Abstract
Introduction
Uveal melanoma (UM) is the most common primary intraocular malignancy in adults, and despite treatment of the primary tumor, approximately 15%–50% of patients will develop metastatic disease. Based on gene expression profiling (GEPs), UM can be categorized as Class 1A (low metastatic risk), Class 1B (intermediate metastatic risk), or Class 2 (high metastatic risk). PReferentially expressed Antigen in MElanoma (PRAME) status is an independent prognostic UM biomarker and a potential target for immunotherapy in metastatic UM. PRAME expression status can be detected in tumors using reverse-transcription polymerase chain reaction (RT-PCR). More recently, immunohistochemistry (IHC) has been developed to detect PRAME protein expression. Here, we employed both techniques to evaluate PRAME expression in 18 UM enucleations.
Methods
Tumor material from the 18 UM patients who underwent enucleation was collected by fine-needle aspiration before or during enucleation and sent for GEP and PRAME analysis by RT-PCR. Histologic sections from these patients were stained with an anti-PRAME monoclonal antibody. We collected patient demographics and tumor characteristics and included this with our analysis of GEP class, PRAME status by RT-PCR, and PRAME status by IHC. PRAME IHC and RT-PCR results were compared.
Results
Twelve males (12/18) and 6 females (6/18) with an average age of 60.6 years underwent enucleation for UM. TNM staging of the UM diagnosed Stage I in 2 patients (2/18), Stage II in 7 patients (7/18), Stage III in 8 patients (8/18), and Stage IV in 1 (1/18). GEP was Class 1A in 6 tumors (6/18), Class 1B in 6 tumors (6/18), and Class 2 in 6 tumors (6/18). PRAME IHC showed diffusely positive labeling of all UM cells in 2/18 enucleations; negative IHC labeling of UM cells in 9/18 enucleations; and IHC labeling of subsets of UM cells in 7/18 enucleations. Eleven of the 17 UMs tested for PRAME by both RT-PCR and IHC had consistent PRAME results. In the remaining 6/17 cases tested by both modalities, PRAME results were discordant between RT-PCR and IHC.
Conclusions
We find that PRAME IHC distinguishes PRAME-positive and PRAME-negative UM tumor cells. Interestingly, IHC reveals focal PRAME expression in subsets of tumor cells consistent with tumor heterogeneity. PRAME RT-PCR and IHC provide concordant results in most of our cases. We suggest that discordance in PRAME results could arise from spatial or temporal variation in PRAME expression between tumor cells. Further studies are required to determine the prognostic implications of PRAME IHC in UM.
Keywords: Uveal melanoma, Preferentially expressed antigen in melanoma, Metastasis, Immunohistochemistry, Reverse-transcription polymerase chain reaction, Prognosis
Introduction
Uveal melanoma (UM) is the most common primary intraocular malignancy in adults [1]. The mean age-adjusted incidence of UMs is 5.1 cases per million each year in the USA with an average age of presentation between the ages of 50 and 70 years [2]. The most common location for UMs is the choroid (90%), followed by the ciliary body (6%) and iris (4%) [3]. Radiation and enucleation are standard treatment methods worldwide. Despite treatment of the primary tumor, metastasis has been reported in 15%–50% of the patients with UM depends on the length of the study and location of the tumor [4, 5, 6, 7]. The liver is the primary location for metastasis of UMs [8]. Metastatic UMs respond poorly to treatment options, including chemotherapy or targeted therapy, and are typically fatal within 1 year [1]. As such, extensive efforts are being made to identify prognostic biomarkers for UM metastasis risk and to identify molecular targets for immunotherapy of metastatic tumors [9].
Risk factors for UM include fair skin, light eye color, inability to tan, ocular or oculodermal melanocytosis, cutaneous or iris or choroidal nevus, and germline BRCA1-associated protein 1 mutation [10]. More recently, molecular tests have been developed to evaluate the metastatic risk of primary UMs. Gene expression profiling (GEP) can classify UMs into Class 1A (low metastatic risk), Class 1B (intermediate metastatic risk), or Class 2 (high metastatic risk) [11, 12, 13, 14, 15] using genetic material extracted from needle biopsies of fresh UM tumor or formalin-fixed paraffin-embedded (FFPE) UM tumor sections. PReferentially expressed Antigen in MElanoma (PRAME) expression in UMs also correlates with increased risk for metastasis [16, 17], and a PRAME gene expression test can be performed using extracted tumor genetic material. However, additional studies are required to confirm the data, but it appears that molecular testing predicts the risk of UM metastasis more precisely, compared to histologic and anatomic features, especially when the three-step GEP combined with PRAME expression are used to create a prognostic model [7, 14, 18, 19].
