Abstract
Antibiotic resistance is one of the major causes of morbidity and mortality, triggered by the adhesion of microbes and to some extent the formation of biofilms. This condition has been quite challenging in the health and industrial sector. Conditions and processes required to foil these infectious and resistance are of much concern. The synthesis of PDA material, inspired by the Mytilus edulis foot protein (MEFP)5 possesses unique characteristics that allow for, adhesion, photothermal therapy, synergistic effects with other materials, biocompatibility process, etc. Therefore, their usage holds great potential for dealing with both the infectious nature and the antibiotic resistance processes. Hence, this review provides an overview of the mechanism involved in accomplishing and eradicating bacteria, the recently harnessed antibacterial effect of the PDA through other properties they possess, a way forward in tapping the benefit embedded in the PDA, and the future perspective.
Keywords: Polydopamine, Antibiotic resistance, Biofilm, Nanoparticle, Antibacterial activity
Graphical abstract
1. Introduction
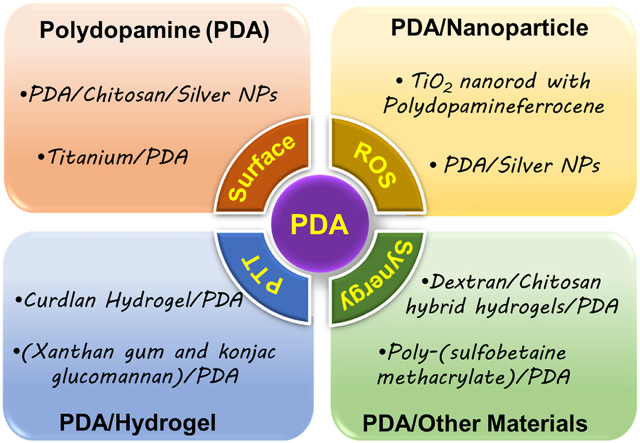
Bacterial infections and antibiotic resistance remain a condition the health sector still battles globally. This poses a serious threat to researchers, food processing, the marine industry, and the health sector, one strategy employed by these bacteria to elude conditions set to eradicate them is by the formation of biofilms. This biofilm can adhere to surfaces, forming a complex form of community that enables them to withstand several forms of harsh conditions. They are capable of being formed on medical implants, catheters, etc. together with other related medical devices [1,2]. Intriguingly, studies reveal that biomedical device-associated infections happen to be a major cause of clinical implant failure, and the idea of dealing with such conditions with a high amount of antibiotics, which seems to be the remedy is even devastating to the human body. Consequently, coating implants with antibiotic eluting materials seems to be one of the best options to prevent the infection and formation of biofilms [[3], [4], [5], [6]]. One mechanism devised to overcome this challenge is the painting of surfaces of devices which can be said to be a traditional means [[7], [8], [9]]. However, this method may not be safe and reliable since any minimal scratch can still provide a surface for the bacteria to thrive. These surfaces a very crucial for usage since some medical devices or implants turned out to lose their functionality immediately after their surfaces are compromised. And so, the material that makes up the surface of the device becomes very crucial for the function and duration of the device since it is the surface that makes contact with the environment. This calls for a means to deal with such conditions, because, improvement of the surface is not to improve its duration alone but to enhance the functionalities [10,11]. Additionally, in the textile industry, the growth of bacteria is known to result in product deterioration and other health-related conditions such as body odor, dermal infections, and other related allergies. Although other agents that are fungicidal and bactericidal are normally employed, these agents exist in chemical forms which are normally toxic and non-biodegradable, they include urea, inorganic salts, onium salts, organometallic, phenols, thiophenols, etc [[12], [13], [14], [15]]. However, advances in nanotechnology have chosen to help adjust the function of these materials and devices with a zero rate of bacteria adherence through the surface coating of these devices with materials of controllable thickness. Dopamine (4-(2-aminoethyl) benzene-1,2-diol), a catecholamine derivative that mimics the adhesive protein inspired by the Mytilus edulis foot protein (MEFP)5, which allows its strong adhesion to solid surfaces [16]. Studies show that melanin possesses some optical and electronic properties [17], and the catalytic process of their generation involves the catalysation of tyrosine hydroxylation to yield 3,4-dihydroxy-l-phenylalanine by the enzyme tyrosinase [18], and dopamine is capable of forming structures that are equivalent to the natural melanin, this brings us to the polydopamine (PDA) inspired by the MEFP5 whose amino acid sequence also reveals the presence of 3,4-dihydroxy-l-phenylalanine (molar fraction of 40%) [[19], [20], [21]]. Nanotechnology development allows the coating of devices and other surfaces with this PDA, materials that possess these catechol groups to mimic the MEFP5 of the mussel, allowing adhesion on both inorganic and organic surfaces through mild conditions, studies show that these dopamine poses hydroxyl groups on their ortho positions which allows them to reduce metals to metal ions [[22], [23], [24]]. Here we outlined the overview of the nature of depositions of the PDA, the mechanism involved in accomplishing and eradicating bacteria, the recently harnessed antibacterial effect, and the future perspective of this material, Fig. 1.
Fig. 1.
Overview of the antibacterial mechanism of PDA and fabrication of PDA nanocomposites.
2. Mechanism of action
It's worth noting that, small molecules of catechol and proteins of amine in the presence of alkaline condition (aqueous) and oxygen, allow dopamine to undergo polymerization to form a polydopamine film that is thin and adherent, this can then be formed in the varying forms of surfaces. PDA which forms on surfaces goes on to prevent the attachment and growth of bacteria through the seizure of the bacteria nutrient supply, blocking the process of growth by acting as a barrier between bacteria and those surfaces [20,21,[25], [26], [27], [28], [29]]. Intriguingly, this antimicrobial activity in most cases has been attributed to the benzene ring of dopamine and the formation of local toxic effects by some active groups formed on the outer membrane of the bacteria cell [30,31]. Which is in keeping with the fact that the negative impact of the active groups on the bacterial outer membrane subsequently affects the permeability of the cell membrane by obstructing components required for the bacteria to survive. Hence a contributing factor to the intrinsic antimicrobial activity of dopamine is its ability to effectually eradicate bacteria, microbes, plankton, or biofilm [30,32,33].
3. Properties
PDA is equipped with several properties, such as hydrophilicity, biodegradability, photothermal conversion capacity, toughness, high stability, surface morphology and roughness, and conditions set for ROS generation, biocompatibility, etc, responsible for their various potentials.
3.1. Hydrophilicity (wettability)
Hydrophilicity will have to do with the sustainability of the liquid-solid surface contact by a liquid through the balance of intermolecular force of interaction. Several studies reveal that after PDA coating, the surface angle of the material reduces indicating a high level of hydrophilicity. Forte et al. reveal a variation in the contact angle after PDA deposition but attributed it to the less homogeneity of the composite sample surface [34,35]. However, the values fall within 50° to 55° when PDA is normally deposited on a hydrophilic substrate [27,36,37]. Next, Ti6A14V coated with PDA and some peptides reveals a low contact angle due to the presence of the PDA [38]. Also, the water contact angle of Ti coated with PDA decreased significantly from 64 ± 2° to 37± 3° [39], similarly, PDA showed a drastic decline in its water contact angle (less than 15°) and was associated with the ability of water droplets to immerse via the gaps between the aggregates of the PDA particles [40,41]. In addition, an uncoated polypropylene (PP) sheet exhibited a water contact angle of 98.3° but decreased to 62° upon coating with PDA similar to PDA coating which resulted in a water contact angle ranging between 50°-70°. The water surface angle does not indicate the hydrophilicity alone but the surface energy of a substrate and could help to ensure the effectual evaluation of a surface modification [[41], [42], [43]].
3.2. Toughness and high stability
PDA-coated material can exhibit stability for a long period, and a PDA composite material exhibited long-lasting stability for more than 30 days. Non shaken PDA deposition resulted in poor stability and conformity as those shaken reveal homogeneously thicken PDA film [43,44]. Based on the thickness and the ability of the coating prepared, a great amount of H2O2 can be generated. As a way of generation of good orthopedic implants, stability turns out to be one of the key properties required [[45], [46], [47], [48]]. Furthermore, the stability condition could be determined by the rate of polymerization which is also dependent on the pH, dopamine concentration, and oxygen concentration [[49], [50], [51]]. Nonetheless, if the stability of PDA is somewhat dependent on the pH, then the examination of this stability can be accomplished, using the surface plasmon resonance, a study revealed that coating stability which was pH-dependent i.e. based on the pH of the aqueous solution, resulted in a ratio of detachment up to 66% and 80% under pH of 1.0 and 14.0 respectively. Therefore, under strongly acidic or strongly alkaline conditions the aqueous solutions' ionic strength could be increased to check the rate of PDA detachment [46,52]. Also, a PDA attained a film thickness of about 62.8 nm (thickness plateau) in 72 h and this was associated with the decrease in oxygen after the formation of the first layer since oxidation will occur while oxygen is present, and polymerization will also occur so far as dopamine is not completely consumed [27,36,49,[53], [54], [55]].
3.3. Surface morphology and roughness
Normally the surface of PDA becomes rough after deposition (where dopamine acts as the building block) and this has been associated with the deposition of an enormous amount of PDA or the film [52,56]. Also, this condition becomes intense when other nanoparticles are used together with the PDA. A metal nanoparticle (NPs)/PDA modified substrate will exhibit a great amount of surface roughness and this process allows for a long-time release of nanoparticles. The relative roughness of Ti-PDA increased by 427% as compared to the pristine Ti and this condition in most cases correlates with water contact angle [57,58]. Secondly, PDA/CS/Ag NPs coating was said to have a smooth or improved surface due to the presence of chitosan (CS) [44]. Lastly, inductively coupled plasma mass spectroscopy results, revealed a continuous silver release for Ag coupled with polydopamine. However, the addition of PEG reduces the rate at which the Ag NPs were released consequently affecting the morphology of the surface as well [59,60].
4. Dissipation of antibacterial/antimicrobial effect
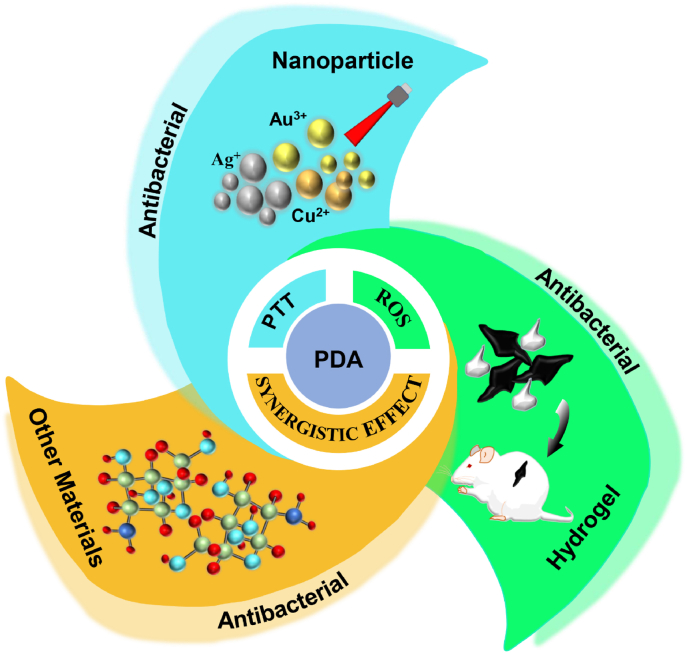
PDAs themselves can alter several conditions to suit their purpose, including pH, temperature, dopamine concentration, reaction time, and more. This just endows them, or the nanoparticles they coat with biocompatibility, hydrophilicity, biodegradability, photothermal conversion capabilities, and the ability to generate reactive oxygen species (ROS), targeting and imaging, chemical processing, light conditions set by dynamic properties (PDT), anti-inflammatory and regenerative properties [[61], [62], [63], [64], [65], [66], [67]]. These outlined conditions allow the PDA to accomplish an antibacterial effect through the employment of (i) Photothermal property (PTT) for antibacterial effect (ii) synergistical/surface modification antibacterial effect (iii) Antibacterial effect from ROS generations. It's worth noting that, these conditions operate hand in hand to ensure great efficiency with minimal side effects.
4.1. PDA
Although over time the Au NPs, silver nanoparticles, carbon nanodot, etc have proven the use of their photothermal property for antibacterial inhibition, however, the PDA has also emerged as a material endowed with such capabilities, high photothermal efficiency with facile functionalization, under local hyperthermia to counter antibiotic resistance bacteria [[68], [69], [70], [71], [72]].
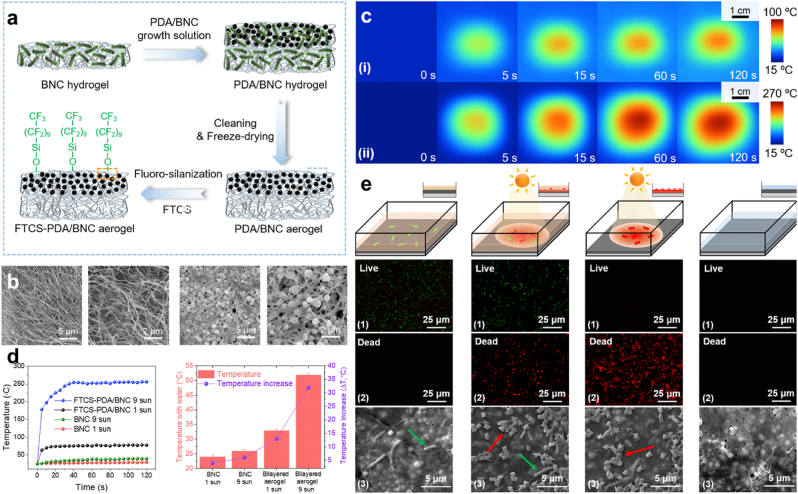
PTT. PDA exhibit some appreciable percentage of photothermal conversion efficiency with an enormous range of absorption to visible light and NIR, the photothermal property is an efficient broad-spectrum antimicrobial approach that can substitute other approaches that trigger antibiotic resistance [73,74]. Hence, this approach has been widely used to replace sterilization methods and other antibacterial methods. Accumulating evidence shows that proteins and enzymes are denatured in the process of killing bacteria at a temperature of 50 °C or higher, also their metabolism is known to be disordered at this point but at a temperature below 50 °C, there is a chance of it being repaired but when temperatures move a bit higher, there is a high possibility of tissues damaged or inflammation [[75], [76], [77], [78], [79]]. Wu et al. engineered a photothermal membrane that was made up of PDA and bacterial nanocellulose (BNC) Fig. 2a. However due to the porous nature of BNC (98%), though possess a high mechanical strength good biocompatibility and non-toxicity [[80], [81], [82]], permitted the employment of the PDA, in addition, owing to the light absorption and photothermal conversion efficiency, biocompatibility, biodegradability and also the low toxicity they exhibit [83]. With the two materials together representing a bi-layer exposed to tridecafluoro-1,1,2, 2-tetrahydrooctyl)-trichlorosilane (FTCS) vapor to give FTCS-PDA/BNC Fig. 2b. Wu et al. proved that the membrane possessed a bactericidal effect following solar illumination, thus a means for anti-bacterial fouling and solar disinfection. Infrared camera images (IR) depicted the rapid increased of the surface temperature of FTCS-PDA/BNC membrane from ∼24 °C to ∼78 °C and ∼256 °C following 10 s and 40 s of exposure to simulated solar light 1 kW m−2 (∼1 sun) and 9 kW m−2 (∼9 suns) respectively Fig. 2c and d. Solar photothermal disinfection examination exhibited the killing of bacteria in water contaminated with E. coli. Analysis of FTCS-PDA/BNC membrane at the unset depicted no inhibition, until the exposure to light irradiation (1 kW m−2). Also, removal of water and continuous exposure to irradiation for 10 min revealed the complete killing of all the bacteria, results were implicated in the SEM images and fluorescence live/dead staining assay with a membrane containing live bacteria as green and the dead as red Fig. 2e [84]. Consequently, the gain of much attention by the PTT of the PDA can be attributed to the low invasiveness, deep tissue penetration, high selectivity, and promising antibacterial strategy in the field of nanomedicine [85].
Fig. 2.
(a) Diagrammatic representation of the preparation process for FTCS-PDA/BNC membrane. (b) SEM images of BNC and FTCS-PDA/BNC membrane. (c) IR Images of FTCS-PDA/BNC membrane following exposure to 1 kW m−2 and 9 kW m−2. (d) Changes in temperature for BNC and FTCS-PDA/BNC membrane with/without water on top. (e) Measurement of the photothermal disinfection activity. With permission, reprinted from Ref. [84]. Copyright 2021 Elsevier.
