Abstract
The ability of marine bacteria to adhere to detrital particulate organic matter and rapidly switch on metabolic genes in an effort to reproduce is an important response for bacterial survival in the pelagic marine environment. The goal of this investigation was to evaluate the relationship between chitinolytic gene expression and extracellular chitinase activity in individual cells of the marine bacterium Pseudoalteromonas sp. strain S91 attached to solid chitin. A green fluorescent protein reporter gene under the control of the chiA promoter was used to evaluate chiA gene expression, and a precipitating enzyme-linked fluorescent probe, ELF-97–N-acetyl-β-d-glucosaminide, was used to evaluate extracellular chitinase activity among cells in the bacterial population. Evaluation of chiA expression and ELF-97 crystal location at the single-cell level revealed two physiologically distinct subpopulations of S91 on the chitin surface: one that was chitinase active and remained associated with the surface and another that was non-chitinase active and released daughter cells into the bulk aqueous phase. It is hypothesized that the surface-associated, non-chitinase-active population is utilizing chitin degradation products that were released by the adjacent chitinase-active population for cell replication and dissemination into the bulk aqueous phase.
Detrital particulate organic matter (POM) in the pelagic marine environment harbors dense microbial assemblages (1). Bacteria associated with POM synthesize ectohydrolytic enzymes to convert insoluble substrates into bacterial biomass. Release of dissolved organic matter (DOM) during ectohydrolytic enzyme degradation of POM is thought to support bacterial production of free-living bacterial populations in the pelagic marine environment (21, 32, 33, 40). This process has been referred to as “sloppy feeding” by POM-associated bacteria. However, due to methodological difficulties, bacterial colonization and enzymatic degradation of POM are difficult to directly observe on these surfaces.
Natural particle surfaces in the marine environment, such as chitin, possess properties that preclude the nondestructive visualization of single cells over time. Chitin is an insoluble homopolymer of β-1,4-linked N-acetyl-β-d-glucosamine and occurs commonly as an exoskeletal and endoskeletal material in many marine organisms including Mollusca, Coelenterata, Protozoa, Fungi, and Crustacea (12, 20, 28). As a result, chitin is a dominant form of particulate carbon, nitrogen, and energy in the pelagic marine environment (27). Bacteria are the principal mediators of chitin degradation, and their ability to promote extracellular hydrolysis of chitin is an essential step in recycling carbon and other nutrients in the pelagic marine environment (34). Unfortunately, chitin displays surface roughness that generally prevents the visualization of single cells on the surface. In addition, chitin fluoresces under epi-illumination, preventing the use of fluorogenic compounds that are used to visualize individual cells. Due to the difficulty in overcoming these properties of chitin, the dynamic relationship between the production of bacterial biomass and chitin hydrolysis has been evaluated only at the population or community level, and never at the single-cell level.
It was the goal of this research to overcome the obstacles associated with natural chitin surfaces by using thin films of pure, spun-cast chitin to visualize attached bacteria during chitin degradation. In addition, a new precipitating enzyme-linked fluorescent (ELF-97) probe was employed to relate chitinase gene expression to extracellular chitinase activity in individual cells of a bacterial population during the degradation of chitin thin films. The new enzyme substrate, ELF-97–N-acetyl-β-d-glucosaminide, is soluble in aqueous solutions, membrane impermeable, and nonfluorescent until cleaved by extracellular chitinase. The resulting fluorophore crystallizes at the site of enzyme action and is used in conjunction with a green fluorescent protein (GFP) reporter of chitinase gene expression. The combination of the chitin thin films, the precipitating enzyme substrate, and the reporter gene construct permitted the direct observation of chitinase gene expression and the activity of the chitinase gene product at the single-cell level during chitin degradation.
MATERIALS AND METHODS
Bacterial strains.
A GFP reporter gene (gfp) was used to visualize chitinase gene expression of Pseudoalteromonas sp. strain S91, during the degradation of solid chitin. Strain S91 was derived from the wild-type strain S9, a chitinolytic marine bacterium isolated from the surface waters of Botany Bay, New South Wales, Australia, in 1981 (22). Strain S91 is streptomycin (SM) resistant and contains the plasmid pDSK519, which confers kanamycin (KM) resistance. The gene for gfp is under the control of the chiA promoter on the plasmid, and a fully functional chiA gene is present on the chromosome (35, 37, 40). The expression of this gene is essential for utilization of high-molecular-weight chitin as a carbon, nitrogen, and energy source (39). The gfp gene is the GFPmut2 derivative of the wild-type gene and confers a 30-fold increase in GFP fluorescence intensity (13).
Bacterial cultivation.
Stock cultures of S91 were grown to exponential phase in MB2216 (Difco, Detroit, Mich.). Approximately 5 μl of this culture was used to inoculate 50 ml of a defined seawater medium. A defined seawater medium consisted of a defined seawater solution to which a specific carbon, nitrogen, and energy source was added to support growth. Defined seawater solution consisted of 402.1 mM NaCl, 4.8 mM H3BO4, 27.5 mM Na2SO4, 2.4 mM NaHCO3, 88.5 mM KCl, 8.4 mM KBr, 54.1 mM MgCl2 · 6H2O, and 1.5 mM SrCl2 · 6H2O suspended in 0.05 M Sigma 7-9 buffer using once-distilled Millipore water and adjusted to pH 8.0. Following autoclave sterilization, 0.008 mM FeCl3, 0.04 mM K2HPO4, and 2.0 mM CaCl2 were each added separately by filter sterilization through a 0.2-μm-pore-size Millipore syringe filter. To ensure retention of the plasmid by the bacterial cells during cultivation, KM was added to all culture media at a final concentration of 600 μg ml−1.
Growth of S91 under conditions of chiA down-expression was achieved by supplementing the defined seawater solution with 0.05 M glutamic acid as the sole carbon, nitrogen, and energy source (39). When expression of the chiA gene was desired during the growth of free-living cells of S91 in batch culture, 0.05 M N-acetyl-β-d-glucosamine (GlcNAc) was added to the defined seawater solution as the sole carbon, nitrogen, and energy source. Starvation cultures of S91 were first cultured on defined seawater medium containing glutamic acid. Upon achieving a cell density of approximately 4.2 × 107 CFU ml−1, the culture was harvested by centrifugation in a Sorvall RC-5B centrifuge at 16,000 × g for 10 min. The resulting cell pellet was washed three times in defined seawater solution, and the final cell pellet was resuspended in 1.0 liter of defined seawater solution in an air-sparged, continuously stirred tank reactor (CSTR) at 20°C. The cells were maintained in this starvation state for 400 h to mimic conditions that free-living bacteria encounter in the pelagic marine environment. KM was added to the defined seawater solution at a final concentration of 600 μg ml−1 to ensure retention of the plasmid by the bacterial cells during starvation. SM was added to the defined seawater solution at a final concentration of 200 μg ml−1 to minimize contamination of the cell suspension during the starvation period. The CSTR (Knick Machining, Bozeman, Mont.) was constructed of a 1.0-liter fluted glass reaction kettle (Lab Glass, Buena, N.J.) and a 304L stainless steel headplate. All reactor ports were constructed of Swagelock gas fittings with Teflon ferrules (Idaho Valve and Fitting, Idaho Falls, Idaho).