PRAME was first identified as a cell surface protein antigen highly expressed by metastatic cutaneous melanoma but not by most normal tissues [20]. Because of its preferential expression in cutaneous melanoma cells, PRAME has emerged as an attractive target for tumor immunotherapy with multiple clinical trials underway [21, 22]. In cutaneous melanomas, immunohistochemically and GEP tests are both used to evaluate PRAME protein and mRNA expression [23, 24, 25, 26]. In UMs, PRAME gene expression testing by reverse-transcription polymerase chain reaction (RT-PCR) is routinely used but less is known about the utility of PRAME immunohistochemistry (IHC) in UMs. Due to PRAMEs importance as a prognostic biomarker for UM metastasis risk, along with growing interest in therapeutic anti-PRAME targeting for UM tumor immunotherapy, it is important to evaluate the use of PRAME IHC in UM samples. We tested 18 UMs for PRAME expression by IHC and compared the findings with PRAME RT-PCR testing performed on the same tumors.
Material and Methods
UM Patient Cases
Eighteen patients underwent enucleation for UM between 2018 and 2021, and the clinicopathologic data were reviewed from our institutional medical record database following IRB-approved protocols. We extracted patients' sex, age, laterality, and UM enucleation tumor characteristics (TNM, margins, metastasis) (Table 1).
Table 1.
Demographic and clinicopathologic characteristics of UM patients
| Case | Sex | Age | Tumor laterality | GEP class | PRAME RT-PCR result | PRAME IHC result | Tumor stage | Category | Histo-morphology |
|---|---|---|---|---|---|---|---|---|---|
| 1 | m | 67 | Left | 1B | Positive | Positive − diffuse | 3a | 3 | Mixed |
| 2 | m | 49 | Left | 1A | Positive | Negative | 2a | 2 | Mixed |
| 3 | m | 80 | Left | 2 | Positive | Negative | 2a | 2 | Epithelioid |
| 4 | m | 50 | Right | 1B | Positive | Positive − focal | 3a | 3 | Spindle |
| 5 | m | 41 | Left | 2 | Positive | Negative | 3d | 3 | Mixed |
| 6 | m | 37 | Left | 1B | Not performed | Positive − diffuse | 1a | 1 | Epithelioid |
| 7 | m | 72 | Right | 1A | Negative | Negative | 3a | 3 | Epithelioid |
| 8 | f | 66 | Left | 1B | Positive | Negative | 2b | 2 | Spindle |
| 9 | m | 66 | Right | 1B | Negative | Negative | 2a | 2 | Spindle |
| 10 | m | 69 | Left | 2 | Positive | Positive − focal | 2b | 2 | Mixed |
| 11 | f | 72 | Right | 2 | Negative | Negative | 2a | 2 | Spindle |
| 12 | f | 40 | Right | 2 | Positive | Positive − focal | 3a | 3 | Epithelioid |
| 13 | m | 72 | Right | 1B | Positive | Negative | 1a | 1 | Spindle |
| 14 | m | 62 | Right | 2 | Negative | Positive − focal | 3a | 3 | Mixed |
| 15 | f | 67 | Left | 1A | Positive | Positive − focal | 4e | 1 | Spindle |
| 16 | M | 38 | Left | 1A | Positive | Positive − focal | 2a | 2 | Epithelioid |
| 17 | f | 87 | Left | 1A | Negative | Negative | 3a | 3 | Mixed |
| 18 | f | 56 | Right | 1A | Positive | Positive − focal | 3a | 3 | Mixed |
PRAME RT-PCR and PRAME IHC
Seventeen out of 18 UM patients had Decision Dx-PRAME testing performed at Castle BioScience Inc. (Friendswood, TX, USA) using samples obtained by fine-needle aspiration (FNA). In brief, Decision Dx-PRAME is a RT-PCR-based gene expression test that analyzes mRNA levels of the PRAME gene relative to the mean of 3 control genes from the UM specimen [16]. The relative expression level of PRAME is compared to a validated threshold that was derived from the PRAME expression values of 958 UM tumors using a LOESS model [16]. Decision Dx-PRAME results follow this protocol to classify the tumor as PRAME-positive or PRAME-negative (Table 1).
IHC staining was performed on FFPE tissue blocks by automation from UM enucleations in all 18 cases. Pupil-optic nerve cross-sections of the UM enucleations were stained for H&E and MelMix (Ki67-MIB-1/HMB45 + A103 + T311; Dako & Ventana M7240/790-4677; predilute) for histopathology diagnosis, staging, and to confirm the melanocytic origin of the tumor cells. PRAME IHC was then performed on a serial adjacent section of the UM using anti-PRAME antibody (EPR20330; Abcam #219650, 1:125 dilution) with appropriate on slide negative and positive controls from skin melanoma samples. UM cells with PRAME-positive IHC expression showed strong uniform nuclear IHC labeling (Fig. 1), whereas UM cells with PRAME-negative IHC expression showed no nuclear labeling (Fig. 2). Melanin pigment in UM cells was easily differentiated from PRAME immunopositivity by the cytoplasmic subcellular location and globular nature of melanin pigments versus the diffuse PRAME labeling in the cell nucleus. Three pathologists reviewed all UM cases individually and, in a group, to establish consensus about PRAME IHC expression in each UM enucleation.
