolytically active nuclear condensates, increase the efficiency of the ubiquitin-proteasome system. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2107321118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kaganovich D., Kopito R., Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zimmerli C.E., Allegretti M., Rantos V., Goetz S.K., Obarska-Kosinska A., Zagoriy I., et al. Nuclear pores dilate and constrict in cellulo. Science. 2021;374 doi: 10.1126/science.abd9776. [DOI] [PubMed] [Google Scholar]
- 80.Lee J., Le L., Kim E., Lee M.J. Formation of non-nucleoplasmic proteasome foci during the late stage of hyperosmotic stress. Cells. 2021;10:2493. doi: 10.3390/cells10092493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mediani L., Guillen-Boixet J., Vinet J., Franzmann T.M., Bigi I., Mateju D., et al. Defective ribosomal products challenge nuclear function by impairing nuclear condensate dynamics and immobilizing ubiquitin. EMBO J. 2019;38 doi: 10.15252/embj.2018101341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tye B.W., Commins N., Ryazanova L.V., Wuhr M., Springer M., Pincus D., et al. Proteotoxicity from aberrant ribosome biogenesis compromises cell fitness. Elife. 2019;8 doi: 10.7554/eLife.43002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Song A., Hazlett Z., Abeykoon D., Dortch J., Dillon A., Curtiss J., et al. Branched ubiquitin chain binding and deubiquitination by UCH37 facilitate proteasome clearance of stress-induced inclusions. Elife. 2021;10 doi: 10.7554/eLife.72798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Albert S., Wietrzynski W., Lee C.W., Schaffer M., Beck F., Schuller J.M., et al. Direct visualization of degradation microcompartments at the ER membrane. Proc. Natl. Acad. Sci. U. S. A. 2020;117:1069–1080. doi: 10.1073/pnas.1905641117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wolf D.H., Stolz A. The Cdc48 machine in endoplasmic reticulum associated protein degradation. Biochim. Biophys. Acta. 2012;1823:117–124. doi: 10.1016/j.bbamcr.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 86.Hao R., Nanduri P., Rao Y., Panichelli R.S., Ito A., Yoshida M., et al. Proteasomes activate aggresome disassembly and clearance by producing unanchored ubiquitin chains. Mol. Cell. 2013;51:819–828. doi: 10.1016/j.molcel.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kopito R.R. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 2000;10:524–530. doi: 10.1016/s0962-8924(00)01852-3. [DOI] [PubMed] [Google Scholar]
- 88.Fabunmi R.P., Wigley W.C., Thomas P.J., DeMartino G.N. Interferon gamma regulates accumulation of the proteasome activator PA28 and immunoproteasomes at nuclear PML bodies. J. Cell Sci. 2001;114:29–36. doi: 10.1242/jcs.114.1.29. [DOI] [PubMed] [Google Scholar]
- 89.Zhao W., Zhou T., Zheng H.Z., Qiu K.P., Cui H.J., Yu H., et al. Yeast polyubiquitin gene UBI4 deficiency leads to early induction of apoptosis and shortened replicative lifespan. Cell Stress Chaperones. 2018;23:527–537. doi: 10.1007/s12192-017-0860-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gu Z.C., Wu E., Sailer C., Jando J., Styles E., Eisenkolb I., et al. Ubiquitin orchestrates proteasome dynamics between proliferation and quiescence in yeast. Mol. Biol. Cell. 2017;28:2479–2491. doi: 10.1091/mbc.E17-03-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saunier R., Esposito M., Dassa E.P., Delahodde A. Integrity of the Saccharomyces cerevisiae Rpn11 protein is critical for formation of proteasome storage granules (PSG) and survival in stationary phase. PLoS One. 2013;8 doi: 10.1371/journal.pone.0070357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Crosas B., Hanna J., Kirkpatrick D.S., Zhang D.P., Tone Y., Hathaway N.A., et al. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell. 2006;127:1401–1413. doi: 10.1016/j.cell.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 93.Hanna J., Meides A., Zhang D.P., Finley D. A ubiquitin stress response induces altered proteasome composition. Cell. 2007;129:747–759. doi: 10.1016/j.cell.2007.03.042. [DOI] [PubMed] [Google Scholar]
- 94.Shin J.Y., Muniyappan S., Tran N.N., Park H., Lee S.B., Lee B.H. Deubiquitination reactions on the proteasome for proteasome versatility. Int. J. Mol. Sci. 2020;21:5312. doi: 10.3390/ijms21155312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hanna J., Waterman D., Boselli M., Finley D. Spg5 protein regulates the proteasome in quiescence. J. Biol. Chem. 2012;287:34400–34409. doi: 10.1074/jbc.M112.390294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moscovitz O., Tsvetkov P., Hazan N., Michaelevski I., Keisar H., Ben-Nissan G., et al. A mutually inhibitory feedback loop between the 20S proteasome and its regulator, NQO1. Mol. Cell. 2012;47:76–86. doi: 10.1016/j.molcel.2012.05.049. [DOI] [PubMed] [Google Scholar]