Substratum preparation.
The physical complexity and chemical heterogeneity found in natural detrital POM were minimized, in this system, by using a substratum consisting of the insoluble biopolymer chitin. Thin films of pure chitin served as the only added sources of carbon, nitrogen, and energy during chitin degradation. Silicon coupons, 1 cm by 1 cm square and 1.5 mm thick (Harrick Scientific, Ossining, N.Y.), were used as nonnutritional controls. The methods used in the preparation of pure chitin thin films were adapted from previously published methods for spin-casting chitin films from chitosan solutions (5, 29, 30). Briefly, the thin films were cast onto the silicon substrates at 4,500 rpm from a 1.5% solution of high-molecular-weight chitosan (Aldrich, Milwaukee, Wis.) in an aqueous solution of 2.0% acetic acid. These thin films of chitosan were N acetylated to chitin using a 20% solution of acetic anhydride in methanol for 18 h at 4°C. The N acetylation of the chitosan film was monitored using Fourier transform, infrared spectroscopy, and the chemical purity and homogeneity were assessed with small-spot X-ray photoelectron spectroscopy (7a). Film thickness and surface coverage were assessed using profilometry (15, 16). The average dry density of these films was calculated to be 2.05 g cm−3 from quartz crystal microbalance mass data and profilometry data. The exact thickness of the thin films used is uncertain, as there was some variability that was dependent on environmental conditions (temperature and humidity) at the time that the films were spun-cast. However, these chitin thin films are approximately 230 nm thick. These chitin thin films were ultrasmooth, continuous, and nonfluorescent. These physical and chemical properties permitted spatial quantification of attached bacteria at the single-cell level during chitin degradation.
Laminar flow cell (LFC) preparation.
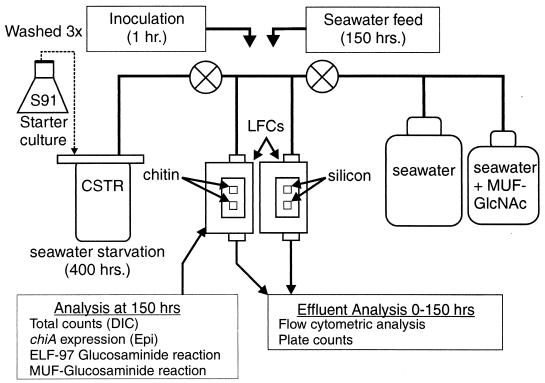
An LFC was used to evaluate bacterial attachment, reproduction, and biofilm development on chitin and silicon surfaces under a constant defined seawater solution flow. Teflon LFCs with glass viewing ports were used to allow direct microscopic examination of the surfaces without disturbing the flowing system. Two separate LFCs were used to monitor surface-associated bacterial activity (Fig. 1). The first LFC contained two chitin thin films cast onto silicon coupons. The second LFC contained clean silicon coupons and served as nonnutritional control surfaces. The LFCs were of a design modified from that used previously to monitor reporter gene expression on surfaces (14). The LFCs were constructed of virgin Teflon (McMaster Carr, Los Angeles, Calif.) and had a flow channel that was 0.8 mm deep, 12 mm wide, and 48 mm long (Knick Machining). Two 1- by 1-cm squares were recessed, in series, into the floor of each LFC to allow the placement of silicon or chitin coupons. A no. 2 coverslip (24 by 60 mm) was used as the viewing window and was sealed against the Teflon using an oversized Viton gasket and an aluminum coverplate. Teflon influent and effluent lines were connected with Chemfluor polytetrafluoroethylene gas fittings (Cole Palmer, Vernon Hills, Ill.). The LFCs were connected with platinum-cured silicon tubing (Cole Palmer) to the air-sparged CSTR and a 20-liter carboy that contained defined seawater solution. Glass flow breaks placed in line between the medium feed and the LFCs prevented back contamination of the sterile defined seawater solution feed. A Buchler 12 roller Multistatic pump (Cole Palmer) was used to transfer the starved-cell inoculum and sterile defined seawater from their respective reservoirs to the LFCs.
FIG. 1.
Schematic representation of the experimental design showing LFCs, CSTR, and peristaltic pumps (⊗). The silicon and chitin LFCs were run in tandem. Epi, epifluorescence.
A strict cleaning procedure was used for all containers and surfaces to which the bulk aqueous phase was exposed in order to minimize the introduction of chemical contaminants that might support bacterial growth. The 20-liter carboy, Teflon LFCs, glass coverslips, and silicon coupons were cleaned in Piranha solution, which consisted of a 70:30 mixture of concentrated sulfuric acid and 30.0% hydrogen peroxide, rinsed in double-distilled water, and baked dry at 120°C prior to assembly of the system. (Be warned that Piranha solution reacts violently, even explosively, with organic materials [18].) All influent lines were cleaned by flushing with 99.8% high-pressure liquid chromatography-grade ethanol at a flow rate of 1 cm3 min−1 for 60 min, followed by double-distilled water to remove residual organic carbon on the surface of the tubing. Any deviation from the cleaning procedure described above resulted in increased bacterial growth on the control surfaces.
The chitin and silicon LFCs were simultaneously inoculated with a continuous flow of a 400-h starved-cell suspension (4.9 × 105 CFU ml−1) from the CSTR over a 1-h period at a flow rate of 0.5 ml min−1. After inoculation, the bulk aqueous phase flowing through the LFCs was switched to sterile defined seawater solution containing KM and SM to select for plasmid retention by cells of S91 and inhibit contamination, respectively, of the solution reservoirs, tubing, and LFCs by other bacteria. The sterile defined seawater solution was maintained at 20°C and pumped through the LFCs at a constant rate of 0.5 ml min−1 for a period of 150 h. The residence time of the defined seawater solution in the flow cells was 55 s, precluding significant replication of free-living cells in the bulk aqueous phase during the time that it resided in the LFCs.
Total chitinase activity.
