Abstract
Mounting clinical evidence suggests that viral infections can lead to detectable changes in an individual’s normal physiologic and behavioral metrics, including heart and respiration rates, heart rate variability, temperature, activity, and sleep prior to symptom onset, potentially even in asymptomatic individuals. While the ability of wearable devices to detect viral infections in a real-world setting has yet to be proven, multiple recent studies have established that individual, continuous data from a range of biometric monitoring technologies can be easily acquired and that through the use of machine learning techniques, physiological signals and warning signs can be identified. In this review, we highlight the existing knowledge base supporting the potential for widespread implementation of biometric data to address existing gaps in the diagnosis and treatment of viral illnesses, with a particular focus on the many important lessons learned from the coronavirus disease 2019 pandemic.
Keywords: biomarker, BioMeT, biometric, COVID-19, FDA, health equity, infectious disease, remote monitoring, wearables
1. INTRODUCTION
Recent advancements in wearable sensors, also known as wearables, have improved their popularity and ability to continuously collect raw physiological parameters that can be used to determine clinically useful health information. The combination of data collected from a range of wearable sensors with advanced data processing algorithms has unearthed the potential for early stage infection detection, a key step to limiting the spread of infectious diseases. The objective of this review is to highlight recent advancements in biometric monitoring technologies (BioMeTs), data processing algorithms, and clinical insights associated with the detection of influenza-like illnesses (ILIs). Particular attention is paid to data quality, machine learning (ML) algorithms, the need for standards, and clinical challenges that remain to be addressed. While much work is still to be done before BioMeTs are routinely used for detection of infections, the potential for continuous monitoring to help limit the spread of infectious disease is great, especially given the recent coronavirus disease 2019 (COVID-19) pandemic.
2. CURRENT AND FUTURE BioMeTs
BioMeTs process mobile sensor–generated data to develop physiological and/or behavioral measures using algorithms (1). These technologies are also referred to as biometric monitoring devices, wearable health devices, smart devices (e.g., smartwatches), wearable sensors, or simply wearables. Form factors include watches/wristbands, rings, earbuds, headbands, clothing, and patches that capture information on movement, heart rate (HR), heart rate variability (HRV), respiratory rate (RR), blood pressure, eye tracking, hydration level, glucose level, skin conductivity, sleep, temperature, posture, brain activity, oxygen level, muscle activity, and gastric activity, among others. BioMeTs have shown promise for promoting general health and wellness by enabling self-monitoring of habits, providing data-driven feedback, and supporting sharing of personal data with healthcare providers and family members.
New wellness features such as displaying fitness and sleep statistics and trends, personalized coaching, and tailored self-monitoring ranging from water intake to women’s health tracking are continuously being added to commercially available consumer BioMeTs as demand for such capabilities increases. The ease of use, cost, efficacy, and ability of BioMeTs to continuously monitor fitness, wellness, and more recently, health, has led to an increase in the number of people using these devices. A study to evaluate the use of wearable devices among US adults found that 30% use wearable devices and 82% are willing to share the health data from their BioMeTs with their care providers (2). Increased access to long-term user data can help healthcare providers understand and monitor individual health trends and concerns and facilitate more personalized treatment paths, highlighting the tremendous potential of BioMeTs to transform health care.
2.1. Intended BioMeT Use
As a result of the types of continuous and longitudinal information collected, BioMeTs hold immense potential for real-time health tracking and illness prediction in free-living and remote conditions. They may also provide insights into disease origin, severity, and progression. Thus, BioMeTs offer a huge advantage over in-clinic measurements at single points in time that do not provide the complete picture of an individual’s health. Due to their capabilities, low cost, and ease of use, BioMeTs now form a major component of digital medicine technologies. Digital medicine is the use of technologies for improving human health through measurement and intervention to support the practice of medicine (3). These technologies include high-quality hardware and software tools that undergo rigorous clinical validation for their impact on disease diagnosis, treatment, recovery, prevention, and health promotion (3, 4).
BioMeTs can be classified into two main designations depending on their intended use (5):
Wellness: nonmedical, low-risk products that promote a healthy lifestyle and are intended by the manufacturer for self-monitoring (6). Examples of consumer wearables in the wellness category include the Xiaomi Mi Band, the Amazon Halo band, and the Garmin smartwatch.
Medical: products intended for the diagnosis or treatment of medical conditions. Medical devices are used primarily for prediction, anomaly detection, and diagnosis support. Examples include the Philips Actiwatch Spectrum Pro, the VitalConnect HealthPatch MD, the Empatica Embrace, and Dexcom’s Continuous Glucose Monitoring System (7).
The reader should note that this is a rapidly evolving climate where existing wellness devices are increasingly being adapted toward medical purposes by improving accuracy and expanding functionality. The US Food and Drug Administration (FDA) defines intended use of medical devices as the “objective intent of the persons legally responsible for the labeling of devices.” The intended use can be shown through “labeling claims, advertising matter, or oral or written statements” (8). However, some BioMeTs simultaneously have separate functionalities, where one feature is intended for wellness and another feature is intended for medical purposes. Thus, a single device can be subject to different sets of FDA regulations, making it difficult for consumers and clinicians to assess the evidence supporting their approval status. For instance, at the time of this writing, the Apple Watch Series 6 electrocardiogram (ECG) is FDA cleared for detecting irregular cardiac rhythms, while the blood oxygen monitor on the same device is advertised by the company for wellness purposes and is therefore not subject to FDA regulations (9).
The FDA’s Center for Devices and Radiological Health (CDRH) regulates companies who “manufacture, repackage, relabel, and/or import medical devices sold in the United States” and establishes requirements for safe and effective medical devices (7, 10). The CDRH’s Digital Health Center of Excellence works toward digital health advancements by fostering responsible innovations in digital health (https://www.fda.gov/medical-devices/digital-health-center-excellence). In a step toward collaboration and interdisciplinary partnership, the Network of Digital Health Experts created by the CDRH comprises scientists, clinicians, and engineers from diverse organizations who advise the FDA staff on matters regarding digital health oversight (7). Similarly, there are collaborative communities led by private- and public-sector stakeholders that often work with and provide guidance to the CDRH to advance digital health.
2.2. BioMeT Accuracy
The accuracy of BioMeTs is a key consideration for their real-world deployment in health research and care. Therefore, standardized best practice guidelines for the systematic evaluation of BioMeTs are essential. Goldsack et al. (1) describe such a framework to evaluate BioMeTs, including verification, analytical validation, and clinical validation (which they refer to as V3). Verification is determining sample-level sensor data accuracy compared with a reference standard (e.g., evaluating an accelerometer using an in-lab vibration plate), and validation is determining the real-world performance for detecting behavior and physiology against predefined criteria (11). The latter is divided into analytic and clinical validation, which are defined respectively as evaluation from the bench to in vivo (e.g., evaluating an accelerometer to detect steps from a human) and evaluation on patients to determine the ability of the technology to distinguish health and disease states. It is also essential to determine whether a BioMeT is fit for purpose; in other words, that its data collection and processing methods are adequate for the BioMeT’s intended purpose and that these functions have been confirmed experimentally. This determination may include assessing other useful BioMeT characteristics such as economic feasibility, security risks, data protections, utility, and usability, all of which are currently understudied areas (11).
BioMeT data processing algorithms may also vary substantially by manufacturer. The lack of transparency around BioMeT data processing by manufacturers is problematic because there is a loss of interpretability for research and clinical decision-making. Wearables can be prone to inaccuracies in measurements due to several factors, including on-body placement, improper wear, and the processing algorithms. For instance, inaccuracies in photoplethysmography (PPG), which is used to derive HR from optical sensing, can arise from diverse skin types, motion artifacts, and signal crossover (12). The accuracy of BioMeTs for measuring steps and HR can vary by manufacturer, device type, activity intensity, and type of exercise or activity being performed (13, 14). To convey such important digital health facts for consumers in an easy-to-understand way, labels for digital health products have been proposed that can detail information about product accuracy, past use, adverse events, data safety, and other important digital health facts, similar to food nutrition labels (15). Transparency, measurement accuracy, reliability, and sensitivity of BioMeTs are some of the factors that need to be carefully considered and reported when including consumer BioMeTs in clinical trials or for healthcare purposes. Product labels with this information could greatly benefit both clinicians and consumers.
2.3. Detecting Infection Using BioMeTs
Infection detection models based on physiological signals can be used for personal infection risk alerts and prescreening assessments to intelligently guide who should be tested for infections and when. If an individual is determined to be at-risk for infection, they can quarantine and get tested promptly, hence reducing further spread. Furthermore, contact tracing using ubiquitous personal technologies such as smartphones can be done to notify people who have been exposed to a pathogen and enable them to take timely precautions and isolate. Hence, individuals can be nudged toward a proactive, data-empowered, and personal-responsibility-driven approach for infection prevention using a network of combined biometric monitoring and digital contact tracing technologies, as shown in Figure 1. This approach can reduce infection transmission while conserving expensive resources. A number of studies using wearables that were implemented to help address care gaps exposed during the COVID-19 pandemic are discussed in detail later in this review.
Figure 1.
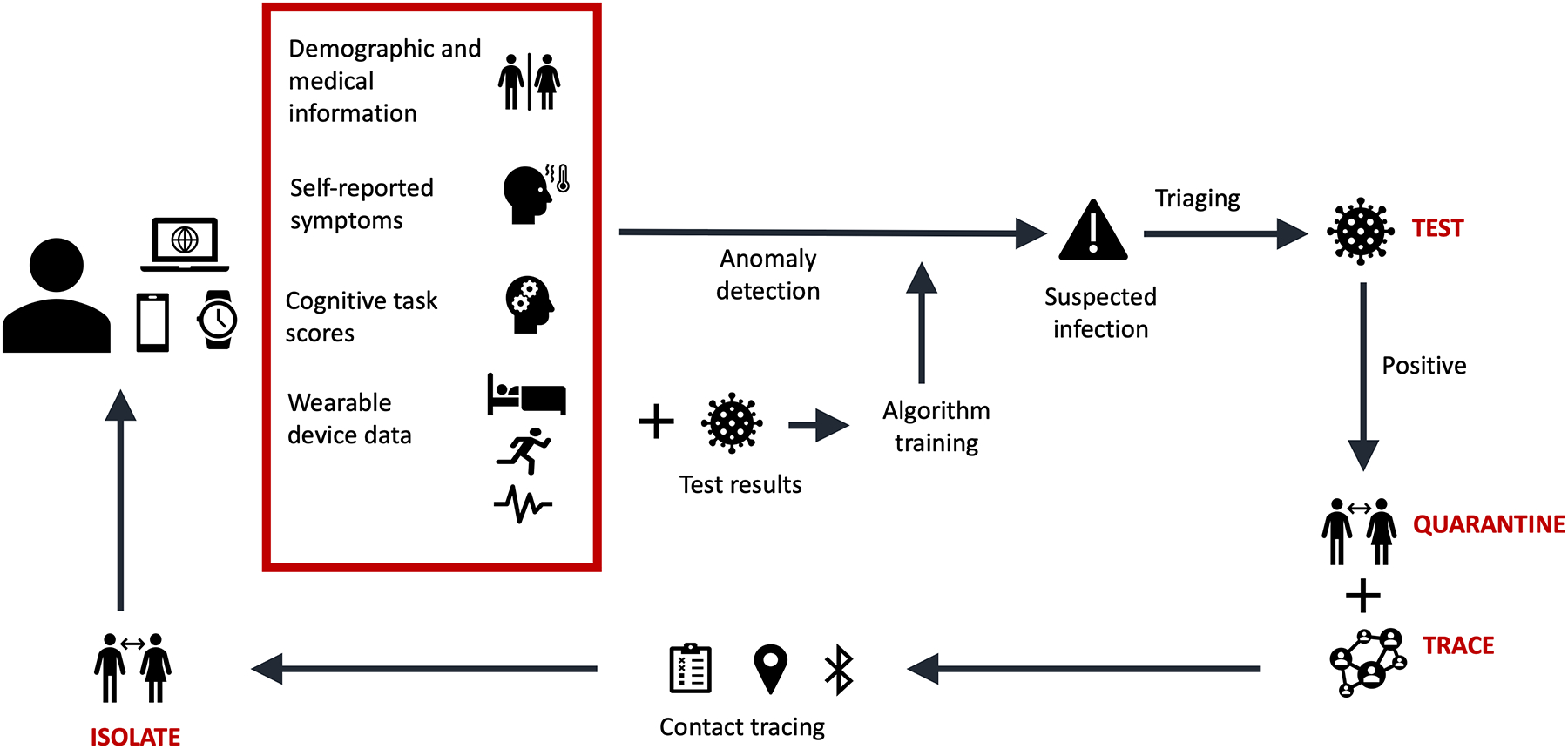
Biometric monitoring and tracking. A conceptual framework is shown for an infectious-disease tracking network using biometric monitoring technologies (BioMeTs) and digital contact tracing tools for rapid test, trace, isolate, and quarantine strategies.
