Abstract
The community structure of sulfate-reducing bacteria (SRB) of a marine Arctic sediment (Smeerenburgfjorden, Svalbard) was characterized by both fluorescence in situ hybridization (FISH) and rRNA slot blot hybridization by using group- and genus-specific 16S rRNA-targeted oligonucleotide probes. The SRB community was dominated by members of the Desulfosarcina-Desulfococcus group. This group accounted for up to 73% of the SRB detected and up to 70% of the SRB rRNA detected. The predominance was shown to be a common feature for different stations along the coast of Svalbard. In a top-to-bottom approach we aimed to further resolve the composition of this large group of SRB by using probes for cultivated genera. While this approach failed, directed cloning of probe-targeted genes encoding 16S rRNA was successful and resulted in sequences which were all affiliated with the Desulfosarcina-Desulfococcus group. A group of clone sequences (group SVAL1) most closely related to Desulfosarcina variabilis (91.2% sequence similarity) was dominant and was shown to be most abundant in situ, accounting for up to 54.8% of the total SRB detected. A comparison of the two methods used for quantification showed that FISH and rRNA slot blot hybridization gave comparable results. Furthermore, a combination of the two methods allowed us to calculate specific cellular rRNA contents with respect to localization in the sediment profile. The rRNA contents of Desulfosarcina-Desulfococcus cells were highest in the first 5 mm of the sediment (0.9 and 1.4 fg, respectively) and decreased steeply with depth, indicating that maximal metabolic activity occurred close to the surface. Based on SRB cell numbers, cellular sulfate reduction rates were calculated. The rates were highest in the surface layer (0.14 fmol cell−1 day−1), decreased by a factor of 3 within the first 2 cm, and were relatively constant in deeper layers.
Marine sediments play a significant role in the global cycling of carbon and nutrients. Organic matter from primary production settles to the sea floor, where a major part is remineralized by microorganisms that colonize the sediments (50). Steep redox potentials provide niches for a wide variety of metabolically diverse microorganisms. Sulfate reduction is the major bacterial process in marine sediments, accounting for up to 50% of the total organic carbon remineralization (3, 17). The sulfate-reducing bacteria (SRB) make up a complex physiological group of organisms that can use a variety of volatile or long-chain fatty acids, alcohols, or aromatic compounds as carbon and energy sources but cannot use polysaccharides or other polymeric substrates. Some SRB are not completely dependent on sulfate; they can also use alternative electron acceptors, such as Fe(III) (4, 24) and nitrate (49), can disproportionate inorganic sulfur compounds (15, 21), or can grow under fermentative conditions (48). Although some SRB have been shown to survive in the presence of oxygen, no growth has been observed under these conditions (5, 9, 22). SRB, therefore, must combine two divergent needs for survival and growth in the sediment. While an input of organic matter is usually provided by sedimentation from the water column, the optimal redox conditions are most likely deeper in the sediment. Studying the occurrence and distribution of SRB along the depth profile combined with measuring metabolic activity can help elucidate the way that SRB cope with these specific challenges.
The study described here was part of an ongoing research project to investigate microbial communities and microbial physiology at permanently low temperatures. During a previous cruise to Svalbard (Arctic Ocean) in 1995, several studies were performed, including sulfate reduction measurements (35), isolation of psychrophilic sulfate reducers (20), determination of prokaryotic abundance, and vertical profiling of SRB rRNA with selected oligonucleotide probes (37). The SRB community was shown to be highly diverse in terms of species richness as determined by cloning of genes encoding 16S rRNA (rDNA) (32).
With the valuable set of data obtained previously in hand, we went back to Svalbard in 1998 and 1999 with several open questions. Could we back up the relevance of psychrophilic sulfate-reducing isolates and prominent clone groups from the previous cruise? And what was the physiological status of these groups with respect to localization along the sediment profile? To answer these questions, we combined fluorescence in situ hybridization (FISH) and rRNA slot blot hybridization to study SRB obtained from Smeerenburgfjorden. To do this, we used previously described and newly developed oligonucleotide probes for different groups of SRB belonging to the delta subgroup of Proteobacteria.
Despite their power, both FISH and rRNA slot blot hybridization have their limitations (for a review see reference 2). FISH may fail due to a low cellular rRNA content of the target organisms, autofluorescence of samples, impermeability of cell walls, and limited accessibility of probe target sites (12). Quantitative slot blot hybridization targets an rRNA pool that depends on both the number of target cells and the rRNA content per cell. Cell numbers cannot be directly inferred from the data, cell morphology and exact localization remain unclear, and rRNA recovery may be influenced by species-dependent differences in the efficiency of cell lysis. In this study we used both methods for quantification of SRB for two reasons. First, many studies have been conducted with one of these two methods (8, 13, 23, 29, 31, 38, 39), but it is still not clear to what degree the limitations of the methods influence the comparability of the data. Second, in order to better understand an organism's role in a given ecosystem, it is important not only to determine the composition of the microbial community but also to combine this information with a measure of the metabolic status. A first step is the calculation of specific rRNA contents for individual groups, which correlates with growth rates under certain circumstances (for a review see reference 26).
MATERIALS AND METHODS
Study site and sampling.
Sediment samples were collected on 28 July 1998 from Smeerenburgfjorden, Svalbard, Arctic Ocean (79°42′815"N, 11°05′189"E; station J). The sediment temperature was 0°C, the surface water temperature was 5°C, and the water depth was 218 m. Sediment samples were obtained with a Haps corer, subsampled, and kept at the in situ temperature during transport. Two parallel cores were sliced. One-half of each slice was frozen in liquid nitrogen for RNA extraction (stored at −80°C), and the other half was fixed for 2 to 3 h with 3% (final concentration) formaldehyde, washed twice with 1× phosphate-buffered saline (PBS) (10 mM sodium phosphate [pH 7.2], 130 mM NaCl), and then stored in 1× PBS–ethanol (1:1) at −20°C. The sediment was characterized by a soft brown silty oxidized surface (upper 2 cm) overlaying a transition zone consisting of darker, black-streaked clayey mud. Below the transition zone (2 to 6 cm) there was a black sulfidic zone. Worm tubes were present in the sediment, as were small shells (diameter, 2 to 3 mm) to depths below 10 cm. In addition to the samples obtained at the main study site at Smeerenburgfjorden, sediment samples were collected at the following three stations off the coast of Svalbard: Magdalenefjorden (station I; 79°34′052"N, 11°03′59"7E; depth, 125 m; temperature, −0.5°C; samples collected on 28 July 1998), Raudfjorden (station K; 79°46′150"N, 12°04.375"E; depth, 154 m; temperature, −1°C; samples collected on 29 July 1998), and Hornsund (76°59′415"N, 15°53′517"E; depth, 176 m; temperature, 1.0°C; samples collected on 16 July 1999).
RNA extraction and slot blot hybridization.
RNA was extracted from 1.5 ml of wet sediment (per layer) by bead beating, phenol extraction, and isopropanol precipitation as described previously (36). The quality of the RNA was checked by polyacrylamide gel electrophoresis. Approximately 50 ng of RNA was blotted onto nylon membranes (Magna Charge; Micron Separations, Westborough, Mass.) in triplicate and hybridized with radioactively labeled oligonucleotide probes as described by Stahl et al. (42). The membranes were washed at different temperatures depending on the dissociation temperature of the probe. The probes used and their dissociation temperatures are shown in Table 1. The dissociation temperatures of the probes were determined as described by Raskin et al. (30), with slight modifications. For dissociation temperature determinations and hybridizations we used washing buffer with a lower sodium dodecyl sulfate (SDS) concentration (1× SSC [150 mM NaCl, 15 mM sodium citrate; pH 7.0]–0.1% SDS). However, for hybridizations with probes Uni1390, EUB338, Sval428, 660, and 221 we used washing buffer containing 1% SDS.
TABLE 1.