At 150 h post-inoculation of the LFCs, chitinase activity was determined by using the fluorogenic substrate 4-methylumbelliferyl–N-acetyl-β-d-glucosaminide dihydrate (MUF-GlcNAc) (Fluka no. 69585). This chitinase substrate is soluble in water and membrane permeable and reacts with both extracellular and periplasmic β-N-acetylhexosaminidase (chitinase) to yield the fluorophore 4-methylumbelliferone (MUF). A stock solution of MUF-GlcNAc was dissolved in dimethyl sulfoxide at a 100 mM concentration. A 50 μM solution of MUF-GlcNAc was prepared by adding 500 μl of the stock solution to 1,000 ml of defined seawater solution. At 150 h postinoculation, the bulk aqueous phase flowing through the LFC was switched from sterile defined seawater solution to sterile defined seawater solution containing MUF-GlcNAc. The flow rate was maintained at 0.5 ml min−1. After 10 min, nine 2.5-ml effluent samples were collected for determination of chitinase activity contributed by surface-associated and bulk aqueous phase enzyme.
The nine effluent samples collected were partitioned into three compartments in an effort to locate chitinase activity. The first three effluent samples were immediately frozen at liquid nitrogen temperature after exiting the LFCs with no additional incubation of MUF-GlcNAc. This set of samples captured primarily that activity contributed by surface-associated chitinase enzyme as the defined seawater solution containing MUF-GlcNAc flowed through the LFCs. The second set of three effluent samples was filtered through 0.2-μm-pore-size Millipore filters to remove whole cells and then incubated for 1 h in the dark at 20°C. After the 1-h incubation, these samples were immediately frozen at liquid nitrogen temperature. This set of samples captured chitinase activity that resulted from surface-associated enzyme and any additional activity contributed by free chitinase liberated into the bulk aqueous phase. The final set of three effluent samples was incubated for 1 h in the dark at 20°C and then frozen at liquid nitrogen temperature. This set of samples captured the combined activity of chitinase associated with the surface, free chitinase associated with the bulk aqueous phase, and chitinase activity associated with detached cells. Prior to freezing of the samples at liquid nitrogen temperatures, 0.5 ml of glycine-OH buffer at pH 10.5 was added to the samples to raise the pH to 10, the pH at which maximum fluorescence emission of MUF occurs. The samples were then stored at −40°C during the 4-week period prior to measurement of fluorescence.
Immediately prior to measurement of sample fluorescence, each sample was thawed and placed in a 3.5-ml optical glass cuvette (Starna Cells Inc., Atascadero, Calif.) with a wavelength range of 320 to 2,500 nm. The cuvette was placed in the analysis chamber of a TD-700 fluorometer (Turner Design Inc., Sunnyvale, Calif.). A 2.5-ml volume of 50 μM MUF-GlcNAc in defined seawater solution adjusted to pH 10.0 with glycine-OH buffer served as a blank. The fluorescence intensity of the samples was determined at an excitation wavelength of 360 nm and an emission wavelength of 430 nm. MUF concentration was determined by relating fluorescence intensity to MUF concentration using a standard curve over the range of 0.01 to 10 μM.
Surface localization of extracellular chitinase activity.
Spatial distribution of extracellular chitinase activity, on the chitin and silicon surfaces, was identified by using a new precipitating fluorescent probe for β-N-acetylhexosaminidase (chitinase) activity. ELF-97–N-acetyl-β-d-glucosaminide (ELF-97–GlcNAc) is soluble in water and cell impermeable and reacts with chitinase to yield the fluorophore ELF-97. This fluorophore is insoluble in water and crystallizes at the site of enzyme action. The methods used in the synthesis of this enzyme substrate were adapted from previously published methods for synthesizing ELF-97–β-d-glucuronide (17). Briefly, ELF-97–GlcNAc was synthesized by oxidatively condensing 4-chloroanthranilamide with 4-chloro-2-formylphenyl-aceto-β-d-glucosaminide. The resulting 4-chloro-2-[2′-(6"-chloro-4(3H)-quinazolinonyl)]-phenyl-aceto-β-d-glucosaminide was deprotected to yield the enzyme substrate 4-chloro-2- [2′-(6"-chloro-4(3H)-quinazolinonyl)]-phenyl-β-d-glucosaminide or ELF-97–GlcNAc. The enzymatic hydrolysis of this substrate with pure Streptomyces griseus chitinase (Fluka no. 22725) or with whole up-expressed cells of Pseudoalteromonas sp. strain S91 yields a bright yellow-green precipitate with the excitation (360 nm) and emission (540 nm) wavelengths of the fluorophore 2-[2′-hydroxy-5′-chlorophenyl-6-chloro-4(3H)-quinazolinone] or ELF-97 (23).
At 150 h postinoculation and after the MUF-GlcNAc analysis, the LFCs were incubated with ELF-97–GlcNAc for 1 h in an effort to spatially resolve chitinase activity with respect to total cells and cells up-expressed for chiA activity on both the chitin and silicon surfaces. ELF-97–GlcNAc was dissolved in dimethyl sulfoxide at a 10 mM concentration. A 50 μM solution of ELF-97–GlcNAc was prepared by adding a 10-μl volume of the 10 mM stock solution to 2.0 ml of defined seawater solution. The mixture was filtered through a 0.2-μm-pore-size polytetrafluoroethylene Millipore membrane to remove any residual ELF-97 crystals and injected into the LFCs. The LFCs were allowed to incubate in the dark at 20°C for 1 h. The ELF-97–GlcNAc–defined seawater solution was then rinsed out of the flow cells, and the ELF-97 activity was assessed using epifluorescence microscopy and image analysis.
Microscopy and image analysis.
