Summary
Although some long noncoding (lnc)RNAs are known since the 1950s, the past 25 years have uncovered myriad lncRNAs with diverse sequences, structures, and functions. The advent of high-throughput and sensitive technologies has further uncovered the vast heterogeneity of lncRNA-interacting molecules and patterns of expressed lncRNAs. We propose a unifying functional theme for the expansive family of lncRNAs. At an elementary level, the genomic program of gene expression is elicited via canonical transcription and post-transcriptional mRNA assembly, turnover, and translation. Building upon this regulation, an epigenomic program refines the basic genomic control by modifying chromatin architecture as well as DNA and RNA chemistry. Superimposed over the genomic and epigenomic programs, lncRNAs create an additional regulatory dimension: by interacting with the proteins and nucleic acids that regulate gene expression in the nucleus and cytoplasm, lncRNAs help establish robust, nimble, and specific transcriptional and post-transcriptional control. We describe our present understanding of lncRNA-coordinated control of protein programs and cell fate, and discuss challenges and opportunities as we embark on the next 25 years of lncRNA discovery.
eTOC blurb
Herman et al. review progress in lncRNA research over the past 25 years. Vast and heterogeneous, lncRNAs interact broadly with gene regulatory machineries. By superimposing a layer of control upon genomic and epigenomic processes, lncRNAs modulate many levels of gene regulation, from transcription to protein modification.
Since the central dogma of molecular biology was proposed by Crick in 1958, we have marveled at the ever-expanding molecular complexity built upon this core process to enable the specific gene expression programs that sustain life. After the discovery of noncoding (nc)RNAs, transfer (t)RNAs, and ribosomal (r)RNAs in the 1950s, messenger (m)RNAs, identified in the 1960s, offered templates for protein synthesis (Brenner et al., 1961; Gros et al., 1961). The ncRNA family grew with the discovery of small nuclear (sn)RNAs and small nucleolar (sno)RNAs in the 1980s (Matera et al., 2007), but it was the discovery of small regulatory RNAs from the 1990s [micro (mi)RNAs, piwi-interacting (pi)RNAs, small interfering (si)RNAs] (Wilson and Doudna, 2013), that set the stage for a rapid escalation in the discovery of regulatory long noncoding (lnc)RNAs over the ensuing 25 years. The advent of genomic tiling arrays and especially high-throughput RNA-sequencing technologies in the mid 2000s fueled this explosion, uncovering a vast, versatile, and rich universe of lncRNAs (>200 nucleotides in length), spanning a large range of sizes, sequences, structures, and functions. These technologies soon revealed that a striking ~75% of the human genome is transcribed, and that only 2% of the genes transcribed encode mRNAs with protein-synthesis potential; the vast majority of transcription produces lncRNAs, as reported by the ENCODE Project Consortium (Dunham et al., 2012).
LncRNAs in this huge family are often classified based on their site of transcription relative to protein-coding genes, and thus there are enhancer lncRNAs, promoter lncRNAs, antisense lncRNAs (transcribed in antisense orientation from protein-coding genes), intergenic lncRNAs, and circular lncRNAs (circRNAs) arising from introns and/or exons that are excised and religated (Box 1). In general, lncRNA primary sequences are less conserved across species than mRNAs sequences, their secondary structures are often linked to their functions, and their expression is highly cell type-specific. The past 25 years have uncovered an astonishingly diverse array of mechanisms whereby lncRNAs influence upon gene expression programs and cell function. Despite their heterogeneity, lncRNA-regulated processes share three key features.
Box 1: The many monikers of lncRNAs.
Throughout the years, lncRNAs have been named depending on features such as the genomic regions from which they originate or their structure, conservation, or function. Below are listed some of the most common types of lncRNAs.
lincRNAs: long intergenic noncoding RNAs
elncRNAs: enhancer-associated lncRNAs
plncRNAs/PROMPTS: promoter-associated lncRNAs
NATs: natural antisense transcripts, antisense lncRNAs
mtlncRNAs: mitochondrial DNA-encoded lncRNAs
snolncRNAs: intron-derived long noncoding RNAs with snoRNA ends
circRNAs: circular lncRNAs
EcircRNAs: circRNAs consisting of exons
IcircRNAs: circRNAs consisting of introns
EIcircRNAs: circRNAs consisting of exons and introns
ceRNAs: competing endogenous lncRNAs
mtcircRNAs: mitochondrial DNA-encoded circRNAs
First, their complex influence is tied to their wide presence across the entire cellular space (Bridges et al., 2021; Carlevaro-Fita et al., 2019; Fazal et al., 2019). Nuclear lncRNAs associate with specialized domains like paraspeckles, nucleoli, and the lamina, as well as with chromosomes, chromatin domains, and gene regions; accordingly, they modulate nuclear processes like chromatin organization, and RNA transcription and splicing. Cytoplasmic lncRNAs interact with membrane-less cytosolic domains like stress granules, processing bodies, ribosomes, and the cytoskeleton, as well as with membranous cytoplasmic structures like the endoplasmic reticulum and mitochondria; accordingly, they regulate mRNA transport, stability, and translation, as well as protein stability, post-translational modification, and function. Second, lncRNA function is closely linked to their relative abundance. Besides their transcription rates, the relative levels of lncRNAs are influenced by their widely different stability; the presence of 5’ end m7G caps and 3’ end poly(A) tails, structured 3’ ends, small nucleolar RNA-protein complexes (snoRNPs) at the ends, and covalent circularization of the 5’ and 3’ ends, all modulate their relative stability in the nucleus and the cytoplasm (Yin et al., 2012; Wu et al., 2017). Third, lncRNA function is directly associated to the molecules with which the lncRNAs interact. Although some lncRNAs have intrinsic catalytic function in the absence of proteins (e.g., ribozymes and riboswitches), and some lncRNAs can be translated in certain instances, the function of most lncRNAs is closely associated to their interaction with other nucleic acids and with RNA-binding proteins (RBPs).