Oligonucleotide probes used in this study
| Probe | Specificity | Sequence (5′→3′) | Positiona | FISH FA concn (%, vol/vol)b | Slot blot Td (°C)c | Reference |
|---|---|---|---|---|---|---|
| Uni1390 | Universal (all organisms) | GACGGGCGGTGTGTACAA | 1390–1407 | 44d | 51 | |
| EUB338 | Bacteria | GCTGCCTCCCGTAGGAGT | 338–355 | 10 | 54d | 1 |
| NON338 | ACTCCTACGGGAGGCAGC | 338–355 | 10 | 47 | ||
| DSR651 | Desulforhopalus spp. | CCCCCTCCAGTACTCAAG | 651–668 | 35 | 62 | 25 |
| DSS658 | Desulfosarcina spp., Desulfofaba sp., Desulfococcus spp., Desulfofrigus spp. | TCCACTTCCCTCTCCCAT | 658–685 | 60 | 58 | 25 |
| DSV698 | Desulfovibrio spp. | GTTCCTCCAGATATCTACGG | 698–717 | 35 | 58 | 25 |
| DSV214 | Desulfomicrobium spp. | CATCCTCGGACGAATGC | 214–230 | 10 | 25 | |
| DSV407 | Desulfovibrio spp. | CCGAAGGCCTTCTTCCCT | 407–424 | 50 | 25 | |
| DSV1292 | Desulfovibrio spp. | CAATCCGGACTGGGACGC | 1292–1309 | 35 | 25 | |
| DSD131 | Desulfovibrio sp. | CCCGATCGTCTGGGCAGG | 131–148 | 20 | 25 | |
| DSMA488 | Desulfarculus sp., Desulfomonile sp., Syntrophus spp. | GCCGGTGCTTCCTTTGGCGG | 488–507 | 60 | 25 | |
| Sval428 | Desulfotalea spp., Desulfofusiis sp. | CCATCTGACAGGATTTTAC | 428–446 | 25 | 52d | 37 |
| 660 | Desulfobulbus spp. | GAATTCCACTTTCCCCTCTG | 660–679 | 60 | 59d | 6 |
| 221 | Desulfobacterium spp. | TGCGCGGACTCATCTTCAAA | 221–240 | 35 | 57d | 6 |
| DSB985 | Desulfobacter spp., Desulfobacula spp. | CACAGGATGTCAAACCCAG | 985–1003 | 20 | 53 | 25 |
| DRM432 | Desulfuromonas spp., Pelobacter spp. | CTTCCCCTCTGACAGAGC | 432–449 | 40 | 62 | This study |
| DSF672 | Desulfofrigus spp., Desulfofaba sp. | CCTCTACACCTGGAATTCC | 672–690 | 45 | This study | |
| DSC193 | Desulfosarcina spp. | AGGCCACCCTTGATCCAA | 193–210 | 35 | This study | |
| DCC209 | Desulfococcus spp. | CCCAAACGGTAGCTTCCT | 209–226 | 25 | This study | |
| DSS225 | Svalbard clone group SVAL1 | TGGTACGCGGGCTCATCT | 225–242 | 40 | This study | |
| cl81-644 | Svalbard clones Sva0081 and Sva0863 | CCCATACTCAAGTCCCTT | 644–661 | 25 | This study |
Position in the 16S rRNA of E. coli.
Formamide (FA) concentration in the hybridization buffer.
Td, dissociation temperature.
Washing buffer containing 1× SSC, and 1% SDS was used.
Quantification.
Hybridization signal intensity was measured with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and was quantified as described previously (38). Reference rRNAs isolated from pure cultures of strain LSv23 (= DSM 13040) (19), strain LSv22 (= DSM 13039) (19), Desulfococcus multivorans DSM 2059, Desulfobulbus elongatus DSM 2908, Desulfobacterium vacuolatum DSM 3385, Desulfovibrio salexigens DSM2638, Desulfobacter latus DSM 3381, Desulfuromonas acetexigens DSM 1397, and Escherichia coli (purchased from Boehringer, Mannheim, Germany) were used as standards for hybridization with the probes shown in Table 1.
FISH.
Samples stored in PBS-ethanol were diluted and treated by mild sonication with a type MS73 probe (Sonopuls HD70; Bandelin, Berlin, Germany) at a setting of 20 s, an amplitude of 42 μm, and <10 W. A 10-μl aliquot of a 1:40 dilution was filtered onto a 0.2-μm-pore-size type GTTP polycarbonate filter (Millipore, Eschborn, Germany). Hybridization and microscopic counting of hybridized and 4′,6′-diamidino-2-phenylindole (DAPI)-stained cells were performed as described previously (39). Means were calculated by using 10 to 20 randomly chosen fields for each filter section, which corresponded to 800 to 1,000 DAPI-stained cells. Counting results were always corrected by subtracting signals observed with probe NON338. The formamide concentrations used are shown in Table 1.
Oligonucleotides.
Oligonucleotides were purchased from Interactiva (Ulm, Germany). For FISH, oligonucleotide probes were synthesized with Cy3 fluorochrome at the 5′ end.
Quantification of cell fluorescence.
To verify the calculated trend observed for the cellular rRNA content of the Desulfosarcina-Desulfococcus group (probe DSS658) along the sediment profile with an independent method, we quantified the mean FISH fluorescence at the single-cell level by confocal laser scanning microscopy. To ensure that the hybridization conditions were the same, sediment samples were hybridized on filters in 20-ml scintillation vials containing 1 ml of hybridization buffer with 1.25 ng of probe ml−1 as described above. The hybridized filters were mounted immediately before microscopy and were analyzed with a confocal laser scanning microscope (Zeiss model LSM510) by using the following settings: pinhole diameter, 176 μm; optical slice thickness, <0.9 μm; and detector gain 822 with an HeNe laser (excitation wavelength, 543 nm; 0.5 mW) and an argon laser (excitation wavelength, 514 nm; 12 mW). Pictures of 20 to 30 randomly selected fields containing a total of approximately 60 probe-targeted cells were used for quantification with the MetaMorph software (version 3.51; Universal Imaging Corp., West Chester, Pa.). Cells were selected manually to determine average cellular gray values and to quantify fluorescence. A mean cell fluorescence value was calculated for each depth; the lowest mean cell fluorescence value was defined as 1, and the mean fluorescence value for cells in each of the other layers was expressed relative to this value.
DNA extraction, PCR amplification, and clone library construction.
Total community DNA was directly extracted from the sediment (Smeerenburgfjorden, station J) according to Zhou et al. (52), with slight modifications as described previously (32). Aliquots (1.5 g) of wet sediment from different sections (depth, 2.25 to 3.75 cm) were used for DNA extraction. Extracted DNAs were then combined. The crude DNA was purified by dialysis. Sterile water (1 ml) was added to a six-well microtiter plate, and a 0.025-μm-pore-size nitrocellulose membrane (Millipore) was placed on the water surface. Approximately 30 μl of the crude DNA was dropped onto the membrane and incubated for 3 h at room temperature, and the purified DNA was removed with a pipette. The volume increased during incubation to roughly 400 μl (a 10- to 15-fold increase).
DNAs that were targeted by probe DSS658 were amplified by a specific PCR. One universal bacterial primer, EUB008 (14), and probe DSS658, as a specific second primer, were used for specific amplification of the target 16S rDNAs from the chromosomal DNA pool. A PCR was performed with a Mastercycler Gradient (Eppendorf, Hamburg, Germany) as follows. A mixture containing 50 pmol of each primer, 2.5 μmol of each deoxyribonucleoside triphosphate, 300 μg of bovine serum albumin, 1× reaction buffer, 1× TaqMaster PCR enhancer, and 1 U of MasterTaq DNA polymerase (Eppendorf) was adjusted to a final volume of 100 μl with sterile water. Template DNA was added to the reaction mixture (preheated to 70°C) to avoid nonspecific annealing of the primers to nontarget DNA. The following cycling conditions were used: one cycle at 70°C for 1 min; 38 cycles at 95°C for 1 min, 52°C for 1 min, and 72°C for 3 min; and one cycle at 72°C for 10 min. The annealing temperature was optimized with a temperature gradient in order to use the highest stringency possible. Control DNAs with one, two, or three mismatches with primer DSS658 were used to determine the stringency of amplification. DNA with more than one mismatch could be discriminated completely, but it was not possible to discriminate DNA with only one mismatch without losing the PCR product of the target DNA. The PCR products were cloned in the vector pGEM-T (Promega, Madison, Wis.), and a clone library was constructed as described previously (32). Forty clones were selected for further analysis. Amplified rDNA restriction analysis (ARDRA) was performed in order to identify clones with different inserts. Digestion with two restriction enzymes (HaeIII and RsaI; Promega) was used to screen the clones as described previously (32).
Sequencing and phylogenetic analysis.
Representatives of most ARDRA pattern groups were used for sequencing. PCR products obtained from selected 16S rDNA clones were sequenced by Taq cycle sequencing performed with vector primers and a model ABI377 sequencer (Applied Biosystems, Inc.). Sequence data were analyzed with the ARB software package (43). Phylogenetic trees were calculated by performing parsimony, neighbor-joining, and maximum-likelihood analyses with different sets of filters. For tree calculation, only full-length sequences were considered. The 650-nucleotide clone sequences were added to the tree after tree reconstruction. The organisms shown in the tree and the accession numbers of their sequences are as follows: Desulfobacterium vacuolatum, M34408; Desulfobacterium autotrophicum, M34409; Desulfobacter postgatei, M26633; Desulfobacula toluolica, X70953; Desulfofaba gelida, AF099063; Desulfofrigus oceanense, AF099064; Desulfofrigus fragile, AF099065; Desulfobacterium indolicum, AJ237607; Desulfonema ishimotoei, U45992; Desulfonema limicola, U45990; Desulfococcus multivorans, M34405; Desulfosarcina cetonicum, AJ237603; Desulfosarcina variabilis, M34407; Desulfobulbus rhabdoformis, U12253; Desulfobulbus elongatus, X95180; Desulfotalea psychrophila, AF099062; Desulfotalea arctica, AF099061; Desulforhopalus vacuolatus, L42613; Desulfofustis glycolicus, X99707; Desulfocapsa sulfoexigens, Y13672; Desulfocapsa thiozymogenes, X95181; Desulfuromonas acetoxidans, M26634; Desulfuromonas acetexigens, U23140; Desulfovibrio gigas, M34400; Desulfovibrio longus, Z24450; Desulfovibrio desulfuricans, M34113; LSv53, AF099058; vadinH60, U81720; Sva0863, AJ240977; Sva0081, AJ240975; S2551, AF177428; str. MMP1991, L06457; AK-01, AF141328; ACE-32, AF142807; CLEAR-29, AF146251; A34, U08389; A52, U08394; RFLP25, AF058007; A01, U85480; DGGE-BS3, AJ011668; and SB-29, AF029047.
