Abstract
Single domain antibodies, including the antigen binding variable domains of the shark immunoglobulin new antigen receptor and the camelid variable region of the heavy chain, are the smallest antigen recognition domains (~15 kDa) and have unique characteristics compared to conventional antibodies. They are capable of binding epitopes that are hard to access for classical antibodies and can also be used for therapeutics or diagnostics, or modular building blocks for multi-domain constructs, antibody-drug conjugates, immunotoxins, or chimeric antigen receptor therapy. This protocol contains detailed procedures for the purification and validation of two single domain antibodies (one shark and one camel), which bind to the S2 subunit of the SARS-CoV-2 spike protein, using both bacterial and mammalian cell expression systems. It provides a comprehensive reference for the production of single domain antibodies with high yield, good quality, and purity.
Keywords: Single domain antibody, shark VNAR, camel VHH, nanobody, Signal peptide, polymyxin B
Introduction
There are two major naturally occurring single domain antibodies: the camelid variable region of the heavy chain, also called nanobody (VHH), and the antigen binding variable domains (VNAR) of the shark immunoglobulin new antigen receptor (IgNAR). Both VHHs and VNARs are the smallest antigen recognition domains found in camels, alpacas, llamas, and sharks (English, Hong, & Ho, 2020; Feng et al., 2019; Flajnik, 2018; Muyldermans, 2013) (see Table 1 for abbreviations). In contrast to mouse and human conventional antibody IgG (150 kDa) and their respective antigen binding domains (50 kDa), single domain antibodies are only 12 to 15 kDa in size (Figure 1), highly stable across a wide range of pH and temperature, display strong binding affinities to target proteins and can recognize concave and hidden epitopes (Ho, 2018; Muyldermans, 2013).
Table 1.
List of abbreviations
| Abbreviations | Full name |
|---|---|
| VNAR | Antigen binding variable domain of the shark immunoglobulin new antigen receptor |
| IgNAR | Shark immunoglobulin new antigen receptor |
| VHH | Camelid variable region of the heavy chain only antibody |
| IgG | Immunoglobulin G |
| 2XYT medium | 2x yeast extract tryptone medium |
| FPLC | Fast protein liquid chromatography |
| IPTG | Isopropyl β- d-1-thiogalactopyranoside |
| PBS | Phosphate-buffered saline |
| SOC medium | Super optimal broth with Catabolite repression |
| HRP | Horseradish peroxidase |
| OD | Optical density |
| SDS-PAGE | Sodium dodecyl sulfate–polyacrylamide gel electrophoresis |
| PEI | Polyethylenimine |
| ELISA | Enzyme-linked immunosorbent assay |
Figure 1.
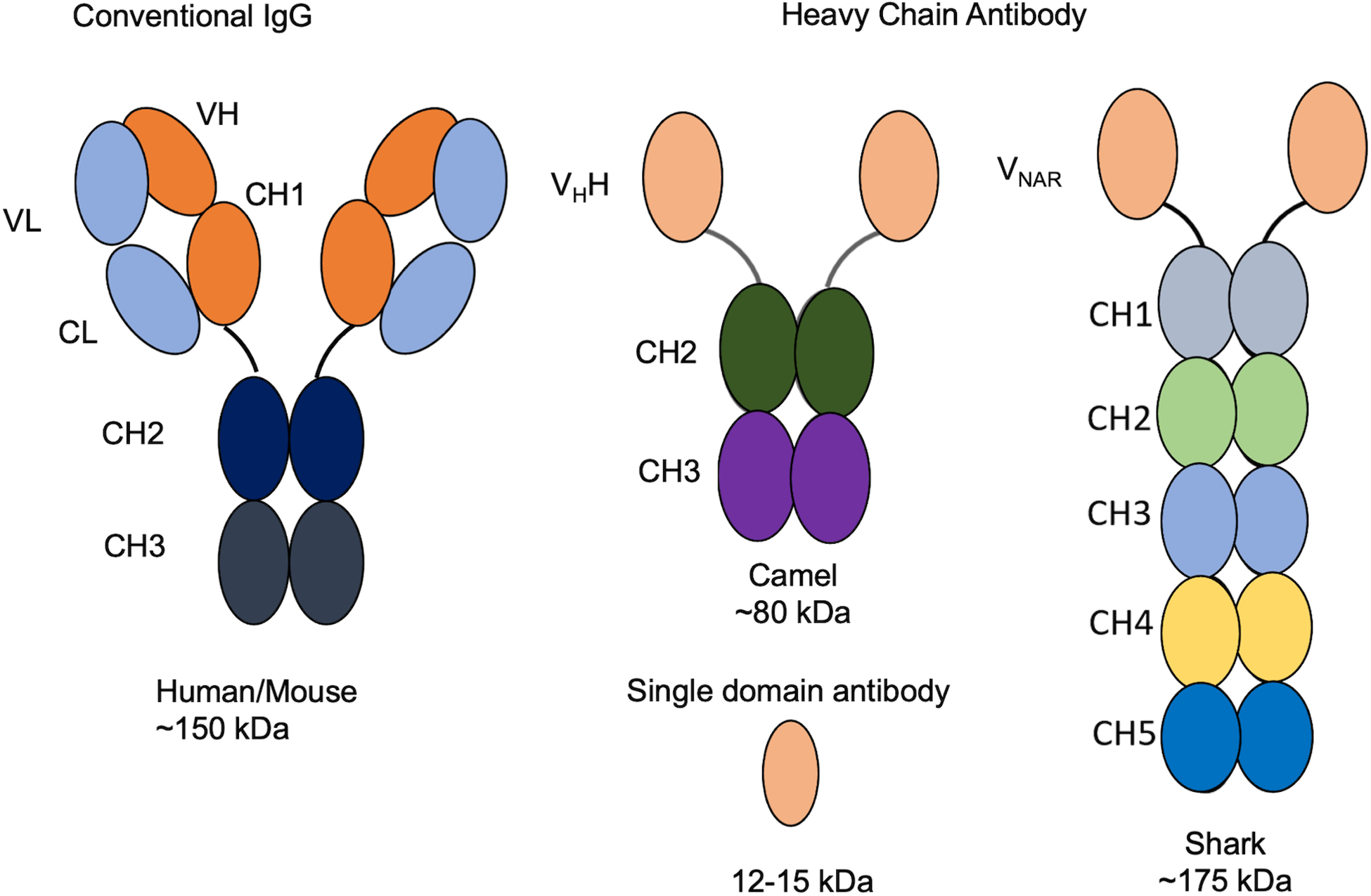
Comparisons of heavy chain antibody, single domain antibody with conventional IgG. Human and mouse antibodies have both heavy and light chains. Camel and shark have heavy chain only antibody and the variable domain of them are so called single domain antibodies. VL: variable domain of light chain; CL: constant domain of light chain; VH: variable domain of heavy chain; CH (1,2,3,4,5): constant domain of heavy chain (1,2,3,4,5); VHH: Camelid variable region of the heavy chain only antibody; VNAR: Antigen binding variable domain of the shark immunoglobulin new antigen receptor.