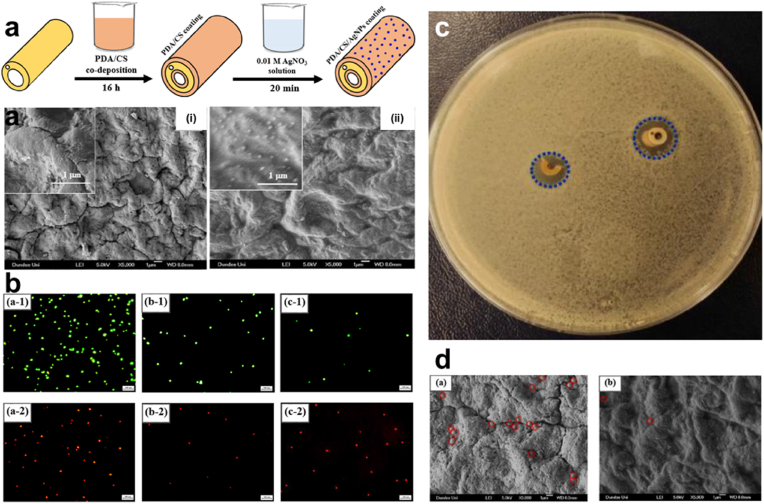
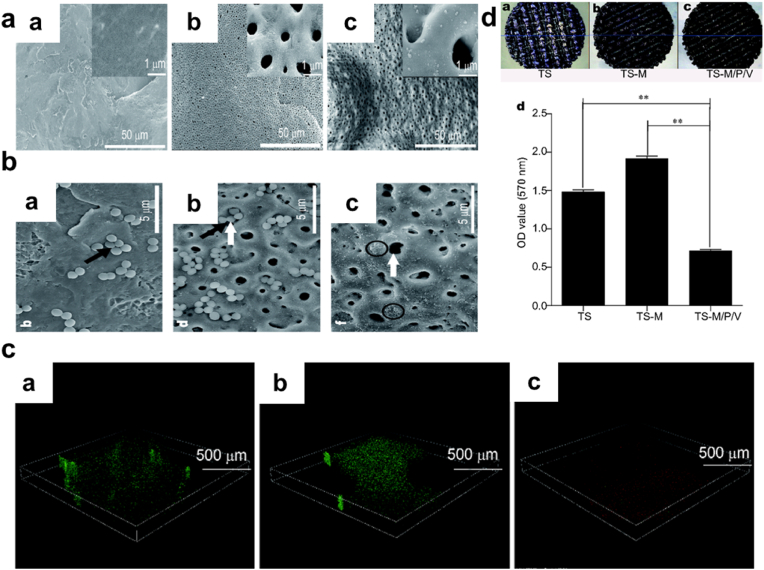
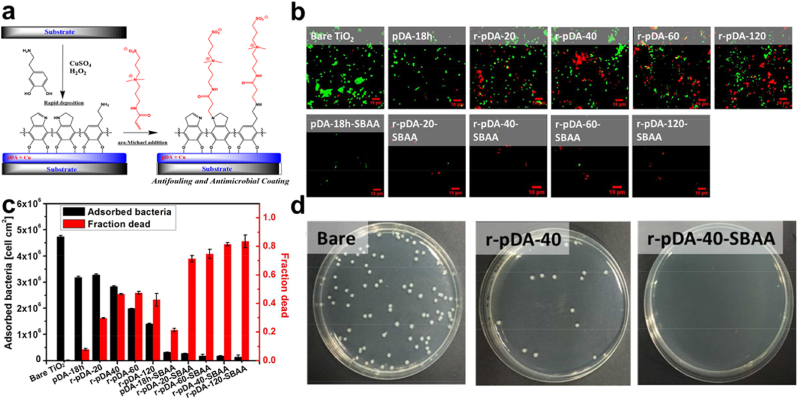
Synergistic effect/Surface Modification. What's more, the act of combining different antibacterial agents that have antibiotics inclusive though has been proven to reduce implant-associated infection, still presents concerns of triggering multidrug resistance, an unsustainable form of antibacterial activity, and even toxicity [[86], [87], [88], [89]]. PDA-coupled with chitosan (CS) and silver nanoparticles (Ag NPs) (PDA/CS/Ag NPs) improved catheters and their antibacterial efficiency, with the successful coating with the PDA, the coated surface exhibited a somewhat smooth nature due to the PDA, Fig. 3a. Also, live/dead fluorescent images implicated the fact that PDA/CS/Ag NPs stepped-up the efficiency for bacterial or biofilm inhibition Fig. 3b, respectively. PDA/CS/Ag NPs catheters prove long-lasting stability with superb antibacterial activity against S. aureus, for more than 30 days in a PBS solution. Additionally, only a few bacteria were able to adhere to the surface made up of PDA coupled with Cs and Ag NPs. SEM images proved an anti-adhesion efficacy compared to Ti substrate that was uncoated Fig. 3c and d [44]. Next, SEM images (8000 × ) of Ti6Al4V scaffold (TS), TS with micro-arc oxidization (MAO) (TS-M), and TS, MOA, PDA, and Vancomycin (V) (TS-M/P/V) Fig. 4a, showed the killing of most numbers of adherent bacterial by TS-M/P/V Fig. 4b, live/dead assay after co-culturing of scaffolds for 24 h with S. aureus (1 × 108 CFU mL−1) showed TS-M/P/V scaffold significantly inhibited bacteria with enormous dead adherent bacteria compared to TS and TS-M, which revealed the presence of many bacteria emphasizing the minimal alteration in the adhering process of the bacteria respectively Fig. 4c. In addition, TS-M/P/V showed a high percentage, 95.66% of antibacterial efficiency, similarly exhibiting a remarkable percentage for a reduction in the mature biofilm biomass (52% & 63%) as compared to the other scaffolds Fig. 4d [48]. Intriguingly studies have revealed that a high percentage (95%) of subcutaneous in vivo implants stand the chance of being infected with S. aureus [90]. And so, the formation of a PDA functional basal layer through deposition triggered by CuSO4/H2O2 with further grafting of zwitterionic sulfobetaine acrylamide (SBAA) (r-pDA-SBAA), proved to be antibacterial effective against E. coli and Staphylococcus epidermis (S. epidermis) contributing to the reduction in the adsorption rate of bacteria Fig. 5a. Following a 3hr incubation of a silicone-based urinary catheter to be tested with a bacterial solution of E. coli, made up of an optical value of OD670 (0.1), live/dead fluorescent images showed the decline in the adsorbed bacteria (E. coli) associated with the time of deposition of PDA Fig. 5b. On the other hand, quantitative data for E. coli and S. epidermis revealed a drastic decrease in the fraction of adherent and dead bacteria in groups treated with r-pDA-SBAA over time Fig. 5c. Hence bacteria adsorption resistance percentage of about 96%, upon examinations of S. epidermidis colonies from urinary catheters modified with or without the PDA and SBAA, indicated the extent to which the PDA can disallow the thriving of bacteria, with r-pDA-40-SBAA suppressing the viability of bacteria. Fig. 5d [91]. Interestingly, reports suggest that PDA possession of antibacterial effect does not make it hazardous, until they are activated via hydration in an aqueous solution to release H2O2, thereby making it easier for transport and even storage [43,92].
Fig. 3.
(a) Diagrammatic illustration of the fabrication process for PDA/CS/Ag NPs for catheter coating and SEM images of uncoated and PDA/CS/Ag NPs-coated catheter (b) Fluorescent images of adhered bacteria on uncoated, PDA/CS coated and PDA/CS/Ag NPs coated Ti surfaces following incubation in S. aureus. (c) S. aureus inhibition halo test (d) SEM images of adhered bacteria on the surfaces of uncoated and PDA/CS/Ag NPs-coated catheters. With permission, reprinted from Ref. [44]. Copyright 2020 Elsevier.
Fig. 4.
(a) SEM images of the surfaces of TS, TS-M, and TS-M/P/V scaffolds (b) SEM images of TS, TS-M, and TS-M/P/V scaffolds with adhered S. aureus ( × 8000 magnification). (c) Live/dead fluorescent images of S. aureus ( × 200 magnification and 500 μm scale bar) (d) Representation of the biomass of bacteria and biofilm component of TS, TS-M, and TS-M/P/V. With permission, reprinted from Ref. [48]. Copyright 2018 Science China Materials.
Fig. 5.
(a) Schematic representation of the coating preparation for dual antifouling and antimicrobial activity. (b) Live/dead fluorescent images of adhered E. coli on unmodified and modified TiO2 (c) Quantitative representation of the number of adsorbed and dead E. coli on the unmodified and modified TiO2 substrate. (d) Plate-counting assay of S. epidermidis from catheters unmodified and those modified with r-pDA-40 and r-pDA-40-SBAA. With permission, reprinted from Ref. [91]. Copyright 2018 American Chemical Society.
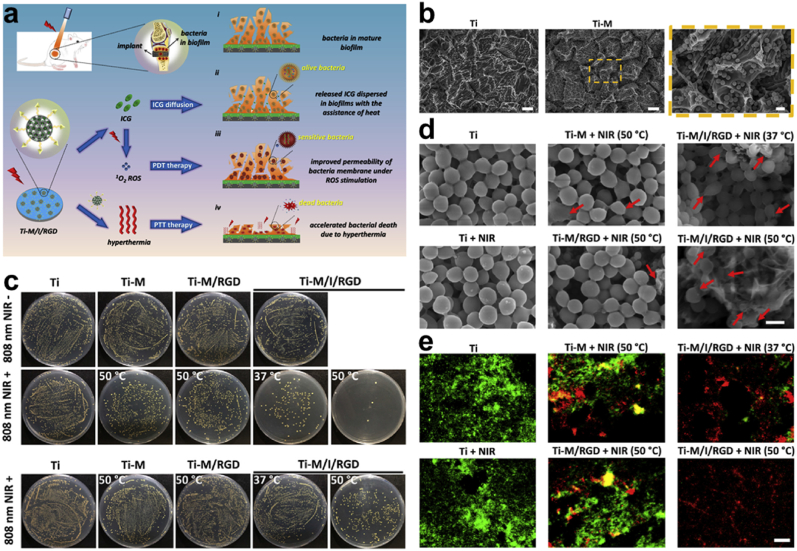
ROS Generation (see Table 1, Table 2, Table 3, Table 4). One of the key mechanisms of combating bacterial infection is via ROS generation of O2•─, H2O2, nitric oxide (NO•), etc. which damages biomolecules of bacterial cells and eventually leads to cell death. Also, the reaction of H2O2 with some redox reactive metals through a Fenton/Haber-Weiss reaction could yield hydroxyl radicals (•OH) that are more reactive and potentially toxic [[93], [94], [95], [96], [97], [98], [99]]. Yuan and co-workers developed a coating on a Ti surface through the immobilization of MPDA Fig. 6a with further conjugation and loading of osteogenic peptides (RGD) and indocyanine green (ICG) to generate, Ti-M/I/RGD. Characterization of the MPDA revealed a concentration-dependent temperature change, further irradiation (808 nm laser) for 600 s exhibited an increase in temperature from 28.0 °C to 61.0 °C Fig. 6b. A clear indication of the PDA photothermal conversion ability. However, evaluation of antibacterial activity in vitro showed a reduction in the number of bacteria treated MPDA samples following exposure to NIR with Ti-M/I/RGD (50 °C) inhibition reaching 99.7% Fig. 5c. Examination of the biofilm efficiency further revealed that the ROS was required with PTT to effectively eradicate the S. aureus biofilm and this was confirmed by the in vivo anti-biofilm data Fig. 6c. Consequently, the morphology of the bacteria in Ti-M/I/RGD (50 °C) depicted the destruction of the structure and cellular integrity Fig. 6d, thereby staining them with red fluorescence under the live/dead staining assay Fig. 6e. This approach of ROS generation for antibacterial activity happens to ensure great efficiency with minimal side effects, and they are capable of impacting an enormous range of pathogens including multidrug resistance bacteria [[100], [101], [102], [103], [104]]..
Table 1.
Antibacterial applications of polydopamine.
| Material | Application | Bacterial Strain | Mechanism | Efficiency (%) | Ref |
|---|---|---|---|---|---|
| PDA | Antibacterial | E. coli | ROS | 100.0% | [43] |
| Antibacterial | S. epidermidis | ROS | 98.90% | [43] | |
| PDA | Antibacterial | E. coli | Polymerization | Effective | [26] |
| PDA | Antibacterial | E. coli | Contact-killing | Effective | [41] |
| Antibacterial | P. aeruginosa | Contact-killing | Effective | [41] | |
| Antibacterial | S. aureus | Contact-killing | Effective | [41] | |
| PDA | Antibacterial | E. coli | PTT | Effective | [84] |
| PDA | Antibacterial/Antifouling | E. coli | Synergism | Effective | [91] |
| Antibacterial/Antifouling | S. epidermidis | Synergism | Effective | [91] | |
| PDA | Antibacterial | S. aureus | ROS | Effective | [104] |
| PDA | Antibacterial | MRSA | Contact-killing | Effective | [105] |
| Antibacterial | P. aeruginosa | Contact-killing | Effective | [105] | |
| Antibacterial | S. aureus | Contact-killing | Effective | [105] |
Table 2.
Antibacterial applications of polydopamine/nanoparticles.
| Material | Application | Bacterial Strain | Mechanism | Efficiency (%) | Ref |
|---|---|---|---|---|---|
| PDA/Cu(II) | Antibacterial | S. aureus | Synergism | Effective | [114] |
| Antibacterial | E. coli | Synergism | Effective | [114] | |
| PDA/Si NPs | Antibacterial | E. coli | Contact-killing | Effective | [124] |
| Ti-Nd-PDA-Fc | Antibacterial | E. coli | PTT/ROS | >92.00% | [134] |
| Antibacterial | MRSA | PTT/ROS | >95.00% | [134] | |
| PDA/Cu NPs | Antibacterial | S. aureus | Cu2+ release | 99.90% | [142] |
| PDA/Ag NPs | Antibacterial | S. aureus | Ag+ release | 91.30% | [143] |
| PDA/Ag NPs | Antibacterial | E. coli | Ag+ release | Effective | [144] |
| Antibacterial | S. mutans | Ag+ release | Effective | [144] | |
| PDA/Ag NPs | Antibacterial | S. mutans | Ag+ release | 99.80% | [145] |
| Antibacterial | E. coli | Ag+ release | 97.90% | [145] | |
| PDA/Ag NPs | Antibacterial | S. mutans | Synergism | Effective | [146] |
| Antibacterial | E. coli | Synergism | Effective | [146] | |
| Ti/PDA/Ag NPs | Antibacterial | S. mutans | Synergism | Effective | [147] |
| Antibacterial | P. gingivalis | Synergism | Effective | [147] | |
| Fe3O4/PDA/Ag NPs | Antibacterial | S. mutans | Synergism | Effective | [148] |
| Antibacterial | E. coli | Synergism | Effective | [148] |
Table 3.
Antibacterial applications of polydopamine/hydrogel.
| Material | Application | Bacterial Strain | Mechanism | Efficiency (%) | Ref |
|---|---|---|---|---|---|
| Hydrogel/PDA | Antibacterial | S. aureus | PTT | Effective | [160] |
| Antibacterial | E. coli | PTT | Effective | [160] | |
| CHX/Curdlan/PDA | Antibacterial | S. aureus | PTT | Effective | [161] |
| Antibacterial | E. coli | PTT | Effective | [161] | |
| PDA/Alg/Fe3O4 | Antibacterial | S. aureus | Contact-killing | Effective | [170] |
| Antibacterial | E. coli | Contact-killing | Effective | [170] | |
| Antibacterial | L. monocytogenes | Contact-killing | Effective | [170] | |
| Antibacterial | S. typhi | Contact-killing | Effective | [170] | |
| PEG-PDA | Antibacterial | MRSA | PTT/ROS | Effective | [177] |
| Antibacterial | S. aureus | PTT/ROS | Effective | [177] | |
| Hydrogel/PDA | Antibacterial | S. aureus | Synergism | Effective | [178] |
| Antibacterial | E. coli | Synergism | Effective | [178] | |
| GT-DA/CS/CNT | Antibacterial | S. aureus | PTT | Effective | [179] |
| Antibacterial | E. coli | PTT | Effective | [179] |
Table 4.