2.4. Reproducibility and Replicability
To improve the generalizability of results from studies using BioMeTs and the utility of those results in clinical settings, it is crucial to address issues that affect replicability (using a new data set that should produce the same results as the original data set) and reproducibility (using the same data set to obtain the same results in a repeated analysis). Various factors that can influence reproducibility and replicability and should be involved in standard reporting include, but are not limited to, protocol design, data cleaning, defining nonwear time and nonadherence, wear location characteristics (e.g., skin tone, freckling/tattoos, skin thickness, hair level, etc.), data provenance/the data supply chain, individual biobehavioral and demographic data, device type, version, sampling rate, known algorithmic changes, etc. (1, 16).
There are other factors that can improve replicability of studies involving BioMeTs. Preregistering studies, an already established practice in clinical trials, can prevent publication bias and define modifications to the original analysis plan. Study groups can report all aspects of the study including the detailed analysis plan and expected outcomes before the data collection process. For bring-your-own-device (BYOD) studies, collaborations between different study sites and groups can increase the power and generalizability of the study. However, BYOD studies can be impacted by unequal BioMeT ownership among different demographic groups, and efforts must be made to ensure a balanced representation from all groups in the study population, such as adoption of demographic improvement guidelines (17). Collaborative and transparent solutions will improve replicability and generalizability of wearable devices studies.
2.5. Opportunities and Challenges When Partnering with Corporate and Healthcare Systems
The increasing footprint of the BioMeT industry has created a unique opportunity for industry partnerships with clinical research teams to explore and expand the usefulness of BioMeTs for health. Preplanning such relationships can facilitate a mutually beneficial and smooth partnership. Maintaining privacy and stringent data security standards while sharing user data is critical. Ensuring that users understand the risks and benefits associated with sharing their data with third parties is another crucial aspect that could be addressed through dedicated digital health counselors. With growing sensing capabilities, the world of personal health is constantly evolving. It is important to ensure that as these capabilities and opportunities grow, so do the requirements for privacy and security standards.
2.6. Multimodal Sensing for Infection Monitoring
Novel and multimodal technologies that passively monitor aspects of human health potentially related to infection are advancing at a rapid pace. Sensors capable of assessing sleep duration and quality exist in smartwatches, rings, and bed sensors (18). Audio signals can be used for cough detection and lung health assessment (19). Wearable eccrine sweat sensors measuring cytokine markers are being explored for real-time infection detection (20). Data collected from these different sources can be combined with an individual’s contextual information to provide a holistic picture of human health and show infection indicators in real time. These data can provide the foundation for the future development of alarm systems to prompt users to seek care at the right time. These personalized alarm systems can be displayed to the users via a cloud-based analytics platform that combines data from multiple sources and uses ML to detect abnormalities that might have been missed from single-source data. For instance, multiple cough detections alone might not trigger the alarm because, for example, they may be a consequence of allergies. This could be verified by geographical, environmental, and smart home sensor data. However, coughing episodes in the absence of environmental allergens with consistent increased resting HR and disturbed sleep patterns might be a cause for concern. These can only be identified by processing multiple sensor data to detect deviations from personal baselines. Such personalized health dashboards can enable individuals to take active charge of their health and give healthcare providers timely access to data to support medical applications (21). Long-term stability, user acceptability, interdevice compatibility, accuracy, and biocompatibility are some of the major aspects that will determine which sensing technologies prove to be the major players in the connected sensor network for passive, holistic human health monitoring.
3. DEVICE ACCESS AND DATA AGGREGATION IN MULTIPLATFORM ENVIRONMENTS
The advent of low-cost consumer BioMeTs has allowed for large-scale physiological data collection without the need for device distribution. This can result in lower-cost BioMeT studies and fewer lost devices as well as further reach to various participant populations. Consumer-grade BioMeTs are often easy to use and connect to familiar smartphone ecosystems. However, consumer BioMeTs are subject to less regulation than validated medical BioMeTs. Given their intended use, consumer BioMeTs also may not have the same durability or battery life as cleared medical devices. A major challenge of multiplatform BioMeT studies is the necessary agglomeration and alignment of disparate data streams. Data sets that are collected across different devices must be eventually reconciled, although they could have been sampled at different rates with separate hardware.
3.1. Current and Proposed Standards
There is no uniform standard currently governing how data from BioMeTs are joined or stored. The pharmaceutical world adopted the Study Data Tabulation Model defined by the Clinical Data Interchange Standards Consortium in 2004 as the FDA sought to streamline the review process for clinical trial data. Accompanying standards such as the Analytical Data Model, the Operational Data Model, and the Clinical Data Acquisition Standards Harmonization model have also risen to meet these needs and are now required components during FDA clinical trial submission (22). Organizations are seeking to maintain health data interoperability through standards such as Fast Healthcare Interoperability Resources and Health Level 7, but these largely concern intrahospital data streams. For data generated and collected in mobile environments, the Open mHealth standard has been proposed.
3.1.1. Data compression.
BioMeTs are often required to transmit freshly collected data in a nearly continuous manner. This is especially true in the context of monitoring viral infections that progress rapidly. In these situations, data compression is necessary to conserve power and maintain data transmission rates. Furthermore, optimizing the rate at which data streams are sampled for the intended use is critical (23). Particularly in limited resource settings, devices may not be recharged for the duration of their wear. Compressive sensing techniques (24) are well suited for BioMeT data compression since they can be implemented using analog-to-digital conversion and digital signal processing (DSP) hardware on the device itself. Data reconstruction is then needed for downstream interpretation of BioMeT signals, but this can be performed centrally after data transmission. The Biosignal Compression Toolbox is the first open-source software to address this need (25).
In the case of ECG-based signals, adaptive linear data prediction methods have performed well (26). Hybrid approaches combining both lossy and lossless compression techniques allow for a reduction in power consumption by nearly 50% (27), even during lossless Bluetooth transmission. Recent findings also suggest that the ECG signal can be successfully compressed and reconstructed using a neural network architecture (28, 29). The particular method of data compression can be further dictated by the arrangement of sensor nodes and the mode of transmission (30). Determining how to maintain data privacy during compression and transmission remains an active area of research (29).
3.1.2. Interplatform harmonization.
Although there is not yet a unifying standard to harmonize BioMeT data, several data- and patient-focused approaches have been proposed (31). The wearable knowledge-as-a-service platform (32) is presented as an extension of the National Institute of Standards and Technology big data model (33) and incorporates data semantics to merge information from different BioMeTs. Other platforms focus on clinical decision support with open BioMeT data warehousing and dashboard tools that integrate across multiple electronic health record (EHR) systems (34, 35). During outbreaks of viral infection, such tools could be markedly useful in drawing public health insights across various healthcare information systems.
The application of the International Organization for Standardization/Institute of Electrical and Electronics Engineers (IEEE) 11073 standard was proposed for connected personal health devices (36) but has not been widely or consistently adopted. More recently, the effort to codify a BioMeT data standard continues with the IEEE P2933 Working Group on the Standard for Clinical Internet of Things Data and Device Interoperability. Focus is placed on integration with existing EHR systems and onboarding of BioMeTs that are not currently interoperable with these platforms. The Open mHealth community has also been leading the development of the now-approved IEEE 1752.1–2021 standard for personal health data collected from sensors and mobile applications. Data privacy and security considerations remain at the forefront of this standard.
3.2. Clinical Trials
Consumer BioMeTs in the wellness category are increasingly being explored for use in clinical trials and telehealth to assess physiological parameters in novel ways and over longer periods. Clinical trials are selectively employing customized digital endpoints, some of which are exemplified in the Digital Medicine Society’s Digital Endpoints Library. In both clinical trials and telehealth, the lines between the medical and wellness FDA categories have been blurred. Current consumer BioMeTs have capabilities to continuously measure both traditional vital signs (5) and additional biosignals to gain more information about evolving physiological state during clinical trials. BioMeTs allow continuous physiological measurements in real-world, ambulatory environments as opposed to single measurements at predefined time points in clinical settings (37). Such flexibility gives researchers and clinicians the ability to assess intervention and treatment outcomes longitudinally as well as avoid confounding effects of circadian variation (38, 39). Mitigating these effects is especially important in times when trial participant mobility is affected, as during the COVID-19 pandemic. However, these devices need to be removed periodically for battery charging and comfort. Hence, reports of problems with adherence where participants lose or regularly forget to wear the device are unsurprising (38). These problems create issues with data missingness that need to be comprehensively reported.
Hundreds of clinical trials have already incorporated BioMeTs into their protocols, and this number continues to grow (40, 41). As of July 2021, ClinicalTrials.gov listed the Fitbit, Apple Watch, Garmin, and Oura ring in 661, 67, 80, and 14 clinical trials, respectively. The trial indications spanned cancer, heart failure, sleep apnea, atrial fibrillation, diabetes, alcohol use disorder, obesity, and depression. These numbers highlight the increasing use of consumer BioMeTs to measure and evaluate outcomes in clinical trials, thus moving them beyond their intended use for wellness purposes.
As novel data types are streamed from BioMeTs and stored as part of clinical trials, an ethical question is how to communicate off-target findings. For instance, it remains unclear how clinical trial operators ought to inform a trial participant of their detected arrhythmia in a noncardiac clinical trial. Additionally, clinical trials of the past have been rife with inequity along race, sex, and age dimensions (42, 43). Time will tell whether digital endpoints and incorporation of BioMeTs aid in exposing clinical trial disparities and reducing these imbalances.
3.3. Health Equity in Bring-Your-Own-Device Studies
In studies supplying BioMeTs, each participant typically receives the same quality of measurement, although this can be subject to wear and compliance. When participants supply their own BioMeTs for a study in a bring-your-own-device (BYOD) configuration, a host of health equity questions come into play. By default, robust conclusions might result only for those participants resourced enough to obtain high-quality devices. It is thus important to consider how to equitably analyze data from devices of differing quality and manufactures.
To obtain generalizable results, an analyzed data set must first be representative of the entire target population (44). However, demographic distributions of BYOD studies in the US frequently do not align with the broader population (Table 1). Even among large studies, minority groups are notably underrepresented. These underrepresented groups, including Black and Latino populations, also experience disproportionately greater COVID-19 mortality rates (56). To improve representation in the CovIdentify (17) study, targeted advertising as well as BioMeT subsidies were pursued. However, BioMeT subsidies or even donations were insufficient at times, since many consumer BioMeTs rely on specific smartphone models for pairing and use.
Table 1.
Racial and ethnic representations in US BYOD studies
| Study name/data | White | Black | Asian | Hispanic/Latino | Other | Reference(s) |
|---|---|---|---|---|---|---|
| All of Us (Fitbit) | 82.4 | 4.5 | 3.0 | 6.4 | 2.4 | 45 |
| All of Us (all participants) | 51.5 | 21.2 | 3.3 | 18.8 | NA | 45 |
| Asthma Health App | 69 | 5 | NA | 14 | 7 | 46 |
| CovIdentify | 87.6 | 3.6 | 3.3 | 4 | NA | 47 |
| MyHeart Counts | 76.1 | 3.3 | 8.8 | 7.3 | 4.44 | 48 |
| MyPHD | 74.9 | 2.9 | 3.9 | 0 | 18.3 | 49 |
| PARADE App | 80.7 | 4 | 2.8 | 10 | NA | 50 |
| Predicting Daily Mood | 57.5 | 16.2 | NA | 15.1 | NA | 51 |
| SleepHealth Mobile App | 77.9 | 2.9 | 5.2 | 11.3 | 3.7 | 52 |
| TemPredict | 81 | 0 | 4 | 17 | 15 | 53 |
| US Census demographics | 60.1 | 13.4 | 5.9 | 18.5 | NA | 54,55 |
| COVID-19 positive cases | 34.8 | 21.8 | 3.9 | 33.4 | NA | 54,55 |
| Deaths in the United States due to COVID-19 | 53.6 | 23.3 | 5.0 | 17.1 | NA | 54,55 |
Studies such as All of Us and Predicting Daily Mood have promoted inclusivity; however, minorities remain routinely underrepresented in US BYOD studies. This includes studies focused on COVID-19, although Black and Hispanic/Latino populations exhibit disproportionately greater mortality rates due to COVID-19.
Abbreviations: BYOD, bring-your-own-device; COVID-19, coronavirus disease 2019.