The production of single domain antibodies is important to test and identify potential candidates for further testing with protein assays. They can be expressed in bacteria, yeast, or mammalian expression systems (Feng et al., 2019; Hong et al., 2022). Bacterial systems are suitable and economic for producing large amounts of single domain antibodies for research purposes. This article describes the production and purification of single domain antibodies from Escherichia coli (E. coli) cells on a large scale by polymyxin B lysis and immobilized metal affinity chromatography with polyhistidine-tag in the Basic Protocol. The antibody gene is in the phagemid pComb3x with a signal peptide derived from the out membrane protein A (ompA) which can lead the antibody to the periplasm of the bacteria (HB2151) (Barbas, Kang, Lerner, & Benkovic, 1991) (Figure 2). In the Alternate Protocol, we describe the use of the mammalian system to express and purify the antibodies with the expression cell Expi293F and plasmid pcDNA3.1 which contains CMV promoter for high level recombinant protein production in mammalian cells (Running Deer & Allison, 2004). The methods of small-scale production of the single domain antibodies and the ELISA to validate them are described in the Support Protocols. We also describe the critical steps in troubleshooting and background information about the single domain antibodies in the Commentary.
Figure 2.
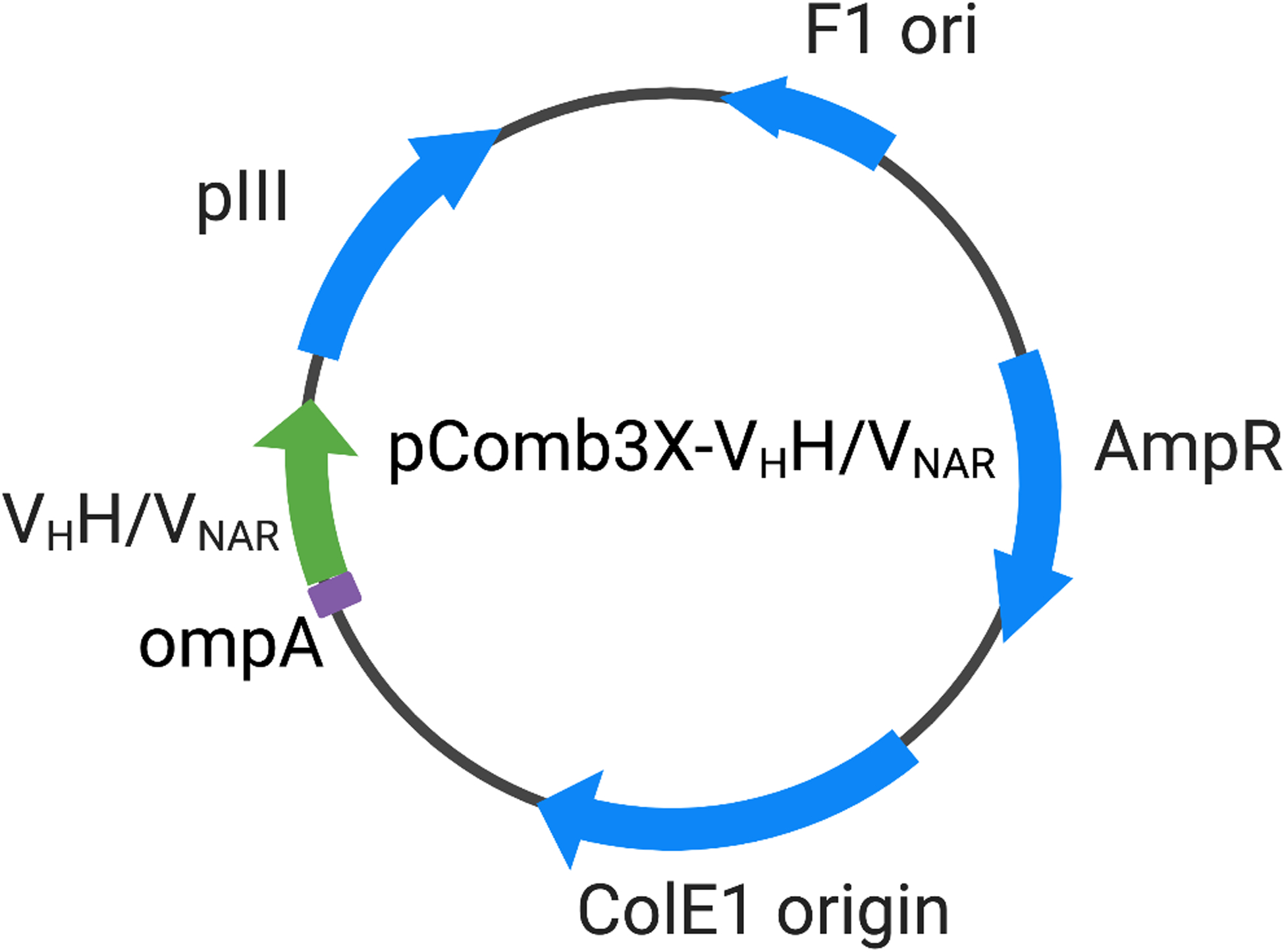
Plasmid map of pComb3X-VHH and pComb3X-VNAR. VHH: Camelid variable region of the heavy chain only antibody; VNAR: Antigen binding variable domain of the shark immunoglobulin new antigen receptor; ompA: the out membrane protein A; pIII: filamentous phage protein III; F1 ori: phage derived ori that allows for the replication and packaging of ssDNA into phage particles; AmpR: ampicillin-resistance gene; ColE1 origin: the ori carries a gene for colicin E1.
Basic protocol: Production of single domain antibodies from E. coli
This protocol describes the methods required for single domain antibody production using FPLC (AKTA explorer), starting from the transformation to the purification. Briefly, the phagemid containing the antibody sequence is transformed into the expression bacteria strain HB2151 and then the cells are cultured with the antibiotics and isopropyl β- d-1-thiogalactopyranoside (IPTG) to induce the protein expression overnight. The cells are collected and lysed with the polymyxin B to release the antibody from the periplasm. The antibody has 6x histidine (His) tag and thus can be purified by the nickel-charged affinity resin column on AKTA FPLC system (Figure 3). The shark VNAR is S3B3 and the camel VHH is CG2G2. Both bind to the antigen S2 subunit of the spike protein of SARS-CoV-2.
Figure 3.
