Summary
Brown adipose tissue (BAT) has a role in maintaining systemic metabolic health in rodents and humans. Here, we show that metabolic stress induces BAT to produce coagulation factors, which then—together with molecules derived from the circulation—promote BAT dysfunction and systemic glucose intolerance. When mice were fed a high-fat diet (HFD), the levels of tissue factor, coagulation Factor VII (FVII), activated coagulation Factor X (FXa), and protease-activated receptor 1 (PAR1) expression increased significantly in BAT. Genetic or pharmacological suppression of coagulation factor-PAR1 signaling in BAT ameliorated its whitening and improved thermogenic response and systemic glucose intolerance in mice with dietary obesity. Conversely, the activation of coagulation factor-PAR1 signaling in BAT caused mitochondrial dysfunction in brown adipocytes and systemic glucose intolerance in mice fed normal chow. These results indicate that BAT produces endogenous coagulation factors that mediate pleiotropic effects via PAR1 signaling under metabolic stress.
Subject areas: Biological sciences, Human Physiology, Human metabolism, Cell biology
Graphical abstract

Highlights
-
•
Coagulation factors and protease-activated receptor 1 (PAR1) are upregulated in brown adipose tissue (BAT) under metabolic stress
-
•
Inhibition of coagulation factor-PAR1 signaling in BAT improves metabolic dysfunction
-
•
Activation of coagulation factor-PAR1 signaling in BAT causes metabolic dysfunction
Biological sciences; Human Physiology; Human metabolism; Cell biology
Introduction
Obesity is increasing rapidly in many societies, and new therapies for obesity-related disorders are urgently required (Ezzati et al., 2017). Brown adipose tissue (BAT) is an active metabolic organ with abundant mitochondria that produce heat through uncoupled respiration (Nedergaard et al., 2011; Rosen and Spiegelman, 2014). Studies have shown that the activation of BAT contributes to the suppression of systemic metabolic disorders in rodents (Bartelt et al., 2011; Shimizu et al., 2014), and there is accumulating evidence that BAT also promotes systemic metabolic health in humans (Ouellet et al., 2012; Yoneshiro et al., 2013). Although it is known that BAT function decreases with obesity and aging (Cypess et al., 2009; van Marken Lichtenbelt et al., 2009), the underlying mechanisms have yet to be defined. Obesity is linked to a pro-coagulant state that contributes to the progression of various obesity-related disorders, including atherosclerotic disease (Borissoff et al., 2010; Spronk et al., 2014). Coagulation factor X (FX) is a component of the coagulation cascade that has become a major target of anticoagulant therapy (Patel et al., 2011). Activated FX (FXa) has pleiotropic biological effects that are mediated through the protease-activated receptor (PAR) signaling (Badeanlou et al., 2011; Ghorpade et al., 2018; Samad and Ruf, 2013; Spronk et al., 2014). In visceral white adipose tissue, tissue factor-PAR2 mediated signaling induced chronic sterile inflammation and systemic insulin resistance (Badeanlou et al., 2011); however, the role of PAR signaling in BAT is yet to be defined. Here we show that PAR1 is a dominant form in BAT, and coagulation factor-PAR1 mediated signaling promotes functional decline of this organ by excessive mitochondrial ROS production, leading to systemic glucose intolerance in a murine model with dietary obesity.
Results
Coagulation factors and protease-activated receptors are upregulated in brown adipose tissue under metabolic stress
Through analysis of GSE64718, a microarray dataset available in a public database (NCBI GEO (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64718)), we found the upregulation of transcripts for PAR1 receptor and several coagulation factors, including factor VII (FVII) and FX, in BAT from obese mice (Figure S1A). These findings led us to investigate whether the coagulation system has a role in the response of BAT to metabolic stress. We generated a murine model of dietary obesity by feeding a high-fat diet (HFD) to C57BL/6NCrSlc mice, which resulted in whitening of BAT, as reported previously (Figures 1A and S1B) (Shimizu et al., 2014). We found a significant increase in tissue factor (Figures 1B, 1C, and S1C), FVII (Figures 1D–1F and S1D), and FX in BAT from obese mice (Figure 1G). FXa also increased in BAT from mice with dietary obesity (Figure 1H). It is generally considered that coagulation factors are mainly synthesized by the liver; however, quantitative PCR studies showed that transcripts for some coagulation factors were detectable in BAT, including transcripts for factor VIII (F8), factor V (F5), and factor XIII A chain (F13a1), while transcripts for factor XII (F12), factor XI (F11), factor IX (F9), factor II (F2), factor I (Fga, Fgb, Fgg), and factor XIII B chain (F13b) were expressed at a very low level or were undetectable (Figures S1E and S1F). It was previously reported that signaling via the tissue factor/protease-activated receptor 2 (PAR2) pathway induces the inflammation of white adipose tissue (WAT) and systemic glucose intolerance (Badeanlou et al., 2011). The PAR family has four members (PAR1 to PAR4), and PARs have been reported to mediate a broad spectrum of biological effects (Spronk et al., 2014; Zhao et al., 2014). We found that PAR2 was predominantly expressed in epididymal WAT (eWAT), whereas PAR1 was the dominant form in BAT or brown adipocytes (Figures 1I, S1G, S1H, and S1I). In BAT, we found that PAR1 expression was increased by dietary obesity (Figures 1I, 1J, S1J, and S1K), whereas those in eWAT or inguinal WAT (iWAT) were comparable (Figure S1L). Transcript F2R increased in BAT of obese individuals (Figure 1K and Table 1). Plasma FXa levels were comparable in lean and obese mice and in lean and obese humans (Figure 1L and Table 1). The level of tissue factor in circulation was higher in our dietary obese model (Figure S1M), but the concentration of coagulation FVII was similar between the groups (Figure S1N).
Figure 1.
Coagulation factors and protease-activated receptors are upregulated in brown adipose tissue under metabolic stress
(A) Hematoxylin and eosin (HE) staining of brown adipose tissue (BAT) from wild-type mice fed a normal chow (NC) or a high-fat diet (HFD). In the HFD group, mice were fed the diet from 4 weeks of age and were analyzed at 19 to 22 weeks of age. Scale bar = 100 μm.
(B and C) Enzyme-linked immunosorbent assay (ELISA; B, n = 10, 10) or immunofluorescent staining (C) for tissue factor in BAT of the indicated mice aged 19-22weeks. Scale bar = 50 μm.
(D-H) Results of quantitative polymerase chain reaction (PCR) for coagulation factor VII (F7; D, n = 8, 7) or coagulation factor X (F10; G, n = 8, 7) in mice aged 19-22weeks. Results of enzyme-linked immunosorbent assay (ELISA; E, H) or immunofluorescent staining (F) for coagulation factor VII (E and F, n = 10, 11) or activated coagulation factor X (Factor Xa; H, n = 9, 9) in BAT of the indicated mice aged 19-22weeks. Scale bar = 50 μm.
(I) Western blot analysis of protease-activated receptor-1 (PAR-1) or PAR-2 expression in epididymal white adipose tissue (eWAT) or BAT from the indicated groups. Right panel indicates the quantification of protease-activated receptor-1 (PAR-1) relative to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) in BAT (n = 5, 5).
(J) Immunofluorescent staining for PAR-1 in BAT from the indicated group. Scale bar = 50 μm.
(K) RNA sequence data analyzing F2R in brown adipose tissue (BAT) from individuals with a body mass index (BMI) < 25 (n = 8) or BMI ≥25 (n = 23). Datasets were taken from Jespersen et al. (https://www.biorxiv.org/content/10.1101/2020.05.07.082057v1).
(L) Enzyme-linked immunosorbent assay (ELISA) for Factor Xa in plasma from NC or HFD mice aged 19-22weeks (left panel; n = 3, 3), and plasma from lean (BMI <22) or obese (BMI >28) human volunteers (right panel; n = 13, 13). All data were analyzed by a 2-tailed Student’s t test. ∗p < 0.05, ∗∗p < 0.01. Values represent the mean ± SEM NS = not significant. All data are from different biological replicates. See also Figure S1.
Table 1.
Characteristics of volunteers enrolled in human studies
| Group | BMI <25 (N = 8) | BMI ≥25 (N = 23) |
|---|---|---|
| Sex (W:M) | n = 7:1 | n = 16:7 |
| Age (years) | 47 (23–67) | 55 (41–74) |
| BMI (kg/m2) | 22.6 (17.6–24.9) | 30.8 (25.1–42.9) |
| WHR | 0.79 (0.69–0.96) | 0.91 (0.81–1.1) |
| Fat% | 31.2 (19.8–41.2) | 44.7 (28.3–54.5) |
| HOMAIR | 1.31 (0.56–4.60) | 2.36 (0.78–5.79) |
| 120 min plasma-glucose (mmol/L) | 5.65 (4.2–13.5) | 7.05 (5.9–11) |
| HBA1C (mmol/mol) | 32 (26–43) | 35 (27–52) |
| T2D (N) | 1 | 3 |
W = women, M = men, BMI = body mass index, WHR = waist-to-hip ratio, Fat% = total body fat percentage analyzed with a dual energy X-ray absorptiometry (DXA) scanner, 120 min plasma glucose = plasma glucose at 120 min after an oral glucose load, HbA1c = glycated hemoglobin A1c, T2D = type 2 diabetes, N = number.