Antibacterial applications of polydopamine/other materials.
| Material | Application | Bacterial Strain | Mechanism | Efficiency (%) | Ref |
|---|---|---|---|---|---|
| PDA/Au-Hap | Antibacterial | E. coli | PTT/ROS | 96.80% | [112] |
| Antibacterial | S. aureus | PTT/ROS | 95.25% | [112] | |
| PNAGA/Au/PDA | Antibacterial | E. coli | PTT | 98.40% | [184] |
| Antibacterial | S. aureus | PTT | 97.60% | [184] | |
| COS/PDA/PU | Antibacterial | S. aureus | Synergism | Effective | [167] |
| PDA/Dextran/Chitosan | Antibacterial | S. aureus | Contact-killing | Effective | [187] |
| PU/PDA-Hep/CMCS | Antibacterial | S. aureus | Synergism | Effective | [199] |
| Antibacterial | E. coli | Synergism | Effective | [199] | |
| CS/PDA | Antibacterial | S. aureus | Synergism | Effective | [200] |
| Antibacterial | E. coli | Synergism | Effective | [200] | |
| PU/PDA/Cu/PSBMA | Antibacterial | S. aureus | Synergism | >90.0% | [190] |
| Antibacterial | E. coli | Synergism | >90.0% | [190] | |
| Cot/PDA/SiO2N+/HDTMS | Antibacterial | S. aureus | Synergism | 99.70% | [201] |
| Antibacterial | E. coli | Synergism | 99.90% | [201] | |
| Polystyrene/PDA/Ag NPs | Antibacterial | S. aureus | Synergism | Effective | [202] |
| Antibacterial | E. coli | Synergism | Effective | [202] | |
| PET/PDA/nHA/Ag | Antibacterial | S. aureus | Ag+ release | Effective | [203] |
| N3P3/PDA/Ag NPs | Antibacterial | S. aureus | Ag+ release | 99.99% | [204] |
| Antibacterial | E. coli | Ag+ release | 99.99% | [204] | |
| Lys/C4H9NO2/PDA | Antibacterial | B. subtilis | Synergism | Effective | [205] |
| Antibacterial | E. coli | Synergism | Effective | [205] | |
| Colistin/PDA/Ag NPs | Antibacterial | E. coli | Synergism | Effective | [206] |
| HA-Cu/PDA | Antibacterial | E. coli | Synergism | Effective | [207] |
| Antibacterial | S. aureus | Synergism | Effective | [207] | |
| PDA/HAP/Nisin | Antibacterial | S. aureus | Synergism | Effective | [208] |
Fig. 6.
(a) Diagrammatic illustration of the process involved in eliminating from Ti-M/I/RGD implant, already-established S. aureus's biofilm. (b) Acid etched Ti, Ti-M SEM images. (c) Images of S. aureus colonies following different treatment Ti, Ti-M, Ti-M/RGD, and Ti-M/I/RGD with the anti-biofilm efficiency. (d) SEM images of the morphology of bacteria treated with Ti, Ti + NIR, Ti-M + NIR (50 °C), Ti-M/RGD + NIR (50 °C), Ti-M/I/RGD + NIR (37 °C), and Ti-M/I/RGD + NIR (50 °C), and (e) the images of the live/dead staining assay for the S. aureus. With permission, reprinted from Ref. [104]. Copyright 2019 Elsevier.
4.2. PDA/nanoparticle
Nanoparticles such as Ag NPs, Au NPs, Cu NPs, Zn NPs, etc. are much considered nano-antibacterial agents, However, under some conditions, these NPs may have their antimicrobial activities reduced, an example is when Ag nanoparticles aggregate in colloidal solution, also other NPs can leach off or fall in the process of filtration, or presented with toxicity and so under these conditions, the NPs are faced with challenges that can affect their effectual actions [106,107]. However, a condition such as the employment of nanocarriers or encapsulation with porous templates presents the nanoparticles with many advantages improving and even presenting them with improved effectiveness, reduced toxicity, and an electrostatic attraction to other surfaces and membranes. Intriguingly, the PDA can present a mesoporous effect and a firm adherence on varying surfaces through strong intermolecular interactions [[108], [109], [110]].
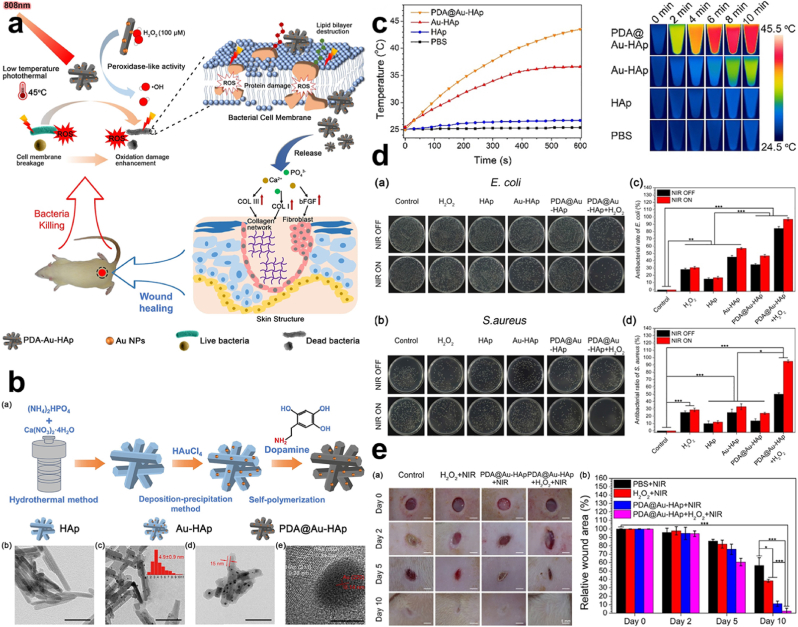
PTT. Dopamine is shown to be a catecholamine of low molecular weight taking the form of an adhesive protein that is capable of undergoing self-polymerization, acting as a binding agent and reductant, PDA coupled with AgNO3 reduced the Ag + to Ag0, which was used to combat bacteria that rapidly grew on a textile surface [111]. Furthermore, Xu et al. showed that PDA exhibits excellent biocompatibility with latent photothermal effect Fig. 7a, after coating hydroxyapatite (HAp) and gold (Au) (Au-HAp) with PDA, (PDA@Au-HAp) through the catalysis of H2O2 Fig. 7b. This proved a great antibacterial efficiency after NIR (808 nm laser) exposure, with a photothermal temperature increase and a conversion efficiency of 13.28% Fig. 7c. PDA@Au-HAp + H2O2 + NIR exhibited inhibition of about 96.8% and 95.2% against Escherichia coli and Staphylococcus aureus respectively Fig. 7d. This also placed some emphasis on the synergy property of photothermal and catalytic effects possessed by the PDA@Au-HAp + H2O2. On the other hand, PDA@Au-HAp showed a percentage of about 46.3% and 23.2% antibacterial effect with Au-HAp exhibiting a weak photothermal effect Fig. 7d. Assessment of a wound infected with S. aureus depicted that, wounds recovered following treatment with PDA@Au-HAp + NIR and PDA@Au-HAp + H2O2 + NIR compared to the other groups, after 10 days Fig. 7e. Nanoparticles with PDA exhibit a remarkable photothermal conversion efficiency by taking advantage of that to be used against Gram-positive and Gram-negative bacteria [112].
Fig. 7.
(a) Schematic representation of the combined activity of photothermal therapy and catalysis for antibacterial effect. (b) Diagrammatic representation of the synthesis of HAp, Au-HAp & PDA@Au-HAp with their respective SEM for characterization. (c) Changes in temperature with the infrared thermography for HAp, Au-HAp & PDA@Au-HAp under 808 nm laser irradiation (d) Images of bacteria colonies and antibacterial rates of E. coli and S. aureus following treatment with H2O2, HAp, Au-HAp, PDA@Au-HAp & PDA@Au-HAp + H2O2. (e) Examination of the S. aureus infected wound healing after exposure to HAp, Au-HAp, PDA@Au-HAp + NIR & PDA@Au-HAp + H2O2 + NIR. With permission, reprinted from Ref. [112]. Copyright 2018 Elsevier.
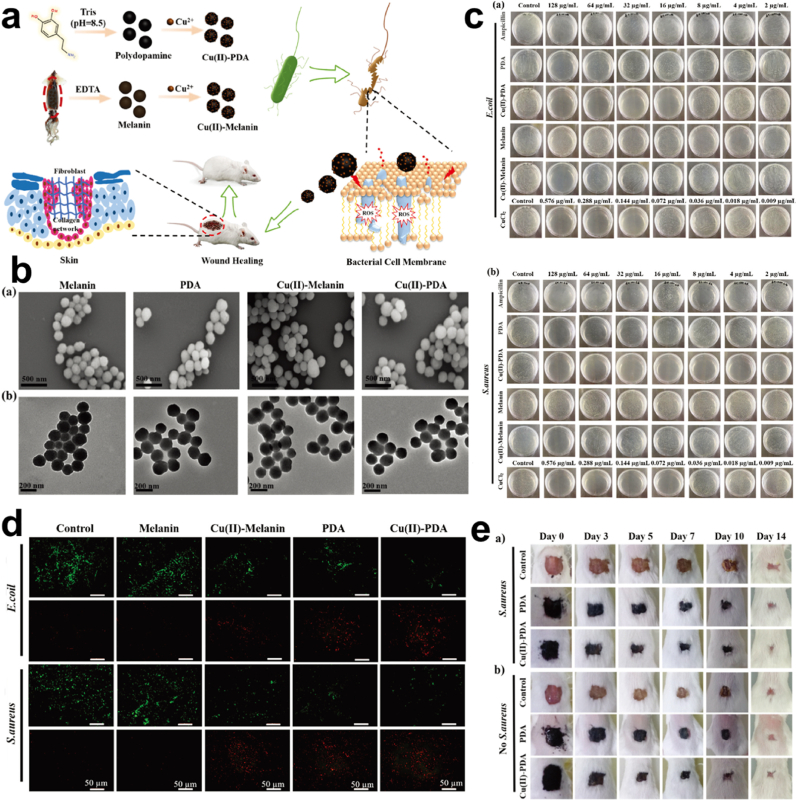
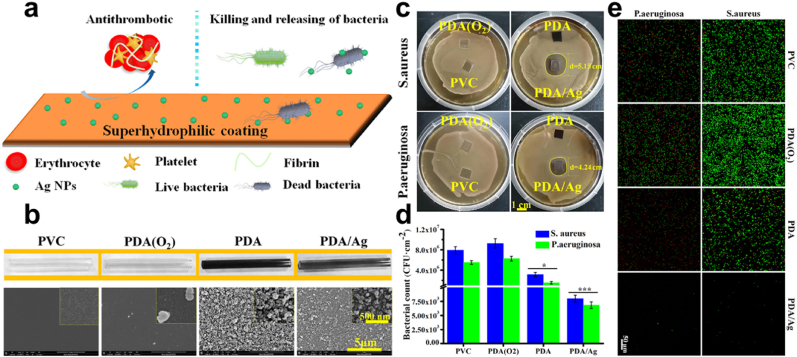
Synergistic effect/Surface Modification. It's worth noting that, as wound healing therapy is to impede the bleeding and infection process so is rapid hemostasis required to step up the closure of wounds and the generation of tissues [113]. However, some inorganic materials used as hemostatic materials induce inflammation and thermal damage due to their highly exothermic nature [114]. Also, clotting of blood that forms in the wound bed can easily be colonized by bacteria and can become quite impermeable as well as rendering the use of antibiotics very ineffective. This calls for materials that are capable of combining antimicrobial properties with wound healing [[115], [116], [117]]. Lin and colleagues developed and studied four nanomaterials; melanin, PDA, Cu(II) loaded melanin, and Cu(II) loaded PDA Fig. 8a and b. Although, soluble copper ion is capable of ROS generation, respiration inhibition, and DNA damage for the killing of bacteria, the great toxicity that they possess calls for a chelating agent to neutralize the toxic effect [[118], [119], [120]]. Using the melanin and PDA to chelate copper ions under acidic conditions, evaluation of the antibacterial efficiency of the various materials reveals the excellent bactericidal effect for the Cu(II) loaded PDA NPs compared to the other NPs with Ampicillin as control, having a minimum bactericidal concentration (MBC) which was as low as 16 μg mL−1, Cu(II) loaded PDA depicted an excellent bactericidal effect Fig. 8c. However, further investigation showed that, although Cu(II) ion had its constituent concentration of Cu(II) loaded PDA (16 μg mL−1) to be 0.072 μg mL−1, the MBC for the Cu(II) ion alone was noticed to be 0.144 μg mL−1, which was more than the Cu(II) ion constituent in the composite structure of Cu(II) loaded PDA, indicating the synergistic bactericidal effect. Accordingly, Cu(II) loaded PDA exhibited a greater bacterial effect in S. aureus compared to E. coli which was attributed to the cell structure. Also, Cu(II) loaded PDA depicted strong red fluorescence indicating an effective antibacterial property although the PDA and Cu(II) loaded melanin also showed some sort of red fluorescence Fig. 8d. Next, SEM images depicted the presence of ruptured membranes in the PDA, Cu(II) loaded melanin and, Cu(II) loaded PDA. Intriguingly, the PDA antibacterial effect is said to be dependent on amine or the hydroxyl groups on their surface, the PDA positive charged amine is capable of causing bacterial lysis upon contacting bacterial cell walls, proving that the nucleophilic hydroxyl groups are capable of destroying bacterial membrane [41,121,122]. On wound healing, Cu(II) loaded PDA exerted much antibacterial effect. After 7 days the reduction in the infected wound size was equivalent to that of an uninfected wound indicating how PDA aided in infection prevention and also wound healing Fig. 8e [114]. Hence PDA improvement of the antimicrobial performance of Cu through a synergistic effect as well as the reduction of the toxicity. The catechol of PDA is capable of being adsorbed during bleeding to cause a sealing effect and capable of activating the coagulation system, preventing bleeding, infections and allowing effective wound healing [123,124]. Next, PDA nanoparticle and Ag nanoparticles coatings were fabricated, endowed with stability and long-term super hydrophilicity effect, exhibiting a great antibacterial effect against S. aureus and P. aeruginosa Fig. 9a and b. Zone of inhibition of PDA/Ag showed a circular bacteria-free zone of diameter (5.15 cm and 4.24 cm) in the S. aureus and P. aeruginosa respectively Fig. 9c, also the live/dead staining images depicted a significant reduction in the adherent bacteria compared to the other surfaces that indicated high susceptibility to the S. aureus and P. aeruginosa Fig. 9d and e [125]. Studies reveal that super-hydrophilic coating can effectively prevent protein adhesion, maintaining adhesion proteins' native conformation thus their good biocompatibility and the low/non-fouling properties [126,127].
Fig. 8.
(a) Schematic illustration of the fabrication of PDA, melanin, Cu(II) loaded PDA, and Cu(II) loaded melanin nanoparticles for antibacterial activity. (b) SEM and TEM images of melanin, PDA, Cu(II) loaded melanin, and Cu(II) loaded PDA nanoparticles. (c) The E. coli and S. aureus inhibition efficiency of ampicillin, PDA, Cu(II) loaded PDA NPs, melanin, Cu(II) loaded melanin NPs and CuCl2, and (d) the corresponding live/dead fluorescent images. (e) Wound healing assessment for S. aureus infected wound, with PDA, Cu(II) loaded PDA nanoparticles within 14 days. With permission, reprinted from Ref. [114]. Copyright 2021 The Royal Society of Chemistry.
Fig. 9.
(a) Schematic illustration of the fabrication of the super hydrophilic coating of PDA with Ag+, with the antithrombotic and anti-infection. (b) Image of the PVC been coated with the PDA before and after with the coating morphology's SEM images. (c) Zone of inhibition for S. aureus and P. aeruginosa. (d) Live/Dead staining of adherent bacteria. (e) Adherent bacteria count for S. aureus and P. aeruginosa. With permission, reprinted from Ref. [125]. Copyright 2020 Elsevier.