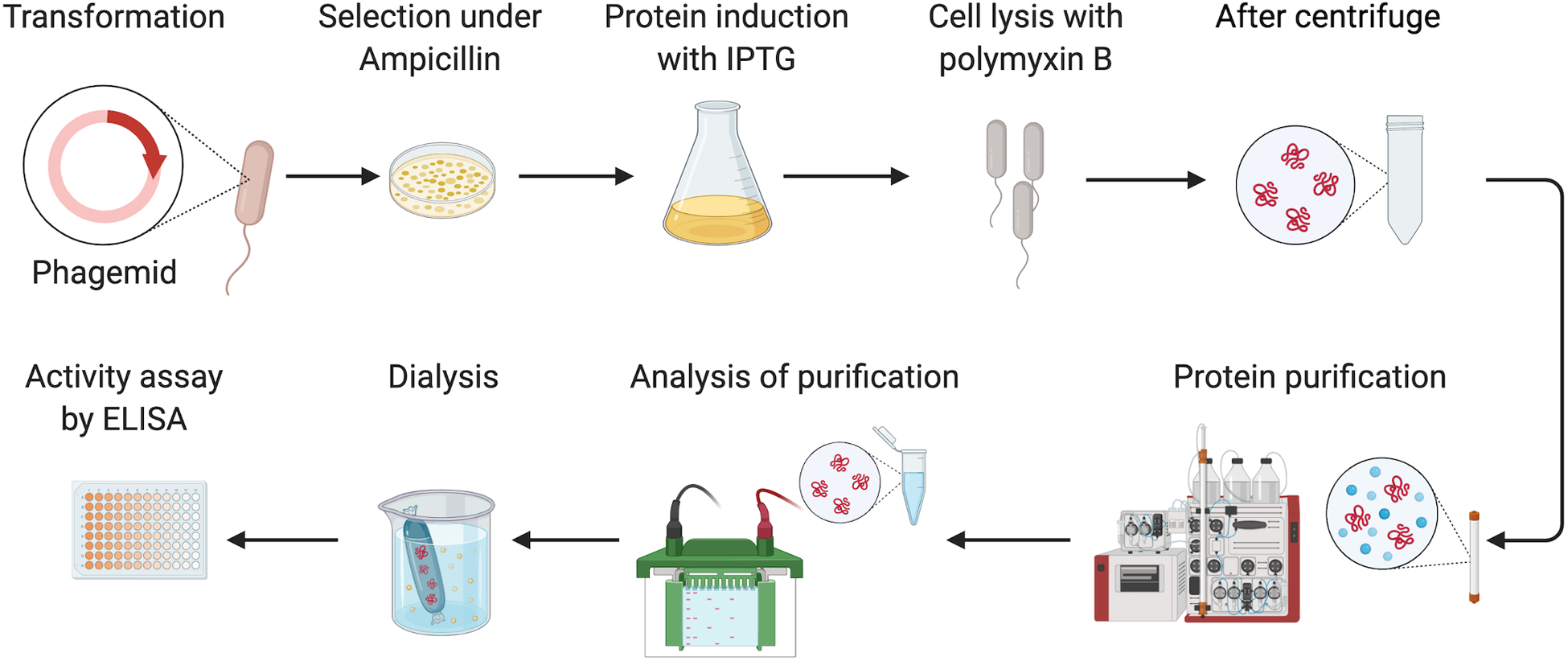
Overview of the basic protocol. Briefly, the phagemid containing the antibody sequence is transformed into HB2151 and the cells are cultured with the antibiotics and isopropyl β- d-1-thiogalactopyranoside (IPTG) to induce the protein expression overnight. The cells are collected and lysed with the polymyxin B to release the antibody from the periplasm. The 6x His tagged antibody can be purified by the nickel-charged affinity resin column on AKTA FPLC system. Fractions containing the antibody are subject to dialysis against PBS. Eventually, ELISA is performed to validate the binding of the antibody to the antigen.
Materials
E. coli strain: HB2151(LifeScienceMarket #S0122)
Phagemids: pComb3X-S3B3; pComb3X-CG2G2; pComb3X (a gift from Dennis Burton at the Scripps Research Institute, La Jolla, CA; available from Addgene #63888)
2XYT medium (Research Products International #L24045)
SOC medium (ThermoFisher Scientific #15544034)
50% (v/v) Glycerol (Sigma-Aldrich #G5516)
100 mg/ml Ampicillin (Sigma-Aldrich #A1593)
2XYT agar plates with 100 μg/ml ampicillin and 2% (w/v) glucose
20% (w/v) glucose solution (Sigma-Aldrich #D9434)
IPTG (Sigma-Aldrich #16758)
Polymyxin B (Sigma-Aldrich #P0972)
Protease inhibitor cocktail 100X (ThermoFisher Scientific #78429)
Buffer A: PBS + 0.5 M NaCl
Buffer B: Buffer A + 200 mM imidazole (Sigma-Aldrich #68268)
Phosphate-buffered saline (PBS) 10x (ThermoFisher Scientific #70011044)
PBST: 1x PBS containing 0.1% (v/v) Tween 20 (Biorad #1610781)
Deionized water
HisTrap HP His tag protein purification column 1 × 1mL (Cytiva #29051021)
AKTAexplorer (GE Healthcare Life Sciences)
SDS-PAGE gel: Novax 4%−20% Tris-Glycine gel (Invitrogen XP04205BOX)
Precision Plus Protein™ All Blue Prestained Protein Standards (Biorad #1610373)
4x Laemmli sample buffer (Biorad #1610747)
Slide-A-Lyzer dialysis cassette 10K (ThermoFisher Scientific #66380)
Nanodrop 1000 spectrophotometer (ThermoFisher Scientific)
TMB Chromogen Solution (ThermoFisher Scientific #002023)
Centrifuge with 96-well microtiter plate adapter (Sorvall)
0.45-μm syringe filter (Millipore #SLHVN33RS)
MaxiSoap 96-well plates (Sigma-Aldrich #M9410)
250-ml Erlenmeyer flask (Sigma-Aldrich #CLS431144)
2 liter Erlenmeyer flask (Sigma-Aldrich #CLS431255)
Incubator shaker (New Brunswick Scientific)
L-shaped cell spreader (CellTreat #229617)
Spectrometer (Amersham GE Healthcare)
Ice
Transformation
-
1
Thaw an aliquot of HB2151 competent cells (around 50 μl) on ice for 20 minutes.
-
2
Add 100 to 500 ng phagemid to cells and gently mix them.
-
3
Place on ice for> 30 minutes and < 1 hour.
-
4
Heat shock cells at 42 °C for 60 seconds using a water bath or a heating block.
-
5
Place cells on ice for 2 minutes.
-
6
Pipette 950 μl SOC media into the tube.
-
7
Place at 37 °C at 250 rpm for 1 to 2 hours.
-
8
Spin cells at 6000 × g for 5 minutes and resuspend pellet in 100 μl supernatant.
-
9
Spread all cells on a 2XYT agar plates with 100 μg/ml ampicillin and 2% (w/v) glucose.
-
10
Place the plate at 37 °C incubator overnight.
Induction of antibody expression
-
11
Take out the plate from the incubator; the colonies should be circular, firm and light color (white) dots.
Stoppable here: the plate can be stored at 4 °C for no more than five days before induction.
-
12
Collect all colonies from the transformed plate by L-shaped cell spreader with 10 ml 2XYT media.
-
13
Inoculate all the colonies into 1L 2XYT with 2% glucose and 100 μg/ml ampicillin.
-
14
Incubate at 37 °C with 250 rpm until OD600 is 0.7 to 0.9, which normally takes about 3 to 5 hours depending on the number of colonies obtained and should be checked often (every half hour at late stage) to make sure the cells are not overgrown.
-
15
Harvest the culture by centrifuge for 20 minutes at 6,000 g and remove old media.
-
16
Resuspend the culture in 1-liter fresh 2XYT with 100 μg/ml ampicillin. Add IPTG to 1 mM final concentration.
-
17
Incubate at 30 °C overnight in a shaker at 250 rpm.
Antibody purification
Both nanobodies (S3B3 and CG2G2) have a 6-histidine tag which can be purified using HisTrap HP column prepacked with Nickel Sepharose by immobilized metal ion affinity chromatography (Figure 2).
-
18
Collect the cell pellet by centrifugation at 6,000 g for 20 min. Pour off the supernatant and drain the residual media from the pellet by inverting the bottle on a paper towel.
Stoppable here: put the bacteria pellet at −80 °C no more than 1 month before the next steps
-
19
Wash pellet with 50 ml cold PBS and centrifuge at 6,000 g for 10 min. Drain residual media from the pellet by inverting the bottle on a paper towel.
-
20
Resuspend pellet in cold PBS + 0.5 M NaCl supplemented with protease inhibitor cocktail (100X). Use 50 ml PBS + 0.5 ml cocktail for the pellet from every liter culture. Vortex the pellet and/or pipet the cells up and down until no clump is visible.
-
21
Add polymyxin B 1 ml (500,000 units/ml) for 50 ml suspension in PBS/NaCl. Let it shake at 250 rpm in the 37 °C shaker incubator for 1 hour. Since the polymyxin B makes pores on the outside membrane and releases the protein from the periplasm, the cells are not completely lysed and chromosomal DNA is not released.
-
22
Centrifuge the lysate at 12,000 g for 30 min at 4 °C.