SRR measurement.
Sulfate reduction rates (SRRs) were measured in whole sediment cores by the radiotracer method (16, 41). Undisturbed sediment cores were injected with 500 kBq of 35S tracer at 1-cm intervals and incubated for 12 h at the in situ temperature in the dark. To stop the reaction, the sediment cores were cut into 1-cm-thick slices that were thoroughly mixed with 20 ml of 20% (wt/vol) zinc acetate and then deep frozen for transport. All samples were distilled with 6 M HCl and chromium(II) chloride in a single-step distillation process to convert reducible sulfur compounds into H2S (11). SRRs were calculated from the ratio of added 35S-sulfate to produced 35S-sulfide.
Nucleotide sequence accession numbers.
The nucleotide sequence data reported in this paper have been deposited in the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession no. AF233491 to AF233500.
RESULTS
FISH detection rates.
In the Smeerenburgfjorden sediment a large fraction of the bacteria living in the top 5 cm could be detected by FISH (Table 2). Up to 73.6% (core A, 57.9%) of the total DAPI cell counts hybridized to eubacterial probe EUB338. Below 10 cm the detection rate with probe EUB338 became too low (<20% of the total DAPI cell counts) for further FISH analysis.
TABLE 2.
Quantification of SRB in Smeerenburgfjorden sediments by FISH and rRNA slot blot hybridization
| Depth (cm) | Absolute cell no. (109) ml−1 | Bacteria (probe EUB338): % DAPI | Concn of prokaryotic RNA (EUB338 + Arch915) (ng ml−1) |
Desulfovibrio spp. (DSV698)
|
Desulforhopalus spp. (DSR651)
|
||||
|---|---|---|---|---|---|---|---|---|---|
| % DAPI | RNA concn (ng ml−1) | % of prokaryotic RNA | % DAPI | RNA concn (ng ml−1) | % of prokaryotic RNA | ||||
| 0.25 | 2.9/2.1a | 57.9/72.9 | 9,865/8,977 | 0.9/0.8 | 220/338 | 2.2/3.8 | 1.1/0.9 | 257/207 | 2.6/2.3 |
| 0.75 | 3.5/3.3 | 55.9/73.6 | 10,036/13,846 | 1.1/1.6 | 264/521 | 2.6/3.8 | 1.5/1.1 | 146/547 | 1.5/4.0 |
| 1.25 | 3.1/3.4 | 56.1/65.4 | 7,321/10,597 | 0/0.8 | 178/349 | 2.4/3.3 | 2.1/1.6 | 284/571 | 3.9/5.4 |
| 1.75 | 4.2/4.2 | 53.1/57.7 | 6,879/8,325 | 0.6/0 | 152/266 | 2.2/3.2 | 2.3/1.3 | 195/202 | 2.8/2.4 |
| 2.25 | 4.1/3.5 | 57.5/48.4 | 7,161/9,605 | 0.4/0 | 171/329 | 2.4/3.4 | 2.2/1.5 | 91/502 | 1.3/5.2 |
| 2.75 | 3.7/3.3 | 47.9/51.2 | 5,037/5,866 | 0 | 119/176 | 2.4/3.0 | 3.2/2.3 | 94/96 | 1.9/1.6 |
| 3.25 | 2.7/3.2 | 43.7/46.6 | 4,724/6,417 | 0 | 118/177 | 2.5/2.8 | 3.0/2.4 | 138/129 | 3.0/2.0 |
| 3.75 | 3.1/3.7 | 42.8/50.0 | 5,212/6,671 | 0 | 128/206 | 2.5/3.1 | 2.9/2.3 | 145/151 | 2.8/2.3 |
| 4.25 | 3.5/3.4 | 43.5/47.3 | 3,613/5,070 | 0 | 94/133 | 2.6/2.6 | 2.7/1.7 | 86/86 | 2.4/1.7 |
| 4.75 | 3.4/3.6 | 28.4/42.2 | 3,605/4,427 | 0 | 107/113 | 3.0/2.5 | 2.7/1.7 | 132/75 | 3.7/1.7 |
| 5.5 | 3.5/4.6 | 26.0/36.0 | 2,616/4,046 | 0 | 77/115 | 3.0/2.9 | 1.8/1.8 | 48/120 | 1.8/3.0 |
| 6.5 | 3.7/4.7 | 24.1/27.4 | 2,984/4,494 | 0 | 90/117 | 3.0/2.6 | 1.9/1.3 | 67/91 | 2.2/2.0 |
| 7.5 | 4.7/3.9 | 23.6/23.6 | 2,662/3,520 | 0 | 77/98 | 2.9/2.8 | 1.5/0.8 | 65/69 | 2.5/2.0 |
| 8.5 | 2.9/3.1 | 28.9/22.6 | 2,869/3,246 | 0 | 82/92 | 2.9/2.8 | 1.5/1.3 | 76/78 | 2.6/2.4 |
| 9.5 | 3.7/3.9 | 22/15.7 | 2,308/2,898 | 0 | 64/83 | 2.8/2.8 | 1.6/1.2 | 89/58 | 3.9/2.0 |
|
Desulfobulbus spp. (660)
|
Desulfotalea spp. (Sval430)
|
Desulfobacter spp. (DSB985)
|
Desulfobacterium spp. (221)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| % DAPI | RNA concn (ng ml−1) | % of prokaryotic RNA | % DAPI | RNA concn (ng ml−1) | % of prokaryotic RNA | % DAPI | RNA concn (ng ml−1) | % of prokaryotic RNA | % DAPI | RNA concn (ng ml−1) |
| 0 | 19/31 | 0.2/0.4 | 0 | 113/35 | 1.2/0.4 | 0 | 45/57 | 0.5/0.7 | 0 | 0 |
| 0 | 19/38 | 0.2/0.3 | 0 | 229/71 | 2.3/0.5 | 0 | 67/67 | 0.7/0.5 | 0 | 0 |
| 0 | 22/30 | 0.3/0.3 | 0/0.8 | 229/59 | 3.1/0.6 | 0 | 76/71 | 1.0/0.7 | 0 | 0 |
| 0 | 19/31 | 0.3/0.4 | 1.3/1.2 | 180/25 | 2.6/0.3 | 0 | 28/128 | 0.4/1.5 | 0 | 0 |
| 0 | 20/22 | 0.3/0.2 | 1.1/1.0 | 256/32 | 3.6/0.3 | 0 | 49/60 | 0.7/0.6 | 0.5/0 | 0 |
| 0 | 15/19 | 0.3/0.3 | 0.8/1.0 | 167/46 | 3.3/0.8 | 0 | 15/45 | 0.3/0.8 | 2.2/0 | 0 |
| 0 | 9/23 | 0.2/0.4 | 0.7/1.4 | 173/71 | 3.7/1.1 | 0 | 5/0 | 0.1/0 | 1.7/0.6 | 0 |
| 0 | 10/28 | 0.2/0.4 | 1.2/1.3 | 293/230 | 5.6/3.4 | 0 | 0 | 0 | 0.7/0 | 0 |
| 0 | 7/19 | 0.2/0.4 | 1.0/1.8 | 114/41 | 3.2/0.8 | 0 | 0 | 0 | 0 | 0 |
| 0 | 7/15 | 0.2/0.4 | 0.6/1.6 | 114/31 | 3.2/0.7 | 0 | 0 | 0 | 0 | 0 |
| 0 | 4/13 | 0.2/0.3 | 1.4/1.5 | 64/35 | 2.5/0.9 | 0 | 0 | 0 | 0 | 0 |
| 0 | 6/10 | 0.2/0.2 | 0.4/1.4 | 61/38 | 2.0/0.9 | 0 | 0 | 0 | 0 | 0 |
| 0 | 5/7 | 0.2/0.2 | 0.6/1.5 | 42/39 | 1.6/1.1 | 0 | 0 | 0 | 0 | 0 |
| 0 | 5/8 | 0.2/0.2 | 1.3/1.0 | 32/29 | 1.1/0.9 | 0 | 0 | 0 | 0 | 0 |
| 0 | 5/6 | 0.2/0.2 | 0.5/0 | 25/21 | 1.1/0.7 | 0 | 0 | 0 | 0 | 0 |
|
Desulfosarcina spp. and Desulfococcus sp. (probe DSS658)
|
Sum for SRB detected
|
DSS clones (probe DSS225): % DAPI | Clones Sva0081 and Sva0863 (probe cl81-644): % DAPI |
Desulfuromonas spp. (probe DRM432)
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| % DAPI | RNA concn (ng ml−1) | % of prokaryotic RNA | % DAPI | RNA concn (ng ml−1) | % of prokaryotic RNA | % DAPI | RNA concn (ng ml−1) | % of prokaryotic RNA | ||
| 1.7/1.8 | 465/555 | 4.7/6.2 | 3.7/3.5 | 1,120/1,224 | 11.4/13.6 | 1.2/0.7 | 0.7/0.7 | 0.7/0.7 | 201/214 | 2.0/2.4 |
| 3.1/3.1 | 562/1,180 | 5.6/8.5 | 5.7/5.7 | 1,286/2,423 | 12.8/17.5 | 2.3/1.8 | 1.0/1.4 | 1.5/1.1 | 182/490 | 1.8/3.5 |
| 4.7/4.5 | 648/1,311 | 8.9/12.4 | 6.8/7.7 | 1,437/2,391 | 19.6/22.6 | 3.3/2.8 | 1.6/1.8 | 1.9/1.5 | 239/535 | 3.3/5.1 |
| 6.5/6.7 | 906/848 | 13.2/10.2 | 10.6/9.3 | 1,481/1,500 | 21.5/18.0 | 4.0/3.9 | 1.6/2.6 | 1.4/1.5 | 283/349 | 4.1/4.2 |
| 8.9/9.5 | 595/1,066 | 8.3/11.1 | 13.2/12.1 | 1,181/2,011 | 15.5/20.9 | 6.3/5.6 | 2.4/2.7 | 1.9/2.2 | 150/434 | 2.1/4.5 |