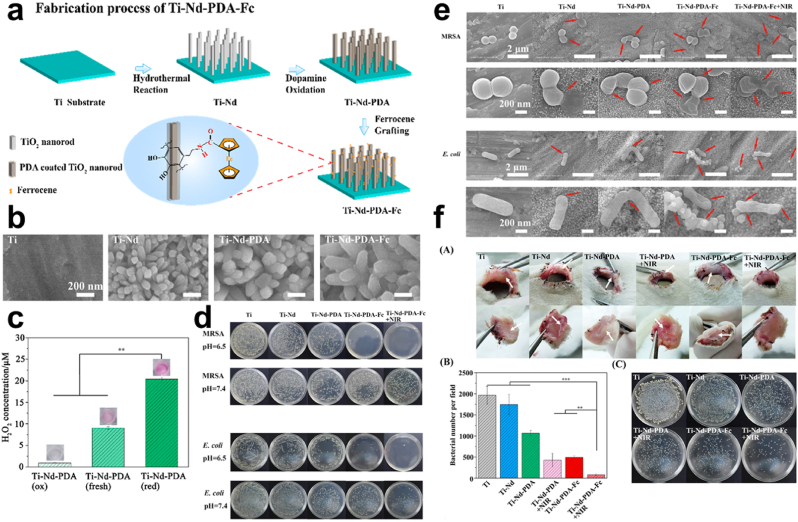
ROS Generation. Oxidation of catechols by air ends up yielding H2O2 and O2•─. This condition enables the PDA to scavenge some radicals' species together with reducing inflammation induced by ROS [[128], [129], [130], [131]]. Also, because the PDA nanoparticles can be redox-active the catechol moieties are capable of donating electrons to O2•─ for H2O2 generation [43,[132], [133], [134]]. Studies reveal that the increment in H2O2 generation was somewhat dependent on the concentration of dopamine after samples with double coated PDA revealed significantly higher forms of H2O2 compared with those with single depositions of dopamine [41,43,131,135,136]. Song and co-workers generated TiO2 nanorod (Ti–Nd) for a nanostructured surface via a hydrothermal approach followed by the coating with PDA, and functionalization with iron (Fe) containing ferrocene (Fc) (Ti-Nd-PDA-Fc), hence polydopamineferrocene (PDA-Fc)-functionalized TiO2 nanorods (Ti-Nd-PDA-Fc) Fig. 10a–b. H2O2 generated from the composite Ti-Nd-PDA measured via an enzymatic colorimetric assay with a dye (Amplex Red) indicated the highest amount of H2O2 concentration which was in keeping with the composite structure's higher donateable electron to oxygen thus their ability to generate more H2O2 Fig. 10c. Substrates contaminated with E. coli and methicillin-resistance Staphylococcus aureus (MRSA) incubated for 8 h showed that substrates treated with Ti-Nd-PDA-Fc resulted in almost a complete eradication of bacteria under mildly acidic conditions. Upon treating the substrates with Ti-Nd-PDA-Fc + NIR, the plate did not show much difference and this was evident in both the E. coli and MRSA Fig. 10d. SEM images implicated the results from the plate after treatment of bacteria with Ti, Ti–Nd, Ti-Nd-PDA, Ti-Nd-PDA-Fc, and Ti-Nd-PDA-Fc + NIR which revealed that treatments with inhibition have bacteria surface damaged compared to those in other groups like the Ti with smooth bacteria surfaces Fig. 10e. Also, biofilms were formed on the substrate following contamination except for Ti-Nd-PDA-Fc + NIR substrates. In vivo antimicrobial assessment indicated the presence of an antimicrobial effect for Ti-Nd-PDA-Fc but, enhanced after irradiation (Ti-Nd-PDA-Fc + NIR) Fig. 10f. In effect, PDA is capable of generating ROS which can be converted to a more reactive radical, its photothermal property allows for temperature increase and rapid generation of •OH [134]. ROS which includes H2O2, NO•, and O2•─ for some time now has drawn attention when it comes to the fight against bacteria since reports indicate that some ROS has high potential with minimal toxicity [103]. Accordingly, some of these ROS can be controlled one way or the other through activation by varying stimuli such as temperature, pH, etc [[137], [138], [139], [140], [141]].
Fig. 10.
(a) Schematic representation of the fabrication process of Ti-Nd-PDA-Fc. (b) SEM images of Ti–Nd, Ti-Nd-PDA, and Ti-Nd-PDA-Fc (c) Graphical representation of H2O2 generations for a specific time under a pH 7.4. (d) Images and quantification of MRSA and E. coli colonies treated with Ti–Nd, Ti-Nd-PDA, Ti-Nd-PDA-Fc, and Ti-Nd-PDA-Fc + NIR under an acid and neutral environment (e) SEM images of the MRSA and E. coli morphology on substrates of Ti, Ti–Nd, Ti-Nd-PDA, Ti-Nd-PDA-Fc, and Ti-Nd-PDA-Fc + NIR. (f) In vivo antibacterial test of MRSA infected wound, quantification and colonies of MRSA after treatment with Ti, Ti–Nd, Ti-Nd-PDA, Ti-Nd-PDA + NIR, Ti-Nd-PDA-Fc, and Ti-Nd-PDA-Fc + NIR. With permission, reprinted from Ref. [134]. Copyright 2020 American Chemical Society.
4.3. PDA/hydrogel
Hydrogels are interconnected polymer chains that form a 3D porous material, with high water content some features which are crucial for their antibacterial activity are their porosity and hydrophilicity characteristics [[149], [150], [151]], however, studies indicate that the careful selection of the monomers and crosslinkers can help make this characteristic desirable. Also, reports indicate that their stability, biocompatibility, biodegradability, mechanical strength, and degree of porosity are all tunable. Although some possess inherent antibacterial property, a synergistic hydrogel can be produced, coupling its characteristics with that of the PDA which equally exhibit effective antimicrobial activity [[152], [153], [154], [155], [156]].
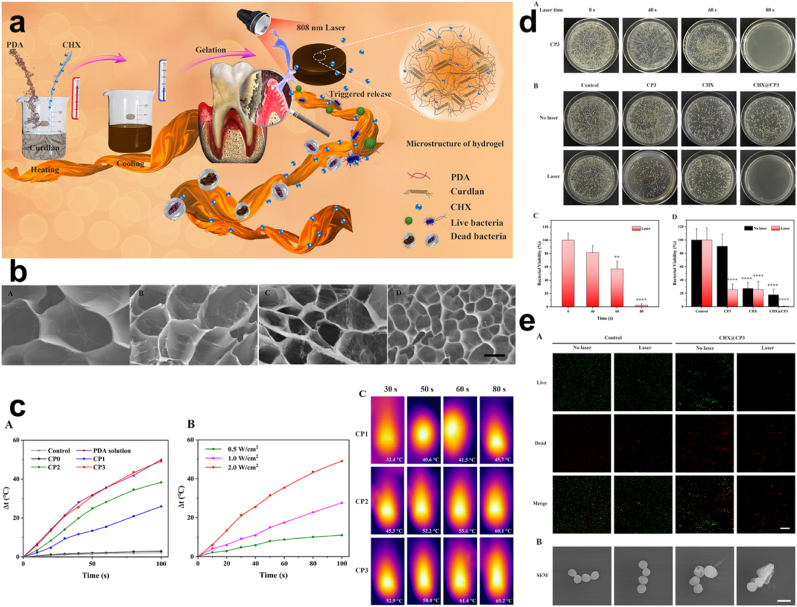
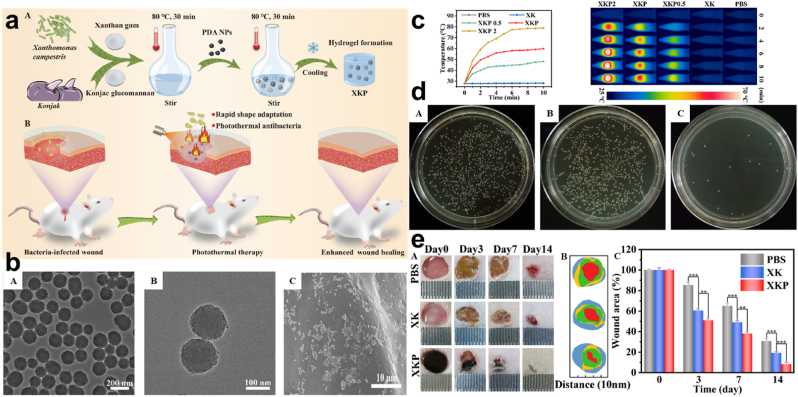
PTT. Although wound dressing serves as a means of promoting wound healing the current state of drug resistance and wide infection doesn't present it as direct means of wound healing. And so, the use of bandages, gauze, cotton, etc may have provided and contributed a lot in terms of wound healing [[157], [158], [159]], over time the rate of bacteria evolution does not allow these materials to be fully relied on as the main material for wound healing. Also, with how they become tightly attached to the surface on which they are used, due to their lack of elasticity, some easily fall off after a short time [160]. This calls for the development of materials that will employ elasticity and dissipate antibacterial resistance conditions. Tong and co-workers synthesized curdlan hydrogel with about 4 mg/mL PDA (CP3) with further additions of acetate CHX (CHX@CP3) Fig. 11a–b, based on the fact that PDA would play a crucial role in photothermal ability, morphology, and physiochemical characteristics improvement. Pure curdlan exhibited no thermal effect, however, the composite hydrogels with PDA displayed heat, recording a highest Δt zenith of more than 40 °C for NIR density (2 W cm−2) Fig. 11c. Also, on the rate of bacterial killing, CHX@CP3 + NIR presented average bacterial cell viability of 0.47%, although CHX@CP3 groups without laser exhibited some inhibition but not as compared to CHX@CP3 laser-treated Fig. 11d. The role of NIR irradiation emphasized the exertion of the bactericidal effect of the CHX@CP3. Fluorescent staining showed an increase in the red fluorescence for the CHX@CP3 as compared to the other treatments which were green. This result was in keeping with the SEM results for the examination of the bacterial S. aureus morphology Fig. 11e [161]. Consequently, all these correspond to the fact that the PDA influenced the composite to some extent in the aspect of stabilized photothermal effect for the improved bactericidal outcome. Also, Zeng and colleagues fabricated a nanocomposite hydrogel (xanthan gum and konjac glucomannan) doped with PDA (XKP) Fig. 12a. Since PDA has an excellent photothermal conversion efficiency and light absorption capacity, good compatibility with easy fabrication. SEM and TEM images showed the relative uniformity of PDA, the smoothed surfaced XK, and the dotted representation of PDA in XKP Fig. 12b [112,162,163]. Fabricated XKP0.5, XKP1 & XKP2 exhibited effective photothermal properties following 10 min exposure to 808 nm NIR laser which revealed an increase in temperature to the concentration of PDA present (0.5, 1, and 2 mg/L) Fig. 12c. The temperature difference based on the concentration of PDA allowed for the selection of XKP since its temperature was 59.9 °C which exceeded the 50 °C consequently proving a chance of bacteria inhibition. Reports indicate that above 50 °C bacteria happen to be killed through protein and enzyme denaturing [164,165]. In vitro antibacterial evaluation of XKP showed inhibition of 98.12% and 96.64% for E. coli and S. aureus respectively following 10 min of NIR irradiation. Examination of bacteria with SYTO 9 and propidium iodide (PI) revealed a red fluorescence for XKP as a result of the presence of dead bacteria, this was implicated by results from SEM which showed disruption and damages in the morphology, hence the ability and possibility of preventing bacterial infections through the photothermal therapy Fig. 12d. Also, results from in vivo were in keeping with what was evident in the in vitro, where a bacteria-infected wound, revealed XKP ability to significantly treat the infected wound without inflammation Fig. 12e [160]. Hence, the PDA effect can easily allow for the dissipation of antimicrobial activity through photothermal NIR irradiations.
Fig. 11.
(a) Schematic representation of the synthesis of CP hydrogel and the synergistic antibacterial effect for periodontal therapy (b) SEM images of CP0, CP1, CP2, and CP3. (c) Photothermal changes, temperature based on NIR densities, and infrared thermography (d) Antibacterial assessment with CP3 alone with laser and CP3, CHX and CHX@CP3 with/without NIR irradiation against S. aureus. (e) Fluorescent staining and SEM images of live and dead S. aureus. With permission, reprinted from Ref. [161]. Copyright 2020 Elsevier.
Fig. 12.
(a) Schematic illustration of the preparation and application of XKP nanocomposite hydrogels. (b) SEM images of XK and XKP with high magnification image of XKP. (c) Photothermal effect and temperature change curves with photothermal images for XK containing varying concentrations of PDA. (d) Images of E. coli and S. aureus colonies following treatment with XK and XKP with the corresponding fluorescent images of the live/dead assay. (e) Representation of bacteria-infected skin wound with the size and area after treatment with PBS, XK, and XKP. With permission, reprinted from Ref. [160]. Copyright 2021 Elsevier.
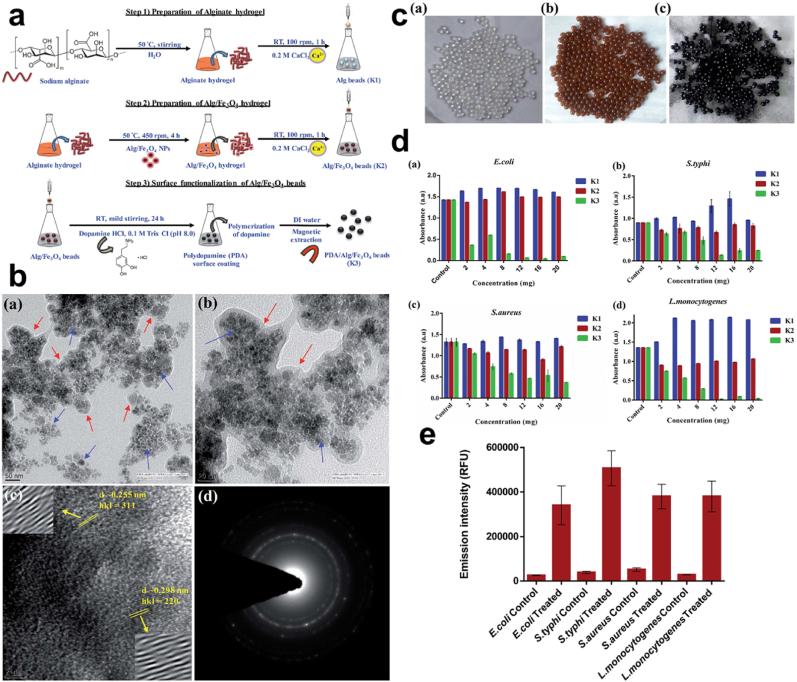
Synergistic effect/Surface Modification. Matai employed a magnetic hydrogel bead which was PDA functionalized through the immersion of the magnetic hydrogel beads in DA (mild alkaline solution) thus the functionalization with PDA Fig. 13a. In comparing PDA/Alg/Fe3O4 beads (K3) to alginate hydrogel (Alg) beads (K1) and Alg/Fe3O4 beads (K2) for their antibacterial activity Fig. 13b–c, K3 tremendously decreased E. coli, S. typhi, S. aureus, and Listeria monocytogenes (L. monocytogenes) growth which was concentration-dependent compared to K1 and K2, with K1 rather allowing for the growth of E. coli as the K2 did not render any change in the bacteria. Consequently, the effectiveness of the K3 was attributed to the PDA, studies indicate that PDA granules are capable of up-surging surface irregularities for attachment, build up, and killing of bacteria [41,166,167]. Information from the bacteria growth kinetics further implicated the action of the K3 beads in the effective killing of bacteria which was time-dependent Fig. 13d. Also, further works reveal the death of bacteria was due to the loss of their membrane integrity after the membrane integrity assay displayed the fluorescent intensity of PI dye following their penetrations into the membrane of damaged and slightly damaged bacteria Fig. 13e. PI is capable of penetrating such membranes other than those unaffected or intact membranes [[168], [169], [170]]. Thus, exposure to K3 resulted in the loss of membrane integrity and subsequently bacterial cell death. PDA can be modified to improve its antioxidant property, although an increased dose of it is capable of inducing toxicity, there can be ways and means to make these conditions and processes possible [[171], [172], [173]].
Fig. 13.
(a) Schematic illustration of the fabrication of PDA/Alg/Fe3O4 (K3) beads. (b) HR-TEM, lattice fringes, and SAED images of PDA/Alg/Fe3O4 and (c) images of the prepared K1, K2, and K3 beads. (d) Bacteria inhibition efficiency based on the concentration of prepared K1, K2, and K3 beads after 12 h. (e) Variation in fluorescent of P·I for varying bacteria and a control. With permission, reprinted from Ref. [170]. Copyright 2019 The Royal Society of Chemistry.
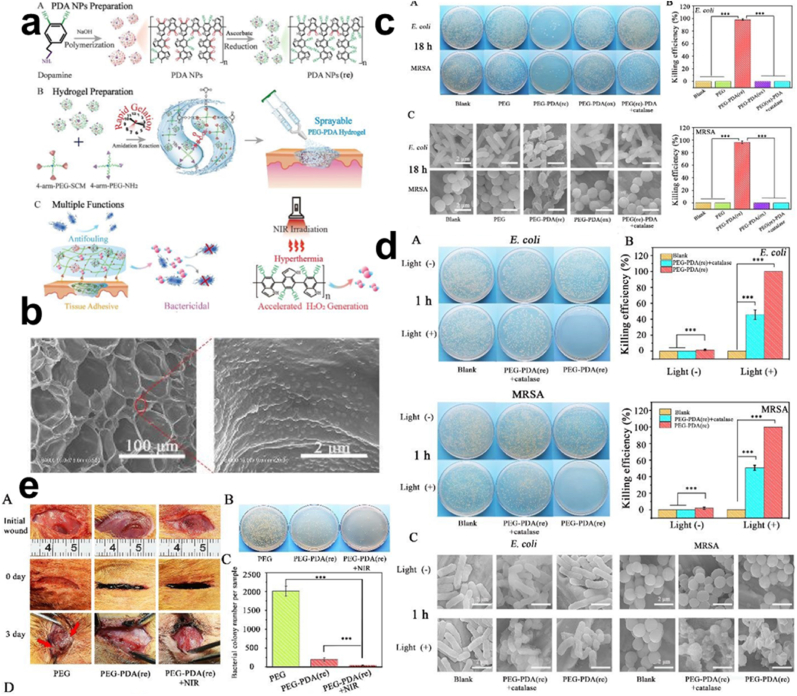
ROS Generation. Pathogens can have their physiological process hampered by ROS, through the exertion of oxidative stress against the cell membrane, consequently yielding to the damage of nucleic acids, protein, or cellular components [174]. For the convenience of the open wound treatment, sprayable hydrogels with some features such as mixing efficiency and adequate atomized droplet to tissue contact etc were examined by Sun, however, another concern was the equipping of the sprayable hydrogel with an inherent antibacterial feature due to inflammation and infection. Eventually, Sun and co-workers fabricated a sprayable antibacterial hydrogel endowed with PDA (PEG-PDA) Fig. 14a–b. As previously mentioned, the PDA is capable of donating an electron to O2 for H2O2 generation, and with their equipped amino acid groups, they easily combine with varying materials exhibiting their biocompatibility and providing an antibacterial effect [175,176]. Characterization of the ROS generated showed that autoxidation of the PDA NPs in an oxygenated environment will subsequently result in the conversion of catechol to quinone, further allowing the transfer of electrons in the polyphenol to the oxygen for H2O2 generation, hence the release of the ROS (H2O2). Also, based on the reduced-PDA (rPDA) in the PEG-PDA hydrogel content, ROS was generated continuously in an increasingly (100, 130, 175) μM manner according to the increasing re-PDA concentration for 18 h compared to those with oxidized PDA, exposure to NIR (808 nm, 1.5 W cm−2) for 10 min indicated a further rise from 8 to 70 μM of H2O2. PEG-PDA (re) hydrogel groups once again, were marked with antibacterial inhibition efficiency of 98.9% and 97.9% against the MRSA and E. coli respectively after 18 h, this action was impeded upon the addition of an H2O2 quenching agent. Hence, outcomes could be attributed to the ROS generations. SEM results depicted the damaged morphology of bacteria which were in keeping with the aforementioned results Fig. 14c. Also, exposure to NIR revealed a moderate killing of bacteria in PEG-PDA (re) groups which was further implicated by the SEM images Fig. 14d. In vivo antibacterial assessment showed a rapid wound healing through rapid gelation and tissue adhesion with a great antibacterial effect for PEG-PDA (re) and much strong for PEG-PDA (re) +NIR following the dilution and plating of fluid from the wound Fig. 14e. Although the level of H2O2 generation ceases to increase after NIR exposure at the varying stages this was attributed to the fact that phenolic hydroxyl groups of the PDA were completely consumed [177].