Here, we review the progress over the past 25 years in learning about the functions of lncRNAs as regulators of gene expression and cell function. Using a few select examples, mostly from the mammalian world, we present an emerging picture in which there is a basic, foundational genomic control of gene expression by proteins regulating transcriptional and post-transcriptional processes that affect mRNA production, turnover, and translation. Superimposed upon this level is an epigenomic program that revises the genomic control by modifying DNA chemistry, RNA chemistry, and chromatin organization. We propose that lncRNAs create an additional dimension of control, superimposed upon genomic and epigenomic layers. In this dimension, lncRNAs form scaffolds to organize DNA regions and modulate transcription, recruit RNAs and cytoplasmic factors to sites of post-transcriptional control, and serve as assembly platforms for multiprotein complexes functionally linked: in effect, they enable a supragenomic layer of protein expression programs and cell fate (Figure 1).
Figure 1. Conceptual summary of supragenomic regulation by lncRNAs.
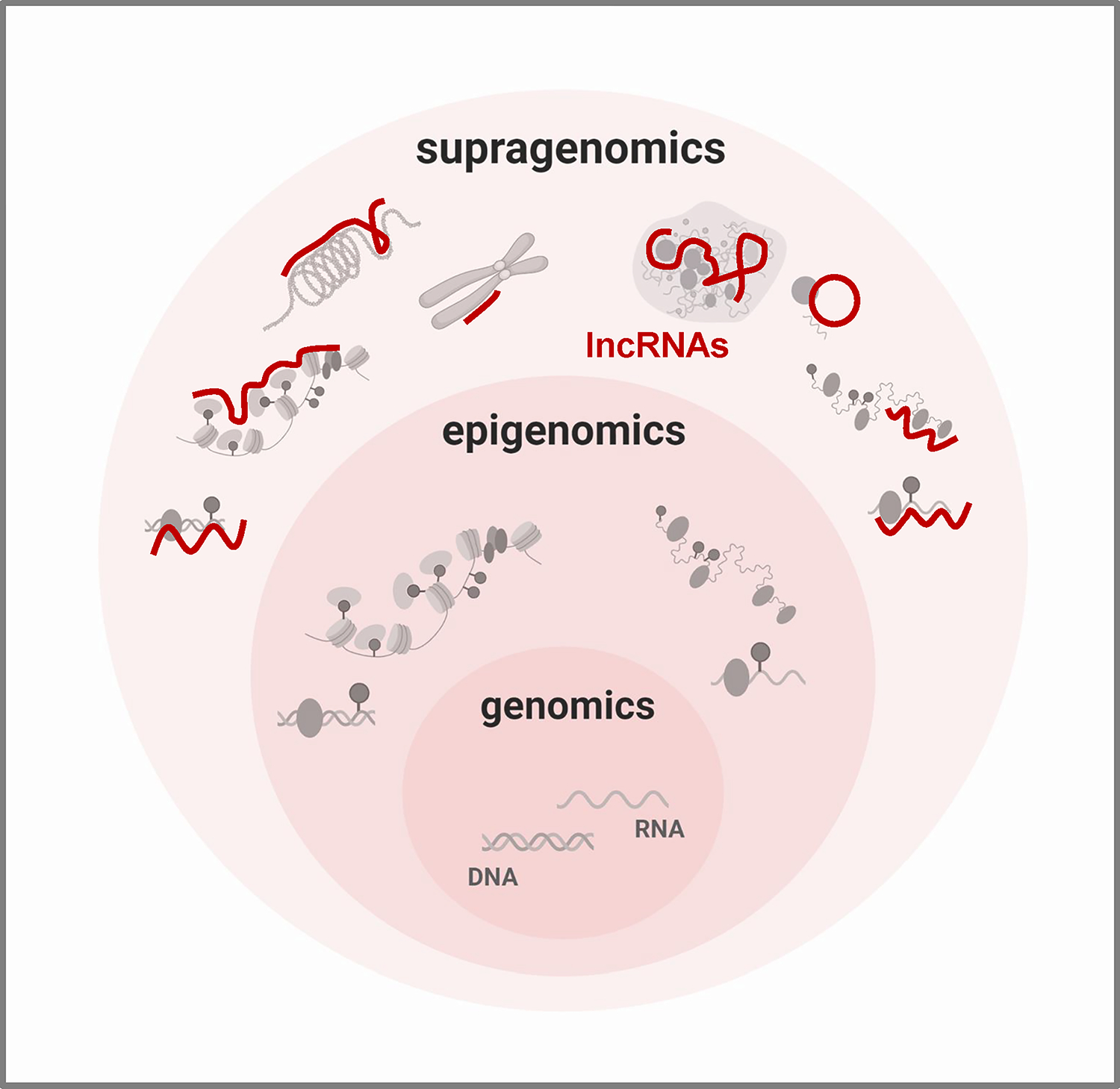
Vast and heterogeneous, lncRNAs interact broadly with gene regulatory machineries. By providing a supragenomic layer of control built upon genomic and epigenomic processes, lncRNAs modulate many levels of gene regulation, from transcription to protein modification. Figure was prepared using BioRender.