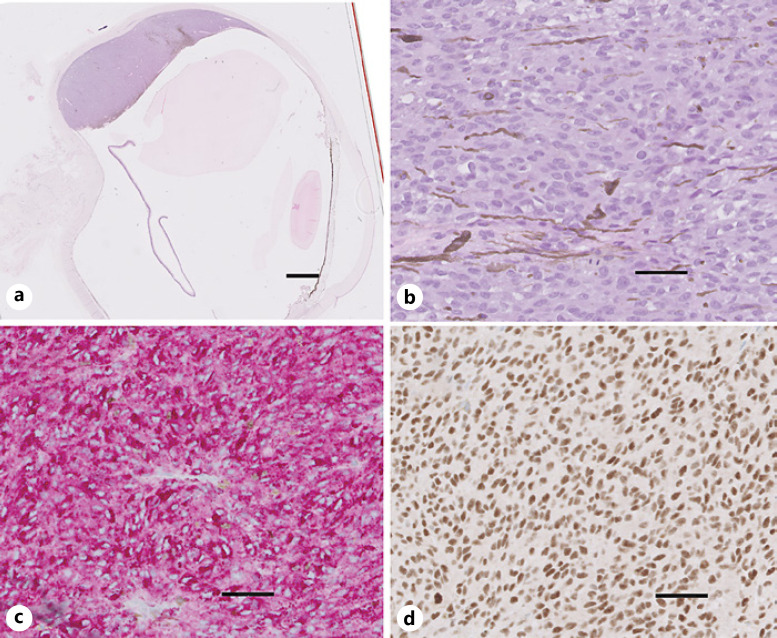
Fig. 1.
UM enucleation (case 1) with diffusely positive PRAME IHC expression. a Cross-section of enucleation at the pupil-optic nerve shows choroidal melanoma tumor toward the superior-posterior pole (H&E stain). b High-power image of the malignant melanoma cells in the tumor (H&E stain). c MART-1 immunolabeling (red cytosolic staining) highlights melanoma cells. d PRAME immunolabeling (brown nuclear staining) with diffuse immunolabeling of tumor cells. Nuclei in b–d are counterstained with hematoxylin (blue). Scale bars, 2 mm (a) and 20 μm (b–d).
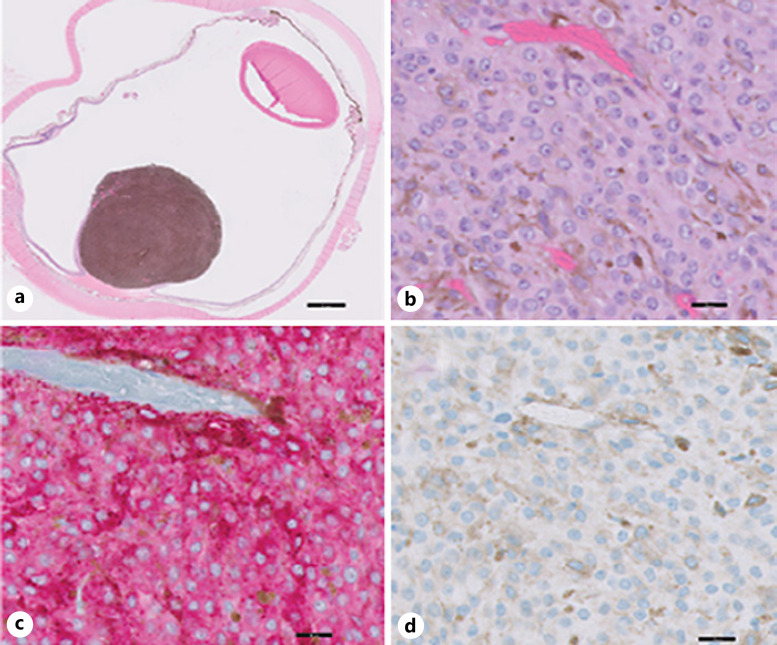
Fig. 2.
UM with negative PRAME IHC expression (case 17). a Cross-section of enucleation shows choroidal melanoma tumor toward the posterior pole (H&E stain). b High-power image of the malignant melanoma cells in the tumor (H&E stain). c MART-1 immunolabeling (red cytosolic staining) highlights melanoma cells. d PRAME immunolabeling (brown nuclear staining) is negative. Nuclei in b–d have been counterstained with hematoxylin (blue). Scale bars, 2 mm (a) and 20 μm (b–d).