Fig. 14.
(a) Schematic representation of the preparation and uses of the PEG-PDA (b) SEM images of PEG-PDA. (c) Images of the E. coli and MRSA colonies following 18 h of incubation with the SEM images representing the bacterial morphology. (d) Images of the E. coli and MRSA colonies following treatment PEG-PDA + catalase and PEG-PDA with/without NIR (808 nm, 6 min) incubated for 1 h with the SEM images of bacterial morphology. (e) In vivo wound treatment with PEG and PEG-PDA, plating of fluid from the wound and the MRSA bacterial colonies count and histogram. With permission, reprinted from Ref. [177]. Copyright 2020 The Royal Society of Chemistry.
4.4. PDA/other materials
The PDA via varying covalent and other non-covalent interactions can be coated or conjugated with organic or inorganic materials which include graphene oxide, magnetic nanoparticles, glass beads, etc. Also, the PDA particles can solely be used in the removal of organic pollutants and heavy metals [[180], [181], [182], [183]].
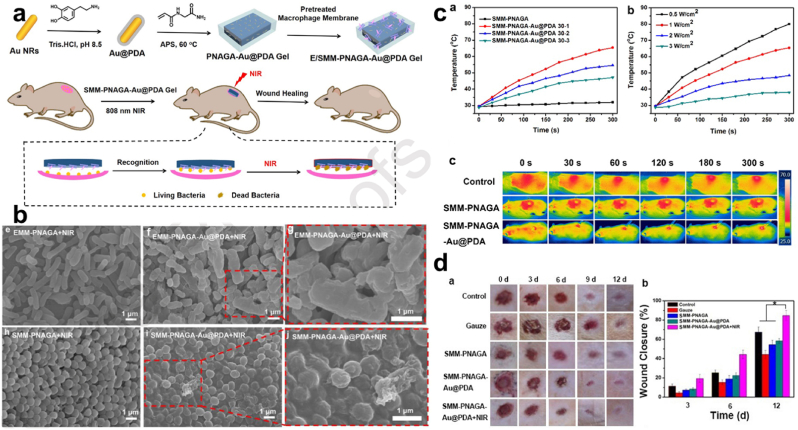
PTT. Furthermore, Li et al. revealed that hardly was there a temperature rise among some hydrogels mounted and therefore attributed that to lack of Au coupled with PDA (Au@PDA), following the synthesis of N-acryloyl glycinamide (NAGA) mixed with Au@PDA (PNAGA-Au@PDA) which has E. coli and S. aureus pre-treated-macrophage membranes coating (E/SMM-PNAGA-Au@PDA) Fig. 15a. The exposure of SMM-PNAGA-Au@PDA-30 to NIR (808 nm, 2 W cm−2) displayed remarkable color change, indicating the presence of excellent photothermal performance Fig. 15b, subsequently, NIR laser irradiation of hydrogel (EMM-PNAGA-Au@PDA & SMM-PNAGA-Au@PDA) resulted in an effectual bacterial killing at a percentage of 98.4% against E. coli and 97.6% against S. aureus respectively Fig. 15c. Meanwhile, their counterpart lacking the Au@PDA couldn't exhibit such an effective antibacterial yield of a near-total bacterial kill. And this phenomenon was equally observed in live/dead staining images of bacteria treated by the various hydrogel containing the Au@PDA. Also, examination of the S. aureus infected wound healing revealed a healing ratio of about 84.7% for SMM-PNAGA-Au@PDA + NIR after 12 days, results after the 3 days showed no serious infections unlike the other groups Fig. 15d [184]. Thus Au@PDA functionally equipped the various hydrogels with a photothermal effect.
Fig. 15.
(a) Diagrammatic illustration of the synthesis of MM-PNAGAAu@ PDA for antibacterial activity. (b) Temperature changes for SMM-PNAGA and SMM-PNAGA + varying concentration of Au@PDA under 808 nm (2 W cm−2) laser irradiation, a test with different laser powers, and the infrared thermography. (c) SEM images of E. coli and S. aureus following exposure to E/SMM-PNAGA + NIR, and E/SMM-PNAGA-Au@PDA + NIR. (d) In vivo wound healing activity assessment for S. aureus infected wound. With permission, reprinted from Ref. [184]. Copyright 2020 Elsevier.
Synergistic effect/Surface Modification. Reports indicate that phenolic quinone regulation with stronger reductants can enhance the antioxidant property of the PDA. Also, the supply of the poly-catechol group with electrons from a stronger reducing agent can be later transferred to the oxidant, in a sense of reduction of the catechol (QH2) for improved antioxidant properties and enhanced wound healing aside from the antibacterial activity [132,185]. Fu and co-workers synthesized a reduced PDA (rPDA) with oxidized dextran/chitosan hybrid hydrogels Fig. 16a. The antibacterial activity revealed an obvious indication of bacterial inhibition exhibited by rPDA incorporated polysaccharides hydrogels depending on the concentration of rPDA present, with GelNP-05 which had the highest concentration of PDA depicting a greater area of bacteria inhibition Fig. 16b. Application of the GelNP-05 on the surface of infected pig skin reduced the S. aureus presence to a small amount compared to the infected skin treated with saline solution Fig. 16c. Also, cells incubated with 50 μM H2O2 and after being treated with PDA containing hydrogel samples showed improved cell viability due to the PDA, since an enormous amount of H2O2 could have resulted in oxidative stress and further destruction of cells Fig. 16d. Next, monitoring of the healing of the infected wound for 15 days showed complete healing with a small scar following the treatment with GelNP-05 with control groups still indicating the presence of a wound. GelNP-05 was able to aid in the accumulation and modification of the collagen fibers together with many vessels around the wound Fig. 16e. Hence, GelNP-05 was said to enable the inhibition of bacterial infection together with the protection of cells at the wound site from oxidative stress damage. Intriguingly, as reports suggest that PDA's antibacterial activity might result from the phenolic hydroxyl group, under this condition ascorbic acid can also, reduce quinone structure to phenolic hydroxyl, subsequently contributing to an excellent antibacterial effect [186,187]. Lastly, the antifouling and antibacterial property of polyurethane (PU) was improved after PDA and poly-(sulfobetaine methacrylate) (PSBMA) co-deposition to give PU/PDA (Cu)/PSBMA Fig. 17a. Antibacterial examination against E. coli and S. aureus showed decreased colonies for PU/PDA (Cu) and PU/PDA (Cu)/PSBMA, unlike other PU materials. Results from the examination of live/dead assay were in keeping with the results evident from the colonies of E. coli and S. aureus, an effective display of anti-fouling and antibacterial activity Fig. 17b. Also, SEM images revealed the dead bacteria in the PU/PDA (Cu) and PU/PDA (Cu)/PSBMA treated groups Fig. 17c. Though PU/PDA (Cu)/PSBMA displayed more antibacterial activity against E. coli compared to the S. aureus which was attributed to the fact that Gram-positive and Gram-negative bacteria wall compositions happen to be different from each other. Results from S. aureus infected wound implicate earlier results from the PU/PDA (Cu) and PU/PDA (Cu)/PSBMA Fig. 17d [[188], [189], [190]]. PDA though able to elicit H2O2 upon activation can equally be combined with other antibacterial agents to kill bacteria on surfaces, considering their durability and biocompatibility, hence providing the active site for another antibacterial agent to function.
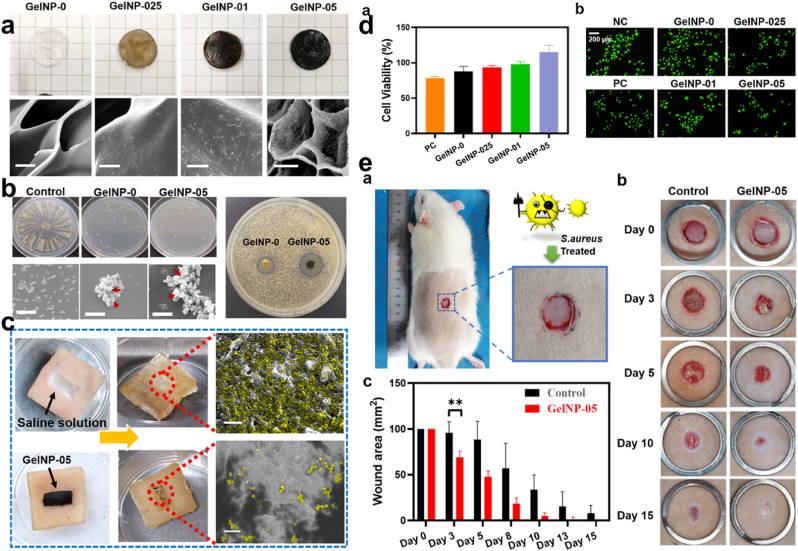
Fig. 16.
(a) Optical and SEM images of GelNP-0, GelNP-025, GelNP-01 & GelNP-05. (b) Antibacterial assessment of GelNP-01 & GelNP-05 with the SEM images of adhered S. aureus. (c) Invitro skin disinfection test, with S. aureus infected skin, treated separately with saline solution and GelNP-05. (d) Cell viability and live/dead staining assay after incubation with 50 μM H2O2 (e) Wound healing assessment for S. aureus infected wound, diameter, and the wound area statistics. With permission, reprinted from Ref. [187]. Copyright 2021 Elsevier.
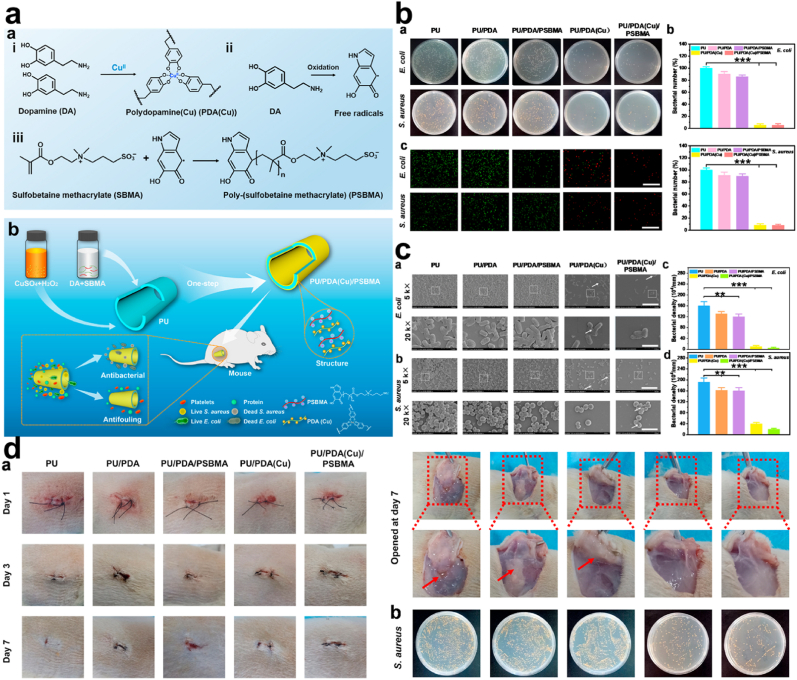
Fig. 17.
(a) Diagrammatic representation of PU/PDA (Cu)/PSBMA antibacterial and antifouling effect. (b) Antibacterial activity against E. coli and S. aureus and the fluorescent images of the live/dead assay following treatment with PU, PU/PDA, PU/PDA/PSBMA, PU/PDA (Cu), and PU/PDA (Cu)/PSBMA (c) SEM images of different magnification showing the anti-adhesive effect of PU, PU/PDA, PU/PDA/PSBMA, PU/PDA (Cu) and PU/PDA (Cu)/PSBMA (d) images S. aureus infected wound treated with PU, PU/PDA, PU/PDA/PSBMA, PU/PDA (Cu) and PU/PDA (Cu)/PSBMA (arrow indicates the pus), with LB plate containing S. aureus present in the pus after treatment with PU, PU/PDA, PU/PDA/PSBMA, PU/PDA (Cu) and PU/PDA (Cu)/PSBMA. With permission, reprinted from Ref. [190]. Copyright 2021 Elsevier.
ROS Generation. PDA's O-quinone and O-hydroquinone groups serve as the elementary point for PDA to be either semi-oxidized or reduced, thus allowing the PDA the ability to scavenge for ROS in its environment [50,191,192]. Although ROS itself can be considered as the by-product of metabolism and also crucial for physiological processes which include intracellular signal transduction etc., an excessive amount is capable of inducing lipids, protein, and cellular damages [[193], [194], [195]]. Xiang and co-workers fabricated an injectable folic-acid conjugated PDA hydrogel with carbon quantum dots/ZnO(C/ZnO) to give DFT-C/ZnO hydrogel Fig. 18a., as the CQDs and PDA exhibit their photothermal and ROS generations properties for rapid bacterial eradication Fig. 18b. Consequently, the photothermal effect and reactive ROS generation assay showed an increase in temperature and even the good temperature stability of DFT and DFT-C/ZnO–hydrogels under 808 nm, 15 min after assessing the cycles Fig. 18c. Although earlier exposure to the red light (606 nm) yielded no effect. Furthermore, an examination of the ROS with, 2,7-dichlorodihydrofluorescein diacetate (DCFH-DA), a ROS fluorescent probe under mixed light irradiation showed DFT-C/ZnO–hydrogels generation of an enormous amount of ROS. Also, in vitro antibacterial test indicated that DFT-C/ZnO–hydrogels under mixed-light, 15 min displayed a near-complete (99.9996% and 99.9998%) eradication of the S. aureus and the E. coli respectively revealing how the composite material was endowed by their photothermal property and ROS generations ability for the bacterial inhibition as this was evident in bacterial morphology from the SEM images Fig. 18d [196]. A clear indication of the fact that NIR irradiation is capable of motivating ROS generation for subsequent destruction of the bacteria membrane integrity and death [104,197,198].
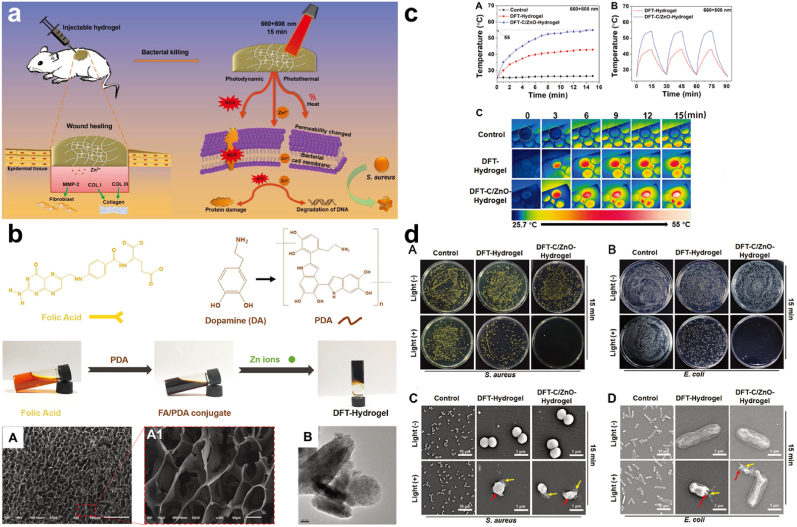
Fig. 18.
