Abstract
Mitochondria are vital organelles that provide energy for muscle function. When these organelles become dysfunctional, they produce less energy as well as excessive levels of reactive oxygen species which can trigger muscle atrophy, weakness and loss of endurance. In this review, molecular evidence is provided to show that exercise serves as a useful therapeutic countermeasure to overcome mitochondrial dysfunction, even when key regulators of organelle biogenesis are absent. These findings illustrate the complexity and compensatory nature of exercise-induced molecular signaling to transcription, as well as to post-transcriptional events within the mitochondrial synthesis and degradation (i.e. turnover) pathways. Beginning with the first bout of contractile activity, exercise exerts a medicinal effect to improve mitochondrial health and whole muscle function.
Keywords: Mitochondria, Muscle, Exercise, Health, Aging, Metabolism, Training
Introduction
Mitochondria are unique organelles that provide the energy (ATP) for cell survival. They consist of approximately 1200 proteins within the matrix, inner and outer membranes, and intermembrane space compartments.1 Most of these proteins are transcribed from the nuclear genome, translated in the cytosol, and imported into existing mitochondria. A small number of mitochondrial proteins (13) are synthesized within the organelle itself, transcribed from a small, compact, circular mitochondrial DNA (mtDNA). These proteins are vital for the structure and function of the electron transport chain (ETC), the series of reactions responsible for the oxidation of reduced coenzymes (NADH2, FADH2) which are products of glucose and fatty acid catabolism. The oxidation of these substrates leads to the formation of an electrochemical gradient across the inner membrane, which is used to power the synthesis of ATP.2 The ETC is composed of five large multi-subunit complexes of protein (Complexes I–V) which perform this task. mtDNA-encoded components are found in all complexes, with the exception of Complex II. During the process of electron transport, excess electrons can be inadvertently donated to oxygen, thereby forming reactive oxygen species (ROS). These molecules are useful when produced in moderation, acting as signaling agents which can affect the transcription of nuclear genes encoding mitochondrial proteins.3, 4, 5, 6 However, when produced in excess, such as during conditions of ETC inhibition, or in the presence of protein subunit mutations, excess electrons can serve to oxidize proteins, lipids and mtDNA, leading to functional defects.7
In skeletal muscle, mitochondria exhibit structurally complex morphologies which are regionally distinct.8,9 Below the sarcolemma, a high concentration of “subsarcolemmal” (SS) mitochondria exist which have relatively simple configurations. In contrast, more diffusely localized organelles exist between the myofibrils (intermyofibrillar, or IMF mitochondria).10 This localized organelle is more reticular in nature, having interconnections between neighbouring mitochondria and forming an irregular network of organelles. SS mitochondria likely serve to provide ATP for membrane transport functions and provide energy support for nuclear processes such as transcription, and nuclear molecule transport. On the other hand, IMF mitochondria support the energy requirements of actin-myosin interactions, and contractile activity per se.11, 12, 13 An interesting note is that these mitochondrial populations possess subtle differences in biochemical characteristics when isolated and studied in vitro. In addition, when they are quantified based on electron microscopy analyses, SS mitochondria occupy about 10–20% of the total organelle population, while IMF mitochondria comprise of the remaining 80–90%.14 These proportions can be altered as a result of adaptations to exercise, disuse, or disease.9,12 In general, the SS mitochondria appear to proliferate more in response to cellular perturbations. The increase in SS mitochondria as a result of training, for example, is about twice the response of IMF mitochondria.9 This increase is further amplified under conditions of mtDNA-driven disease conditions, in which a remarkable increase in SS mitochondria produces a morphology termed ragged-red fibers.15 This adaptation appears to be the result of a compensatory nuclear gene expression response to a mitochondrial DNA defect.
The adaptive response of muscle mitochondrial content to regularly performed exercise (i.e. training) was first convincingly demonstrated by John Holloszy in his classic publication in 1967.16 He demonstrated that endurance exercise training performed at an appropriately prescribed intensity, duration, and frequency per week, in combination with high intensity intervals, could produce a remarkable 100% increase in the level of oxidative enzymes per gram of muscle. Modest changes in mitochondrial composition favouring more densely packed electron transport chain proteins were also observed. Subsequently he and his research group showed that this adaptation was muscle fibre type-dependent, and that the physiological consequences of this increase in mitochondria was more favourable lipid oxidation, accompanied by reduced glycolytic flux and glycogen utilization. These physiological adaptations, along with reduced lactic acid production, were shown to contribute meaningfully to the overall enhancement of endurance performance observed as a result of training. Indeed, a number of studies have confirmed that the best correlation between any physiological parameter and endurance performance is the mitochondrial content of the muscle. Gollnick et al. subsequently showed that these adaptive changes also took place in human muscle subject to endurance training.17
These studies led to a remarkable growth in interest for the adaptive plasticity of muscle. While these adaptations have now repeatedly been observed as a result of endurance training, other forms of exercise also produce changes in muscle mitochondrial content. For example, repeated interval training bouts at high intensity force the recruitment of all muscle fibers within the muscle, and while the duration of the exercise is short, convincing changes in mitochondrial content have been observed.18, 19, 20 In contrast, mitochondrial adaptations to resistance training have shown inconsistent results. Original studies suggested that, while the stimulus invoked by such training produced large changes in myofibrillar protein levels and muscle fiber hypertrophy, little to no change in mitochondrial content was observed, resulting in a “dilution” of the mitochondrial content within the fiber.21,22 This adaptation is physiologically disadvantageous, because mitochondrial content dilution increases the diffusion distance between the capillary and the mitochondrial location, with potential impairments in endurance performance. More recent work has suggested that despite a lack of change in organelle content, some improvements in mitochondrial function may take place.23 Further, changes in mitochondria gene expression may be most evident in muscle possessing a lower mitochondrial content, such as aged muscle. Indeed this adaptation appears to be a general principle: the lower the mitochondrial content to begin with, the greater the adaptive potential observed.24 The greater adaptation observed under these conditions is most likely the result of a larger metabolic response to the exercise stress. The higher levels of metabolites, such as AMP, lactic acid or others, provoke a greater nuclear gene expression response by activating transcription factors, leading to a stronger mitochondrial adaptation.
These longstanding results regarding the effects of exercise on mitochondrial adaptations were largely based on observations in young, healthy skeletal muscle. These findings led to the realization that, if mitochondrial content and function were impaired under certain conditions (eg. disease, aging), exercise might serve as a form of “behavioural medicine” that could ameliorate the pathophysiology of that condition. However, the molecular basis for such adaptations remains to be fully resolved. In the remaining sections of this review, we highlight several important protein regulators and processes that serve to maintain mitochondrial content and function in muscle, the absence of which leads to muscle pathology. In addition, we describe the evidence that exercise can serve to compensate for these deficiencies and lead to healthier muscle, thereby illustrating the function of exercise as “medicine” to attenuate muscle pathophysiology.
Important signaling proteins for biogenesis
PGC-1α
Likely the most important regulators of the transcription of nuclear genes encoding mitochondrial proteins (NuGEMPs) is the peroxisome proliferator-activated receptor gamma coactivator-1 (PGC-1) family of transcriptional coactivators. Among the three members, PGC-1α is widely regarded as the master regulator of mitochondrial biogenesis. As a co-activator, PGC-1α does not directly interact with the genome, but rather binds other transcription factors to exert its function.7,25,26 Through its LXXLL motif, PGC-1α is capable of binding a host of transcription factors including PPARs, NRFs, and ERRs. In response to a bout of exercise, a series of signaling events lead to the nuclear translocation and activation of PGC-1α to initiate mitochondrial adaptations through mitochondrial biogenesis27 (Fig. 1). Signals such as influxes of cytosolic calcium, elevations in reactive oxygen species (ROS) and increases in the AMP to ATP ratio all occur following an acute bout of exercise, and serve to promote PGC-1α activation through their respective kinases.28 The convergence of these various signals on this transcriptional co-activator illustrates the importance of PGC-1α activation in the maintenance and expansion of the mitochondrial reticulum.
