Abstract
The concentration of Ca2+ in the endoplasmic reticulum (ER) is critically important for maintaining its oxidizing environment as well as for maintaining luminal ATP levels required for chaperone activity. Therefore, local luminal Ca2+ concentrations and the dynamic Ca2+ flux between the different subcellular compartments are tightly controlled. Influx of Ca2+ into the ER is enabled by a reductive shift, which opens the sarcoendoplasmic reticulum calcium transport ATPase pump, building the Ca2+ gradient across the ER membrane required for ATP import. Meanwhile, Ca2+ leakage from the ER has been reported to occur via the Sec61 translocon following protein translocation. In this review, we provide an overview of the complex regulation of Ca2+ homeostasis, Ca2+ flux between subcellular compartments, and the cellular stress response (the unfolded protein response) induced upon dysregulated luminal Ca2+ metabolism. We also provide insight into the structure and gating mechanism at the Sec61 translocon and examine the role of ER-resident cochaperones in assisting the central ER-resident chaperone BiP in the control of luminal Ca2+ concentrations.
Keywords: translocon, chaperone, J-proteins, DnaJ, thioredoxin, stress, BiP/Grp78/HspA5, diabetes, UPR
Abbreviations: AXER, ATP/ADP exchanger in the ER membrane; CRAC, Ca2+-release activated Ca2+ channel; ER, endoplasmic reticulum; ERAD, ER-associated degradation; IP3R, inositol 1,4,5-trisphosphate receptor; JDPs, J-domain proteins; MAM, mitochondria-associated ER membrane; NBD, nucleotide-binding domain; PDI, protein disulfide isomerase; SBD, substrate-binding domain; SERCA, sarcoplasmic/endoplasmic reticulum calcium ATPase; SOCE, store-operated Ca2+ entry; UPR, unfolded protein response; VDCA1, voltage-dependent anion channel 1
The endoplasmic reticulum (ER) is the major store for calcium ions (Ca2+) in the cell. Under physiological conditions, a steep concentration gradient exists between the ER (100 μM up to 1 mM [Ca2+]) and the cytosol (about 100 nM [Ca2+]) (1). Influx of Ca2+ into the ER is mainly achieved by the energy-consuming ATPase SERCA2b (sarcoplasmic/endoplasmic reticulum calcium ATPase) (2) (Fig. 1). On the other hand, Ca2+ efflux from the ER into the cytosol mainly occurs via inositol 1,4,5-trisphosphate receptors (IP3Rs) and ryanodine receptors (3, 4, 5) but also via the translocon Sec61 (6). The IP3R is activated by binding of cytosolic IP3, which is generated by splicing of the membrane anchored phosphoinositol-2-phosphate upon activation of receptors at the plasma membrane by external stimuli such as neurotransmitters, hormones, and growth factors (7). IP3, thus, is the cytosolic messenger, which links signals at the plasma membrane to the release of Ca2+ from intracellular stores. At the Sec61 channel, which is required for the import of secretory and membrane proteins into the ER, Ca2+ efflux from the ER to the cytosol occurs at the end of protein translocation, a process depicted as “leakiness” of Ca2+ (6).
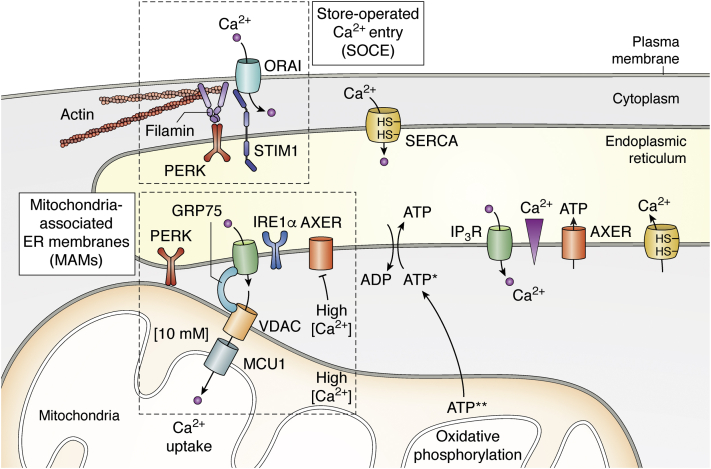
Figure 1.
Schematic illustration of the Ca2+and ATP flux across membranes. The ER is the major Ca2+ store. Ca2+ is transported against a steep concentration gradient into the lumen of the ER via the SERCA pump. Upon ligand stimulation, Ca2+ are released via the IP3R and RyR (here not shown) into the cytosol to activate Ca2+-dependent proteins. ATP import via AXER requires a Ca2+ gradient across the ER membrane and is inhibited in the presence of [Ca2+]cyt > 500 nM. ATP, generated at the inner mitochondrial membrane by oxidative phosphorylation (ATP∗∗), is preferentially transported into the ER. In the presence of high [Ca2+]cyt, transport of ATP (ATP∗) into the ER is discussed to be mediated by a yet unknown transporter. Misfolded proteins in the ER lead to ER-stress, which stimulates Ca2+ transfer from the ER to mitochondria. It is not clear whether Ca2+ uptake in mitochondria is coupled to ATP release. The refilling of intracellular Ca2+ stores by extracellular Ca2+ occurs via SOCE by close apposition of the transmembrane proteins STIM and ORAI. Close apposition of the mitochondrial and ER membrane occurs at MAM sites, in which high [Ca2+] prevail, a prerequisite for Ca2+ uptake into mitochondria. Further details are addressed in the text. AXER, ATP/ADP exchanger in the ER membrane; ER, endoplasmic reticulum; IP3R, inositol 1,4,5-trisphosphate receptor; MAM, mitochondria-associated ER-membrane; RyR, ryanodine receptor; SERCA, sarcoplasmic/endoplasmic reticulum calcium ATPase; SOCE, store-operated Ca2+ entry.
Apart from Ca2+ flux between the ER and the cytosol, there are also Ca2+ flux between the ER and mitochondria. This Ca2+ transfer from the ER to mitochondria is closely linked to mitochondrial ATP production and cell survival (8, 9) and requires a close apposition of the ER membrane and the outer mitochondrial membrane thereby forming microdomains. These microdomains are named mitochondria-associated ER membranes (MAMs) (Fig. 1). MAM contacts have a width of 10 to 25 nm and are optimal for Ca2+ transfer, so called “Ca2+-hot spots”. In these zones, the concentration of Ca2+ is very high reaching levels of more than 10 μM (see review (8)). Efficient Ca2+ flux from the ER to mitochondria is mediated via IP3R, located within the ER membrane, the voltage-dependent anion channel 1 (VDAC1), which is located at the outer mitochondrial membrane, and mitochondrial Ca2+ uniporter 1, located at the inner mitochondrial membrane (8). Interaction of IP3R with VDAC1 is facilitated by the chaperone GRP75, which is enriched in MAMs and associated with both, IP3R and VDAC1 (9) (Fig. 1). Although mitochondrial Ca2+ levels are closely linked to mitochondrial ATP synthesis, Ca2+ influx into and prevailing [Ca2+] within mitochondria need to be carefully surveyed, since excessive Ca2+ accumulation within mitochondria leads to oxidative stress through production of reactive oxygen species and to subsequent apoptosis by releasing cytochrome c and caspase activation (8, 10).
Replenishment of intracellular Ca2+ stores is achieved by a mechanism known as store-operated Ca2+ entry (SOCE) (11, 12) (Fig. 1). SOCE is also known to be triggered by Ca2+ efflux from the ER to establish a rapid and sustained elevation of Ca2+ levels in the cytosol. Elevated Ca2+ levels are required to initiate signaling pathways by regulating Ca2+-dependent proteins, such as kinases and phosphatases (13). SOCE depends on two proteins, STIM, located at the ER membrane, and ORAI, at the plasma membrane, which is a structural component of the Ca2+-release activated Ca2+ channel (CRAC). Two isoforms STIM1 and STIM2 exist, which differentially sense ER Ca2+ levels. Depletion of ER luminal [Ca2+] below its resting levels (of about 400 μM) is sensed by STIM1. While STIM1 senses significant store depletion and activates SOCE, STIM2 senses mild store depletion to maintain basal Ca2+ homeostasis (14). Activated STIM1 relocalizes to ER/plasma membrane junctions to bind to ORAI1 at the plasma membrane and to induce opening of CRAC channels, which triggers Ca2+ entry into the cell (15). After cessation of the stimulus, Ca2+ is pumped into the ER by SERCA. STIM1 is deactivated and releases ORAI1, which results in closing of the CRAC channels (15). Having depicted the most important Ca2+ flux between the ER and the cytosol as well as mitochondria, we will now delineate the interconnection of luminal Ca2+ levels, ER redox state, and ATP levels.
Luminal calcium ion levels affect ER redox equilibrium and luminal ATP levels
The concentration of Ca2+ in the ER is linked to the prevailing redox conditions. Low luminal Ca2+ concentrations induce a reductive shift, which is required for reducing disulfide bonds in the SERCA channel thereby enabling Ca2+ import into the ER (Fig. 2). When luminal Ca2+ levels are restored, oxidation of the respective cysteine residues arrests further Ca2+ import and Ca2+ levels decrease due to the Ca2+ “leakage” at the translocon (16, 17). In order to experimentally study the effects of Ca2+-depletion, the activity of SERCA can be inhibited by the chemical compound thapsigargin (18). Ca2+ depletion in thapsigargin-treated cells results in a reductive shift as shown in HEK293 cells transfected with an ‘ER-tuned GFP redox-responsive’ probe (roGFPiE), which enables fluorescent lifetime imaging in the ER lumen of mammalian cells (19, 20). This reductive shift was shown to be associated with a decrease in protein disulfide isomerase (PDI) mobility, discussed to be due to complexing of PDI with calreticulin (20) (Fig. 2). However, downregulation of PDI using shRNA only resulted in a slight reduction of the fluorescent lifetime of roGFPiE in HEK293 cells (20) indicating that PDI does not play a major role in providing the reductive shift required for the opening of the SERCA channel. Recently, it was shown that the reductive shift—resulting from Ca2+ depletion—is largely independent of the Ca2+-binding calreticulin but associated with increased import of GSH into the ER (21) (Fig. 2). The GSH system with the reduced GSH and the oxidized GSSG redox pair provides an important thiol-disulfide buffer system in the ER (22). Since GSSG cannot be reduced in the ER, maintenance of the ER redox state likely depends on GSH import and GSSG export (23) (Fig. 2). Since thapsigargin-treated cells were shown to have increased GSH levels in the ER lumen, it is now suspected that Ca2+-depletion drives GSH transport through a yet unknown Ca2+-sensitive GSH transporter in the ER membrane (21).
