Abstract
Rapid analysis of microbial communities has proven to be a difficult task. This is due, in part, to both the tremendous diversity of the microbial world and the high complexity of many microbial communities. Several techniques for community analysis have emerged over the past decade, and most take advantage of the molecular phylogeny derived from 16S rRNA comparative sequence analysis. We describe a web-based research tool located at the Ribosomal Database Project web site (http://www.cme.msu.edu/RDP/html/analyses.html) that facilitates microbial community analysis using terminal restriction fragment length polymorphism of 16S ribosomal DNA. The analysis function (designated TAP T-RFLP) permits the user to perform in silico restriction digestions of the entire 16S sequence database and derive terminal restriction fragment sizes, measured in base pairs, from the 5′ terminus of the user-specified primer to the 3′ terminus of the restriction endonuclease target site. The output can be sorted and viewed either phylogenetically or by size. It is anticipated that the site will guide experimental design as well as provide insight into interpreting results of community analysis with terminal restriction fragment length polymorphisms.
Many microbial communities have proven to be complex assemblages of different phylotypes and physiologies. For example, the number of species in soil is estimated to be more than 4,000 species/30 g of soil (17), and estimates of the number of bacterial species are enormous (6). It has been only within the last 20 years that we have begun to recognize the phylogenetic diversity of the microbial world. During this time 16S rRNA has emerged as one of the premier phylogenetic markers (7, 16, 19), providing a landmark for comparative analyses of isolated and uncultured strains as well as microbial communities.
Comparative community analysis provides an accelerated approach to understanding community structure and function. It allows for the identification of unique or numerically dominant strains or groups under defined or controlled conditions. Thus, one can begin to dissect the trophic complexity of a community by changing nutrient patterns and observing the resulting changes in community structure. A number of rapid techniques to screen communities have been developed (2, 3, 8, 9, 13, 14), and many of those described to date take advantage of the 16S rRNA phylogenetic marker and are culture-independent approaches (1). Preliminary steps include the isolation of community DNA and the PCR amplification of a phylogenetic marker from the community DNA template. It is at this point that the various approaches differ in how the PCR products are separated. Terminal restriction fragment length polymorphism (T-RFLP) takes advantage of the high resolution and throughput of automated sequencing technologies to separate the polymorphic terminal fragments after restriction digestion. Because the polymorphism is based solely on the length of the fragment, direct reference can be made to the sequence database (9, 11).
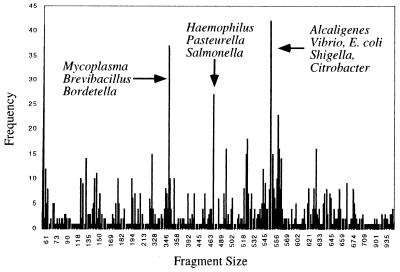
The potential effectiveness of distinguishing phylotypes by T-RFLP is presented in Fig. 1. The frequency distribution of terminal fragment sizes derived from an in silico digestion with TspEI (AATT) of all sequences in release 7 of the Ribosomal Database Project (RDP) is plotted, with fragment size on the abscissa. Of the 1,663 nearly complete sequences from release 7, 1,200 are recognized by both the 27F primer and restriction endonuclease TspEI. Among the 1,200 restriction products, there are 349 unique terminal fragment sizes. The ability to resolve 29% of the phylotypes from the current database is significant, given the skewed composition (e.g., duplicated sequences and emphasis on medically important organisms) of the database. The terminal fragment sizes with the greatest frequency are indicated.
FIG. 1.
Frequency distribution profile of terminal fragments derived from an in silico digestion of the RDP-II with the restriction enzyme TspEI. The phylotypes with the highest frequency found in the database are labeled.
To increase the effectiveness of T-RFLP as a tool for community analysis, knowledge of the distribution of restriction sites on the 16S ribosomal DNA (rDNA) and of the relationship of terminal fragment size to phylogeny is required. To that end we have developed a web site that integrates the most recent release of the Ribosomal Database Project (10), including the phylogenetic tree, with a pattern-searching algorithm. The site provides the investigator with a rapid way to determine optimal primer and restriction enzyme combinations for community analysis. Moreover, it permits an approach to tentatively identify experimentally determined phylotypes from cognate phylotypes in the database.
Brief description of T-RFLP.