-
23
Using a 35 ml syringe to collect the supernatant and filter it through the 0.45 μm filter. The sample is ready for purification now.
-
24
Before loading the sample, the 1 ml HisTrap column is cleaned with water and equilibrated with 5 ml buffer A.
-
25
Load the supernatant into the column at a flow rate of 1 ml/min through a loading pump.
-
26
Connect the loaded column to AKTA Explorer.
-
27
Wash the column with 15 column volumes (CV) of Buffer A (PBS + 0.5 M NaCl).
-
28
Elute the antibody with gradient Buffer B targeting 100% in 15 minutes and maintain 100% for another 15 minutes (Figure 4). The collection volume of fractions will be 1 ml.
Figure 4.
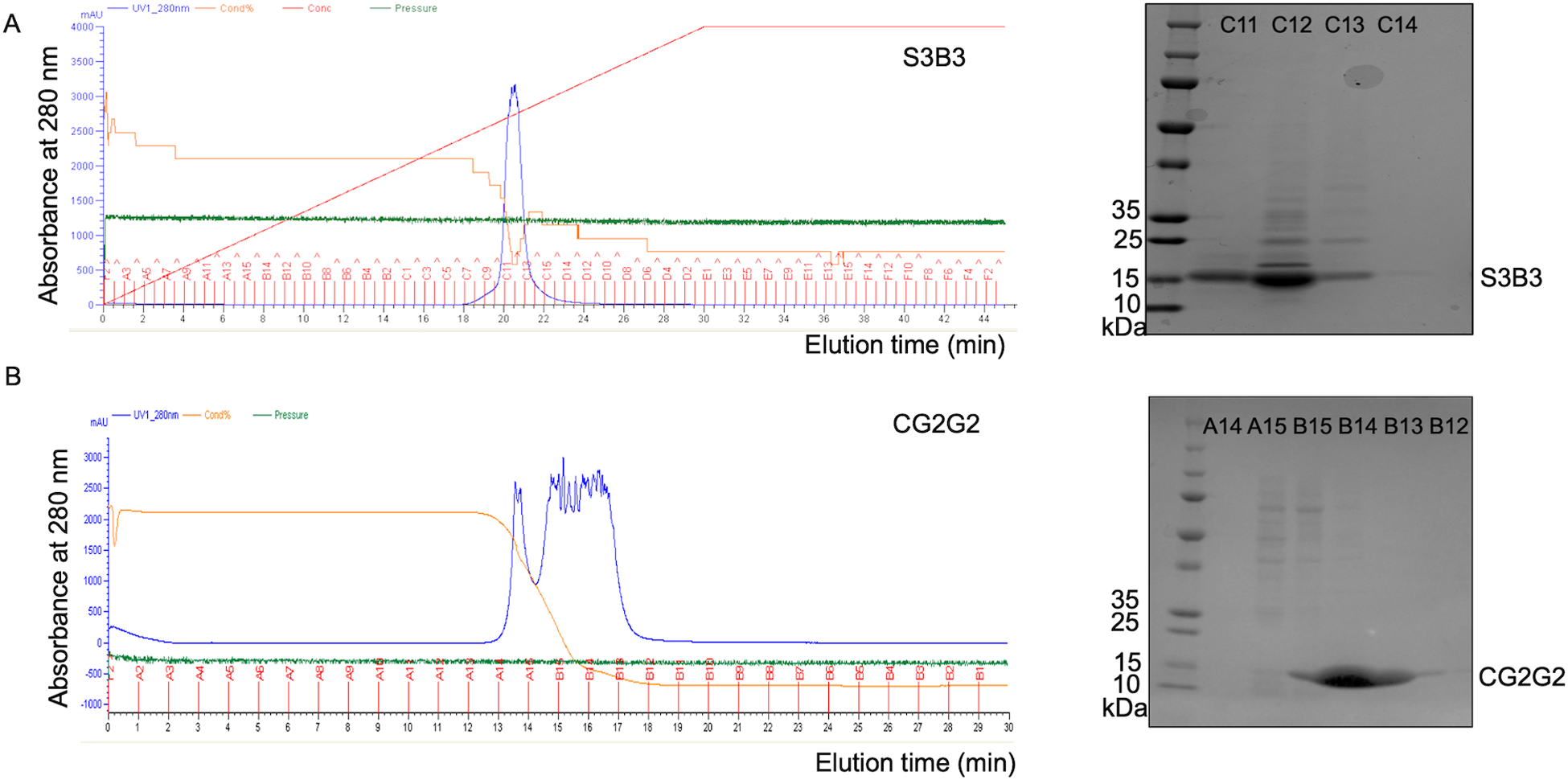
The purification of S3B3 and CG2G2 on AKTA FPLC system. Through the 6x Histidine-tag, A) S3B3 and B) CG2G2 can be purified by Ni-NTA column which is an immobilized metal ion affinity chromatography on FPLC-AKTA. After extensive wash with buffer A, the antibodies were eluted with gradient buffer B starting from 0 minute and targeting 100% in 30 (A) or 15(B) minutes. The fractions would be subjected to SDS-PAGE before combining and dialysis. The general column pressure for the purification is around 0.1 to 0.2 Mpa.
Buffer exchange
-
29
Run the fractions on an SDS-PAGE gel to check protein purity and recovery (Figure 4). 18 μl of the 1ml fraction mixed with 6 μl the 4x loading dye are added into each lane of the gel. A prestained protein ladder (10–250 kDa; 10 μl) is needed to monitor the gel status.
-
30
Pool the fractions containing the antibody protein which is around 15 kDa.
-
31
Dialyze the pooled fractions with a dialysis cassette (10 kDa cutoff) against PBS overnight at 4 °C.
-
32
Harvest the antibody protein from the dialysis cassette and measure the protein concentration using a Nanodrop spectrophotometer (A280).
-
33
Aliquot small volumes (50 to 200 μl each) of the antibody immediately, and store them at −80 °C.
Alternate protocol: Production of single domain antibodies using mammalian cell Expi293F
Single domain antibodies can be expressed in bacteria, yeast, or mammalian cells. The advantage of mammalian cell expression is that it can bring the post-translational modifications, especially glycosylation, on the antibody (Frenzel, Hust, & Schirrmann, 2013; Ho, Nagata, & Pastan, 2006). The antibody gene (S3B3 or CG2G2) with a 6x His and Flag tag is cloned into a mammalian expression vector (pcDNA3.1 or similar vector) with a signal peptide (MGWSCIILFLVATATGVHS), which can lead to the secretion of the antibody from the cells (Figure 5). With the multiple His tag, the protein can be purified with the nickel column on the AKTA FPLC system.
Figure 5.
