Abstract
Target site inaccessibility represents a significant problem for fluorescence in situ hybridization (FISH) of 16S rRNA with oligonucleotide probes. Here, unlabeled oligonucleotides (helpers) that bind adjacent to the probe target site were evaluated for their potential to increase weak probe hybridization signals in Escherichia coli DSM 30083T. The use of helpers enhanced the fluorescence signal of all six probes examined at least fourfold. In one case, the signal of probe Eco474 was increased 25-fold with the use of a single helper probe, H440-2. In another case, four unlabeled helpers raised the FISH signal of a formerly weak probe, Eco585, to the level of the brightest monolabeled oligonucleotide probes available for E. coli. The temperature of dissociation and the mismatch discrimination of probes were not significantly influenced by the addition of helpers. Therefore, using helpers should not cause labeling of additional nontarget organisms at a defined stringency of hybridization. However, the helper action is based on sequence-specific binding, and there is thus a potential for narrowing the target group which must be considered when designing helpers. We conclude that helpers can open inaccessible rRNA regions for FISH with oligonucleotide probes and will thereby further improve the applicability of this technique for in situ identification of microorganisms.
Over the past 10 years fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes has become a much-used tool in microbial ecology (e.g., reference 1). In situ identification of individual cells by FISH may be hindered by limited cell wall permeability (3), low cellular ribosome contents (4, 9), and differences in the in situ accessibility of probe target sites (10). In a recent systematic study, about one-third of the 200 probes examined yielded signals of <20% of the maximum fluorescence conferred by a monolabeled oligonucleotide on the 16S rRNA of Escherichia coli (10). Presumably, these differences in probe binding are due to the higher-order structure of the ribosome, which includes rRNA-rRNA interactions as well as interactions of the rRNAs with ribosomal proteins (2, 6). Unfortunately, the inaccessible regions encompass some of the most variable sites of the 16S rRNA, which would be highly valuable for the design of probes at the genus to species level (10).
Here, we report results obtained with unlabeled oligonucleotides, so-called helpers, in an attempt to open inaccessible regions on the 16S rRNA of E. coli for fluorescently labeled oligonucleotides. This work was inspired by publications of O'Meara et al. (14) and Niemeyer et al. (13), who observed an enhanced binding of oligonucleotide probes on isolated nucleic acids when using oligonucleotides complementary to regions neighboring the probe target site.
MATERIALS AND METHODS
FISH.
E. coli K-12 DSM 30083T and Azospirillum brasilense DSM 1690T (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany) were grown according to the DSM catalogue of strains and subsequently fixed for FISH with 4% paraformaldehyde as described previously (10). The 5′ carboxyfluorescein-labeled oligonucleotide probes (Interactiva, Ulm, Germany) were those used in the study of Fuchs et al. (10). In the new Eco603x, one central A-C mismatch was introduced. The unlabeled helper oligonucleotides were identical in sequence to the corresponding probes (e.g., H119 has the same sequence as probe Eco119 (10), with the following exceptions: helper H440-2 was shortened by 2 nucleotides to prevent overlap with probe H455, H621azo was altered from H621 to fully match the sequence of A. brasilense (EMBL accession number Z29617), and H621L and H621azoL were 21-nucleotide-long versions of the regular 18mer helpers. The sequences of all probes and helpers used in this study are listed in Table 1. Oligonucleotide concentrations and quality of labeling were analyzed spectrophotometrically (10). FISH of suspended cells was done as previously described in the standard hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 7.2], 0.01% sodium dodecyl sulfate) (17). Probes and helpers were added at final concentrations of 2.5 ng μl−1, assuming that an optical density at 260 nm of 1 is equivalent to 20 μg ml−1. Dissociation studies used hybridization temperatures of 37, 41, and 46°C; higher theoretical temperatures were achieved by adding formamide to the hybridization buffer, assuming an increase in the effective hybridization temperature of 0.5°C per 1% of added formamide (15).
TABLE 1.