Inhibition of FactorXa prevents whitening and dysfunction of brown adipose tissue
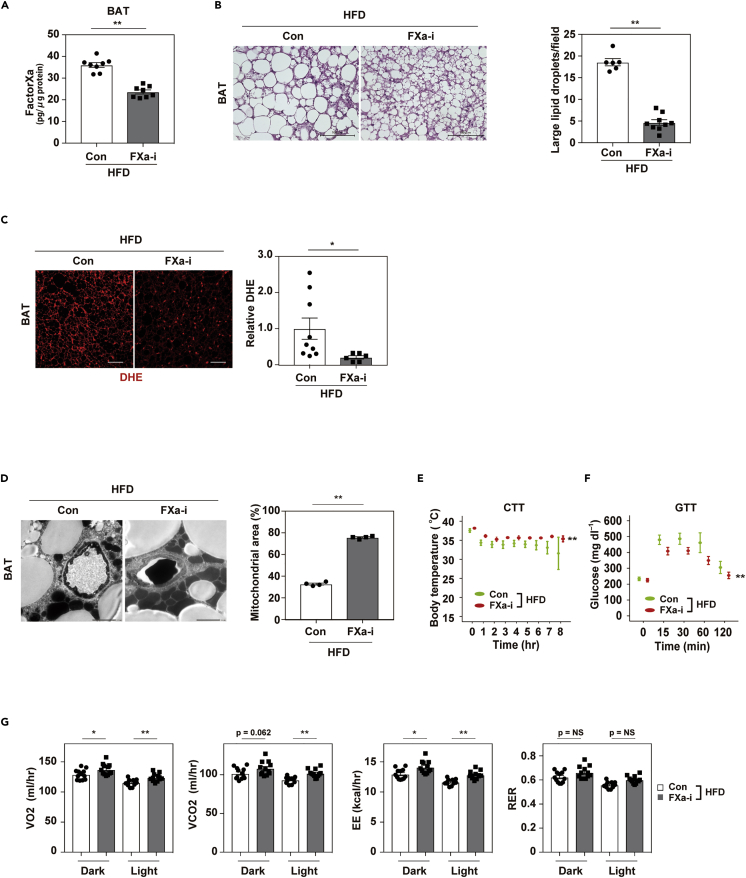
To further investigate the role of coagulation factors and PARs in BAT, we suppressed FXa by the administration of a FXa inhibitor (FXa-i) anticoagulant to our mice with dietary obesity. FXa-i treatment significantly reduced the FXa level in BAT (Figure 2A) and ameliorated its whitening (Figure 2B). In BAT, the level of oxidative stress markers including dihydroethidium (DHE) declined after FXa-i administration (Figure 2C), and mitochondrial rarefaction and fragmentation were reduced in brown adipocytes (Figure 2D). These changes were associated with an increase in the thermogenic response to acute cold exposure (Figure 2E). Moreover, FXa-i treatment improved systemic glucose intolerance (Figure 2F), along with an increase in energy expenditure (Figure 2G). Body weight and food intake were comparable between the control group and the FXa-i group, but BAT weight was reduced by FXa-i administration (Figures S2A–S2C).
Figure 2.
Inhibition of FactorXa prevents whitening and dysfunction of brown adipose tissue
Mice were fed a high-fat diet (HFD) from 4 weeks of age with or without the administration of an FXa inhibitor (FXa-i). Physiological studies were performed at age 13 to 19 weeks for FXa-i treatment, and samples were collected at age 18 to 22 weeks.
(A) Enzyme-linked immunosorbent assay study of Factor Xa in brown adipose tissue (BAT) from mice aged 19-22 weeks fed a high-fat diet (HFD; Con HFD) or HFD + FXa inhibitor (FXa-i HFD; n = 8, 8).
(B) Hematoxylin and eosin (HE) staining of BAT from the indicated mice. Scale bar = 100 μm. Right panel indicates the quantification of large lipid droplets in the indicated mice (n = 6, 9). Large lipid droplets were defined as droplets with a surface area >1000 μm2.
(C) Staining with dihydroethidium (DHE) in brown adipose tissue (BAT) from mice fed the HFD (Con HFD) or HFD + FXa-i (FXa-i HFD). Scale bar = 50 μm. Right panels indicate quantification shown as relative dihydroethidium (DHE) level (DHE area of FXa-i HFD/Con HFD, n = 9, 6).
(D) Findings on transmission electron microscopy in the indicated mice (scale bar = 2 μm). Right panels indicate mitochondrial area (%; analyzed as mitochondrial area/[non-capillary and non-lipid area]) in the pericapillary area of respective groups (n = 4, 4).
(E and F) Cold tolerance test (CTT; E, n = 7, 7) or glucose tolerance test (GTT; F, n = 14, 17) in the indicated mice aged 13 weeks for CTT, and 12 weeks for GTT.
(G) Oxygen consumption (VO2), CO2 production (VCO2), energy expenditure (EE), and respiratory exchange ratio (RER) in the indicated groups aged 14-16 weeks (n = 12, 12, 12, 12). Data were analyzed by a 2-tailed Student’s t test (A, B, C, D, G) or 2-way repeated measures ANOVA (E, F). ∗p < 0.05, ∗∗p < 0.01. Values represent the mean ± SEM NS = not significant. All data are from different biological replicates. See also Figure S2.
Warfarin does not ameliorate brown adipose tissue dysfunction upon dietary obesity
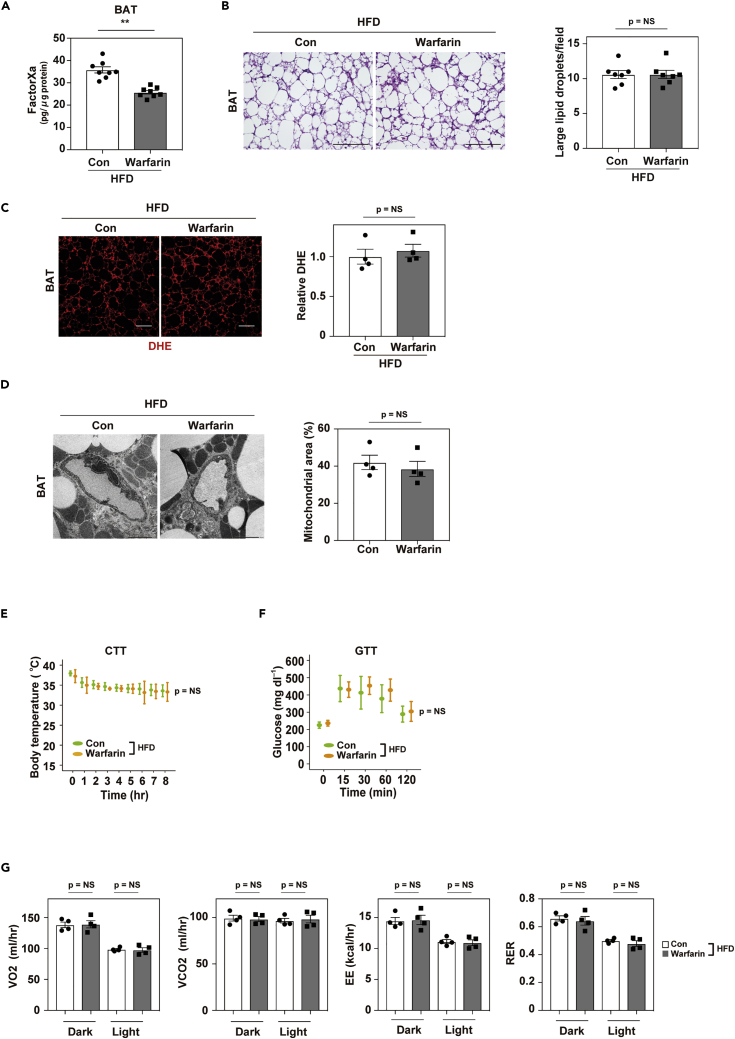
Next, we investigated whether another anticoagulant (Deykin, 1970), warfarin, had any effect on BAT in obese mice. We found that warfarin reduced the FXa level in BAT (Figure 3A) but did not induce re-browning of BAT or reduce DHE levels in obese mice (Figures 3B and 3C). Electron microscopy also showed no reversal of mitochondrial rarefaction and fragmentation in brown adipocytes after warfarin treatment (Figure 3D). Furthermore, warfarin did not improve the thermogenic response (Figure 3E) and had no effect on systemic metabolism (Figures 3F and 3G). Body weight, food intake, and BAT weight were comparable between control mice and warfarin-treated mice (Figures S3A–S3C). Antithrombotic activity was assessed from the tail vein bleeding time and was significantly higher in mice receiving warfarin or FXa-i than in the control groups but was comparable in the FXa-i and warfarin groups (Figure S3D). Thrombin activity in plasma was also suppressed to a similar extent in the warfarin and FXa-i groups (Figure S3E). In the clinical settings, both warfarin and FXa-i are used as anticoagulants; however, our results suggest that these compounds mediate different biological effects in BAT.
Figure 3.
Warfarin does not ameliorate brown adipose tissue dysfunction on dietary obesity
Mice were fed a high-fat diet (HFD) from 4 weeks of age with or without the administration of warfarin. Physiological studies were performed at age 12 to 17 weeks for warfarin treatment, and samples were collected at age 17 weeks.
(A) Enzyme-linked immunosorbent assay study analyzing Factor Xa in brown adipose tissue (BAT) from mice aged 17weeks fed a high-fat diet (HFD; Con HFD) or HFD + Warfarin (Warfarin HFD; n = 8, 8).
(B) Hematoxylin and eosin (HE) staining of BAT from the indicated mice. Scale bar = 100 μm. Right panel indicates the number of large lipid droplets per field in BAT from the indicated mice. Large lipid droplets were defined as droplets with a surface area >1000 μm2 (n = 7, 7).