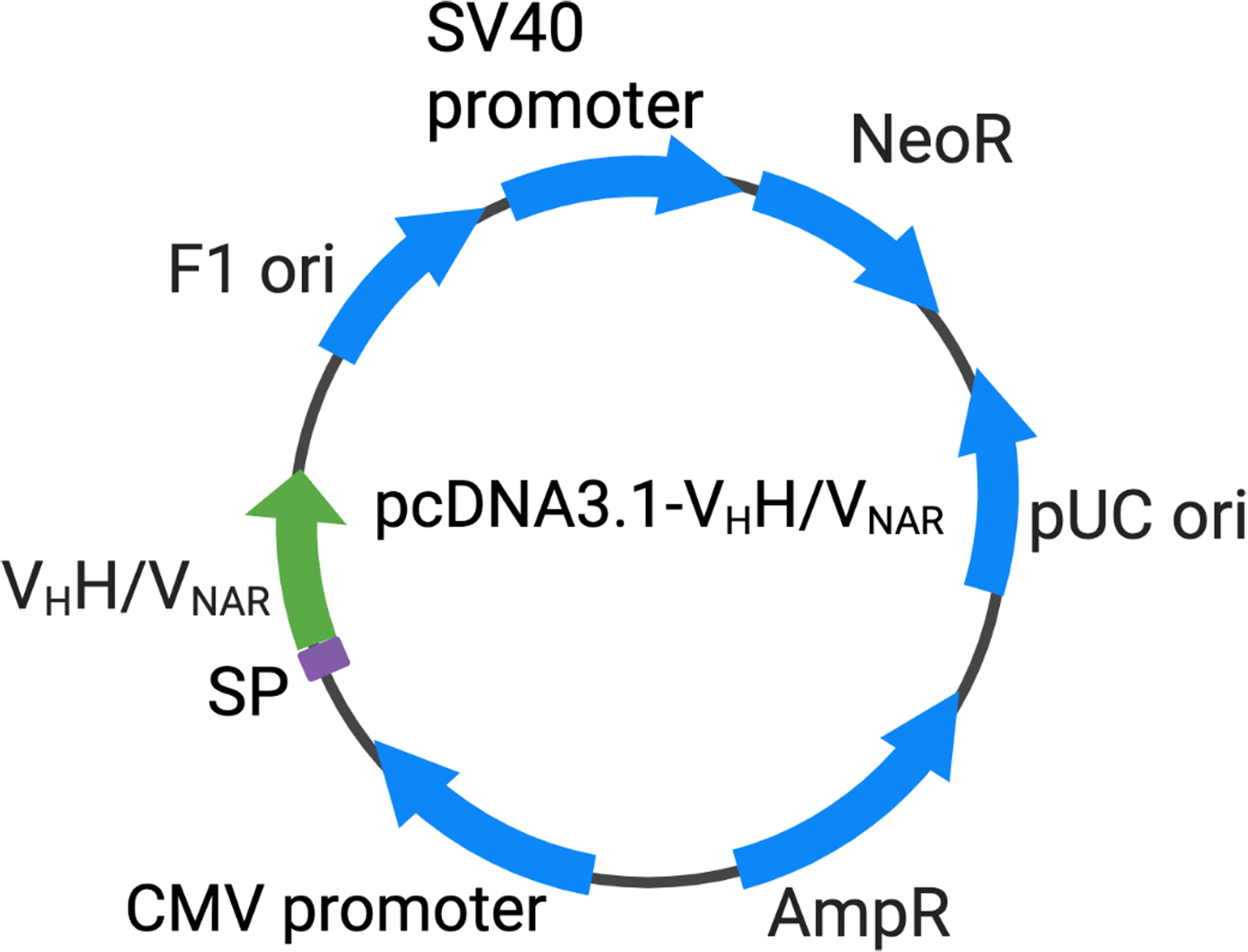
Plasmid map of pcDNA3.1-VHH and pcDNA3.1-VNAR. VHH: Camelid variable region of the heavy chain only antibody; VNAR: Antigen binding variable domain of the shark immunoglobulin new antigen receptor; SP: signal peptide; F1 ori: phage derived ori that allows for the replication and packaging of ssDNA into phage particles; SV40 promoter: the early promoter of the simian virus 40; NeoR: neomycin-resistance gene; pUC ori: pUC origin of replication; AmpR: ampicillin-resistance gene; CMV promoter: the human cytomegalovirus promoter.
Materials
pcDNA3.1-S3B3 or pcDNA3.1-CG2G2 (pcDNA3.1 can be obtained from Invitrogen #V790–20)
Expi293F cells (ThermoFisher Scientific #A14527)
Expi293 expression medium (ThermoFisher Scientific #A1435101)
125 ml shaker flask (Sigma-Aldrich #TMO4115)
PEI (Sigma-Aldrich #408727)
Phosphate-buffered saline (PBS) 10x (ThermoFisher Scientific #70011044)
Buffer A: PBS + 0.5 M NaCl
Buffer B: Buffer A + 200 mM imidazole (Sigma-Aldrich #68268)
HisTrap HP His tag protein purification column 1 × 1mL (Cytiva #29051021)
AKTAexplorer (GE Healthcare Life Sciences)
Shaker in Reach-In CO2 incubator (ThermoFisher Scientific)
SDS-PAGE gel: Novax 4%−20% Tris-Glycine gel (Invitrogen XP04205BOX)
Precision Plus Protein™ All Blue Prestained Protein Standards (Biorad #1610373)
4x Laemmli sample buffer (Biorad #1610747)
Slide-A-Lyzer dialysis cassette 10K (ThermoFisher Scientific #66380)
Nanodrop 1000 spectrophotometer (ThermoFisher Scientific)
The mammalian cell used here is Expi293F, which is a suspension growing cell line adapted to high-density growth conditions with a doubling time of around 24 hours during log phase growth. Frozen Expi293F cells are thawed directly into 35 ml pre-warmed Expi293 expression medium in a 125 ml shaker flask. The cells are then shaking at 120 rpm in the 37 °C incubator filled with 8% CO2. Typically, the cells will be passed when the density reaches 3–5 × 106 viable cells/mL.
PEI Transfection with Expi293F cells
-
1
One day before the transfection, seed the Expi293F cells with an appropriate inoculum to reach 3×106 cells/mL on the day of transfection.
-
2
Before transfection, spin down cells with a 50 ml conical tube at 250 g for 5 minutes. Remove the supernatant and resuspend the cell pellet in 30 ml fresh culture medium. The cell culture should have high viability (>95%).
-
3
Add 2 pg DNA/cell to the culture and gently mix the suspension. DNA should be prepared with high quality and endotoxin-free.
-
4
Add PEI stock solution in a 1:2 to 1:4 (w/w) DNA: PEI ratio and gently mix.
Optimal DNA: PEI ratio may need to be determined in advance.
-
5
Incubate cells under normal cultivation conditions mentioned above for 2–4 hours.
-
6
Spin down cells with a 50 ml conic tube at 250 g for 5 minutes. Remove the supernatant and resuspend the cell pellet in 35 ml fresh culture medium.
-
7
Culture for at least 7 days. At day 3, 5 and 7, spin down cells with a 50 ml conic tube at 250 g for 5 minutes. Collect the supernatant and put it in fridge. Resuspend the cell pellet in 35 ml fresh culture medium.
-
8
Pool all supernatant in a 250 ml centrifuge bottle and spin for 20 min at 4500 g to remove cell debris.
-
9
Filtrate the supernatant through a 0.45 μm filter before loading it through the column.
Antibody purification
-
10
Before loading the sample, the 1 ml HisTrap column is cleaned with water and equilibrated with 5 ml buffer A.
-
11
Load the supernatant into the column at a flow rate of 1 ml/min through a loading pump
-
12
Connect the loaded column to AKTA.
-
13
Wash the column with 15 CV of Buffer A (PBS + 0.5 M NaCl).
-
14
Elute the antibody with gradient Buffer B targeting 100% in 30 minutes for S3B3 and 15 minutes for CG2G2 and maintain 100% for another 15 minutes. The collection volume of fractions will be 1 ml.
Buffer exchange
-
15
Run the fractions on an SDS-PAGE gel to check protein purity and recovery. 18 μl of the 1ml fraction mixed with 6 μl the 4x loading dye are added into each lane of the gel. A prestained protein ladder (10–250 kDa; 10 μl) is needed to monitor the gel status.
-
16
Pool the fractions containing antibody protein which is around 15 kDa.