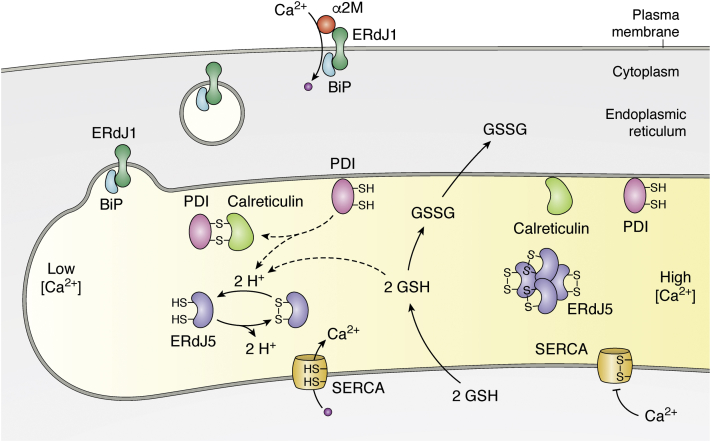
Figure 2.
Schematic illustration of the function of ERdJ1 and ERdJ5 in controlling Ca2+influx. ERdJ1 targets the chaperone BiP to the plasma membrane where it functions as a receptor for α2-macroglobulin (α2M) to enable Ca2+ influx across the plasma membrane into the cytosol. Within the ER, the cochaperone ERdJ5 regulates Ca2+ influx by reducing disulfide bonds of SERCA thereby activating SERCA, which pumps Ca2+ into the ER. The color shading of the ER lumen symbolizes different Ca2+ concentrations in the ER with low [Ca2+] at the left and high [Ca2+] at the right. Under normal Ca2+ concentrations, PDI1A prevails in its reduced and ERdJ5 in its oxidized form, in which it forms large protein complexes. The reductive milieu, required to reduce ERdJ5, is presumed to be provided by PDI, bound to calreticulin and by GSH, which is transported into the ER when Ca2+ concentrations in the ER decrease. Reduced ERdJ5 increases the reductive power to reduce and activate the ATPase activity of the Ca2+ channel SERCA, which—in its reduced state—can pump Ca2+ into the ER. ER, endoplasmic reticulum; PDI, protein disulfide isomerase; SERCA, sarcoplasmic/endoplasmic reticulum calcium ATPase.
Taken together, the low [Ca2+]ER induces a reductive shift in the ER, which leads to the opening of the SERCA pump enabling Ca2+ import. In the presence of high luminal [Ca2+], the oxidizing milieu is readjusted, which is required for protein folding, and further [Ca2+] import is inhibited due to the oxidized state of the SERCA pump.
Besides the function of the ER to be the major cellular Ca2+ store enabling rapid cellular responses, high luminal Ca2+ levels are also mandatory for maintaining a Ca2+ gradient across the ER membrane, which is required for ATP import (24, 25, 26) (Fig. 1). As shown above for GSH, ATP cannot be synthesized in the ER but solely stems from glycolysis (in the cytosol) and oxidative phosphorylation (at the inner mitochondrial membrane) (25) and, therefore, also needs to be transported into the ER (Fig. 1). The corresponding ATP/ADP carrier, named AXER (ATP/ADP exchanger in the ER membrane) has been identified only recently by screening databases of solute carriers (26). In thapsigargin-treated HeLa cells, Ca2+ depletion in the ER results in transiently increased ATP levels, an effect that largely decreased when AXER was downregulated (26). This transient elevation in [ATP]ER is followed by a drastic decline in luminal ATP levels (24, 25, 26). The reason for the transient increase in luminal ATP upon Ca2+ depletion is not known. It is discussed to be due to downregulation of ATP-consuming activities within the ER and could serve to promote chaperoning activity during the initial phase of ER-stress sensed by accumulating misfolded proteins (as outlined further below). The subsequent downregulation of luminal ATP upon Ca2+ depletion has been shown to be due to corresponding increases in cytosolic Ca2+ levels (24). High cytosolic Ca2+ concentrations (ranging between 500 nM to 2 μM)—sensed by a yet unidentified Ca2+ sensing component—block AXER activity and inhibit ATP import into the ER (Fig. 1). The regulation of ATP import by cytosolic Ca2+ levels has been termed CaATiER (Ca2+ antagonizing transport into ER) by Yong et al. (24). This group further elaborated that a Ca2+ gradient across the ER membrane (with [Ca2+]ER > [Ca2+]cyt) is required for ATP import into the ER (Fig. 1). Furthermore, they could show that the ATP pumped into the ER stems from mitochondrial oxidative phosphorylation (24). Since mitochondrial and ER membranes are closest at MAM sites, efficient transport of mitochondrial-derived ATP is most likely to occur at these sites. Accordingly, a yet unresolved question evolves since at these sites, such high Ca2+ concentrations (10 μM) prevail that AXER activity is inhibited. It must be assumed that ATP transport and cytosolic Ca2+ hotspots are either temporally or spatially separated. Alternatively, another not yet identified ATP transporter might mediate the ATP import in the presence of high [Ca2+] at MAMs. Yong et al (24) postulate the existence of a Ca2+ sensing component, which attenuates the ATP transport in the presence of high [Ca2+]cyt enabling the transport of ATP into the ER only when the [Ca2+]cyt is at physiological levels.
Dysregulated calcium homeostasis induces the unfolded protein response
As discussed above, Ca2+ levels within the ER have to be tightly controlled in order to ensure appropriate luminal ATP levels. Furthermore, Ca2+ and ATP are required for the activity of molecular chaperones and folding enzymes.
Therefore, it is not surprising that depletion of Ca2+ from the ER and subsequent energy deficiency lead to ER-stress due to impaired folding capacity and subsequent accumulation of misfolded or unfolded proteins within the ER lumen (2, 27, 28, 29). Likewise, agents that impair the ER redox homeostasis such as DTT or thapsigargin cause ER-stress (30). During ER-stress, a stress response is induced to reestablish ER homeostasis and to minimize cell damage. This complex stress response is named the unfolded protein response (UPR). The UPR succeeds in transiently arresting protein synthesis by inducing transcriptional and translational upregulation of chaperones in an attempt to refold proteins or at least to keep unfolded and misfolded proteins in a soluble conformation to enable restoration of proteostasis. Simultaneously, irreversibly misfolded proteins are subjected to ER-associated degradation (ERAD). Prolonged ER-stress eventually induces apoptosis (29). During the UPR, ER-stress can mainly be sensed by three ER membrane proteins: protein kinase RNA-like ER kinase (PERK), inositol-requiring enzyme 1 α (IRE1α) and activating transcription factor 6 (ATF6) (29, 31).
With regard to IRE1α, autophosphorylation of its ribonuclease domain activates the endoribonuclease domain of IRE1α, which catalyzes the splicing of prevailing mRNA of X-Box binding protein 1 (Xbp1) resulting in spliced Xbp1 mRNA (Xbp1s) (32). Xbp1s is efficiently translated into XBP1 protein, which travels to the nucleus to act as a transcription factor for several genes associated with the ERAD pathway and the folding machinery in the ER lumen (33). Apart from upregulating a subset of genes that are important for restoration of ER homeostasis, the activated endoribonuclease domain of IRE1α degrades a subset of coding and noncoding cytosolic mRNAs to stop ongoing protein synthesis. This mechanism is called regulated IRE1α-dependent decay of mRNA (34). Termination of IRE1α signaling occurs via dephosphorylation of IRE1α and its monomerization (29, 35). If ER homeostasis cannot be restored, apoptosis is induced at late stages of ER-stress. Interestingly, besides its classical function as ER-stress transducer, IRE1α is found to be located at MAM sites (36, 37) (Fig. 1). Experiments with different IRE1α mutants revealed that the function of IRE1α at MAM sites is independent of its enzymatic activities but due to its ability to form a stable complex with IP3R (36). At MAM sites, IRE1α was shown to control Ca2+ transport from the ER to mitochondria i) by serving as a scaffold keeping the optimal distance between ER and mitochondrial membranes and ii) by directing IP3R to MAMs (36).
Autophosphorylation of PERK results in phosphorylation of the transcription factor NRF2 and the eukaryotic initiation factor 2α (eIF2α) (31, 38). Phosphorylation of eIF2α inhibits general protein synthesis in the cytosol but simultaneously stimulates preferential synthesis of specific proteins including the activating transcription factor 4 (ATF4) (29). ATF4 increases transcription of genes involved in transport of amino acids, synthesis of GSH, and maintenance of redox homeostasis (31, 39, 40). Upon prolonged ER-stress, ATF4 induces the transcription of the transcription factor CHOP, which can induce subsequent apoptosis. Furthermore, ATF4 can activate the transcription of GADD34, which dephosphorylates p-eIF2α to restore general protein synthesis (29). Phosphorylation of NRF2, on the other hand, results in dissociation of the protein from its binding partner KEAP1 in the cytosol, which enables trafficking of the transcription factor to the nucleus (31). NRF2 target genes include many detoxifying enzymes as well as cellular transporters, proteins involved in protein folding, as well as genes of proteasomal subunits (40). Besides these classical ER-stress transducer functions, PERK is also found to be enriched at MAMs, where it is required to maintain ER-mitochondrial juxtapositions, a function that is independent of the kinase domain but—similar to the findings for IRE1α (see above)—requires the cytosolic domain (Fig. 1). PERK-deficient cells were shown to contain considerable weaker MAMs than WT cells (41), see also the recent review (37). The drastic structural modifications at MAM sites and the reduced ER-stress–induced mitochondria-mediated apoptotic rate in PERK-deficient cells indicate the necessity of PERK for propagation of ER-stress to the mitochondria to induce apoptotic signaling (41). An additional, UPR-independent function could be assigned to PERK by the identification of the actin-binding protein Filamin A (FLNA) as a novel PERK-interaction partner. This interaction seems to be crucial for the formation of juxtapositions of the ER membrane with the plasma membrane, the close vicinity of the membranes is a prerequisite for SOCE-mediated Ca2+ influx (42) (Fig. 1).
The third sensor of ER-stress, activating transcription factor 6 (ATF6), which is also silenced by its binding to BiP and activated upon its dissociation (29), differs in its activation pattern from PERK and IRE1α. When activated, ATF6 travels to the Golgi complex where the cytosolic N-terminus of ATF6 (ATF6N) is cleaved off by regulated intramembrane proteolysis (29). ATF6N travels to the nucleus to function as transcription factor to upregulate transcription of BiP and other chaperones (43). ATF6N has also been reported to dimerize with the transcription factor XBP1 to increase upregulation of several ERAD components and cochaperones (43). There is no evidence to date that ATF6 contributes to the formation, stabilization, or architecture of membrane juxtapositions.