3.4. Developing World
BioMeTs have proven even more difficult to obtain in low- and middle-income countries, although they are perhaps more needed in these settings to monitor infectious diseases outside of the direct care of medical professionals. Low-power BioMeTs such as the MultiSense patient sensor (57) have demonstrated utility during Ebola treatment in Sierra Leone. This device, equipped with single-lead ECG capability, also wirelessly transmits skin temperature, RR, actigraphy, and oxygen saturation information for more frequent touchpoints than the typical 8-h intervals healthcare workers could provide. The disposable device with a Band-Aid form factor (Figure 2) resulted in measurements that correlated well with known references (58). A networking hub is still necessary to receive transmitted data, but such equipment for centralized hubs could be strategically distributed to treatment centers more easily than costly BioMeTs for each patient. Low-cost BioMeTs have also been proposed for remote hemoglobin monitoring (59), interferometry (60), and pulse oximetry (61). In settings with limited resources, remote sensing via satellite imagery can reveal mosquito activity (62) or ecological change (63, 64) related to vector-borne infectious diseases.
Figure 2.
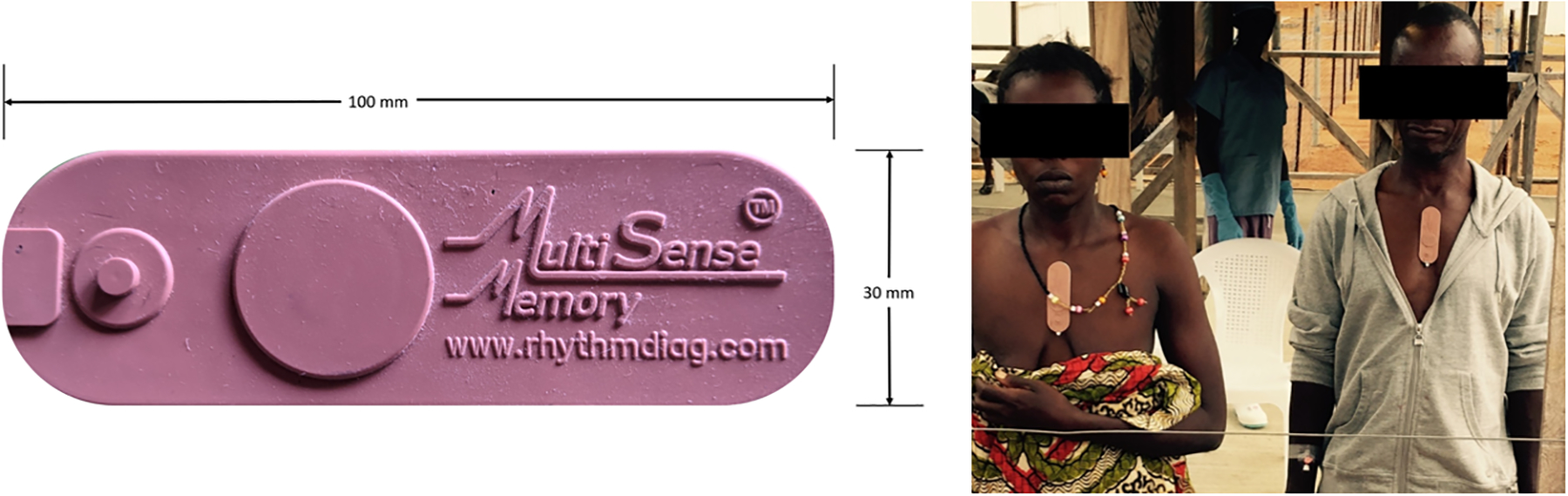
MultiSense patch for remote sensing of vital signs. The wireless, low-power sensor transmitted multiple biometrics from patients in an Ebola treatment center in Sierra Leone. Figure adapted from Reference 57 (CC BY 4.0).
More recently, the developing world has felt extreme impact from the COVID-19 pandemic. Internet of medical things (IoMT)-embedded applications (65) and even BioMeTs (66, 67) were proposed for quarantine and contact tracing purposes in India. Newly designed BioMeT remote monitoring solutions (68, 69) for COVID-19 patient management included HR, RR pulse oximetry, and temperature sensing. If integrated, such remote monitoring systems could bolster geography-specific epidemiological models (70, 71) predicting hospital utilization and supply needs. However, many of the aforementioned challenges surrounding multiplatform BioMeTs’ interoperability and real-time processing have remained major impediments to large-scale BioMeT integration during COVID-19 outbreaks in India and much of the developing world (72).
4. ANALYTICS AND MACHINE LEARNING FOR BIOMETRIC DATA
The development of ML techniques has created a number of possibilities for improved mining, searching, and analyzing of large amounts of BioMeT data. As BioMeT data is typically considered big data made up of a growing number of physiologic metrics, automated and intelligent ML approaches have become increasingly necessary (73). With that in mind, ML is typically defined as computational approaches that can extract desired information from data via a variety of learning paradigms using probabilistic frameworks. A variety of ML techniques for predicting patient outcomes or detecting events have become common in biomedical research and healthcare (74). For example, ML algorithms have shown promise in rapidly and robustly characterizing the relationship between predictor BioMeT data and clinical outcomes (75–77). Established methods include principal component analysis, decision trees, k-nearest neighbors, and random forests, with a greater current interest in neural networks (78) (Figure 3). As technology has advanced and larger and more complex data sets are collected, consideration has also been given to (a) where the data analysis occurs (i.e., on a device or in a cloud-based platform), (b) data quality and availability, and (c) the ultimate ability of users and clinicians to detect clinically important events. Advancements in wearable technology in particular have opened possibilities both for the integration of BioMeT data into clinical decision-making and for BioMeT data to serve as a direct source for ML algorithms to help identify changes that may be early signs of ILIs (79). The following subsections highlight recent advancements concerning how data analytics and ML tools are being used to obtain clinically useful information from BioMeTs.
Figure 3.
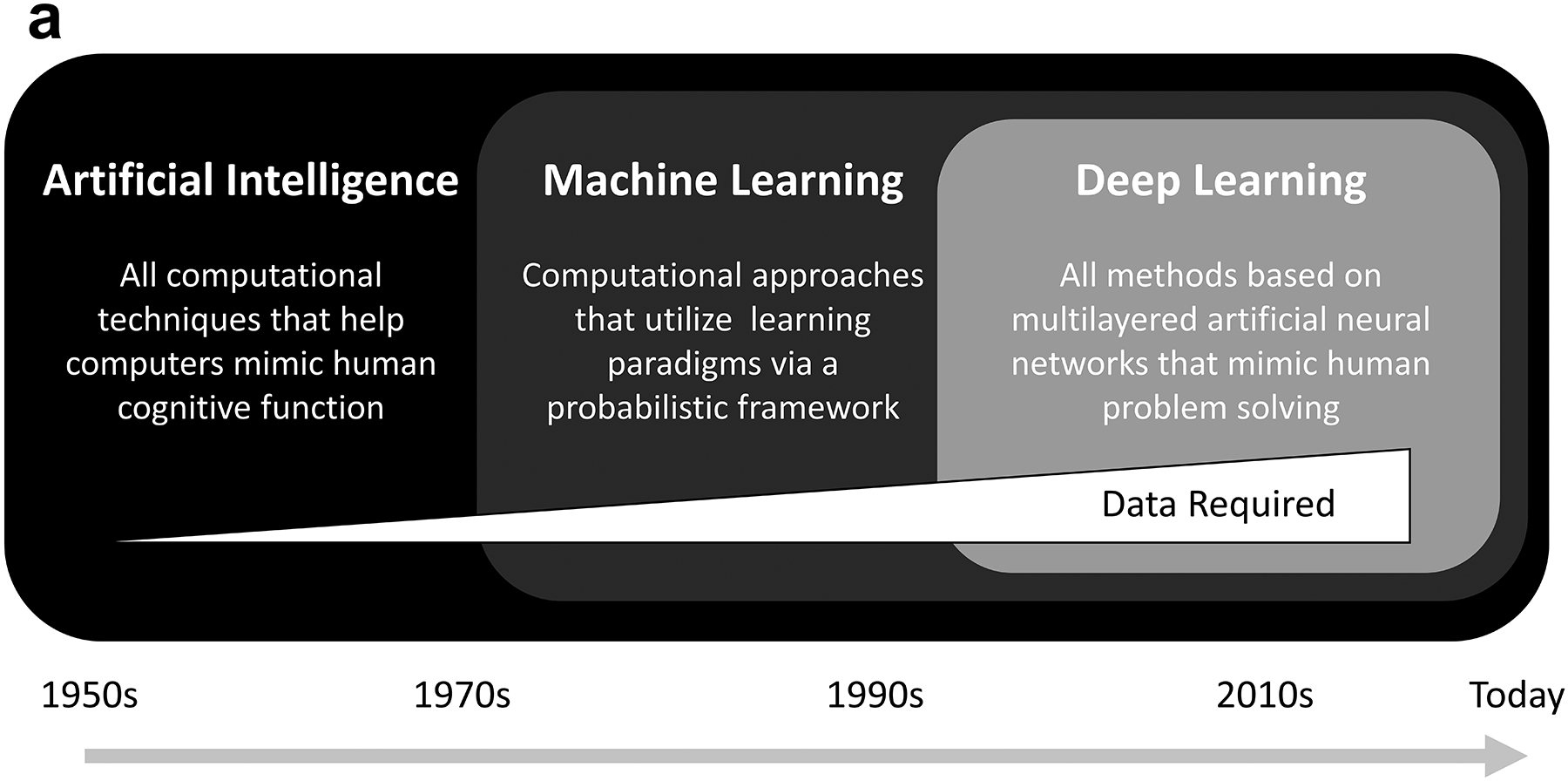
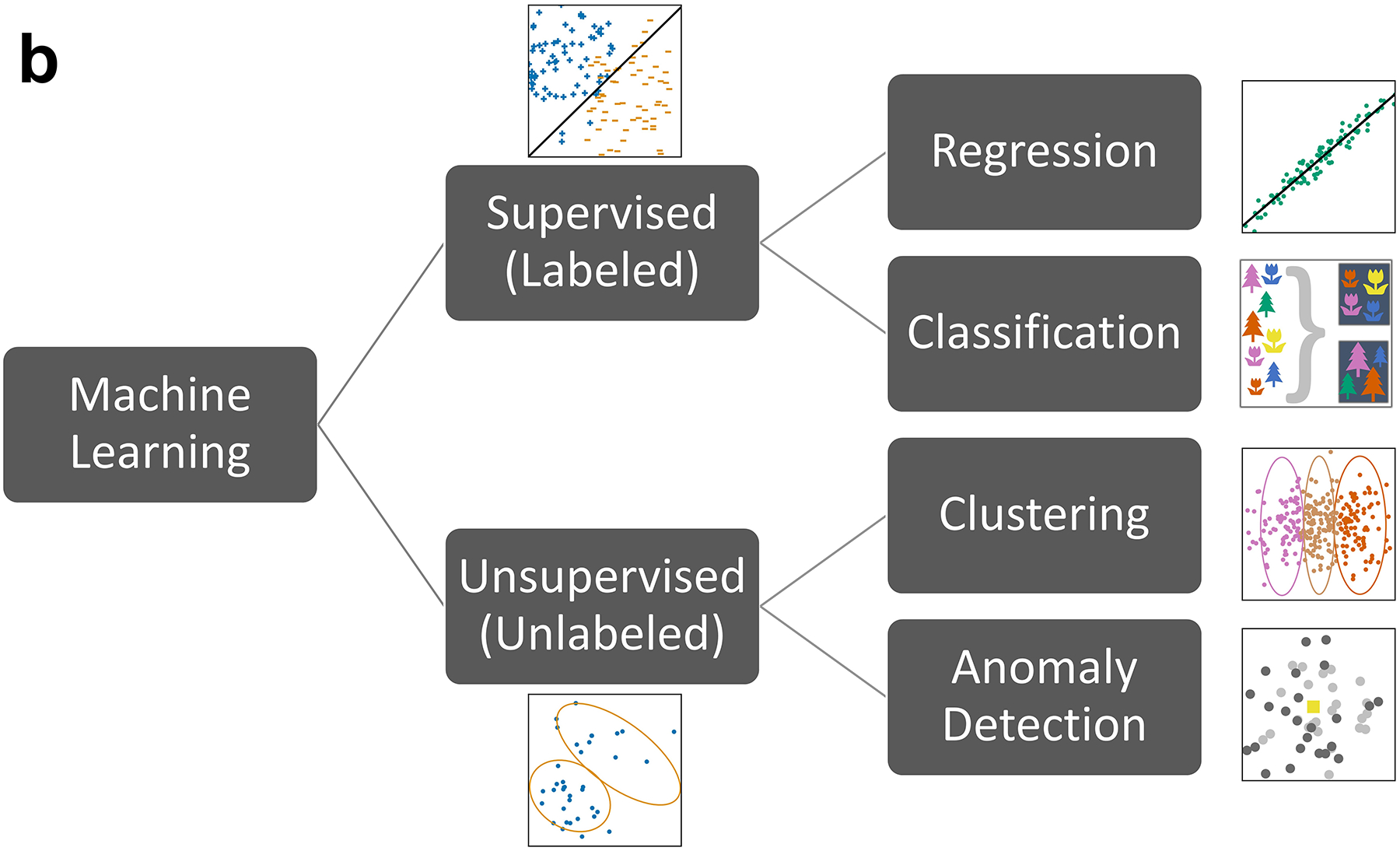
Timeline and breakdown of various machine learning tasks. (a) Timeline of artificial intelligence developing into deep learning since the 1950s. (b) Machine learning tasks related to data analytics.