This proposed supragenomic layer of control globally involves lncRNAs, but it does not occur in isolation. Instead, as we discuss here, it is carried out through the association of lncRNAs with individual proteins and protein complexes, with DNA and chromatin in different states, with RNAs coding and noncoding, and with machineries that control transcription, splicing, translation, phase-separation states, and more. While other ncRNAs like miRNAs, siRNAs, piRNAs, and snoRNAs, are functionally associated to lncRNAs, here we focus primarily on lncRNAs.
Supragenomic control of nuclear functions by lncRNAs
The past 25 years have firmly established that nuclear lncRNAs can influence many processes related to DNA replication, chromatin organization, and gene transcription (Figure 2). As the first functional roles for lncRNAs were in chromatin metabolism, it was generalized early on that lncRNAs had predominantly nuclear functions, although many lncRNA functions were identified in the cytoplasm soon afterwards. Here, we discuss key examples of the supragenomic control of gene expression by nuclear lncRNAs (Figure 3).
Figure 2. Timeline of lncRNA discoveries over the past 25 years.
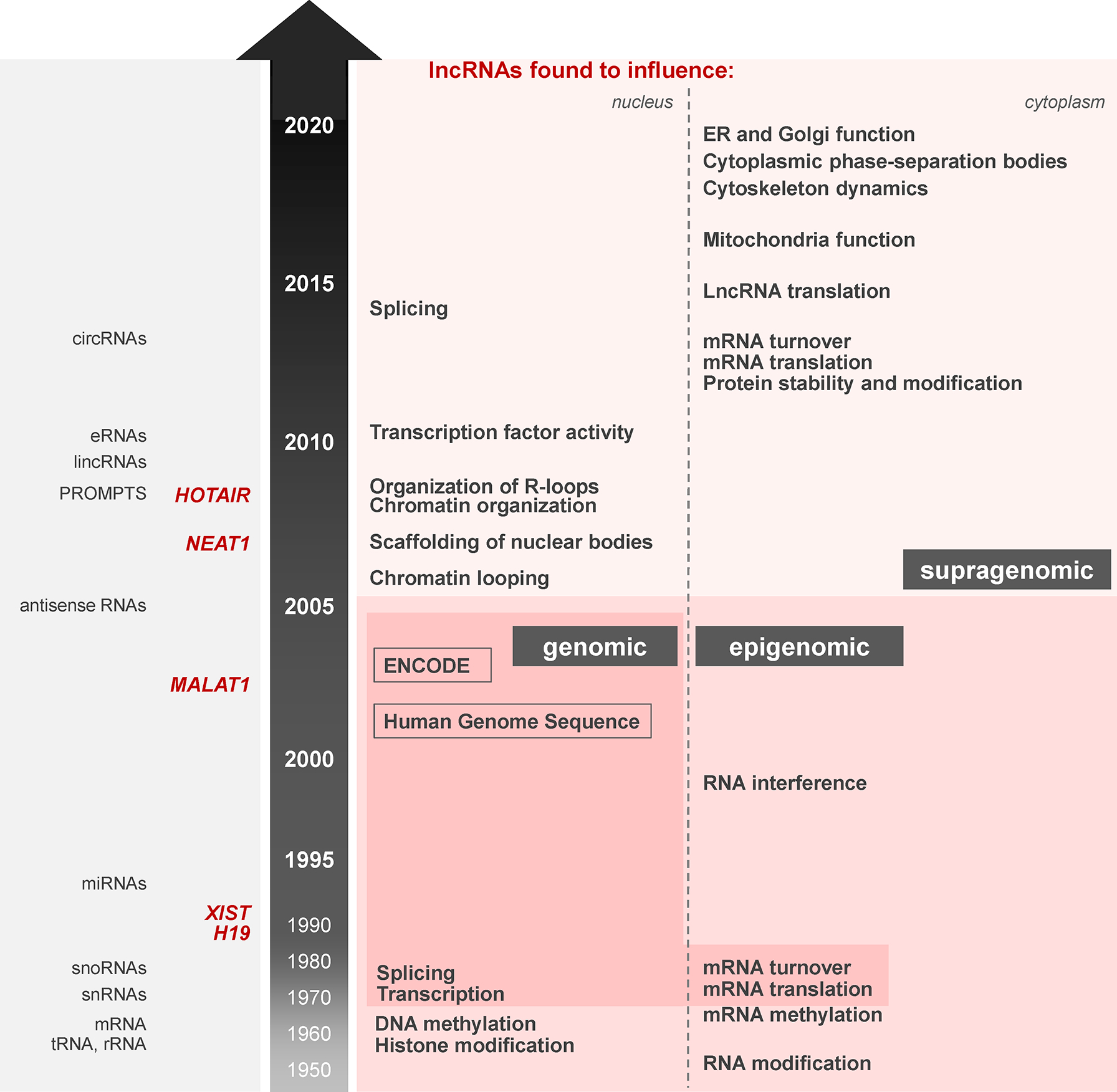
Brief timeline of gene expression discoveries with a focus on the most recent 25 years. Right. Breakthroughs in genomic regulation [transcription, mRNA turnover, translation (darker shade)] and epigenomic changes [modifications of DNA, RNA, and histones (medium shade)] are highlighted at the bottom of the schematic. Superimposed upon genomic and epigenomic processes, a supragenomic array of regulatory schemes mediated by lncRNAs (light shade) in the nucleus and cytoplasm has emerged over the past 25 years, as discussed in this article. Left, key lncRNAs (red font) and RNA families (black font) are listed.
Figure 3. Emerging supragenomic functions of nuclear lncRNAs.
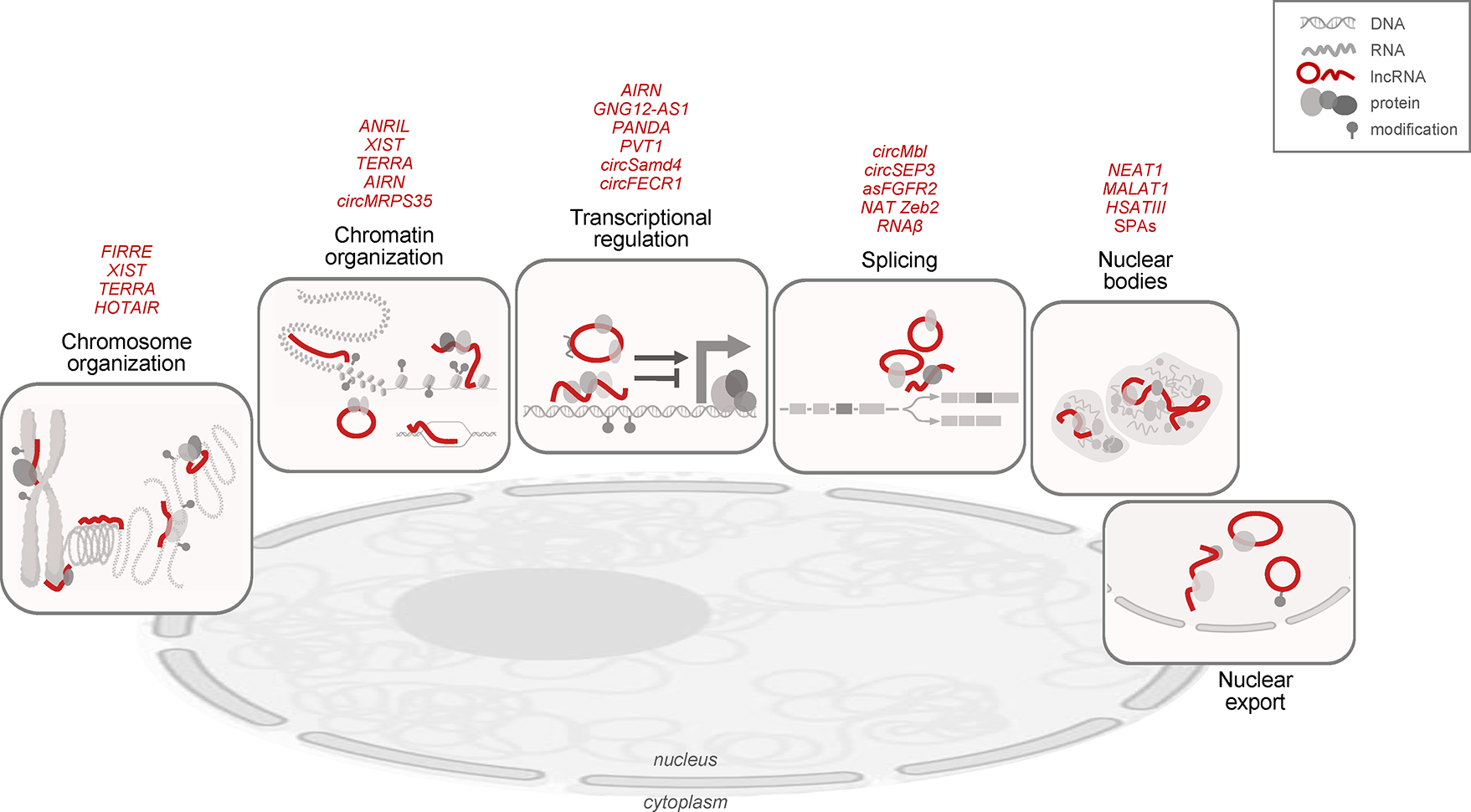