Activation of the UPR is mediated by the ER-resident Hsp70 chaperone BiP, which is the central protein for chaperoning activity in the ER. It has been shown that the binding of BiP to the luminal domains of IRE1α, PERK, and ATF6 keeps them in a silenced state (29, 31, 44, 45). BiP has two distinct domains: a substrate-binding domain (SBD) and a nucleotide-binding domain (NBD). Under control conditions, BiP is bound to luminal domains of the transmembrane proteins IRE1α and PERK by its NBD site (44) (see Fig. 3). When misfolded proteins accumulate in the ER, their binding to the SBD induces the release of BiP from IRE1α and PERK thereby activating them and enabling their oligomerization and autophosphorylation (31, 38, 46, 47) (Fig. 3B). In addition to this longstanding model, an extended model based on structural and mutational analyses in yeast was presented by Credle et al (48). They showed that unfolded proteins can activate and enhance the UPR by directly binding into the highly conserved groove of the Ire1 (45, 48) (Fig. 3C). It is, thus, possible that BiP release is not a requirement for Ire1 activation, but that it might provide a regulatory role under extreme activation conditions when the pool of free BiP becomes severely depleted. Since the Ire1 mutant, defective in binding BiP, is mildly constitutively active, BiP binding to Ire1 might dampen activation of Ire1 under conditions of mild accumulation of unfolded protein, thereby reducing ‘‘noise’’ in UPR signaling (48). Since the luminal domains of IRE1α and PERK are highly homologous, this model can also be extended to the activation mechanism of PERK by unfolded proteins (48) (Fig. 3C).
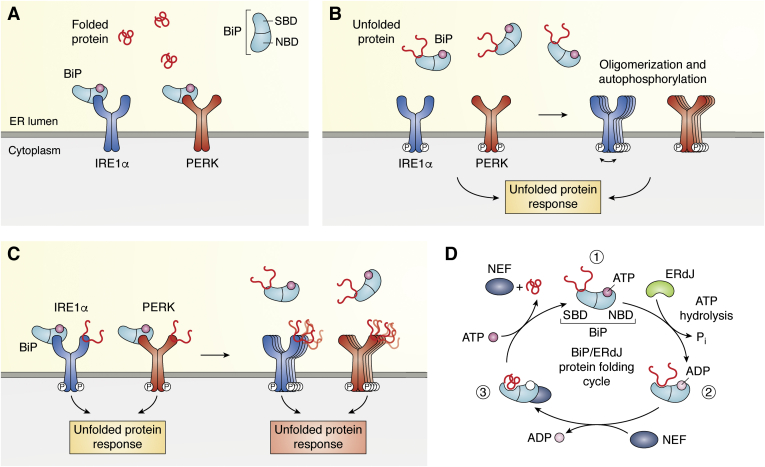
Figure 3.
Induction of ER-stress by misfolded proteins.A, BiP has a substrate-binding domain (SBD) and a nucleotide-binding domain (NBD). IRE and PERK are transmembrane proteins with high homologous luminal domains, which are silenced by BiP-NBD binding under normal conditions, when nascent proteins are folded properly (red coiled lines). B, in ER-stressed cells, misfolded proteins accumulate and engage luminal BiP for refolding. By binding of the misfolded proteins to the SBD, BiP dissociates from the luminal domains, enabling the oligomerization and autophosphorylation of the transmembrane proteins thereby activating the UPR. C, according to the extended model, misfolded proteins bind directly to the luminal domains of the transmembrane proteins, enabling their activation and stimulation of the UPR. Under more severe stress conditions, even BiP, which is usually bound to the transmembrane domains, binds unfolded protein and dissociates from the respective luminal domains thereby enforcing the UPR. D, misfolded proteins, bound by BiP, are subjected to a cycle to enable their refolding. This refolding, ATP-consuming chaperoning is assisted by ERdJ proteins and nucleotide exchange factors (NEF). In a first step (1), BiP is shown in its ATP-bound state, in which binding affinity to the substrate (unfolded, native protein or misfolded protein) is low. In a second step (2), ERdJ proteins, as cochaperones, bind to BiP and stimulate ATP hydrolysis. In the ADP-bound state, the affinity of BiP to the substrate is increased. When protein folding has been completed, ADP is exchanged for ATP by the nucleotide exchange factor (NEF) resulting in release of the substrate protein (3). ER, endoplasmic reticulum; UPR, unfolded protein response.
Apart from its role in silencing the transmembrane proteins IRE1α, PERK, and ATF6 and in transmitting ER-stress via activation of the UPR, BiP has manifold additional functions. BiP mediates proper protein folding and refolding of misfolded proteins, processes which depend on the availability of ATP. Due to its Ca2+-binding capacity, BiP is a major Ca2+ store in the ER (49) and it also controls [Ca2+]ER efflux at the Sec61 translocon and at IP3R. At the Sec61 translocon, BiP is required to keep it in a closed state (50), while at the IP3R (type 1) channel, BiP was shown to activate it and to promote [Ca2+] efflux (51). The chaperoning and gating functions of BiP are ATP dependent as shown for IP3R1 (51) and outlined below for its function at the Sec61 translocon. Chaperoning activity of BiP is known to be assisted by cochaperones, which stimulate—due to their highly conserved J-domain—the ATPase activity of BiP. These cochaperones are also known by now to exert diverse functions. In the following section, we will briefly describe the chaperone cycle of BiP and its cochaperones and then consider the role of these individual cochaperones in regulating the Ca2+ flux across membranes.
The ATP-consuming chaperoning activity of the Hsp70 protein BiP is based on two conformationally distinct structures of BiP depending on its binding to ADP or ATP at its NBD (Fig. 3D). When bound to ATP, the affinity of BiP-SBD to substrate is low, while in the ADP-bound state, substrate affinity is high. Changes in the affinity of BiP to substrates are mediated by conformational changes following ATP hydrolysis. As soon as folding of a substrate protein has been accomplished, BiP is released from the substrate. This release is mediated by so called nucleotide exchange factors, which mediate exchange of ADP for ATP (52) (Fig. 3D). A group of proteins, the J-domain proteins (JDPs), has been identified to cooperate with Hsp70 chaperones to stimulate their ATPase activity and thus enhance affinity of Hsp70 proteins for substrates. Binding to Hsp70 and stimulation of the ATPase activity is mediated by the J-domain, a highly conserved sequence characteristic for all members of the JDP family (53). As these proteins are no client proteins of Hsp70 themselves but stimulate the function of Hsp70 chaperones, these proteins are also termed cochaperones (54). A specific set of cochaperones resides within the ER lumen, the so called ERdJ proteins. While the J-domain is highly conserved and required for their function as cochaperones, some JDPs contain additional features such as a glycine/phenylalanine rich region (G/F region) as well as a zinc finger domain. It was proposed that apart from the J-domain, the G/F region is also important for binding of JDPs to Hsp70 chaperones (55). The G/F rich region was further shown to be essential for interaction with target proteins as was shown for the ERdJ4 (56, 57, 58, 59). Up to now, eight members of the ER-resident JDPs, ERdJ1 – ERdJ8, are characterized that stimulate ATPase activity of BiP and assist in maintaining ionic homeostasis and proteostasis in the ER (52, 60). Apart from their function in assisting BiP in folding of target proteins, BiP and four of the eight ER-resident cochaperones are also known to be involved in the regulation of Ca2+ homeostasis. Of these, ERdJ3 and ERdJ6 are operating at the Sec61 translocon (Fig. 4), ERdJ1 and ERdJ5 at the plasma membrane or acting as oxidoreductases, respectively (Fig. 2). No evidence is currently available pointing to a contribution of ERdJ2 and ERdJ4 in controlling or regulating Ca2+ homeostasis. Little is yet known about the function of the recently detected proteins ERdJ7 and ERdJ8. Kaz Nagata showed that ERdJ8 (DNAJC16) is an ER-membrane protein with both, the J and a thioredoxin domain, facing the luminal side of the ER. Due to its location in ER membrane subdomains, which are in close vicinity to mitochondrial membranes, ERdJ8 is presumed to be localized at MAMs (61). Yet, knockdown experiments indicate that ERdJ8 regulates autophagosomal degradation of mitochondria and controls the size of newly formed autophagosomes (60). In the following, we will briefly summarize recent understandings of the Sec61 translocon and the impact of BiP in gating and inhibiting Ca2+ efflux at the translocon, before we discuss cochaperone-mediated mechanisms, which regulate the luminal Ca2+ homeostasis.
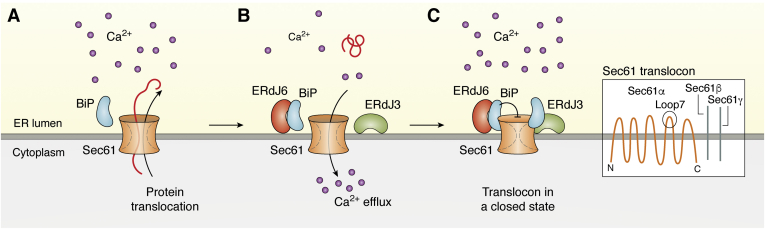
Figure 4.
Schematic illustration of the function of ERdJ3 and ERdJ6 in controlling Ca2+efflux.A, protein translocation across the ER membrane is mediated via the Sec61 translocon. B, at the end of the translocation process, the Sec61 channel mediates efflux of Ca2+ from the ER to the cytosol resulting in loss of Ca2+ within the ER. C, translocon closure is achieved by binding of BiP to loop7 of the Sec61α subunit and by binding of ERdJ3 and ERdJ6. Translocon closure results in upregulation of [Ca2+]ER levels. The exact mechanism how ERdJ3 and ERdJ6 gate the translocon and whether interaction with BiP is required for this function has still to be elucidated. Inset: The translocon Sec61 is composed of three subunits (α, β, γ). Sec61 β and Sec61 γ have one transmembrane domain. Sec61α has 10 transmembrane domains. Loop7, which is located between the transmembrane domains 7 and 8, has been shown to be required for interaction with BiP. ER, endoplasmic reticulum.
BiP and cochaperone-mediated, Sec61-dependent regulation of calcium ion homeostasis
The Sec61 translocon constitutes the main channel for protein translocation into the ER in mammalian cells (62). Around one third of all cellular proteins have to transit this channel to enter the ER where they undergo maturation, quality control, and—in case of enduring misfolding—their delivery to the ERAD pathway. The processes at the Sec61 translocon are highly demanding since the translocon must allow for passage of nascent proteins while maintaining the permeability barrier between the cytosol and ER lumen. Leakiness of this channel for Ca2+ has been shown in experiments using planar lipid (6), and in Sec61A1-deficient cells, live cell Ca2+ imaging experiments revealed a significant decline in [Ca2+]ER efflux in the presence of thapsigargin (6). These results strongly indicated that experimental inhibition of the SERCA pump results in Ca2+ depletion of the ER lumen that is not based on an active IP3R- or ryanodine receptor–mediated Ca2+ burst but based on a Ca2+ efflux along the Ca2+ gradient at the end of protein translocation across the Sec61 channel (63, 64, 65, 66, 67, 68) (Fig. 4). The structure of Sec61 pore and the mechanism engaged in gating this channel are depicted in the following section.