-
17
Dialyze the pooled fractions with a dialysis cassette (10 kDa cutoff) against PBS overnight at 4 °C.
-
18
Harvest the antibody protein from the dialysis cassette and measure the protein concentration using a Nanodrop spectrophotometer (A280).
-
19
Aliquot the antibody immediately with small volume (50 to 200 μl each) , and store them at −80 °C.
Support protocol 1: Production and purification of single domain antibodies on small scale with the polymyxin B method
When many antibodies are identified and proteins are needed for further validation, small-scale purification of antibodies is helpful to screen the potential binders with less effort before the large-scale production. This protocol describes the small-scale production of single domain antibodies which is performed with a nickel spin column and needs only small bacteria culture (30 to 50 ml). The protocol includes a transformation that is identical to the process in the Basic Protocol, protein induction, purification by nickel spin column. The transformation part is skipped and can be found in the Basic Protocol. The yield of small-scale production for shark VNAR is normally 0.1 to 0.5 mg/40 ml culture and for camel VHH is 0.1 to 1 mg/40ml culture.
Materials
E. coli strain: HB2151(LifeScienceMarket #S0122)
Phagemids: pComb3X-S3B3; pComb3X-CG2G2
2XYT medium (Research Products International #L24045)
SOC medium (ThermoFisher Scientific #15544034)
50% (v/v) Glycerol (Sigma-Aldrich #G5516)
100 mg/ml Ampicillin (Sigma-Aldrich #A1593)
2XYT agar plates with 100 μg/ml ampicillin and 2% (w/v) glucose
20% (w/v) glucose solution (Sigma-Aldrich #D9434)
IPTG (Sigma-Aldrich #16758)
Polymyxin B (Sigma-Aldrich #P0972)
Ni-NTA spin column (Qiagen #31314)
Phosphate-buffered saline (PBS) 10x (ThermoFisher Scientific #70011044)
Buffer A: PBS + 0.5M NaCl
Equilibration buffer: Buffer A + 10 mM imidazole (Sigma-Aldrich #68268)
Washing buffer: Buffer A + 20 mM imidazole
Elution buffer: Buffer A + 500 mM imidazole
SDS-PAGE gel: Novax 4%−20% Tris-Glycine gel (Invitrogen XP04205BOX)
Precision Plus Protein™ All Blue Prestained Protein Standards (Biorad #1610373)
4x Laemmli sample buffer (Biorad #1610747)
Slide-A-Lyzer dialysis cassette 10K (ThermoFisher Scientific #66380)
Nanodrop 1000 spectrophotometer (ThermoFisher Scientific)
Protein induction
-
1
Take out the transformation plates from the incubator.
Stoppable here: put plates at 4 °C for less than five days before induction
-
2
Scrape all colonies from transformed plates with a L-shaped scrapper with 10 ml 2XYT with 2% glucose and 100 μg/ml ampicillin.
-
3
Add 40 ml 2XYT with 2% glucose and 100 μg/ml ampicillin to the 10 ml culture above and split it into two 50 ml conical tube evenly.
-
4
Incubate at 37 °C with 250 rpm shaking until OD600 is 0.7 to 0.9.
-
5
Spin down the cells at 6,000 g for 20 minutes.
-
6
Remove old media. Resuspend each pellet in 25 ml fresh 2XYT with 100 μg/ml ampicillin. Add IPTG to 1 mM final concentration.
-
7
Incubate the flask at 30 °C overnight at 250 rpm.
Antibody purification
-
8
Collect cell pellet by centrifugation at 7,000 g for 20 min. Pour off supernatant. Drain the residual media from the pellet by inverting the bottle on a paper towel.
Stoppable here: put the bacteria pellet at −80 °C before the next steps
-
9
Wash pellet with 25 ml cold PBS and centrifuge at 6,000 g for 20 min. Drain residual media from the pellet by inverting the bottle on a paper towel.
-
10
Resuspend pellet in 1.2 ml cold PBS + 0.5 M NaCl. Pipet up and down till no clumps are visible and transfer to a 1.5 ml centrifuge tube.
-
11
Add polymyxin B 30 μl (500,000 units/ml). Incubate at 37 °C for 1 hour.
-
12
Centrifuge lysate at 12,000 g for 30 min at 4°C. Collect supernatant.
-
13
Equilibrate the Ni-NTA spin column with 600 μl equilibration buffer. Centrifuge for 2 min at 890 g. Centrifuge with open lid.
-
14
Load up to 600 μl of cleared lysate containing the 6xHis-tagged protein onto the pre-equilibrated Ni-NTA spin column. Centrifuge for 5 min at 270 g and collect flow-through. Centrifuge with open lid.
-
15
Wash the Ni-NTA spin column twice with 600 μl washing buffer. Centrifuge for 2 min at 890 g.
-
16
Elute the protein twice with 300 μl elution buffer. Centrifuge for 2 min at 890 g and collect the eluate.
-
17
Run SDS-PAGE gel to confirm size.
Buffer exchange
-
18
Dialyze the 600 μl with a dialysis cassette (10 kDa cutoff) against PBS overnight at 4 °C.
-
19
Harvest the antibody protein from the dialysis cassette and measure the protein concentration using a Nanodrop spectrophotometer (A280).
-
20
Aliquot the antibody immediately with small volume (50 to 200 μl each), and store them at −80 °C.
Support protocol 2: Validation of single domain antibodies by ELISA
The characterization and validation of purified single domain antibodies is performed by ELISA. Both antibodies have a Flag tag at the C terminal which can be used for ELISA testing. The antigen is coated on 96-well plates and the antibody at various concentrations was added to wells to bind to the antigen. The binding signal is detected by probing with HRP conjugated anti-flag antibodies (Figure 6).
Figure 6.