Results
Clinicopathologic Characteristics of Primary UM Cases
The clinicopathologic characteristics of 18 UM patients were retrospectively reviewed in this IRB-approved study. These included 12 (67%) males and 6 (33%) females, with an average age of 60.6 years at the time of diagnosis (range 37–87). All patients underwent enucleations from 2018 to 2021. The UM enucleation histopathologic findings regarding TNM stage, category, and histologic cell type (epithelioid, spindle) are listed in Table 1. The mean follow-up period following enucleation was 18 months. Liver metastases were identified in 2/18 cases during follow-up. Case 6 developed liver metastasis 43 months after the initial diagnosis of UM with a GEP Class 1B (Table 1). Case 11 also developed liver metastasis 16 months after diagnosis of UM with a GEP Class 2 (Table 1). PRAME IHC was performed on FFPE sections from all 18 UM enucleations.
UMs with Diffusely Positive PRAME IHC
Two of the 18 UM enucleations, cases 1 and 6, showed strong PRAME nuclear expression in virtually all tumor cells (Fig. 1; Table 1) and were classified as Class 1B by GEP (Table 1). The UM tumor from case 1 was PRAME-positive by RT-PCR with no liver metastasis during the follow-up period of this study. The UM tumor from case 6 developed liver metastasis shortly after diagnosis and was not tested for PRAME by RT-PCR (Table 1).
UMs with Negative PRAME IHC
Nine of the 18 UM enucleations showed no PRAME nuclear immunohistochemical labeling in melanoma cells (Fig. 2; Table 1). Four of the 9 PRAME IHC-negative UMs (cases 7, 9, 11, and 17) were also PRAME-negative by RT-PCR (Table 1). Cases 7 and 19 were Class 1A by GEP; case 9 was Class 1B; and case 11 was GEP Class 2 (Table 1). Case 11 developed liver metastasis during the follow-up period of this study.
The other 5 of the 9 PRAME IHC-negative tumors (cases 2, 3, 5, 8, and 13) were PRAME-positive by RT-PCR (Table 1). Case 2 showed a Class 1A GEP; cases 3 and 5 were Class 2 GEP; and cases 8 and 13 were Class 1B GEP (Table 1).
UMs with Focally Positive PRAME IHC
Seven of the 18 UM enucleations in our current study (cases 4, 10, 12, 14, 15, 16, and 18) showed focal melanoma cell labeling by PRAME IHC where PRAME IHC-positive and PRAME IHC-negative UM cells were found within the same tumor (Fig. 3, 4). In some of these UMs with focal PRAME antibody immunopositivity, the PRAME IHC-positive and PRAME IHC-negative UM cells form separate clusters which are spatially recognizable areas (Fig. 3). In other UMs, the PRAME-positive and PRAME-negative cells were more intermixed and do not form spatially recognizable areas (Fig. 4). Expression of the melanoma marker (MART-1) confirmed all PRAME-positive and PRAME-negative neoplastic cells were of melanocytic origin (Fig. 3, 4). We described PRAME expression in these UM cases as being variably positive with some showing separate populations of positive cells and some showing intermixed individual positive cells (Table 1).
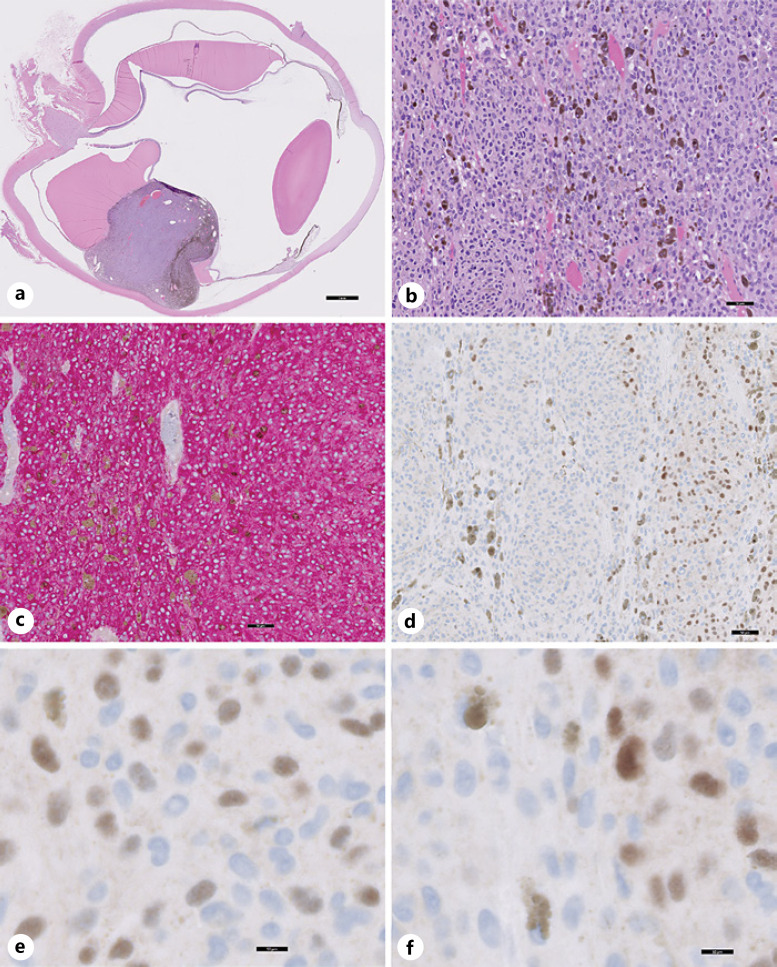
Fig. 3.