The translocon is composed of three subunits (α, β, and γ) (69). The Sec61α subunit is the largest subunit with 10 transmembrane domains that create a pore whereas the smaller β and γ subunits span the ER membrane only once (69) (Fig. 4). A lateral gate enables the insertion of transmembrane domains of translocated proteins. At the cytosolic side, the translocon is sealed by tight ribosome-membrane junctions (70). At the luminal side, a plug-like structure is formed by Sec61α that keeps the translocon closed in the absence of protein translocation and opens the translocon upon initiation of translocation (69, 71, 72).
In addition, BiP was shown to be necessary to seal the translocon during early stages of translocation as well as repetitively during transmembrane domain insertion of emerging proteins (70, 73, 74). BiP was also shown to be required for sealing nontranslocating translocons (70). The sealing of the translocon by BiP occurs in a stochiometric, noncatalytic manner (70). By using hydrophilic collisional quenching agents in isolated microsomes, reconstituted with purified BiP, it was shown that BiP-mediated sealing of the translocon closely resembles the BiP ATPase cycle described above (see also Fig. 3D). BiP associates with the translocon in its ATP-bound state. When ATP is hydrolyzed to ADP, affinity of BiP to the translocon increases so that the translocon is sealed. Exchange of ADP for ATP then induces a conformational change in BiP so that the translocon is opened (74). Even though it was shown that BiP has to be in the ADP-bound state to seal the translocon during protein translocation (74), recently, the necessity of ATP to close the Sec61 channel in order to limit Ca2+ efflux was verified by downregulation of the ATP carrier AXER, which resulted in increased Ca2+ leakage from the ER (26). Experiments with reconstituted microsomes further revealed that a membrane-bound JDP is required for translocon closure (74). BiP-mediated closure of the pore seems to depend on a functional interaction of BiP with a membrane-bound JDP at or near the translocon (70, 74). Interestingly, since it is the ADP-bound conformation of BiP that is required for sealing the translocon, the interaction of BiP and ERdJ protein is important for something other than stimulation of the ATPase activity of BiP (74). According to these results, a model was proposed in which an ERdJ protein is located within the ER membrane near the translocon, which recruits ADP-BiP to the translocon to close it in a plug-like way. According to their localization near the translocon, the transmembrane proteins ERdJ1 and ERdJ2 are good candidate J-proteins to fulfill such a required function, which comprises possibly the targeting of ADP-BiP to the translocon. Yet, neither downregulation of ERdj1, ERdj2, nor ERdj7 in HeLa cells did increase Ca2+ efflux from the ER (75). This draws the attention to two other cochaperones ERdJ3 and ERdJ6, both of them being known to be involved in the degradation of proteins. ERdJ3 was shown to deliver substrate proteins to BiP to facilitate their retrograde translocation within the ERAD pathway (76, 77, 78, 79, 80). Furthermore, secreted ERdJ3 was shown to bind to aggregation-prone proteins in the extracellular environment (81). ERdJ6 has different subcellular locations and functions (as reviewed (82)). It is known to mediate protein degradation via a cotranslocational degradation pathway (83). Recently, it was shown that ERdJ6 is required for Golgi-independent trafficking of BiP to the cell surface (84). This unconventional traffic of BiP to the cell surface has been reported in lung and colon cancer cell lines (84). Here, ER-derived vesicles translocate to the cell surface through early and recycling endosomes. The loading into the ER-derived vesicles was shown to require ERdJ6, which is upregulated by PERK via the PERK–Akt–mTOR axis in response to ER-stress (84). Both, ERdJ3 and ERdJ6 are known to be soluble proteins in the ER lumen (85, 86) but a membrane-bound pool of both cochaperones has been shown to exist (see recent review (82)). ERdJ3 and ERdJ6 were both shown to interact with BiP and the Sec61 translocon at the ER membrane (76, 83, 85, 86, 87) (Fig. 4). Knockdown experiments revealed that downregulation of either ERdJ3 or ERdJ6 increases Ca2+ efflux from the ER. This effect could be abolished when silencing the expression of Sec61 itself pointing to the ability of both cochaperones to survey Ca2+ efflux at the translocon (75). It is of interest to note that downregulation of ERdJ6 leads to a compensatory upregulation in ERdJ3 protein levels, the increased levels are, however, not able to complement for downregulated ERdJ6 levels. Downregulation of both cochaperones even increased Ca2+ leakage from the ER (75), suggesting that both cochaperones cooperate during translocon closure and that they do not have overlapping functions in this respect. An even increased Ca2+ efflux from the ER was reported to occur upon simultaneous downregulation of ERdJ3 or ERdJ6 and BiP (75).
Since knocking down of ERdJ3 or ERdJ6 could possibly result in a general disturbance of ER proteostasis, which induces ER-stress and subsequent Ca2+ efflux (88), it is of interest to note that neither silencing of ERdJ3 nor ERdJ6 resulted in activation of the UPR (75). These data further manifest that Ca2+ efflux in ERdJ3/ERdJ6 knockdowns is not a consequence of ER-stress but that Ca2+ efflux is prevented by interaction of both cochaperones with BiP at the translocon. Nothing is yet known about the binding dynamics of ERdJ3 and ERdJ6 to BiP with regard to gating the Sec61 translocon. It would be of major interest to investigate the effect of ERdJ3/ERdJ6 mutants that cannot bind to BiP on Ca2+ efflux to investigate whether interaction with BiP or even stimulation of the ATPase activity of BiP by ERdJ3 or ERdJ6 activity is required for closing the translocon and whether ERdJ3 and ERdJ6 act simultaneously or sequentially.
It is also not known whether ADP-BiP or the complex ADP-BiP–J-protein serve as a plug for sealing the translocon or whether the underlying mechanism for translocon closure is based on a conformational change induced by the interaction of ADP-BiP or the complex ADP-BiP–J-protein with Sec61. Yet, the binding site of BiP to the translocon could be specified using different Sec61 mutants. The Y344H Sec61 mutant carrying a point mutation within loop7—located between transmembrane domain 7 and 8 of Sec61α—showed impaired binding to BiP and increased Ca2+ leakage revealing that loop7 of the Sec61 translocon is central for translocon closure (50). ATP and J-domain dependency for gating the translocon points to the participation of cochaperones in stimulating the ATPase activity of BiP and furthermore clearly shows that the Sec61 translocon is in fact a gated Ca2+ channel, regulated by the activity of BiP.
Taken together, ERdJ3 and ERdJ6 are good candidates in cooperating with BiP at the translocon since a subpopulation of both has been shown to be membrane bound and both assist BiP in sealing the translocon. Since ERdJ3 and ERdJ6 cannot compensate each other’s function in limiting Ca2+ efflux, they affect translocon closure by distinct mechanisms.
Having specific functions in translocon closure to limit Ca2+ efflux, ERdJ3 and ERdJ6 are expected to be upregulated in response to Ca2+ depletion within the ER to prevent further Ca2+ efflux. This assumption could be confirmed by showing that indeed, thapsigargin treatment, which depletes ER Ca2+ stores by blocking SERCA2b, leads to upregulation of ERdj3 (78) and ERdj6 (89, 90).
Considering the experimental data outlined above, a regulatory feedback loop can be postulated linking ATP homeostasis to Ca2+ homeostasis in the ER lumen, which enables the cell to regain proteostasis in the ER at the early onset of ER-stress. Since chaperoning and closing the Sec61 translocon mediated by BiP require energy (26), changes in cellular microenvironment challenge the maintenance of the ionic and protein homeostasis and more ATP is required to meet the increasing chaperoning demand for proper folding of translocated proteins. Eventually, the ER-localized ATP pool will decline. Since the closure of the Sec61 translocon requires ATP (74), it cannot be kept in a closed state (26), resulting in Ca2+ depletion from the ER via the Sec61 translocon. The initial increase in [ATP]ER observed in response to Ca2+ depletion (25, 26) could be a mechanism to restore ER Ca2+ homeostasis by closing the Sec61 translocon as well as by enabling BiP to continue its chaperoning function. In case, the temporary rise in ATP is not sufficient to restore ER homeostasis (e.g., due to high levels of ER-stress, and an ATP demand that cannot be met by the rise in ATP), Ca2+ leakage into the cytosol would progress resulting in abrogation of the Ca2+ gradient across the ER membrane and increased cytosolic Ca2+ concentrations, conditions which inhibit AXER and ATP transport into the ER (24). This scenario could be resolved by regaining physiological [Ca2+]cyt levels either by pumping Ca2+ across the plasma membrane into the extracellular space or by refilling luminal Ca2+ stores via SOCE and SERCA activity.
This model is in accordance with another hypothetical model presented by Ushioda et al., who discuss that occasional Ca2+ depletion could clear the ER of misfolded proteins (16). This hypothesis is based on the finding that Ca2+ depletion can activate the UPR signaling cascade, which results in an arrest of general mRNA translation and increased degradation of ERAD substrates. Thus, occasional periods of Ca2+ depletion could be exploited to clear the ER of misfolded cargo while preventing degradation of nascent proteins due to translational arrest. Ca2+ homeostasis could then be regained by the presented regulatory feedback mechanism.
Diabetic phenotype in ERdJ6-deficient mice
Since the dynamics in Ca2+ flux across membranes and the prevailing Ca2+ levels within every subcellular compartment are key positions for regulating the cellular metabolism, disturbances in Ca2+ homeostasis have been shown to be associated with metabolic diseases, such as diabetes mellitus (9). A diabetic phenotype was reported in mice carrying the Y344H mutation in loop7 of the Sec61 translocon, which was found to abolish BiP binding and resulted in an increased Ca2+ efflux from the ER (50, 91). Considering the suggested role of ERdJ6 in Sec61 channel closure, it is interesting that a diabetic phenotype was also obtained in ERdj6 KO mice (92). ERdj6 KO mice present with glucosuria, hyperglycaemia, and hypoinsulinemia. Furthermore, there is a reduced amount of pancreatic beta cells in these mice, presumably due to increased proapoptotic signaling (92). Increased apoptotic rates could be due to [Ca2+]ER depletion, resulting in general ER-stress and subsequent apoptosis by UPR signaling pathways. Furthermore, [Ca2+]ER depletion could prevent Ca2+ flux between the ER and mitochondria. Increased mitochondrial Ca2+ concentrations are important for mitochondrial ATP production, which in turn is required for insulin secretion (9), when cells are stimulated with glucose to produce around one million molecules of proinsulin per minute. In addition, downregulation of ERdJ6 might increase PERK signaling and PERK-induced apoptosis due to lack of inhibition through ERdJ6 (92).