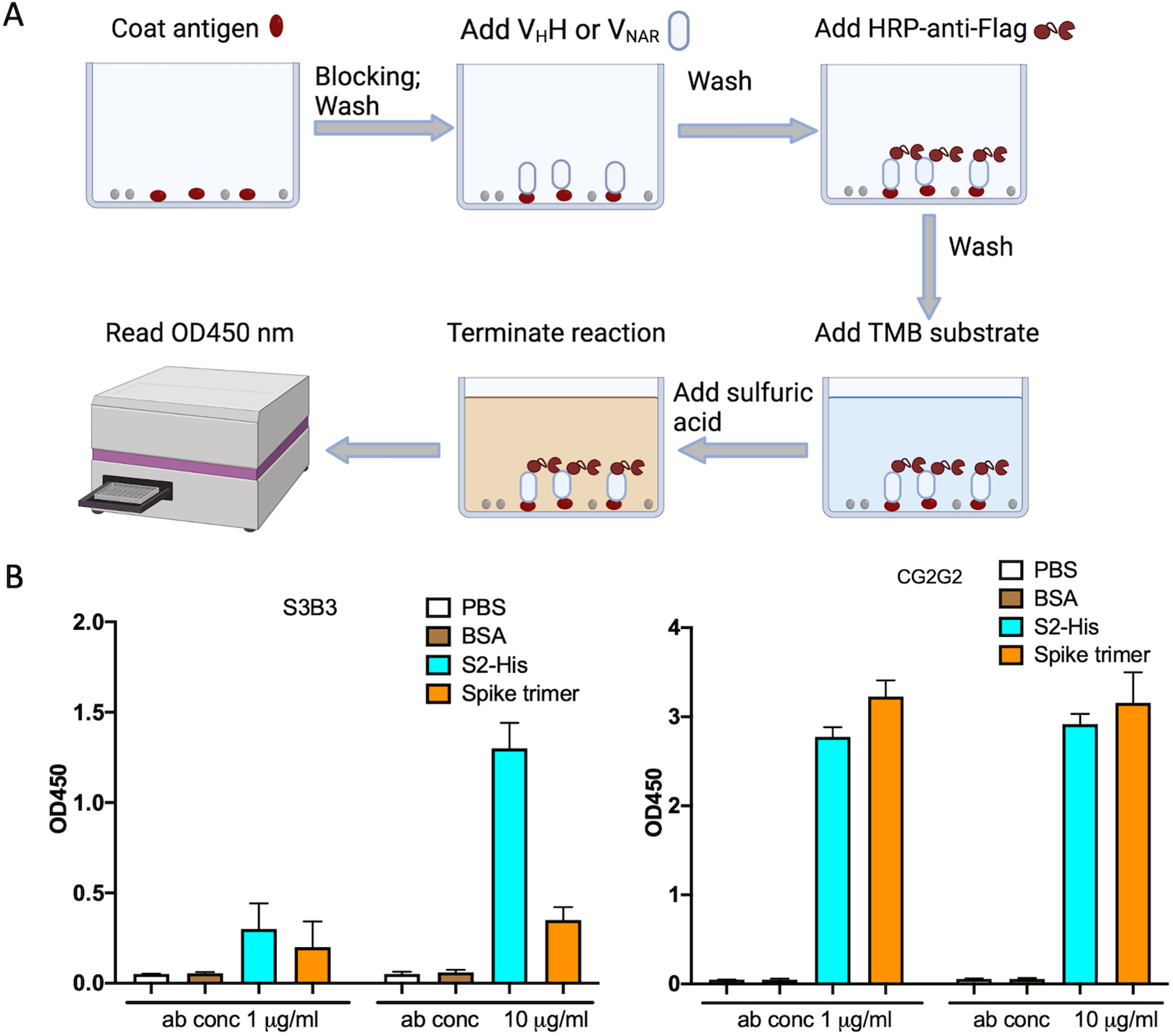
ELISA testing of S3B3 and CG2G2 binding to S2 subunit and spike trimer of SARS-CoV-2. A) The diagram of the ELISA. B) The antigens (5 μg/ml in PBS) were coated on ELISA plate and two concentrations of S3B3 and CG2G2 were used to test the binding. The signal was read on the plate reader at the OD450 nm. The graph was made by GraphPad Prism.
Materials
Phosphate-buffered saline (PBS) 10x (ThermoFisher Scientific #70011044)
PBST: 1x PBS containing 0.1% (v/v) Tween 20 (Biorad #1610781)
MaxiSoap 96-well plates (Sigma-Aldrich #M9410)
Superblock (ThermoFisher Scientific #37515)
HRP-anti-Flag 2nd antibody (Sigma-Aldrich #A8592)
TMB Chromogen Solution (ThermoFisher Scientific #002023)
1 M sulfuric acid (Sigma-Aldrich #84736)
Microplate reader (Molecular Devices)
ELISA
Coat the maxiSoap 96-well plate with the antigens (SARS-CoV-2 Spike trimer and S2 subunit) or controls (PBS and BSA). Pipet 50 μl of the antigen (5 μg/ml) into each well. Put the plate in fridge overnight.
Wash the plate three times with 200 μl PBST each time with 30 seconds waiting time.
Incubate with 100 μl blocking buffer (Superblock) for 1 hr at room temperature (RT).
Dilute the antibody with the dilution buffer (PBST+10% superblock) at various concentrations (1, 5, 10, and 20 μg/ml), which should give low to high reading signals at the end; add 50 μl of all diluted antibodies separately into the wells and incubate for 1 hr at RT.
Discard the solution and wash the plate three times with PBST.
Add 50 μl of 1:5000 dilution of HRP-anti-Flag 2nd antibody and incubate for 1hr at RT.
Wash the plate three times with PBST.
Add 50 μl TMB substrate solution to each well and incubate for 2 to 15 min at RT.
Stop the reaction by adding 25 μl of 1 M sulfuric acid.
Read the OD at 450 nm.
COMMENTARY
Background information
Single domain antibodies are the smallest single domain antigen binding proteins identified so far (about 15 kDa), whereas mouse and human antibody domains are about 50 kDa in size. They can be derived from the variable domain (VHH) of variant heavy chain-only IgGs (HCAb) found in camelids (e.g. llamas, alpacas, and camels), or the antigen binding variable domain (VNAR) of the shark immunoglobulin new antigen receptor (IgNAR) (English et al., 2020; Feng et al., 2019). Although very small, single domain antibodies still retain full antigen specificity and can bind to antigens with very high affinities. They can bind epitopes that are not normally accessible to conventional antibodies because of their extended complementarity determining regions (CDRs) (Ho, 2018; Stanfield, Dooley, Flajnik, & Wilson, 2004). In addition, camel nanobodies can be readily humanized and show low immunogenicity in recent clinical trials (Scully et al., 2019), and also shark IgNARs can be humanized (Steven et al., 2017). Another major advantage of single domain antibodies is the flexibility in drug delivery. For instance, these antibodies can be directly delivered into the lungs by nebulization because of their small size and good thermal stability (Ho, 2020), which is very useful against respiratory viruses like SARS-CoV-2.
Single domain antibodies are highly soluble, stable, lack glycans and are readily cloned and expressed in bacteria (Muyldermans, 2013). To date, people mainly utilize genetic engineering to add the signal sequence and specific tags including multiple histidines or maltose binding protein to the N or C terminus of antibodies, and then purify them with affinity chromatography (Pardon et al., 2014). The common method to release the protein into the periplasm is the osmotic shock (Pardon et al., 2014), which is less efficient than the polymyxin B treatment and generally has moderate to poor yield. In addition, small scale purification is useful if multiple binders need to be screened against the antigen. If several good antibodies have been identified, they can be purified by large scale FPLC purification, especially if large amounts of high-quality antibodies are important for further studies. Overall, we found that the yield for both camel and shark antibodies from this proposal is sufficient. The final product of antibodies from small- and large-scale production are biologically functional.
Critical parameters
The signal peptide is critical for antibody purification. It is a sequence of amino acids attached to a protein and directs the protein to the bacterial periplasm, where the sequence is removed by a signal peptidase (Steiner, Forrer, Stumpp, & Pluckthun, 2006). Protein secretion can increase the stability of the cloned gene products. It was shown that the half-life of the recombinant proinsulin is increased 10-fold when the protein is secreted to the periplasmic space (Sockolosky & Szoka, 2013). The commonly used signal peptides are MKYLLPTAAAGLLLLAAQPAMA derived from periplasmic pectate lyase(pelB), and MKKTAIAIAVALAGFATVAQAA derived from the outer membrane protein A (ompA), which is used in our constructs. The other important factor for protein expression is the control of the lac promoter. Glucose suppresses lacZ promoter activity and decreases protein leaking expression. Glucose can shorten the shaking time for the bacteria to reach desired density. It is important to remove the glucose before adding the IPTG which will remove the repressor of the lac operon and induce gene expression.
In mammalian system, the signal peptide which can lead the protein to be secreted is also critical. The signal sequence used here is MGWSCIILFLVATATGVHS. The other important factor is the transfection efficiency with mammalian cells. Generally, the efficiency will need to be optimized in advance with the fine testing of different ratio of PEI to DNA. PEI is economic and cheap. If the antibody has very low yield from PEI, it might be useful to try different mammalian cells or transfection reagents.
Troubleshooting
Polymyxin B treatment will destabilize the outer membrane of the bacteria (Zavascki, Goldani, Li, & Nation, 2007) and release the proteins in the periplasm. This is a very important step in the protocol. If there is not enough polymyxin B, the lysis of the cell membranes will be insufficient, and the yield of the antibody will be affected. More tips on troubleshooting are in Table 2.