UM with focally positive PRAME expression (case 16). a Cross-section of enucleation shows choroidal melanoma tumor toward the inferior pole (H&E stain). b Low-power image of the malignant melanoma cells in the tumor (H&E stain). c MART-1 immunolabeling (red cytosolic staining) highlights the tumor cells. d–f Focally positive PRAME immunolabeling with intermixed PRAME-positive (brown nuclear staining) and PRAME-negative tumor cells (no brown nuclear staining) present in the same enucleation. Nuclei in (b–f) have been counterstained with hematoxylin (blue). Scale bars, 2 mm (a), 50 μm (b–d), and 10 μm (e, f).
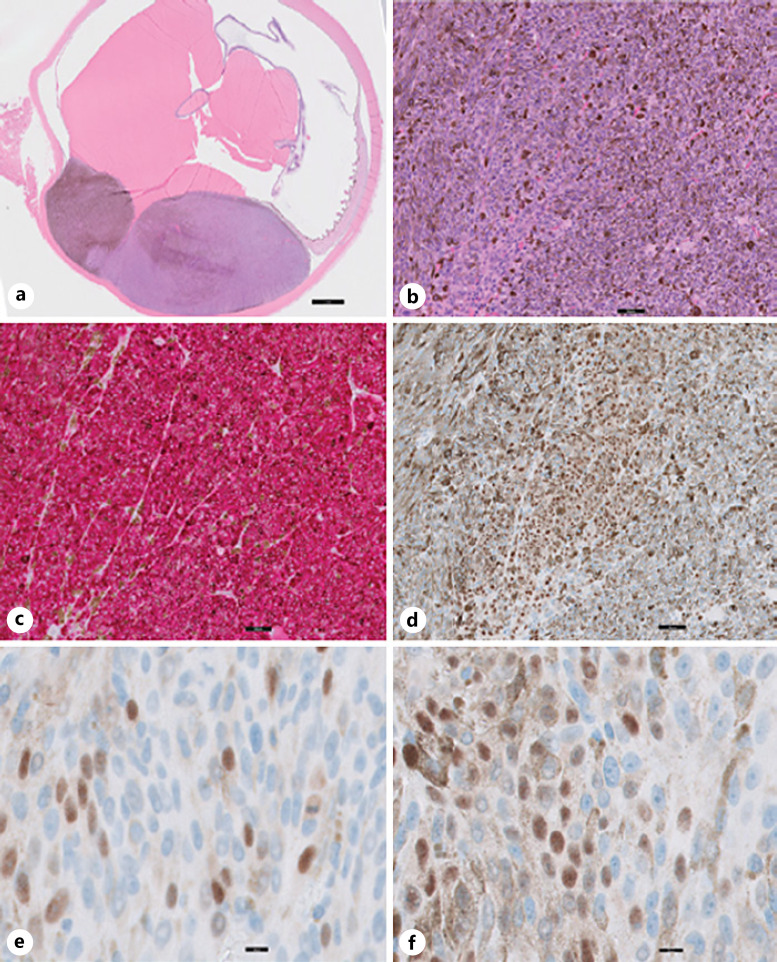
Fig. 4.
UM with focally positive PRAME expression (case 14). a Cross-section of enucleation shows choroidal melanoma tumor toward the inferior pole (H&E stain). b Low-power image of the malignant melanoma cells in the tumor (H&E stain). c MART-1 immunolabeling (red cytosolic staining) highlights the tumor cells. d–f Focally positive PRAME immunolabeling (brown nuclear staining) with intermixed PRAME-positive and PRAME-negative tumor cells in the same enucleation. Nuclei in (b–f) have been counterstained with hematoxylin (blue). Scale bars, 2 mm (a), 50 μm (b–d), and 10 μm (e, f).