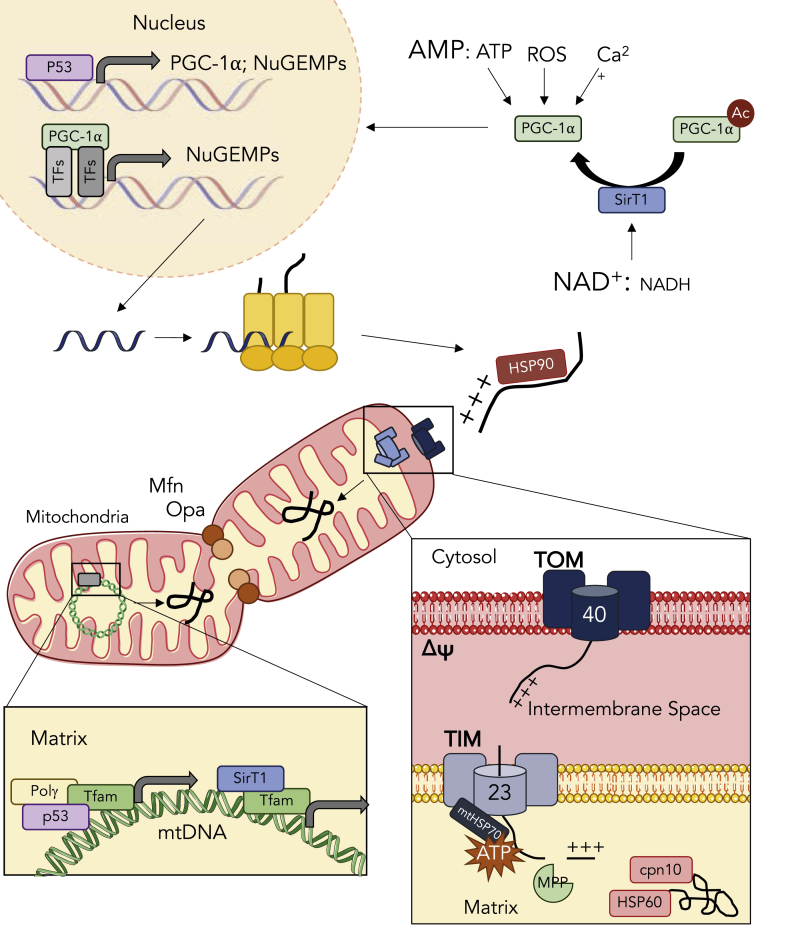
Fig. 1.
Exercise-induced activation of mitochondrial biogenesis. An acute bout of exercise elicits signaling events that promote the expansion of the mitochondrial reticulum. These signals, including increases in cytosolic calcium, elevations in reactive oxygen species, and declines in the energy status converge on the activation of PGC-1α. Concomitantly, increases in the NAD+: NADH ratio activate SirT1, a deacetylase that activates PGC-1α thus allowing its nuclear translocation. p53 also transcriptionally regulates PGC-1α and various nuclear encoded mitochondrial proteins (NuGEMPs). PGC-1α coactivates a number of transcription factors to promote the expression of NuGEMPs, which are subsequently translated in the cytosol. To facilitate mitochondrial localization, NuGEMPs contain a mitochondrial targeting sequence (MTS) that is recognized by cytosolic chaperones such as HSP90, that unfold preproteins and bring them to the translocase of the outer membrane (TOM) complex. Receptors on the TOM complex recognize the MTS and initiate the translocation of the preprotein through Tom40. Preproteins destined for the mitochondrial matrix are shuttled to the translocase of the inner membrane (TIM) complex, and actively pulled through the Tim23 channel into the matrix via mtHSP70. Once in the matrix, mitochondrial processing peptidase (MPP) cleaves the MTS, thus allowing chaperones such as cpn10 and HSP60 to refold the protein into its mature conformation. The expansion of the mitochondrial reticulum also relies on the mitochondrial genome. Nuclear-encoded mitochondrial transcription factor A (TFAM) is a major regulator of the expression of mtDNA. TFAM has also been found to form complexes with Sirt1, p53 and Polγ to stabilize and maintain the integrity of the mitochondrial genome. Expansion of the mitochondrial reticulum can also be achieved through events of fusion in which mitofusin 1/2 (Mfn) and optical atrophy 1/2 (Opa) tether and ligate the outer and inner membranes, respectively, to fuse the joining organelle into the network. As discussed in the text, defects in several of these processes can be overcome with exercise.
Within skeletal muscle, overexpression of PGC-1α is sufficient to induce a more oxidative metabolic profile.29 Mitochondrial content increases, and there is a shift to a greater proportion of type I fibers.30 These adaptations contribute to improving exercise performance and fatigue resistance. In stark contrast, PGC-1α null animals have lower mitochondrial content, reduced state 3 and state 4 respiration,31 elevated ROS production and are typically described as exercise intolerant. Similarly, following periods of inactivity or muscle atrophy, such as aging, declines in PGC-1α are observed.32 Thus, a strong positive correlation exists between PGC-1α and mitochondrial content, demonstrating the importance of this co-activator in the maintenance of these organelles. Indeed, a single bout of exercise initiates the transcription of PGC-1α, and promotes the subsequent accumulation of mitochondrial proteins.33,34
Despite the value of PGC-1α in determining muscle phenotype, a number of studies have now shown that exercise training-induced improvements in mitochondrial content and function can proceed in the absence of PGC-1α.31,35,36 For example, the mitochondrial respiratory defects observed in PGC-1α null animals are completely rescued by a period of endurance training,31 suggesting the presence of other compensatory transcriptional proteins that are activated to compensate for PGC-1α. However, this compensation appears to diminish with age, as the exercise training benefits are diminished in older animal models.37 Thus, exercise serves a medicinal function to improve muscle health, even in the absence of PGC-1α.
p53
p53 is widely regarded as a tumor suppressor protein as it is heavily involved in apoptosis as well as in DNA damage and repair. The absence of p53 leads to cancer, and mutations in this protein are found in approximately 50% of human cancers.38,39 However, p53 also plays a notable role in the regulation of mitochondria, as the loss of p53 results in a decline in mitochondrial content and functional deficiencies.40, 41, 42, 43 p53 can regulate mtDNA and is imported into the mitochondrial matrix through CHCHD4, a component of the protein import machinery (PIM).44 p53 controls mtDNA in two ways, first by directly interacting with the D-Loop region to upregulate the transcription of mtDNA.45,46 p53 can also interact with mitochondrial transcription factor A, Tfam, and DNA polymerase γ to promote its binding to mtDNA46, 47, 48 (Fig. 1). This binding is important for the transcriptional regulation of the mitochondrial genome, but also to maintain mtDNA integrity.