Table 2.
Troubleshooting
| Problem | Possible cause | Solution |
|---|---|---|
| No colonies on the transformation plates | Wrong antibiotics in the plate Bad competent cells Wrong plasmid |
Make fresh plates Try fresh cells Sequence the plasmid |
| No cell growth in the flask | Too many antibiotics Failed shaker incubator |
Add accurate amount of antibiotics Check incubator |
| No protein expression | Glucose inhibition Bad IPTG |
Remove glucose before induction Make fresh IPTG |
| Low yield | Insufficient Polymyxin B | Make fresh stocks and add enough amount |
| Bad HisTrap column | Try new column | |
| Low purity | Not enough washing stringency | Increase washing stringency by increasing imidazole concentration (less then 50 mM) in the washing step |
| No ELISA signal | Bad TMB substrate or secondary antibodies | Use positive controls and test the secondary antibody with the TMB substrate directly |
| High background on ELISA | Insufficient blocking or washing | Increase blocking time or Including milk or BSA in the blocking buffer Wash more |
Understanding Results
If the purification is successful, the antibodies should be the major band (around 15 kDa) on an SDS-PAGE gel. Sometimes, the antibody might form a dimer which is around 30 kDa. Generally, VHHs and VNARs are quite stable. Protease cleavage might occur if the antibodies are exposed to proteases. The purity of the protein can be estimated on the gel too. The activity of the purified antibodies could be tested by ELISA and a strong signal indicates good binding to the antigens.
Time Considerations
Time considerations are listed in Table 3. Generally, it will take 5 to 7 days to finish the large-scale purification and ELISA validation. The yield of S3B3 is around 6 mg/L and CG2G2 is around 20 mg/L. Overall, the yield range for shark VNAR is 2 to 15 mg/L and for camel VHH is 5 to 40 mg/L according to our experiences.
Table 3.
Time considerations
| Days | Time | Procedure |
|---|---|---|
| 1 | 4 hrs | Transformation |
| 2 | 2 days | Protein induction |
| 3 | 6 hrs | Protein purification |
| 4 | 1 day | Buffer exchange |
| 5 | 2 days | ELISA |
Acknowledgement
This project was supported by the Intramural Research Program of NIH, Center for Cancer Research (CCR) National Cancer Institute (NCI), NCI CCR Antibody Engineering Program (ZIC BC 011891 to M.H.) and NIH Intramural Targeted Anti-COVID-19 (ITAC) Program (ZIA BC 011943 to M.H.). The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. We thank the NIH Fellows Editorial Board and NIH library for editorial assistance.
The VHH and VNAR nanobodies targeting SARS-CoV-2 spike protein are the subject of a pending patent application and are available for license in certain fields of use to qualified candidates. Please contact the corresponding leading author Dr. Mitchell Ho at homi@mail.nih.gov if you are interested in pursuing a license.
Literature Cited
- Barbas CF 3rd, Kang AS, Lerner RA, & Benkovic SJ (1991). Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc Natl Acad Sci U S A, 88(18), 7978–7982. doi: 10.1073/pnas.88.18.7978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English H, Hong J, & Ho M (2020). Ancient species offers contemporary therapeutics: an update on shark VNAR single domain antibody sequences, phage libraries and potential clinical applications. Antib Ther, 3(1), 1–9. doi: 10.1093/abt/tbaa001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng M, Bian H, Wu X, Fu T, Fu Y, Hong J, … Ho M (2019). Construction and next-generation sequencing analysis of a large phage-displayed V(NAR) single-domain antibody library from six naïve nurse sharks. Antib Ther, 2(1), 1–11. doi: 10.1093/abt/tby011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flajnik MF (2018). A cold-blooded view of adaptive immunity. Nat Rev Immunol, 18(7), 438–453. doi: 10.1038/s41577-018-0003-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenzel A, Hust M, & Schirrmann T (2013). Expression of recombinant antibodies. Front Immunol, 4, 217. doi: 10.3389/fimmu.2013.00217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M (2018). Inaugural Editorial: Searching for Magic Bullets. Antib Ther, 1(1), 1–5. doi: 10.1093/abt/tby001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M (2020). Perspectives on the development of neutralizing antibodies against SARS-CoV-2. Antib Ther, 3(2), 109–114. doi: 10.1093/abt/tbaa009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho M, Nagata S, & Pastan I (2006). Isolation of anti-CD22 Fv with high affinity by Fv display on human cells. Proc Natl Acad Sci U S A, 103(25), 9637–9642. doi: 10.1073/pnas.0603653103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J, Kwon HJ, Cachau R, Chen CZ, Butay KJ, Duan Z, … Ho M (2022). Dromedary camel nanobodies broadly neutralize SARS-CoV-2 variants. Proc Natl Acad Sci U S A, 119(18), e2201433119. doi: 10.1073/pnas.2201433119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyldermans S (2013). Nanobodies: natural single-domain antibodies. Annu Rev Biochem, 82, 775–797. doi: 10.1146/annurev-biochem-063011-092449 [DOI] [PubMed] [Google Scholar]
- Pardon E, Laeremans T, Triest S, Rasmussen SG, Wohlkonig A, Ruf A, … Steyaert J, (2014). A general protocol for the generation of Nanobodies for structural biology. Nat Protoc, 9(3), 674–693. doi: 10.1038/nprot.2014.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Running Deer J, & Allison DS (2004). High-level expression of proteins in mammalian cells using transcription regulatory sequences from the Chinese hamster EF-1alpha gene. Biotechnol Prog, 20(3), 880–889. doi: 10.1021/bp034383r [DOI] [PubMed] [Google Scholar]
- Scully M, Cataland SR, Peyvandi F, Coppo P, Knobl P, Kremer Hovinga JA, … Investigators H (2019). Caplacizumab Treatment for Acquired Thrombotic Thrombocytopenic Purpura. N Engl J Med, 380(4), 335–346. doi: 10.1056/NEJMoa1806311 [DOI] [PubMed] [Google Scholar]
- Sockolosky JT, & Szoka FC (2013). Periplasmic production via the pET expression system of soluble, bioactive human growth hormone. Protein Expr Purif, 87(2), 129–135. doi: 10.1016/j.pep.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanfield RL, Dooley H, Flajnik MF, & Wilson IA (2004). Crystal structure of a shark single-domain antibody V region in complex with lysozyme. Science, 305(5691), 1770–1773. doi: 10.1126/science.1101148 [DOI] [PubMed] [Google Scholar]
- Steiner D, Forrer P, Stumpp MT, & Pluckthun A (2006). Signal sequences directing cotranslational translocation expand the range of proteins amenable to phage display. Nat Biotechnol, 24(7), 823–831. doi:nbt1218 [pii] 10.1038/nbt1218 [DOI] [PubMed] [Google Scholar]
- Steven J, Muller MR, Carvalho MF, Ubah OC, Kovaleva M, Donohoe G, … Barelle CJ, (2017). In Vitro Maturation of a Humanized Shark VNAR Domain to Improve Its Biophysical Properties to Facilitate Clinical Development. Front Immunol, 8, 1361. doi: 10.3389/fimmu.2017.01361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavascki AP, Goldani LZ, Li J, & Nation RL (2007). Polymyxin B for the treatment of multidrug-resistant pathogens: a critical review. J Antimicrob Chemother, 60(6), 1206–1215. doi: 10.1093/jac/dkm357 [DOI] [PubMed] [Google Scholar]