PRAME RT-PCR testing of these cases with focal PRAME-positive IHC labeling showed PRAME-positive by RT-PCR in 6 of 7 cases (Table 1). Case 14 was the only focal PRAME IHC-positive case where the RT-PCR result was PRAME-negative (Table 1). None of the UM cases with focal positive PRAME IHC expression developed metastases in the follow-up period.
Discussion
Since PRAME is an important prognostic molecular biomarker for aggressive UMs [18, 27], it is important to characterize the best way to determine PRAME expression. PRAME status is commonly determined by RT-PCR testing of genetic material extracted from UM tumor cells. The status of PRAME and other molecular mutations found in UMs are being evaluated in >1,300 UM patients in the ongoing Collaborative Ocular Oncology Group Study 2 to stratify UM patient risk profiles and to assign optimal UM clinical treatment regimens based on status of the biomarkers (2021 Schefler IOVS ARVO2021). In addition to its prognostic utility, PRAME is also being intensively studied as a therapeutic target for cancer immunotherapies [17]. Several clinical trials are testing anti-PRAME immunotherapy regimens in UM, cutaneous melanoma, acute myeloid leukemia, and non-small-cell lung cancer based on targeting tumor cells expressing PRAME protein (clinicaltrials.gov).
In this study, we examined PRAME protein expression in tumor cells by immunohistochemical staining of enucleation sections collected from 18 UM patients. We found negative PRAME nuclear expression in 9 cases where no UM tumor cells expressed PRAME nuclear expression by IHC (Fig. 2). One of these PRAME IHC-negative cases (Table 1, case 11) was GEP Class 2 and developed liver metastasis. We found diffusely positive PRAME expression in 2 cases where virtually all UM tumor cells expressed PRAME by IHC (Fig. 1). One of these diffusely PRAME IHC-positive cases (Table 1, case 6) also developed liver metastasis. Interestingly, in the remaining 7 enucleations of our study, we found focal PRAME nuclear expression by IHC in subsets of UM cells within the same enucleation suggestive of tumor heterogeneity. None of these cases with focal PRAME IHC positivity developed metastases. In our study, the mean age at the time of diagnosis was 60.6, similar to prior studies [2, 3]. However, the male-to-female ratio was 2:1, in contrast to other studies which showed a 1:1 ratio [2, 3], and the small sample size was considered as the cause of this difference between this study and others. We did not find a correlation or a pattern between PRAME positivity by IHC or RT-PCR with patients' age, gender, tumor laterality, histology (epithelioid vs. spindle), or tumor stage in our cohort of UMs. Prior studies showed there is a significant association between PRAME-positive status and largest basal diameter of the tumor as well as GEP class [18, 28]. However, a recent study showed GEP was the only statistically significant factor to predict metastatic-free survival and melanoma-specific survival in multivariate analysis [19].
In this study, we also compared the results of PRAME testing performed by RT-PCR and by IHC. In the majority of UM cases (11/17), RT-PCR and IHC provided the same PRAME results with both testing modalities. However, in 6/17 UM cases, RT-PCR and IHC PRAME testing gave discordant results (e.g., PRAME-negative by RT-PCR and PRAME-positive by IHC or vice versa). One potential reason for this discordance could be heterogeneity in PRAME expression within the UM, and the subset of tumor cells collected by FNA for RT-PCR PRAME testing may not have reflected the heterogeneity of PRAME expression within the UM. A second potential reason for the difference could arise from changes (positive or negative) in PRAME expression in the UM between the time of the initial FNA biopsy for RT-PCR PRAME testing and the time of the enucleation on which the PRAME IHC was performed (e.g., the UM was initially PRAME-positive at the time of needle biopsy but became PRAME-negative by the time of enucleation after primary eye treatment). A third potential reason for discordance could be the higher sensitivity of the RT-PCR compared to IHC. We suggest in these cases, the PRAME alteration present at the RNA level is detected by RT-PCR but not at the protein level by IHC. If the targeting of PRAME becomes a possible treatment modality, it remains an important question if response to treatment can be predicted by protein expression (PRAME detected by IHC) or PRAME active transcription by GEP.
In sum, we show that PRAME IHC can reveal heterogeneity in PRAME expression by tumor cells within the same UM. The major limitation of our study is the small cohort of UMs (n = 18) with both PRAME IHC and RT-PCR data for comparison, which limits the value of statistical analysis. Additionally, a more extensive scale study would be helpful to elucidate the cause of discordance between the two methods and define the methodologic and prognostic implications of PRAME IHC in UM. Tracking PRAME IHC status in UM is another potential diagnostic tool to determine the prognosis and risk stratification of patients in combination with other biomarkers. PRAME IHC alone or in combination with RT-PCR may be especially useful for identifying the PRAME-positive UM tumor cells that can be targeted by anti-PRAME immunotherapies.