At 150 h postinoculation, total cells, chiA up-expressed cells, and sites of ELF-97 activity were directly enumerated and spatially related on both the chitin and silicon surfaces. Images were acquired using an Olympus B-Max 60 microscope (Olympus Optical Co., Tokyo, Japan) employing both reflected differential interference contrast (DIC) and epifluorescence optics. All images were acquired using a Nikon infinity-corrected, 40×, water-immersion objective (Nikon Inc., Torrance, Calif.) and a mercury lamp (Chiu Technical Corporation, Kings Park, N.Y.). Digital images were gathered using a Photometrics Imagepoint cooled charge-coupled device camera (Photometrics, Tucson, Ariz.). Fluorescence images of chiA-gfp reporter gene expression were acquired using an excitation wavelength of 481 nm and an emission wavelength of 507 nm (13). Fluorescence images of ELF-97 were acquired using an excitation wavelength of 360 nm and an emission wavelength of 540 nm. All images were analyzed with Image-Pro Plus software (Media Cybernetics, Silver Spring, Md.). Image manipulation of the DIC images consisted of a background correction, adjustment of the gray-scale contrast, and one pass of a three-by-three sharpening filter. The images of gfp expression were pseudocolored green, and the images of ELF-97 activation were pseudocolored red. Total cells, chiA up-expressed cells, and sites of ELF-97 activity were counted using triple image overlays. Individual cells were counted as chitinase active if sites of ELF-97 activity were touching the cell. Four physiological states were established at the single-cell level using these image overlays: cells that were chiA down-expressed and non-chitinase active (chiA negative-chitinase negative), cells that were chiA up-expressed and non-chitinase active (chiA positive-chitinase negative), cells that were chiA up-expressed and chitinase active (chiA positive-chitinase positive), and cells that were chiA down-expressed and chitinase active (chiA negative-chitinase positive). Chitinase-active sites not associated with cells (cell negative-chitinase positive) were also located and enumerated. The software was used to count individual cells and individual sites of ELF-97 activity in all images using manually adjusted threshold values for each individual image. Four random fields were counted in each of 10 images on the chitin surface, and three random fields were counted in each of eight images on the silicon surface.
Flow cytometric analysis.
A total of 17 LFC effluent samples were gathered throughout the time course of the 150-h experiment and analyzed using a Becton Dickinson FACSCalibur fluorescence-activated cell sorter. chiA expression was monitored by measuring GFP fluorescence upon excitation at 481 nm and emission at 507 nm. A maximum of 50,000 counts were acquired for each sample. Batch cultures of S91 grown on glutamic acid and GlcNAc were used as controls for chiA down- and up-expression, respectively, in cells. Detached cells were partitioned on the basis of their fluorescence intensity using ranges defined by the batch culture controls.
RESULTS
chiA expression in glutamate-grown and starved cells of S91.
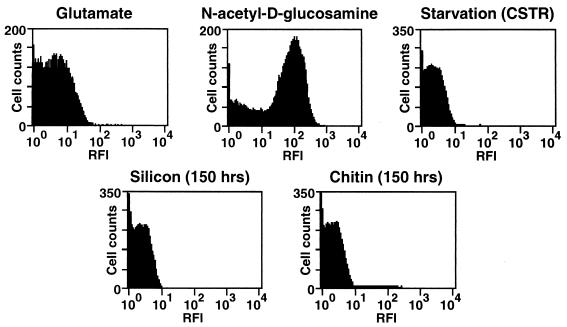
gfp expression and product fluorescence reported the level of expression of chiA, a gene involved in chitin degradation in S91. When S91 was cultured in defined seawater medium with glutamate as the sole added carbon, nitrogen, and energy source, glutamate being an amino acid previously determined to support cell growth but unable to induce detectable chitinase activity (35), a range of levels of chiA expression from 1 to 11 relative fluorescence intensity (RFI) units was displayed by cells in the population, based on flow cytometric analysis, a level of chiA expression resulting in an RFI of 6 being the most common (Fig. 2). In contrast, a population of S91 cells grown in the presence of GlcNAc, the subunit of chitin that is a strong inducer of chitinase activity, yielded a range of fluorescence intensities from 1 to 1,000 RFI units, the most common RFI displayed being 140 RFI units (Fig. 2).
FIG. 2.
Histograms of RFI based on chiA gene activity in a population of cells from a GlcNAc-grown batch culture, a glutamate-grown batch culture, a 400-h starved culture, or the effluent of LFCs containing a silicon or chitin substratum at 150 h postinoculation. Cells displaying RFIs between 1 and 11 were defined as down-expressed for chiA, while cells displaying RFIs between 12 and 1,000 were defined as up-expressed for chiA. Cell counts refer to the number of cells in the population displaying a particular RFI.
Following resuspension of the glutamate-grown culture in defined seawater solution and exposure to starvation conditions in the CSTR for 400 h, the RFIs displayed by the population of cells shifted to a lower range, an RFI of 1 being displayed by the largest number of cells under these conditions (Fig. 2). Thus, cells used to inoculate the surfaces in the LFCs displayed a very low level of chiA expression, one that was undetectable in individual cells by epifluorescence microscopy.
Surface colonization.
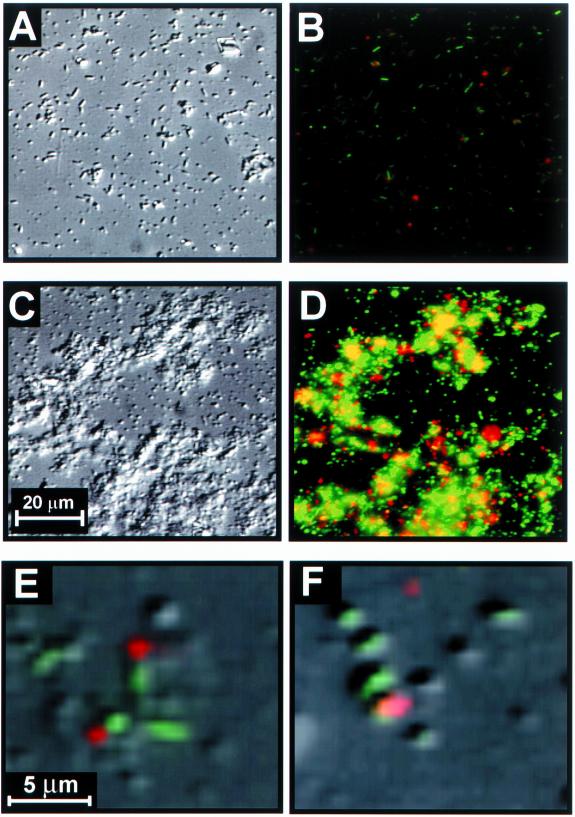
The use of thin films of the natural biopolymer chitin permitted the assessment of chiA gene expression in single cells attached to surfaces without interference from the autofluorescence of natural forms of chitin. Cells that had been starved for 400 h rapidly colonized the silicon and chitin surfaces. Following the switch to sterile defined seawater solution, surface-associated cells increased in size and proliferated across each surface. DIC images of the chitin and silicon surfaces at 150 h postinoculation revealed higher cell densities on the chitin surface than on the silicon surface (Fig. 3A and C). The mean cell densities ± standard errors on the chitin and silicon surfaces were 7.6 × 106 ± 1.5 × 105 and 3.4 × 106 ± 6.4 × 104 cells cm−2, respectively. These results demonstrate that surfaces, regardless of their inherent nutritional value, stimulate the growth of starved cells.
FIG. 3.