| 9.3/11.0 | 743/498 | 14.8/8.5 | 15.6/14.3 | 1,152/880 | 22.9/15.0 | 6.6/7.0 | 2.6/2.9 | 1.7/1.7 | 212/236 | 4.2/4.0 |
| 10.0/11.7 | 727/522 | 15.4/8.1 | 15.4/16.1 | 1,169/922 | 24.8/14.4 | 7.4/6.1 | 2.3/1.7 | 1.3/1.4 | 235/247 | 5.0/3.8 |
| 8.8/7.4 | 929/836 | 17.8/12.5 | 13.6/10.9 | 1,504/1,449 | 28.9/21.7 | 5.4/4.6 | 1.2/2.3 | 1.2/0.7 | 291/351 | 5.6/5.3 |
| 8.8/8.1 | 608/450 | 16.8/8.9 | 12.5/11.6 | 910/729 | 25.2/14.4 | 6.4/3.9 | 2.5/2.7 | 0.8/0.5 | 205/149 | 5.7/2.9 |
| 6.0/6.2 | 559/590 | 15.5/13.3 | 9.2/9.5 | 918/823 | 25.5/18.6 | 3.8/4.1 | 1.6/0.6 | 1.3/1.0 | 229/191 | 6.4/4.3 |
| 6.6/5.2 | 305/651 | 11.7/16.1 | 9.8/8.5 | 499/933 | 19.1/23.1 | 3.7/3.6 | 0.9/3.1 | 1.3/1.0 | 104/246 | 4.0/6.1 |
| 7.1/6.5 | 378/725 | 12.7/16.1 | 9.4/9.1 | 601/981 | 20.1/21.8 | 5.2/3.4 | 1.9/1.8 | 0.9/1.0 | 86/191 | 2.9/4.3 |
| 5.2/5.6 | 378/519 | 14.2/14.7 | 7.3/7.9 | 567/732 | 21.3/20.8 | 3.3/2.8 | 1.8/1.3 | 1.1/0.7 | 68/129 | 2.6/3.7 |
| 4.4/4.2 | 442/492 | 15.4/15.2 | 7.3/6.4 | 637/698 | 22.2/21.5 | 2.4/2.6 | 0.5/0.7 | 0.8/0.6 | 91/126 | 3.2/3.9 |
| 3.0/3.6 | 393/350 | 17.0/12.1 | 5.1/4.7 | 575/519 | 24.9/17.9 | 1.9/1.0 | 1.0/1.5 | 0.5/0.2 | 93/136 | 4.0/4.7 |
Core A value/core B value.
SRB community structure.
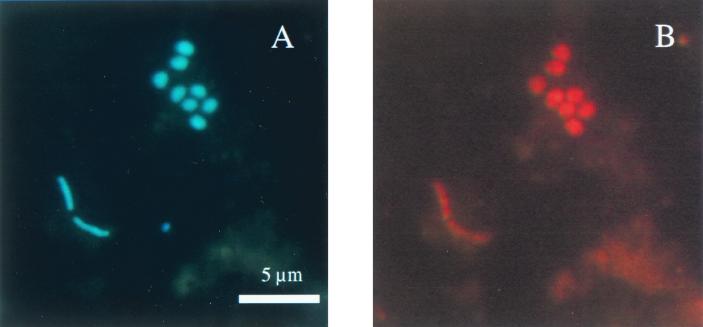
The emphasis of this study was on the SRB community structure in Smeerenburgfjorden sediment. SRB were quantified by both FISH and rRNA slot blot hybridization. The profiles of the individual groups of SRB in duplicate cores revealed comparable trends and abundances, indicating that there was horizontal homogeneity within the sediment at the level of our experimental resolution. The SRB community was dominated by complete oxidizers: the monophyletic group of Desulfosarcina spp., Desulfococcus spp., Desulfofrigus spp., Desulfofaba sp., and related clone sequences. This group is targeted by probe DSS658 and is referred to as the Desulfosarcina-Desulfococcus group below. Almost 12% of the DAPI cell counts were detected with this probe by FISH (Table 2). The highest abundance occurred at a depth of 2.25 cm, where 3.7 × 108 cells ml of sediment−1 accounted for 73% of the total SRB detected. Typically, the DSS658-positive cells had sarcinalike cell morphology (Fig. 1). Approximately 80% of the cells were irregularly shaped cocci that were about 1 μm in diameter and occurred in sarcinalike tetrads, in large clusters consisting of 10 or more cells, or (very often) as diplococci. About 20% of the cells detected were rods (0.5 by 1 to 3 μm).
FIG. 1.
Epifluorescence micrographs of bacteria in sediment samples from Smeerenburgfjorden, Svalbard, Arctic Ocean. (A) DAPI staining. (B) FISH performed with probe DSS658 specific for the Desulfosarcina-Desulfococcus group (same microscopic field).
Similar quantitative results were obtained when we used rRNA hybridization, which also showed that DSS658 rRNA was the most abundant rRNA. The vertical profile had a broad peak at depths between 3.75 and 9 cm, with 14 to 15% of prokaryotic rRNA (Table 2), while the absolute rRNA yields were highest at depths between 1.25 and 3.75 cm, where the mean maximum concentration was 1,000 ng of rRNA ml of wet sediment−1. At depths below 3.75 cm the rRNA yield decreased with depth by a factor of approximately 1.5 to 2.0. The second dominant group, which was present at much lower abundance (one-third the level of the Desulfosarcina-Desulfococcus group), was Desulforhopalus spp. (Table 2). Probe DSR651 detected a maximum of 3.2% of all cells when FISH was used (1.2 × 108 cells ml−1) and 5.4% of the prokaryotic rRNA. In general, the rRNA yield decreased with depth (Table 2). Members of the genus Desulfotalea, a newly described genus of psychrophilic SRB (20), could be detected in numbers of up to 6.9 × 107 cells ml of sediment−1 (1.0 to 1.8% of the DAPI cell count) in the depth profile. There was no clear maximum visible by FISH. In vertical profiles for Desulfotalea sp. rRNA the values were constant at almost all depths, with a clear maximum (4.4% of the prokaryotic rRNA, mean of two cores) at 3.75 cm.
For quantification of members of the frequently cultivated genus Desulfovibrio we used several probes designed by Manz et al. (25). Only with the most general probe, probe DSV698, could cells be detected, and they were detected only in the upper layers (surface layer to a depth of 2.25 cm); the maximum value obtained was only 1.6% of the DAPI cell count (5.2 × 107 cells ml−1). The cell morphology was not vibriolike as expected for most Desulfovibrio species; the cells were short or long thin rods, and a few cells were coccoidal. This could have been due to a lack of probe specificity, but there are also rod-shaped Desulfovibrio species (e.g., Desulfovibrio piger and Desulfovibrio carbinolicus) (48). When rRNA hybridization was used, we detected a constant level of Desulfovibrio sp. rRNA (approximately 3% of the prokaryotic rRNA) throughout the vertical profile. The recovered rRNA yield decreased with depth.
Other probe target groups, like Desulfomicrobium spp. (probe DSV214) and Desulfarculus spp.-Desulfomonile spp.-Syntrophus spp. (probe DSMA488), were below the detection limit. Members of the genus Desulfobacterium (probe 221), which are completely oxidizing bacteria, were detected only at depths between 2.25 and 3.75 cm (up to 2.2% of the DAPI cell count). The level of RNA of this group was below the detection limit for slot blot hybridization. Desulfobulbus spp. (probe 660) and Desulfobacter spp.-Desulfobacula spp. (probe DSB985) were not detected by FISH, but small amounts of rRNAs of these organisms were found. Constant small amounts of Desulfobulbus sp. rRNAs were detected (0.2 to 0.4% of the prokaryotic rRNA throughout the vertical profile). Desulfobacter sp. rRNA was recovered down to a depth of 3.25 cm, and the maximum value was 1.5% of the prokaryotic RNA.