(C) Staining with dihydroethidium (DHE) in brown adipose tissue (BAT) from mice fed the HFD (Con HFD) or HFD + warfarin (Warfarin HFD). Scale bar = 50 μm. Right panels indicate quantification shown as relative dihydroethidium (DHE) level (DHE area of Warfarin HFD/Con HFD; n = 4, 4).
(D) Findings on transmission electron microscopy in the indicated mice (scale bar = 2 μm). Right panel indicate the mitochondrial area (%; analyzed as mitochondrial area/[non-capillary and non-lipid area]) in the pericapillary area of respective groups (n = 4, 4).
(E and F) Cold tolerance test (CTT; E, n = 7, 7) or glucose tolerance test (GTT; F, n = 7, 6) in the indicated mice aged 13 weeks for CTT, and 12 weeks for GTT.
(G) Oxygen consumption (VO2), CO2 production (VCO2), energy expenditure (EE), and respiratory exchange ratio (RER) in the indicated groups aged 14-16 weeks (n = 4, 4, 4, 4). Data were analyzed by a 2-tailed Student’s t test (A, B, C, D, G) or 2-way repeated measures ANOVA (E, F). ∗p < 0.05, ∗∗p < 0.01. Values represent the mean ± SEM NS = not significant. All data are from different biological replicates. See also Figure S3.
Brown adipose tissue-specific protease-activated receptor-1 depletion ameliorates brown adipose tissue dysfunction
To assess the role of PAR1 signaling in these processes, we generated a BAT-specific PAR1 knockout model by crossing Ucp1-Cre+/− mice with PAR1fl/fl mice (Ucp1-Cre+/−; PAR1fl/fl (BAT-PAR1 KO)) (Figure S4A). Upon a normal chow diet, BAT-PAR1 KO mice showed better thermogenic capacity than littermate control (PAR1fl/fl) mice (Figure S4B), but the results of GTT were similar between the genotypes (Figure S4C). Compared with control mice, BAT-PAR1 KO mice on a HFD were resistant to the development of a whitening BAT phenotype (Figure 4A), along with a reduction of DHE level (Figure 4B) and improvement of mitochondrial rarefaction and fragmentation in brown adipocytes (Figure 4C). These mice also showed improvement in the thermogenic response and systemic glucose metabolism (Figures 4D and 4E). FXa-i administration had no synergistic effect in this model of BAT-specific PAR1 depletion (Figures 4A-4E). Next, we compared other coagulation factors and metabolic profiles in littermate control and BAT-PAR1 KO mice. Transcripts for PAR-2 (F2rl1), tissue factor (F3), FVII (F7), and factor X (F10) were comparable, and body weight, food intake and BAT weight did not differ between the two groups (Figures S4D-S4H). Autophagy has a critical role in lipid metabolism and mitochondrial quality control, but the expression of autophagy-related molecules in BAT was comparable between the two groups (Figures S4I and S4J). Next, we analyzed interscapular skin temperature with an infrared camera and found that upon acute cold exposure an area in HFD BAT-PAR1 KO mice showed a higher temperature than in the littermate control mice, indicating the enhanced thermogenic capacity of BAT in the KO group (Figure S4K).
Figure 4.
Brown adipose tissue-specific protease-activated receptor-1 depletion ameliorates brown adipose tissue dysfunction
Brown adipose tissue (BAT)-specific protease-activated receptor-1 (PAR1) knockout (KO) mice (UCP1-Cre+/−; PAR1flox/flox; BAT PAR1 KO) were fed a high-fat diet (HFD) from 4 weeks of age and physiological studies were performed after 12 to 13 weeks. Tissues were harvested at 18-21 weeks of age.
(A, B, and C) Hematoxylin and eosin (HE) staining (A; scale bar = 100 μm), dihydroethidium (DHE) staining (B; scale bar = 50 μm), and transmission electron microscopy (C; scale bar = 2 μm) of BAT from littermate control mice (PAR1flox/flox; Con), BAT PAR1 KO, and BAT PAR1 KO mice treated with an FXa inhibitor (BAT PAR1 KO + FXa-i). Right panels in Figure 4A indicate the quantification of large lipid droplets (n = 4, 3, 4), and Figure 4B indicates the quantification of dihydroethidium (DHE) staining (n = 5, 5, 5) of BAT in littermate control mice (PAR1flox/flox; Con), BAT PAR1 KO, and BAT PAR1 KO + FXa-i mice. Right panel in Figure 4C indicates mitochondria area in % (analyzed as mitochondrial area/[non-capillary and non-lipid area]) in the pericapillary area of the respective groups (n = 4, 4, 4). (D, E) Cold tolerance test (CTT; D, n = 16, 8, 5) and glucose tolerance test (GTT; E, n = 14, 11, 11) in the indicated mice aged 13 weeks for CTT, and 12 weeks for GTT. ∗ indicates Con vs. BAT PAR1 KO, ## indicates Con vs. BAT PAR1 KO + FXa-i. Data were analyzed by 2-way ANOVA followed by Tukey’s multiple comparison test (A, B, C) or by 2-way repeated-measures ANOVA followed by Tukey’s multiple comparison test (D, E). ∗p < 0.05, ∗∗p < 0.01, ##p < 0.01. Values represent the mean ± SEM NS = not significant. All data are from different biological replicates. See also Figure S4.
Introduction of tissue factor and protease-activated receptor 1 in brown adipose tissue promotes functional decline of this organ
To further characterize coagulation factor-PAR1 signaling in BAT, we generated a gain-of-function model by the direct injection of adeno-associated virus (AAV) encoding PAR1 (AAV-F2r) or tissue factor (AAV-F3) into the BAT of mice maintained on normal chow. Co-administration of these AAVs led to a significant increase in tissue factor and PAR1 expression in BAT, as well as a significant elevation of FXa (Figures 5A–5C and S5A–S5C). Although the introduction of the AAVs into BAT did not change body weight or food intake (Figures S5D and S5E), whitening of this tissue increased (Figures 5D and S5F). BAT weight was comparable between treated and control mice (Figure S5G), but AAV introduction increased DHE level in BAT (Figure 5E) and promoted mitochondrial rarefaction in brown adipocytes (Figure 5F). Furthermore, AAV introduction reduced the thermogenic response to acute cold exposure and promoted systemic glucose intolerance (Figures 5G and 5H).
Figure 5.
Introduction of tissue factor and PAR-1 in brown adipose tissue promotes functional decline of this organ
All experiments with Adeno-associated virus (AAV) were performed in mice fed normal chow (NC). AAVs were injected at 10 weeks of age. Physiological studies were performed 10 to 14 days after AAV injection, and tissues were harvested at 14 weeks of age.
(A-C) Enzyme-linked immunosorbent assay (ELISA) for tissue factor (A, n = 10, 9) or FactorXa (C, n = 8, 8) in BAT from mice injected with control AAV (Mock) or with both AAV encoding F3 and AAV encoding F2r (AAV F3+F2r) aged 14 weeks. (B) Western blot analysis of PAR-1 in BAT from the indicated groups. Right panel indicates the quantification of PAR1 relative to glyceraldehyde 3-phosphate dehydrogenase (GAPDH; n = 9,9).
(D, E, and F) Hematoxylin and eosin (HE) staining (D; scale bar = 100μm), dihydroethidium (DHE) staining (E; scale bar = 50 μm), and transmission electron microscopy (F; scale bar = 10 μm for low magnification [Low Mag] and 2 μm for high magnification [High Mag]) of BAT from the indicated mice. Right panel in Figure 5E indicates the quantification of dihydroethidium (DHE) analyzed as relative DHE signal (DHE area of AAV F3+F2r/Mock) of indicated mice (n = 4, 5). Right panel in Figure 5F indicates mitochondria area (%; analyzed as mitochondrial area/[non-capillary and non-lipid area]) in the pericapillary area of the respective groups (n = 4, 4).
(G and H) Cold tolerance test (CTT; G, n = 8, 8) and glucose tolerance test (GTT; H, n = 6, 7) in the indicated mice aged 13 weeks for CTT, and 12 weeks for GTT. Data were analyzed by a 2-tailed Student’s t test (A, B, C, E, F) or by 2-way repeated measures ANOVA (G and H). ∗p < 0.05, ∗∗p < 0.01. Values represent the mean ± SEM NS = not significant. All data are from different biological replicates. See also Figure S5.
These findings in our loss- and gain-of-function models indicated a causal role of coagulation factor/PAR1-mediated signaling in dietary BAT dysfunction. Although vitamin K-dependent coagulation factors (including FVII and FX) are presumed to be mainly synthesized in the liver (Hassan et al., 1990), extrahepatic production has also been reported (Wilcox et al., 2003). In addition to the microarray (GSE64718) data, the results of these in vivo studies indicate that BAT produces components of the endogenous coagulation system. In association with coagulation factors delivered to BAT through the circulation, these would enhance pleiotropic coagulation factor/PAR1 signaling in this tissue, promoting the functional decline in BAT.