Reflected DIC (A and C) and epifluorescence (B and D) micrographs of silicon (A and B) and chitin (C and D) surfaces at 150 h postinoculation. Shown are low-magnification (B and D) and high-magnification (E and F) reflected DIC-epifluorescence image overlays showing the tight association between chiA up-expressed cells (green) and chitinase activity as reported by cleavage of the ELF-97–N-acetyl-β-d-glucosaminide enzyme substrate (red) at the single-cell level. Overlap of chiA gene activity (green) and chitinase activity (red) appears as yellow.
Expression of chiA by surface-associated cells.
Since it was difficult to normalize the levels of fluorescence obtained by flow cytometry and epifluorescence microscopy, up-expression of chiA by surface-associated cells was defined in this study as any level of fluorescence detected in a single cell by epifluorescence microscopy. On the chitin surface, 3.2 × 106 ± 1.5 × 105 cells cm−2 or 41.5% of the total cell population was up-expressed for chiA at 150 h postinoculation. On the silicon surface, 1.8 × 106 ± 4.9 × 104 cells cm−2 or 53.8% of the total cell population was up-expressed for chiA at 150 h postinoculation. Thus, a significant fraction of attached cells become up-expressed for chiA regardless of the presence of chitin.
Chitinolytic activity of the surface-associated population.
Quantification by image analysis of the area of the chitin and silicon surfaces displaying ELF-97 fluorescence allowed a comparison of chitinase activity by cells colonizing these surfaces. Of the total exposed surface area, 11.6% ± 1.5% displayed fluorescence 150 h postinoculation on the chitin surface and 2.9% ± 0.3% displayed fluorescence on the silicon surface.
Relationship between expression of chiA gene and chitinase activity of surface-associated populations.
The combined use of the gfp reporter for chiA gene expression and the precipitating enzyme substrate ELF-97–N-acetyl-β-d-glucosaminide permitted the evaluation of the relationship between chitinase gene expression and gene product activity at the single-cell level. While it was difficult to resolve individual cells in some areas of the chitin surface containing high cell densities, other areas of the surface contained low-enough cell densities to resolve individual cells. These areas of lower cell density were therefore used to evaluate phenotypic differences among cells associated with the chitin and silicon surfaces. Image overlays revealed chitinase activity both directly adjacent to (Fig. 3E) and directly associated with (Fig. 3F) chiA up-expressed cells. When chitinase activity was observed, it was associated with chiA up-expressed cells on both chitin and silicon surfaces (Fig. 3B and D). However, many chiA up-expressed cells were not associated with chitinase-active sites. Furthermore, many cells on both the chitin and silicon surfaces exhibited no chiA activity.
Four physiological states were resolved for the surface-associated cell population based on chiA gene expression and chitinase activity (Table 1). The data show that (25.2 ± 7.1)% and (20.8 ± 4.6)% of the total population are chiA positive-chitinase positive on the chitin and silicon surfaces, respectively. Thus, some cells of S91 become chiA up-expressed and excrete active chitinase enzyme upon attachment to a surface regardless of whether the surface contains chitin. In addition, (39.0 ± 12.4)% and (32.7 ± 6.1)% of the total population are chiA negative-chitinase negative on the chitin and silicon surfaces, respectively. Furthermore, a large fraction of chiA up-expressed cells displayed no detectable chitinase activity (chiA positive-chitinase negative) (Table 1). The ELF-97 enzyme substrate also identified a very small percentage of free enzyme activity not associated with cells (cell negative-chitinase positive) and enzyme-active sites containing cells down-expressed for chiA (chiA negative-chitinase positive). The results suggest that chitinase activity, when present, closely correlated with up-expression of chiA by the cells.
TABLE 1.
Contribution of subpopulations to total cell population associated with chitin and silicon surfaces
| Surfacea | % Subpopulation (mean ± SD)
|
||||
|---|---|---|---|---|---|
| chiA positive-chitinase positive | chiA positive-chitinase negative | chiA negative-chitinase negative | chiA negative-chitinase positive | Cell negative-chitinase positive | |
| Chitin | 25.2 ± 7.1 | 31.8 ± 8.7 | 39.0 ± 12.4 | 1.33 ± 0.19 | 1.61 ± 0.29 |
| Silicon | 20.8 ± 4.6 | 44.2 ± 8.2 | 32.7 ± 6.1 | 1.25 ± 0.27 | 0.88 ± 0.26 |
A total of 1,780 and 1,149 cells were counted on the chitin and silicon surfaces, respectively.
Expression of chiA by detached cells.
Analysis of chiA expression by cells that were displaced from the surfaces into the effluent of the LFCs was determined by flow cytometry. Cells that detached from either the chitin or the silicon surface displayed comparably low levels of chiA expression relative to that of up-expressed cells grown in GlcNAc (Fig. 2). Whereas an RFI of 140 was displayed by the greatest number of cells grown in GlcNAc, an RFI of 3 was displayed by the greatest number of cells that had detached from either the chitin or the silicon surface (Fig. 2). Interestingly, the level of expression of chiA in the population of cells that detached from the chitin and silicon surfaces was comparable to that of the starved-cell population used to inoculate the LFCs (Fig. 2).
Throughout the 150-h time course of the experiment, only (0.2 ± 0.03)% (n = 17) and (0.1 ± 0.02)% (n = 17) of the total cells that detached from the chitin and silicon surfaces, respectively, displayed RFI levels of 12 or greater, the minimum RFI for a cell considered to be up-expressed for chiA. By comparison, (71.5 ± 3.4)% of the total cells in a GlcNAc-grown batch culture, (3.3 ± 0.8)% of the total cells in a glutamate-grown batch culture, and (0.8 ± 0.05)% of the total cells in the 400-h starvation culture used to inoculate the LFCs were up-expressed for chiA. Thus, the fraction of detached cells that are up-expressed for chiA is even lower than that of a starved-cell population.
Distribution of chitinase activity.
To assess the distribution of chitinase activity between the surface and bulk aqueous phase, chitinase activity was determined with MUF-GlcNAc, a soluble, membrane-permeable analog to the ELF-97–N-acetyl-β-d-glucosaminide substrate (Table 2). Using the same molar concentration for MUF-GlcNAc as for ELF-97–N-acetyl-β-d-glucosaminide, enzyme activities in the effluent from the LFCs containing chitin and silicon were 0.47 ± 0.05 and 0.24 ± 0.03 μmol liter−1 h−1, respectively, when no further incubation was carried out. When the effluent samples were incubated for an additional 60-min period, no additional chitinase activity was detected, suggesting that there was no significant activity contributed by free enzyme or enzyme associated with detached, free-living cells. Therefore, the majority of chitinase activity in this system was surface associated. The lack of chitinase activity among detached free-living bacteria indicates that this subpopulation maintains the chiA-negative–chitinase-negative phenotype.