(a) Diagrammatic illustration of antibacterial effect of DFT-C/ZnO-hydrogel. (b) Images and SEM images of DFT-hydrogel, TEM images of C/ZnO. (c)Photothermal curve with the temperature cooling profile and thermographic images of the various hydrogels. (d) Representation of the antibacterial activity of DFT-hydrogel, DFT-C/ZnO-hydrogel against S. aureus and E. coli, with/without 15 min of mixed light irradiation and SEM images of the bacteria. With permission, reprinted from Ref. [196]. Copyright 2019 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
5. Impact of coating and shaking on the PDA antibacterial effect
PDA can be prepared by shaking and this has proven to result in nanoscale surface roughness thus providing the surface area for oxidization and release of H2O2 further enabling the atmospheric exchange of oxygen [36,41]. Also, shaken PDA samples have been shown to possess higher content of (Nitrogen) N, hence a lower N/C ratio compared to their unshaken counterpart with equivalent PDA composition [27,43]. Conversely, coating with PDA can be done either in one step which involves a single layer, or a two-step which results in a double layer. Studies indicate that the initial step enables the stabilization of the base layer while the second or last step enables the adhesion of dopamine to previous PDA which can further enable the effective generation of H2O2 [41,43,209]. The antibacterial effect is greatly influenced by the combined effect of shaking and double layer coating. Forooshani reported that shaken two-step coated (20-2S) exhibited a high log reduction value (LRV) completely eradicating E. coli after 24 h. This phenomenon of 20–2S was evident against other forms of bacteria compared to the other forms of PDA formulations. Hence double coating and shaking conditions pose to be more effective in the killing of the bacteria [41,43]. Though studies reveal H2O2 didn't impact the gram-positive bacteria that much yet, enough time for H2O2 release was capable of completely killing such bacteria. Shaking in itself allows the oxidation of catechol which further allows for many depositions to create a much thicker film. Batul et al. further emphasized that the rough nature of a PDA ensured maximum contact with a great bactericidal activity which can be attributed to the vigorous shaking process [131,135,136]. Also, one-step coating resulted in a PDA thickness of about 34 nm and that of the two steps yielded a PDA of a thickness of 72 nm. Which explains the fact that the base layer of the two-step coating allows PDA deposition in the second step [27,210,211]. Finally, two-step coating can yield a large amount of H2O2 compared to those that are single coated though the coating alone does not guarantee enough increment in H2O2 provided other conditions meant for the purpose are set in place [212].
6. Cytotoxicity and biocompatibility
For a feasible biomedical application of any material, nanoparticle, or drug, the evaluation of the toxicity and biocompatibility is very key and of much concern. Although studies show the PDA is capable of generating semiquinone radicals in solutions and under aerobic conditions produces O2•─ which can later lead to the formation of •OH through the Haber Weiss and Fenton reaction [[213], [214], [215]]. PDAs have been proven to possess negligible toxicity when compared to metals, and also good biocompatibility which is confirmed by their nature, that can be likened to melanin [50,216]. HA-Cu/PDA exhibited good biocompatibility enabling collagen deposition and granulation formation for wound healing [207]. Also, colistin-loaded PDA with silver displayed good biocompatibility and negligible toxicity. Proving that, apart from PDA's easy modification, excellent quenching effect, high photothermal conversion efficiency, etc. they are biocompatible, and for this reason, they are employed in numerous composite materials [206,217,218].
7. Future perspective and conclusion
This article gives an overview of the nature, properties, and mechanisms of polydopamine for antibacterial inhibition, the recent approaches or modes of inhibiting bacteria, and their infections. PDAs are known for their adhesion property but apart from this well-known ability, the PDA has been utilized and employed in several ways together with their combined utilization with other materials such as the nanoparticle, through the employment of some conditions such as photothermal therapy with other microbial applications, etc. Consequently, portraying great potential wielded by this material and a promising outcome for the future as far as antibiotic resistance bacteria and organisms are concerned. Much investigation and research into this material will facilitate and provide an understanding of their nature, together with all the conditions that surround their synthesis process. Further research will also bring into light the complete harnessing of all the potentials held within this material, together with the toxicologically related issues concerning its application and usage in humans. These will allow for maximum usage and the ensuring of a cost-effective approach for the complete eradication of antibiotic-resistant organisms.
Declaration of competing interest
All authors agree to be published. All authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was financially supported by the National Natural Science Foundation of China (21977081), Zhejiang Provincial Natural Science of Foundation of China (LZ19H180001), University of Chinese Academy of Science (WIBEZD2017001-03), Wenzhou Science and Technology Plan Project (ZG2021035), Wenzhou Medical University (KYYW201901). Zhejiang Qianjiang Talent Plan (QJD20020224), J. Yoon thanks to Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (2021R1A6A1A10039823).
Footnotes
All authors have approved the final version of this manuscript.
Contributor Information
Jianliang Shen, Email: shenjl@wiucas.ac.cn.
Juyoung Yoon, Email: jyoon@ewha.ac.kr.
References
- 1.Hall-Stoodley L., Costerton J.W., Stoodley P. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 2.Zou Y., Zhang Y., Yu Q., Chen H. J. Mater. Sci. Technol. 2021;70:24–38. [Google Scholar]
- 3.Amin Yavari S., Loozen L., Paganelli F.L., Bakhshandeh S., Lietaert K., Groot J.A., Fluit A.C., Boel C., Alblas J., Vogely H.C. ACS Appl. Mater. Interfaces. 2016;8:17080–17089. doi: 10.1021/acsami.6b03152. [DOI] [PubMed] [Google Scholar]
- 4.Busscher H.J., van der Mei H.C., Subbiahdoss G., Jutte P.C., van den Dungen J.J., Zaat S.A., Schultz M.J., Grainger D.W. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3004528. 153rv110. [DOI] [PubMed] [Google Scholar]
- 5.Jia Z., Xiu P., Xiong P., Zhou W., Cheng Y., Wei S., Zheng Y., Xi T., Cai H., Liu Z. ACS Appl. Mater. Interfaces. 2016;8:28495–28510. doi: 10.1021/acsami.6b10473. [DOI] [PubMed] [Google Scholar]
- 6.Lopez-Heredia M.A., Sohier J., Gaillard C., Quillard S., Dorget M., Layrolle P. Biomaterials. 2008;29:2608–2615. doi: 10.1016/j.biomaterials.2008.02.021. [DOI] [PubMed] [Google Scholar]
- 7.Cosnier S., Holzinger M. Wiley VCH; Weinheim: 2010. Electropolymerization; p. 189. [Google Scholar]
- 8.Nguyen V.-N., Zhao Z., Tang B.Z., Yoon J. Chem. Soc. Rev. 2022;51:3324. doi: 10.1039/d1cs00647a. [DOI] [PubMed] [Google Scholar]
- 9.Wu X., Yang M., Kim J.S., Wang R., Kim G., Ha J., Kim H., Cho Y., Nam K.T., Yoon J. Angew. Chem. Int. Ed. 2022;134 doi: 10.1002/anie.202200808. [DOI] [PubMed] [Google Scholar]
- 10.Langer R. Science. 2001;293:58–59. doi: 10.1126/science.1063273. [DOI] [PubMed] [Google Scholar]
- 11.Norde W. Marcel Dekker; New York: 2000. [Google Scholar]
- 12.Gao Y., Cranston R. Textil. Res. J. 2008;78:60–72. [Google Scholar]
- 13.Gorjanc M., Kovač F., Gorenšek M. Textil. Res. J. 2012;82:62–69. [Google Scholar]
- 14.Hou A., Zhou M., Wang X. Carbohydr. Polym. 2009;75:328–332. [Google Scholar]
- 15.Raza Z.A., Rehman A., Mohsin M., Bajwa S.Z., Anwar F., Naeem A., Ahmad N. J. Clean. Prod. 2015;101:377–386. [Google Scholar]
- 16.Bell E., Gosline J. Oceanogr. Lit. Rev. 1998;6:987. [Google Scholar]
- 17.McGinness J., Corry P., Proctor P. Science. 1974;183:853–855. doi: 10.1126/science.183.4127.853. [DOI] [PubMed] [Google Scholar]
- 18.Simon J.D., Peles D., Wakamatsu K., Ito S. Pigment Cell Melanoma Res. 2009;22:563–579. doi: 10.1111/j.1755-148X.2009.00610.x. [DOI] [PubMed] [Google Scholar]
- 19.Gademann K. Chembiochem. 2005;6:913–919. doi: 10.1002/cbic.200400343. [DOI] [PubMed] [Google Scholar]
- 20.Lee H., Scherer N.F., Messersmith P.B. Proc. Natl. Acad. Sci. USA. 2006;103:12999–13003. doi: 10.1073/pnas.0605552103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waite J.H., Tanzer M.L. Science. 1981;212:1038–1040. doi: 10.1126/science.212.4498.1038. [DOI] [PubMed] [Google Scholar]
- 22.Hossain M.M., Herrmann A.S., Hegemann D. Plasma Process. Polym. 2006;3:299–307. [Google Scholar]
- 23.Stokes A.H., Hastings T.G., Vrana K.E. J. Neurosci. Res. 1999;55:659–665. doi: 10.1002/(SICI)1097-4547(19990315)55:6<659::AID-JNR1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 24.Sureshkumar M., Siswanto D.Y., Chen Y.C., Lee C.K., Wang M.J. J. Polym. Sci. B Polym. Phys. 2013;51:303–310. [Google Scholar]
- 25.Gan D., Xu T., Xing W., Ge X., Fang L., Wang K., Ren F., Lu X. Adv. Funct. Mater. 2019;29 [Google Scholar]
- 26.Iqbal Z., Lai E.P., Avis T.J. J. Mater. Chem. 2012;22:21608–21612. [Google Scholar]
- 27.Lee H., Dellatore S.M., Miller W.M., Messersmith P.B. Science. 2007;318:426–430. doi: 10.1126/science.1147241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waite J.H. Integr. Comp. Biol. 2002;42:1172–1180. doi: 10.1093/icb/42.6.1172. [DOI] [PubMed] [Google Scholar]
- 29.Zhou J., Wang P., Wang C., Goh Y.T., Fang Z., Messersmith P.B., Duan H. ACS Nano. 2015;9:6951–6960. doi: 10.1021/acsnano.5b01138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji Y., Ji T., Liang K., Zhu L. J. Mater. Sci. Mater. Med. 2016;27:30. doi: 10.1007/s10856-015-5649-2. [DOI] [PubMed] [Google Scholar]
- 31.Sushomasri M., Maji Himangshu S., Pranabesh C., Sujata G.D. Int. J. Pharm. Biomed. Res. 2010;4:70–75. [Google Scholar]
- 32.Fu Y., Yang L., Zhang J., Hu J., Duan G., Liu X., Li Y., Gu Z. Mater. Horiz. 2021;8:1618–1633. doi: 10.1039/d0mh01985b. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Y.-W., Sun Y.-J., Wang J.-L., Yu B.-R. Rare Met. 2022;41:499–518. [Google Scholar]
- 34.Forte L., Torricelli P., Bonvicini F., Boanini E., Gentilomi G.A., Lusvardi G., Della Bella E., Fini M., Nepita E.V., Bigi A. J. Inorg. Biochem. 2018;178:43–53. doi: 10.1016/j.jinorgbio.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 35.Mallinson D., Mullen A.B., Lamprou D.A. J. Mater. Sci. 2018;53:3198–3209. doi: 10.1007/s10853-017-1806-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernsmann F., Ponche A., Ringwald C., Hemmerle J., Raya J., Bechinger B., Voegel J.-C., Schaaf P., Ball V. J. Phys. Chem. C. 2009;113:8234–8242. [Google Scholar]
- 37.Zhu L.-P., Jiang J.-H., Zhu B.-K., Xu Y.-Y. Colloids Surf., B: Bionterfaces. 2011;86:111–118. doi: 10.1016/j.colsurfb.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 38.Wei Q., Li B., Yi N., Su B., Yin Z., Zhang F., Li J., Zhao C. J. Biomed. Mater. Res. 2011;96:38–45. doi: 10.1002/jbm.a.32956. [DOI] [PubMed] [Google Scholar]
- 39.He S., Zhou P., Wang L., Xiong X., Zhang Y., Deng Y., Wei S. J. R. Soc. Interface. 2014;11 doi: 10.1098/rsif.2014.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang C.-J., Hung S.-T. J. Nanosci. Nanotechnol. 2010;10:4674–4678. doi: 10.1166/jnn.2010.1691. [DOI] [PubMed] [Google Scholar]
- 41.Su L., Yu Y., Zhao Y., Liang F., Zhang X. Sci. Rep. 2016;6:1–8. doi: 10.1038/srep24420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding Y., Weng L.-T., Yang M., Yang Z., Lu X., Huang N., Leng Y. Langmuir. 2014;30:12258–12269. doi: 10.1021/la5026608. [DOI] [PubMed] [Google Scholar]
- 43.Kord Forooshani P., Polega E., Thomson K., Bhuiyan M., Akram S., Pinnaratip R., Trought M., Kendrick C., Gao Y., Perrine K.A. Front. Chem. 2019;7:631. doi: 10.3389/fchem.2019.00631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang B.-B., Quan Y.-H., Xu Z.-M., Zhao Q. Prog. Org. Coating. 2020;149 [Google Scholar]
- 45.Liu Y., Zheng Y., Hayes B. Sci. China Mater. 2017;60:377–391. [Google Scholar]
- 46.Xu N., Wei F., Liu X., Jiang L., Cai H., Li Z., Yu M., Wu F., Liu Z. Spine. 2016;41:E50–E54. doi: 10.1097/BRS.0000000000001179. [DOI] [PubMed] [Google Scholar]
- 47.Yang J., Cai H., Lv J., Zhang K., Leng H., Sun C., Wang Z., Liu Z. Spine. 2014;39:E486–E492. doi: 10.1097/BRS.0000000000000211. [DOI] [PubMed] [Google Scholar]
- 48.Zhang T., Zhou W., Jia Z., Wei Q., Fan D., Yan J., Yin C., Cheng Y., Cai H., Liu X. Sci. China Mater. 2018;61:579–592. [Google Scholar]
- 49.Jiang L., Jin G., Kang J., Yu L., Yoon W., Lim M., Lee M., Jin D. J. Wuhan Univ. Technol.-Materials Sci. Ed. 2014;29:197–200. [Google Scholar]