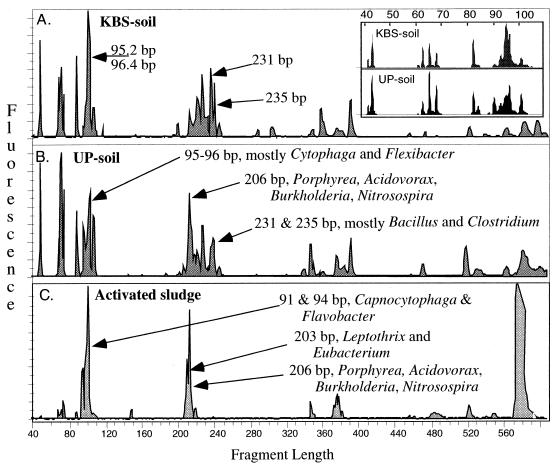
The initial steps of community analysis protocols vary only in detail. Briefly, community DNA is extracted directly from the environment by any of the techniques that are efficient for the particular community (15, 20). The 16S rDNAs from phylotypes present in the community are then PCR amplified using primers targeted to conserved regions of the gene. Primers can be designed to be nondiscriminating, amplifying nearly all 16S rDNAs, or selective, targeting specific domains or groups. The 5′ primer is fluorescently labeled to tag the products. The amplification products are then digested with restriction endonucleases, usually 4-base cutters, and the primer-proximal products (hence, the use of the descriptor “terminal”) are sized on a sequencing gel (2, 3, 5, 9, 11). Three T-RFLP electrophoretic profiles from an HhaI digestion are presented in Fig. 2. The two profiles from soil communities (Fig. 2A and B) are similar to one another and decidedly different from the profile of activated sludge (Fig. 2C). Presented in Fig. 2 are terminal fragments ranging from 35 to 600 bp long. The insert in Fig. 2A presents an expanded view of the soil community profile (with terminal fragments from 35 to 120 base pairs long) as a demonstration of the resolving power of the method. While longer fragments are possible, they are not accurately sized by the combination of gel parameters and size markers employed in this experiment.
FIG. 2.
T-RFLP profile of three bacterial communities. Terminal fragments were generated from an HhaI digestion of 16S rDNAs amplified from total community DNA using a fluorescently tagged 27F primer and an unlabeled 1392R primer. The terminal fragments were sized on an ABI 373 automated sequencer using internal size standards (ABI TAMRA 2500). (A) Agricultural soil from Kellogg's Biological Station (KBS), Michigan State University; (B) uncultivated soil from the upper peninsula (UP) of Michigan; (C) activated sludge. The insert presents an expanded view of the soil profile for fragments from 40 to 110 bp long.
T-RFLP analysis program (TAP) web site.
We have developed a web site (located at http://www.cme.msu.edu/RDP/trflp/#program) that allows the investigator to answer the following initial questions in community analysis using T-RFLP. (i) What restriction enzyme(s) will provide the most discriminating activity for estimates of population diversity? (ii) What enzyme(s) will provide the best resolution for the phylogenetic group(s) of interest? (iii) What primer-enzyme combination will be optimal for the community under investigation? In the final analysis, each specific community may have its own set of optima that are empirically defined. However, initial general directions regarding appropriate enzyme(s) to use, as well as phylogenetic insights, can be gained by examining an in silico digestion of the 16S rRNA database (4, 9).
TAP gives its users the ability to simulate the T-RFLP procedure with the entire RDP as the surrogate community. The required user input includes a forward or reverse primer sequence that may contain non-Watson-Crick International Union of Biochemistry (IUB) characters and a restriction enzyme target sequence(s). In addition, the maximum number of base mismatches allowed within a specified number of bases from the 5′ end of the primer can be specified. In the event that more than one restriction enzyme is selected, the operator has the additional option of performing multiple single digests or a single multiple enzyme digest.