Further to its roles in regulating and maintaining mtDNA, p53 can also regulate mitochondria by modulating the expression of NuGEMPs, as well as that of PGC-1α, as the promoter of PGC-1α contains multiple putative p53 binding sites49,50 (Fig. 1). p53 null mice exhibit lower levels of PGC-1α, which contributes to the decline in mitochondrial content. p53 can also directly transcriptionally regulate the ETC assembly factor SCO2, resulting in improper complex assembly of the electron transport chain (ETC).51 Impaired COX holoenzyme formation may contribute to the functional declines seen in the absence of p53. These changes in mitochondrial content are reflected in reduced muscle endurance performance in p53 null mice,40 as well a higher blood lactate level during a progressive exercise test to exhaustion.43
Despite the reduced mitochondrial content and function in the absence of p53, endurance training was able to increase mitochondrial content and ameliorate blood lactate levels during an exercise test. In addition, PGC-1α and Tfam were restored to normal or higher levels despite the absence of p53, while the elevated ROS levels evident in muscle specific KO (mKO) muscle was markedly reduced with exercise training.43 In addition, training also attenuated the elevated basal cytochrome c release evident in isolated subsarcolemmal mitochondria, and reduced pro-apoptotic Bax expression in p53 mKO animals.43 Thus, exercise training effectively reversed the mitochondrial defects brought about by the absence of p53. This suggests that muscle in patients with p53 loss-of-function mutations could potentially adapt to exercise as effectively as healthy individuals, to restore muscle function and metabolism.
Sirt1
Sirt1 is a deacetylase that participates in the regulation of mitochondria through both mitochondrial biogenesis and mitophagy. Sirt1 is responsive to the NAD+/NADH ratio, as NAD + serves as a coenzyme for Sirt1, thus making Sirt1 sensitive to both the energy and redox status of the cell. Deacetylation of PGC-1α by Sirt1 promotes PGC-1α activity, while also indirectly increasing its transcription52 (Fig. 1). Similar to p53, Sirt1 appears to translocate into the mitochondrion where it interacts with Tfam to regulate the transcription of mtDNA.53,54 Therefore, Sirt1 participates in the coordination of both the nuclear and mitochondrial genome in the expansion of the mitochondrial reticulum. Alternatively, Sirt1 also regulates autophagy through various mechanisms, ultimately leading to an increase in mitochondrial degradation. Through deacetylation, Sirt1 activates TSC2 preventing the inhibition of autophagy by mTOR55 (Fig. 2). Sirt1 also activates FOXO3, a transcription factor that regulates a number of genes including BNIP and LC3, which are both involved in mitophagy.56,57 Thus, Sirt1 is involved in the optimization of the mitochondrial pool by stimulating the synthesis of new organelles and the clearance of damaged ones.
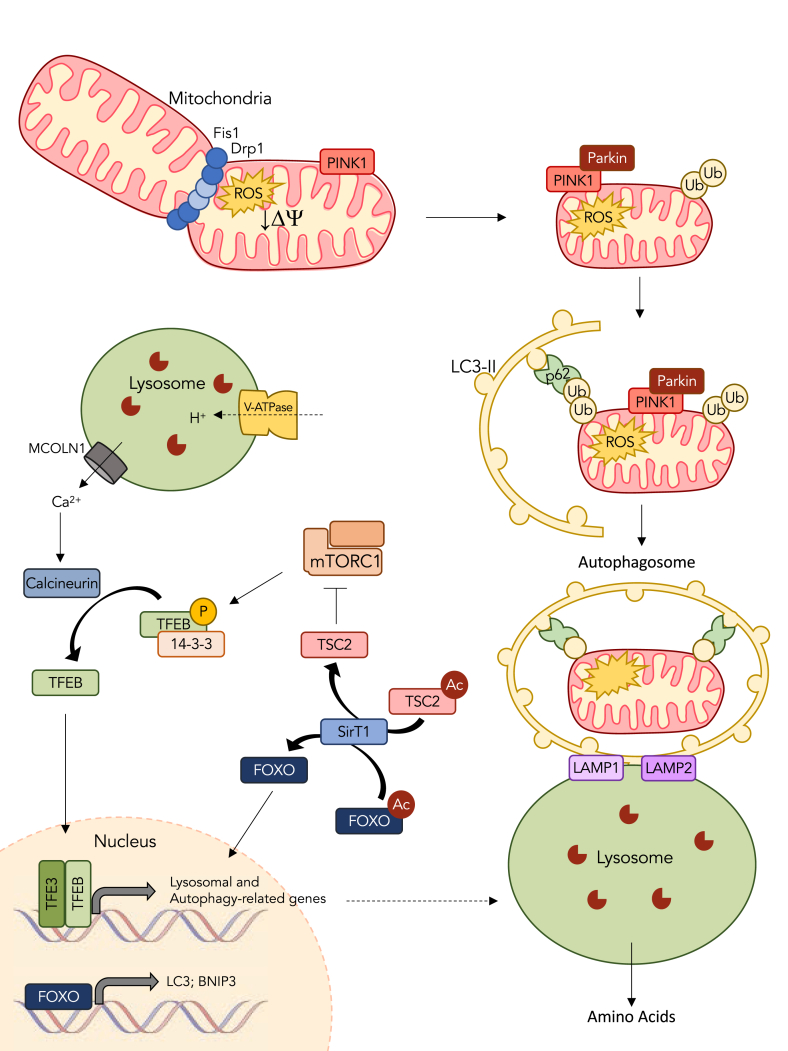
Fig. 2.
Exercise promotes the turnover of mitochondria through mitophagy. Damaged organelles exhibit classic signs of dysfunction including the loss of membrane potential and elevations in reactive oxygen species emission. These organelles are removed from the mitochondrial reticulum through events of fission, carried out by Drp1 and Fis1, which constrict the outer and inner membranes to excise portions of the network. Mitochondrial depolarization inhibits the basal import of PINK1, causing it to accumulate on the outer membrane. Pink1 then recruits Parkin, an E3 ligase capable of ubiquitinating various proteins on the outer membrane. These ubiquitin chains serve as a signal to target the engulfment of the organelle in an autophagosomal membrane to be degraded by the lysosome. p62 links the ubiquitin chains on the dysfunctional cargo to LC3-II present in the phagophore. Once fully encapsulated, the autophagosome travels along microtubules to fuse with the lysosome with the aid of lysosome associated membrane proteins (LAMP1 and LAMP2). The lysosome then degrades the contents of the autophagosome via resident proteolytic enzymes and releases amino acids to support future protein synthesis. In resting muscle, mTOR inhibits TFEB and TFE3 translocation into the nucleus through phosphorylation that promotes the binding of chaperone 14-3-3, thereby preventing nuclear translocation. In response to exercise, mTOR is inhibited, in part by the deacetylation and activation of TSC2 by SirT1. At the same time, calcium is released from the lysosome through mucolipin-1 (MCOLN1), which activates calcineurin to dephosphorylate TFEB and TFE3 and allow their nuclear translocation. Once in the nucleus, TFEB and TFE3 transcriptionally regulate numerous lysosomal and autophagy-related genes. In conjunction to its role in inhibiting mTOR, SirT1 also activates FOXO, thereby allowing it to localize to the nucleus and transcribe components of the autophagy/mitophagy pathway including LC3 and BNIP3. Exercise also stimulates lysosomal biogenesis to increase the capacity of the muscle for organelle turnover.
Since Sirt1 regulates both arms of mitochondrial turnover, it is no surprise that genetic and pharmacological manipulation of Sirt1 has dramatic effects on these organelles. Sirt1 null animals exhibit lower mitochondrial content in skeletal muscle, which contributes to poor exercise endurance performance.58 In addition, the absence of Sirt1 led to a marked increase in mitochondrial ROS emission and reductions in both state 3 and state 4 respiration.58 Alternatively, pharmacological activation of Sirt1 through Resveratrol partially restored mitochondrial content and function. However, when combined with endurance training, the effects on mitochondrial content were synergistic improvements, compared to either treatment alone. These data suggest that exercise training, particularly in combination with Resveratrol form a potent combination to restore mitochondrial health in skeletal muscle.