Sulfur-reducing and fermenting bacteria.
Members of Desulfuromonas spp., which are sulfur-reducing bacteria, and of Pelobacter spp., which are strictly anaerobic fermenting bacteria, have been shown to constitute a dominant group in our Svalbard sediment clone library (32). Therefore, we investigated the abundance of these organisms to assess their potential contribution to the sulfur cycle in the sediments studied. This group was targeted by probe DRM432 (Table 2). Members of this group accounted for up to 2.2% of the DAPI cell counts and up to 6.4% of the total prokaryotic RNA, and thus this group's contribution to the sulfur cycle may be important and deserves further attention.
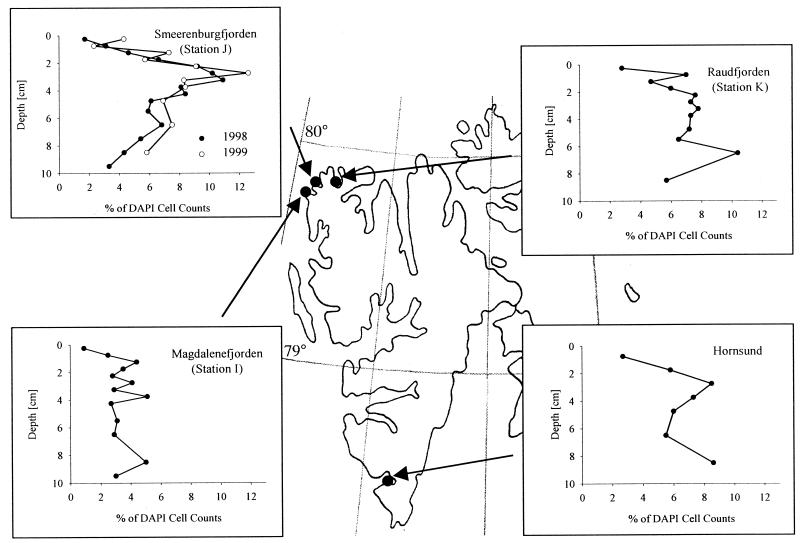
Predominance of the Desulfosarcina-Desulfococcus group at various stations along the coast of Svalbard.
Sediment samples from three other sampling sites off the coast of Svalbard and from Smeerenburgfjorden, which was sampled again 1999, were investigated with FISH to determine if the predominance of the Desulfosarcina-Desulfococcus group is a common feature at these stations. The vertical profile for DSS658-targeted cells from 1999 Smeerenburgfjorden sediment was almost identical to the profile obtained with samples collected in 1998 (Fig. 2). The highest percentage of cells detected in 1999 was 12.6% at a depth of 2.75 cm (11.7% at a depth of 3.25 cm depth in 1998). Members of the Desulfosarcina-Desulfococcus group were also found in high abundance in sediment samples obtained from Raudfjorden (station K) and from Hornsund (Fig. 2); both profiles exhibited a maximum at a depth of 2.75 cm depth (7.5 and 8.5% of the DAPI cell counts, respectively). The sediment profile for station K had another maximum (10.4% of the DAPI cell counts) in a deeper layer (depth, 6.5 cm). Only at Magdalenefjorden (station I) was this group detected in lower numbers (4 to 5% of the DAPI cell counts). This lower abundance was not due to a lower rate of detection of eubacterial cells at this station. DSS658-targeted cells accounted for 13% of the EUB338-detected cells at station I, compared to up to 30% at the other stations.
FIG. 2.
Detection and quantification of the Desulfosarcina-Desulfococcus group at various stations along the coast of Svalbard by FISH (probe DSS658).
In-depth analysis of the Desulfosarcina-Desulfococcus group.
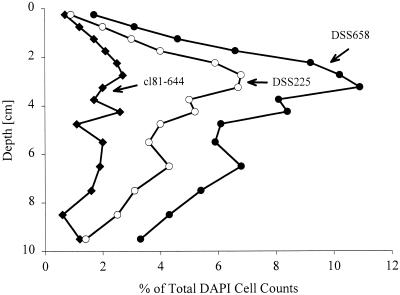
The existence of a prominent group of SRB at several stations prompted us to analyze this group in greater detail. We developed new specific probes for the different genera targeted by DSS658 on the basis of the available 16S rDNA data set (Table 1). Probe DSC193 was specific for Desulfosarcina spp., probe DCC209 was specific for Desulfococcus spp., probe DSF672 was specific for Desulfofaba sp. and Desulfofrigus spp., and probe cl81-644 was specific for 16S rDNA clones from Hornsund sediment (32). All of the probes could also be used for FISH. The hybridization conditions were adjusted by using several reference strains. When we used these probes for known and cultivated genera with Smeerenburgfjorden sediment samples collected in 1998, we did not detect any cells. However, when we used probe cl81-644, which was specific for Svalbard clones Sva0081 and Sva0863, almost 3% of the DAPI-stained cell counts were detected (Fig. 3). The targeted cells had a rod-shaped morphology (0.5 by 1 to 3 μm). Nevertheless, the very abundant sarcinalike cells could not be affiliated with cultivated genera or 16S rDNA clone sequences in the databases.
FIG. 3.
Depth profile for subgroups of the Desulfosarcina-Desulfococcus group as detected by FISH (Smeerenburgfjorden sediment). Symbols: ●, probe DSS658 targeting the Desulfosarcina-Desulfococcus group; ○, probe DSS225 targeting group SVAL1; ⧫, probe cl81-644 targeting previously cloned 16S rDNA sequences from the same habitat.
Search for the identity of sarcinalike cells.
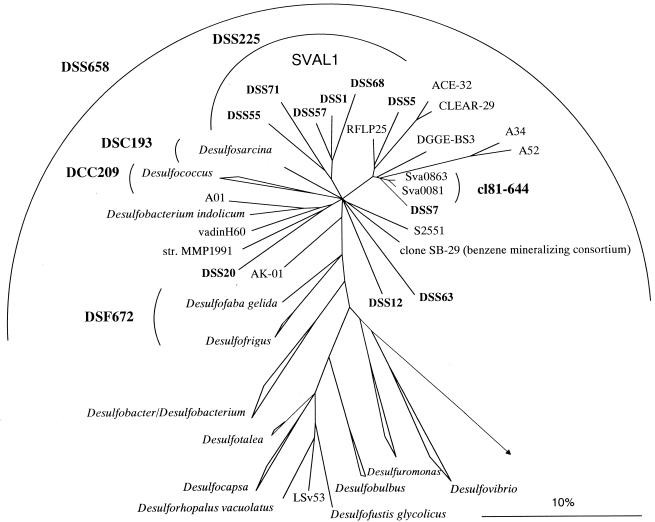
A new strategy was needed to further identify the sarcinalike cells in the DSS658 target group. We used probe DSS658 as a specific primer in combination with a universal eubacterial primer for specific amplification of the DSS658 target 16S rDNAs. To verify that the amplification was specific, we performed parallel PCR with reference DNAs with one to three mismatches with DSS658. Targets with more than one mismatch could be distinguished. A clone library was set up, and 40 clones with an insert of the correct size, 650 bp, were screened by ARDRA. Fourteen different patterns were found after digestion with two restriction enzymes. Sequence analysis of representatives of all of the patterns showed that all of the clone sequences except two gamma-proteobacterial sequences fell in the Desulfosarcina-Desulfococcus group (Fig. 4). The highest sequence similarity was 96.9% between clone DSS7 and DGGE BS3, a sequence retrieved from Black Sea sediment (34). Desulfosarcina variabilis was the closest relative of the most frequent clone group (19 of 40 clones), which was designated the SVAL1 group (91.2%). On the basis of this new sequence data we developed a probe (DSS225) for SVAL1. Desulfosarcina variabilis and Desulfofaba gelida exhibited one weak central mismatch with the probe (G = U) and therefore could not be discriminated. The inclusion of Desulfosarcina spp. and Desulfofaba sp. was not relevant for this study because no members of the genera Desulfosarcina and Desulfofaba were detected in our samples. Using new probe DSS225, we detected very high numbers of cells with sarcinalike morphology (Fig. 3) and very few rods. The distributions of DSS225- and DSS658-targeted cells were almost identical, and maxima occurred at the same depth. Probe DSS658 detected both DSS225 and cl81-644 target cells. By adding the detection rates for the individual more specific probes we could recover roughly 100% of the DSS658-detected cells along the vertical profile.
FIG. 4.
Phylogenetic tree showing the affiliations of 16S rDNA clone sequences with selected reference sequences of members of the delta subclass of the Proteobacteria. The tree was calculated by using maximum-likelihood analysis and was corrected with filters which considered only 50% conserved regions of the 16S rRNAs of members of the delta subclass of the Proteobacteria. The DSS clones, as well as clone sequences A01, SB-29, RFLP25, ACE-32, CLEAR-29, A52, A34, and DGGE-BS3, are not full-length sequences (length, 650 to 900 bp) and therefore were added to the existing tree by using a special algorithm included in the ARB software without allowing changes in the tree topology based on almost complete sequences. Different calculations of phylogenetic trees did not result in a stable branching order for some subgroups. Consequently, the phylogenetic affiliations of these subgroups are shown as multifurcations. New cloned 16S rDNA sequences are indicated by boldface type. The group consisting of clone sequences DSS1, DSS5, DSS55, DSS71, and DSS68 was designated SVAL1. Bar = 10% estimated phylogenetic divergence.