4.1. Data Availability and Computational Modalities
Biosensor data collected by BioMeTs drive the derivation of physiological parameters. Generally, there are two data collection schemes used with wearable devices: BioMeTs that acquire and store data on board for later offload and those that acquire data and transfer them in real time (or near real time) to secondary processing systems. Patient-monitoring applications where clinical outcomes are highly dependent on early intervention require real-time data collection, whereas research involving the collection of observational study data may not. That said, even in the observational case, having real-time knowledge of participant compliance may be important to ensure that BioMeTs data intended to be collected per a study protocol actually are. The ubiquity of cellular networks has enabled real-time, continuous monitoring using BioMeTs. Additionally, the cost and data bandwidth of cellular networks have continued to improve over recent years. These improvements have enabled the acquisition and wireless transfer of high-resolution, 24–7 biosensor data during activities of daily living. Raw biosensor data have become more commonly available; this availability has in turn enabled the development and deployment of advanced signal processing and ML techniques applied directly to raw sensor data, replacing reliance on typically available onboard calculated physiological features such as average HR, step counts, and activity level summaries. As the availability of raw sensor data continues to increase, the development of advanced algorithms will continue to progress as well, enhancing the clinical and actionable insights that BioMeTs can provide (80, 81).
The physiological parameters that can be derived from, and the ML techniques that can be applied to, data collected from BioMeTs are entirely dependent on the embedded sensors, data collection schemes, and computational resources available to process sensor data. There are several common computational modalities used with BioMeTs today, each with advantages and disadvantages as summarized in Table 2. Often a combination of these modalities is utilized. As cellular technology and coverage, low-power electronics, and wearable sensor technologies have continued to evolve, more and more raw sensor data are being captured, uploaded, and processed using cloud computing resources that support the application of advanced ML techniques to those data. This evolution has enabled BioMeT-driven early detection of clinically significant health issues not only related to viral infection but also found in other areas as far ranging as heart failure exacerbation detection and safety monitoring of patients undergoing cancer therapy (82–84).
Table 2.
Computational modalities of biometric monitoring technologies: advantages and disadvantages
| Computational modality | Advantages | Disadvantages |
|---|---|---|
| Onboard microcontroller units/embedded firmware | Low power utilization Longer battery life Enables continuous data collection for long periods of time Smaller data transfer size |
Limited computational capabilities; low random access memory (RAM) Loss of raw biosensor data Limited support for new algorithm development Real-time access to results often limited Uncommon to have direct access to Internet Limited data storage |
| Smartphone | Higher computational capabilities Ubiquity of smartphones Good battery life Advanced data displays Real-time display of results Access to Internet Waveform data viewable |
Competing with other apps; limits available RAM Many different phone types; testing and user support can be difficult Real-time display requires radio connection to device Limited data storage |
| Cloud processing | Very high computational capabilities Deep learning algorithms are feasible Virtually unlimited data storage Enables development of advanced machine learning algorithms Raw data can be reprocessed Waveform data viewable |
High bandwidth requirement for data transfer from device Lower device battery life Requires data storage host Higher end-to-end complexity |
4.2. Signal Processing, Quality, Availability, and Implementation
Physiological features can be extracted from a variety of BioMeTs, including ECG, PPG, ballistocardiograms, electrodermal activity sensors, accelerometers, magnetometers, and gyroscopes. However, a schism exists in use cases for BioMeTs between the benefit of cardiorespiratory plus actigraphy versus actigraphy-only monitoring (40). Cardiorespiratory BioMeTs are sensitive to motion artifacts, electromyographic interference, and baseline wander (85, 86). Extracting physiological features from these inherently noisy signals can lead to false metrics, incorrect conclusions, or missed symptomology (87, 88). Therefore, to mitigate the possibility of false feature extraction, DSP and quality assessments are necessary. For the purposes of evaluating infectious diseases, we focus only on those sensors capable of continuous ambulatory cardiorespiratory monitoring (ECG and PPG) when discussing quality, availability, and usability.
4.2.1. Quality.
Quality assessment determines the usability of a signal by identifying fiducial points, amplitude, duration, and/or frequency (89, 90). Traditional algorithms for determining signal quality index (SQI) include perfusion, skewness, kurtosis, entropy, zero-crossing rate, signal-to-noise ratio, matching of multiple systolic wave detection, and relative power (91–93). However, as more BioMeT data become readily available, more computationally expensive methods such as long short-term memory, least-squares support vector machine, and convolutional neural networks will become more commonplace (94–97).
4.2.2. Filtering.
While ECG and PPG signals utilize different filtering approaches, both aim to reduce noise to amplify the signal. These filtering approaches rely on specific frequencies and sampling rates related to the desired output. For example, filtering for an HR algorithm may utilize a passband between 0.5 and 5 Hz (30–300 bpm), whereas an RR algorithm may utilize one between 0.08 and 0.7 Hz (4.8–42 breaths/min), (98). Popular ECG and PPG filtering approaches implement wavelet transforms or DSP filters such as low-pass, high-pass, band-pass, adaptive, Chebyshev, Kalman, and notch (70, 94, 98–101). However, in addition to traditional PPG filtering methods, it is becoming more common to leverage accelerometer signals in tandem with PPG in a multimodal approach to account for motion artifacts to enable monitoring during higher activity periods (102–104). This multimodal approach tends to work best during periodic motion (e.g., walking, cycling, or running), rather than during sporadic, nonperiodic motion (102). While there have been significant advancements in this field, effective DSP approaches are constantly being developed in an effort to optimize data yield as their implementation, in combination with an SQI, has drastic implications for downstream feature extraction, including improved accuracy and decreased data availability that may introduce biases (105, 106).
4.2.3. Aggregate comparisons.
To better characterize how SQI implementation can impact downstream feature extraction, algorithmic input requirements must be defined. Similar to variations in band-pass filters, the minimum continuity of quality data is dependent on features being extracted. For example, HR algorithms can require anywhere from 8 to 60 s to generate an output (107–109), while an RR algorithm may require 20 to 30 s of data (110) and HRV can require more than 5 min (111). It is hypothesized that the continuity of ECG signals frequently meets these requirements; however, PPG, as a more sensitive signal, is less likely to have long contiguous segments of quality data. Current literature only reports the accuracy of SQI algorithms and aggregate levels of good data/availability. ECG availability is typically greater than 90% while PPG falls between 20–50% (112–114), with a bias toward low activity and sleep (12, 115, 116). To date, however, no studies have reported on the intermittency of signal quality and how often quality segments of varying lengths occur for each signal; nor has it been reported how mixing high- and low-quality data to meet algorithmic requirements impacts accuracy. Definitions of acceptable accuracy are varied (117), though these can be dependent upon the use case. For example, monitoring the frequency of tachycardia throughout a day versus a daily HR may have different thresholds of validity. Further characterization of a signal’s SQI as a function of accuracy is required to better evaluate possible applications for continuous monitoring.
4.2.4. Data completeness.
To analytically benefit from continuous monitoring, continuous data streams must be present, and this is only possible with participant compliance (PC). Factors directly impacting PC include long-term comfort, complexity of setup, and user interface (118, 119). Additionally, various factors impact these data streams, including seasonality, diurnal variation, menstrual cycles, fitness regimes, and daily schedules (120, 121). Other factors substantially impacting downstream analyses include definitions of a day of data and clinical implementation. Currently, there is no consensus as to how many minutes are needed to summarize a day of data. However, there have been numerous studies demonstrating the minimal duration of monitoring to accurately characterize physical activity (122–124), although this research is lacking for cardiorespiratory monitoring. Additionally, current ML applications focus on data science’s definitions of success (i.e., area under the curve, sensitivity, specificity, etc.) rather than a clinical definition. To better validate the success of an algorithm in clinical practice, a decision curve analysis could be leveraged (125). However, more research into these approaches is required as ML becomes more prevalent in today’s medical space.
4.3. Symptom Tracking via Machine Learning and BioMeTs
Disease symptoms can be thought of as biological features that are indicative of sickness. While symptom detection via BioMeTs is not likely to be able to diagnose illness with any real-level certainty in the near future, identification of early changes in BioMeT data via ambulatory monitoring does have the potential to signal that a patient may be experiencing the initial stages of infection (79). This type of event or persistent change in information could help with early testing and quarantine efforts designed to limit the spread of infectious diseases but will require continued efforts to characterize algorithm performance, verify and validate underlying code, and implement event criteria that can benefit clinical decisions. A key question then is what constitutes baseline BioMeT data and what period of time is needed to robustly characterize a patient’s physiological state (38). While still in the relatively early stages of development and testing, the multiparameter aspects of BioMeTs combined with advanced ML techniques have the potential to greatly improve both sensitivity and specificity of event detection related to infections (126).
5. REAL-WORLD EXPERIENCE
The collection and analysis of individual biometric data via BioMeTs to help diagnose and manage infectious diseases such as COVID-19 are still in their earliest stages of clinical use, with a great deal to learn. While the COVID-19 pandemic has rapidly accelerated research in this area, related work addressing other infectious diseases had been ongoing in the preceding years (127). Those experiences, in addition to the knowledge being gained through multiple programs addressing COVID-19, have helped identify some early successes along with challenges and knowledge gaps. Building off these early real-world experiences will help move the field beyond just research programs and toward large-scale, global implementation.
5.1. Hospitalized Patients
Prior to the recent availability of consumer BioMeTs, hospitalized patients were the only people whose physiologic vital signs were routinely monitored serially and, occasionally, continuously. As concerns for serious infections such as pneumonia and sepsis often lead to hospitalizations, and because infections are a common complication in people hospitalized for other reasons, the ability to promptly recognize someone with an infection, or at risk for decompensating from one, has long been recognized as a critical need.
5.1.1. Heart rate variability.
Multiple investigators have evaluated a single metric, HRV, as a measure of severe infection or to identify a concerning trajectory toward decompensation (128). In a study of 17 bone-marrow transplant patients who underwent continuous HR monitoring beginning the day prior to transplant, 14 patients developed sepsis requiring antibiotic therapy (129). By initiating monitoring prior to transplant, the investigators were able to explore changes in an individual’s baseline well HRV to identify the earliest changes prior to the development of sepsis. They found that the majority (12 out of 14) of the people who developed sepsis experienced a 25% reduction in several measures of HRV, whereas none of the three people who did not develop sepsis experienced a decrease. For infected individuals, wavelet HRV decreased to the 25% threshold, on average, 35 h prior to the clinical diagnosis of sepsis. Despite decades of research and encouraging early results, HRV as a predictor of infection remains primarily a focus of research rather than a valuable clinical tool. Challenges to clinical implementation of HRV tracking include the plurality of analytic techniques available to determine HRV and their inconsistent application across studies (130).
5.1.2. Multiparametric prediction algorithms.
The early prediction of decompensation of hospitalized patients due to sepsis has also been evaluated using multiparametric vital signs (blood pressure, HR, temperature, RR, and oxygen saturation), age, and additional selected lab inputs when they were available, often with the use of ML techniques. In a small, prospective, randomized trial, 142 eligible adults admitted to an intensive care unit were randomized to either standard care or the ML-based predictive algorithm (131). Use of the predictive algorithm was associated with a decrease in both mortality and length of stay. In a real-world analysis of the impact of implementation of a predictive algorithm for severe sepsis, outcomes in more than 17,700 sepsis-related patients hospitalized in nine hospitals across the United States were compared with historical controls (132). After implementation of the predictive algorithm into routine clinical care, significant reductions of in-hospital mortality (39.5%), length of stay (32.3%), and 30-day readmission (22.7%) were seen relative to preimplementation. Nonetheless, the true value of widespread implementation of early sepsis prediction algorithms remains unclear, as a recent external validation study of the Epic Sepsis Model, already used in hundreds of healthcare systems, found dismal performance for early warning with a sensitivity of 33% and a positive predictive value of only 12% (133).