Hypoinsulinemia in ERdJ6-deficient mice could also be based on the resultant reductive shift due to [Ca2+]ER depletion (20). For folding and sorting of insulin in the ER, three intramolecular disulfide bonds have to be formed by PDI (9). Accordingly, defective insulin folding and maturation could be based on the lack of PDI, which is sequestered by calreticulin upon [Ca2+]ER depletion (20), thereby affecting redox homeostasis (as further outlined in the following section).
While ERdJ3 and ERdJ6 assist in closing the Sec61 translocon to limit Ca2+ leakage from the ER, ERdJ1 and ERdJ5 have been found to regulate cellular Ca2+ homeostasis by Sec61-independent mechanisms.
Cochaperone mediated, Sec61-independent regulation of calcium ion homeostasis
ERdJ5 activates SERCA2b upon [Ca2+]ER depletion and regulates mitochondrial dynamics
ERdJ5 is an ER-resident reductase, which contains a J-domain at its N-terminus and four thioredoxin-like domains with redox-active motifs (Cys-X-X-Cys, CXXC) (93). It is known to cleave disulfide bonds of misfolded proteins to target them to BiP for retrotranslocation via the ERAD pathway (94, 95). Within the ER, ERdJ5 maintains luminal Ca2+ stores by controlling Ca2+ influx in response to [Ca2+]ER depletion (16) (Fig. 2). In HEK293T cells, it was shown that ERdJ5 is normally present in its oxidized form without reductase activity. However, upon decreasing Ca2+ concentrations within the ER, an increasing fraction of ERdJ5 becomes reduced (Fig. 2). Thus, upon Ca2+ depletion, the reductive power of ERdJ5 increases while the oxidizing PDI1A is sequestered by calreticulin thereby increasing the reductive potential of ERdJ5 in the ER (16). This reductive power is required for activating SERCA. ERdJ5 can interact with the Ca2+ pump SERCA2b, as shown in HeLa and HEK293T cell, and increases its ATPase activity (16). This effect is lost when all four thioredoxin domains of ERdJ5 were mutated (16). Since ATPase activity was shown to positively correlate with the amount of reduced SERCA2b, it is assumed that ERdJ5 activates SERCA2b by reduction of an intramolecular disulfide bond under low Ca2+ concentrations (16). In line with this is the observation that ERdj5 KO MEFs had decreased [Ca2+]ER levels, a defect that could be compensated by overexpression of ERdj5 (16). Interestingly, compensation was also obtained by overexpression of an ERdJ5 mutant that could not bind to BiP, indicating that the interaction of ERdJ5 with BiP is not required for controlling Ca2+ influx. The ability to control Ca2+ influx into the ER thus seems to be solely due to the reductive potential of ERdJ5. Taking into account that ERdJ5 regulates SERCA activity and Ca2+ influx in response to [Ca2+]ER depletion (16), ERdj5 should be upregulated upon Ca2+ depletion. Indeed, transcription of ERdj5 was shown to be upregulated in response to treatment with thapsigargin and Ca2+ ionophore, which depletes [Ca2+]ER stores by promoting Ca2+ efflux from the ER (96).
Though interaction with BiP is not required for the function of ERdJ5 to control Ca2+ influx into the ER, BiP has been shown to modulate ERdJ5 motility and its ability to bind to SERCA2b in a [Ca2+]- dependent way (16). While Ca2+ depletion did not affect ERdJ5 mobility, high Ca2+ concentrations promoted oligomeric assembly of ERdJ5 within the ER keeping the complex less mobile as shown by coimmunoprecipitation studies using FLAG- and myc-tagged ERdJ5 constructs as well as by cellular fractionation experiments (16, 20). Monomerization and mobility of ERdJ5 is regained in the presence of BiP as indicated by two independent experiments: While overexpression of BiP facilitated binding of ERdJ5 to SERCA2b even under higher Ca2+ concentrations, in the absence of BiP, ERdJ5 is found in higher molecular weight fractions having less binding affinity (16). This BiP-controlled availability of ERdJ5 points to the presence of an additional mechanism that allows fine-tuning of Ca2+ import. All in all, the data point to an important role of ERdJ5 reductase activity in maintaining the physiological Ca2+ influx into the ER (Fig. 2). By means of ERdj5 KO cells, it was recently shown that ERdJ5 is not only required for Ca2+ homeostasis in the ER but also for the Ca2+ homeostasis in the cytosol (97). Interestingly, mitochondrial Ca2+ levels were unaffected in ERdJ5-deficient cells although fragmentation of mitochondria was observed, and ERdJ5-deficient cells proved to be more sensitive to apoptosis (97). The observed mitochondrial fission in ERdJ5-deficient cells was shown to be due to aberrant phosphorylation of the cytosolic GTPase DRP1 (dynamin-related protein 1), which is caused by the increase in cytosolic [Ca2+] (97). The exact mechanism how ERdJ5 affects cytosolic [Ca2+] levels is not fully understood, yet the measured elevation in [Ca2+]cyt is very likely due to the inhibition of SERCA2b activity. The impact on DRP1 phosphorylation is very interesting, since the phosphorylation pattern of DRP1 has been shown to be a key driver of metabolic diseases and inhibition of DRP1 phosphorylation, by overexpression of the phosphorylation mutant of S600A, was shown to improve glucose tolerance and to protect against diet-induced glucose intolerance (98). Whether ERdJ5-deficiency directs the metabolism toward diabetes is yet not known.
Taken together, the presented data point to another regulatory feedback loop linking redox homeostasis with Ca2+ homeostasis within the ER lumen. As previously discussed, Ca2+ depletion in the ER results in depletion of ATP (26) and an accompanying reductive shift within the ER either by sequestration of PDI, or increasing influx of GSH, or both (16, 19, 21). This reductive shift should result in increased breaking of disulfide bridges, which is important for the retrotranslocation of misfolded proteins into the cytosol for their degradation. Activation of the UPR by Ca2+ depletion and accompanying upregulation of ERAD components could further aid in clearing the ER of misfolded cargo. At the same time, the reductive shift within the ER would result in SERCA activation by ERdJ5, increasing Ca2+ influx into the ER and establishing the Ca2+ gradient required for AXER activity and ATP transfer into the ER. The rise in ER luminal ATP levels would result in closing the translocon and subsequent restoration of ER homeostasis.
ERdJ1 mediates BiP translocation to the cell surface and regulates intracellular Ca2+ levels
ERdJ1 is a transmembrane protein with its J-domain in the ER lumen and a cytosolic C-terminus (99). The cytosolic C-terminus of ERdJ1 can associate with ribosomes (99), thereby arresting protein synthesis, an arrest that is released upon binding of BiP (99, 100, 101). ERdJ1 and BiP, which are primarily localized in the ER compartment, have also been shown to form a complex at the plasma membrane of macrophages (102, 103, 104). At the plasma membrane, BiP functions as a cell surface receptor for various ligands including α2-macroglobulin (104, 105) (Fig. 2). Under normal conditions, this leads to an increase of the intracellular [Ca2+], probably via G-protein–mediated signaling (102, 103, 104). Coimmunoprecipitation experiments in peritoneal macrophages revealed that downregulation of ERdJ1 by RNA interference reduced the binding of BiP to the plasma membrane and the binding of α2-macroglobulin to the cell surface. Consequently, downregulation of ERdJ1 inhibited the α2-macroglobulin–mediated increase of the intracellular Ca2+ concentrations (103). These results indicate that—in the presence of low cellular Ca2+ levels—ERdJ1 plays a crucial role in targeting BiP to the plasma membrane of cells to regulate the Ca2+ influx across the plasma membrane (103), possibly by inducing SOCE, which had been shown to be activated by ER Ca2+ depletion (Fig. 1). As ERdJ1 stimulates Ca2+ influx into the cytosol, it is reasonable to assume that ERdJ1 is upregulated upon Ca2+ depletion. Yet, presently no data are published with respect to the regulation of ERdJ1 upon treatment of cells with thapsigargin or ionophore.
The mechanism how BiP enables Ca2+ influx is yet not fully understood. As a receptor of α2-macroglobulin, BiP could assist and control the formation and opening of the CRAC channel. Studies in 1-LN cells implicated that BiP colocalizes with plasma membrane–located VDAC channels and that binding of ligands to BiP induces a rise in intracellular Ca2+ through VDAC (106).
Another interesting aspect is the question how BiP is transported to the plasma membrane, since BiP contains the KDEL sequence at its C-terminus, which redirects it to the ER compartment. The phenomenon itself, that BiP is localized at the plasma membrane, was observed as early as in 1997, when a “highly homologous protein to BiP” was reported to be localized to the cell surface of lymphoma cells of malignant cutaneous T cell lymphoma (107). One year later, it was shown that cell surface expression of BiP was induced by thapsigargin in human rhabdomyosarcoma cells (108). Also, in NG108-15 glioblastoma cells, cell surface expression of BiP was detected (109). In NG 108-15, it was found that BiP translocation to the plasma membrane was blocked after treatment with brefeldin A, suggesting that BiP translocation is mediated through the Golgi apparatus (109), which was confirmed in liver and pancreatic cell lines (84). By now, the underlying mechanism has been elaborated. KDEL receptors at the Golgi apparatus are normally responsible for binding to proteins carrying a KDEL sequence (KDEL proteins) that have translocated from the ER to the Golgi. Upon binding of these proteins to the KDEL receptors, the proteins are retrotranslocated to the ER resulting in the retention of KDEL proteins in the ER (110). The translocation of KDEL receptors from the Golgi apparatus to the ER is due to the activation of tyrosinkinase SRC, which is localized at Golgi membranes (111). In ER-stressed HeLa cells, it was shown that IRE1α binds to SRC and induces a dispersion of KDEL receptors in the Golgi apparatus, enabling BiP to escape from ER retention and to traffic to the plasma membrane (112). The exact mechanism on how ERdJ1 is involved in mediating BiP transport to the plasma membrane is yet not known.