Applying probe DSS225 on selected layers of sediment samples obtained from the other stations (stations I and K and Hornsund), very high cell numbers and relative proportions of DSS658-targeted cells were detected (data not shown). Up to 94% of DSS658-detected cells were targeted by specific probe DSS225 in Hornsund sediment (station K, 80%; station I, 33%).
Total SRB and SRRs.
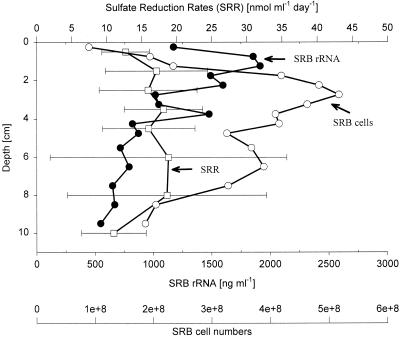
Adding up the number of cells from the individual groups of SRB, as well as the rRNA recovered from these groups, gave an overview of the detectable SRB population along the depth profile (Fig. 5). Up to 5.2 × 108 SRB ml−1 (15% of DAPI cell counts) and up to 25.3% prokaryotic rRNA were detected at depths of 2.25 and 2.75 cm, respectively. The highest SRB rRNA yield, however, was obtained at a 1.25 cm depth with (1,914 ng ml of sediment−1). Due to the high standard deviation the sulfate reduction rates (SRR) had to be considered essentially constant along the vertical profile.
FIG. 5.
Depth profiles for SRB abundance, SRB rRNA concentrations, and SRRs. Numbers of SRB cells (○) and rRNA concentrations (●) were determined by adding the values for the groups targeted by probes DSS658 (Desulfosarcina-Desulfococcus group), DSR651 (Desulforhopalus spp.), DSV698 (Desulfovibrio spp.), Sval428 (Desulfotalea spp.), DSB985 (Desulfobacter spp.), 221 (Desulfobacterium spp.), and 660 (Desulfobulbus spp.); means based on the values for two cores are shown. The SRRs (□) are mean values based on the values for three cores.
Cell-specific SRRs.
Based on SRB cell numbers, average cellular SRRs were calculated. The highest rate per cell was found in the uppermost layer (0.14 fmol cell−1 day−1). This rate decreased by a factor of 3.5 within the first 3 cm and was relatively constant in deeper layers.
Cell-specific rRNA content.
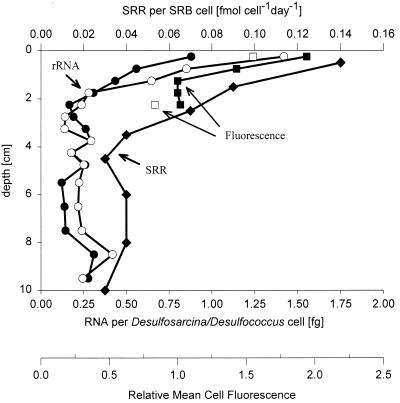
A combination of two methods, FISH and slot blot hybridization, allowed us to calculate specific cellular rRNA contents for individual groups. The calculated average cellular rRNA content for the Desulfosarcina-Desulfococcus group, the most abundant group in this study, exhibited a trend similar to that of the vertical profile of SRB cell-specific SRRs. The RNA content was highest in the first 5 mm of the sediment; the values obtained were 0.9 fg of RNA per cell (core A) and 1.4 fg of RNA per cell (core B) (Fig. 6). In the vertical profile there was a strong decrease in the cellular rRNA content in the first 1.75 cm; in cores A and B the rRNA content decreased by factors of 3 and 6, respectively. In deeper layers the cellular rRNA content remained low. To verify these findings with an independent method, we quantified the average cell fluorescence at the single-cell level with confocal laser scanning microscopy (Fig. 6). We found that the cells in the uppermost layer had an approximately twofold brighter signal with probe DSS658 than the cells in deeper layers. This finding supports the calculated trend since a brighter FISH signal is related to a higher cellular rRNA content.
FIG. 6.
Depth profiles for specific rRNA contents and mean cell fluorescence for DSS658 (Desulfosarcina-Desulfococcus group)-targeted cells and SRRs per SRB cell. The average cellular rRNA contents were determined by combining FISH and rRNA hybridization data (●, core A; ○, core B). The mean fluorescence of hybridized cells was quantified by laser scanning microscopy in the following way. For each depth a mean cell fluorescence was calculated; the lowest mean cell fluorescence value was defined as 1, and the mean fluorescence values for cells in the other layers were expressed relative to this value (■, core A; □, core B). SRRs per SRB cell (⧫) were based on SRB abundance.
DISCUSSION
SRB community structure.
The major group of SRB identified was the Desulfosarcina-Desulfococcus group. Between 49 and 73% of FISH-detected SRB and between 44 and 70% of total SRB rRNA belonged to this group of completely oxidizing sulfate reducers. The predominance of the Desulfosarcina-Desulfococcus group in the Smeerenburgfjorden sediment was confirmed with sediment samples taken 1 year later. This group was also found to be the dominant group at three other sampling sites off the coast of Svalbard. Thus, members of the Desulfosarcina-Desulfococcus group seem to be able to survive under various conditions. This conclusion is supported by the fact that high abundances of members of the Desulfosarcina-Desulfococcus group have been shown previously in different habitats. Sahm et al. (38) found between 71.7 and 85% SRB rRNA in a coastal sediment, Rooney-Varga et al. (33) detected up to 15.5% of bacterial rRNA with probes specific for 16S rDNA clones affiliated with Desulfosarcina variabilis and Desulfococcus multivorans, and Edgcomb et al. (10) estimated high cell numbers based on probe-detected rDNAs in salt marsh sediments. Furthermore, Mußmann and Llobet Brossa found high levels of members of the Desulfosarcina-Desulfococcus group in Wadden Sea sediment (personal communication). Desulfosarcina spp. and Desulfococcus spp. are known to be nutritionally versatile with respect to potential electron donors and are capable of complete oxidation of organic carbon to CO2 (48). Some strains and 16S rDNA clone sequences in this group (Fig. 6) have been isolated from contaminated sites; strain S2552 (accession no. AF177428) was isolated from an oil reservoir, clone RFLP25 (accession no. AF058007) was derived from a polychlorinated biphenyl-dechlorinating culture (27), and strain AK-01 (accession no. AF141328) was isolated from an estuarine sediment with a history of chronic petroleum contamination (40).
The predominance of members of the Desulfosarcina-Desulfococcus group may reflect the availability of a variety of complex organic matter rather than the input of one specific substrate as an electron donor. This seems only reasonable in a natural habitat, where a diverse community of prokaryotes might produce a wide range of carbon sources in the food chain. Using bag incubations of sediment slurries, Purdy et al. (28) demonstrated that the availability of a single substrate potentially favors other groups; e.g., propionate supported the growth of Desulfobulbus spp. The nutritional versatility of the Desulfosarcina-Desulfococcus group could also be advantageous in case of competition for limited carbon sources in this extreme habitat. Other species, like Desulfovibrio spp., can use only a few simple organic acids, hydrogen, and (in some cases) ethanol as an electron donor. They do not grow well in the presence of low substrate concentrations but were found to be favored by higher substrate concentrations (44, 45, 48).
In previous studies on the microbial community of Svalbard sediments, Sahm and coworkers quantified selected groups of SRB by slot blot hybridization and found that rRNA of members of the Desulfovibrionaceae was dominant (37). However, due to results obtained with a clone library established by using the same sediment samples (32), they assumed that the rRNA detected might have come from organisms belonging to the Geobacteraceae group because a significant portion of the clone sequences in the Svalbard sediment clone library (32) gave positive signals with the same probe (probe 687). All sequences were sequences of members of the family Geobacteraceae and were most closely related to the Desulfuromonas palmitatis sequence. The fact that we detected high levels of Desulfuromonas rRNA in the present study supports this conclusion. The newly isolated genus Desulfotalea was the second most abundant group of SRB in the studies of Sahm et al.; the relative abundance was 0.6 to 4.4% of the prokaryotic rRNA at the relevant depth. In the present study we obtained similar results; Desulfotalea spp. accounted for 0.3 to 5.6% of the prokaryotic rRNA. However, detection of additional groups of SRB (e.g., the Desulfosarcina-Desulfococcus group) showed that there are other groups that are present at even higher abundance. Sahm et al. did not find significant amounts of Desulfosarcina-Desulfococcus rRNA (37). A possible explanation for the failure to detect Desulfococcus-Desulfosarcina rRNA is that these organisms were not targeted by the probe used (probe 804) (6). We cannot resolve this discrepancy yet, since the sequence data for the dominant subgroup of uncultured sarcinalike cells which we describe in this paper does not contain the target position for probe 804.
Resolution of the Desulfosarcina-Desulfococcus group.