TABLE 2.
Distribution of chitinase activity among various compartments at 150 h postinoculation
| Surface | Rate of MUF-GlcNAc hydrolysis (μmol liter−1 h−1) (mean ± SD)
|
||
|---|---|---|---|
| Surface associated | Surface associated + free enzyme + free cells | Surface associated + free enzyme (<0.2-μm-pore-size filtrate) | |
| Chitin | 0.47 ± 0.05 | 0.50 ± 0.04 | 0.46 ± 0.05 |
| Silicon | 0.24 ± 0.03 | 0.21 ± 0.02 | 0.22 ± 0.02 |
DISCUSSION
Up-expression of chiA following attachment of starved cells of S91 to chitin is consistent with other studies that have shown chitin utilization by marine bacteria to be a complex and highly regulated process (20). Bassler et al. (6, 7) have shown chitin utilization by Vibrio furnissii to be a multistep process, involving a chemotactic response and production of different enzymes that hydrolyze the various degradation products of the chitin polymer. That cells became up-expressed for chiA when attached to the silicon surface in the absence of chitin was unanticipated and suggests that expression of this gene was a surface-controlled response, independent of the inherent nutritional value of the surface. While it is now known that expression of numerous genes is controlled by cell attachment to a surface (8), little is known about those properties of the surface that are involved in the process. Yu et al. (42) showed that attachment of V. furnissii to a surface was dependent on the surface displaying glycosides containing either GlcNAc, d-mannose, or d-glucose. Cell adhesion to these sugars was mediated by a Ca2+-dependent cell surface lectin that interacted directly with these sugar subunits. Since the silicon surface contained no sugars, the mechanism of adhesion of cells of S91 is not likely to involve sugar-lectin interactions.
In addition, attachment to a surface, irrespective of the presence of chitin, promoted synthesis and excretion of active chitinase. We hypothesize that the cells produce a “sensing” amount of the enzyme to continuously monitor availability of chitin on the surface, analogous to the nutrient sensorium proposed for the adhesion-deadhesion apparatus in V. furnissii (42).
A new method for localizing extracellular chitinase activity confirmed the presence of four subpopulations during chitin degradation: two subpopulations (chiA positive-chitinase positive and chiA negative-chitinase positive) that actively synthesized and excreted extracellular chitinase enzyme and two other subpopulations (chiA negative-chitinase negative and chiA positive-chitinase negative) that did not produce extracellular chitinase enzyme. The detection of chiA-positive–chitinase-negative and chiA-negative–chitinase-positive subpopulations was unexpected. Cells displaying the chiA-positive–chitinase-negative phenotype may have recently initiated chitinase synthesis, with not enough active enzyme excreted to convert a threshold amount of enzyme substrate to a detectable fluorescent product. We cannot exclude the possibility that the chiA-positive–chitinase-negative subpopulation produced an endochitinase that may not have reacted with ELF-97–N-acetyl-β-d-glucosaminide. It is possible using the current synthesis method for ELF-97–N-acetyl-β-d-glucosaminide to synthesize the corresponding ELF-97–N,N′-diacetyl-β-d-chitobioside and -N,N′,N"-triacetyl-β-d-chitotrioside enzyme substrates to distinguish different chitinase enzymes. In combination with the MUF analogs, these substrates should enable the distinction of different chitinase activities at the single-cell level.
Detection of the chiA-negative–chitinase-positive phenotype, on the other hand, suggests that cells are capable of liberating an ELF-97–N-acetyl-β-d-glucosaminide-hydrolyzing enzyme that is encoded by a gene that operates independently of the chiA promoter. Many chitinolytic bacteria, including S91, possess multiple chitinase genes controlled by different promoters (24, 36, 38, 41). The small contribution made by this phenotype to the total population of S91 cells, however, suggests that up-expression of the chiA gene was highly correlated with chitinase activity in the population, as reported with ELF-97–N-acetyl-β-d-glucosaminide under the conditions of this study. The small contribution of the chiA-negative–chitinase-positive phenotype among cells of the total surface-associated population also suggests that the chiA promoter-gfp construct is relatively stable in the S91 population under the conditions employed in this study, as loss of the reporter gene would give rise to this phenotype.
The detection of such a large subpopulation that displayed the chiA-negative–chitinase-negative phenotype was also unexpected. Others have suggested that POM-associated bacteria generate more DOM from the hydrolysis of POM than they can utilize, resulting in the release of DOM into the bulk aqueous phase for utilization by free-living bacteria (3, 4, 11). It is hypothesized that, in our model system, the chiA-positive–chitinase-positive subpopulation was responsible for the generation of excess chitin hydrolysate during growth and replication on the chitin surface and that the excess hydrolysate was utilized by the chitin surface-associated chiA-negative–chitinase-negative subpopulation for growth and replication. An alternative hypothesis is that a portion of the chiA-positive–chitinase-positive subpopulation changes to the chiA-negative–chitinase-negative phenotype while associated with the surface. This may be the case when cells displaying the chiA-positive–chitinase-positive phenotype deplete their surroundings of chitin or produce adequate amounts of soluble chitin degradation products for their metabolic needs. Either of these conditions may promote down-expression of chiA, causing the cells to transiently display the chiA-negative–chitinase-positive phenotype before the extracellular chitinase produced previously by the cell is degraded or diffuses away, which would then result in a chiA-negative–chitinase-negative phenotype. Support for the alternative hypothesis therefore requires the presence of the transient chiA-negative–chitinase-positive phenotype, since conversion to the chiA-negative phenotype would not cause the precipitated, fluorescent ELF-97 product around the cell to disappear simultaneously but instead yield a chiA-negative–chitinase-positive phenotype. That this phenotype was displayed by only 1% of the total population suggests that conversion of cells from the chiA-positive–chitinase-positive to the chiA-negative–chitinase-negative phenotype was not significant among cells attached to either the chitin or the silicon surface.