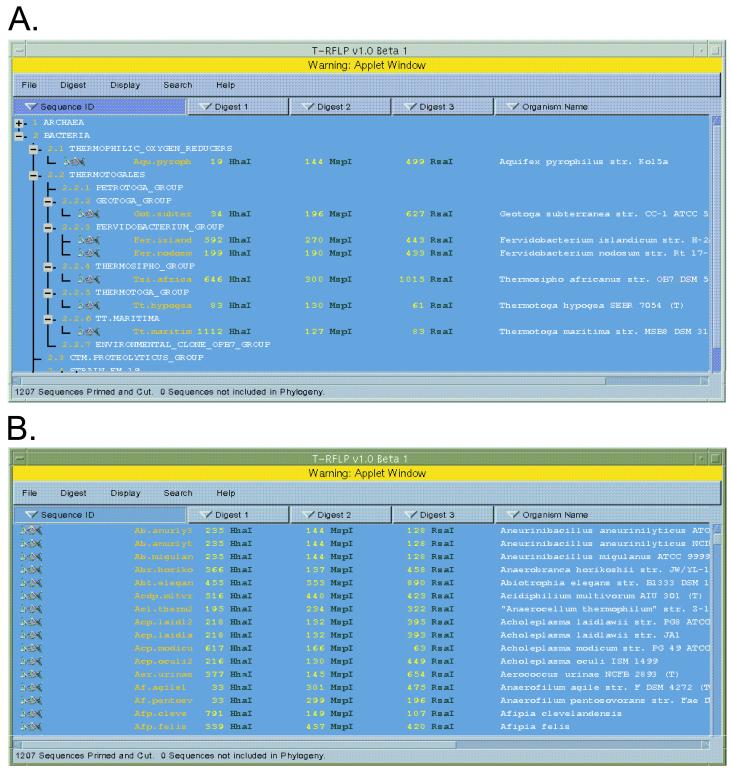
Upon submitting a digest request, the program accesses the most recent release of the RDP prokaryote database, extracts a name and a sequence, and attempts to prime each sequence with the supplied primer sequence under the specified conditions. Each sequence that is successfully recognized by the primer sequence is digested by the specified enzyme(s). If the user selects multiple single digestions, the program will display each resulting terminal fragment size and enzyme with the organism's name and short ID (SID) (Fig. 3). If the user selects a multienzyme digestion, the shortest fragment size and corresponding enzyme is displayed with the name and SID. In the event that a sequence is successfully primed but no restriction site is found, “NA” is entered in the data set at the appropriate site.
FIG. 3.
Phylogenetic and sorted display windows of TAP. (A) Organism RDP identifiers, fragment sizes, selected restriction enzymes, and organism names are displayed within the RDP-II phylogenetic hierarchy. Organism names and RDP phylogenetic indices are displayed adjacent to expansion status boxes. A plus image indicates the presence of undisplayed subheadings, as with Archaea, and a minus image indicates that the level is fully expanded. (B) Organisms are displayed sorted in according to the selected column header buttons. In this instance, the organisms are sorted alphabetically in ascending order by their RDP sequence identifiers.
The resultant digest data, including the organism's name, its RDP identifier, and one or more fragment sizes and enzyme pairs, can be displayed in two configurations. The default configuration places the data within the RDP's prokaryotic phylogenetic hierarchy (Fig. 3). For each phylogenetic group, the display shows the group's level and name and an image indicating whether any levels and/or sequences are collapsed underneath. If a phylogenetic group contains subnodes, the indicator can be selected to toggle the display between expanded and collapsed forms.
In addition to the phylogenetic hierarchy display, TAP can display the digest data in a sorted order. The user can sort the data by sequence name, by SID, or by a digest's restriction fragment size. The data is ordered alphabetically if sorted by the organism's name or SID and is ordered numerically if sorted by a digest's restriction fragment size. The program determines what information to sort by the selection of a header at the top of each column of data.
An additional function the program offers is the ability to highlight specific organisms of interest. The highlight status of an organism is retained between the phylogenetic and sorted displays. The user can toggle the highlight status of an organism by selecting it. In addition, the highlight status can be toggled by performing a search. The program's search engine will examine the column with the selected header for the word, or set of fragment sizes, supplied by the user. By default, the search engine adds to those already highlighted organisms that meet the user-specified criteria. This allows the user to perform a Boolean “OR” search. Alternately, the user can search only the previously highlighted organisms for the specified word or fragment size(s), removing the highlighting from any organisms that do not match the search parameters. This allows the user to construct a Boolean “AND” search. Furthermore, when searching a digest column for particular restriction fragment sizes, the user can specify a base-pair tolerance.