Statement of Ethics
This study was performed with the ethical standards per the World Medical Association Declaration of Helsinki. The study protocol was reviewed and approved by Stanford University Panel on Human Subjects in Medical Research, IRB approval number #54760. The subjects provided their written informed consent before surgery or FNA biopsy.
Conflict of Interest Statement
All authors: no reported conflicts.
Funding Sources
There are no funding sources to declare.
Author Contributions
Saman S. Ahmadian, Ian J. Dryden, and Jonathan H. Lin contribute to study conception and design, interpretation and analysis of the data, and writing the paper manuscript. Andrea Naranjo, Prithvi Mruthyunjaya, and Ryanne A. Brown contribute to the interpretation and analysis of data. All the authors discussed the results and contributed to the final manuscript.
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further inquiries can be directed to the corresponding author.
References
- 1.Jager MJ, Shields CL, Cebulla CM, Abdel-Rahman MH, Grossniklaus HE, Stern MH, et al. Uveal melanoma. Nat Rev Dis Primers. 2020 Apr 9;6((1)):24. doi: 10.1038/s41572-020-0158-0. [DOI] [PubMed] [Google Scholar]
- 2.Singh AD, Turell ME, Topham AK. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology. 2011 Sep;118((9)):1881–5. doi: 10.1016/j.ophtha.2011.01.040. [DOI] [PubMed] [Google Scholar]
- 3.Shields CL, Kaliki S, Furuta M, Mashayekhi A, Shields JA. Clinical spectrum and prognosis of uveal melanoma based on age at presentation in 8,033 cases. Retina. 2012 Jul;32((7)):1363–72. doi: 10.1097/IAE.0b013e31824d09a8. [DOI] [PubMed] [Google Scholar]
- 4.Manschot WA, Van Peperzeel HA. Uveal melanoma: location, size, cell type, and enucleation as risk factors in metastasis. Hum Pathol. 1982 Dec;13((12)):1147–8. doi: 10.1016/s0046-8177(82)80258-x. [DOI] [PubMed] [Google Scholar]
- 5.Kujala E, Mäkitie T, Kivelä T. Very long-term prognosis of patients with malignant uveal melanoma. Invest Ophthalmol Vis Sci. 2003 Nov;44((11)):4651–9. doi: 10.1167/iovs.03-0538. [DOI] [PubMed] [Google Scholar]
- 6.Rietschel P, Panageas KS, Hanlon C, Patel A, Abramson DH, Chapman PB. Variates of survival in metastatic uveal melanoma. J Clin Oncol. 2005 Nov 1;23((31)):8076–80. doi: 10.1200/JCO.2005.02.6534. [DOI] [PubMed] [Google Scholar]
- 7.Cai L, Paez-Escamilla M, Walter SD, Tarlan B, Decatur CL, Perez BM, et al. Gene expression profiling and PRAME status versus tumor-node-metastasis staging for prognostication in uveal melanoma. Am J Ophthalmol. 2018 Nov;195:154–60. doi: 10.1016/j.ajo.2018.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diener-West M, Reynolds SM, Agugliaro DJ, Caldwell R, Cumming K, Earle JD, et al. Development of metastatic disease after enrollment in the COMS trials for treatment of choroidal melanoma: Collaborative Ocular Melanoma Study Group Report No. 26. Arch Ophthalmol. 2005 Dec;123((12)):1639–43. doi: 10.1001/archopht.123.12.1639. [DOI] [PubMed] [Google Scholar]
- 9.Kaliki S, Shields CL, Shields JA. Uveal melanoma: estimating prognosis. Indian J Ophthalmol. 2015 Feb;63((2)):93–102. doi: 10.4103/0301-4738.154367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaliki S, Shields CL. Uveal melanoma: relatively rare but deadly cancer. Eye. 2017 Feb;31((2)):241–57. doi: 10.1038/eye.2016.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Gils W, Lodder EM, Mensink HW, Kilic E, Naus NC, Bruggenwirth HT, et al. Gene expression profiling in uveal melanoma: two regions on 3p related to prognosis. Invest Ophthalmol Vis Sci. 2008 Oct;49((10)):4254–62. doi: 10.1167/iovs.08-2033. [DOI] [PubMed] [Google Scholar]