5.2. Population Data for Viral Illness Epidemiology
The initial description of the potential value of recognizing changes in individuals’ normal patterns of activity, sleep, and HR via a consumer wearable as a tool for better identifying population trends in influenza was in 2018 (127). By identifying trends in deviations from normal in ~64,000 Fitbit users in various regions of the United States, Radin and colleagues were able to demonstrate a strong correlation with weekly ILI rates from the US Centers for Disease Control and Prevention. Since then, multiple groups across the globe have explored the use of BioMeTs to improve surveillance for influenza and, more recently, COVID-19 (Supplemental Table 1). The size of these studies, most notably one from China including data from more than 1 million smartwatch/activity tracker users (134), highlights the potential for incorporating these unique data from willing volunteers to help guide future routine epidemiological surveillance.
These population-based studies provide early, encouraging results supporting how BioMeT data from large, geographically dispersed populations could potentially offer valuable insights into the location, timing, and trajectory of future infectious diseases. While many of these analyses were conducted without the active consent and knowledge of the participants, the Corona Data Donation project is an exception and an example of peoples’ willingness to voluntarily donate their deidentified BioMeT data to help address COVID-19 (https://corona-datenspende.de/science/en/). In just a month after launching, several hundred thousand people had enrolled, and the project eventually attracted more than half a million participants, leading to the development of fever maps for all of Germany. The example of using BioMeT data to demonstrate the behavioral and physiologic impact of various degrees of quarantine across different countries also highlights the usefulness of these data to track the impact of different disease prevention measures at a population level (135). For epidemiological purposes, the use of BioMeT data in linkage with other surveillance methods such as Internet searches, social media, and wastewater surveillance might be especially powerful (136).
5.3. BioMeT Data for Individual Diagnosis and Prognosis
As part of a wide-ranging, multisensor, long-term monitoring study from Stanford University, one participant noticed he had an unusual, small increase in his expected resting HR and skin temperature (137). He later developed a fever and was subsequently diagnosed with Lyme disease, likely marking the first time a BioMeT provided an early indication of an infectious disease. Shortly after, investigators from Evidation were able to identify individuals with influenza-like symptoms during the 2017–2018 influenza season via sleep and HR changes tracked by BioMeTs (138). Since that time, with the increasing ubiquity of multiparametric BioMeTs and the rapid acceleration of implementation of large research programs due to the COVID-19 pandemic, the body of literature describing the use of BioMeTs as novel diagnostic and prognostic tools has rapidly expanded. Within the initial weeks of the enormity of the COVID-19 pandemic being recognized, multiple studies were rapidly set up to determine if individual-level data from BioMeTs could enable the earlier diagnosis of COVID-19 infection, or subsequent decompensation. To date, the findings of nearly a dozen of these studies have been published (Supplemental Table 2). Most reported studies have focused on data from wrist-based wearables, but at least one has used a chest patch sensor and another a ring sensor. All the studies have provided novel information supporting the potential for BioMeT data to contribute valuable diagnostic and prognostic information in the setting of infection. For example, some studies described the diagnostic value in a single parameter, such as RR (139), resting HR (140), peripheral temperature (141), or HRV (142). Other studies evaluated individual changes in multiple parameters, typically resting HR, sleep, and activity (79, 126, 143, 144). Several studies even incorporated interaction features between measured parameters, such as HR and/or RR during activity, in their predictive models (126, 145).
The potential value of prospective implementation of real-time alerting based solely on individual changes in BioMeT-generated physiologic data was demonstrated in a study of more than 2,000 individuals (140). Alerts were sent to volunteers if their resting HR was elevated above their expected normal for two consecutive nights. Alerts were generated in 78% of all participants who tested positive for COVID-19 during the study—both asymptomatic and symptomatic—with the alert being sent a median of 3 days prior to symptom onset in presymptomatic individuals. In addition, one wrist-wearable device has recently received regulatory approval in Europe for the early detection of an acute respiratory infection, although no data are yet available supporting its performance (146).
While the limited results available are encouraging, there is still a great deal to learn about the value of real-world implementation of BioMeTs for individual illness detection. One especially important concern, discussed earlier, is that since many of the studies described were initiated rapidly and without external funding, the majority depended on enrolling individuals who already owned their own BioMeT, potentially introducing important biases through the limited inclusion of populations most impacted by COVID-19 (17).
5.4. BioMeT Data in Complications and Treatments of Viral Infections
Beyond the capability of BioMeTs to enable the detection of the acute physiologic and behavioral changes secondary to an acute infection, knowledge of an individual’s healthy baseline can also be of unique value in understanding an individual’s trajectory of recovery and their response to specific therapeutic interventions. Two examples of that capability from the COVID-19 pandemic are the inter-individual variability in long-term recovery from acute COVID-19 and the detection of individual response to vaccination.
5.4.1. Postacute sequelae of severe acute respiratory syndrome coronavirus 2 infection.
More than 70% of people who have recovered from an acute episode of COVID-19 report continuing to experience at least one symptom beyond 60 days after their initial diagnosis (147). Studies evaluating HR changes with COVID-19 infection have consistently shown a characteristic HR pattern in the acute setting with an initial spike, followed by a several-week transient bradycardia (143). One study has looked beyond the first weeks of recovery in 234 COVID-19-positive individuals relative to 641 symptomatic COVID-19-negative individuals (147). They found that individuals with COVID-19, but not those with acute respiratory infection symptoms who were COVID-19 negative, experienced a relative tachycardia that did not return to baseline, on average, until 79 days after symptom onset. Additionally, variable trajectories of recovery were identifiable on the basis of how long resting HR remained above a person’s pre-COVID normal, with a subset of ~10% whose resting HR remained >2 standard deviations above their preinfection value for >130 days from infection (Figure 4). More work is needed to explore the correlation between these objective changes and subjective symptoms.
Figure 4.
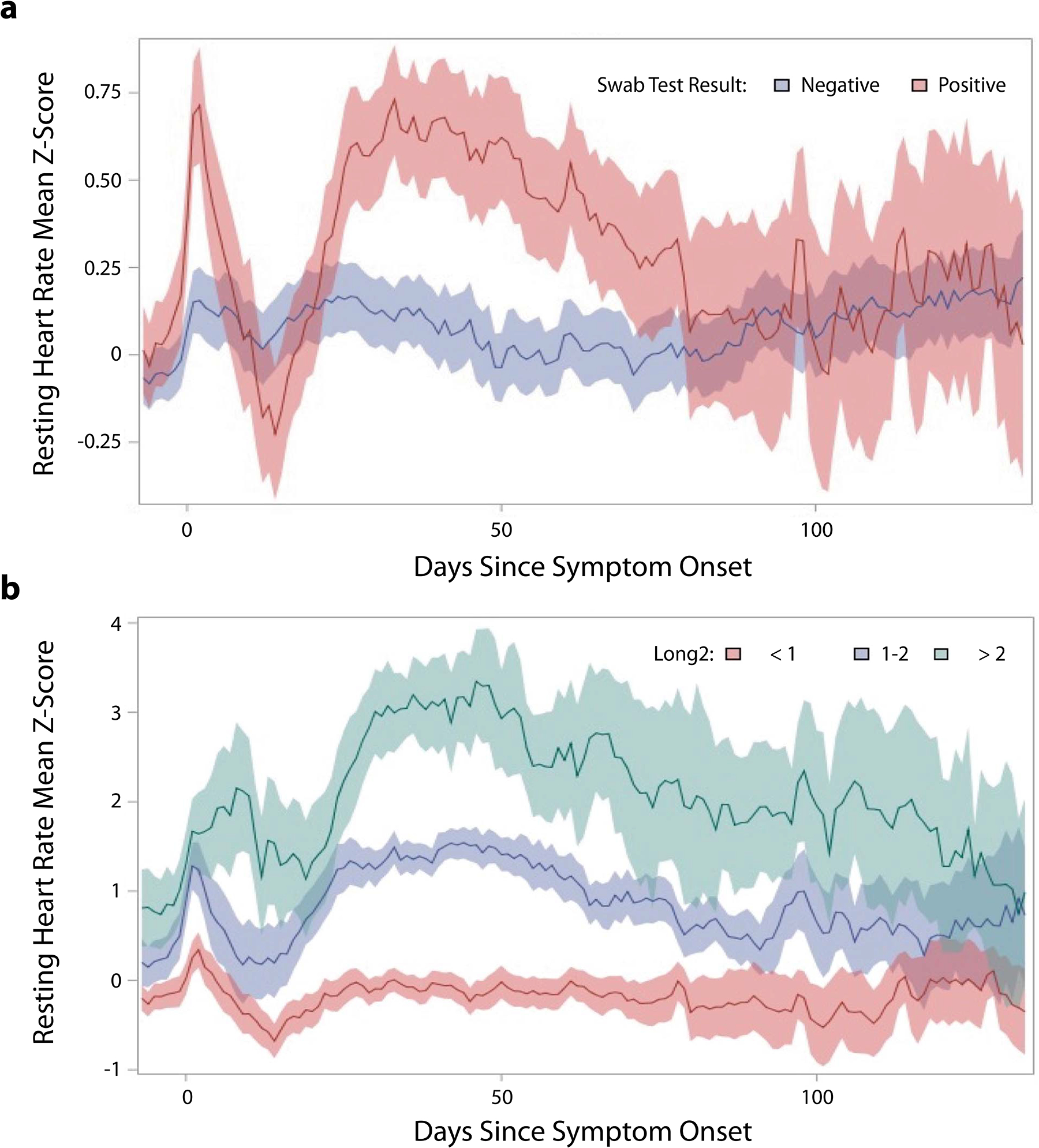
Change in resting heart rate (HR) by Z-score, relative to peoples’ normal, preinfection mean, during recovery from symptomatic acute respiratory infection. (a) Data based on whether the patient tested positive or negative for coronavirus disease 2019 (COVID-19). (b) Trajectories in resting HR change in subsets of COVID-19-positive individuals based on degree of abnormality in Z-score relative to their preinfection normal in the second month after symptom onset. Figure adapted from Reference 147 (CC BY 4.0).
5.4.2. Response to vaccination.
Vaccination has long been the key to controlling seasonal influenza and, most recently, has proven to be remarkably effective in helping control the COVID-19 pandemic (148). While most people undergoing vaccination appear to receive protection, no vaccine is 100% effective. The ability to determine if someone has had the expected immune system activation following vaccination could help guide next steps, such as the need for an additional booster shot. It is possible that BioMeT data might be able to provide some indication of that. Two early studies from individuals receiving a COVID-19 vaccination have shown that the majority of people experience a detectable increase in their normal HR in the days following the vaccine, with the degree of change influenced by the vaccine type, prior infection status, and age (149, 150). Future work correlating physiologic changes with measures of immune system activation and specific humoral and T cell responses may lead to the tracking of physiologic response after vaccination becoming a key component of any vaccination program.
6. CONCLUSIONS AND FUTURE DIRECTIONS
Wrist-based fitness devices that can measure HR have been available to consumers for less than a decade, but already more than one-third of people in the United States use one. Over this short period of time, data types and quality have continuously improved, while the form factors available for BioMeTs keep simultaneously expanding. As more and better consumer devices reach the shelves, the questions about how to incorporate such devices in clinical studies, and more importantly in clinical care, will become even more important. One could imagine multiplatform IoMT ecosystems in which smartwatches, continuous glucose monitors, implanted devices, and even ambient sensing systems could all stream to the cloud irrespective of a manufacturer. Privacy and security will need to be maintained, with data managed in a patient-centric way.
These are still the very early days of BioMeTs, continuous remote monitors, and advanced individualized big-data analytics. The COVID-19 pandemic has rapidly accelerated the implementation of these technologies in research and even in healthcare. While this experience has established BioMeTs as being central to a future care paradigm based on the recognition of subtle individual changes, there remains a great deal to learn. Nonetheless, the time is now right to start taking advantage of what BioMeTs make possible to improve the treatment of individuals at risk for infectious diseases by developing innovative systems of care that enable earlier, individualized alerting at the earliest sign of possible infection, possibly triggering home diagnostic testing followed by, if appropriate, early isolation and more intensive remote monitoring. With the ongoing collaboration of engineers, clinicians, researchers, and manufacturers, healthcare can be transformed around BioMeTs to extend better care to people everywhere.
Supplementary Material
SUMMARY POINTS.
Biometric monitoring technologies (BioMeTs) hold the potential for remote and continuous physiological monitoring. They have increasingly been utilized in clinical trials, and their use is being explored for infectious disease detection monitoring.
Clinical trials and research studies on infectious disease now include both medical- and consumer-grade BioMeTs. The challenge of reconciling data from different BioMeTs and ensuring equitable participation among historically underrepresented populations, especially those in the developing world, continues to grow as more devices become available.
The ubiquity of wireless networks along with the computational capabilities of BioMeT platforms has enabled real-time, continuous monitoring of individuals for a variety of clinical applications including viral infection detection and severity monitoring. The artificial intelligence and machine learning (ML) algorithms that rely on data produced by BioMeTs continue to advance as well, thus improving the physiological monitoring capabilities that BioMeTs can provide. Determining best clinical monitoring practices for selecting the type of BioMeTs and their computational modalities, in addition to establishing consistent data collection requirements (i.e., amount of data per day, minimum participant compliance, and definition of accuracy), are imperative for ensuring continued advancements.