Cardiolipin metabolism
Cardiolipin is a unique phospholipid located predominantly within the mitochondrial inner membrane. Cardiolipin is known as the “glue” of the electron transport chain because of its important interactions with resident proteins, and its requirement for ETC activity.59 The absence of cardiolipin as a result of a mutation in Taffazin, a key enzyme in the cardiolipin synthesis pathway, causes Barth Syndrome, a disease characterized by cardiomyopathy and muscle weakness. Cardiolipin synthesis is reduced by muscle disuse60,61 and may contribute to the mitochondrial dysfunction observed.62 However, exercise can reverse this decline. Experiments using a chronic contractile activity model of exercise have shown the rapid augmentation of cardiolipin in muscle,63 along with an increase in gene expression of important enzymes that contribute to cardiolipin synthesis.61 Thus, exercise can serve to maintain the appropriate phospholipid environment essential for electron transport chain function in muscle.
Protein import
Mitochondria are comprised of ∼1200 proteins that are encoded by both the nuclear and mitochondrial genome (mtDNA), although, mtDNA encodes for merely 13 proteins.1 Thus, the vast majority of the mitochondrial proteome is encoded by nuclear DNA, translated on cytosolic ribosomes as precursor proteins and imported into the mitochondrion. Nascent polypeptides destined for the mitochondrion typically contain a mitochondrial targeting sequence (MTS) that dictates mitochondrial-localization.64 Several MTS variations that specify their sub-localization within the mitochondrion exist. Regardless of their final destination, all precursor proteins destined for the mitochondrion are recognized and transported by cytosolic chaperones to the translocase of the outer membrane (TOM) complex (Fig. 1).
Once bound to a receptor of the TOM complex, unfolded preproteins transit through the OM via the Tom40 channel. Precursor proteins destined for the mitochondrial matrix are brought to the TIM complex, where polypeptide chains translocate through the IM via Tim23, the major channel of the TIM complex. Translocation through the IM requires ΔΨ and ATP, as transiting proteins are actively pulled into the matrix by ATP-driven action of mitochondrial Hsp70 (mtHsp70). Once in the matrix, the MTS is cleaved by mitochondrial processing peptidase (MPP), and refolded by resident chaperones such as Hsp60 and chaperonin10 (cpn10) into their mature conformation28,65 (Fig. 1).
Mitochondrial protein import is a highly dynamic and energetically-responsive mechanism, as the rate of import can be modified to match the needs of the cell. For example, chronic exercise has been shown to increase the expression of several components of the protein import machinery (PIM), and this increases coincides with increased rates of protein translocation into the mitochondrion.66 Various key components of the import machinery such as Tom20, Tom22, Tim23, Hsp90 and MSF have been shown to increase following chronic contractile activity, a model of endurance training. A similar drive in the expression of mitochondrial chaperones Hsp60, Hsp70 and cpn10 is also observed following training. Thus, the upregulation of import components is vital to support the augmented drive in mitochondrial biogenesis that occurs with chronic exercise.
In contrast, in experimental models of muscle disuse such as denervation, protein import is reduced and is strongly correlated with the decline in mitochondrial content under these conditions.62 However, exercise has the potential to reverse this import defect. For example, in animals lacking the pro-apoptotic proteins BAX and BAK, a surprising impairment in protein import was observed. Interestingly, a program of voluntary wheel running upregulated the expression of various PIM components and normalized their import rate.67 Thus, the beneficial effects of exercise on the protein import process is critical for improving skeletal muscle mitochondrial content and function.
Fission-fusion
Mitochondria exist in complex reticular structures that are highly dynamic and constantly undergo morphological changes. The morphology of these organelles is important for their function, as a highly interconnected network is able to share mtDNA, metabolites and substrates, and distribute energy throughout the muscle.68 In contrast, small fragmented mitochondria typically produce more ROS and exhibits signs of dysfunction. The dynamic nature of the mitochondrial reticulum is mediated by events of fusion and fission, through which mitochondria are constantly being added or removed from the network.69 Fusion supports the expansion of the network, where mitofusin 1/2 (mfn1 and mfn2) and optical atrophy 1/2 (Opa1 and Opa2) tether and ligate the outer and inner membranes, respectively, to facilitate the addition of the mitochondrion to the reticulum (Fig. 1). In contrast, segments of the reticulum can be removed through events of fission, in which dynamin-related protein 1 (Drp1), mitochondrial fission factor (Mff) and fission protein 1 (Fis1) actively constrict around the outer mitochondrial membrane to facilitate separation and excision from the network.70,71 Fission is critical for the clearance of dysfunctional segments of the mitochondrial network through mitophagy.4 The selective removal promotes the optimization of the organelle pool and ensure that damaging byproducts such as ROS produced by adjacent dysfunctional segments do not compromise the network as a whole.
Both aging and chronic periods of muscle disuse result in an increase in fission, relative to fusion, regulatory proteins.72 This imbalance results in mitochondrial populations that are smaller and more fragmented, and tend to produce more ROS. These smaller mitochondrial fragments serve as substrates for mitophagy (see below) which is elevated basally under these conditions. Chronic exercise can reverse the imbalance in these fission-fusion proteins, and favour the formation of a more physiologically efficient organelle reticulum.72
Mitophagy
Mitochondrial content at any given time is a byproduct of two opposing process, the expansion of the network through mitochondrial biogenesis, and the degradation of dysfunctional mitochondria through mitophagy. Mitophagy is a selective form of autophagy, in which dysfunctional portions of the mitochondrial reticulum are excised through events of fission and tagged for recycling73 (Fig. 2). Dysfunctional organelles are then engulfed in an autophagosomal membrane and brought to the lysosome, where the two will fuse to degrade the contents of the autophagosome. Lysosomes contain a highly acidic lumen and a plethora of catalytic enzymes capable of degrading cellular material into its basic amino acids, which are then released into the cytosol to support future protein synthesis.