FactorXa/protease-activated receptor 1 signaling promotes the dysfunction of brown adipocytes
Next, we performed in vitro characterization of this signaling pathway by studying differentiated brown adipocytes in culture. FXa increased the mitochondrial superoxide indicator level and reduced the mitochondrial membrane potential, as analyzed by the MitoSox or MitoRed signal, and these changes were ameliorated by the co-administration of a PAR-1 inhibitor (Figures 6A and 6B) but not with a PAR-2 inhibitor (Figure S6A). MitoTEMPO is a mitochondrial-targeted superoxide dismutase mimetic that is known to mediate antioxidant effects in this organelle. MitoTEMPO treatment reduced the MitoSox level and increased the MitoRed signal in brown adipocytes administered FXa (Figures 6C and 6D). To further explore downstream molecules mediating PAR1 signaling, we focused on mitochondrial calcium (Ca2+). It is well known that Ca2+ has roles in enhancing mitochondrial ATP production by regulating the tricarboxylic acid (TCA) cycle; however, excessive mitochondrial Ca2+ overload is known to increase oxidative stress in this organelle and induce mitochondrial dysfunction (Arruda et al., 2014; Brookes et al., 2004). We found that FXa administration increased the mitochondrial Ca2+ level and that this increase was suppressed by PAR1 inhibition (Figure 6E). Inhibition of mitochondrial calcium uniporter (MCU), a transporter involved in the passage of Ca2+ into mitochondria, reduced mitochondrial Ca2+, leading to a reduction of MitoSox signals and an increase of MitoRed signals (Figures 6F-6H). These results indicated that FXa/PAR1 signaling induces Ca2+ overload and oxidative stress in mitochondria, resulting in a functional decline in this organelle.
Figure 6.
FactorXa/PAR1 signaling promotes the dysfunction of brown adipocytes
(A-H) Nivo Multimode Microplate Reader study analyzing signal of mitochondrial ROS (MitoSox; A, n = 5, 5, 5; C, n = 8, 8, 8; G, n = 8, 8, 8), mitochondrial membrane potential (MitoRed; B, n = 8, 8, 8; D, n = 20, 20, 20; H, n = 8, 8, 8), mitochondrial calcium (Ca2+; Rhod2; E, n = 8, 8, 8; F, n = 8, 8, 8) in the indicated groups. For studies of MitoSox and MitoRed, recombinant FXa protein (10nM) was administered for a total of 3 h and other compounds were administered for 1 h before the administration of FXa at the following concentrations: PAR1 inhibitor (PAR1-i), 1μM; MitoTEMPO, 10μM; MCU inhibitor (250nM). For studies of Rhod2, recombinant FXa was administered for a total of 30 min, with or without the reagents described above.
(I) Results of qPCR for expression of coagulation factor VII (F7; n = 5, 6), factor X (F10; n = 6, 6), tissue factor (F3; n = 4, 5), and F2r (PAR-1 transcript; n = 6, 6) in differentiated brown adipocytes incubated with control adenovirus (Mock) or adenovirus encoding constitutively activated Hif-1a (Ad-Hif-1a; 10 MOI, 48 h for F7, F10, and F2r; 30 MOI, 24 h for F3).
(J) Enzyme-linked immunosorbent assay (ELISA) for tissue factor (n = 10, 10), coagulation factor VII (FVII; n = 9, 14), and FXa (n = 11, 9) in conditioned medium from differentiated brown adipocytes incubated with control adenovirus (Mock) or Ad-Hif-1a.
(K and L) MitoSox/MitoGreen ratio (K, n = 8, 7, 7) and MitoRed/MitoGreen ratio (L, n = 8, 8, 8) in differentiated brown adipocytes incubated with conditioned medium (CM) obtained from brown adipocytes infected with Mock (Mock-CM), Ad-Hif-1a (Ad-Hif-1a-CM), or Ad-Hif-1a-CM + PAR1-i. Data were analyzed by a 2-tailed Student’s t test (I-L), or by 2-way ANOVA followed by Tukey’s multiple comparison test (A and C-H), or non-parametric Kruskal Wallis test (B). ∗p < 0.05, ∗∗p < 0.01. Values represent the mean ± SEM NS = not significant. All data are from different biological replicates. See also Figure S6.
It was previously reported that metabolic stress induces hypoxia of BAT, resulting in the activation of hypoxia-inducible factor 1α (Hif1α) signaling (Shimizu et al., 2014). The results of in silico analysis [DBTSS (http://dbtss.hgc.jp); JASPAR (http://jaspar.binf.ku.dk)] predicted that transcripts for FVII (F7), FX (F10), tissue factor (F3), and PAR1 (F2r) have Hif1α binding sites in their putative promoter regions (Figure S6B). We detected F7, F10, F3, and F2r transcripts in brown adipocytes, and exposure to hypoxia or adenovirus-mediated introduction of constitutively activated Hif1α (Ad-Hif1α) led to an increase in the expression of all these genes (Figures 6I and S6C-S6F). The levels of tissue factor, FVII, and FXa were increased in a conditioned medium of brown adipocytes infected with Ad-Hif1α (Figure 6J). When untreated brown adipocytes were exposed to a conditioned medium obtained from Ad-Hif1α-infected brown adipocytes, there was a marked increase in mitochondrial ROS production and a decrease in the mitochondrial membrane potential (Figures 6K and 6L). These changes were ameliorated by co-treatment of the cells with a PAR1 inhibitor (PAR1-i) (Figures 6K and 6L).
Discussion
In the present study, we demonstrated that coagulation factors produced by brown adipocytes or delivered to BAT through the systemic circulation mediate biological effects via PAR1 receptor signaling in an autocrine, paracrine, or endocrine manner, leading to excessive mitochondrial ROS production, BAT dysfunction, and systemic glucose intolerance.
Our studies showed a clear difference in responses against BAT between warfarin and FXa-i. Warfarin affects coagulation by post-translational modification of the gamma-carboxyglutamic acid (Gla)-rich domain of vitamin K-dependent coagulation factors. FXa synthesized without the Gla domain is described as protein induced by vitamin K antagonism or absence Xa (PIVKA-Xa) and is known to retain the capacity to activate PAR-mediated signaling without activating the coagulation cascade (Spronk et al., 2014). The observation of a different phenotype between FXa-i and warfarin indicated that PIVKA-Xa (which was suppressed by FXa-i but not by warfarin) may have enhanced PAR1 signaling in these anti-coagulation models. Therefore, these results suggest that the activation of coagulation factor-PAR1 signaling, rather than an increase of coagulation activity per se, was involved in BAT dysfunction associated with dietary obesity.
In our study, body weight showed comparable results in the HFD and HFD + FXa-inhibitor models and in the WT HFD and BAT PAR1 KO HFD models. Healthy BAT did not lead to BW reduction in the HFD + FXa-inhibitor model or BAT PAR1 KO HFD model compared with the respective control. This puzzling but interesting finding suggests the existence of a feedback loop between BAT and systemic metabolic homeostasis that lets BW remain constant. A thrombin inhibitor, dabigatran, was previously reported to reduce BW under a HFD in an analysis of WT mice on a C57BL/6J background (Kopec et al., 2014, 2017). Although the mouse strain and diet were different from those in our study (we used C57BL/6NCrSlc male mice), a potential difference between thrombin and FXa inhibition needs to be explored.
In conclusion, our finding that coagulation factors can mediate biological effects in BAT provides a new avenue toward understanding the functions of this tissue. Further human studies would be required to examine the role of coagulation factors in patients with metabolic disease.
Limitations of the study
Coagulation factors are in general considered to be predominantly produced by the liver. We did not generate or analyze tissue-specific coagulation factor models and could not show quantitative data for BAT in the production of these factors under dietary obesity. We cannot exclude the possibility that FXa-i may also affect the activity of downstream proteases and PAR2 signaling. The role of PAR1-mediated signaling in human BAT remains to be explored. Animals on HFD were treated with a FXa inhibitor or warfarin from the beginning of the study (before the establishment of the disease), and we have not examined the effects of these compounds on metabolic dysfunction after the establishment of the disease.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-GAPDH antibody | Cell Signaling | #2118 RRID: AB_561053 |
| anti-PAR1 antibody | Novus Biologicals | NBP1-71770 RRID: AB_11027203 |
| anti-PAR2 antibody | Abcam | ab180953 |
| anti-Tissue factor antibody | Abcam | ab189483 |
| anti-Factor VII antibody | Abcam | ab97614 |
| anti-p62 antibody | Abcam | ab219581 |
| anti-LC3A/B antibody | Cell Signaling | #4108 RRID:AB_2137703 |
| Hoechst | Life Technologies | 33258 |
| horseradish peroxidase-conjugated anti-rabbit immunoglobulin G | Jackson Immunoresearch | #111-035-003 RRID: AB_2313567 |
| horseradish peroxidase-conjugated goat anti-mouse IgG(H + L) | Jackson Immunoresearch | #115-035-003 RRID: AB_10015289 |