TAP was developed for multiplatform usage through the web. The priming and digest calculations and access to the Ribosomal Database Project RDP-II database are handled by a web server using common gateway interface programs at the RDP-II web site. The user interface is implemented as an applet in the Java 2 programming language. Because browser support for Java is not uniform, we have designed the applet to work with a Java plug-in available from Sun Microsystem's web site (http://java.sun.com/products/plugin) for Solaris and Windows operating systems or from Mozilla Organization's web site (http://www.mozilla.org/oji/MRJPlugin.html) for the Macintosh operating system. Some Macintosh users may also need to obtain the most recent version of the Macintosh Runtime for Java which is available from Apple Computer's web site (http://www.apple.com/java). All aforementioned software packages are available free of charge. Links to these packages can be found on the TAP web page.
Fragment lengths and phylogeny.
The power of the technique rests in the high throughput and resolution of terminal fragment lengths with nucleic acid sequencing technology. The fact that in silico digestions of the RDP indicate that a significant fraction of the current database can be distinguished on the basis of terminal restriction fragment length in no way is meant to imply that one can positively identify phylogenetic groups or species based upon terminal fragment length. However, the disposition of restriction sites along the length of the 16S rDNA molecule does reflect phylogeny at some level. Table 1 presents the terminal fragments measured from 5′ Escherichia coli position 8 of three in silico digests (HhaI, MspI, and RsaI) of the RDP. The fragments were sorted first by the length of the HhaI fragment and then by the lengths of the MspI and RsaI fragments, respectively. Only HhaI digestion products between 1,098 and 1,105 bp long are presented, along with the subgroup affiliation from release 7.1 of the RDP. Several points are revealed by the list in this table. First, note that the table lists 26 members from group 2.28.3, 4 from group 2.30.8, and 1 from group 2.15.1. Hence, 84% of the sequences from this range of HhaI terminal fragment sizes are from the gamma subdivision of the Proteobacteria. Second, coherent groupings based upon one, two, or three digestions are indicated. All nine of the phylogenetic subgroups represented can be resolved from the remaining database with two or three digestions. Clusters of species from subgroups 2.28.3.26.17, 2.28.3.26.18, 2.28.3.26.11, 2.28.3.26.12 cannot be distinguished with three enzymes. These are, of course, phylogenetically quite close to one another, accounting for the conservation of restriction site positions. The Buchnera subgroup is resolved below the species level. Third, the HhaI fragments from these phylogenetic groups have terminal fragment sizes greater than 1,000 bp. Terminal fragments of sizes greater than 600 bp are resolved poorly, if at all, by gel systems. While capillary electrophoresis systems offer longer sequence reads (up to 1,000 bp under optimal conditions) and fewer electrophoretic anomalies, coverage of the entire molecule with one labeled primer is still impossible. Hence a single-label profile, even under optimal conditions, may not reveal or track all potentially resolvable populations of a community. An inspection of the digested database with TAP will quickly reveal if there are any known phylogenetic groups that would be out of range of a single labeled primer with a specified enzyme. This underscores the need for several primer sets or multiplexed fluorescently labeled primers when dissecting a complex community by T-RFLP.
TABLE 1.
Derivation of phylogenic inference from TAPa
| Fragment length (bp)
|