The most commonly studied pathway in the context of skeletal muscle involves the proteins PINK1 and Parkin. PINK1 is a cytosolic kinase that is regularly imported into the mitochondrion and degraded by PARL.74 While this process seems energy-inefficient, it allows the constant monitoring of the mitochondria as the import process requires a viable membrane potential and ATP. In the event of mitochondrial dysfunction, the import of PINK1 is arrested, causing it to accumulate on the outer membrane. The presence of PINK1 on the outer mitochondrial membrane recruits Parkin, an E3 ubiquitin ligase, that ubiquitinates various outer mitochondrial membrane proteins75 (Fig. 2). These ubiquitin chains serve as a flag to recruit the phagophore to engulf the organelle. The phagophore is a membranous structure that is scavenged from other membranes in the cell and contains LC3-II, the lipidated form of LC3-I. p62 serves as an adaptor protein to dock the ubiquitin chains on the mitochondria to the LC3-II embedded within the phagophore membrane. Once the organelle is fully engulfed, creating an autophagosome, it will travel along microtubule tracts to the lysosome. Here the two vesicles will fuse with the aid of lysosomal membrane bound protein LAMP1 and LAMP2, to expose the contents of the autophagosome to the catabolic environment of the lysosome.76
Exercise serves a major role as mitochondrial medicine because even a single bout of exercise activates mitophagy above the basal levels observed at rest.77, 78, 79 Alongside this enhanced removal, signals to increase the capacity for cellular recycling are also initiated following an acute bout of exercise. TFEB and its family member TFE3, key regulators of lysosomal and autophagy related genes, translocate to the nucleus in response to exercise, thereby promoting lysosomal biogenesis.80,81 Basally, TFEB and TFE3 are sequestered in the cytosol via phosphorylation by mTOR82,83 (Fig. 2). Phosphorylation of TFEB and TFE3 promote the binding of chaperone 14-3-3 which effectively masks the nuclear localization sequence of these transcription factors.84 However, in response to an acute bout of exercise, mTOR is inhibited. Concomitantly, calcium is released from the lysosome through mucolipin-1 (MCOLN1), thereby generating an influx of cytosolic calcium capable of activating Calcineurin, a calcium-dependent phosphatase.85 Dephosphorylation of TFEB and TFE3 by Calcineurin, promotes the release of 14-3-3 and allows the nuclear translocation of these transcription factors (Fig. 2). This series of reactions facilitates the subsequent removal of dysfunctional mitochondria and matches the timing of signaling toward mitochondrial biogenesis (see PGC-1 discussion, above). The fact that exercise initiates this organelle “turnover” with one bout is an important mechanism to maintain a healthy pool of mitochondria within muscle.86
Repeated bouts of exercise in the form of training lead to an enhancement of mitochondrial content, as well as subtle improvements in mitochondrial quality.16,87 If the mitochondria within muscle undergo adaptive qualitative improvements, a reduced signaling toward biogenesis as well as mitophagy is provoked. This reduction is evident when muscle with a high mitochondrial content is subjected to exercise; a lower kinase activation is observed compared to the same exercise stimulus imposed on a muscle with a lower mitochondrial content.88 Thus, repeated bouts of exercise lead to a reduced activation of turnover pathways because of a higher quality of the organelle pool. Interestingly, this exercise-induced mitophagy signaling is absent in Parkin-null animals, thereby illustrating a fundamentally important function for this protein in muscle.79 Chronic exercise also increases the level of lysosomal enzymes within muscle, and leads to lysosomal biogenesis.89 This altered biogenesis may have a beneficial role to help increase the capacity of muscle for autophagic degradation when lysosome dysfunction is apparent, as in aging muscle (see below), or in lysosomal storage diseases. More research in this exciting area is warranted.
Effect of age on mitochondria
With age a gradual and progressive loss of muscle mass and function known as sarcopenia can occur. Sarcopenia is often accompanied and preceded by changes in mitochondrial content and function.90, 91, 92, 93 Declines in PGC-1α mRNA and protein have been observed with aging likely contributing to the age-associated losses in mitochondrial content.33 Age-related reductions in the transcriptional drive for PGC-1α are likely due to increased DNA methylation and altered expression of PGC-1α regulators such as NRF2, YY1 and ATF2 resulting in declines in PGC-1α promoter activity. In addition, there is an increase in PGC-1α mRNA instability that likely contributes to the decline in PGC-1α expression seen with age, and is an important contributor to the decrease mitochondrial biogenesis with age.33
Muscle mitochondria exhibit signs of dysfunction with age, including elevations in ROS and losses in membrane potential, both of which are triggers for mitophagy.94 Enhanced targeting for mitophagy has been observed in aged skeletal muscle, as shown by elevations of p62 and LC3-II on the mitochondrial fraction and elevations in Parkin expression.79,95 Recent studies have confirmed that aged skeletal muscle exhibits enhanced mitochondrial engulfment by autophagosomes, thus suggesting enhanced mitophagy with age. Despite the drive for enhanced mitophagy, mitochondrial dysfunction remains a common characteristic of the aging phenotype suggesting that although the clearance of damaged mitochondria is elevated with age, this is insufficient to maintain mitochondrial function. Furthermore, with age lysosomes begin to accumulate lipofuscin, also known as the “aging pigment”, an index of lysosomal dysfunction.95,96 Thus, despite having enhanced targeting and encapsulation, a decrement in the final clearance of these autophagosomes at the level of the lysosome may exist, again contributing to the age-related decline in function.
In response to exercise, aging muscle responds with a lower amplitude of signaling activation toward gene expression.97 The consequence of this change is a lower mitochondrial adaptive response to long-term exercise.24 Despite this, chronic exercise can rescue, in part, the mitochondrial declines observed with age. Exercise restores mitochondrial content back toward levels observed in younger individuals,94,95 and at the same time reduces mitochondrially-produced ROS and apoptotic signaling.24 Exercise also reverses the decline in PGC-1α transcription observed with age, while increasing the expression of the coactivator at the mRNA and protein level.33,98 Exercise training helps to restore mitochondrial health in aging muscle.
Conclusion: exercise is mitochondrial medicine for muscle
Well established for many years, regularly performed exercise has beneficial effects on muscle endurance through the stimulation of mitochondrial biogenesis. The applied consequence of this adaptation on athletic performance is well recognized. Less established is the role that exercise can play to improve muscle health and metabolism under disease conditions, or with age when mitochondrial function is compromised. Further, the molecular basis of this potential utility of exercise as “medicine” to treat metabolic and genetic disorders to restore mitochondrial function is poorly understood. In this review, we have provided fundamental evidence illustrating that 1) exercise can initiate organelle turnover with each bout of contractile activity to refresh the organelle pool, 2) exercise can trigger this response, in whole or in part, even with the genetic loss-of-function of important metabolic regulators, 3) exercise ameliorates mitochondrial dysfunction through multiple compensatory pathways that are not all fully characterized, and 4) exercise serves as a therapeutic modality, under behavioural control, that can improve muscle and mitochondrial health, and ultimately quality of life.
Conflict of interest
The authors are not aware of any affiliations, memberships, funding or financial holdings that might be perceived as affecting the objectivity of this review.
Submission statement
This manuscript has not been published and is not under consideration for publication elsewhere.
Each authors’ contributions
D.A.H. and A.N.O. wrote the manuscript. D.A.H. edited the final document.
Acknowledgements
The work was funded by Natural Sciences and Engineering Research Council (NSERC) and Canadian Institutes of Health Research (CIHR) grants to D.A.H. A.N.O. is the recipient of an NSERC CGS-D. D.A.H. is the recipient of a Canada Research Chair in Cell Physiology.