Representative functions of linear and circular lncRNAs in the nucleus (red lines), typically carried out by interacting with proteins and DNA, and modulating their activity. From left to right, lncRNAs can control chromosome organization at a full scale (e.g., X-chromosome inactivation) and at telomeres, in condensed and open chromatin, in chromatin loops, and in contacts across chromosomes. LncRNAs can also influence chromatin organization at a smaller scale by altering local DNA methylation and histone acetylation and/or methylation at nucleosomes, as well as by forming R-loops. LncRNAs directly influence transcription through proteins that bind enhancers or promoters and either promote or suppress the transcription machinery. LncRNAs can influence pre-mRNA splicing by affecting the presence of splicing factors to modulate intron inclusion or exclusion, and can assemble proteins and RNAs in nuclear bodies such as paraspeckles. At the far right, canonical and specialized proteins contribute to exporting linear and circular lncRNAs through nucleopores. Some lncRNAs representing these functions are named above the vignettes; molecules are not drawn at scale. Figure was prepared using BioRender.
LncRNAs implicated in chromatin dynamics
The packaging of DNA and organization into three-dimensional (3D) structures are critical for enabling the carefully orchestrated interactions within and among chromosomes that ensure tight gene expression patterns and genetic transmission during cell division. DNA wraps around histones to form nucleosomes, which then cluster to form loops organized into topologically associated domains (TADs); these domains in turn aggregate into compartments that occupy chromosome territories across the nuclear space. The chromatin must have a stable organization, but it must also be capable of changing to meet the needs of the cell (Wachsmuth et al., 2008). This organization began to be investigated over a century ago and was known to comprise DNA, proteins, and RNA (reviewed by Nickerson et al., 1986; Olins and Olins, 2003). The past 25 years have uncovered many diverse and unexpected ways in which lncRNAs contribute to chromatin regulation.