Real-world experience using wearable devices and biometric data to address infectious diseases is limited but encouraging. Further work in the setting of infectious diseases and other noninfectious stressors will be critical for refining the diagnostic and prognostic capabilities of these technologies.
FUTURE ISSUES.
Accuracy and reliability of BioMeT data are important factors to consider for inclusion in clinical trials and infectious disease monitoring. Coherence between intended use of BioMeTs and regulatory oversight needs to be evaluated when employing such devices in clinical trials.
Uniform standards and data compression methods will be required to meaningfully harmonize data streams from different BioMeTs. As consumer BioMeTs become more popular in research studies, it is important to ensure equitable access to BioMeTs across populations and geography.
As BioMeT-based artificial intelligence and ML algorithms continue to evolve, it is crucial that these algorithms are properly validated, repeatable, and replicable; otherwise, clinical adoption is unlikely.
As the types and quality of wearable sensors continue to increase and data analytic techniques consistently improve, implementation of these technologies as meaningful solutions to address infectious diseases will require a focus on all aspects of usability, especially the return of value to users.
ACKNOWLEDGMENTS
Work described in this article is funded, in part, by grant UL1TR002550 from the National Center for Advancing Translational Sciences at the National Institutes of Health (S.R.S.).
DISCLOSURE STATEMENT
M.J.T. and S.W.W. are employees of physIQ. S.R.S. is an employee of physIQ, has served as a consultant for Tempus, and received research funding from Janssen Pharmaceuticals. K.S. is part of the research team for the CovIdentify study at Duke University.
LITERATURE CITED
- 1.Goldsack J, Coravos A, Bakker JP, Bent B, Dowling AV, et al. 2020. Verification, analytical validation, and clinical validation (V3): the foundation of determining fit-for-purpose for biometric monitoring technologies (BioMeTs). NPJ Digit. Med 3:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandrasekaran R, Katthula V, Moustakas E. 2020. Patterns of use and key predictors for the use of wearable health care devices by US adults: insights from a national survey. J. Med. Internet Res 22(10):e22443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Digit. Med. Soc. 2021. Defining digital medicine. Digital Medicine Society. https://www.dimesociety.org/about-us/defining-digital-medicine/ [Google Scholar]
- 4.Elenko E, Underwood L, Zohar D. 2015. Defining digital medicine. Nat. Biotechnol 33(5):456–61 [DOI] [PubMed] [Google Scholar]
- 5.Dias D, Paulo Silva Cunha J. 2018. Wearable health devices—vital sign monitoring, systems and technologies. Sensors 18(8):2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cent. Devices Radiol. Health. 2019. General wellness: policy for low risk devices. Guidance Doc., US Food Drug Admin., Silver Spring, MD [Google Scholar]
- 7.Cent. Devices Radiol. Health. 2020. Network of digital health experts. Memo, US Food Drug Admin., Silver Spring, MD. https://www.fda.gov/medical-devices/digital-health-center-excellence/network-digital-health-experts [Google Scholar]
- 8.US Food Drug Admin. 2020. CFR—Code of Federal Regulations Title 21. Database, US Food Drug Admin., Silver Spring, MD [Google Scholar]
- 9.Bent B, Dunn J. 2020. Wearables in the SARS-CoV-2 pandemic: What are they good for? JMIR Mhealth Uhealth 8(12):e25137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.US Food Drug Admin. 2020. Overview of device regulation. U.S. Food & Drug Administration. https://www.fda.gov/medical-devices/device-advice-comprehensive-regulatory-assistance/overview-device-regulation [Google Scholar]
- 11.Manta C, Jain SS, Coravos A, Mendelsohn D, Izmailova ES. 2020. An evaluation of biometric monitoring technologies for vital signs in the era of COVID-19. Clin. Transl. Sci 13(6):1034–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bent B, Goldstein BA, Kibbe WA, Dunn JP. 2020. Investigating sources of inaccuracy in wearable optical heart rate sensors. NPJ Digit. Med 3:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuller D, Colwell E, Low J, Orychock K, Tobin M, et al. 2020. Reliability and validity of commercially available wearable devices for measuring steps, energy expenditure, and heart rate: systematic review. JMIR Mhealth Uhealth 8(9):e18694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillinov S, Etiwy M, Wang R, Blackburn G, Phelan D, et al. 2017. Variable accuracy of wearable heart rate monitors during aerobic exercise. Med. Sci. Sports Exerc 49(8):1697–703 [DOI] [PubMed] [Google Scholar]
- 15.Cohen A, Mathews S, Dorsey E, Bates D, Safavi K. 2020. Direct-to-consumer digital health. Lancet Digit. Health 2(4):e163–65 [DOI] [PubMed] [Google Scholar]
- 16.Nelson BW, Low CA, Jacobson N, Arean P, Torous J, Allen NB. 2020. Guidelines for wrist-worn consumer wearable assessment of heart rate in biobehavioral research. NPJ Digit. Med 3:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cho PJ, Yi JJ, Ho E, Dinh YH, Patil A, et al. 2021. The demographic improvement guideline to reduce bias resulting from bring-your-own-device study design. JMIR preprint 23/04/2021:29510. https://preprints.jmir.org/preprint/29510 [Google Scholar]
- 18.Hendriks M, van Lotringen JH, Vos-van der Hulst M, Keijsers N. 2021. Bed sensor technology for objective sleep monitoring within the clinical rehabilitation setting: observational feasibility study. JMIR Mhealth Uhealth 9(2):e24339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller E, Banerjee N, Zhu T. 2021. Smart homes that detect sneeze, cough, and face touching. Smart Health 19:100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jagannath B, Lin K, Pali M, Sankhala D, Muthukumar S, Prasad S. 2021. Temporal profiling of cytokines in passively expressed sweat for detection of infection using wearable device. Bioeng. Transl. Med 6:e10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Snyder M, Zhou W. 2019. Big data and health. Lancet Digit. Health 1(6):e252–54 [DOI] [PubMed] [Google Scholar]
- 22.Wood F, Guinter T. 2008. Evolution and implementation of the CDISC Study Data Tabulation Model (SDTM). Pharm. Program 1(1):20–27 [Google Scholar]
- 23.Bent B, Dunn J. 2021. Optimizing sampling rate of wrist-worn optical sensors for physiologic monitoring. J. Clin. Transl. Sci 5(1):e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang C, Lu W, Narayanan M, Redmond S, Lovell N. 2015. Low-power technologies for wearable telecare and telehealth systems: a review. Biomed. Eng. Lett 5(1):1–9 [Google Scholar]
- 25.Bent B, Lu B, Kim J, Dunn J. 2021. Biosignal compression toolbox for digital biomarker discovery. Sensors 21(2):516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deepu CJ, Lian Y. 2014. A joint QRS detection and data compression scheme for wearable sensors. IEEE Trans. Biomed. Eng 62(1):165–75 [DOI] [PubMed] [Google Scholar]
- 27.Deepu C, Heng C, Lian Y. 2016. A hybrid data compression scheme for power reduction in wireless sensors for IoT. IEEE Trans. Biomed. Circuits Syst 11(2):245–54 [DOI] [PubMed] [Google Scholar]
- 28.Diamant N, Reinertsen E, Song S, Aguirre A, Stultz C, Batra P. 2021. Patient contrastive learning: a performant, expressive, and practical approach to ECG modeling. arXiv:2104.04569 [cs.LG] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ibaida A, Abuadbba A, Chilamkurti N. 2021. Privacy-preserving compression model for efficient IoMT ECG sharing. Comput. Commun 166:1–8 [Google Scholar]
- 30.Imtiaz S, Casson AJ, Rodriguez-Villegas E. 2013. Compression in wearable sensor nodes: impacts of node topology. IEEE Trans. Biomed. Eng 61(4):1080–90 [DOI] [PubMed] [Google Scholar]
- 31.Bent B, Sim I, Dunn J. 2021. Digital medicine community perspectives and challenges: survey study. JMIR Mhealth Uhealth 9(2):e24570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mezghani E, Exposito E, Drira K, Da Silveira M, Pruski C. 2015. A semantic big data platform for integrating heterogeneous wearable data in healthcare. J. Med. Syst 39(12):185. [DOI] [PubMed] [Google Scholar]
- 33.Chang W, Grady N. 2019. NIST big data interoperability framework. NIST Special Pub. 1500–1r2, Natl. Inst. Stand. Technol., Gaithersburg, MD [Google Scholar]
- 34.Jayaratne M, Nallaperuma D, De Silva D, Alahakoon D, Devitt B, et al. 2019. A data integration platform for patient-centered e-healthcare and clinical decision support. Future Gener. Comput. Syst 92:996–1008 [Google Scholar]
- 35.Budd J, Miller BS, Manning E, Lampos V, Zhuang M, et al. 2020. Digital technologies in the public-health response to COVID-19. Nat. Med 26(8):1183–92 [DOI] [PubMed] [Google Scholar]
- 36.Santos D, Perkusich A, Almeida H. 2014. Standard-based and distributed health information sharing for mHealth IoT systems. In 2014 IEEE 16th International Conference on e-Health Networking, Applications and Services (Healthcom), pp. 94–98. New York: IEEE [Google Scholar]
- 37.Possamai C, Ravaud P, Ghosn L, Tran V. 2020. Use of wearable biometric monitoring devices to measure outcomes in randomized clinical trials: a methodological systematic review. BMC Med. 18(1):310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrison D, Marshall P, Berthouze N, Bird J. 2014. Tracking physical activity: problems related to running longitudinal studies with commercial devices. In Proceedings of the 2014 ACM International Joint Conference on Pervasive and Ubiquitous Computing: Adjunct Publication, pp. 699–702. New York: ACM [Google Scholar]
- 39.Dunn J, Kidzinski L, Runge R, Witt D, Hicks J, et al. 2021. Wearable sensors enable personalized predictions of clinical laboratory measurements. Nat. Med 27(6):1105–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Izmailova E, Wagner J, Perakslis E. 2018. Wearable devices in clinical trials: hype and hypothesis. Clin. Pharmacol. Therapeut 104(1):42–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cox S, Lane A, Volchenboum S. 2018. Use of wearable, mobile, and sensor technology in cancer clinical trials. JCO Clin. Cancer Informat 2:1–11 [DOI] [PubMed] [Google Scholar]
- 42.Geller S, Koch A, Pellettieri B, Carnes M. 2011. Inclusion, analysis, and reporting of sex and race/ethnicity in clinical trials: have we made progress? J. Women’s Health 20(3):315–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murthy V, Krumholz H, Gross C. 2004. Participation in cancer clinical trials: race-, sex-, and age-based disparities. JAMA 291(22):2720–26 [DOI] [PubMed] [Google Scholar]
- 44.Panch T, Mattie H, Atun R. 2019. Artificial intelligence and algorithmic bias: implications for health systems. J. Glob. Health 9(2):010318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Natl. Inst. Health. 2019. All of Us research program expands data collection efforts with Fitbit. News Release, January 16. https://allofus.nih.gov/news-events-and-media/announcements/all-us-research-program-expands-data-collection-efforts-fitbit
- 46.Chan YFY, Bot BM, Zweig M, Tignor N, Ma WP, et al. 2018. The asthma mobile health study, smartphone data collected using ResearchKit. Sci. Data 5:180096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Univ Duke. 2020. CovIdentify–a Duke University study. Duke University. https://covidentify.covid19.duke.edu [Google Scholar]
- 48.Hershman SG, Bot BM, Shcherbina A, Doerr M, Moayedi Y, et al. 2019. Physical activity, sleep and cardiovascular health data for 50,000 individuals from the MyHeart Counts Study. Sci. Data 6:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mishra T, Wang M, Metwally AA, Bogu GK, Brooks AW, et al. 2020. Pre-symptomatic detection of COVID-19 from smartwatch data. Nat. Biomed. Eng 4:1208–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crouthamel M, Quattrocchi E, Watts S, Wang S, Berry P. 2018. Using a ResearchKit smartphone app to collect rheumatoid arthritis symptoms from real-world participants: feasibility study. JMIR Mhealth Uhealth 6(9):e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pratap A, Atkin DC, Renn BN, Tanana MJ, Mooney SD, et al. 2019. The accuracy of passive phone sensors in predicting daily mood. Depress. Anxiety 36(1):72–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deering S, Pratap A, Suver C, Borelli AJ Jr., Amdur A, et al. 2020. Real-world longitudinal data collected from the SleepHealth mobile app study. Sci. Data 7:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smarr BL, Aschbacher K, Fisher SM, Chowdhary A, Dilchert S, et al. 2020. Feasibility of continuous fever monitoring using wearable devices. Sci. Rep 10:21640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.US Census Bur. 2021. Table: United States. QuickFacts: United States, US Census Bureau, Washington, DC, updated May 18. https://www.census.gov/quickfacts/fact/table/US/RHI125219 [Google Scholar]