RDP sequence | RDP phylogenetic subgroup | ||
|---|---|---|---|---|
| HhaI | MspI | RsaI | ||
| 1,098 | 136 | 626 | Eubacterium biforme | 2.30.8.2.9 EUB.CYLINDROIDES_SUBGROUP |
| 1,098 | 139 | 1,221 | Eubacterium cylindroides | 2.30.8.2.9 EUB.CYLINDROIDES_SUBGROUP |
| 1,098 | 496 | 427 | Buchnera aphidicola | 2.28.3.27.1 BUCHNERA_SUBGROUP |
| 1,099 | 495 | 223 | Vibrio sp. | 2.28.3.20.4 MRT.MARINA_SUBGROUP |
| 1,099 | 496 | 74 | Actinobacillus capsulatus | 2.28.3.26.17 ACB.PLEUROPNEUMONIAE_SUBGROUP |
| 1,099 | 496 | 650 | Pasteurella haemolytica | 2.28.3.26.18 MNH.HAEMOLYTICA_SUBGROUP |
| 1,099 | 496 | 650 | Pasteurella sp. | 2.28.3.26.18 MNH.HAEMOLYTICA_SUBGROUP |
| 1,099 | 496 | 650 | Actinobacillus suis | 2.28.3.26.17 ACB.PLEUROPNEUMONIAE_SUBGROUP |
| 1,099 | 496 | 650 | Actinobacillus equuli | 2.28.3.26.17 ACB.PLEUROPNEUMONIAE_SUBGROUP |
| 1,099 | 496 | 650 | Actinobacillus capsulatus | 2.28.3.26.17 ACB.PLEUROPNEUMONIAE_SUBGROUP |
| 1,099 | 496 | 650 | Actinobacillus hominis | 2.28.3.26.17 ACB.PLEUROPNEUMONIAE_SUBGROUP |
| 1,099 | 496 | 741 | Pasteurella sp. | 2.28.3.26.18 MNH.HAEMOLYTICA_SUBGROUP |
| 1,099 | 496 | 882 | Pasteurella sp. | 2.28.3.26.18 MNH.HAEMOLYTICA_SUBGROUP |
| 1,099 | 496 | 882 | Pasteurella haemolytica | 2.28.3.26.18 MNH.HAEMOLYTICA_SUBGROUP |
| 1,099 | 496 | 882 | Haemophilus paracuniculus | 2.28.3.26.12 H.PARACUNICULUS_SUBGROUP |
| 1,099 | 496 | 882 | Haemophilus parasuis | 2.28.3.26.11 H.PARASUIS_SUBGROUP |
| 1,100 | 138 | 630 | Eubacterium cylindroides | 2.30.8.2.9 EUB.CYLINDROIDES_SUBGROUP |
| 1,101 | 97 | 459 | Porphyromonas levii | 2.15.1.2.7 PPM.MACACAE_SUBGROUP |
| 1,101 | 283 | 755 | Thiobacillus hydrothermalis | 2.28.3.7 DICHELOBACTER_GROUP |
| 1,101 | 496 | 224 | Vibrio sp. | 2.28.3.20.4 MRT.MARINA_SUBGROUP |
| 1,101 | 498 | 652 | Actinobacillus pleuropneumoniae | 2.28.3.26.17 ACB.PLEUROPNEUMONIAE_SUBGROUP |
| 1,101 | 498 | 652 | Actinobacillus lignieresii | 2.28.3.26.17 ACB.PLEUROPNEUMONIAE_SUBGROUP |
| 1,101 | 498 | 884 | Haemophilus parasuis | 2.28.3.26.11 H.PARASUIS_SUBGROUP |
| 1,101 | 606 | 224 | Vibrio sp. | 2.28.3.20.4 MRT.MARINA_SUBGROUP |
| 1,102 | 140 | 592 | Streptococcus pleomorphus | 2.30.8.2.9 EUB.CYLINDROIDES_SUBGROUP |
| 1,102 | 496 | 224 | Environmental isolate | 2.28.3.20.4 MRT.MARINA_SUBGROUP |
| 1,102 | 496 | 884 | Environmental isolate | 2.28.3.20.4 MRT.MARINA_SUBGROUP |
| 1,104 | 499 | 430 | Buchnera aphidicola | 2.28.3.27.1 BUCHNERA_SUBGROUP |
| 1,104 | 501 | 432 | Buchnera aphidicola | 2.28.3.27.1 BUCHNERA_SUBGROUP |
| 1,105 | 496 | 224 | Vibrio sp. | 2.28.3.20.4 MRT.MARINA_SUBGROUP |
Two additional separate enzymatic digestions on the group defined by an HhaI terminal fragment size of between 1,098 and 1,105 bp were carried out. The values in bold indicate coherent groups of terminal fragment phylotypes and the corresponding phylogenetic subgroups (determined with RDP release 7.1)
Pitfalls of T-RFLP.
T-RFLP analysis of microbial communities is gaining increased usage in the scientific community because it is rapid and has high resolution. It is, however, subject to all of the caveats routinely applied to molecular approaches that are dependent on efficient extraction of community DNA and PCR amplification of a target gene. These difficulties have been discussed previously in considerable detail (15, 18, 20) and include, primarily, concerns regarding preferential extraction of genomic DNAs and amplification bias during PCR. In addition, care must be taken to assure that the restriction digests are complete and specific. This can be monitored by including the amplified product from a well-characterized isolate in representative digestions. If this control product is amplified with a primer labeled with a different fluor, it can easily be distinguished from fragments derived from the community profile.