The hypothesized dependence of the surface-associated chiA-negative–chitinase-negative subpopulation on chitin degradation products produced by the chiA-positive–chitinase-positive subpopulation may suggest cell-cell interaction. Intraspecies cooperation may result in more efficient utilization of the carbon, nitrogen, and energy available in chitin by the surface-associated population as a whole and allow for two energy-intensive cellular activities (chitinase production and cell replication) to occur simultaneously. Phenotypic variation within a bacterial population is generally recognized when individual traits manifest themselves through distinguishable morphological features (9). In one of the best-known examples, vegetative cells of Myxococcus xanthus aggregate and undergo morphogenesis to form fruiting bodies that differentiate into myxospores (25, 31). Likewise, as Streptomyces colonies age, cells differentiate into aerial filaments called sporophores that give rise to spores called conidia that germinate to form new vegetative cells (10). Caulobacter crescentus is known to attach to surfaces, form a stalk that serves as an adhesive holdfast, and undergo cell division that results in a flagellated swarmer cell (19, 26). In all of these examples, morphological differentiation distinguishes members of a population with different life histories that work cooperatively to enhance the survival of the population. However, unlike the above examples, differentiation among members of S91 was not manifested as a gross change in morphology. Rather, the differentiation occurred at the level of gene expression and extracellular enzyme production. Considering the possibilities of differential gene expression within any genome, it is likely that this type of phenotypic variation is more common than that which results in gross changes in morphology.
That detachment of cells was observed from both the chitin and silicon surfaces is consistent with the behavior described previously for the chitin-degrading marine bacterium V. furnissii (42). Yu et al. (42) reported that the first progeny of adherent cells of V. furnissii continued to bind to glycoside-coated beads but the population gradually shifted over six cell divisions to a large fraction of free-swimming cells that represented 80 to 90% of the total population. It was proposed that the adhesion-deadhesion apparatus is used by the bacterium to continuously monitor the nutrient status of the environment, prevent overcrowding, and permit colonization of more favorable environments.
The chiA-negative–chitinase-negative phenotype displayed by the bulk of the cells that detached from both chitin and silicon surfaces suggests that the cells that detach from these surfaces are physiologically more homogeneous than those cells remaining on the surfaces. Since the residence time of the bulk liquid in the system is less than the generation time of the bacteria, and the half-life of the non-protease-sensitive GFP used in the cells of this study is more than the 45 min to several hours reported for the less stable protease-sensitive GFP (2), the biomass associated with the chiA-negative–chitinase-negative free-living cell population must have originated from the surface-associated population of the same phenotype. In this scenario, chiA up-expressed cells still remain associated with the surface, while detached cells are derived almost exclusively from the subpopulation displaying the chiA-negative–chitinase-negative phenotype. The basis for this strong partitioning of chitinase-producing cells to remain associated with the surface and the detachment of only cells displaying the chiA-negative–chitinase-negative phenotype remains to be determined.
In summary, evaluation of microbial processes at the single-cell level may be necessary in order to resolve pathways of carbon flux in the marine environment. We have demonstrated that synthesis of extracellular chitinase enzyme, like the up-regulation of chitinase genes in surface-associated bacteria, varies among individual cells within a population exposed to apparently homogeneous environmental conditions. The ability of a population to coordinate chitinase activity, cell reproduction, and surface detachment among different cells on a surface should enhance their ability to locate new sources of nutrients in the pelagic marine environment.
ACKNOWLEDGMENTS
We gratefully acknowledge Sandra Kurk at the Department of Veterinary Molecular Biology at Montana State University for her help in flow cytometric analysis. We also recognize Samuel Hudson at North Carolina State University for sharing his expertise in the preparation of chitin films.
Part of this work was supported by The Flinders University of South Australia and the Australian Research Council. Somkiet Techkarnjanaruk was supported by a Royal Thai Government Scholarship. This work was sponsored by the National Science Foundation under grant OCE 9720151 to Gill Geesey and the National Institutes of Health under grant S10RR11877 to Mark Jutila and under the National Science Foundation cooperative agreement EEC 8907039.
REFERENCES
- 1.Alldredge A L, Youngbluth M. The significance of macroscopic aggregates (marine snow) as sites for heterotrophic bacterial production in the mesopelagic zone of the subtropical Atlantic. Deep Sea Res. 1985;32:1445–1456. [Google Scholar]
- 2.Anderson J B, Sternberg C, Poulsen L K, Bjorn S P, Givskov M, Molin S. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl Environ Microbiol. 1998;64:2240–2246. doi: 10.1128/aem.64.6.2240-2246.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azam F. Microbial control of oceanic carbon flux: the plot thickens. Science. 1998;280:694–696. [Google Scholar]
- 4.Azam F, Cho B C. Bacterial utilization of organic matter in the sea. In: Fletcher M, Gray T R G, Jones J G, editors. Ecology of microbial communities. New York, N.Y: Cambridge University Press; 1987. pp. 261–281. [Google Scholar]
- 5.Bae H, Hudson S M. The cooperative binding behavior of sodium dodecyl sulfate to crosslinked chitosan films. J Appl Polymer Sci: Part A Polymer Chem. 1997;35:3755–3765. [Google Scholar]
- 6.Bassler B L, Gibbons P J, Yu C, Roseman S. Chitin utilization by marine bacteria. Chemotaxis to chitin oligosaccharides by Vibrio furnissii. J Biol Chem. 1991;266:24268–24275. [PubMed] [Google Scholar]
- 7.Bassler B L, Yu C, Lee C, Roseman S. Chitin utilization by marine bacteria. Degradation and catabolism of chitin oligosaccharides by Vibrio furnissii. J Biol Chem. 1991;266:2476–2486. [PubMed] [Google Scholar]