Conclusion
Protein and ionic homeostasis in ER are essential for cellular metabolism and survival. Disturbance of this finely balanced homeostasis results in metabolic diseases such as diabetes. Within the ER, the Hsp70 chaperone BiP and its ER-resident cochaperones have manifold functions. Besides their role in controlling cotranslational and posttranslational translocation of proteins, in assisting in protein folding and promoting the degradation of mutated or misfolded proteins, four of the eight ER-resident cochaperones are known to be involved in regulating Ca2+ flux along cellular membranes. ERdJ3 and ERdJ6 control Ca2+ leakage from the ER by closing the Sec61 channel at the end of protein translocation. Two other ER-resident cochaperones, ERdJ1 and ERdJ5, control Ca2+ homeostasis by Sec61-independent mechanisms. ERdJ1 plays a central role in targeting BiP to the plasma membrane. Here, the ERdJ1–BiP complex seems to control Ca2+ influx with BiP acting as a receptor for alpha2-macroglobulin. ERdJ5 has a central role in controlling Ca2+ influx into the ER. Due to its reductive potential, it has the ability to reduce and open the SERCA to enable Ca2+ influx and to maintain luminal Ca2+ stores. In this review, we present the current knowledge on the impact of these ERdJ proteins in controlling the cellular Ca2+ metabolism, yet the elaboration of the underlying mechanisms is at its very onset.
Conflict of interest
The authors declare that they have no conflict of interest with the contents of this article.
Acknowledgments
Author contributions
L. F. D. and F. P. investigation; L. F. D. writing–original draft; L. F. D. and F. P. writing–review and editing; L .F. D. and F. P. methodology.
Edited by Ronald Wek
References
- 1.Case R.M., Eisner D., Gurney A., Jones O., Muallem S., Verkhratsky A. Evolution of calcium homeostasis: from birth of the first cell to an omnipresent signalling system. Cell Calcium. 2007;42:345–350. doi: 10.1016/j.ceca.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 2.Mekahli D., Bultynck G., Parys J.B., De Smedt H., Missiaen L. Endoplasmic-reticulum calcium depletion and disease. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amador F.J., Stathopulos P.B., Enomoto M., Ikura M. Ryanodine receptor calcium release channels: lessons from structure-function studies. FEBS J. 2013;280:5456–5470. doi: 10.1111/febs.12194. [DOI] [PubMed] [Google Scholar]
- 4.Raffaello A., Mammucari C., Gherardi G., Rizzuto R. Calcium at the center of cell signaling: interplay between endoplasmic reticulum, mitochondria, and lysosomes. Trends Biochem. Sci. 2016;41:1035–1049. doi: 10.1016/j.tibs.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyazaki S. IP3 receptor-mediated spatial and temporal Ca2+ signaling of the cell. Jpn. J. Physiol. 1993;43:409–434. doi: 10.2170/jjphysiol.43.409. [DOI] [PubMed] [Google Scholar]
- 6.Lang S., Erdmann F., Jung M., Wagner R., Cavalie A., Zimmermann R. Sec61 complexes form ubiquitous ER Ca2+ leak channels. Channels (Austin) 2011;5:228–235. doi: 10.4161/chan.5.3.15314. [DOI] [PubMed] [Google Scholar]
- 7.Berridge M.J. The inositol trisphosphate/calcium signaling pathway in health and disease. Physiol. Rev. 2016;96:1261–1296. doi: 10.1152/physrev.00006.2016. [DOI] [PubMed] [Google Scholar]
- 8.Marchi S., Patergnani S., Pinton P. The endoplasmic reticulum-mitochondria connection: one touch, multiple functions. Biochim. Biophys. Acta. 2014;1837:461–469. doi: 10.1016/j.bbabio.2013.10.015. [DOI] [PubMed] [Google Scholar]
- 9.Madec A.M., Perrier J., Panthu B., Dingreville F. Role of mitochondria-associated endoplasmic reticulum membrane (MAMs) interactions and calcium exchange in the development of type 2 diabetes. Int. Rev. Cell Mol. Biol. 2021;363:169–202. doi: 10.1016/bs.ircmb.2021.06.001. [DOI] [PubMed] [Google Scholar]
- 10.Arruda A.P., Hotamisligil G.S. Calcium homeostasis and organelle function in the pathogenesis of obesity and diabetes. Cell Metab. 2015;22:381–397. doi: 10.1016/j.cmet.2015.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Venkatachalam K., van Rossum D.B., Patterson R.L., Ma H.T., Gill D.L. The cellular and molecular basis of store-operated calcium entry. Nat. Cell Biol. 2002;4:E263–272. doi: 10.1038/ncb1102-e263. [DOI] [PubMed] [Google Scholar]
- 12.Parekh A.B., Penner R. Store depletion and calcium influx. Physiol. Rev. 1997;77:901–930. doi: 10.1152/physrev.1997.77.4.901. [DOI] [PubMed] [Google Scholar]
- 13.Prakriya M., Lewis R.S. Store-operated calcium channels. Physiol. Rev. 2015;95:1383–1436. doi: 10.1152/physrev.00020.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y.F., Chen L.H., Shen M.R. The distinct role of STIM1 and STIM2 in the regulation of store-operated Ca(2+) entry and cellular function. J. Cell Physiol. 2019;234:8727–8739. doi: 10.1002/jcp.27532. [DOI] [PubMed] [Google Scholar]
- 15.Lewis R.S. Store-operated calcium channels: from function to structure and back again. Cold Spring Harb. Perspect. Biol. 2020;12 doi: 10.1101/cshperspect.a035055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ushioda R., Miyamoto A., Inoue M., Watanabe S., Okumura M., Maegawa K.I., et al. Redox-assisted regulation of Ca2+ homeostasis in the endoplasmic reticulum by disulfide reductase ERdj5. Proc. Natl. Acad. Sci. U. S. A. 2016;113:E6055–E6063. doi: 10.1073/pnas.1605818113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dremina E.S., Sharov V.S., Davies M.J., Schoneich C. Oxidation and inactivation of SERCA by selective reaction of cysteine residues with amino acid peroxides. Chem. Res. Toxicol. 2007;20:1462–1469. doi: 10.1021/tx700108w. [DOI] [PubMed] [Google Scholar]
- 18.Lytton J., Westlin M., Hanley M.R. Thapsigargin inhibits the sarcoplasmic or endoplasmic reticulum Ca-ATPase family of calcium pumps. J. Biol. Chem. 1991;266:17067–17071. [PubMed] [Google Scholar]
- 19.Avezov E., Cross B.C., Kaminski Schierle G.S., Winters M., Harding H.P., Melo E.P., et al. Lifetime imaging of a fluorescent protein sensor reveals surprising stability of ER thiol redox. J. Cell Biol. 2013;201:337–349. doi: 10.1083/jcb.201211155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Avezov E., Konno T., Zyryanova A., Chen W., Laine R., Crespillo-Casado A., et al. Retarded PDI diffusion and a reductive shift in poise of the calcium depleted endoplasmic reticulum. BMC Biol. 2015;13:2. doi: 10.1186/s12915-014-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lizak B., Birk J., Zana M., Kosztyi G., Kratschmar D.V., Odermatt A., et al. Ca(2+) mobilization-dependent reduction of the endoplasmic reticulum lumen is due to influx of cytosolic glutathione. BMC Biol. 2020;18:19. doi: 10.1186/s12915-020-0749-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Appenzeller-Herzog C. Glutathione- and non-glutathione-based oxidant control in the endoplasmic reticulum. J. Cel. Sci. 2011;124:847–855. doi: 10.1242/jcs.080895. [DOI] [PubMed] [Google Scholar]
- 23.Banhegyi G., Lusini L., Puskas F., Rossi R., Fulceri R., Braun L., et al. Preferential transport of glutathione versus glutathione disulfide in rat liver microsomal vesicles. J. Biol. Chem. 1999;274:12213–12216. doi: 10.1074/jbc.274.18.12213. [DOI] [PubMed] [Google Scholar]
- 24.Yong J., Bischof H., Burgstaller S., Siirin M., Murphy A., Malli R., et al. Mitochondria supply ATP to the ER through a mechanism antagonized by cytosolic Ca(2) Elife. 2019;8 doi: 10.7554/eLife.49682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vishnu N., Jadoon Khan M., Karsten F., Groschner L.N., Waldeck-Weiermair M., Rost R., et al. ATP increases within the lumen of the endoplasmic reticulum upon intracellular Ca2+ release. Mol. Biol. Cell. 2014;25:368–379. doi: 10.1091/mbc.E13-07-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein M.C., Zimmermann K., Schorr S., Landini M., Klemens P.A.W., Altensell J., et al. AXER is an ATP/ADP exchanger in the membrane of the endoplasmic reticulum. Nat. Commun. 2018;9:3489. doi: 10.1038/s41467-018-06003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kania E., Pajak B., Orzechowski A. Calcium homeostasis and ER stress in control of autophagy in cancer cells. Biomed. Res. Int. 2015;2015:352794. doi: 10.1155/2015/352794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krebs J., Agellon L.B., Michalak M. Ca(2+) homeostasis and endoplasmic reticulum (ER) stress: an integrated view of calcium signaling. Biochem. Biophys. Res. Commun. 2015;460:114–121. doi: 10.1016/j.bbrc.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Read A., Schroder M. The unfolded protein response: an overview. Biology (Basel) 2021;10:384. doi: 10.3390/biology10050384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Z., Zhang L., Zhou L., Lei Y., Zhang Y., Huang C. Redox signaling and unfolded protein response coordinate cell fate decisions under ER stress. Redox Biol. 2019;25:101047. doi: 10.1016/j.redox.2018.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schroder M., Kaufman R.J. ER stress and the unfolded protein response. Mutat. Res. 2005;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 32.Minakshi R., Rahman S., Jan A.T., Archana A., Kim J. Implications of aging and the endoplasmic reticulum unfolded protein response on the molecular modality of breast cancer. Exp. Mol. Med. 2017;49:e389. doi: 10.1038/emm.2017.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee A.H., Iwakoshi N.N., Glimcher L.H. XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol. Cell Biol. 2003;23:7448–7459. doi: 10.1128/MCB.23.21.7448-7459.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maurel M., Chevet E., Tavernier J., Gerlo S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem. Sci. 2014;39:245–254. doi: 10.1016/j.tibs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 35.Li X., Sun S., Appathurai S., Sundaram A., Plumb R., Mariappan M. A molecular mechanism for turning off IRE1alpha signaling during endoplasmic reticulum stress. Cell Rep. 2020;33:108563. doi: 10.1016/j.celrep.2020.108563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carreras-Sureda A., Jana F., Urra H., Durand S., Mortenson D.E., Sagredo A., et al. Non-canonical function of IRE1alpha determines mitochondria-associated endoplasmic reticulum composition to control calcium transfer and bioenergetics. Nat. Cell Biol. 2019;21:755–767. doi: 10.1038/s41556-019-0329-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar V., Maity S. ER stress-sensor proteins and ER-mitochondrial crosstalk-signaling beyond (ER) stress response. Biomolecules. 