| goat anti-rabbit IgG H&L (Cy5) | Abcam | ab97077 RRID: AB_10679461 |
| anti-mouse IgG H&L (Cy5) | Abcam | ab6563 RRID: AB_955068 |
| wheat germ agglutinin-conjugated Alexa Fluor 488 | Thermo Fisher Scientific | W11261 |
| dihydroethidium (DHE) | WAKO | 041-28251 |
| Bacterial and Virus Strains | ||
| AAV-DJ Helper Free Expression System | Cell Biolabs Inc | VPK-410-DJ |
| Adeno-X Expression System | Clontech | 631513 |
| Biological Samples | ||
| Human plasma samples | Biobank in Niigata University | N/A |
| Human BAT samples | Biobank in Zealand University Hospital Køge, Denmark | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| HFD32 | CLEA Japan | N/A |
| Rivaroxaban | Bayer | N/A |
| Warfarin | Sigma-Aldrich | A2250 |
| MitoSox | Thermo Fisher Scientific | M36008 |
| MitoTracker Green FM | Thermo Fisher Scientific | M7514 |
| MitoTracker Red CM-H2Xros | Thermo Fisher Scientific | M7513 |
| Rhod-2 AM | Thermo Fisher Scientific | R1245MP |
| human FactorXa protein | Abcam | ab62229 |
| SCH79797 | Santa Cruz | sc-203693A |
| Mito TEMPO | Sigma-Aldrich | SML0737 |
| KB-R7943 | TOCRIS Bioscience | 1244 |
| FSLLRY-NH2 | R&D Systems | 4751/1 |
| Critical Commercial Assays | ||
| Mouse Coagulation factor Xa ELISA Kit | My BioSource | MBS726968 |
| Human Coagulation factor Xa ELISA Kit | My BioSource | MBS721605 |
| Mouse Tissue Factor ELISA Kit | My BioSource | MBS016365 |
| Factor VII ELISA Kit | My BioSource | MBS944525 |
| Deposited Data | ||
| DNA microarray | Gene2015 Jul 1; 565(1):15–21. | GSE64718 |
| Experimental Models: Cell Lines | ||
| Brown pre-adipocyte cell line | Gifted from Dr. C. Ronald Kahn (Joslin Diabetes Center and Harvard Medical School) | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: Adipoq-Cre+: Pgam1flox/flox Mouse: Ucp1-Cre+/−; PAR1fl/fl (BAT-PAR1 KO) | UCP1-Cre; Rosen and Spiegelman (2014). floxed Par1; Generated in our lab | N/A |
| Oligonucleotides | ||
| Forward primer for mouse Actb; 5′- CTAAGGCCAACCGTGAAAAG-3′ |
This paper | N/A |
| Reverse primer for mouse Actb; 5′-ACCAGAGGCATACAGGGACA-3′ | This paper | N/A |
| Forward primer for mouse Rplp0; 5′-GATGCCCAGGGAAGACAG-3′ | This paper | N/A |
| Reverse primer for mouse Rplp0; 5′-ACAATGAAGCATTTTGGATAA-3′ | This paper | N/A |
| Forward primer for mouse Atg5; 5′-AAGTCTGTCCTTCCGCAGTC-3′ | This paper | N/A |
| Reverse primer for mouse Atg5; 5′-TGAAGAAAGTTATCTGGGTAGCTCA-3′ | This paper | N/A |
| Forward primer for mouse Atg7; 5′-CCGGTGGCTTCCTACTGTTA-3′ | This paper | N/A |
| Reverse primer for mouse Atg7; 5′-AAGGCAGCGTTGATGACC-3′ | This paper | N/A |
| Forward primer for mouse Bnip3; 5′-CCTGTCGCAGTTGGGTTC-3′ | This paper | N/A |
| Reverse primer for mouse Bnip3; 5′- GAAGTGCAGTTCTACCCAGGAG-3′ |
This paper | N/A |
| Forward primer for mouse F2r;; 5′-G TCTTCCCGCGTCCCTAT-3′ |
This paper | N/A |
| Reverse primer for mouse F2r;; 5′- GGGTTCACCGTAGCATCTGT-3′ |
This paper | N/A |
| Forward primer for mouse F2rl1; 5′- GGACCGAGAACCTTGCAC-3′ |
This paper | N/A |
| Reverse primer for mouse F2rl1; 5′- GGAACCCCTTTCCCAGTG-3′ |
This paper | N/A |
| Forward primer for mouse F10; 5′-C CATCTTGAATGAGTTCTACATCCT-3′ |
This paper | N/A |
| Reverse primer for mouse F10; 5′-TT CTCTGTGTTCCGATCACCT-3′ |
This paper | N/A |
| Forward primer for mouse F7; 5′-CG CTACTGGGGAAACATCAC-3′ |
This paper | N/A |
| Reverse primer for mouse F7; 5′-CC CCATCCTTCTCACTGAAG-3′ |
This paper | N/A |
| Forward primer for mouse F3; 5′-G CTCAAGCACGGGAAAGA-3′ |
This paper | N/A |
| Reverse primer for mouse F3; 5′-G CTTGCACAGAGATATGGACAG-3′ |
This paper | N/A |
| Forward primer for mouse F12; 5′-T GGGGTCACCAGTTCGAG-3′ |
This paper | N/A |
| Reverse primer for mouse F12; 5′-A GGGAACCTGTGCCTCCT-3′ |
This paper | N/A |
| Forward primer for mouse F11 5′-GGTGCTTGCTCTTCACGTT-3′ |
This paper | N/A |
| Reverse primer for mouse F11 5′-TCAGGATGCAGGCAAACC-3′ |
This paper | N/A |
| Forward primer for mouse F9; 5′-C GCTACTGGGGAAACATCAC-3′ |
This paper | N/A |
| Reverse primer for mouse F9; 5′-C CCCATCCTTCTCACTGAAG-3′ |
This paper | N/A |
| Forward primer for mouse F8; 5′-A GATACACTTACCCTGTTCCCATT-3′ |
This paper | N/A |
| Reverse primer for mouse F8; 5′-A CCCCAAGACCCATAGACCT-3′ |
This paper | N/A |
| Forward primer for mouse F5; 5′-G GACCAGAGGGGTGTACAGA-3′ |
This paper | N/A |
| Reverse primer for mouse F5; 5′-T GTTCTCGTCAAACACAGCAA-3′ |
This paper | N/A |
| Forward primer for mouse F2; 5′-G CAGTGTCCCTGTCTGTGG-3′ |
This paper | N/A |
| Reverse primer for mouse F2; 5′-T TGTCCTTGGAACCTCCAGA-3′ |
This paper | N/A |
| Forward primer for mouse Fga; 5′-G CAGGAAGCGAAGCTCAC-3′ |
This paper | N/A |
| Reverse primer for mouse Fga; 5′-T GGAGGACATCATCACAGTCTC-3′ |
This paper | N/A |
| Forward primer for mouse Fgb; 5′-G CCGATGATGACTACGATGA-3′ |
This paper | N/A |
| Reverse primer for mouse Fgb; 5′-A GGTCTTAGGCTAGGAGGCTCT-3′ |
This paper | N/A |
| Forward primer for mouse Fgg; 5′-G GCCACCTCAATGGAGTTTA-3′ |
This paper | N/A |
| Reverse primer for mouse Fgg; 5′-C ATGGTGGTTTCCTTCATAGAAT-3′ |
This paper | N/A |
| Forward primer for mouse F13a1; 5′-T TCAGAGTGGAATATGTCATTGGT-3′ |
This paper | N/A |
| Reverse primer for mouse F13a1; 5′-A CAGGCACAGGGATGTAGGT-3′ |
This paper | N/A |
| Forward primer for mouse F13b; 5′-A CTGCCCCCTGAGTGTGTT-3′ |
This paper | N/A |
| Reverse primer for mouse F13b; 5′-A ACAACCACACCGTTTGCTA-3′ |
This paper | N/A |
| Forward primer for mouse Map1c3a; 5′-C CCCACCAAGATCCCAGT-3′ |
This paper | N/A |
| Reverse primer for mouse Map1c3a; 5′- CGCTCATGTTCACGTGGT-3′ |
This paper | N/A |
| Recombinant DNA | ||
| pCMV6-Kan/Neo-mouse F3 | OriGENE | MC200642 |
| pCMV6-Entry-mouse F2r | OriGENE | MR206862 |
| Software and Algorithms | ||
| SPSS version 24 software | IBM | N/A |
| KEGG mapper | KEGG (Kyoto Encyclopedia of Genes and Genomes) | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Tohru Minamino at t.minamino@juntendo.ac.jp.
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human samples
The human data were obtained from a subcohort of patients undergoing an operation for benign thyroid disease in whom RNA sequencing of human deep neck BAT was performed (Jespersen et al. bioRxiv 2020.05.07.082057). The full dataset and detailed methodology is available online at Biorxiv (https://www.biorxiv.org/content/10.1101/2020.05.07.082057v1). Briefly, patients were recruited at the outpatient clinics of the Otorhinolaryngology, Head and Neck Surgery, and Audiology Departments of Rigshospitalet and Gentofte Hospitals, Copenhagen, Denmark, and the Department of Otorhinolaryngology and Maxillofacial Surgery, Zealand University Hospital, Køge, Denmark. Apart from thyroid malignancy and inability to provide informed consent, there were no other specific exclusion criteria if patients were considered eligible for surgery. The patients of the subcohort underwent thorough metabolic characterization before the surgical procedure, including an oral glucose tolerance test and a dual energy X-ray absorptiometry (DXA) scan to assess body composition. The clinical characteristics of the included patients can be found in Table 1. Most patients were diagnosed with non-toxic goiter, although 4 had Grave’s disease and 2 had myxedema; however, all underwent the operation and, importantly, all had thyroid hormone levels (T3 and T4) within the normal range. Participants were recruited over the course of a year. We found no correlation between UCP1 levels and the mean or minimal outdoor temperature and no systematic effect of the season (Jespersen et al. bioRxiv 2020.05.07.082057). All participants provided written informed consent before participation. The Scientific-Ethics Committees of the Capital Region of Denmark approved the study protocol and amendments (approval number H-1-2014-015), and the study was performed in accordance with the Declaration of Helsinki. Depending on the specific type of surgical procedure used, the supraclavicular deep neck biopsy was collected from the deep neck fatty tissue either at the lymph node level 4, between the 2 heads of the sternocleidomastoid muscle, or at lymph node level 6, from in front of the thyroid gland. Tissue used for RNA sequencing was immediately frozen in liquid nitrogen and stored at −80°C until analyses were performed. In the current study, the level of transcript F2R was compared between lean individuals (BMI <25; n = 8) and overweight or obese individuals (BMI ≥25; n = 23). Blood samples and clinical data were collected from patients of Niigata University Hospital. All subjects provided written informed consent prior to participation in these studies. The Scientific-Ethics Committees of Niigata University approved the protocols of all the studies (protocol number G2015-0837), and each investigation was performed in accordance with the Declaration of Helsinki.