References
- 1.Calvo S.E., Clauser K.R., Mootha V.K. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44(D1):D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 3.Petrosillo G., Ruggiero F.M., Paradies G. Role of reactive oxygen species and cardiolipin in the release of cytochrome c from mitochondria. FASEB J. 2003;17(15):2202–2208. doi: 10.1096/fj.03-0012com. [DOI] [PubMed] [Google Scholar]
- 4.Frank M., Duvezin-Caubet S., Koob S., et al. Mitophagy is triggered by mild oxidative stress in a mitochondrial fission dependent manner. Biochim Biophys Acta Mol Cell Res. 2012;1823(12):2297–2310. doi: 10.1016/j.bbamcr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Heusch P., Canton M., Aker S., et al. The contribution of reactive oxygen species and p38 mitogen-activated protein kinase to myofilament oxidation and progression of heart failure in rabbits. Br J Pharmacol. 2010;160(6):1408–1416. doi: 10.1111/j.1476-5381.2010.00793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barbieri E., Sestili P. Reactive oxygen species in skeletal muscle signaling. J Signal Transduct. 2012;2012:982794. doi: 10.1155/2012/982794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lowell B.B., Spiegelman B.M. Towards a molecular understanding of adaptive thermogenesis. Nature. 2000;404(6778):652–660. doi: 10.1038/35007527. [DOI] [PubMed] [Google Scholar]
- 8.Kirkwood S.P., Munn E.A., Brooks G.A. Mitochondrial reticulum in limb skeletal muscle. Am J Physiol. 1986;251(3 Pt 1):C395–C402. doi: 10.1152/ajpcell.1986.251.3.C395. [DOI] [PubMed] [Google Scholar]
- 9.Hoppeler H. Exercise-induced ultrastructural changes in skeletal muscle. Int J Sports Med. 1986;07(04):187–204. doi: 10.1055/s-2008-1025758. [DOI] [PubMed] [Google Scholar]
- 10.Hood D.A. Contractile activity-induced mitochondrial biogenesis in skeletal muscle. J Appl Physiol. 2001;90(3):1137–1157. doi: 10.1152/jappl.2001.90.3.1137. [DOI] [PubMed] [Google Scholar]
- 11.Cogswell A.M., Stevens R.J., Hood D.A. Properties of skeletal muscle mitochondria from subsarcolemmal and intermyofibrillar isolated regions. Am J Physiol. 1993;264(Pt 1):C383–C389. doi: 10.1152/ajpcell.1993.264.2.C383. [DOI] [PubMed] [Google Scholar]
- 12.Picard M., White K., Turnbull D.M. Mitochondrial morphology, topology, and membrane interactions in skeletal muscle: a quantitative three-dimensional electron microscopy study. J Appl Physiol. 2013 doi: 10.1152/japplphysiol.01096.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferreira R., Vitorino R., Alves R.M.P., et al. Subsarcolemmal and intermyofibrillar mitochondria proteome differences disclose functional specializations in skeletal muscle. Proteomics. 2010;10(17):3142–3154. doi: 10.1002/pmic.201000173. [DOI] [PubMed] [Google Scholar]
- 14.Vincent A.E., White K., Davey T., et al. Quantitative 3D mapping of the human skeletal muscle mitochondrial network. Cell Rep. 2019;26(4):996–1009. doi: 10.1038/nature22814.Trans-kingdom. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Molnar M., Kovacs G. Mitochondrial diseases. Handb Clin Neurol. 2017;145:147–155. doi: 10.1016/B978-0-12-802395-2.00010-9. [DOI] [PubMed] [Google Scholar]
- 16.Holloszy J.O. Biochemical adaptations in muscle. J Biol Chem. 1967;242(9):2278–2282. [PubMed] [Google Scholar]
- 17.Gollnick P.D., Armstrong R.B., Saltin B., Saubert C.W., Sembrowich W.L., Shepherd R.E. Effect of training on enzyme activity and fiber composition of human skeletal muscle. J Appl Physiol. 1973;34(1):107–111. doi: 10.1152/jappl.1973.34.1.107. [DOI] [PubMed] [Google Scholar]
- 18.Burgomaster K.A., Howarth K.R., Phillips S.M., et al. Similar metabolic adaptations during exercise after low volume sprint interval and traditional endurance training in humans. J Physiol. 2008;5861:151–160. doi: 10.1113/jphysiol.2007.142109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartlett J.D., Hwa Joo C., Jeong T.-S., et al. Matched work high-intensity interval and continuous running induce similar increases in PGC-1 mRNA, AMPK, p38, and p53 phosphorylation in human skeletal muscle. J Appl Physiol. 2012;112(7):1135–1143. doi: 10.1152/japplphysiol.01040.2011. [DOI] [PubMed] [Google Scholar]
- 20.MacInnis M.J., Gibala M.J. Physiological adaptations to interval training and the role of exercise intensity. J Physiol. 2017;595(9):2915–2930. doi: 10.1113/JP273196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Groennebaek T., Vissing K. Impact of resistance training on skeletal muscle mitochondrial biogenesis, content, and function. Front Physiol. 2017;15(8):713. doi: 10.3389/fphys.2017.00713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.MacDougall J., Sale D., Moroz J., Elder G., Sutton J., Howald H. Mitochondrial volume density in human skeletal muscle following heavy resistance training. Med Sci Sport. 1979;11(2):164–166. [PubMed] [Google Scholar]
- 23.Porter C., Reidy P.T., Bhattarai N., Sidossis L.S., Rasmussen B.B. Resistance exercise training alters mitochondrial function in human skeletal muscle. Hum Skelet Muscle Med Sci Sport Exerc. 2015;47(9):1922–1931. doi: 10.1249/MSS.0000000000000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ljubicic V., Joseph A.-M., Adhihetty P.J., et al. Molecular basis for an attenuated mitochondrial adaptive plasticity in aged skeletal muscle. Aging (Albany NY) 2009;1(9):818–830. doi: 10.18632/aging.100083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puigserver P., Adelmant G., Wu Z., et al. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286(5443):1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 26.Lin J., Handschin C., Spiegelman B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metabol. 2005;1(6):361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 27.Hock M.B., Kralli A. Transcriptional control of mitochondrial biogenesis and function. Annu Rev Physiol. 2009;71(1):177–203. doi: 10.1146/annurev.physiol.010908.163119. [DOI] [PubMed] [Google Scholar]
- 28.Hood D.A., Memme J.M., Oliveira A.N., Triolo M. Maintenance of skeletal muscle mitochondria in health, exercise, and aging. Annu Rev Physiol. 2019;81(1) doi: 10.1146/annurev-physiol-020518-114310. annurev-physiol-020518-114310. [DOI] [PubMed] [Google Scholar]
- 29.Calvo J.A., Daniels T.G., Wang X., et al. Muscle-specific expression of PPARgamma coactivator-1alpha improves exercise performance and increases peak oxygen uptake. J Appl Physiol. 2008;104(5):1304–1312. doi: 10.1152/japplphysiol.01231.2007. [DOI] [PubMed] [Google Scholar]
- 30.Lin J., Wu H., Tarr P.T., et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418(6899):797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 31.Adhihetty P.J., Uguccioni G., Leick L., Hidalgo J., Pilegaard H., Hood D.A. The role of PGC-1 on mitochondrial function and apoptotic susceptibility in muscle. AJP Cell Physiol. 2009;297(1):C217–C225. doi: 10.1152/ajpcell.00070.2009. [DOI] [PubMed] [Google Scholar]
- 32.Chabi B., Ljubicic V., Menzies K.J., Huang J.H., Saleem A., Hood D a. Mitochondrial function and apoptotic susceptibility in aging skeletal muscle. Aging Cell. 2008;7(1):2–12. doi: 10.1111/j.1474-9726.2007.00347.x. [DOI] [PubMed] [Google Scholar]
- 33.Carter H.N., Pauly M., Tryon L.D., Hood D.A. Effect of contractile activity on PGC-1α transcription in young and aged skeletal muscle. J Appl Physiol. 2018;124(6):1605–1615. doi: 10.1152/japplphysiol.01110.2017. [DOI] [PubMed] [Google Scholar]