Probe and helper sequences and location of target site on E. coli 16S rRNA positions (5)
| Probea | Sequence (5′->3′)b | E. coli target position |
|---|---|---|
| H48 | TCGACTTGCATGTGTTAG | 48–65 |
| H66 | GCTGCTTCCTGTTACCGT | 66–83 |
| Eco84 | TCGTCAGCAAAGCAGCAA | 84–101 |
| H102 | ACTCACCCGTCCGCCAC | 102–118 |
| H119 | GGCAGTTTCCCAGACATT | 119–136 |
| H422 | AAAGTACTTTACAACCCG | 422–439 |
| H440-2 | TCCCTTCCTCCCCGC | 440–454 |
| H455 | AAAGGTATTAACTTTACTC | 455–473 |
| Eco474 | CGGGTAACGTCAATGAGC | 474–491 |
| H492 | TAGCCGGTGCTTCTTCTG | 492–509 |
| H567 | CTGCGTGCGCTTTACGCC | 567–584 |
| Eco585, H585 | TCTGACTTAACAAACCGC | 585–602 |
| Eco603, H603 | GAGCCCGGGGATTTCACA | 603–620 |
| Eco603x | GAGCCCGGAGATTTCACA | 603–620 |
| Eco621, H621 | AGATGCAGTTCCCAGGTT | 621–638 |
| H621L | ATCAGATGCAGTTCCCAGGTT | 621–641 |
| H621azo | AAATGCAGTTCCCAGGTT | 621–638 |
| H621azoL | ATCAAATGCAGTTCCCAGGTT | 621–641 |
| Eco639, H639 | CTCAAGCTTGCCAGTATC | 639–656 |
| H657 | CTACCCCCCTCTACGAGA | 657–674 |
| Eco711 | TCGCCACCGGTATTCCTC | 711–728 |
| H729 | GTCCAGGGGGCCGCCT | 729–744 |
| H745 | ACCTGAGCGTCAGTCTTC | 745–762 |
EcoXXX, probes labeled with carboxyfluorescein; HXXX, unlabeled helper oligonucleotides.
Sequences modified from reference 10. Modifications are shown in boldface.
Flow cytometric analysis.
A FACStar Plus flow cytometer (Becton Dickinson, Mountain View, Calif.) was used to quantify the fluorescence intensities of hybridized cells. Probe-conferred fluorescence is given relative to that of the brightest probe of a former study, Eco1482 (10). Settings of the flow cytometer and data acquisition and processing have been described previously (10). Green fluorescent polystyrene beads (0.5-μm diameter) (catalogue no. 17152; Polysciences, Warrington, Pa.) were used to check the optical alignment of the flow cytometer and to standardize fluorescence measurements. All measurements were done as fully independent triplicates for which means were calculated. The coefficient of variation of the triplicates was in all cases <10%. Temperatures of dissociation were determined by sigmoidal fittings with the software package Origin (Microcal, Northampton, Mass.).
RESULTS
Helper effect.
The influence of unlabeled helper oligonucleotides on probe-conferred fluorescence was tested in three variable regions, the helices 6, 18, and 22-23 (Fig. 1). We selected six probes targeting sites with low in situ accessibility which were, according to the brightness classes defined by Fuchs et al. (10), either of class V (20 to 6% maximum fluorescence; Eco84 and Eco603) or VI (5 to 0% maximum fluorescence; Eco474, Eco585, Eco621, and Eco639) (Fig. 1).
FIG. 1.
Fluorescence conferred by probes and probe-helper combinations (probes are in boldface; addition of helper is indicated by +Hxxx) on whole fixed cells of E. coli DSM 30083T. Relative fluorescence intensities are standardized to the brightest probe, Eco1482. The secondary structure according to Gutell (11) of the respective target region on the 16S rRNA is shown. Different colors indicate the different brightness classes I through VI (10). The binding sites of helpers are shown in gray.
While probe Eco84 without helpers yielded only 8% of the fluorescence signal of the brightest probe, the signal could be enhanced roughly sixfold to 45% by simultaneous hybridization with the helper H66, targeting the 5′ adjacent region of Eco84, and fivefold to 38% with the 3′ adjacent H102 (Table 2). The helper effect was less pronounced with the more distantly located helper H119 (twofold to 17%) and absent with H48 (6%). The simultaneous use of the two adjacent helper probes H66 and H102 resulted in an 11-fold increase of the fluorescence conferred by probe Eco84 to 87%. With all four helper probes, Eco84 showed 70% of the fluorescence of Eco1482.
TABLE 2.