Although only a few strains of the DSS658 target group have been cultivated so far, perhaps due to the use of substrates that are too simple (such as lactate or propionate), molecular biological studies have revealed a very high diversity in this group. In the last few years the diversity of the Desulfosarcina-Desulfococcus group has been greatly extended by 16S rDNA cloning (7, 27, 33, 46), denaturing gradient gel electrophoresis analysis (34), and cultivation with complex substrates (40). The sequences often exhibit only 90% sequence similarity to their closest relative or to a cultivated strain. Cultivation of this major group of marine SRB should be a goal for future studies.
In our study none of the Desulfosarcina-Desulfococcus cells detected could be affiliated with known genera. The closest relative of the most abundant clone group, group SVAL1, was Desulfosarcina variabilis, with 92% sequence similarity. The newly designed probe DSS225, which is specific for group SVAL1, detected up to three-quarters of the DSS658-targeted cells and produced an almost identical vertical profile. However, the phylogenetic distance between Desulfosarcina spp. and the clone sequences is so large (8%) that we can only speculate on the physiological properties of the organisms. The ability to oxidize substrates completely to CO2 and nutritional versatility are features that are common to almost all species belonging to the Desulfosarcina-Desulfococcus group; thus, we assume that the bacteria detected also have these physiological characteristics. Attempts to perform directed cultivation of SRB from the same habitat are under way. Additional studies on the presence and abundance of the new SVAL1 group in other habitats, like antarctic sediments or temperate environments, should show whether the dominance and ecological significance of this group are restricted to Svalbard sediments.
Specific cellular rRNA content and specific SRRs.
In this study we combined FISH and rRNA hybridization data to calculate average cellular rRNA contents of Desulfosarcina-Desulfococcus cells. The calculated average cellular rRNA content of these cells was greatest in the upper 5 mm of sediment and decreased steeply within the first 2 cm. The ribosome content and with that the rRNA content are directly connected to the growth rate in steady-state cultures (for a review see reference 26). Molin and Givskow, however, have cautioned to use cellular rRNA measurements on cells growing in a complex environment under changing nutritional conditions to address cellular growth activities. To translate the measured cellular rRNA contents into absolute growth rates, pure-culture experiments performed with specific strains are needed. Even then, in a natural habitat different biological and nonbiological factors interfere with each other and might activate different global-transcriptional control networks in the cell, thereby influencing the direct correlation between growth and rRNA synthesis. We would like to add that species heterogeneity within a probe target group might further complicate the picture. In our study, however, the major target group of SRB was dominated by one group of closely related organisms throughout the whole depth profile, and the cellular rRNA content was consistent with independently determined cellular SRRs. This makes us confident that we obtained useful information about the physiological state of the SRB detected. These results suggest that although growth rates might be generally low in the natural habitat, they change along the depth profile. Closeness to the sediment surface guarantees the availability of different substrates and could therefore explain why the highest cellular rRNA contents are in the first layer.
In this paper we describe, to our knowledge for the first time, cellular SRRs and the corresponding vertical profile obtained for total SRB cells, as quantified by a cultivation-independent method. The cellular SRRs, which were calculated from numbers of SRB cells as detected by FISH, were highest at the sediment surface, where they were 0.14 fmol of SO4 per day, and decreased steeply with depth to 0.02 fmol of SO4 per day. They were lower, by factors of 5 to 50, than the specific SRRs of mesophilic SRB that were grown in pure cultures at 4°C (19). For Desulfosarcina variabilis and Desulfococcus niacini SRRs of 0.7 ± 0.4 and 1.2 ± 0.05 fmol of SO4 cell−1 day−1, respectively, were obtained. Nevertheless, our calculated rates seem to be in a reasonable range for natural, substrate-limited environments. The general finding that the cellular SRRs were much higher in the first 5 mm than in the suboxic or anoxic zones might even be more pronounced since SRRs probably are underestimated rather than overestimated in oxidized layers (18).
Combination of FISH and rRNA slot blot hybridization for quantification of bacteria—methodological considerations.
Quantification of SRB by FISH and quantification of SRB by slot blot hybridization gave comparable results. This comparability is encouraging. A comparison of studies based on FISH and studies based on slot blot hybridization is possible almost without reservation, although the two methods have different drawbacks (2). Despite the different methodological constraints, only detection of groups at levels just above the detection limit resulted in minor discrepancies in this study. For example, Desulfobacter sp. rRNA could be detected in some layers, but no cells were detected by FISH, suggesting that the rRNA detected was distributed over a relatively large fraction of probably less active cells with low cellular rRNA contents.
A combination of the two methods allowed us to calculate the specific cellular rRNA contents. In this study we found a good correlation between the cellular SRR and the cellular rRNA content of SRB. It would be rewarding to investigate other natural systems for this correlation between cellular activity and rRNA content.
ACKNOWLEDGMENTS
We thank Marc Mußmann for the 1999 sampling and fixation of Hornsund and Smeerenburgfjorden sediments. Annelie Hentschke and Armin Gieseke are acknowledged for introducing us to confocal laser scanning microscopy, and we thank Falk Warnecke for assistance with RNA extraction.
This work was supported by the Max Planck Society.
REFERENCES
- 1.Amann R I, Binder B J, Olson R J, Chisholm S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canfield D E, Jørgensen B B, Fossing H, Glud R, Gundersen J, Ramsing N B, Thamdrup B, Hansen J W, Nielsen L P, Hall P O J. Pathways of organic carbon oxidation in three continental margin sediments. Mar Geol. 1993;113:27–40. doi: 10.1016/0025-3227(93)90147-n. [DOI] [PubMed] [Google Scholar]
- 4.Coleman M L, Hedrick D B, Lovley D R, White D C, Pye K. Reduction of Fe(III) in sediments by sulphate-reducing bacteria. Nature. 1993;361:436–438. [Google Scholar]
- 5.Dannenberg S, Kroder M, Dilling W, Cypionka H. Oxidation of H2, organic compounds and inorganic sulfur compounds coupled to reduction of O2 or nitrate by sulfate-reducing bacteria. Arch Microbiol. 1992;158:93–99. [Google Scholar]
- 6.Devereux R, Kane M D, Winfrey J, Stahl D A. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst Appl Microbiol. 1992;15:601–609. [Google Scholar]
- 7.Devereux R, Mundfrom G W. A phylogenetic tree of 16S rRNA sequences from sulfate-reducing bacteria in sandy marine sediment. Appl Environ Microbiol. 1994;60:3437–3439. doi: 10.1128/aem.60.9.3437-3439.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devereux R, Winfrey M R, Winfrey J, Stahl D A. Depth profile of sulfate-reducing bacterial ribosomal RNA and mercury methylation in an estuarine sediment. FEMS Microbiol Ecol. 1996;20:23–31. [Google Scholar]
- 9.Dilling W, Cypionka H. Aerobic respiration in sulfate-reducing bacteria. FEMS Microbiol Lett. 1990;71:123–128. [Google Scholar]
- 10.Edgcomb V P, McDonald J H, Devereux R, Smith D W. Estimation of bacterial cell numbers in humic acid-rich salt marsh sediments with probes directed to 16S ribosomal DNA. Appl Environ Microbiol. 1999;65:1516–1523. doi: 10.1128/aem.65.4.1516-1523.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fossing H, Jørgensen B B. Measurements of bacterial sulfate reduction in sediments: evaluation of a single-step chromium reduction method. Biogeochemistry. 1989;8:205–222. [Google Scholar]
- 12.Fuchs B M, Wallner G, Beisker W, Schwippl I, Ludwig W, Amann R. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 1998;64:4973–4982. doi: 10.1128/aem.64.12.4973-4982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glöckner F O, Amann R, Alfreider A, Pernthaler J, Psenner R, Trebesius K, Schleifer K-H. An in situ hybridization protocol for the detection and identification of planktonic bacteria. Syst Appl Microbiol. 1996;63:4237–4242. [Google Scholar]
- 14.Hicks R E, Amann R I, Stahl D A. Dual staining of natural bacterioplankton with 4′,6-diamidino-2-phenylindole and fluorescent oligonucleotide probes targeting kingdom-level 16S rRNA sequences. Appl Environ Microbiol. 1992;58:2158–2163. doi: 10.1128/aem.58.7.2158-2163.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Janssen P H, Schuhmann A, Bak F, Liesack W. Disproportionation of inorganic sulfur compounds by the sulfate-reducing bacterium Desulfocapsa thiozymogenes gen. nov., sp. nov. Arch Microbiol. 1996;166:184–192. [Google Scholar]