Inasmuch as the power of this technique lies in comparative community analysis, considerable attention must be paid to standardizing all parameters during the processing of the samples. That having been done, any differences detected in community profiles can be attributed to differences in community structure rather than to differences in sample preparation. It should also be noted that a terminal restriction fragment profile is a quantitative and detailed view of the PCR product pool derived from a community. It is not, however, a quantitative view of the structure of the native community, primarily because of possible PCR bias during amplification and the diversity of rRNA operon copy numbers seen within bacterial genomes (18).
Evolving web site.
The future directions for this site will depend in part upon suggestions from users. There are several new features currently being considered. First, we will extend the analysis function to the 18S and large subunit database. Second, we hope to develop a data analysis function that would provide rapid identification of species in the database that match a submitted T-RFLP profile. Third, we will further enhance the methodologies for rapid comparisons of T-RFLP profiles with an eye to identifying pandemic as well as endemic populations among the communities being compared.
ACKNOWLEDGMENTS
This research was supported by the Center for Microbial Ecology through NSF DEB-9120006 and funding from the DOE (DE-FG02-99ER62848) to the Ribosomal Database Project. T.L. Marsh is supported, in part, by the DOE (DE-FG02-97ER62477).
REFERENCES
- 1.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avaniss-Aghajani E, Jones K, Chapman D, Brunk C. A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences. BioTechniques. 1994;17(1):144–149. [PubMed] [Google Scholar]
- 3.Bruce K D. Analysis of mer gene subclasses within bacterial communities in soils and sediments resolved by fluorescent-PCR-restriction fragment length polymorphism profiling. Appl Environ Microbiol. 1997;63:4914–4919. doi: 10.1128/aem.63.12.4914-4919.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunk C F, Avaniss-Aghajani E, Brunk C A. A computer analysis of primer and probe hybridization potential with bacterial small-subunit rRNA sequences. Appl Environ Microbiol. 1996;62:872–879. doi: 10.1128/aem.62.3.872-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clement B G, Kehl L E, DeBord K L, Kitts C L. Terminal restriction fragment patterns [TRFPs], a rapid, PCR-based method for the comparison of complex bacterial communities. J Microbiol Methods. 1998;31:135–142. [Google Scholar]
- 6.Dykhuizen D E. Santa Rosalia revisited: why are there so many species of bacteria? Antonie Leeuwenhoek. 1998;73:25–33. doi: 10.1023/a:1000665216662. [DOI] [PubMed] [Google Scholar]
- 7.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee D-H, Zo Y-G, Kim S-J. Nonradioactive method to study genetic profiles of natural bacterial communities by PCR–single-strand-conformation polymorphism. Appl Environ Microbiol. 1996;62:3112–3120. doi: 10.1128/aem.62.9.3112-3120.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maidak B L, Cole J R, Parker C T, Jr, Garrity G M, Larsen N, Li B, Lilburn T G, McCaughey M J, Olsen G J, Overbeek R, Pramanik S, Schmidt T M, Tiedje J M, Woese C R. A new version of the RDP [Ribosomal Database Project] Nucleic Acids Res. 1999;27:171–173. doi: 10.1093/nar/27.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marsh T L. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplicons. Curr Opin Microbiol. 1999;2:323–327. doi: 10.1016/S1369-5274(99)80056-3. [DOI] [PubMed] [Google Scholar]
- 12.Marsh T L, Liu W-T, Forney L J, Cheng H. Beginning a molecular analysis of the eukaryal community in activated sludge. Water Sci Technol. 1998;37:455–460. [Google Scholar]
- 13.Massol-Deya A A, Odelson D A, Hickey R F, Tiedje J M. Bacterial community fingerprinting of amplified 16S and 16-23S ribosomal DNA gene sequences and restriction endonuclease analysis [ARDRA] In: Akkermans A D, et al., editors. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 1–8. [Google Scholar]
- 14.Muyzer G A, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogram A. Isolation of nucleic acids from environmental samples. In: Burlage R S, Atlas R, Stahl D, Geesey G, Sayler G, editors. Techniques in microbial ecology. New York, N.Y: Oxford University Press; 1998. [Google Scholar]
- 16.Pace N R, Stahl D A, Lane D J, Olsen G J. The analysis of natural microbial populations by ribosomal RNA sequences. Adv Microbial Ecol. 1986;9:1–55. [Google Scholar]
- 17.Torsvik V, Goksoyr J, Daae F L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wintzingerode F V, Göbel U B, Satackebrandt E. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol Rev. 1997;21:213–229. doi: 10.1111/j.1574-6976.1997.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 19.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]