Relative fluorescence intensities of probes and probe-helper combinations
| Probe | Relative probe fluorescencea | Increase | Brightness classb |
|---|---|---|---|
| Eco84 | 8 | V | |
| Eco84 + 48 | 6 | 0.7 | V |
| Eco84 + 66 | 45 | 5.7 | III |
| Eco84 + 102 | 38 | 4.8 | IV |
| Eco84 + 119 | 17 | 2.1 | V |
| Eco84 + 66 + 102 | 87 | 11.1 | I |
| Eco84 + all | 70 | 8.9 | II |
| Eco474 | 2 | VI | |
| Eco474 + 422 | 2 | 1.0 | VI |
| Eco474 + 440-2 | 43 | 25.1 | III |
| Eco474 + 455 | 6 | 3.3 | V |
| Eco474 + 492 | 34 | 19.8 | IV |
| Eco474 + 455 + 492 | 47 | 27.4 | III |
| Eco474 + all | 75 | 43.2 | II |
| Eco585 | 5 | VI | |
| Eco585 + 567 | 47 | 9.3 | III |
| Eco585 + 603 | 41 | 8.1 | III |
| Eco585 + 621 | 13 | 2.5 | V |
| Eco585 + 639 | 28 | 5.5 | IV |
| Eco585 + all | 103 | 20.3 | I |
| Eco603 | 9 | V | |
| Eco603 + 567 | 19 | 2.1 | V |
| Eco603 + 585 | 39 | 4.1 | IV |
| Eco603 + 621 | 49 | 5.2 | III |
| Eco603 + 639 | 30 | 3.2 | IV |
| Eco603 + all | 78 | 8.4 | II |
| Eco621 | 3 | VI | |
| Eco621 + 585 | 17 | 5.2 | V |
| Eco621 + 603 | 42 | 12.6 | III |
| Eco621 + 639 | 11 | 3.3 | V |
| Eco621 + 657 | 8 | 2.3 | V |
| Eco621 + all | 60 | 17.9 | III |
| Eco639 | 5 | VI | |
| Eco639 + 585 | 12 | 2.4 | V |
| Eco639 + 603 | 15 | 2.9 | V |
| Eco639 + 621 | 9 | 1.8 | V |
| Eco639 + 657 | 39 | 7.9 | IV |
| Eco639 + all | 70 | 14.2 | II |
Fluorescence intensities are expressed as percentages of that of the brightest probe, Eco1482.
Probes were grouped according to their relative fluorescences into six classes of brightness: class I (100 to 81% relative fluorescence of Eco1482), class II (80 to 61%), class III (60 to 41%), class IV (40 to 21%), class V (20 to 6%), and class VI (5 to 0%).
Probe Eco474, targeting a site in helix 18, gave signals at the background level (2%). With the helpers H440-2 and H492, signals could be improved to 43 and 34% of maximum fluorescence, respectively. In contrast, H422 showed no effect (2%) and H455 showed little effect (6%). Combination of the two adjacent helper probes H455 and H492 with Eco474 resulted in 47% of maximum fluorescence. The use of all four helper probes raised the signal to 75%.
Helper effects on in situ accessibility of helix 22 were studied in detail on probes Eco585, Eco603, Eco621, and Eco639. Without helpers, all these probes had low signals (3 to 9%). The pair Eco585-H567 showed 47% of maximum fluorescence, and the pair Eco585-H603 showed 41%. The latter value almost matched that of the pair Eco603-H585 (39%). Obviously, the signal achieved with a given oligonucleotide pair is about the same, independent of which of the two oligonucleotides is labeled. This also applied for the probe helper pairs Eco603-H621 and Eco621-H603, which yielded relative signals of 49 and 42%, respectively, as well as for the pairs Eco621-H639 (11%) and Eco639-H621 (9%). H657 increased the signal of probe Eco639 to 39%.
For each of the probes tested, the signal achieved with four adjacent helpers was higher than the effect of a single helper (Fig. 1). With four helpers, all probes reached at least 60% of the maximum fluorescence (Table 2).
Influence of helper oligonucleotides on the Td.
The influence of helpers on the dissociation temperature (Td) of probes was initially explored with Eco603. The Tds of Eco603, Eco603-H621, and Eco603-H621L were 64, 61, and 62°C, respectively (Fig. 2A). All Tds were within a narrow temperature range. The helper nucleotides caused no increase in the Td. Interestingly, the change in the length of the helper H621L to a 21mer that is 3 nucleotides longer than H621 had no significant effect on the Td but caused a significant increase in relative fluorescence.