- 16.Jørgensen B B. A comparison of methods for the quantification of bacterial sulfate reduction in coastal marine sediments. I. Measurements with radiotracer techniques. Geomicrobiol J. 1978;1:11–27. [Google Scholar]
- 17.Jørgensen B B. Mineralization of organic matter in the sea bed—the role of sulphate reduction. Nature. 1982;296:643–645. [Google Scholar]
- 18.Jørgensen B B, Bak F. Pathways and microbiology of thiosulfate transformations and sulfate reduction in a marine sediment (Kattegat, Denmark) Appl Environ Microbiol. 1991;57:847–856. doi: 10.1128/aem.57.3.847-856.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knoblauch C, Jørgensen B B, Harder J. Community size and specific sulfate reduction rates of psychrophilic sulfate-reducing bacteria in arctic marine sediments: evidence for high activity at low temperature. Appl Environ Microbiol. 1999;65:4230–4233. doi: 10.1128/aem.65.9.4230-4233.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knoblauch C, Sahm K, Jørgensen B B. Psychrophilic sulfate-reducing bacteria isolated from permanently cold Arctic marine sediments: description of Desulfofrigus oceanense gen. nov., sp. nov., Desulfofrigus fragile sp. nov., Desulfofaba gelida gen. nov., sp. nov., Desulfotalea psychrophila gen. nov., sp. nov. and Desulfotalea arctica sp. nov. Int J Syst Bacteriol. 1999;49:1631–1643. doi: 10.1099/00207713-49-4-1631. [DOI] [PubMed] [Google Scholar]
- 21.Krämer M, Cypionka H. Sulfate formation via ATP sulfurylase in thiosulfate- and sulfite-disproportionating bacteria. Arch Microbiol. 1989;151:232–237. [Google Scholar]
- 22.Krekeler D, Teske A, Cypionka H. Strategies of sulfate-reducing bacteria to escape oxygen stress in a cyanobacterial mat. FEMS Microbiol Ecol. 1998;25:89–96. [Google Scholar]
- 23.Llobet-Brossa E, Rossello-Mora R, Amann R. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl Environ Microbiol. 1998;64:2691–2696. doi: 10.1128/aem.64.7.2691-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lovley D R, Phillips E J P. Novel processes for anaerobic sulfate production from elemental sulfur by sulfate-reducing bacteria. Appl Environ Microbiol. 1994;60:2394–2399. doi: 10.1128/aem.60.7.2394-2399.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manz W, Eisenbrecher M, Neu T R, Szewzyk U. Abundance and spatial organization of gram negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol Ecol. 1998;25:43–61. [Google Scholar]
- 26.Molin S, Givskov M. Application of molecular tools for in situ monitoring of bacterial growth activity. Environ Microbiol. 1999;1:383–391. doi: 10.1046/j.1462-2920.1999.00056.x. [DOI] [PubMed] [Google Scholar]
- 27.Pulliam Holoman T R, Elberson M A, Cutter L A, May H D, Sowers K R. Characterization of a defined 2,3,5,6-tetrachlorobiphenyl-ortho-dechlorinating microbial community by comparative sequence analysis of genes coding for 16S rRNA. Appl Environ Microbiol. 1998;64:3359–3367. doi: 10.1128/aem.64.9.3359-3367.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Purdy K J, Nedwell D B, Embley T M, Takii S. Use of 16S rRNA-targeted oligonucleotide probes to investigate the occurrence and selection of sulfate-reducing bacteria in response to nutrient addition to sediment slurry microcosms from a Japanese estuary. FEMS Microbiol Ecol. 1997;24:221–234. [Google Scholar]
- 29.Ramsing N, Fossing H, Ferdelman T G, Andersen F, Thamdrup B. Distribution of bacterial populations in a stratified fjord (Mariager Fjord, Denmark) quantified by in situ hybridization and related to chemical gradients in the water column. Appl Environ Microbiol. 1996;62:1391–1404. doi: 10.1128/aem.62.4.1391-1404.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raskin L, Poulsen L K, Noguera D R, Rittmann B E, Stahl D A. Quantification of methanogenic groups in anaerobic biological reactors by oligonucleotide probe hybridization. Appl Environ Microbiol. 1994;60:1241–1248. doi: 10.1128/aem.60.4.1241-1248.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raskin L, Stromley J M, Rittmann B E, Stahl D A. Group-specific 16S rRNA hybridization probes to describe natural communities of methanogens. Appl Environ Microbiol. 1994;60:1232–1240. doi: 10.1128/aem.60.4.1232-1240.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ravenschlag K, Sahm K, Pernthaler J, Amann R. High bacterial diversity in permanently cold marine sediments. Appl Environ Microbiol. 1999;65:3982–3989. doi: 10.1128/aem.65.9.3982-3989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rooney-Varga J N, Devereux R, Evans R S, Hines M E. Seasonal changes in the relative abundance of uncultivated sulfate-reducing bacteria in a salt marsh sediment and in the rhizosphere of Spartina alterniflora. Appl Environ Microbiol. 1997;63:3895–3901. doi: 10.1128/aem.63.10.3895-3901.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rossello-Mora R, Thamdrup B, Schaefer H, Weller R, Amann R. The response of the microbial community of marine sediments to organic carbon input under anaerobic conditions. Syst Appl Microbiol. 1999;22:237–248. doi: 10.1016/S0723-2020(99)80071-X. [DOI] [PubMed] [Google Scholar]
- 35.Sagemann J, Jørgensen B B, Greeff O. Temperature dependence and rates of sulfate reduction in cold sediments of Svalbard, Arctic Ocean. Geomicrobiol J. 1998;15:85–100. [Google Scholar]
- 36.Sahm K, Berninger U-G. Abundance, vertical distribution, and community structure of benthic prokaryotes from permanently cold marine sediments (Svalbard, Arctic Ocean) Mar Ecol Prog Ser. 1998;165:71–80. [Google Scholar]
- 37.Sahm K, Knoblauch C, Amann R. Phylogenetic affiliation and quantification of psychrophilic sulfate-reducing isolates in marine arctic sediments. Appl Environ Microbiol. 1999;65:3976–3981. doi: 10.1128/aem.65.9.3976-3981.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahm K, MacGregor B J, Jørgensen B B, Stahl D A. Sulfate reduction and vertical distribution of sulfate-reducing bacteria quantified by rRNA slot-blot hybridization in a coastal marine sediment. Environ Microbiol. 1999;1:65–74. doi: 10.1046/j.1462-2920.1999.00007.x. [DOI] [PubMed] [Google Scholar]
- 39.Snaidr J, Amann R, Huber I, Ludwig W, Schleifer K H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl Environ Microbiol. 1997;63:2884–2896. doi: 10.1128/aem.63.7.2884-2896.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.So C M, Young L Y. Isolation and characterization of a sulfate-reducing bacterium that anaerobically degrades alkanes. Appl Environ Microbiol. 1999;65:2969–2976. doi: 10.1128/aem.65.7.2969-2976.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorokin D Y, Teske A, Robertson L A, Kuenen J G. Anaerobic oxidation of thiosulfate to tetrathionate by obligately heterotrophic bacteria, belonging to the Pseudomonas stutzeri group. FEMS Microbiol Ecol. 1999;30:113–123. doi: 10.1111/j.1574-6941.1999.tb00640.x. [DOI] [PubMed] [Google Scholar]
- 42.Stahl D A, Flesher B, Mansfield H R, Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988;54:1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strunk O, Gross O, Reichel B, May M, Hermann S, Stuckman N, Nonhoff B, Lenke M, Ginhart A, Vilbig A, Ludwig T, Bode A, Schleifer K-H, Ludwig W. ARB: a software environment for sequence data. http://www.mikro.biologie.tu-muenchen.de/pub/ARB. Munich, Germany: Department of Microbiology Technische Universität München; 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor J, Parkes R J. Identifying different populations of sulfate reducing bacteria within marine sediment systems, using fatty acid biomarkers. J Gen Microbiol. 1985;131:631–642. [Google Scholar]
- 45.Trimmer M, Purdy K J, Nedwell D B. Process measurement and phylogenetic analysis of the sulfate reducing bacterial communities of two contrasting benthic sites in the upper estuary of the Great Ouse, Norfolk, UK. FEMS Microbiol Ecol. 1997;24:333–342. [Google Scholar]
- 46.Urakawa H, Kita-Tsukamota K, Ohwada K. Microbial diversity in marine sediments from Sagami Bay and Tokyo Bay, Japan, as determined by 16S rRNA gene analysis. Microbiology. 1999;145:3305–3315. doi: 10.1099/00221287-145-11-3305. [DOI] [PubMed] [Google Scholar]
- 47.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganism. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 48.Widdel F, Bak F. Gram-negative mesophilic sulfate-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. Vol. 1. New York, N.Y: Springer-Verlag; 1992. pp. 3352–3378. [Google Scholar]
- 49.Widdel F, Hansen T. The dissimilatory sulfate- and sulfur-reducing bacteria. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The Prokaryotes. 2nd ed. Vol. 1. New York, N.Y: Springer-Verlag; 1992. pp. 583–624. [Google Scholar]
- 50.Wollast R. The coastal organic carbon cycle: fluxes, sources, and sinks. In: Mantoura R F C, Martin J-M, Wollast R, editors. Ocean margin processes in global change. New York, N.Y: John Wiley & Sons; 1991. pp. 365–381. [Google Scholar]
- 51.Zheng D, Alm E W, Stahl D A, Raskin L. Characterization of universal small-subunit rRNA hybridization probes for quantitative molecular microbial ecology studies. Appl Environ Microbiol. 1996;62:4314–4317. doi: 10.1128/aem.62.12.4504-4513.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou J, Brunns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]