- 50.Ju K.-Y., Lee Y., Lee S., Park S.B., Lee J.-K. Biomacromolecules. 2011;12:625–632. doi: 10.1021/bm101281b. [DOI] [PubMed] [Google Scholar]
- 51.Postma A., Yan Y., Wang Y., Zelikin A.N., Tjipto E., Caruso F. Chem. Mater. 2009;21:3042–3044. [Google Scholar]
- 52.Yang W., Liu C., Chen Y. Langmuir. 2018;34:3565–3571. doi: 10.1021/acs.langmuir.7b03143. [DOI] [PubMed] [Google Scholar]
- 53.Jiang J., Zhu L., Zhu L., Zhu B., Xu Y. Langmuir. 2011;27:14180–14187. doi: 10.1021/la202877k. [DOI] [PubMed] [Google Scholar]
- 54.Lynge M.E., van der Westen R., Postma A., Städler B. Nanoscale. 2011;3:4916–4928. doi: 10.1039/c1nr10969c. [DOI] [PubMed] [Google Scholar]
- 55.Zhang W., Yang F.K., Han Y., Gaikwad R., Leonenko Z., Zhao B. Biomacromolecules. 2013;14:394–405. doi: 10.1021/bm3015768. [DOI] [PubMed] [Google Scholar]
- 56.Asha A.B., Chen Y., Narain R. Chem. Soc. Rev. 2021;50:11668–11683. doi: 10.1039/d1cs00658d. [DOI] [PubMed] [Google Scholar]
- 57.Aslan S., Loebick C.Z., Kang S., Elimelech M., Pfefferle L.D., Van Tassel P.R. Nanoscale. 2010;2:1789–1794. doi: 10.1039/c0nr00329h. [DOI] [PubMed] [Google Scholar]
- 58.Rodriguez-Valverde M.A., Ruiz-Cabello F.J.M., Cabrerizo-Vilchez M.A. Soft Matter. 2011;7:53–56. [Google Scholar]
- 59.Li J., Tan D., Zhang X., Tan H., Ding M., Wan C., Fu Q. Colloids Surf. B Biointerfaces. 2010;78:343–350. doi: 10.1016/j.colsurfb.2010.03.027. [DOI] [PubMed] [Google Scholar]
- 60.Sileika T.S., Kim H.-D., Maniak P., Messersmith P.B. ACS Appl. Mater. Interfaces. 2011;3:4602–4610. doi: 10.1021/am200978h. [DOI] [PubMed] [Google Scholar]
- 61.Chen D., Wang C., Nie X., Li S., Li R., Guan M., Liu Z., Chen C., Wang C., Shu C. Adv. Funct. Mater. 2014;24:6621–6628. [Google Scholar]
- 62.Hu D., Liu C., Song L., Cui H., Gao G., Liu P., Sheng Z., Cai L. Nanoscale. 2016;8:17150–17158. doi: 10.1039/c6nr05502h. [DOI] [PubMed] [Google Scholar]
- 63.Jiang T., Li Y.-M., Lv Y., Cheng Y.-J., He F., Zhuo R.-X. Colloids Surf. B Biointerfaces. 2013;111:542–548. doi: 10.1016/j.colsurfb.2013.06.054. [DOI] [PubMed] [Google Scholar]
- 64.Malinovskaya Y., Melnikov P., Baklaushev V., Gabashvili A., Osipova N., Mantrov S., Ermolenko Y., Maksimenko O., Gorshkova M., Balabanyan V. Int. J. Pharm. 2017;524:77–90. doi: 10.1016/j.ijpharm.2017.03.049. [DOI] [PubMed] [Google Scholar]
- 65.Ponzio F., Bertani P., Ball V. J. Colloid Interface Sci. 2014;431:176–179. doi: 10.1016/j.jcis.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 66.Xi J., Da L., Yang C., Chen R., Gao L., Fan L., Han J. Int. J. Nanomed. 2017;12:3331. doi: 10.2147/IJN.S132270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zheng W., Fan H., Wang L., Jin Z. Langmuir. 2015;31:11671–11677. doi: 10.1021/acs.langmuir.5b02757. [DOI] [PubMed] [Google Scholar]
- 68.Fang J., Chen Y.-C. Curr. Pharmaceut. Des. 2013;19:6622–6634. doi: 10.2174/1381612811319370006. [DOI] [PubMed] [Google Scholar]
- 69.Wu M.-C., Deokar A.R., Liao J.-H., Shih P.-Y., Ling Y.-C. ACS Nano. 2013;7:1281–1290. doi: 10.1021/nn304782d. [DOI] [PubMed] [Google Scholar]
- 70.Li Y., Liu X., Tan L., Cui Z., Yang X., Zheng Y., Yeung K.W.K., Chu P.K., Wu S. Adv. Funct. Mater. 2018;28 [Google Scholar]
- 71.Hong S., Na Y.S., Choi S., Song I.T., Kim W.Y., Lee H. Adv. Funct. Mater. 2012;22:4711–4717. [Google Scholar]
- 72.Gao G., Jiang Y.-W., Jia H.-R., Wu F.-G. Biomaterials. 2019;188:83–95. doi: 10.1016/j.biomaterials.2018.09.045. [DOI] [PubMed] [Google Scholar]
- 73.Liu Y., Ai K., Liu J., Deng M., He Y., Lu L. Adv. Mater. 2013;25:1353–1359. doi: 10.1002/adma.201204683. [DOI] [PubMed] [Google Scholar]
- 74.Zou Y., Chen X., Yang P., Liang G., Yang Y., Gu Z., Li Y. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abb4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ju E., Dong K., Liu Z., Pu F., Ren J., Qu X. Adv. Funct. Mater. 2015;25:1574–1580. [Google Scholar]
- 76.Lei W., Ren K., Chen T., Chen X., Li B., Chang H., Ji J. Adv. Mater. Interfac. 2016;3 [Google Scholar]
- 77.Liu Y., Ai K., Lu L. Chem. Rev. 2014;114:5057–5115. doi: 10.1021/cr400407a. [DOI] [PubMed] [Google Scholar]
- 78.Natan M., Edin F., Perkas N., Yacobi G., Perelshtein I., Segal E., Homsy A., Laux E., Keppner H., Rask-Andersen H. Adv. Funct. Mater. 2016;26:2473–2481. [Google Scholar]
- 79.Sun J.-Y., Zhao X., Illeperuma W.R., Chaudhuri O., Oh K.H., Mooney D.J., Vlassak J.J., Suo Z. Nature. 2012;489:133–136. doi: 10.1038/nature11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fu L., Zhang J., Yang G. Carbohydr. Polym. 2013;92:1432–1442. doi: 10.1016/j.carbpol.2012.10.071. [DOI] [PubMed] [Google Scholar]
- 81.Kang Y.J., Chun S.-J., Lee S.-S., Kim B.-Y., Kim J.H., Chung H., Lee S.-Y., Kim W. ACS Nano. 2012;6:6400–6406. doi: 10.1021/nn301971r. [DOI] [PubMed] [Google Scholar]
- 82.Klemm D., Kramer F., Moritz S., Lindström T., Ankerfors M., Gray D., Dorris A. Angew. Chem. Int. Ed. 2011;50:5438–5466. doi: 10.1002/anie.201001273. [DOI] [PubMed] [Google Scholar]
- 83.Jiang Q., Derami H.G., Ghim D., Cao S., Jun Y.-S., Singamaneni S. J. Mater. Chem. 2017;5:18397–18402. [Google Scholar]
- 84.Wu X., Cao S., Ghim D., Jiang Q., Singamaneni S., Jun Y.-S. Nano Energy. 2021;79 [Google Scholar]
- 85.Cheng L., Wang C., Feng L., Yang K., Liu Z. Chem. Rev. 2014;114:10869–10939. doi: 10.1021/cr400532z. [DOI] [PubMed] [Google Scholar]
- 86.Fielding G.A., Roy M., Bandyopadhyay A., Bose S. Acta Biomater. 2012;8:3144–3152. doi: 10.1016/j.actbio.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stanić V., Dimitrijević S., Antonović D.G., Jokić B.M., Zec S.P., Tanasković S.T., Raičević S. Appl. Surf. Sci. 2014;290:346–352. [Google Scholar]
- 88.Chen W., Liu Y., Courtney H.S., Bettenga M., Agrawal C.M., Bumgardner J.D., Ong J.L. Biomaterials. 2006;27:5512–5517. doi: 10.1016/j.biomaterials.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 89.Mitrano D.M., Rimmele E., Wichser A., Erni R., Height M., Nowack B. ACS Nano. 2014;8:7208–7219. doi: 10.1021/nn502228w. [DOI] [PubMed] [Google Scholar]
- 90.Zimmerli W., Waldvogel F.A., Vaudaux P., Nydegger U.E. J. Infect. Dis. 1982;146:487–497. doi: 10.1093/infdis/146.4.487. [DOI] [PubMed] [Google Scholar]
- 91.Fan Y.J., Pham M.T., Huang C.J. Langmuir. 2019;35:1642–1651. doi: 10.1021/acs.langmuir.8b01730. [DOI] [PubMed] [Google Scholar]
- 92.McDonnell G. 2009. PATAI'S Chemistry of Functional Groups; pp. 1–34. [Google Scholar]
- 93.Wang Y., Branicky R., Noë A., Hekimi S. J. Cell Biol. 2018;217:1915–1928. doi: 10.1083/jcb.201708007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Abreu I.A., Cabelli D.E. Biochim. Biophys. Acta, Proteins Proteomics. 2010;1804:263–274. doi: 10.1016/j.bbapap.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 95.Afanas' ev I. Curr. Med. Chem. 2005;12:2731–2739. doi: 10.2174/092986705774462941. [DOI] [PubMed] [Google Scholar]
- 96.Inoue M., Sato E.F., Park A.-M., Nishikawa M., Kasahara E., Miyoshi M., Ochi A., Utsumi K. Free Radic. Res. 2000;33:757–770. doi: 10.1080/10715760000301281. [DOI] [PubMed] [Google Scholar]
- 97.Radi R. J. Biol. Chem. 2013;288:26464–26472. doi: 10.1074/jbc.R113.472936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Obeng E., Ding F., He X., Shen J. Dyes Pigments. 2021 [Google Scholar]
- 99.Wu S., Xu C., Zhu Y., Zheng L., Zhang L., Hu Y., Yu B., Wang Y., Xu F.J. Adv. Funct. Mater. 2021;31 [Google Scholar]
- 100.Dryden M. Curr. Opin. Infect. Dis. 2017;30:143–149. doi: 10.1097/QCO.0000000000000350. [DOI] [PubMed] [Google Scholar]
- 101.Sies H. Redox Biol. 2017;11:613–619. doi: 10.1016/j.redox.2016.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Y., Wan J., Miron R.J., Zhao Y., Zhang Y. Nanoscale. 2016;8:11143–11152. doi: 10.1039/c6nr01114d. [DOI] [PubMed] [Google Scholar]
- 103.Xie X., Mao C., Liu X., Tan L., Cui Z., Yang X., Zhu S., Li Z., Yuan X., Zheng Y. ACS Cent. Sci. 2018;4:724–738. doi: 10.1021/acscentsci.8b00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yuan Z., Tao B., He Y., Mu C., Liu G., Zhang J., Liao Q., Liu P., Cai K. Biomaterials. 2019;223 doi: 10.1016/j.biomaterials.2019.119479. [DOI] [PubMed] [Google Scholar]
- 105.Dhand C., Ong C.Y., Dwivedi N., Varadarajan J., Halleluyah Periayah M., Jianyang Lim E., Mayandi V., Goh E.T.L., Najjar R.P., Chan L.W., Beuerman R.W., Foo L.L., Loh X.J., Lakshminarayanan R. ACS Biomater. Sci. Eng. 2020;6:3162–3173. doi: 10.1021/acsbiomaterials.0c00229. [DOI] [PubMed] [Google Scholar]
- 106.Ben-Sasson M., Zodrow K.R., Genggeng Q., Kang Y., Giannelis E.P., Elimelech M. Environ. Sci. Technol. 2014;48:384–393. doi: 10.1021/es404232s. [DOI] [PubMed] [Google Scholar]
- 107.Yin J., Deng B. J. Membr. Sci. 2015;479:256–275. [Google Scholar]
- 108.Xu K., Wu J., Fang Q., Bai L., Duan J., Li J., Xu H., Hui A., Hao L., Xuan S. J. Membr. Sci. 2020;398 [Google Scholar]
- 109.He W., Guo X., Zheng J., Xu J., Hayat T., Alharbi N.S., Zhang M. Inorg. Chem. 2019;58:7255–7266. doi: 10.1021/acs.inorgchem.9b00288. [DOI] [PubMed] [Google Scholar]
- 110.Cai R., Tao G., He H., Guo P., Yang M., Ding C., Zuo H., Wang L., Zhao P., Wang Y. Materials. 2017;10:967. doi: 10.3390/ma10080967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Xu H., Shi X., Lv Y., Mao Z. Textil. Res. J. 2013;83:321–326. [Google Scholar]
- 112.Xu X., Liu X., Tan L., Cui Z., Yang X., Zhu S., Li Z., Yuan X., Zheng Y., Yeung K.W.K. Acta Biomater. 2018;77:352–364. doi: 10.1016/j.actbio.2018.07.030. [DOI] [PubMed] [Google Scholar]
- 113.Ul-Islam M., Khan T., Khattak W.A., Park J.K. Cellulose. 2013;20:589–596. [Google Scholar]
- 114.Lin Z., Liu L., Wang W., Jia L., Shen Y., Zhang X., Ge D., Shi W., Sun Y. Biomater. Sci. 2021;9:5951–5964. doi: 10.1039/d1bm00622c. [DOI] [PubMed] [Google Scholar]
- 115.Baym M., Lieberman T.D., Kelsic E.D., Chait R., Gross R., Yelin I., Kishony R. Science. 2016;353:1147–1151. doi: 10.1126/science.aag0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Alekshun M.N., Levy S.B. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 117.Bing W., Chen Z., Sun H., Shi P., Gao N., Ren J., Qu X. Nano Res. 2015;8:1648–1658. [Google Scholar]
- 118.Xu X.X., Nie F.L., Wang Y.B., Zhang J.X., Zheng W., Li L., Zheng Y.F. Acta Biomater. 2012;8:886–896. doi: 10.1016/j.actbio.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 119.Xiao J., Chen S., Yi J., Zhang H., Ameer G.A. Adv. Funct. Mater. 2017;27 doi: 10.1002/adfm.201604872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Guo L., Panderi I., Yan D.D., Szulak K., Li Y., Chen Y.T., Ma H., Niesen D.B., Seeram N., Ahmed A., Yan B., Pantazatos D., Lu W. ACS Nano. 2013;7:8780–8793. doi: 10.1021/nn403202w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Karkhanechi H., Takagi R., Matsuyama H. Desalination. 2014;336:87–96. [Google Scholar]
- 122.Patel K., Singh N., Yadav J., Nayak J.M., Sahoo S.K., Lata J., Chand D., Kumar S., Kumar R. Phys. Chem. Chem. Phys. 2018;20:5744–5755. doi: 10.1039/c7cp08406d. [DOI] [PubMed] [Google Scholar]
- 123.Wu W., Yang Y., Wang L., Xu T., Wang R. RSC Adv. 2021;11:18252–18258. doi: 10.1039/d0ra10249k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Liu C., Yao W., Tian M., Wei J., Song Q., Qiao W. Biomaterials. 2018;179:83–95. doi: 10.1016/j.biomaterials.2018.06.037. [DOI] [PubMed] [Google Scholar]
- 125.Li L., Yang L., Liao Y., Yu H., Liang Z., Zhang B., Lan X., Luo R., Wang Y. Chem. Eng. J. 2020;402 [Google Scholar]
- 126.Cao Z., Jiang S. Nano Today. 2012;7:404–413. [Google Scholar]
- 127.Ishizaki T., Saito N., Takai O. Langmuir. 2010;26:8147–8154. doi: 10.1021/la904447c. [DOI] [PubMed] [Google Scholar]
- 128.Alonso C., Rubio L., Touriño S., Martí M., Barba C., Fernández-Campos F., Coderch L., Parra J.L. Free Radic. Biol. Med. 2014;75:149–155. doi: 10.1016/j.freeradbiomed.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 129.Bao X., Zhao J., Sun J., Hu M., Yang X. ACS Nano. 2018;12:8882–8892. doi: 10.1021/acsnano.8b04022. [DOI] [PubMed] [Google Scholar]
- 130.Jodko-Piórecka K., Litwinienko G. Free Radic. Biol. Med. 2015;83:1–11. doi: 10.1016/j.freeradbiomed.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 131.Kord Forooshani P., Lee B.P. J. Polym. Sci.: Polym. Chem. 2017;55:9–33. doi: 10.1002/pola.28368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Liu H., Qu X., Tan H., Song J., Lei M., Kim E., Payne G.F., Liu C. Acta Biomater. 2019;88:181–196. doi: 10.1016/j.actbio.2019.02.032. [DOI] [PubMed] [Google Scholar]
- 133.Meng H., Forooshani P.K., Joshi P.U., Osborne J., Mi X., Meingast C., Pinnaratip R., Kelley J., Narkar A., He W. Acta Biomater. 2019;83:109–118. doi: 10.1016/j.actbio.2018.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Song J., Liu H., Lei M., Tan H., Chen Z., Antoshin A., Payne G.F., Qu X., Liu C. ACS Appl. Mater. Interfaces. 2020;12:8915–8928. doi: 10.1021/acsami.9b22339. [DOI] [PubMed] [Google Scholar]
- 135.Batul R., Tamanna T., Khaliq A., Yu A. Biomater. Sci. 2017;5:1204–1229. doi: 10.1039/c7bm00187h. [DOI] [PubMed] [Google Scholar]