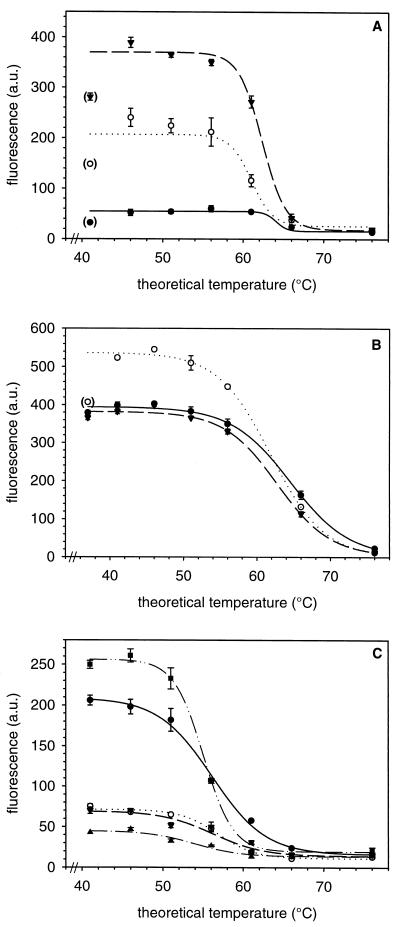
FIG. 2.
Influence of helper oligonucleotides on probe binding at different theoretical temperatures. The lines show the sigmoid melting curves calculated from the respective data points. The values in parentheses are excluded from the fitting procedure. a.u., arbitrary units. The error bars indicate standard deviations calculated from triplicate measurements. (A) E. coli hybridized with Eco603 (solid circles and solid line), Eco603-H621 (open circles and dotted line), and Eco603-H621L (triangles and dashed line). (B) E. coli hybridized with Eco711 (solid circles and solid line), Eco711-H729 (open circles and dotted line), and Eco711-H745 (triangles and dashed line). (C) A. brasilense hybridized with Eco603-H621azo (inverted triangles and dashed line) and Eco603-H621 (open circles and dotted line) and E. coli hybridized with Eco603x-H621L (triangles and dashed-dotted line), Eco603-H621azo (solid circles and solid line), and Eco603-H621azoL (squares and dashed–double-dotted line).
The effect of helpers on the Td was further evaluated on a probe targeting a more accessible site, Eco711, for which the Td without helpers could be determined more accurately. In this case, only the 3′ adjacent helper, H729, had a significant effect by raising the signal from 58 to 83% of maximum fluorescence. The more distant H745 did not change the fluorescence signal significantly (60%). The Tds for Eco711, Eco711-H729, and Eco711-H745 were 65, 62, and 63°C, again very similar (Fig. 2B).
Mismatch discrimination.
A possible influence of helpers on the specificity of hybridization was evaluated with probe Eco603 on A. brasilense, which has one weak mismatch (G instead of U at position 610) within the 16S rRNA target sequence. Whereas Tds on E. coli were 61 to 64°C, the midpoint of dissociation of Eco603-H621 from A. brasilense cells was at 57°C and that of Eco603-H621azo was at 56°C (Fig. 2C). In the latter case, a weak G-U mismatch of H621 to A. brasilense at position 637 was compensated for so that H621azo fully matched the helper site in A. brasilense. In contrast, when probe Eco603x, with a central A-C mismatch, was hybridized to E. coli, the Td with the long full matching helper H621L was 54°C (Fig. 2C). Interestingly, this Td is almost identical to the Tds of probe Eco603-helper pairs on E. coli, where the helpers had one strong mismatch (Fig. 2C). H621azo and H621azoL both have a strong A-C mismatch to E. coli at position 637. Tds of Eco603-H621azo and Eco603-H621azoL were 56 and 55°C, respectively. At low hybridization stringencies, these helpers increased the in situ accessibility almost as much as perfect helpers, but they did not work at higher stringencies (Fig. 2C).
DISCUSSION
Limited accessibility of some 16S rRNA target sites is a major problem when using FISH. We show here that unlabeled helper oligonucleotides can significantly improve FISH of six oligonucleotide probes to three different helices. The graphic presentation of the results (Fig. 1) suggests that the most effective signal enhancement is achieved by directly adjacent helper oligonucleotides (e.g., Eco585) and by helpers targeting the region complementary to the probe target site (e.g., Eco474). In many experiments, these two effects could not be separated, since the adjacent oligonucleotides also targeted the region opposing the probe target site. The helper effect was very specific, as shown by several examples in which there was no (H48 on Eco84; H422 on Eco474) or very little effect of close but not adjacent oligonucleotides. Our results are in good agreement with those of other groups. O'Meara et al. observed good capture of single-stranded DNA on a microchip only after using additional oligonucleotides targeting regions adjacent to the target site of the capture probe (14). Niemeyer et al. (13) suggest a mechanism in which the helper binds to the denatured RNA during hybridization and prevents the reestablishment of the native secondary structure, thereby keeping the probe target site open.