- 34.Pilegaard H., Saltin B., Neufer P.D. Exercise induces transient transcriptional activation of the PGC-1$α$ gene in human skeletal muscle. J Physiol. 2003;546(3):851–858. doi: 10.1113/jphysiol.2002.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rowe G.C., El-Khoury R., Patten I.S., Rustin P., Arany Z. PGC-1 a is dispensable for exercise-induced mitochondrial biogenesis in skeletal muscle. PLoS One. 2012;7(7) doi: 10.1371/journal.pone.0041817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leick L., Wojtaszewski J.F.P., Johansen S.J., et al. PGC-1α is not mandatory for exercise-and training-induced adaptive gene responses in mouse skeletal muscle. Am J Physiol Endocrinol Metab. 2008;294(2):463–474. doi: 10.1152/ajpendo.00666.2007. [DOI] [PubMed] [Google Scholar]
- 37.Leick L., Lyngby S.S., Wojtasewski J.F., Pilegaard H. PGC-1α is required for training-induced prevention of age-associated decline in mitochondrial enzymes in mouse skeletal muscle. Exp Gerontol. 2010;45(5):336–342. doi: 10.1016/J.EXGER.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 38.Lane D.P. Cancer. p53, guardian of the genome. Nature. 1992;358(6381):15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 39.Beyfuss K., Hood D.A. A systematic review of p53 regulation of oxidative stress in skeletal muscle. Redox Rep. 2018;23(1):100–117. doi: 10.1080/13510002.2017.1416773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saleem A., Adhihetty P.J., Hood D.A. Role of p53 in mitochondrial biogenesis and apoptosis in skeletal muscle. Physiol Genom. 2009;3:58–66. doi: 10.1152/physiolgenomics.90346.2008. [DOI] [PubMed] [Google Scholar]
- 41.Ljubicic V., Joseph A.-M., Saleem A., et al. Transcriptional and post-transcriptional regulation of mitochondrial biogenesis in skeletal muscle: effects of exercise and aging. Biochim Biophys Acta Gen Subj. 2010;1800(3):223–234. doi: 10.1016/j.bbagen.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 42.Matoba S., Kang J.-G., Patino W.D., et al. p53 regulates mitochondrial metabolism. Science. 2006;312(5780):1650–1653. doi: 10.1126/science.1126863. 80- [DOI] [PubMed] [Google Scholar]
- 43.Beyfuss K., Erlich A.T., Triolo M., Hood D.A. The role of p53 in determining mitochondrial adaptations to endurance training in skeletal muscle. Sci Rep. 2018;8(1):14710. doi: 10.1038/s41598-018-32887-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhuang J., Kamp W.M., Li J., et al. Forkhead Box O3A (FOXO3) and the mitochondrial disulfide relay carrier (CHCHD4) regulate p53 protein nuclear activity in response to exercise. J Biol Chem. 2016;291(48):24819–24827. doi: 10.1074/jbc.M116.745737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heyne K., Mannebach S., Wuertz E., Knaup K.X., Mahyar-Roemer M., Roemer K. Identification of a putative p53 binding sequence within the human mitochondrial genome. FEBS Lett. 2004;578:198–202. doi: 10.1016/j.febslet.2004.10.099. [DOI] [PubMed] [Google Scholar]
- 46.Park J.-H., Zhuang J., Li J., Hwang P.M. p53 as guardian of the mitochondrial genome. FEBS Lett. 2016;590(7):924–934. doi: 10.1016/j.physbeh.2017.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Achanta G., Sasaki R., Feng L., et al. Novel role of p53 in maintaining mitochondrial genetic stability through interaction with DNA Pol gamma. EMBO J. 2005;24(19):3482–3492. doi: 10.1038/sj.emboj.7600819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Safdar A., Khrapko K., Flynn J.M., et al. Exercise-induced mitochondrial p53 repairs mtDNA mutations in mutator mice. Skelet Muscle. 2016;6(7) doi: 10.1186/s13395-016-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Park J.-Y., Wang P.-Y., Matsumoto T., et al. p53 improves aerobic exercise capacity and augments skeletal muscle mitochondrial DNA content. Circ Res. 2009;105(7):705–712. doi: 10.1161/CIRCRESAHA.109.205310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saleem A., Hood D.A. Acute exercise induces tumour suppressor protein p53 translocation to the mitochondria and promotes a p53-Tfam-mitochondrial DNA complex in skeletal muscle. J Physiol. 2013;591(14):3625–3636. doi: 10.1113/jphysiol.2013.252791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saleem A., Iqbal S., Zhang Y., Hood D.A. Effect of p53 on mitochondrial morphology, import and assembly in skeletal muscle. Am J Physiol Cell Physiol. 2015;308:C319–C329. doi: 10.1152/ajpcell.00253.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mouchiroud L., Houtkooper R.H., Moullan N., et al. The NAD(+)/Sirtuin pathway modulates longevity through activation of mitochondrial UPR and FOXO signaling. Cell. 2013;154(2):430–441. doi: 10.1016/j.cell.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aquilano K., Vigilanza P., Baldelli S., Pagliei B., Rotilio G., Ciriolo M.R. Peroxisome proliferator-activated receptor γ co-activator 1 α (PGC-1α) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J Biol Chem. 2010;285(28):21590–21599. doi: 10.1074/jbc.M109.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang B.L. Sirt1 and the mitochondria. Mol Cells. 2016;39(2):87–95. doi: 10.14348/molcells.2016.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghosh H.S., McBurney M., Robbins P.D. SIRT1 negatively regulates the mammalian target of rapamycin. PLoS One. 2010;5(2):1–8. doi: 10.1371/journal.pone.0009199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ng F., Tang B.L. Sirtuins' modulation of autophagy. J Cell Physiol. 2013;228(12):2262–2270. doi: 10.1002/jcp.24399. [DOI] [PubMed] [Google Scholar]
- 57.Hariharan Nirmala, Maejima Yasuhiro, Nakae Jun, Paik Jihye, DePinho Ronald A., S J. Deacetylation of FoxO by Sirt1 Plays an Essential Role in Mediating Starvation-Induced Autophagy in Cardiac Myocytes. 2013;185:974–981. doi: 10.1038/mp.2011.182. 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Menzies K.J., Singh K., Saleem A., Hood D.A. Sirtuin 1-mediated effects of exercise and resveratrol on mitochondrial biogenesis. J Biol Chem. 2013;288(10):6968–6979. doi: 10.1074/jbc.M112.431155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dudek J. Role of cardiolipin in mitochondrial signaling pathways. Front Cell Dev Biol. 2017;5(90):1–17. doi: 10.3389/fcell.2017.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wicks K.L., Hood D.A. Mitochondrial adaptations in denervated muscle: relationship to muscle performance. Am J Physiol Cell Physiol. 1991;260(29):C841–C850. doi: 10.1152/ajpcell.1991.260.4.C841. [DOI] [PubMed] [Google Scholar]
- 61.Ostojic O., O'Leary M.F.N., Singh K., Menzies K.J., Vainshtein A., Hood D.A. The effects of chronic muscle use and disuse on cardiolipin metabolism. J Appl Physiol. 2013;114(4):444–452. doi: 10.1152/japplphysiol.01312.2012. [DOI] [PubMed] [Google Scholar]
- 62.Singh K., Hood D.A. Effect of denervation-induced muscle disuse on mitochondrial protein import. Am J Cell Physiol. 2011;300:C138–C145. doi: 10.1152/ajpcell.00181.2010. [DOI] [PubMed] [Google Scholar]
- 63.Takahashi M., Hood D.A. Chronic stimulation-induced changes in mitochondria and performance in rat skeletal muscle. J Appl Physiol. 1993;74(2):934–941. doi: 10.1152/jappl.1993.74.2.934. [DOI] [PubMed] [Google Scholar]
- 64.Backes S., Herrmann J.M. Protein translocation into the intermembrane space and matrix of mitochondria: mechanisms and driving forces. Front Mol Biosci. 2017;4:83. doi: 10.3389/fmolb.2017.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wiedemann N., Pfanner N. Mitochondrial machineries for protein import and assembly. Annu Rev Biochem. 2017;86(1):685–714. doi: 10.1146/annurev-biochem-060815-014352. [DOI] [PubMed] [Google Scholar]