Chromatin organization.
Many examples have emerged of lncRNAs providing important tiers of control for chromatin assembly. Numerous lncRNAs help to organize chromatin into active and inactive domains by interacting with major chromatin-modifying proteins like polycomb repressive complex 2 (PRC2), Yin Yang 1 (YY1), and CCCTC-binding factor (CTCF) (Belak and Ovsenek, 2007; Beltran et al., 2016; Cifuentes-Rojas et al., 2014; Hendrickson et al., 2016; Sun et al., 2013; Zhao et al., 2008). One of the first lncRNAs reported, XIST, provides scaffolding for chromatin-modifying enzymes like SMCHD1 to drive X-chromosome inactivation (Engreitz et al., 2013; Wang et al., 2018), while telomeric repeat-containing RNAs (TERRA) recruit chromatin-modifying proteins TRF2 and PRC2 to support heterochromatin formation at telomeres (Deng et al., 2009; Montero et al., 2018), and lncRNA ANRIL regulates neighboring transcription of CDKN2A and CDKN2B mRNAs by recruiting PRC1 and PRC2 to specific gene promoters in senescent cells (Yap et al., 2010). In other examples, transcription of Igf2r non-protein coding RNA (Airn) helps to spread polycomb complexes across chromatin, and HOTAIR may facilitate chromosome condensation and gene silencing at least in part by interacting with epigenetic regulators PRC2 and LSD1 (Gupta et al., 2010; Latos et al., 2012; Rinn et al., 2007; Tsai et al., 2010). A few circRNAs were also found to modulate transcription in related ways; circMRPS35 recruited an acetyltransferase to gene promoters, whereas circFECR1, circAFG1, and circLRP6 recruited methylating enzymes to inactivate gene promoters (Chen et al., 2018; Jie et al., 2020; Wang and Li, 2020; Zheng et al., 2019).
Chromatin looping.