- 136.Chang C.C., Kolewe K.W., Li Y., Kosif I., Freeman B.D., Carter K.R., Schiffman J.D., Emrick T. Biomater. Sci. 2016;3 doi: 10.1002/admi.201500521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jia R., Tian W., Bai H., Zhang J., Wang S., Zhang J. Adv. Healthc. Mater. 2019;8 doi: 10.1002/adhm.201801591. [DOI] [PubMed] [Google Scholar]
- 138.Liu X., Yan Z., Zhang Y., Liu Z., Sun Y., Ren J., Qu X. ACS Nano. 2019;13:5222–5230. doi: 10.1021/acsnano.8b09501. [DOI] [PubMed] [Google Scholar]
- 139.Mao C., Xiang Y., Liu X., Zheng Y., Yeung K.W.K., Cui Z., Yang X., Li Z., Liang Y., Zhu S. ACS Appl. Mater. Interfaces. 2019;11:17902–17914. doi: 10.1021/acsami.9b05787. [DOI] [PubMed] [Google Scholar]
- 140.Sun H., Gao N., Dong K., Ren J., Qu X. ACS Nano. 2014;8:6202–6210. doi: 10.1021/nn501640q. [DOI] [PubMed] [Google Scholar]
- 141.Tong W., Xiong Y., Duan S., Ding X., Xu F.-J. Biomater. Sci. 2019;7:1905–1918. doi: 10.1039/c8bm01483c. [DOI] [PubMed] [Google Scholar]
- 142.Yeroslavsky G., Richman M., Dawidowicz L.-o., Rahimipour S. Chem. Commun. 2013;49:5721–5723. doi: 10.1039/c3cc37762h. [DOI] [PubMed] [Google Scholar]
- 143.Jia Z., Xiu P., Roohani-Esfahani S.-I., Zreiqat H., Xiong P., Zhou W., Yan J., Cheng Y., Zheng Y. ACS Appl. Mater. Interfaces. 2019;11:4447–4469. doi: 10.1021/acsami.8b20429. [DOI] [PubMed] [Google Scholar]
- 144.Raza Z.A., Rehman A., Anwar F., Usman A. Bull. Mater. Sci. 2016;39:391–396. [Google Scholar]
- 145.Gao C., Wang Y., Han F., Yuan Z., Li Q., Shi C., Cao W., Zhou P., Xing X., Li B. J. Mater. Chem. B. 2017;5:9326–9336. doi: 10.1039/c7tb02436c. [DOI] [PubMed] [Google Scholar]
- 146.Wu C., Zhang G., Xia T., Li Z., Zhao K., Deng Z., Guo D., Peng B. Mater. Sci. Eng. C. Mater. Biol. Appl. 2015;55:155–165. doi: 10.1016/j.msec.2015.05.032. [DOI] [PubMed] [Google Scholar]
- 147.Choi S.-H., Jang Y.-S., Jang J.-H., Bae T.-S., Lee S.-J., Lee M.-H. J. Appl. Biomater. Funct. Mater. 2019;17 doi: 10.1177/2280800019847067. [DOI] [PubMed] [Google Scholar]
- 148.Fang Q., Xu K., Zhang J., Xiong Q., Duan J., Xuan S. Materials. 2020;13 doi: 10.3390/ma13173872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Konishi M., Tabata Y., Kariya M., Hosseinkhani H., Suzuki A., Fukuhara K., Mandai M., Takakura K., Fujii S. J. Contr. Release. 2005;103:7–19. doi: 10.1016/j.jconrel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 150.Dragan E.S., Humelnicu D., Dinu M.V., Olariu R.I. Chem. Eng. J. 2017;330:675–691. [Google Scholar]
- 151.Ganguly K., Chaturvedi K., More U.A., Nadagouda M.N., Aminabhavi T.M. J. Contr. Release. 2014;193:162–173. doi: 10.1016/j.jconrel.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 152.Liu L., Feng X., Pei Y., Wang J., Ding J., Chen L. Mater. Sci. Eng. 2018;82:25–28. doi: 10.1016/j.msec.2017.08.045. [DOI] [PubMed] [Google Scholar]
- 153.Zheng Y., Cheng Y., Chen J., Ding J., Li M., Li C., Wang J.-c., Chen X. ACS Appl. Mater. Interfaces. 2017;9:3487–3496. doi: 10.1021/acsami.6b15245. [DOI] [PubMed] [Google Scholar]
- 154.Li S., Dong S., Xu W., Tu S., Yan L., Zhao C., Ding J., Chen X. Adv. Sci. 2018;5 doi: 10.1002/advs.201700527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Caliari S.R., Burdick J.A. Nat. Methods. 2016;13:405–414. doi: 10.1038/nmeth.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Maleki A., He J., Bochani S., Nosrati V., Shahbazi M.-A., Guo B. ACS Nano. 2021;15:18895–18930. doi: 10.1021/acsnano.1c08334. [DOI] [PubMed] [Google Scholar]
- 157.Li S., Chen A., Chen Y., Yang Y., Zhang Q., Luo S., Ye M., Zhou Y., An Y., Huang W. Chem. Eng. J. 2020;402 [Google Scholar]
- 158.Han L., Lu X., Liu K., Wang K., Fang L., Weng L.T., Zhang H., Tang Y., Ren F., Zhao C., Sun G., Liang R., Li Z. ACS Nano. 2017;11:2561–2574. doi: 10.1021/acsnano.6b05318. [DOI] [PubMed] [Google Scholar]
- 159.Wang Y., Zhou P., Xiao D., Zhu Y., Zhong Y., Zhang J., Sui X., Feng X., Xu H., Mao Z. Carbohydr. Polym. 2019;221:202–208. doi: 10.1016/j.carbpol.2019.05.082. [DOI] [PubMed] [Google Scholar]
- 160.Zeng Q., Qian Y., Huang Y., Ding F., Qi X., Shen J. Bioact. Mater. 2021;6:2647–2657. doi: 10.1016/j.bioactmat.2021.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tong X., Qi X., Mao R., Pan W., Zhang M., Wu X., Chen G., Shen J., Deng H., Hu R. Carbohydr. Polym. 2020;245 doi: 10.1016/j.carbpol.2020.116585. [DOI] [PubMed] [Google Scholar]
- 162.Qi X., Zeng Q., Tong X., Su T., Xie L., Yuan K., Xu J., Shen J. J. Hazard Mater. 2021;402 doi: 10.1016/j.jhazmat.2020.123359. [DOI] [PubMed] [Google Scholar]
- 163.Gan D., Xing W., Jiang L., Fang J., Zhao C., Ren F., Fang L., Wang K., Lu X. Nat. Commun. 2019;10:1487. doi: 10.1038/s41467-019-09351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Liang Y., Zhao X., Hu T., Chen B., Yin Z., Ma P.X., Guo B. Small. 2019;15 doi: 10.1002/smll.201900046. [DOI] [PubMed] [Google Scholar]
- 165.Hsiao C.W., Chen H.L., Liao Z.X., Sureshbabu R., Hsiao H.C., Lin S.J., Chang Y., Sung H.W. Adv. Funct. Mater. 2015;25:721–728. [Google Scholar]
- 166.Quirynen M., van der Mei H.C., Bollen C.M., Schotte A., Marechal M., Doornbusch G.I., Naert I., Busscher H.J., van Steenberghe D. J. Dent. Res. 1993;72:1304–1309. doi: 10.1177/00220345930720090801. [DOI] [PubMed] [Google Scholar]
- 167.Luo C., Liu W., Luo B., Tian J., Wen W., Liu M., Zhou C. Carbohydr. Polym. 2017;156:235–243. doi: 10.1016/j.carbpol.2016.09.036. [DOI] [PubMed] [Google Scholar]
- 168.Weeks M.E., Nebe von Caron G., James D.C., Smales C.M., Robinson G.K. J. Microbiol. Methods. 2006;66:43–55. doi: 10.1016/j.mimet.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 169.Sträuber H., Müller S. Cytometry. 2010;77:623–634. doi: 10.1002/cyto.a.20920. [DOI] [PubMed] [Google Scholar]
- 170.Matai I., Garg M., Rana K., Singh S. RSC Adv. 2019;9:13444–13457. doi: 10.1039/c9ra00623k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Hu J., Yang L., Yang P., Jiang S., Liu X., Li Y. Biomater. Sci. 2020;8:4940–4950. doi: 10.1039/d0bm01070g. [DOI] [PubMed] [Google Scholar]
- 172.Zhang J., Fu Y., Yang P., Liu X., Li Y., Gu Z. Adv. Mater. Interfac. 2020;7 [Google Scholar]
- 173.Zhang S., Hou J., Yuan Q., Xin P., Cheng H., Gu Z., Wu J. Chem. Eng. J. 2020;392 [Google Scholar]
- 174.Freitas M., Lima J.L., Fernandes E. Anal. Chim. Acta. 2009;649:8–23. doi: 10.1016/j.aca.2009.06.063. [DOI] [PubMed] [Google Scholar]
- 175.Lim C., Huang J., Kim S., Lee H., Zeng H., Hwang D.S. Angew. Chem. Int. Ed. 2016;55:3342–3346. doi: 10.1002/anie.201510319. [DOI] [PubMed] [Google Scholar]
- 176.Li J., Wu F., Zhang K., He Z., Zou D., Luo X., Fan Y., Yang P., Zhao A., Huang N. ACS Appl. Mater. Interfaces. 2017;9:30343–30358. doi: 10.1021/acsami.7b07444. [DOI] [PubMed] [Google Scholar]
- 177.Sun J., Tan H., Liu H., Jin D., Yin M., Lin H., Qu X., Liu C. Biomater. Sci. 2020;8:6946–6956. doi: 10.1039/d0bm01213k. [DOI] [PubMed] [Google Scholar]
- 178.Liu G., Wang L., He Y., Wang L., Deng Z., Liu J., Peng D., Ding T., Lu L., Ding Y., Zhang J., Liu P., Cai K. Adv. Healthc. Mater. 2021 doi: 10.1002/adhm.202101476. [DOI] [PubMed] [Google Scholar]
- 179.Liang Y., Zhao X., Hu T., Han Y., Guo B. J. Colloid Interface Sci. 2019;556:514–528. doi: 10.1016/j.jcis.2019.08.083. [DOI] [PubMed] [Google Scholar]
- 180.Zhang Q., Yang Q., Phanlavong P., Li Y., Wang Z., Jiao T., Peng Q. ACS Sustain. Chem. Eng. 2017;5:4161–4170. [Google Scholar]
- 181.Rodríguez A.R.C., Saiz-Poseu J., García-Pardo J., García B., Lorenzo J., Ojea-Jiménez I., Komilis D., Sedó J., Busqué F., Sánchez A. RSC Adv. 2016;6:40058–40066. [Google Scholar]
- 182.Dong Z., Zhang F., Wang D., Liu X., Jin J. J. Solid State Chem. 2015;224:88–93. [Google Scholar]
- 183.Nematollahzadeh A., Seraj S., Mirzayi B. Chem. Eng. J. 2015;277:21–29. [Google Scholar]
- 184.Li J., Wang Y., Yang J., Liu W. Chem. Eng. J. 2020 [Google Scholar]
- 185.Kang M., Kim E., Temoçin Z.l., Li J., Dadachova E., Wang Z., Panzella L., Napolitano A., Bentley W.E., Payne G.F. Chem. Mater. 2018;30:5814–5826. [Google Scholar]
- 186.Papuc C., Goran G.V., Predescu C.N., Nicorescu V., Stefan G. Compr. Rev. Food Sci. Food Saf. 2017;16:1243–1268. doi: 10.1111/1541-4337.12298. [DOI] [PubMed] [Google Scholar]
- 187.Fu Y., Zhang J., Wang Y., Li J., Bao J., Xu X., Zhang C., Li Y., Wu H., Gu Z. Carbohydr. Polym. 2021;257 doi: 10.1016/j.carbpol.2020.117598. [DOI] [PubMed] [Google Scholar]
- 188.Pillai P.P., Kowalczyk B., Kandere-Grzybowska K., Borkowska M., Grzybowski B.A. Angew. Chem., Int. Ed. Engl. 2016;55:8610–8614. doi: 10.1002/anie.201602965. [DOI] [PubMed] [Google Scholar]
- 189.Zhang X., Chen X., Yang J., Jia H.R., Li Y.H., Chen Z., Wu F.G. Adv. Funct. Mater. 2016;26:5958–5970. [Google Scholar]
- 190.Zhu Z., Gao Q., Long Z., Huo Q., Ge Y., Vianney N., Daliko N.A., Meng Y., Qu J., Chen H., Wang B. Bioact. Mater. 2021;6:2546–2556. doi: 10.1016/j.bioactmat.2021.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wang Q., Zhang R., Lu M., You G., Wang Y., Chen G., Zhao C., Wang Z., Song X., Wu Y. Biomacromolecules. 2017;18:1333–1341. doi: 10.1021/acs.biomac.7b00077. [DOI] [PubMed] [Google Scholar]
- 192.Gao C., Cheng H., Xu N., Li Y., Chen Y., Wei Y., Gao B., Fu J., Huo K., Xiong W. Nanomedicine. 2019;14:803–818. doi: 10.2217/nnm-2018-0131. [DOI] [PubMed] [Google Scholar]
- 193.Fu P.P., Xia Q., Hwang H.-M., Ray P.C., Yu H. J. Food Drug Anal. 2014;22:64–75. doi: 10.1016/j.jfda.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Ito K., Suda T. Nat. Rev. Mol. Cell Biol. 2014;15:243–256. doi: 10.1038/nrm3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kao H., Chen C.-C., Huang Y.-R., Chu Y.-H., Csík A., Ding S.-J. Surf. Coat. 2019;378 [Google Scholar]
- 196.Xiang Y., Mao C., Liu X., Cui Z., Jing D., Yang X., Liang Y., Li Z., Zhu S., Zheng Y. Small. 2019;15 doi: 10.1002/smll.201900322. [DOI] [PubMed] [Google Scholar]
- 197.Zhu Y., Xu C., Zhang N., Ding X., Yu B., Xu F.J. Adv. Funct. Mater. 2018;28 [Google Scholar]
- 198.Sun L., Jiang W., Zhang H., Guo Y., Chen W., Jin Y., Chen H., Du K., Dai H., Ji J. ACS Appl. Mater. Interfaces. 2018;11:2302–2316. doi: 10.1021/acsami.8b19522. [DOI] [PubMed] [Google Scholar]
- 199.Jin Y., Zhu Z., Liang L., Lan K., Zheng Q., Wang Y., Guo Y., Zhu K., Mehmood R., Wang B. Appl. Surf. Sci. 2020;528 [Google Scholar]
- 200.Zhang X., Sun G.H., Tian M.P., Wang Y.N., Qu C.C., Cheng X.J., Feng C., Chen X.G. Int. J. Biol. Macromol. 2019;138:321–333. doi: 10.1016/j.ijbiomac.2019.07.052. [DOI] [PubMed] [Google Scholar]
- 201.Song J., Chen P., Liu W. Fibers Polym. 2019;20:1380–1386. [Google Scholar]
- 202.Cong Y., Xia T., Zou M., Li Z., Peng B., Guo D., Deng Z. J. Mater. Chem. B. 2014;2:3450–3461. doi: 10.1039/c4tb00460d. [DOI] [PubMed] [Google Scholar]
- 203.Wu Y., Zhang Y., Zhang R., Chen S. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.630745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Li Y., Wang B., Sui X., Xie R., Xu H., Zhang L., Zhong Y., Mao Z. Appl. Surf. Sci. 2018;435:1337–1343. [Google Scholar]
- 205.Patel K., Kushwaha P., Kumar S., Kumar R. ACS Appl. Bio Mater. 2019;2:5799–5809. doi: 10.1021/acsabm.9b00794. [DOI] [PubMed] [Google Scholar]
- 206.Ran H.-H., Cheng X., Gao G., Sun W., Jiang Y.-W., Zhang X., Jia H.-R., Qiao Y., Wu F.-G. ACS Appl. Bio Mater. 2020;3:2438–2448. doi: 10.1021/acsabm.0c00163. [DOI] [PubMed] [Google Scholar]
- 207.Tao B., Lin C., Guo A., Yu Y., Qin X., Li K., Tian H., Yi W., Lei D., Chen L. J. Ind. Eng. Chem. 2021;104:345–355. [Google Scholar]
- 208.Wang N., Yu X., Kong Q., Li Z., Li P., Ren X., Peng B., Deng Z. Colloid Surf. A-Physicochem. Eng. Asp. 2020;602 [Google Scholar]
- 209.Yang F.K., Zhang W., Han Y., Yoffe S., Cho Y., Zhao B. Langmuir. 2012;28:9562–9572. doi: 10.1021/la301388e. [DOI] [PubMed] [Google Scholar]
- 210.Bernsmann F., Frisch B., Ringwald C., Ball V. J. Colloid Interface Sci. 2010;344:54–60. doi: 10.1016/j.jcis.2009.12.052. [DOI] [PubMed] [Google Scholar]
- 211.Shalev T., Gopin A., Bauer M., Stark R.W., Rahimipour S. J. Mater. Chem. 2012;22:2026–2032. [Google Scholar]
- 212.Jia L., Han F., Wang H., Zhu C., Guo Q., Li J., Zhao Z., Zhang Q., Zhu X., Li B. J. Orthop. Translat. 2019;17:82–95. doi: 10.1016/j.jot.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Bothner-By A.A. J. Am. Chem. Soc. 1953;75:728–730. [Google Scholar]
- 214.Mrówczyński R., Coy L.E., Scheibe B.e., Czechowski T., Augustyniak-Jabłokow M., Jurga S., Tadyszak K. J. Phys. Chem. B. 2015;119:10341–10347. doi: 10.1021/acs.jpcb.5b01524. [DOI] [PubMed] [Google Scholar]
- 215.Auten R.L., Davis J.M. Pediatr. Res. 2009;66:121–127. doi: 10.1203/PDR.0b013e3181a9eafb. [DOI] [PubMed] [Google Scholar]
- 216.Yang S.H., Kang S.M., Lee K.-B., Chung T.D., Lee H., Choi I.S. J. Am. Chem. Soc. 2011;133:2795–2797. doi: 10.1021/ja1100189. [DOI] [PubMed] [Google Scholar]
- 217.Saleh B., Dhaliwal H.K., Portillo-Lara R., Shirzaei Sani E., Abdi R., Amiji M.M., Annabi N. Small. 2019;15 doi: 10.1002/smll.201902232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Chien H.-W., Chiu T.-H. Eur. Polym. J. 2020;130 [Google Scholar]