The different efficiencies of adjacent helpers may be due to differences in the accessibilities of the helper sites. A good example of this is probe Eco639, whose target site is presumably blocked by an rRNA-protein interaction at position 642-643 (8, 12). The helping effect of the 5′ adjacent H621 was low. There was an increase only from 5 to 9%, whereas that of the 3′ adjacent H657 was significant, 5 to 39%. The fluorescence conferred on these two sites in an earlier study by carboxyfluorescein-labeled oligonucleotides of identical sequence was 2 (Eco621) and 19% (Eco657) of the maximum signal (10). Only a helper that binds can help. In situ accessibility of rRNA in FISH is influenced by rRNA-rRNA and rRNA-protein interactions of various kinds and strengths. Even though individual helpers sometimes had a profound positive effect, we found the best signals for all probes with several adjacent helper oligonucleotides. Our current data suggest that by the joint action of multiple adjacent helper oligonucleotides, every site on the rRNA can be opened for FISH. However, we cannot rule out the possibility that there might be tightly closed regions on which the helper approach fails.
Can helper oligonucleotides change the specificities of probes?
O'Meara et al. (14) reported higher melting temperatures of probe-helper pairs, which they attributed to a cooperative base-stacking effect. In contrast, we found no evidence for significant Td changes between probes and probe-helper pairs in probes Eco603 and Eco711. Furthermore, the Td of probe Eco474 with four helpers (Eco474–H422–H440-2–H455–H492) was almost identical at 57°C to that of Eco474–H440-2 (59°C; data not shown). In this case, due to lack of signal, no Td could be determined for probe Eco474 alone. In our experimental setup, helper and probe seem to act as independent molecules.
Our examination of single-mismatch discrimination with and without helpers corroborates this view. Despite the fact that probe Eco603 has only a weak A-G mismatch to A. brasilense, dissociation from A. brasilense cells in the presence of helpers occurs at a temperature about 5°C lower than that from E. coli cells. This destabilizing effect of the single mismatch is within the normal range. When we introduced a mismatch in probe Eco603, the resulting probe, Eco603x, had a similar dissociation midpoint at 54°C. When a strong mismatch of probe Eco711 on Burkholderia cepacia LMG 1222T (Laboratorium voor Microbiologie, Universiteit Gent, Belgium) was examined, the presence of H729 did not change the melting behavior (data not shown). At the moment, we have no evidence for an influence of helpers on the potential for single-mismatch discrimination. However, we recommend that the discrimination of target and nontarget cells by probe-helper combinations be examined experimentally.
The data discussed so far suggest little influence of a helper(s) on probe specificity. At first glance, the quite dramatic influence of a single mismatch in the helper, H621azo versus H621 and H621L versus H621azoL, on the signal obtained with E. coli cells by probe Eco603 appears to contradict this assumption. The apparent Td of Eco603 with the single-mismatch helpers H621azo and H621azoL is much lower (56 and 55°C) than those of Eco603-H621, Eco603-H621L, and Eco603, which range between 61 and 64°C. The most likely explanation for the phenomenon where Tds are identical with and without helpers but different with a single-mismatch helper is as follows. At about 55°C, the single-mismatch helper dissociates from its binding site, causing an immediate strong decrease in the accessibility of the probe binding site. Our data (Fig. 2) suggest that a closure of this site could reduce the probe signal from over 200 to about 60 units of relative fluorescence. This would fully mask the subsequent true Td of Eco603, which melts at about 64°C, from 60 units of relative fluorescence to a background value of 30 units. We recommend that helper oligonucleotides be designed so that the Td of the helper is at least as high as that of the probe. This is most easily achieved by selecting longer helper oligonucleotides. One of our experiments (Fig. 2A) even suggests that the stronger binding of a longer oligonucleotide, the 21mer H621L versus the 18mer H621, further improved the probe-conferred fluorescence.
The use of helper oligonucleotides for FISH of environmental samples.