Following the discovery that transcriptional activity assists with chromatin topology and nuclear compartmentalization, transcription of lncRNAs was likewise found to influence chromatin architecture and looping (Hubner et al., 2013; Mao et al., 2011). A ‘cat’s cradling’ model was postulated in which transcribing lncRNAs successively opened chromatin forming ‘grip holds’ to guide looping interactions (Mele and Rinn, 2016). Enhancer lncRNAs and enhancer-associated lncRNAs (eRNAs, elncRNAs) were also implicated in chromatin topology; for example, transcription of lncRNA ThymoD in T-cells triggered local demethylation at CTCF sites, creating a loop that brought together the enhancer and promoter regions of Bcl11b during T-cell fate determination (Isoda et al., 2017). In keeping with earlier results that LINoCR transcription repositioned nucleosomes and expelled CTCF complexes (Lefevre et al., 2008), genome-wide studies found that RNA polymerase II (pol II) transcription displaced CTCF-anchored chromatin loops and remodeled local architecture (Heinz et al., 2018). Interestingly, CTCF itself interacts with many lncRNAs (Kuang and Wang, 2020) that likely influence its activity. Other lncRNAs involved in transcription-enabled chromatin looping include Airn and Lockd (Paralkar et al., 2016; Sleutels et al., 2002), and a full class of trait-relevant long-intergenic ncRNAs (TR-lincRNAs) (Tan et al., 2017) and topological anchor point RNAs (tapRNAs) (Amaral et al., 2018). In sum, superimposed on previously known paradigms of chromatin looping, lncRNAs are now found to perform additional regulatory tiers that influence transcription.
R-loops.
Although proteins mediate many interactions between lncRNAs and chromatin, lncRNAs also directly contact DNA, forming hybrid structures that modify chromatin accessibility. Molecular RNA-DNA-DNA hybrids (R-loops), driven by sequence complementarity, are postulated to be widespread and necessary for lncRNA functions, particularly by contributing to gene activation or silencing (Blank-Giwojna et al., 2019; Kuo et al., 2019; Martianov et al., 2007; Schmitz et al., 2010). A recent genome-wide analysis concluded that lncRNA:DNA triplex-forming structures are enriched in TADs and may predict TAD formation areas (Soibam and Zhamangaraeva, 2021). Many lncRNAs form R-loops in cis, with local biological impact; for instance, the TCF21 antisense lncRNA TARID forms a loop at the TCF21 promoter that induces TCF21 transcription (Arab et al., 2019). In an interesting example from plants, the lncRNA APOLO forms local R-loops in cis but also distant R-loops in trans, jointly coordinating the expression of a subset of genes responsive to the plant hormone Auxin (Ariel et al., 2020). In addition to controlling the transcription of protein-coding genes, R-loops also play a large role in the transcription of lncRNAs; moreover, R-loops appear to function as intrinsic pol II promoters to drive transcription, and ablating R-loop formation decreased the transcription of antisense lncRNAs (Tan-Wong et al., 2019). CircRNAs may also form R-loop structures to alter transcription elongation, as shown for circSMARCA5, which forms an R-loop that pauses transcription at SMARCA5 exon 15 and reduces SMARCA5 production (Xu et al., 2020).
Transcriptional regulation by lncRNAs
The past two decades have revealed that lncRNAs also influence transcriptional programs by interacting directly with the transcriptional machinery and repressing or activating it. Examples of transcriptional repression include Airn, which caused transcriptional pausing at the Igf2r promoter (Latos et al., 2012), and antisense lncRNA GNG12-AS1, which interfered with the transcription of protein-coding DIRAS3 mRNA in the sense direction (Stojic et al., 2016). Examples of transcriptional activation include production of the heart development factor HAND2, which was transcriptionally enhanced by two nearby lncRNAs, Uph and Hdn (Anderson et al., 2016; Han et al., 2019; Ritter et al., 2019). In fact, a global cis function for lncRNAs promoting transcription has been proposed, as genes encoding chromatin-remodeling and transcription factors are preferentially located near sites of lncRNA transcription, pointing to a cooperative role for lncRNAs to produce transcription factors (Ponjavic et al., 2009). LncRNAs directly binding transcription factors to influence gene transcription include lncRNA PANDA, derived from the CDKN1A promoter, which binds nuclear transcription factor Y subunit α (NF-YA) in senescent cells (Hung et al., 2011), lncRNA PVT1, whose functions include blocking phosphorylation and degradation of the transcription factor MYC (Tseng et al., 2014), and LincRNAp21, induced by p53 and capable of binding HNRNPK in the nucleus to repress transcription (Huarte et al., 2010).
In recent years, nuclear circRNAs modulating gene transcription have also been identified. Numerous circRNAs were found associated with pol II, many of them exon-intron circular RNAs (EIciRNAs) like circEIF3J and circPAIP2, which interact with snRNA U1 and promote the transcription of the respective parent genes (Li et al., 2015). Other circRNAs regulate transcription by interacting with transcription factors, as shown for circHuR and circSamd4, or by inducing promoter DNA demethylation, as shown for FECR1 circRNA (Chen et al., 2018; Pandey et al., 2020; Yang et al., 2019). Interestingly, intronic circRNAs (ciRNAs) appear to regulate transcription elongation, a step not typically controlled by linear lncRNAs (Faust et al., 2012; Zhang et al., 2013).