- 97.Solomon H., Brauning B., Fainer I., Ben-Nissan G., Rabani S., Goldfinger N., et al. Post-translational regulation of p53 function through 20S proteasome-mediated cleavage. Cell Death Differ. 2017;24:2187–2198. doi: 10.1038/cdd.2017.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mittag T., Parker R. Multiple modes of protein-protein interactions promote RNP granule assembly. J. Mol. Biol. 2018;430:4636–4649. doi: 10.1016/j.jmb.2018.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Aufderheide A., Beck F., Stengel F., Hartwig M., Schweitzer A., Pfeifer G., et al. Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc. Natl. Acad. Sci. U. S. A. 2015;112:8626–8631. doi: 10.1073/pnas.1510449112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li J., Hochstrasser M. Microautophagy regulates proteasome homeostasis. Curr. Genet. 2020;66:683–687. doi: 10.1007/s00294-020-01059-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li J., Breker M., Graham M., Schuldiner M., Hochstrasser M. AMPK regulates ESCRT-dependent microautophagy of proteasomes concomitant with proteasome storage granule assembly during glucose starvation. PLoS Genet. 2019;15 doi: 10.1371/journal.pgen.1008387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Peters L.Z., Karmon O., Miodownik S., Ben-Aroya S. Proteasome storage granules are transiently associated with the insoluble protein deposit in Saccharomyces cerevisiae. J. Cell Sci. 2016;129:1190–1197. doi: 10.1242/jcs.179648. [DOI] [PubMed] [Google Scholar]
- 103.Patel A., Malinovska L., Saha S., Wang J., Alberti S., Krishnan Y., et al. ATP as a biological hydrotrope. Science. 2017;356:753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- 104.Munder M.C., Midtvedt D., Franzmann T., Nuske E., Otto O., Herbig M., et al. A pH-driven transition of the cytoplasm from a fluid- to a solid-like state promotes entry into dormancy. Elife. 2016;5 doi: 10.7554/eLife.09347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Peters L.Z., Hazan R., Breker M., Schuldiner M., Ben-Aroya S. Formation and dissociation of proteasome storage granules are regulated by cytosolic pH. J. Cell Biol. 2013;201:663–671. doi: 10.1083/jcb.201211146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cheung T.H., Rando T.A. Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol. 2013;14:329–340. doi: 10.1038/nrm3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bingol B., Schuman E.M. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 2006;441:1144–1148. doi: 10.1038/nature04769. [DOI] [PubMed] [Google Scholar]
- 108.Turker F., Cook E.K., Margolis S.S. The proteasome and its role in the nervous system. Cell Chem. Biol. 2021;28:903–917. doi: 10.1016/j.chembiol.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ramachandran K.V., Margolis S.S. A mammalian nervous-system-specific plasma membrane proteasome complex that modulates neuronal function. Nat. Struct. Mol. Biol. 2017;24:419–430. doi: 10.1038/nsmb.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ramachandran K.V., Fu J.M., Schaffer T.B., Na C.H., Delannoy M., Margolis S.S. Activity-dependent degradation of the nascentome by the neuronal membrane proteasome. Mol. Cell. 2018;71:169–177.e6. doi: 10.1016/j.molcel.2018.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Guo Q., Bin H., Cheng J., Seefelder M., Engler T., Pfeifer G., et al. The cryo-electron microscopy structure of huntingtin. Nature. 2018;555:117–120. doi: 10.1038/nature25502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guo Q., Lehmer C., Martinez-Sanchez A., Rudack T., Beck F., Hartmann H., et al. In situ structure of neuronal C9orf72 poly-GA aggregates reveals proteasome recruitment. Cell. 2018;172:696–705.e12. doi: 10.1016/j.cell.2017.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gierisch M.E., Giovannucci T.A., Dantuma N.P. Reporter-based screens for the ubiquitin/proteasome system. Front. Chem. 2020;8:64. doi: 10.3389/fchem.2020.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sakata E., Eisele M.R., Baumeister W. Molecular and cellular dynamics of the 26S proteasome. Biochim. Biophys. Acta Proteins Proteom. 2021;1869:140583. doi: 10.1016/j.bbapap.2020.140583. [DOI] [PubMed] [Google Scholar]
- 115.Schweitzer A., Aufderheide A., Rudack T., Beck F., Pfeifer G., Plitzko J.M., et al. Structure of the human 26S proteasome at a resolution of 3.9 A. Proc. Natl. Acad. Sci. U. S. A. 2016;113:7816–7821. doi: 10.1073/pnas.1608050113. [DOI] [PMC free article] [PubMed] [Google Scholar]