Animal models
All of the animal experiments were conducted in compliance with protocols reviewed by the Institutional Animal Care and Use Committee of Niigata University, Juntendo University and approved by the Presidents of both Universities. C57BL/6NCrSlc male mice were purchased from SLC Japan (Shizuoka, Japan), and were maintained on a high-fat diet (HFD; HFD32, CLEA Japan; https://www.clea-japan.com/en/products/general_diet/item_d0080) for 8–18 weeks, starting from 4 weeks of age, or a chow diet (CE-2; https://www.clea-japan.com/en/products/general_diet/item_d0030). In some HFD groups, a Factor Xa inhibitor (rivaroxaban; Bayer) was added to the diet at a concentration of 12 mg/kg (BW)/day from 4 weeks of age or warfarin (Sigma-Aldrich A2250) was added to the drinking water at 0.2 mg/kg (BW)/day from 4 weeks of age. Ucp1-Cre+/−; PAR1fl/fl (BAT-PAR1 KO) mice were generated as follows. C57BL/6 BAC genomic clones (RP23-335L17 and 435L15) containing the Par1 gene (F2r) were isolated from a RP23 mouse genomic BAC library (Advanced GenoTechs, Tsukuba, Japan). The Quick and Easy BAC modification Kit (Gene Bridges, Dresden, Germany) was used for vector construction. An 11.4 kb DNA fragment containing exon 2 of F2r was subcloned into the vector with a MC1 promoter-driven diphtheria toxin gene. To construct a Par1-flox targeting vector, a DNA fragment containing the loxP sequence and pgk/gb2 promoter-driven Neo-poly(A) flanked by two frt sites was inserted 206 bp upstream of exon 2, while the other loxP sequence was introduced just behind the stop codon. The resulting construct contained the 4.3 kb upstream and 6.0 kb downstream genomic sequences, as well as the 1.4 kb floxed sequence containing exon 2. To establish the Par1-flox mouse line, we performed electroporation of the linearized targeting vector into RENKA cells, a C57BL/6N-derived ES cell line, and then selected recombinant clones by incubation in medium containing G418 (175 μg/mL). Culture of ES cells was performed as described previously (Mishina and Sakimura, 2007). Target clones were confirmed by Southern blot analysis using 5′, 3′ and Neo probes. Generation of chimeric mice was performed as described previously (Mishina and Sakimura, 2007). Briefly, after microinjection of target clones into 8-cell embryos of CD-1 mice, the chimeric embryos developed to the blastocyst stage by incubation for at least 24 h and then were transferred into pseudopregnant CD-1 mice. Germline chimeras were crossed with C57BL/6N female mice to establish the Par1-flox mouse line. DNA was extracted from mouse tail tips with a REDExtract-N-Amp tissue PCR kit (Sigma Aldrich XNAT-100RXN). Genotyping was done with Taq DNA Polymerase (Roche 11 435 094 001) and the following primers: GCCAGCTGATGCCGAGTAAA (F2R forward) and GACTAATGGGATTCCCCGC (F2R reverse). The PCR protocol was as follows: denaturation at 94°C for 1 min, followed by 30 cycles of 94°C for 30 s/68°C for 30 s/74°C for 30 s, and final holding at 4°C. Homozygous male mice were fed a high-fat diet for at least 8 weeks from 4 weeks of age before investigation. The tail vein bleeding time was measured as reported previously (Zhou et al., 2011). Briefly, 50mL conical tubes were filled with saline and kept in a 37°C water bath. Mice were anesthetized and placed above a conical tube with the stretched tail immersed in saline for 1min. The tip of the tail was cut 5mm from the distal end, and the tail was promptly reimmersed into the conical tube. The tail vein bleeding time was defined as the interval from cutting of the tail until the tip of the tail stopped bleeding for 20s.
Method details
Systemic metabolic parameters
Mice were housed individually for one week before the glucose tolerance test. On the day of the test, mice were fasted for 6 h and then glucose was injected intraperitoneally at a dose of 1 g/kg in the early afternoon. Blood glucose levels were measured with a glucose analyzer (Sanwa Kagaku Kenkyusho) at 15, 30, 60, and 120 min after glucose injection.
Acute cold exposure
Body temperature was assessed by subcutaneous implantation of a sterile biocompatible microchip transponder (IPTT-300 Extended Accuracy Calibration; Bio Medic Data Systems) in the scapular region according to the manufacturer’s instructions. Animals were subjected to the cold tolerance test (CTT) at 4°C and body temperature was measured at hourly intervals for 8 h.
Physiological analysis
Oxygen consumption of mice was assessed with an O2/CO2 metabolic measurement system (Columbus Instruments) according to the manufacturer’s instructions. An infrared camera (testo 885-2, Testo SE & Co) was used to detect skin temperature according to the manufacturer’s instructions. Before temperature was measured, mouse hair was removed from the back with a depilatory cream. Skin temperature was analyzed at baseline and 10 min after putting the tail tip (the last 5 mm of the tail) into ice-cold water. Areas of high temperature were defined as areas of orange or higher in the temperature range of the infrared camera (30°C–40°C, and measured with ImageJ).
Histological examination
Interscapular BAT samples were harvested, fixed overnight in 10% formalin, embedded in paraffin, and sectioned for immunofluorescence or hematoxylin-eosin (HE) staining. Lipid droplets and fluorescence signals were quantified with ImageJ software at a magnification of x400, with four fields being randomly selected in each section. Large lipid droplets were defined as those >1000 μm2 in size. The following antibodies and fluorescent dyes were used: anti-PAR1 antibody (NOVUS Biologicals, NBP1-71770), anti-Factor VII antibody (Abcam, ab97614), anti-tissue factor antibody (Abcam, ab189483), wheat germ agglutinin-conjugated Alexa Fluor 488 for staining cell membranes (Thermo Fisher Scientific, W11261), and Hoechst (Life Technologies, 33258). The secondary antibody was goat anti-rabbit IgG H&L (Cy5) (Abcam, ab97077) or anti-mouse IgG H&L (Cy5) (Abcam, ab6563). All primary and secondary antibodies were used at a dilution of 1:50, except for Hoechst (1:1000). Stained sections were photographed under a confocal microscope (FV1200, Olympus, or C2, Nicon), and 4 or 5 randomly selected views were recorded for quantification of the fluorescence signals with ImageJ software. In some studies, ROS were evaluated with dihydroethidium (DHE) staining (WAKO, 041-28251). For electron microscopy, brown adipose tissue was fixed in 2.5% glutaraldehyde. Grids for electron microscopy were prepared by Masaaki Nameta at the electron microscope core facility of Niigata University, and electron microscopy was performed at Niigata University Medical Campus with a JEM1400 transmission electron microscope. Mitochondrial area was analyzed in randomly selected pericapillary areas as mitochondrial area/(non-capillary plus non-lipid area).
RNA analysis
Total RNA (1 μg) was isolated from tissue samples with RNA-Bee (TEL-TEST Inc.). Then, qPCR was performed with a Light Cycler 480 (Roche) with the Universal Probe Library and Light Cycler 480 Probes Master (Roche) according to the manufacturer’s instructions. To quantify the copy number, we used Light Cycler 480 software version 1.5 (Roche) and performed a calculation by the Fit Points method according to the manufacturer’s instructions.
The following primers were employed, with Actb or Rplp0 as the internal control:
Actb; 5′-CTAAGGCCAACCGTGAAAAG-3′, 5′-ACCAGAGGCATACAGGGACA-3′
Atg5; 5′-AAGTCTGTCCTTCCGCAGTC-3′, 5′-TGAAGAAAGTTATCTGGGTAGCTCA-3′
Atg7; 5′-CCGGTGGCTTCCTACTGTTA-3′, 5′-AAGGCAGCGTTGATGACC-3′
Bnip3; 5′-CCTGTCGCAGTTGGGTTC-3′, 5′-GAAGTGCAGTTCTACCCAGGAG-3′
F2r; 5′-GTCTTCCCGCGTCCCTAT-3′, 5′-GGGTTCACCGTAGCATCTGT-3′
F2rl1; 5′-GGACCGAGAACCTTGCAC-3′, 5′-GGAACCCCTTTCCCAGTG-3′
F10; 5′-CCATCTTGAATGAGTTCTACATCCT-3′, 5′-TTCTCTGTGTTCCGATCACCT-3′
F7; 5′-CGCTACTGGGGAAACATCAC-3′, 5′-CCCCATCCTTCTCACTpGAAG-3′
F3; 5′-GCTCAAGCACGGGAAAGA-3′, 5′-GCTTGCACAGAGATATGGACAG-3′
F12; 5′-TGGGGTCACCAGTTCGAG-3′, 5′-AGGGAACCTGTGCCTCCT-3′
F11; 5′-GGTGCTTGCTCTTCACGTT-3′, 5′-TCAGGATGCAGGCAAACC-3′
F9; 5′-CGCTACTGGGGAAACATCAC-3′, 5′-CCCCATCCTTCTCACTGAAG-3′
F8; 5′-AGATACACTTACCCTGTTCCCATT-3′, 5′-ACCCCAAGACCCATAGACCT-3′
F5; 5′-GGACCAGAGGGGTGTACAGA-3′, 5′-TGTTCTCGTCAAACACAGCAA-3′
F2; 5′-GCAGTGTCCCTGTCTGTGG-3′, 5′-TTGTCCTTGGAACCTCCAGA-3′
Fga; 5′-GCAGGAAGCGAAGCTCAC-3′, 5′-TGGAGGACATCATCACAGTCTC-3′
Fgb; 5′-GCCGATGATGACTACGATGA-3′, 5′-AGGTCTTAGGCTAGGAGGCTCT-3′
Fgg; 5′-GGCCACCTCAATGGAGTTTA-3′, 5′-CATGGTGGTTTCCTTCATAGAAT-3′
F13a1; 5′-TTCAGAGTGGAATATGTCATTGGT-3′, 5′-ACAGGCACAGGGATGTAGGT-3′
F13b; 5′-ACTGCCCCCTGAGTGTGTT-3′, 5′-AACAACCACACCGTTTGCTA-3′
Map1c3a; 5′-CCCCACCAAGATCCCAGT-3′, 5′-CGCTCATGTTCACGTGGT-3′
Rplp0; 5′-GATGCCCAGGGAAGACAG-3′, 5′-ACAATGAAGCATTTTGGATAA-3′
Western blot analysis
Whole-cell lysates were prepared in lysis buffer (10mM Tris-HCl, pH 8, 140mM NaCl, 5mM EDTA, 0.025% NaN3, 1% Triton X-100, 1% deoxycholate, 0.1% SDS, 1mM PMSF, 5 μg ml−1 leupeptin, 2 μg ml−1 aprotinin, 50mM NaF, and 1mM Na2VO3). Then the lysates (40–50 μg) were resolved by SDS-PAGE. Proteins were transferred to a PVDF membrane (Millipore) that was incubated with following primary antibodies: anti-PAR1 antibody (NOVUS Biologicals, NBP1-71770), anti-PAR2 antibody (Abcam, ab180953), anti-p62 antibody (Abcam, ab219581), LC3A/B antibody (Cell Signaling #4108), GAPDH antibody (Proteintech, 10494-1-AP). All primary antibodies were used at a dilution of 1:1000. Subsequently, incubation was done with horseradish peroxidase-conjugated goat anti-rabbit IgG(H + L) (light chain specific) (Jackson Immunoresearch, #115-035-174) at a dilution of 1:5000, except that horseradish peroxidase-conjugated goat anti-mouse IgG(H + L) (Jackson Immunoresearch, #115-035-003) was used as the secondary antibody for anti-PAR1 antibody. After incubation, proteins were detected by enhanced chemiluminescence (GE).