Splicing control by lncRNAs
The complex process of splicing is traditionally known to involve short, cis-regulatory elements in pre-mRNA and trans-acting splicing factors. Over the past two decades, lncRNAs have been found to superimpose key layers of regulation upon splicing. Because both canonical splicing and backsplicing to generate circRNAs largely use the same splicing machinery, it was postulated early on that circRNAs might alter pre-mRNA splicing and mRNA production. In fact, it was proposed that the canonical splicing machinery and the backsplicing machinery compete for shared factors, such that there is a balance between pre-mRNA splicing and circRNA backsplicing (Liang et al., 2017). An example of this balance is the MBL locus, which encodes the splicing factor muscleblind (MBL). MBL promotes circularization to yield circMbl, and interestingly, circMbl binds and sequesters MBL; thus, low MBL levels favors splicing to generate mature MBL mRNA, while high MBL levels favors backsplicing to generate instead circMbl (Ashwal-Fluss et al., 2014). On the other hand, circRNAs may also promote alternative splicing of the host transcript. As an example, circSEP3, arising from exon 6 of SEP3 DNA, forms an R-loop, in turn slowing down transcription and promoting splicing of mature SEP3 mRNA (Conn et al., 2017). Instances of circRNAs adding tiers of control on splicing will likely grow, given their innate partnership with the splicing machinery.
The role of linear lncRNAs in splicing is less intuitive, but interesting evidence is emerging. The strong correlation between alternative splicing and the transcription of antisense RNAs has led to the hypothesis that the two processes are connected and evolutionarily conserved (Morrissy et al., 2011). In this scenario, natural antisense transcripts (NATs) transcribed from the opposite strand can form RNA-RNA hybrids with sense pre-mRNAs to modulate the production of splice isoforms (Bardou et al., 2011); for example, lncRNA asFGFR2 regulates alternative splicing of FGFR2 mRNA by interacting with the chromatin-modifying proteins PRC2 and KDM2a and thus creating a splicing-specific chromatin signature (Gonzalez et al., 2015). Conversely, transcription of antisense linear lncRNAs can alter pre-mRNA splicing by masking the splice position and inhibiting further processing. An example of such regulation is NAT Zeb2, which prevents splicing to maintain a 5’UTR Zeb2 intron encoding an internal ribosome entry site (IRES) necessary for translation (Beltran et al., 2008). Others function by attenuating pol II transcriptional elongation or by triggering premature termination to affect isoform expression, as is the case for antisense RNAβ (Stork et al., 2007). Further regulation of splicing factors by lncRNAs is linked to paraspeckles and nuclear bodies (below). Through these actions, lncRNAs help to establish and refine patterns of alternative splicing and protein isoform production.
LncRNAs in nuclear bodies
Responsible for igniting early interest in the functions of nuclear lncRNAs in the 2000s, membrane-less nuclear bodies such as nucleoli, nuclear speckles, and paraspeckles exist both constitutively and in response to changing cell states (Mao et al., 2011). We highlight distinct roles of linear lncRNAs as architectural (arc)RNAs and backbones of nuclear condensates, and discuss their unique ability to anchor to specific nuclear regions and coordinate the assembly of many RNA- and DNA-binding proteins (Chujo et al., 2016).
NEAT1 is an essential component of paraspeckles, where it forms a scaffold for protein binding (Chen and Carmichael, 2009; Clemson et al., 2009; Hutchinson et al., 2007; Sasaki et al., 2009; Sunwoo et al., 2009). In particular, the 3’ end of a long NEAT1 isoform has subdomains that recruit core paraspeckle proteins NONO and SFPQ to initiate liquid-liquid phase separation (Yamazaki et al., 2018). NEAT1 can alter splicing by sequestering splicing factors [SR (serine/arginine-rich) proteins] in the condensates to help balance mRNA isoforms (Cooper et al., 2014; Jiang et al., 2009). The pioneering discovery of a lncRNA with various important roles in the subnuclear space set the stage for identifying other types of RNAs that localize to paraspeckles (Fox et al., 2018; Prasanth et al., 2005).
The abundant lncRNA MALAT1 localizes to nuclear speckles (Hutchinson et al., 2007; Wilusz et al., 2012). Unlike NEAT1, MALAT1 is not essential for nuclear speckle formation, but does orchestrate the complex layering of these nuclear bodies: MALAT1 is positioned near the periphery while splicing factors SON and SC35 are located centrally (Fei et al., 2017). Like NEAT1, MALAT1 also regulates pre-mRNA splicing by modulating the availability of splicing factors like SR proteins and transcriptional repressors like PC2 (Engreitz et al., 2014; Tripathi et al., 2012; Yang et al., 2011). Techniques such as RNA in situ conformation sequencing (RIC-seq) have provided deep insight supporting a function for MALAT1 as an “RNA hub” for other nuclear RNAs including like NEAT1 and U1 (Cai et al., 2020).