- 55.Cent. Dis. Control Prev. 2020. COVID-19. Centers for Disease Control and Prevention. https://www.cdc.gov/coronavirus/2019-ncov/index.html [Google Scholar]
- 56.Andrasfay T, Goldman N. 2021. Reductions in 2020 US life expectancy due to COVID-19 and the disproportionate impact on the Black and Latino populations. PNAS 118(5):e2014746118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steinhubl S, Marriott M, Wegerich S. 2015. Remote sensing of vital signs: a wearable, wireless “Band-Aid” sensor with personalized analytics for improved Ebola patient care and worker safety. Glob. Health Sci. Pract 3(3):516–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Steinhubl S, Feye D, Levine A, Conkright C, Wegerich S, Conkright G. 2016. Validation of a portable, deployable system for continuous vital sign monitoring using a multiparametric wearable sensor and personalised analytics in an Ebola treatment centre. BMJ Glob. Health 1(1):e000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pavithra K, Mary A, Rajasekaran K, Jegan R. 2017. Low cost non-invasive medical device for measuring hemoglobin. In 2017 International Conference on Innovations in Electrical, Electronics, Instrumentation and Media Technology (ICEEIMT), pp. 197–200. New York: IEEE [Google Scholar]
- 60.Duperron M, Carroll L, Rensing M, Collins S, Zhao Y, et al. 2017. Hybrid integration of laser source on silicon photonic integrated circuit for low-cost interferometry medical device. Proc. SPIE 10109:1010915 [Google Scholar]
- 61.Tomlinson S, Behrmann S, Cranford J, Louie M, Hashikawa A. 2018. Accuracy of smartphone-based pulse oximetry compared with hospital-grade pulse oximetry in healthy children. Telemed. e-Health 24(7):527–35 [DOI] [PubMed] [Google Scholar]
- 62.Chuang T, Henebry G, Kimball J, VanRoekel-Patton D, Hildreth M, Wimberly M. 2012. Satellite microwave remote sensing for environmental modeling of mosquito population dynamics. Remote Sens. Environ 125:147–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ledien J, Sorn S, Hem S, Huy R, Buchy P, et al. 2017. Assessing the performance of remotely-sensed flooding indicators and their potential contribution to early warning for leptospirosis in cambodia. PLOS ONE 12(7):e0181044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Palaniyandi M, Anand P, Pavendar T. 2017. Environmental risk factors in relation to occurrence of vector borne disease epidemics: remote sensing and GIS for rapid assessment, picturesque, and monitoring towards sustainable health. Int. J. Mosq. Res 4(3):9–20 [Google Scholar]
- 65.Kumar S, Maheshwari V, Prabhu J, Prasanna M, Jayalakshmi P, et al. 2020. Social economic impact of COVID-19 outbreak in India. Int. J. Pervasive Comput. Commun 16:309–19 [Google Scholar]
- 66.Tripathy A, Mohapatra A, Mohanty S, Kougianos E, Joshi A, Das G. 2020. Easyband: a wearable for safety-aware mobility during pandemic outbreak. IEEE Consum. Electron. Mag 9(5):57–61 [Google Scholar]
- 67.Singh V, Chandna H, Kumar A, Kumar S, Upadhyay N, Utkarsh K. 2020. IoT-Q-band: a low cost internet of things based wearable band to detect and track absconding COVID-19 quarantine subjects. EAI Endorsed Trans. Internet Things 6(21):4 [Google Scholar]
- 68.Mondal M, Roy K, Sarkar S. 2020. Design and development of wearable remote temperature monitoring device for smart tracking of COVID-19 fever. In Proceedings of the 2nd International Conference on IoT, Social, Mobile, Analytics & Cloud in Computational Vision & Bio-Engineering (ISMAC-CVB 2020). Rochester, NY: SSRN. 10.2139/ssrn.3735919 [DOI] [Google Scholar]
- 69.Tayal M, Mukherjee A, Chauhan U, Uniyal M, Garg S, et al. 2020. Evaluation of remote monitoring device for monitoring vital parameters against reference standard: a diagnostic validation study for COVID-19 preparedness. Indian J. Community Med 45(2):235–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chatterjee K, Chatterjee K, Kumar A, Shankar S. 2020. Healthcare impact of COVID-19 epidemic in India: a stochastic mathematical model. Med. J. Armed Forces India 76(2):147–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arora P, Kumar H, Panigrahi B. 2020. Prediction and analysis of COVID-19 positive cases using deep learning models: a descriptive case study of India. Chaos Solitons Fractals 139:110017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krishnamurthi R, Gopinathan D, Kumar A. 2021. Wearable devices and Covid-19: state of the art, framework, and challenges. In Emerging Technologies for Battling Covid-19: Applications and Innovations, ed. Al-Turjman F, Devi A, Nayyar A, pp. 157–80. Cham, Switz.: Springer [Google Scholar]
- 73.Witt D, Kellogg R, Snyder M, Dunn J. 2019. Windows into human health through wearables data analytics. Curr. Opin. Biomed. Eng 9:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang F, Jiang Y, Zhi H, Dong Y, Li H, et al. 2017. Artificial intelligence in healthcare: past, present and future. Stroke Vasc. Neurol 2(4):230–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Turkki R, Byckhov D, Lundin M, Isola J, Nordling S, et al. 2019. Breast cancer outcome prediction with tumour tissue images and machine learning. Breast Cancer Res. Treatment 177(1):41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang H, Hsu W, Lee M, Weng H, Chang S, et al. 2019. Automatic machine-learning-based outcome prediction in patients with primary intracerebral hemorrhage. Front. Neurol 10:910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang C, Gardiner L, Wang H, Hueman M, Chen D. 2019. Creating prognostic systems for well-differentiated thyroid cancer using machine learning. Front. Endocrinol 10:288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.LeCun Y, Bengio Y, Hinton G. 2015. Deep learning. Nature 521(7553):436–44 [DOI] [PubMed] [Google Scholar]
- 79.Quer G, Radin J, Gadaleta M, Baca-Motes K, Ariniello L, et al. 2020. Wearable sensor data and self-reported symptoms for COVID-19 detection. Nat. Med 27(1):73–77 [DOI] [PubMed] [Google Scholar]
- 80.Ren H, Shen J, Tang X, Feng T. 2020. 5G healthcare applications in COVID-19 prevention and control. In 2020 ITU Kaleidoscope: Industry-Driven Digital Transformation, pp. 1–4. New York: IEEE [Google Scholar]
- 81.Gupta N, Kumar Juneja P, Sharma S, Garg U. 2021. Future aspect of 5G-IoT architecture in smart healthcare system. In Proceedings 5th International Conference on Intelligent Computing and Control Systems, ICICCS 2021, pp. 406–11. New York: IEEE [Google Scholar]
- 82.Stehlik J, Schmalfuss C, Bozkurt B, Nativi-Nicolau J, Wohlfahrt P, et al. 2020. Continuous wearable monitoring analytics predict heart failure hospitalization: the LINK-HF multicenter study. Circ. Heart Fail 13(3):e006513. [DOI] [PubMed] [Google Scholar]
- 83.Low CA. 2020. Harnessing consumer smartphone and wearable sensors for clinical cancer research. NPJ Digit. Med 3:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Iqbal S, Mahgoub I, Du E, Leavitt M, Asghar W. 2021. Advances in healthcare wearable devices. NPJ Flex. Electron 5:9 [Google Scholar]
- 85.Nikolaev N, Gotchev A, Egiazarian K, Nikolov Z. 2001. Suppression of electromyogram interference on the electrocardiogram by transform domain denoising. Med. Biol. Eng. Comput 39(6):649–55 [DOI] [PubMed] [Google Scholar]
- 86.D’Aloia M, Longo A, Rizzi M. 2019. Noisy ECG signal analysis for automatic peak detection. Information 10(2):35 [Google Scholar]
- 87.Naeini E, Azimi I, Rahmani A, Liljeberg P, Dutt N. 2019. A real-time PPG quality assessment approach for healthcare internet-of-things. Procedia Comput. Sci 151:551–58 [Google Scholar]
- 88.Whiting S, Moreland S, Costello J, Colopy G, McCann C. 2018. Recognising cardiac abnormalities in wearable device photoplethysmography (PPG) with deep learning. arXiv:1807.04077 [eess.SP] [Google Scholar]
- 89.Satija U, Ramkumar B, Sabarimalai Manikandan M. 2018. A review of signal processing techniques for electrocardiogram signal quality assessment IEEE Rev. Biomed. Eng 11:36–52 [DOI] [PubMed] [Google Scholar]
- 90.Karimipour A, Homaeinezhad M. 2014. Real-time electrocardiogram P-QRS-T detection-delineation algorithm based on quality-supported analysis of characteristic templates. Comput. Biol. Med 52:153–65 [DOI] [PubMed] [Google Scholar]
- 91.Krishnan R, Natarajan B, Warren S. 2010. Two-stage approach for detection and reduction of motion artifacts in photoplethysmographic data. IEEE Trans. Biomed. Eng 57(8):1867–76 [DOI] [PubMed] [Google Scholar]
- 92.Selvaraj N, Mendelson Y, Shelley K, Silverman DG, Chon K. 2011. Statistical approach for the detection of motion/noise artifacts in Photoplethysmogram. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, pp. 4972–75. New York: IEEE; [DOI] [PubMed] [Google Scholar]
- 93.Elgendi M 2016. Optimal signal quality index for photoplethysmogram signals. Bioengineering 3(4):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao Z, Liu C, Li Y, Li Y, Wang J, et al. 2019. Noise rejection for wearable ECGs using modified frequency slice wavelet transform and convolutional neural networks. IEEE Access 7:34060–67 [Google Scholar]
- 95.Fu F, Xiang W, An Y, Liu B, Chen Z, et al. 2021. Comparison of machine learning algorithms for the quality assessment of wearable ECG signals via Lenovo H3 devices. J. Med. Biol. Eng 41(2):231–40 [Google Scholar]
- 96.Sabeti E, Reamaroon N, Mathis M, Gryak J, Sjoding M, Najarian K. 2019. Signal quality measure for pulsatile physiological signals using morphological features: applications in reliability measure for pulse oximetry. Inform. Med. Unlocked 16:100222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang Q, Fu L, Gu L. 2019. A cascaded convolutional neural network for assessing signal quality of dynamic ECG. Comput. Math. Methods Med 2019:7095137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fleming S, Tarassenko L. 2007. A comparison of signal processing techniques for the extraction of breathing rate from the photoplethysmogram. Int. J. Med. Health Biomed. Bioeng. Pharm. Eng 1:366–70 [Google Scholar]
- 99.Weaver C, Cole C, Lawrence R, von der Groeben J, Fitzgerald J, et al. 1968. Digital filtering with applications to electrocardiogram processing. IEEE Trans. Audio Electroacoust 16(3):350–91 [Google Scholar]
- 100.Deric Tang SK, Sebastian Goh Y, Dennis Wong ML, Eileen Lew Y. 2017. PPG signal reconstruction using a combination of discrete wavelet transform and empirical mode decomposition. In International Conference on Intelligent and Advanced Systems, ICIAS 2016. New York: IEEE [Google Scholar]
- 101.Liang Y, Elgendi M, Chen Z, Ward R. 2018. An optimal filter for short photoplethysmogram signals. Sci. Data 5:180076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lee J, Kim M, Park H, Kim I. 2020. Motion artifact reduction in wearable photoplethysmography based on multi-channel sensors with multiple wavelengths. Sensors 20(5):1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Majeed I, Jos S, Arora R, Choi K, Bae S. 2019. Motion artifact removal of photoplethysmogram (PPG) signal. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, pp. 5576–80. New York: IEEE; [DOI] [PubMed] [Google Scholar]
- 104.Seok D, Lee S, Kim M, Cho J, Kim C. 2021. Motion artifact removal techniques for wearable EEG and PPG sensor systems. Front. Electron 2:4 [Google Scholar]
- 105.Sukor J, Redmond S, Lovell N. 2011. Signal quality measures for pulse oximetry through waveform morphology analysis. Physiol. Meas 32(3):369–84 [DOI] [PubMed] [Google Scholar]
- 106.Masinelli G, Dell’agnola F, Valdés A, Atienza D. 2021. SPARE: a spectral peak recovery algorithm for PPG signals pulsewave reconstruction in multimodal wearable devices. Sensors 21(8):2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Saquib N, Tarikul Islam Papon M, Ahmad I, Rahman A. 2015. Measurement of heart rate using photoplethysmography. In 2015 International Conference on Networking Systems and Security, pp. 1–6. New York: IEEE [Google Scholar]
- 108.Melvin Johnson A, Jegan R, Anitha Mary X. 2017. Performance measures on blood pressure and heart rate measurement from PPG signal for biomedical applications. In 2017 International Conference on Innovations in Electrical, Electronics, Instrumentation and Media Technology (ICIEEIMT), pp. 311–15. New York: IEEE [Google Scholar]
- 109.Reiss A, Indlekofer I, Schmidt P, Van Laerhoven K. 2019. Deep PPG: large-scale heart rate estimation with convolutional neural networks. Sensors 19(14):3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Charlton P, Birrenkott D, Bonnici T, Pimentel M, Johnson A, et al. 2018. Breathing rate estimation from the electrocardiogram and photoplethysmogram: a review. IEEE Rev. Biomed. Eng 11:2–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bourdillon N, Schmitt L, Yazdani S, Vesin J, Millet G. 2017. Minimal window duration for accurate HRV recording in athletes. Front. Neurosci 11:456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Eerikainen L, Bonomi A, Schipper F, Dekker L, Vullings R, et al. 2018. Comparison between electrocardiogram- and photoplethysmogram-derived features for atrial fibrillation detection in free-living conditions. Physiol. Meas 39:084001. [DOI] [PubMed] [Google Scholar]