- 66.Takahashi M., Chesley A., Freyssenet D., Hood D.A. Contractile activity-induced adaptations in the mitochondrial protein import system. Am J Physiol. 1998;274(5 Pt 1):C1380–C1387. doi: 10.1152/ajpcell.1998.274.5.C1380. [DOI] [PubMed] [Google Scholar]
- 67.Zhang Y., Iqbal S., O'Leary M.F.N., et al. Altered mitochondrial morphology and defective protein import reveal novel roles for Bax and/or Bak in skeletal muscle. AJP Cell Physiol. 2013;305(5):C502–C511. doi: 10.1152/ajpcell.00058.2013. [DOI] [PubMed] [Google Scholar]
- 68.Glancy B., Hartnell L.M., Combs C.A., et al. Power grid protection of the muscle mitochondrial reticulum. Cell Rep. 2017 doi: 10.1016/j.celrep.2017.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mishra P., Chan D.C. Metabolic regulation of mitochondrial dynamics. J Cell Biol. 2016;212(4):379–387. doi: 10.1083/jcb.201511036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chan D.C. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46(1):265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 71.Losón O.C., Song Z., Chen H., Chan D.C. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell. 2013;24(5):659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iqbal S., Ostojic O., Singh K., Joseph A.-M., Hood D.A. Expression of mitochondrial fission and fusion regulatory proteins in skeletal muscle during chronic use and disuse. Muscle Nerve. 2013;48(6):963–970. doi: 10.1002/mus.23838. [DOI] [PubMed] [Google Scholar]
- 73.Pickles S., Vigié P., Youle R.J. Mitophagy and quality control mechanisms in mitochondrial maintenance. Curr Biol. 2018;28(4):R170–R185. doi: 10.1016/J.CUB.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jin S.M., Lazarou M., Wang C., Kane L.A., Narendra D.P., Youle R.J. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010;191(5):933–942. doi: 10.1083/jcb.201008084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Narendra D.P., Jin S.M., Tanaka A., et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8(1) doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mizushima N., Yoshimori T., Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27(1):107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- 77.Vainshtein A., Tryon L.D., Pauly M., Hood D.A. Role of PGC-1α during acute exercise-induced autophagy and mitophagy in skeletal muscle. Am J Physiol Cell Physiol. 2015;308(9):C710–C719. doi: 10.1152/ajpcell.00380.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Drake J.C., Laker R.C., Wilson R.J., Zhang M., Yan Z. Exercise-induced mitophagy in skeletal muscle occurs in the absence of stabilization of Pink1 on mitochondria. Cell Cycle. 2019;18(1):1–6. doi: 10.1080/15384101.2018.1559556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen C.C.W., Erlich A.T., Crilly M.J., Hood D.A. Parkin is required for exercise-induced mitophagy in muscle: impact of aging. Am J Physiol Metab. 2018;315(3):E404–E415. doi: 10.1152/ajpendo.00391.2017. [DOI] [PubMed] [Google Scholar]
- 80.Mansueto G., Armani A., Viscomi C., et al. Transcription factor EB controls metabolic flexibility during exercise. Cell Metabol. 2017;25(1):182–196. doi: 10.1016/j.cmet.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Pastore N., Vainshtein A., Klisch T.J., et al. TFE3 regulates whole-body energy metabolism in cooperation with TFEB. EMBO Mol Med. 2017;9(5):605–621. doi: 10.15252/emmm.201607204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Roczniak-Ferguson A., Petit C.S., Froehlich F., et al. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5(228):ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Settembre C., Zoncu R., Medina D.L., et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 2012;31(5):1095–1108. doi: 10.1038/emboj.2012.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Napolitano G., Ballabio A. TFEB at a glance. J Cell Sci. 2016;129:2475–2481. doi: 10.1242/jcs.146365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Medina D.L., Di Paola S., Peluso I., et al. Lysosomal calcium signalling regulates autophagy through calcineurin and TFEB. Nat Cell Biol. 2015;17(3):288–299. doi: 10.1038/ncb3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guan Y., Drake J.C., Yan Z. Exercise-induced mitophagy in skeletal muscle and heart. Exerc Sport Sci Rev. 2019;47(3):151–156. doi: 10.1249/JES.0000000000000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim Y., Triolo M., Erlich A.T., Hood D.A. Regulation of autophagic and mitophagic flux during chronic contractile activity-induced muscle adaptations. Pflüg Arch - Eur J Physiol. October 2018:1–10. doi: 10.1007/s00424-018-2225-x. [DOI] [PubMed] [Google Scholar]
- 88.Ljubicic V., Hood D.A. Specific attenuation of protein kinase phosphorylation in muscle with a high mitochondrial content. Am J Physiol Metab. 2009;297(3):E749–E758. doi: 10.1152/ajpendo.00130.2009. [DOI] [PubMed] [Google Scholar]
- 89.Kim Y., Hood D.A. Regulation of the autophagy system during chronic contractile activity-induced muscle adaptations. Phys Rep. 2017;5(14) doi: 10.14814/phy2.13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Alway S.E., Mohamed J.S., Myers M.J. Mitochondria initiate and regulate sarcopenia. Exerc Sport Sci Rev. 2017;45(2):58–69. doi: 10.1249/JES.0000000000000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Calvani R., Joseph A.-M., Adhihetty P.J., et al. Mitochondrial pathways in sarcopenia of aging and disuse muscle atrophy. Biol Chem. 2013;394(3):393–414. doi: 10.1515/hsz-2012-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hepple R.T. Mitochondrial involvement and impact in aging skeletal muscle. Front Aging Neurosci. 2014;6:211. doi: 10.3389/fnagi.2014.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Romanello V., Sandri M. Mitochondrial quality control and muscle mass maintenance. Front Physiol. 2016;6(422):1–21. doi: 10.3389/fphys.2015.00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Carter H.N., Chen C.C.W., Hood D.A. Mitochondria, muscle health, and exercise with advancing age. Physiology. 2015;30(3):208–223. doi: 10.1152/physiol.00039.2014. [DOI] [PubMed] [Google Scholar]
- 95.Carter H.N., Kim Y., Erlich A.T., Zarrin-khat D., Hood D.A. Autophagy and mitophagy flux in young and aged skeletal muscle following chronic contractile activity. J Physiol. July 2018 doi: 10.1113/JP275998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.O'Leary M.F., Vainshtein A., Iqbal S., Ostojic O., Hood D a. Adaptive plasticity of autophagic proteins to denervation in aging skeletal muscle. Am J Physiol Cell Physiol. 2013;304(5):C422–C430. doi: 10.1152/ajpcell.00240.2012. [DOI] [PubMed] [Google Scholar]
- 97.Ljubicic V., Hood D.A. Diminished contraction-induced intracellular signaling towards mitochondrial biogenesis in aged skeletal muscle. Aging Cell. 2009;8(4):394–404. doi: 10.1111/j.1474-9726.2009.00483.x. [DOI] [PubMed] [Google Scholar]
- 98.Koltai E., Hart N., Taylor A.W., et al. Age-associated declines in mitochondrial biogenesis and protein quality control factors are minimized by exercise training. Am J Physiol Integr Comp Physiol. 2012;303(2):R127–R134. doi: 10.1152/ajpregu.00337.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]