Environmental samples often have background fluorescence and contain cells with low rRNA contents. Single-cell identification by FISH, therefore, relies strongly on bright signals. Helpers were shown here to improve the signals of weakly binding probes. There might, however, be cases in which the cellular rRNA contents are so low that helpers alone are insufficient and more sensitive approaches, like multilabeled polynucleotide probes, are required (7, 16). Unfortunately, these probe types lack the tailored specificity typical of oligonucleotides. If strong probe signals depend on helper oligonucleotides, obviously only those cells will be detected that bind both probe and helper. Therefore, when designing a helper, care should be taken that it is complementary to all members of the probe target group. In an ideal world, the helper should have the exact specificity of the probe, thereby assuring the accuracy of identification. In reality, because the helper is placed either adjacent to the probe target site or opposing it, these ideal requirements are hard to fulfill. It should be kept in mind that besides the use of more general helpers, which might in practice require wobble positions in the helper sequence, one could accept the narrowing of the target group by the helper. If applied cautiously, the helper approach will reduce the time and effort lost in searching for open target sites and thereby further improve the applicability of FISH to environmental samples.
ACKNOWLEDGMENTS
The expert technical assistance of Daniela Lange and Torben Stührmann is acknowledged. Special thanks are due to Nicole Dubilier for critically reading the manuscript.
This work was supported by a grant of the Deutsche Forschungsgemeinschaft (Am73/2-4) and by the Max-Planck-Society.
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ban N, Nissen P, Hansen J, Capel M, Moore P B, Steitz T A. Placement of protein and RNA structures into a 5 A-resolution map of the 50S ribosomal subunit. Nature. 1999;400:841–847. doi: 10.1038/23641. [DOI] [PubMed] [Google Scholar]
- 3.Bidnenko E, Mercier C, Tremblay J, Tailliez P, Kulakauskas S. Estimation of the state of the bacterial cell wall by fluorescent in situ hybridization. Appl Environ Microbiol. 1998;64:3059–3062. doi: 10.1128/aem.64.8.3059-3062.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Binder B J, Liu Y C. Growth rate regulation of rRNA content of a marine Synechococcus (Cyanobacterium) strain. Appl Environ Microbiol. 1998;64:3346–3351. doi: 10.1128/aem.64.9.3346-3351.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 6.Clemons W M, May J L C, Wimberly B T, McCutcheon J P, Capel M S, Ramakrishnan V. Structure of a bacterial 30S ribosomal subunit at 5.5 A resolution. Nature. 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- 7.DeLong E, Taylor L, Marsh T, Preston C. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol. 1999;65:5554–5563. doi: 10.1128/aem.65.12.5554-5563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehresmann B, Ehresmann C, Romby P, Mougel M, Baudin F, Westhof E, Ebel J-P. Detailed structures of rRNAs: new approaches. In: Hill W E, Dahlberg A, Garrett R A, Moore P B, Schlessinger D, Warner J R, editors. The Ribosome: structure, function, and evolution. Washington, D.C.: American Society for Microbiology; 1990. pp. 148–159. [Google Scholar]
- 9.Fegatella F, Lim J, Kjelleberg S, Cavicchioli R. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain Rb2256. Appl Environ Microbiol. 1998;64:4433–4438. doi: 10.1128/aem.64.11.4433-4438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs B M, Wallner G, Beisker W, Schwippl I, Ludwig W, Amann R. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl Environ Microbiol. 1998;64:4973–4982. doi: 10.1128/aem.64.12.4973-4982.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutell R R. Collection of small subunit (16S- and 16S-like) ribosomal RNA structures: 1994. Nucleic Acids Res. 1994;22:3502–3507. doi: 10.1093/nar/22.17.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malhorta A, Harvey S C. A quantitative model of the Escherichia coli 16S RNA in the 30S ribosomal subunit. J Mol Biol. 1994;240:308–340. doi: 10.1006/jmbi.1994.1448. [DOI] [PubMed] [Google Scholar]
- 13.Niemeyer C M, Bürger W, Peplies J. Covalent DNA-streptavidin conjugates as building blocks for novel biometallic nanostructures. Angew Chem Int Ed. 1998;37:2265–2268. doi: 10.1002/(SICI)1521-3773(19980904)37:16<2265::AID-ANIE2265>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 14.O'Meara D, Nilsson N, Nygren P, Uhlén M, Lundeberg J. Capture of single-stranded DNA assisted by oligonucleotide modules. Anal Biochem. 1998;255:195–203. doi: 10.1006/abio.1997.2472. [DOI] [PubMed] [Google Scholar]
- 15.Stahl D A, Amann R. Development and application of nucleic acid probes. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1991. pp. 205–248. [Google Scholar]
- 16.Trebesius K, Amann R, Ludwig W, Mühlegger K, Schleifer K-H. Identification of whole fixed bacterial cells with nonradioactive 23S rRNA-targeted polynucleotide probes. Appl Environ Microbiol. 1994;60:3228–3235. doi: 10.1128/aem.60.9.3228-3235.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]