- 12.Onken MD, Worley LA, Harbour JW. Association between gene expression profile, proliferation and metastasis in uveal melanoma. Curr Eye Res. 2010 Sep;35((9)):857–63. doi: 10.3109/02713683.2010.493265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onken MD, Worley LA, Tuscan MD, Harbour JW. An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn. 2010 Jul;12((4)):461–8. doi: 10.2353/jmoldx.2010.090220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onken MD, Worley LA, Char DH, Augsburger JJ, Correa ZM, Nudleman E, et al. Collaborative Ocular Oncology Group report number 1: prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology. 2012 Aug;119((8)):1596–603. doi: 10.1016/j.ophtha.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berry DE, Schefler AC, Seider MI, Materin M, Stinnett S, Mruthyunjaya P, et al. Correlation of gene expression profile status and American Joint Commission on cancer stage in uveal melanoma. Retina. 2020 Feb;40((2)):214–24. doi: 10.1097/IAE.0000000000002385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Field MG, Decatur CL, Kurtenbach S, Gezgin G, van der Velden PA, Jager MJ, et al. PRAME as an independent biomarker for metastasis in uveal melanoma. Clin Cancer Res. 2016 Mar 1;22((5)):1234–42. doi: 10.1158/1078-0432.CCR-15-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu Y, Zou R, Wang J, Wang ZW, Zhu X. The role of the cancer testis antigen PRAME in tumorigenesis and immunotherapy in human cancer. Cell Prolif. 2020 Mar;53((3)):e12770. doi: 10.1111/cpr.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schefler AC, Koca E, Bernicker EH, Correa ZM. Relationship between clinical features, GEP class, and PRAME expression in uveal melanoma. Graefes Arch Clin Exp Ophthalmol. 2019 Jul;257((7)):1541–5. doi: 10.1007/s00417-019-04335-w. [DOI] [PubMed] [Google Scholar]
- 19.Aaberg TM, Covington KR, Tsai T, Shildkrot Y, Plasseraud KM, Alsina KM, et al. Gene expression profiling in uveal melanoma: five-year prospective outcomes and meta-analysis. Ocul Oncol Pathol. 2020 Oct;6((5)):360–7. doi: 10.1159/000508382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda H, Lethe B, Lehmann F, van Baren N, Baurain JF, de Smet C, et al. Characterization of an antigen that is recognized on a melanoma showing partial HLA loss by CTL expressing an NK inhibitory receptor. Immunity. 1997 Feb;6((2)):199–208. doi: 10.1016/s1074-7613(00)80426-4. [DOI] [PubMed] [Google Scholar]
- 21.Pujol JL, De Pas T, Rittmeyer A, Vallieres E, Kubisa B, Levchenko E, et al. Safety and immunogenicity of the PRAME cancer immunotherapeutic in patients with resected non-small cell lung cancer: a phase I Dose Escalation Study. J Thorac Oncol. 2016 Dec;11((12)):2208–17. doi: 10.1016/j.jtho.2016.08.120. [DOI] [PubMed] [Google Scholar]
- 22.Gezgin G, Luk SJ, Cao J, Dogrusoz M, van der Steen DM, Hagedoorn RS, et al. PRAME as a potential target for immunotherapy in metastatic uveal melanoma. JAMA Ophthalmol. 2017 Jun 1;135((6)):541–9. doi: 10.1001/jamaophthalmol.2017.0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lezcano C, Jungbluth AA, Nehal KS, Hollmann TJ, Busam KJ. PRAME expression in melanocytic tumors. Am J Surg Pathol. 2018 Nov;42((11)):1456–65. doi: 10.1097/PAS.0000000000001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Raghavan SS, Wang JY, Kwok S, Rieger KE, Novoa RA, Brown RA. PRAME expression in melanocytic proliferations with intermediate histopathologic or spitzoid features. J Cutan Pathol. 2020 Dec;47((12)):1123–31. doi: 10.1111/cup.13818. [DOI] [PubMed] [Google Scholar]
- 25.Raghavan SS, Wang JY, Toland A, Bangs CD, Rieger KE, Novoa RA, et al. Diffuse PRAME expression is highly specific for malignant melanoma in the distinction from clear cell sarcoma. J Cutan Pathol. 2020 Dec;47((12)):1226–8. doi: 10.1111/cup.13812. [DOI] [PubMed] [Google Scholar]
- 26.Kangas-Dick AW, Greenbaum A, Gall V, Groisberg R, Mehnert J, Chen C, et al. Evaluation of a gene expression profiling assay in primary cutaneous melanoma. Ann Surg Oncol. 2021 Aug;28((8)):4582–9. doi: 10.1245/s10434-020-09563-7. [DOI] [PubMed] [Google Scholar]
- 27.Field MG, Durante MA, Decatur CL, Tarlan B, Oelschlager KM, Stone JF, et al. Epigenetic reprogramming and aberrant expression of PRAME are associated with increased metastatic risk in class 1 and class 2 uveal melanomas. Oncotarget. 2016 Sep 13;7((37)):59209–19. doi: 10.18632/oncotarget.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Correa ZM. Assessing prognosis in uveal melanoma. Cancer Control. 2016 Apr;23((2)):93–8. doi: 10.1177/107327481602300202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further inquiries can be directed to the corresponding author.