2021;11:173. doi: 10.3390/biom11020173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McQuiston A., Diehl J.A. Recent insights into PERK-dependent signaling from the stressed endoplasmic reticulum. F1000Res. 2017;6:1897. doi: 10.12688/f1000research.12138.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harding H.P., Zhang Y., Zeng H., Novoa I., Lu P.D., Calfon M., et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 40.Cullinan S.B., Diehl J.A. Coordination of ER and oxidative stress signaling: the PERK/Nrf2 signaling pathway. Int. J. Biochem. Cell Biol. 2006;38:317–332. doi: 10.1016/j.biocel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 41.Verfaillie T., Rubio N., Garg A.D., Bultynck G., Rizzuto R., Decuypere J.P., et al. PERK is required at the ER-mitochondrial contact sites to convey apoptosis after ROS-based ER stress. Cell Death Differ. 2012;19:1880–1891. doi: 10.1038/cdd.2012.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Vliet A.R., Giordano F., Gerlo S., Segura I., Van Eygen S., Molenberghs G., et al. The ER stress sensor PERK coordinates ER-plasma membrane contact site formation through interaction with filamin-A and F-actin remodeling. Mol. Cell. 2017;65:885–899.e886. doi: 10.1016/j.molcel.2017.01.020. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto K., Sato T., Matsui T., Sato M., Okada T., Yoshida H., et al. Transcriptional induction of mammalian ER quality control proteins is mediated by single or combined action of ATF6alpha and XBP1. Dev. Cell. 2007;13:365–376. doi: 10.1016/j.devcel.2007.07.018. [DOI] [PubMed] [Google Scholar]
- 44.Kopp M.C., Larburu N., Durairaj V., Adams C.J., Ali M.M.U. UPR proteins IRE1 and PERK switch BiP from chaperone to ER stress sensor. Nat. Struct. Mol. Biol. 2019;26:1053–1062. doi: 10.1038/s41594-019-0324-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karagoz G.E., Acosta-Alvear D., Walter P. The unfolded protein response: detecting and responding to fluctuations in the protein-folding capacity of the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 2019;11 doi: 10.1101/cshperspect.a033886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kopp M.C., Nowak P.R., Larburu N., Adams C.J., Ali M.M. In vitro FRET analysis of IRE1 and BiP association and dissociation upon endoplasmic reticulum stress. Elife. 2018;7 doi: 10.7554/eLife.30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carrara M., Prischi F., Nowak P.R., Kopp M.C., Ali M.M. Noncanonical binding of BiP ATPase domain to Ire1 and Perk is dissociated by unfolded protein CH1 to initiate ER stress signaling. Elife. 2015;4 doi: 10.7554/eLife.03522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Credle J.J., Finer-Moore J.S., Papa F.R., Stroud R.M., Walter P. On the mechanism of sensing unfolded protein in the endoplasmic reticulum. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18773–18784. doi: 10.1073/pnas.0509487102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lievremont J.P., Rizzuto R., Hendershot L., Meldolesi J. BiP, a major chaperone protein of the endoplasmic reticulum lumen, plays a direct and important role in the storage of the rapidly exchanging pool of Ca2+ J. Biol. Chem. 1997;272:30873–30879. doi: 10.1074/jbc.272.49.30873. [DOI] [PubMed] [Google Scholar]
- 50.Schauble N., Lang S., Jung M., Cappel S., Schorr S., Ulucan O., et al. BiP-mediated closing of the Sec61 channel limits Ca2+ leakage from the ER. EMBO J. 2012;31:3282–3296. doi: 10.1038/emboj.2012.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Higo T., Hamada K., Hisatsune C., Nukina N., Hashikawa T., Hattori M., et al. Mechanism of ER stress-induced brain damage by IP(3) receptor. Neuron. 2010;68:865–878. doi: 10.1016/j.neuron.2010.11.010. [DOI] [PubMed] [Google Scholar]
- 52.Pobre K.F.R., Poet G.J., Hendershot L.M. The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: getting by with a little help from ERdj friends. J. Biol. Chem. 2019;294:2098–2108. doi: 10.1074/jbc.REV118.002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu X.B., Shao Y.M., Miao S., Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol. Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caplan A.J. What is a co-chaperone? Cell Stress Chaperones. 2003;8:105–107. doi: 10.1379/1466-1268(2003)008<0105:wiac>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheetham M.E., Caplan A.J. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Daverkausen-Fischer L., Prols F. The function of the co-chaperone ERdj4 in diverse (patho-)physiological conditions. Cell Mol. Life Sci. 2021;79:9. doi: 10.1007/s00018-021-04082-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Amin-Wetzel N., Saunders R.A., Kamphuis M.J., Rato C., Preissler S., Harding H.P., et al. A J-protein Co-chaperone recruits BiP to monomerize IRE1 and repress the unfolded protein response. Cell. 2017;171:1625–1637.e1613. doi: 10.1016/j.cell.2017.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sun F., Liao Y., Qu X., Xiao X., Hou S., Chen Z., et al. Hepatic DNAJB9 drives anabolic biasing to reduce steatosis and obesity. Cell Rep. 2020;30:1835–1847.e1839. doi: 10.1016/j.celrep.2020.01.043. [DOI] [PubMed] [Google Scholar]
- 59.Dong M., Bridges J.P., Apsley K., Xu Y., Weaver T.E. ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. Mol. Biol. Cell. 2008;19:2620–2630. doi: 10.1091/mbc.E07-07-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yamamoto Y.H., Kasai A., Omori H., Takino T., Sugihara M., Umemoto T., et al. ERdj8 governs the size of autophagosomes during the formation process. J. Cell Biol. 2020;219 doi: 10.1083/jcb.201903127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kampinga H.H., Andreasson C., Barducci A., Cheetham M.E., Cyr D., Emanuelsson C., et al. Function, evolution, and structure of J-domain proteins. Cell Stress Chaperones. 2019;24:7–15. doi: 10.1007/s12192-018-0948-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gemmer M., Forster F. A clearer picture of the ER translocon complex. J. Cel. Sci. 2020;133 doi: 10.1242/jcs.231340. [DOI] [PubMed] [Google Scholar]
- 63.Van Coppenolle F., Vanden Abeele F., Slomianny C., Flourakis M., Hesketh J., Dewailly E., et al. Ribosome-translocon complex mediates calcium leakage from endoplasmic reticulum stores. J. Cel. Sci. 2004;117:4135–4142. doi: 10.1242/jcs.01274. [DOI] [PubMed] [Google Scholar]
- 64.Ong H.L., Liu X., Sharma A., Hegde R.S., Ambudkar I.S. Intracellular Ca(2+) release via the ER translocon activates store-operated calcium entry. Pflugers Arch. 2007;453:797–808. doi: 10.1007/s00424-006-0163-5. [DOI] [PubMed] [Google Scholar]
- 65.Lomax R.B., Camello C., Van Coppenolle F., Petersen O.H., Tepikin A.V. Basal and physiological Ca(2+) leak from the endoplasmic reticulum of pancreatic acinar cells. second messenger-activated channels and translocons. J. Biol. Chem. 2002;277:26479–26485. doi: 10.1074/jbc.M201845200. [DOI] [PubMed] [Google Scholar]
- 66.Giunti R., Gamberucci A., Fulceri R., Banhegyi G., Benedetti A. Both translocon and a cation channel are involved in the passive Ca2+ leak from the endoplasmic reticulum: a mechanistic study on rat liver microsomes. Arch. Biochem. Biophys. 2007;462:115–121. doi: 10.1016/j.abb.2007.03.039. [DOI] [PubMed] [Google Scholar]
- 67.Flourakis M., Van Coppenolle F., Lehen'kyi V., Beck B., Skryma R., Prevarskaya N. Passive calcium leak via translocon is a first step for iPLA2-pathway regulated store operated channels activation. FASEB J. 2006;20:1215–1217. doi: 10.1096/fj.05-5254fje. [DOI] [PubMed] [Google Scholar]
- 68.Amer M.S., Li J., O'Regan D.J., Steele D.S., Porter K.E., Sivaprasadarao A., et al. Translocon closure to Ca2+ leak in proliferating vascular smooth muscle cells. Am. J. Physiol. Heart Circ. Physiol. 2009;296:H910–916. doi: 10.1152/ajpheart.00984.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Van den Berg B., Clemons W.M., Jr., Collinson I., Modis Y., Hartmann E., Harrison S.C., et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 70.Hamman B.D., Hendershot L.M., Johnson A.E. BiP maintains the permeability barrier of the ER membrane by sealing the lumenal end of the translocon pore before and early in translocation. Cell. 1998;92:747–758. doi: 10.1016/s0092-8674(00)81403-8. [DOI] [PubMed] [Google Scholar]
- 71.Rapoport T.A., Li L., Park E. Structural and mechanistic insights into protein translocation. Annu. Rev. Cell Dev. Biol. 2017;33:369–390. doi: 10.1146/annurev-cellbio-100616-060439. [DOI] [PubMed] [Google Scholar]