ELISA
The following kits were used for ELISA according to the instructions of the manufacturer: Mouse Coagulation factor Xa ELISA Kit (My BioSource, MBS726968), Human Coagulation factor Xa ELISA Kit (My BioSource, MBS721605), Mouse Tissue Factor ELISA Kit (My BioSource, MBS016365), and Factor VII ELISA Kit (My BioSource, MBS944525).
In-vitro studies and molecular probe
The brown pre-adipocyte cell line was a kind gift from Dr. C. Ronald Kahn (Joslin Diabetes Center and Harvard Medical School, Section on Integrative Physiology and Metabolism, Boston, USA) (Klein et al., 1999). This cell line was established from wild-type FVB mice, and was immortalized by infection with the pBabe retroviral vector encoding SV40T antigen. Cells were cultured in high-glucose Dulbecco’s modified minimal essential medium with 10% fetal bovine serum and 100 U/mL P/S, and differentiation was induced as described previously (Fasshauer et al., 2001). Fully differentiated brown adipocytes were used for analyses after 10 days of culture. The cells were stained with the following mitochondrial molecular probes according to the manufacturer’s instructions and were analyzed under a confocal microscope (FV1200, Olympus): MitoSox (1 μM, 10 min, Thermo Fisher Scientific, M36008), MitoTracker Green FM (MitoGreen) (200 nM, 45 min, Thermo Fisher Scientific, M7514), MitoTracker Red CM-H2Xros (MitoRed) (200 nM, 45 min, Thermo Fisher Scientific, M7513) and Rhod-2 AM (5μM, 30 min, Thermo Fisher Scientific, R1245MP). In some experiments, differentiated brown adipocytes were cultured with the full-length human FactorXa protein (ab62229) at 10 nM (6 h for MitoSox and 24 h for MitoTracker Red CM-H2Xros). In other experiments, a PAR1 antagonist (SCH79797) (Santa Cruz, sc-203693A, 1μM), Mito TEMPO (Sigma, SML0737, 10μM), MCU inhibitor (KB-R7943; TOCRIS Bioscience, 1244, 250 nM), and PAR-2 antagonist (FSLLRY-NH2, R&D Systems, 4751/1, 1 μM) were added 1 h before administration of full-length human FactorXa protein. Hypoxic stress was induced by incubating the cells under 1% O2 for 1 h in a hypoxia chamber (Stem Cell Technologies) according to the manufacturer’s instructions, and analyses were done after returning the cells to normoxia for 5 h. For studies using conditioned medium, an adenoviral vector encoding constitutively expressed Hif-1α (Ad Hif-1a, 10 MOI for 48 h) or the Mock vector (10 MOI for 48 h) was introduced into differentiated brown adipocytes incubated in 24-well culture dishes with 500 μL of medium per well. Then the medium was replaced with fresh differentiation medium and cells were incubated for an additional 24 h, followed by collection of conditioned medium. Fully differentiated brown adipocytes were incubated in wells containing 200 μL of conditioned medium mixed with 300 μL of normal differentiation medium (total volume: 500 μL), with or without pretreatment using a PAR1 antagonist (SCH79797) (Santa Cruz, sc-203693A) for 1 h at 1μM. Cells were cultured with conditioned medium for a total of 24 h for either the MitoRed/MitoGreen or MitoSox/MitoGreen studies, and signals were analyzed with a Nivo Multimode Microplate Reader (PerkinElmer) according to the instructions of the manufacturer.
Adenovirus and adeno-associated virus
Adenovirus encoding ODD-deleted Hif-1α (the constitutively expressed activated form of Hif-1) (Koshiji et al., 2004) was generated with the Adeno-X Expression System (Clontech) according to the manufacturer’s instructions. For experiments with adeno-associated virus (AAV), F3 mouse cDNA (Origene MC200642) or F2r mouse cDNA (Origene MR206862) was subcloned into the pAAV-MCS expression vector (AAV-DJ Helper Free Expression System, Cell Biolabs Inc., VPK-410-DJ). Then pAAV-F3, pAAV-F2r, or pAAV-GFP (control) was co-transfected into HEK293 cells with pAAV-DJ and pHelper using X-tremeGENE9 DNA Transfection Reagent (Roche, 06365809001), according to the manufacturer’s instructions. AAV was harvested from the cells by freeze-thaw disruption, and purified with a ViraBind AAV Purification Kit (Cell Biolabs Inc., VPK-140). The titer of purified AAV was quantified with a QuickTiter AAV Quantitation Kit (Cell Biolabs Inc., VPK-145). Then a single injection of AAV was administered into BAT under direct visual inspection in 10-week-old mice fed a normal chow diet, as described previously (Shimizu et al., 2014). The AAV titer was 1 × 109 GC/mouse for AAV-F3 or AAV-F2r and 2 × 109 GC/mouse for AAV-GFP. Physiological studies were performed on days 10–14 after injection, and tissues were collected for further analyses at 4 weeks after injection.
Microarray analysis
GSE64718, a microarray dataset available in a public database (NCBI GEO: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE64718), was analyzed and the results were displayed with the KEGG mapper (https://www.genome.jp/kegg/tool/map_pathway3.html), focusing on the coagulation cascade.
Quantification and statistical analysis
Statistical analysis
Statistical analyses were performed with SPSS software version 24. All values were included in the analyses. If analyses did not reach statistical significance, in some cases outliers and abnormal values were detected by SPSS boxplot analyses (boxplots show the upper whisker, upper quartile, median, lower quartile, and lower whisker) and excluded from further analyses Non-significant (NS) values in the figure indicate that these analyses included or excluded outlier and/or abnormal values and still did not reach statistical significance. All data reported are from different biological replicates. Differences between groups were examined by a 2-tailed Student’s t test or two-way ANOVA, followed by Tukey’s multiple comparison test, a non-parametric Kruskal Wallis test, or Dunnett’s test for comparisons between more than two groups. Data from some experiments were analyzed by 2-way repeated measures ANOVA, followed by Tukey’s multiple comparison test. In all analyses, p < 0.05 was considered significant.
Acknowledgments
This work was supported by a Grant-in-Aid for Scientific Research (A) (20H00533) from MEXT, AMED-CREST under Grant Number JP20gm1110012, Moonshot Research and Development Program (21zf0127003s0201), MEXT Supported Program for the Scleroderma Research Foundation at Private Universities Japan, Private University Research Branding Project, and Leading Initiative for Excellent Young Researchers, and grants from the Science Research Promotion Fund, the Takeda Medical Research Foundation, the Vehicle Racing Commemorative Foundation, Ono Medical Research Foundation, and the Suzuken Memorial Foundation (to T.M.). This research was also supported by grants from the Uehara Memorial Foundation, Kowa Life Science Foundation, SENSHIN Medical Research Foundation, Ono Medical Research Foundation, The Nakajima Foundation, Suzuken Memorial Foundation, as well as a Grant for Basic Science Research Projects from The Sumitomo Foundation, a Grant-in-Aid for Encouragement of Young Scientists (A) (JSPS KAKENHI Grant Number 16H06244), grants from the Japan Cardiovascular Research Foundation, a Grant-in-Aid for Scientific Research (B) (19H03650), AMED Project for Elucidating and Controlling Mechanisms of Aging and Longevity under Grant Number JP21gm5010002, and The Japan Geriatrics Society (to I.S.). Furthermore, support was provided by a Grant-in-Aid for Scientific Research (C) (19K08974), Yujin Memorial Grant, Sakakibara Memorial Research Grant from The Japan Research Promotion Society for Cardiovascular Diseases, TERUMO Life Science Foundation, Kanae Foundation (to Y.Y.), and by a grant from Bourbon (to T.M., I.S., and Y.Y.) and the joint research funds from Bayer (to T.M.). NZ.J. is supported by a research grant from the Danish Diabetes Academy, which is funded by the Novo Nordisk Foundation, grant number; NNF17SA0031406.