- 7a.Baty A M, III, Eastburn C C, Techkarnjanaruk S, Goodman A E, Geesey G G. Spatial and temporal variations in chitinolytic gene expression and bacterial biomass production during chitin degradation. Appl Environ Microbiol. 2000;66:3574–3585. doi: 10.1128/aem.66.8.3574-3585.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belas R, Simon M, Silverman M. Regulation of lateral gene transcription in Vibrio parahaemolyticus. J Bacteriol. 1986;167:210–218. doi: 10.1128/jb.167.1.210-218.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-Jacob E, Cohen I, Gutnick D L. Cooperative organization of bacterial colonies: from genotype to morphotype. Annu Rev Microbiol. 1998;52:779–806. doi: 10.1146/annurev.micro.52.1.779. [DOI] [PubMed] [Google Scholar]
- 10.Chater K F. Genetics of differentiation in Streptomyces. Annu Rev Microbiol. 1993;47:685–713. doi: 10.1146/annurev.mi.47.100193.003345. [DOI] [PubMed] [Google Scholar]
- 11.Cho B C, Azam F. Major role of bacteria in biogeochemical fluxes in the ocean's interior. Nature. 1988;332:441–443. [Google Scholar]
- 12.Clarke A. The biochemical composition of krill, Euphausia superba Dana, from South Georgia. J Exp Mar Biol Ecol. 1980;43:221–236. [Google Scholar]
- 13.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 14.Davies D G, Geesey G G. Regulation of the alginate biosynthesis gene algC in Pseudomonas aeruginosa during biofilm development in continuous culture. Appl Environ Microbiol. 1995;61:860–867. doi: 10.1128/aem.61.3.860-867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deck L, de Groot P. High speed non-contact profiler based on scanning white light interferometry. Appl Optics. 1995;33:7334–7338. doi: 10.1364/AO.33.007334. [DOI] [PubMed] [Google Scholar]
- 16.Demarest F C. High resolution, high speed, low data age uncertainty, heterodyne displacement measuring interferometer electronics. Meas Sci Technol. 1998;9:1024–1030. [Google Scholar]
- 17.Diwu Z, Lu Y, Upson R H, Zhou M, Klaubert D H, Haugland R P. Fluorescent molecular probes I: the synthesis and biological properties of an ELF β-glucuronidase substrate that yields fluorescent precipitates at the enzymatic activity sites. Tetrahedron. 1997;53:7159–7164. [Google Scholar]
- 18.Dobbs D A, Bergman R G, Theopold K H. Piranha solution explosion (H2SO4/H2O2) Chem Eng News. 1990;68:2. [Google Scholar]
- 19.Gober J W, Alley M R, Shapiro L. Positional information during Caulobacter cell differentiation. Curr Opin Genet Dev. 1991;3:324–329. doi: 10.1016/s0959-437x(05)80295-3. [DOI] [PubMed] [Google Scholar]
- 20.Gooday G W. The ecology of chitin degradation. Adv Microb Ecol. 1990;11:387–430. [Google Scholar]
- 21.Hollibaugh J T, Azam F. Microbial degradation of dissolved proteins in seawater. Limnol Oceanogr. 1983;28:1104–1116. [Google Scholar]
- 22.Humphrey B, Kjelleberg S, Marshall K C. Responses of marine bacteria under starvation conditions at a solid-water interface. Appl Environ Microbiol. 1983;45:43–47. doi: 10.1128/aem.45.1.43-47.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larison K D, BeMiller R, Wells S, Clements I, Haugland R P. Use of a new fluorogenic phosphatase substrate in immunohistochemical applications. J Histochem Cytochem. 1995;43:77–83. doi: 10.1177/43.1.7822768. [DOI] [PubMed] [Google Scholar]
- 24.Miyashita K, Fujii T, Sawada Y. Molecular cloning and characterization of chitinase genes from Streptomyces lividans 66. J Gen Microbiol. 1991;137:2065–2072. [Google Scholar]
- 25.Munoz-Dorado J, Arias J M. The social behavior of Myxobacteria. Microbiologia. 1995;11:429–438. [PubMed] [Google Scholar]
- 26.Newton A. Temporal and spatial regulation of differentiation in Caulobacter crescentus. Microbiol Sci. 1987;11:338–341. [PubMed] [Google Scholar]
- 27.Place A R. The biochemical basis and ecological significance of chitin digestion. In: Muzzarelli R A A, editor. Chitin enzymology. Vol. 2. Grottammare, Italy: Atec Edizioni; 1996. pp. 39–54. [Google Scholar]
- 28.Purchase E R, Braun C E. d-Glucosamine hydrochloride. Org Synth. 1946;26:36–37. doi: 10.1002/0471264180.os026.12. [DOI] [PubMed] [Google Scholar]
- 29.Qin Y. The chelating properties of chitosan fibers. J Appl Polymer Sci. 1993;49:727–731. [Google Scholar]
- 30.Rathke T D, Hudson S M. Review of chitin and chitosan as fiber and film formers. J. Mater. Sci.: Rev. Macromol Chem Phys. 1994;C34:375–437. [Google Scholar]
- 31.Shimkets L J. Control of morphogenesis in Myxobacteria. Crit Rev Microbiol. 1987;14:195–227. doi: 10.3109/10408418709104439. [DOI] [PubMed] [Google Scholar]
- 32.Simon M, Alldredge A L, Azam F. Bacterial carbon dynamics on marine snow. Mar Ecol Prog Ser. 1990;51:201–213. [Google Scholar]
- 33.Smith D C, Simon M, Alldredge A L, Azam F. Intense hydrolytic enzyme activity on marine aggregates and implications for rapid particle dissolution. Nature. 1992;359:139–141. [Google Scholar]
- 34.Smucker R A, Kim C K. Microbial extracellular enzyme activity: a new key parameter in aquatic ecology. In: Chrost R J, editor. Microbial enzymes in aquatic environments. New York, N.Y: Springer-Verlag; 1991. pp. 60–83. [Google Scholar]
- 35.Stretton S, Techkarnjanaruk S, McLennan A M, Goodman A E. Use of green fluorescent protein to tag and investigate gene expression in marine bacteria. Appl Environ Microbiol. 1998;64:2554–2559. doi: 10.1128/aem.64.7.2554-2559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svitil A L, Ni Chadhain S M, Moore J A, Kirchman D L. Chitin degradation proteins produced by the marine bacterium Vibrio harveyi growing on different forms of chitin. Appl Environ Microbiol. 1997;63:408–413. doi: 10.1128/aem.63.2.408-413.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Techkarnjanaruk S. Genetic investigation of the chitinase system of a marine bacterium. Ph.D. thesis. Adelaide, Australia: The Flinders University of South Australia; 1998. [Google Scholar]
- 38.Techkarnjanaruk S, Goodman A E. Multiple genes involved in chitin degradation from the marine bacteria Pseudoalteromonas sp. strain S91. Microbiology. 1999;145:925–934. doi: 10.1099/13500872-145-4-925. [DOI] [PubMed] [Google Scholar]
- 39.Techkarnjanaruk S, Pongpattanakitshote S, Goodman A E. Use of a promoterless lacZ gene insertion to investigate chitinase gene expression in the marine bacterium Pseudoalteromonas sp. strain S9. Appl Environ Microbiol. 1997;63:2898–2996. doi: 10.1128/aem.63.8.2989-2996.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vetter Y A, Deming J W. Growth rates of marine bacterial isolates on particulate organic substrates solubilized by freely released extracellular enzyme. Microb Ecol. 1999;37:86–94. doi: 10.1007/s002489900133. [DOI] [PubMed] [Google Scholar]
- 41.Watanabe T, Oyanagi W, Suzuki K, Tanaka H. Chitinase system of Bacillus circulans WL-12 and importance of chitinase A1 in chitin degradation. J Bacteriol. 1990;172:4017–4022. doi: 10.1128/jb.172.7.4017-4022.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu C, Lee A M, Bassler B L, Roseman S. Chitin utilization by marine bacteria. A physiological function for bacterial adhesion to immobilized carbohydrates. J Biol Chem. 1991;266:24260–24267. [PubMed] [Google Scholar]