- 113.Bonomi A, Schipper F, Eerikainen L, Margarito J, Aarts R, et al. 2016. Atrial fibrillation detection using photo-plethysmography and acceleration data at the wrist. In Computing in Cardiology, pp. 277–80. New York: IEEE [Google Scholar]
- 114.Solosenko A, Petrenas A, Paliakaite B, Sörnmo L, Marozas V. 2019. Detection of atrial fibrillation using a wrist-worn device. Physiol. Meas 40(2):025003. [DOI] [PubMed] [Google Scholar]
- 115.Lazaro J, Reljin N, Hossain M, Noh Y, Laguna P, Chon K. 2020. Wearable armband device for daily life electrocardiogram monitoring. IEEE Trans. Biomed. Eng 67(12):3464–73 [DOI] [PubMed] [Google Scholar]
- 116.Spierer D, Rosen Z, Litman L, Fujii K. 2015. Validation of photoplethysmography as a method to detect heart rate during rest and exercise. J. Med. Eng. Technol 39(5):264–71 [DOI] [PubMed] [Google Scholar]
- 117.Sartor F, Papini G, Elisabeth Cox L, Cleland J. 2018. Methodological shortcomings of wrist-worn heart rate monitors validations J. Med. Internet Res 20(7):e10108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Galarnyk M, Quer G, McLaughlin K, Ariniello L, Steinhubl S. 2019. Usability of a Wrist-worn smartwatch in a direct-to-participant randomized pragmatic clinical trial. Digit. Biomark 3(3):176–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yilmaz T, Foster R, Hao Y. 2010. Detecting vital signs with wearable wireless sensors. Sensors 10(12):10837–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pierson E, Althoff T, Thomas D, Hillard P, Leskovec J. 2021. Daily, weekly, seasonal and menstrual cycles in women’s mood, behaviour and vital signs. Nat. Hum. Behav 5(6):716–25 [DOI] [PubMed] [Google Scholar]
- 121.Roussel B, Buguet A. 1982. Changes in human heart rate during sleep following daily physical exercise. Eur. J. Appl. Physiol. Occupat. Physiol 49(3):409–16 [DOI] [PubMed] [Google Scholar]
- 122.Hart T, Swartz A, Cashin S, Strath S. 2011. How many days of monitoring predict physical activity and sedentary behaviour in older adults? Int. J. Behav. Nutr. Phys. Activ 8:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Prescott S, Traynor J, Shilliday I, Zanotto T, Rush R, Mercer T. 2020. Minimum accelerometer wear-time for reliable estimates of physical activity and sedentary behaviour of people receiving haemodialysis. BMC Nephrol. 21(1):230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Leger D, Tonetti L, Gauriau C, Faraut B, Elbaz M, et al. 2019. A study on the optimal length of actigraphic recording in narcolepsy type 1. Clin. Neurophysiol. Pract 4:114–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rousson V, Zumbrunn T. 2011. Decision curve analysis revisited: overall net benefit, relationships to ROC curve analysis, and application to case-control studies. BMC Med. Informat. Decis. Making 11(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mishra T, Wang M, Metwally A, Bogu G, Brooks A, et al. 2020. Pre-symptomatic detection of COVID-19 from smartwatch data. Nat. Biomed. Eng 4(12):1208–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Radin J, Queer G, Steinhubl S. 2018. Can wearable sensors detect influenza epidemics? Correlation of anomalous Fitbit data with influenza surveillance data. Emerg. Infect. Dis 24(9):47 [Google Scholar]
- 128.Wee B, Lee J, Mok Y, Chong S. 2020. A narrative review of heart rate and variability in sepsis. Ann. Transl. Med 8(12):768–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ahmad S, Ramsay T, Huebsch L, Flanagan S, McDiarmid S, et al. 2009. Continuous multi-parameter heart rate variability analysis heralds onset of sepsis in adults. PLOS ONE 4(8):e6642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ahmad S, Tejuja A, Newman K, Zarychanski R, Seely A. 2009. Clinical review: a review and analysis of heart rate variability and the diagnosis and prognosis of infection. Crit. Care 13:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shimabukuro D, Barton C, Feldman M, Mataraso S, Das R. 2017. Effect of a machine learning-based severe sepsis prediction algorithm on patient survival and hospital length of stay: a randomised clinical trial. BMJ Open Respirat. Res 4(1):e000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Burdick H, Pino E, Gabel-Comeau D, McCoy A, Gu C, et al. 2020. Effect of a sepsis prediction algorithm on patient mortality, length of stay and readmission: a prospective multicentre clinical outcomes evaluation of real-world patient data from US hospitals. BMJ Health Care Informat. 27(1):e100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wong A, Otles E, Donnelly J, Krumm A, McCullough J, et al. 2021. External Validation of a widely implemented proprietary sepsis prediction model in hospitalized patients. JAMA Intern. Med 181:1065–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhu G, Li J, Meng Z, Yu Y, Li Y, et al. 2020. Learning from large-scale wearable device data for predicting epidemics trend of COVID-19. Discrete Dyn. Nat. Soc 2020:6152041 [Google Scholar]
- 135.Ong J, Lau T, Karsikas M, Kinnunen H, Chee M. 2021. A longitudinal analysis of COVID-19 lockdown stringency on sleep and resting heart rate measures across 20 countries. Sci. Rep 11(1):14413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kogan N, Clemente L, Liautaud P, Kaashoek J, Link N, et al. 2021. An early warning approach to monitor COVID-19 activity with multiple digital traces in near real time. Sci. Adv 7(10):eabd6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li X, Dunn J, Salins D, Zhou G, Zhou W, et al. 2017. Digital health: tracking physiomes and activity using wearable biosensors reveals useful health-related information. PLOS Biol. 15(1):e2001402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bradshaw B, Konty K, Ramirez E, Lee W, Signorini A, Foschini L. 2019. Influenza surveillance using wearable mobile health devices. Online J. Public Health Inform 11(1):e249 [Google Scholar]
- 139.Miller D, Capodilupo J, Lastella M, Sargent C, Roach G, et al. 2020. Analyzing changes in respiratory rate to predict the risk of COVID-19 infection. PLOS ONE 15(12):e0243693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Alavi A, Bogu G, Wang M, Rangan E, Brooks A, et al. 2021. Real-time alerting system for COVID-19 using wearable data. medRxiv 2021.06.13.21258795. 10.1101/2021.06.13.21258795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Smarr B, Aschbacher K, Fisher S, Chowdhary A, Dilchert S, et al. 2020. Feasibility of continuous fever monitoring using wearable devices. Sci. Rep 10:21640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hirten R, Danieletto M, Tomalin L, Choi K, Zweig M, et al. 2021. Use of physiological data from a wearable device to identify SARS-CoV-2 infection and symptoms and predict COVID-19 diagnosis: observational study. J. Med. Internet Res 23(2):e26107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Natarajan A, Su H, Heneghan C. 2020. Assessment of physiological signs associated with COVID-19 measured using wearable devices. NPJ Digit. Med 3:156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shapiro A, Marinsek N, Clay I, Bradshaw B, Ramirez E, et al. 2021. Characterizing COVID-19 and influenza illnesses in the real world via person-generated health data. Patterns 2(1):100188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Larimer K, Wegerich S, Splan J, Chestek D, Prendergast H, Van den Hoek T. 2021. Personalized analytics and a wearable biosensor platform for early detection of COVID-19 decompensation (DeCODe): protocol for the development of the COVID-19 decompensation index. JMIR Res. Protoc 10(5):e27271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Empatica. 2021. Aura receives first of its kind European CE mark for early detection of COVID-19. News Release, March 9. https://www.empatica.com/blog/aura-and-care-receive-ce-mark-for-early-detection-of-covid-19-and-other-respiratory-infections.html
- 147.Radin J, Quer G, Ramos E, Baca-Motes K, Gadaleta M, et al. 2021. Assessment of prolonged physiological and behavioral changes associated With COVID-19 infection. JAMA Netw. Open 4(7):e2115959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Haas E, Angulo F, McLaughlin J, Anis E, Singer S, et al. 2021. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: an observational study using national surveillance data. Lancet 397(10287):1819–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Presby D, Capodilupo E. 2021. Objective and subjective COVID-19 vaccine reactogenicity by age and vaccine manufacturer. medRxiv 2021.04.29.21256255 [Google Scholar]
- 150.Quer G, Gadaleta M, Radin J, Andersen K, Baca-Motes K, et al. 2021. The physiologic response to COVID-19 vaccination. medRxiv 2021.05.03.21256482. 10.1101/2021.05.03.21256482 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Biosensor data collected by BioMeTs drive the derivation of physiological parameters. Generally, there are two data collection schemes used with wearable devices: BioMeTs that acquire and store data on board for later offload and those that acquire data and transfer them in real time (or near real time) to secondary processing systems. Patient-monitoring applications where clinical outcomes are highly dependent on early intervention require real-time data collection, whereas research involving the collection of observational study data may not. That said, even in the observational case, having real-time knowledge of participant compliance may be important to ensure that BioMeTs data intended to be collected per a study protocol actually are. The ubiquity of cellular networks has enabled real-time, continuous monitoring using BioMeTs. Additionally, the cost and data bandwidth of cellular networks have continued to improve over recent years. These improvements have enabled the acquisition and wireless transfer of high-resolution, 24–7 biosensor data during activities of daily living. Raw biosensor data have become more commonly available; this availability has in turn enabled the development and deployment of advanced signal processing and ML techniques applied directly to raw sensor data, replacing reliance on typically available onboard calculated physiological features such as average HR, step counts, and activity level summaries. As the availability of raw sensor data continues to increase, the development of advanced algorithms will continue to progress as well, enhancing the clinical and actionable insights that BioMeTs can provide (80, 81).
The physiological parameters that can be derived from, and the ML techniques that can be applied to, data collected from BioMeTs are entirely dependent on the embedded sensors, data collection schemes, and computational resources available to process sensor data. There are several common computational modalities used with BioMeTs today, each with advantages and disadvantages as summarized in Table 2. Often a combination of these modalities is utilized. As cellular technology and coverage, low-power electronics, and wearable sensor technologies have continued to evolve, more and more raw sensor data are being captured, uploaded, and processed using cloud computing resources that support the application of advanced ML techniques to those data. This evolution has enabled BioMeT-driven early detection of clinically significant health issues not only related to viral infection but also found in other areas as far ranging as heart failure exacerbation detection and safety monitoring of patients undergoing cancer therapy (82–84).
Table 2.
Computational modalities of biometric monitoring technologies: advantages and disadvantages
| Computational modality | Advantages | Disadvantages |
|---|---|---|
| Onboard microcontroller units/embedded firmware | Low power utilization Longer battery life Enables continuous data collection for long periods of time Smaller data transfer size |
Limited computational capabilities; low random access memory (RAM) Loss of raw biosensor data Limited support for new algorithm development Real-time access to results often limited Uncommon to have direct access to Internet Limited data storage |
| Smartphone | Higher computational capabilities Ubiquity of smartphones Good battery life Advanced data displays Real-time display of results Access to Internet Waveform data viewable |
Competing with other apps; limits available RAM Many different phone types; testing and user support can be difficult Real-time display requires radio connection to device Limited data storage |
| Cloud processing | Very high computational capabilities Deep learning algorithms are feasible Virtually unlimited data storage Enables development of advanced machine learning algorithms Raw data can be reprocessed Waveform data viewable |
High bandwidth requirement for data transfer from device Lower device battery life Requires data storage host Higher end-to-end complexity |