Author contributions
I.S. and T.M. designed the studies and wrote the article. Y.H., Y.Y., and I.S. performed the majority of the experiments. R.I., M.S., G.K., and K.N. provided technical assistance for the in vivo studies. M.A., K.S., and K.W. contributed to the generation of genetic mouse models. S.O. contributed to bioinformatic analysis. T.H. contributed to protein studies. S.F. and K.O. contributed to the human study. NZ.J., S.N. and C.S. analyzed the human bat biopsies.
Declaration of interests
The authors except for T.M. disclose no conflicts of interest. T.M. discloses the joint research funds and the remuneration for a lecture from Bayer; however, this company did not play a role in the study design, data collection and analysis, decision to publish, or preparation of the article.
Published: July 15, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.104547.
Contributor Information
Ippei Shimizu, Email: ippeishimizu@yahoo.co.jp.
Tohru Minamino, Email: t.minamino@juntendo.ac.jp.
Supplemental information
Data and code availability
All data reported in this paper will be shared by the lead contact upon request. Gene expression data are available through the Gene Expression Omnibus database (GSE64718). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Arruda A.P., Pers B.M., Parlakgul G., Guney E., Inouye K., Hotamisligil G.S. Chronic enrichment of hepatic endoplasmic reticulum-mitochondria contact leads to mitochondrial dysfunction in obesity. Nat. Med. 2014;20:1427–1435. doi: 10.1038/nm.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badeanlou L., Furlan-Freguia C., Yang G., Ruf W., Samad F. Tissue factor-protease-activated receptor 2 signaling promotes diet-induced obesity and adipose inflammation. Nat. Med. 2011;17:1490–1497. doi: 10.1038/nm.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartelt A., Bruns O.T., Reimer R., Hohenberg H., Ittrich H., Peldschus K., Kaul M.G., Tromsdorf U.I., Weller H., Waurisch C., et al. Brown adipose tissue activity controls triglyceride clearance. Nat. Med. 2011;17:200–205. doi: 10.1038/nm.2297. [DOI] [PubMed] [Google Scholar]
- Borissoff J.I., Heeneman S., Kilinc E., Kassak P., Van Oerle R., Winckers K., Govers-Riemslag J.W., Hamulyak K., Hackeng T.M., Daemen M.J., et al. Early atherosclerosis exhibits an enhanced procoagulant state. Circulation. 2010;122:821–830. doi: 10.1161/circulationaha.109.907121. [DOI] [PubMed] [Google Scholar]
- Brookes P.S., Yoon Y., Robotham J.L., Anders M.W., Sheu S.S. Calcium, ATP, and ROS: a mitochondrial love-hate triangle. Am. J. Physiol. Cell Physiol. 2004;287:C817–C833. doi: 10.1152/ajpcell.00139.2004. [DOI] [PubMed] [Google Scholar]
- Cypess A.M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A.B., Kuo F.C., Palmer E.L., Tseng Y.H., Doria A., et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 2009;64:519–520. doi: 10.1097/ogx.0b013e3181ac8aa2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deykin D. Warfarin therapy. 1. N. Engl. J. Med. 1970;283:691–694. doi: 10.1056/NEJM197009242831307. [DOI] [PubMed] [Google Scholar]
- Ezzati M., Bentham J., Di Cesare M., Bilano V., Bixby H., Zhou B., Stevens G.A., Riley L.M., Taddei C., Hajifathalian K., et al. Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: a pooled analysis of 2416 population-based measurement studies in 128.9 million children, adolescents, and adults. Lancet. 2017;390:2627–2642. doi: 10.1016/S0140-6736(17)32129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer M., Klein J., Kriauciunas K.M., Ueki K., Benito M., Kahn C.R. Essential role of insulin receptor substrate 1 in differentiation of brown adipocytes. Mol. Cell Biol. 2001;21:319–329. doi: 10.1128/mcb.21.1.319-329.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghorpade D.S., Ozcan L., Zheng Z., Nicoloro S.M., Shen Y.F., Chen E., Hen E.C., Bluher M., Czech M.P., Tabas I. Hepatocyte-secreted DPP4 in obesity promotes adipose inflammation and insulin resistance. Nature. 2018;555:673–677. doi: 10.1038/nature26138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan H.J., Leonardi A., Chelucci C., Mattia G., Macioce G., Guerriero R., Russo G., Mannucci P.M., Peschle C. Blood-coagulation factors in human embryonic-fetal development - preferential expression of the FVII-tissue factor pathway. Blood. 1990;76:1158–1164. doi: 10.1182/blood.v76.6.1158.1158. [DOI] [PubMed] [Google Scholar]
- Klein J., Fasshauer M., Ito M., Lowell B.B., Benito M., Kahn C.R. β3-Adrenergic stimulation differentially inhibits insulin signaling and decreases insulin-induced glucose uptake in Brown adipocytes. J. Biol. Chem. 1999;274:34795–34802. doi: 10.1074/jbc.274.49.34795. [DOI] [PubMed] [Google Scholar]
- Kopec A.K., Abrahams S.R., Thornton S., Palumbo J.S., Mullins E.S., Divanovic S., Weiler H., Owens A.P., Mackman N., Goss A., et al. Thrombin promotes diet-induced obesity through fibrin-driven inflammation. J. Clin. Invest. 2017;127:3152–3166. doi: 10.1172/jci92744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec A.K., Joshi N., Towery K.L., Kassel K.M., Sullivan B.P., Flick M.J., Luyendyk J.P. Thrombin inhibition with dabigatran protects against high-fat diet-induced fatty liver disease in mice. J. Pharmacol. Exp. Therapeut. 2014;351:288–297. doi: 10.1124/jpet.114.218545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiji M., Kageyama Y., Pete E.A., Horikawa I., Barrett J.C., Huang L.E. HIF-1α induces cell cycle arrest by functionally counteracting Myc. EMBO J. 2004;23:1949–1956. doi: 10.1038/sj.emboj.7600196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishina M., Sakimura K. Conditional gene targeting on the pure C57BL/6 genetic background. Neurosci. Res. 2007;58:105–112. doi: 10.1016/j.neures.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Nedergaard J., Bengtsson T., Cannon B. New powers of brown fat: fighting the metabolic syndrome. Cell Metabol. 2011;13:238–240. doi: 10.1016/j.cmet.2011.02.009. [DOI] [PubMed] [Google Scholar]
- Ouellet V., Labbe S.M., Blondin D.P., Phoenix S., Guerin B., Haman F., Turcotte E.E., Richard D., Carpentier A.C. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. J. Clin. Invest. 2012;122:545–552. doi: 10.1172/jci60433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel M.R., Mahaffey K.W., Garg J., Pan G., Singer D.E., Hacke W., Breithardt G., Halperin J.L., Hankey G.J., Piccini J.P., et al. Rivaroxaban versus warfarin in nonvalvular atrial fibrillation. N. Engl. J. Med. 2011;365:883–891. doi: 10.1056/NEJMoa1009638. [DOI] [PubMed] [Google Scholar]
- Rosen E.D., Spiegelman B.M. What we talk about when we talk about fat. Cell. 2014;156:20–44. doi: 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad F., Ruf W. Inflammation, obesity, and thrombosis. Blood. 2013;122:3415–3422. doi: 10.1182/blood-2013-05-427708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu I., Aprahamian T., Kikuchi R., Shimizu A., Papanicolaou K.N., MacLauchlan S., Maruyama S., Walsh K. Vascular rarefaction mediates whitening of brown fat in obesity. J. Clin. Invest. 2014;124:2099–2112. doi: 10.1172/jci71643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spronk H.M.H., de Jong A.M., Crijns H.J., Schotten U., Van Gelder I.C., ten Cate H. Pleiotropic effects of factor Xa and thrombin: what to expect from novel anticoagulants. Cardiovasc. Res. 2014;101:344–351. doi: 10.1093/cvr/cvt343. [DOI] [PubMed] [Google Scholar]
- van Marken Lichtenbelt W.D., Vanhommerig J.W., Smulders N.M., Drossaerts J.M.A.F.L., Kemerink G.J., Bouvy N.D., Schrauwen P., Teule G.J.J. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009;360:1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Wilcox J.N., Noguchi S., Casanova J. Extrahepatic synthesis of factor VII in human atherosclerotic vessels. Arterioscler. Thromb. Vasc. Biol. 2003;23:136–141. doi: 10.1161/01.atv.0000043418.84185.3c. [DOI] [PubMed] [Google Scholar]
- Yoneshiro T., Aita S., Matsushita M., Kayahara T., Kameya T., Kawai Y., Iwanaga T., Saito M. Recruited brown adipose tissue as an antiobesity agent in humans. J. Clin. Invest. 2013;123:3404–3408. doi: 10.1172/jci67803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P.S., Metcalf M., Bunnett N.W. Biased signaling of protease-activated receptors. Front. Endocrinol. 2014;5 doi: 10.3389/fendo.2014.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Schwarting S., Illanes S., Liesz A., Middelhoff M., Zorn M., Bendszus M., Heiland S., van Ryn J., Veltkamp R. Hemostatic therapy in experimental intracerebral hemorrhage associated with the direct thrombin inhibitor dabigatran. Stroke. 2011;42:3594–3599. doi: 10.1161/strokeaha.111.624650. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request. Gene expression data are available through the Gene Expression Omnibus database (GSE64718). This paper does not report original code. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.