- 72.Voorhees R.M., Hegde R.S. Toward a structural understanding of co-translational protein translocation. Curr. Opin. Cell Biol. 2016;41:91–99. doi: 10.1016/j.ceb.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 73.Haigh N.G., Johnson A.E. A new role for BiP: closing the aqueous translocon pore during protein integration into the ER membrane. J. Cell Biol. 2002;156:261–270. doi: 10.1083/jcb.200110074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Alder N.N., Shen Y., Brodsky J.L., Hendershot L.M., Johnson A.E. The molecular mechanisms underlying BiP-mediated gating of the Sec61 translocon of the endoplasmic reticulum. J. Cell Biol. 2005;168:389–399. doi: 10.1083/jcb.200409174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schorr S., Klein M.C., Gamayun I., Melnyk A., Jung M., Schauble N., et al. Co-chaperone specificity in gating of the polypeptide conducting channel in the membrane of the human endoplasmic reticulum. J. Biol. Chem. 2015;290:18621–18635. doi: 10.1074/jbc.M115.636639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo F., Snapp E.L. ERdj3 regulates BiP occupancy in living cells. J. Cel. Sci. 2013;126:1429–1439. doi: 10.1242/jcs.118182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jin Y., Zhuang M., Hendershot L.M. ERdj3, a luminal ER DnaJ homologue, binds directly to unfolded proteins in the mammalian ER: identification of critical residues. Biochemistry. 2009;48:41–49. doi: 10.1021/bi8015923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shen Y., Hendershot L.M. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP's interactions with unfolded substrates. Mol. Biol. Cell. 2005;16:40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Buck T.M., Kolb A.R., Boyd C.R., Kleyman T.R., Brodsky J.L. The endoplasmic reticulum-associated degradation of the epithelial sodium channel requires a unique complement of molecular chaperones. Mol. Biol. Cell. 2010;21:1047–1058. doi: 10.1091/mbc.E09-11-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tan Y.L., Genereux J.C., Pankow S., Aerts J.M., Yates J.R., 3rd, Kelly J.W. ERdj3 is an endoplasmic reticulum degradation factor for mutant glucocerebrosidase variants linked to Gaucher's disease. Chem. Biol. 2014;21:967–976. doi: 10.1016/j.chembiol.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Genereux J.C., Qu S., Zhou M., Ryno L.M., Wang S., Shoulders M.D., et al. Unfolded protein response-induced ERdj3 secretion links ER stress to extracellular proteostasis. EMBO J. 2015;34:4–19. doi: 10.15252/embj.201488896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Daverkausen-Fischer L., Prols F. Dual topology of co-chaperones at the membrane of the endoplasmic reticulum. Cell Death Discov. 2021;7:203. doi: 10.1038/s41420-021-00594-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Oyadomari S., Yun C., Fisher E.A., Kreglinger N., Kreibich G., Oyadomari M., et al. Cotranslocational degradation protects the stressed endoplasmic reticulum from protein overload. Cell. 2006;126:727–739. doi: 10.1016/j.cell.2006.06.051. [DOI] [PubMed] [Google Scholar]
- 84.Van Krieken R., Tsai Y.L., Carlos A.J., Ha D.P., Lee A.S. ER residential chaperone GRP78 unconventionally relocalizes to the cell surface via endosomal transport. Cell Mol. Life Sci. 2021;78:5179–5195. doi: 10.1007/s00018-021-03849-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yu M., Haslam R.H., Haslam D.B. HEDJ, an Hsp40 co-chaperone localized to the endoplasmic reticulum of human cells. J. Biol. Chem. 2000;275:24984–24992. doi: 10.1074/jbc.M000739200. [DOI] [PubMed] [Google Scholar]
- 86.Rutkowski D.T., Kang S.W., Goodman A.G., Garrison J.L., Taunton J., Katze M.G., et al. The role of p58IPK in protecting the stressed endoplasmic reticulum. Mol. Biol. Cell. 2007;18:3681–3691. doi: 10.1091/mbc.E07-03-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bies C., Blum R., Dudek J., Nastainczyk W., Oberhauser S., Jung M., et al. Characterization of pancreatic ERj3p, a homolog of yeast DnaJ-like protein Scj1p. Biol. Chem. 2004;385:389–395. doi: 10.1515/BC.2004.043. [DOI] [PubMed] [Google Scholar]
- 88.Lebeau P.F., Platko K., Byun J.H., Austin R.C. Calcium as a reliable marker for the quantitative assessment of endoplasmic reticulum stress in live cells. J. Biol. Chem. 2021;296:100779. doi: 10.1016/j.jbc.2021.100779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yan W., Frank C.L., Korth M.J., Sopher B.L., Novoa I., Ron D., et al. Control of PERK eIF2alpha kinase activity by the endoplasmic reticulum stress-induced molecular chaperone P58IPK. Proc. Natl. Acad. Sci. U. S. A. 2002;99:15920–15925. doi: 10.1073/pnas.252341799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van Huizen R., Martindale J.L., Gorospe M., Holbrook N.J. P58IPK, a novel endoplasmic reticulum stress-inducible protein and potential negative regulator of eIF2alpha signaling. J. Biol. Chem. 2003;278:15558–15564. doi: 10.1074/jbc.M212074200. [DOI] [PubMed] [Google Scholar]
- 91.Lloyd D.J., Wheeler M.C., Gekakis N. A point mutation in Sec61alpha1 leads to diabetes and hepatosteatosis in mice. Diabetes. 2010;59:460–470. doi: 10.2337/db08-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ladiges W.C., Knoblaugh S.E., Morton J.F., Korth M.J., Sopher B.L., Baskin C.R., et al. Pancreatic beta-cell failure and diabetes in mice with a deletion mutation of the endoplasmic reticulum molecular chaperone gene P58IPK. Diabetes. 2005;54:1074–1081. doi: 10.2337/diabetes.54.4.1074. [DOI] [PubMed] [Google Scholar]
- 93.Ushioda R., Hoseki J., Araki K., Jansen G., Thomas D.Y., Nagata K. ERdj5 is required as a disulfide reductase for degradation of misfolded proteins in the ER. Science. 2008;321:569–572. doi: 10.1126/science.1159293. [DOI] [PubMed] [Google Scholar]
- 94.Ushioda R., Hoseki J., Nagata K. Glycosylation-independent ERAD pathway serves as a backup system under ER stress. Mol. Biol. Cell. 2013;24:3155–3163. doi: 10.1091/mbc.E13-03-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hagiwara M., Maegawa K., Suzuki M., Ushioda R., Araki K., Matsumoto Y., et al. Structural basis of an ERAD pathway mediated by the ER-resident protein disulfide reductase ERdj5. Mol. Cell. 2011;41:432–444. doi: 10.1016/j.molcel.2011.01.021. [DOI] [PubMed] [Google Scholar]
- 96.Cunnea P.M., Miranda-Vizuete A., Bertoli G., Simmen T., Damdimopoulos A.E., Hermann S., et al. ERdj5, an endoplasmic reticulum (ER)-resident protein containing DnaJ and thioredoxin domains, is expressed in secretory cells or following ER stress. J. Biol. Chem. 2003;278:1059–1066. doi: 10.1074/jbc.M206995200. [DOI] [PubMed] [Google Scholar]
- 97.Yamashita R., Fujii S., Ushioda R., Nagata K. Ca(2+) imbalance caused by ERdj5 deletion affects mitochondrial fragmentation. Sci. Rep. 2021;11:20772. doi: 10.1038/s41598-021-99980-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Valera-Alberni M., Joffraud M., Miro-Blanch J., Capellades J., Junza A., Dayon L., et al. Crosstalk between Drp1 phosphorylation sites during mitochondrial remodeling and their impact on metabolic adaptation. Cell Rep. 2021;36:109565. doi: 10.1016/j.celrep.2021.109565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dudek J., Volkmer J., Bies C., Guth S., Muller A., Lerner M., et al. A novel type of co-chaperone mediates transmembrane recruitment of DnaK-like chaperones to ribosomes. EMBO J. 2002;21:2958–2967. doi: 10.1093/emboj/cdf315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dudek J., Greiner M., Muller A., Hendershot L.M., Kopsch K., Nastainczyk W., et al. ERj1p has a basic role in protein biogenesis at the endoplasmic reticulum. Nat. Struct. Mol. Biol. 2005;12:1008–1014. doi: 10.1038/nsmb1007. [DOI] [PubMed] [Google Scholar]
- 101.Behnke J., Mann M.J., Scruggs F.L., Feige M.J., Hendershot L.M. Members of the Hsp70 family recognize distinct types of sequences to execute ER quality control. Mol. Cell. 2016;63:739–752. doi: 10.1016/j.molcel.2016.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Misra U.K., Pizzo S.V. Heterotrimeric Galphaq11 co-immunoprecipitates with surface-anchored GRP78 from plasma membranes of alpha2M∗-stimulated macrophages. J. Cell. Biochem. 2008;104:96–104. doi: 10.1002/jcb.21607. [DOI] [PubMed] [Google Scholar]
- 103.Misra U.K., Gonzalez-Gronow M., Gawdi G., Pizzo S.V. The role of MTJ-1 in cell surface translocation of GRP78, a receptor for alpha 2-macroglobulin-dependent signaling. J. Immunol. 2005;174:2092–2097. doi: 10.4049/jimmunol.174.4.2092. [DOI] [PubMed] [Google Scholar]
- 104.Misra U.K., Gonzalez-Gronow M., Gawdi G., Hart J.P., Johnson C.E., Pizzo S.V. The role of Grp 78 in alpha 2-macroglobulin-induced signal transduction. Evidence from RNA interference that the low density lipoprotein receptor-related protein is associated with, but not necessary for, GRP 78-mediated signal transduction. J. Biol. Chem. 2002;277:42082–42087. doi: 10.1074/jbc.M206174200. [DOI] [PubMed] [Google Scholar]
- 105.Gonzalez-Gronow M., Gopal U., Austin R.C., Pizzo S.V. Glucose-regulated protein (GRP78) is an important cell surface receptor for viral invasion, cancers, and neurological disorders. IUBMB Life. 2021;73:843–854. doi: 10.1002/iub.2502. [DOI] [PubMed] [Google Scholar]
- 106.Gonzalez-Gronow M., Kaczowka S.J., Payne S., Wang F., Gawdi G., Pizzo S.V. Plasminogen structural domains exhibit different functions when associated with cell surface GRP78 or the voltage-dependent anion channel. J. Biol. Chem. 2007;282:32811–32820. doi: 10.1074/jbc.M703342200. [DOI] [PubMed] [Google Scholar]
- 107.Berger C.L., Dong Z., Hanlon D., Bisaccia E., Edelson R.L. A lymphocyte cell surface heat shock protein homologous to the endoplasmic reticulum chaperone, immunoglobulin heavy chain binding protein BIP. Int. J. Cancer. 1997;71:1077–1085. doi: 10.1002/(sici)1097-0215(19970611)71:6<1077::aid-ijc26>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 108.Delpino A., Piselli P., Vismara D., Vendetti S., Colizzi V. Cell surface localization of the 78 kD glucose regulated protein (GRP 78) induced by thapsigargin. Mol. Membr. Biol. 1998;15:21–26. doi: 10.3109/09687689809027514. [DOI] [PubMed] [Google Scholar]
- 109.Xiao G., Chung T.F., Pyun H.Y., Fine R.E., Johnson R.J. KDEL proteins are found on the surface of NG108-15 cells. Brain Res. Mol. Brain Res. 1999;72:121–128. doi: 10.1016/s0169-328x(99)00188-6. [DOI] [PubMed] [Google Scholar]
- 110.Capitani M., Sallese M. The KDEL receptor: new functions for an old protein. FEBS Lett. 2009;583:3863–3871. doi: 10.1016/j.febslet.2009.10.053. [DOI] [PubMed] [Google Scholar]
- 111.Bard F., Mazelin L., Pechoux-Longin C., Malhotra V., Jurdic P. Src regulates Golgi structure and KDEL receptor-dependent retrograde transport to the endoplasmic reticulum. J. Biol. Chem. 2003;278:46601–46606. doi: 10.1074/jbc.M302221200. [DOI] [PubMed] [Google Scholar]
- 112.Tsai Y.L., Ha D.P., Zhao H., Carlos A.J., Wei S., Pun T.K., et al. Endoplasmic reticulum stress activates SRC, relocating chaperones to the cell surface where GRP78/CD109 blocks TGF-beta signaling. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E4245–E4254. doi: 10.1073/pnas.1714866115. [DOI] [PMC free article] [PubMed] [Google Scholar]