Additional scaffolding of nuclear bodies has been found to be mediated by lncRNAs such as SPAs (5’ snoRNA-capped and 3’ polyadenylated lncRNAs) (Wu et al., 2016; Yin et al., 2012), whereas nuclear stress bodies were assembled by lncRNAs such as HSATIII, through the retention of SR proteins, transcription factors, and scaffold attachment proteins (Jolly et al., 1999). The interactions among these lncRNAs and RBPs were linked to a range of diseases associated with alternative splicing of key transcripts (Yap et al., 2018). Although lncRNAs organizing higher-order nuclear architecture remain to be identified, functional intergenic repeating RNA element (FIRRE), expressed from the X chromosome, was capable of bringing together different chromosomes (Hacisuleyman et al., 2014).
Mechanisms of lncRNA nuclear export and retention
We have discussed the functions of many nuclear-enriched lncRNAs, but few lncRNAs are exclusively nuclear or cytoplasmic, and the determinants of their subcellular distribution are not fully known. Early studies suggested that nuclear lncRNAs lacked a default nuclear export pathway (Palazzo and Lee, 2018). However, a recent survey identified NXF1 and TREX as required components for lncRNA nuclear export, with NXF1 being particularly important for the export of long RNAs with high A/U content and few exons, two features often found in lncRNAs (Zuckerman et al., 2020; Zuckerman and Ulitsky, 2019). This discovery linked exon organization and sequence composition as features driving nuclear export for some lncRNAs. Although fewer studies have focused on circRNA export, a recent study found that circRNA length influenced nuclear export, with short and long circRNAs utilizing helicases UAP56 and URH49, respectively, to exit the nucleus (Huang et al., 2018). Soon afterwards, another study found that circRNA N6-methyladenosine (m6A) modification promoted its nuclear export (Chen et al., 2019). The compartmentalization and transport of circRNAs remain areas of active study.
Likewise, elucidating the nuclear retention mechanisms for lncRNAs is critical for understanding their function. Over the years, a number of nuclear retention motifs in lncRNA primary sequences have been identified that drive their nuclear localization (Carlevaro-Fita et al., 2019; Lubelsky and Ulitsky, 2018; Shukla et al., 2018; Zhang et al., 2014). Some nuclear retention signals like repeat insertion domains of lncRNAs (RIDLs) evolved from transposable elements (Carlevaro-Fita et al., 2019; Chillon and Pyle, 2016; Nguyen et al., 2020), while others include repeating RNA domains (RRDs) that direct intracellular localization, as is the case with FIRRE, which uses RRDs to interact with hnRNPU to localize to chromatin (Hacisuleyman et al., 2016). These retention elements bind trans-acting factors to prevent translocation and maintain nuclear enrichment (Guo et al., 2020b; Schiene-Fischer, 2015). Besides primary sequences, lncRNA secondary structures and post-transcriptional modifications provide another layer of complexity for nuclear retention. A recent study assessing lncRNA features like splicing, architecture, chromatin modifications, and sequence motifs to predict subcellular distribution found that pol II pausing and chromatin marks significantly influenced lncRNA localization (Zuckerman and Ulitsky, 2019). Inefficient splicing of lncRNAs may also contribute to nuclear retention due to poor export efficiency (Guo et al., 2020b). As mentioned earlier, intron-containing circRNAs may remain nuclear to modulate processes like transcriptional elongation, but subsets of exonic circRNAs are also predominantly nuclear (Wang et al., 2019; Yang et al., 2017a; Yang et al., 2017c). For example, exonic circAmotl1 increased nuclear retention of the proto-oncoprotein MYC and promoted MYC binding to several target promoters (Yang et al., 2017a) and associated with STAT3 to enable its nuclear translocation (Yang et al., 2017c). We are likely just beginning to scratch the surface on the functions of linear and circular lncRNAs in the nucleus.
Supragenomic control of cytoplasmic functions by lncRNAs
Although lncRNA function initially appeared restricted to the nucleus, work over the past 25 years has uncovered many ways in which cytoplasmic lncRNAs superimpose critical regulatory tiers of protein production and function (Figure 2). After export to the cytosol, lncRNAs associate with RBPs and/or nucleic acids, and may be directed to specific cytosolic domains (e.g., processing bodies, stress granules, or polysomes) or organelles (endoplasmic reticulum or mitochondria). As discussed here, cytoplasmic lncRNAs contribute critical layers of refinement, strength, and specificity to canonical cytoplasmic processes such as mRNA turnover and transport, as well as protein translation, stability, and assembly, mitochondrial function, cytoskeletal dynamics, and cell-cell interactions (Figure 4).
Figure 4. Emerging supragenomic functions of cytoplasmic lncRNAs.
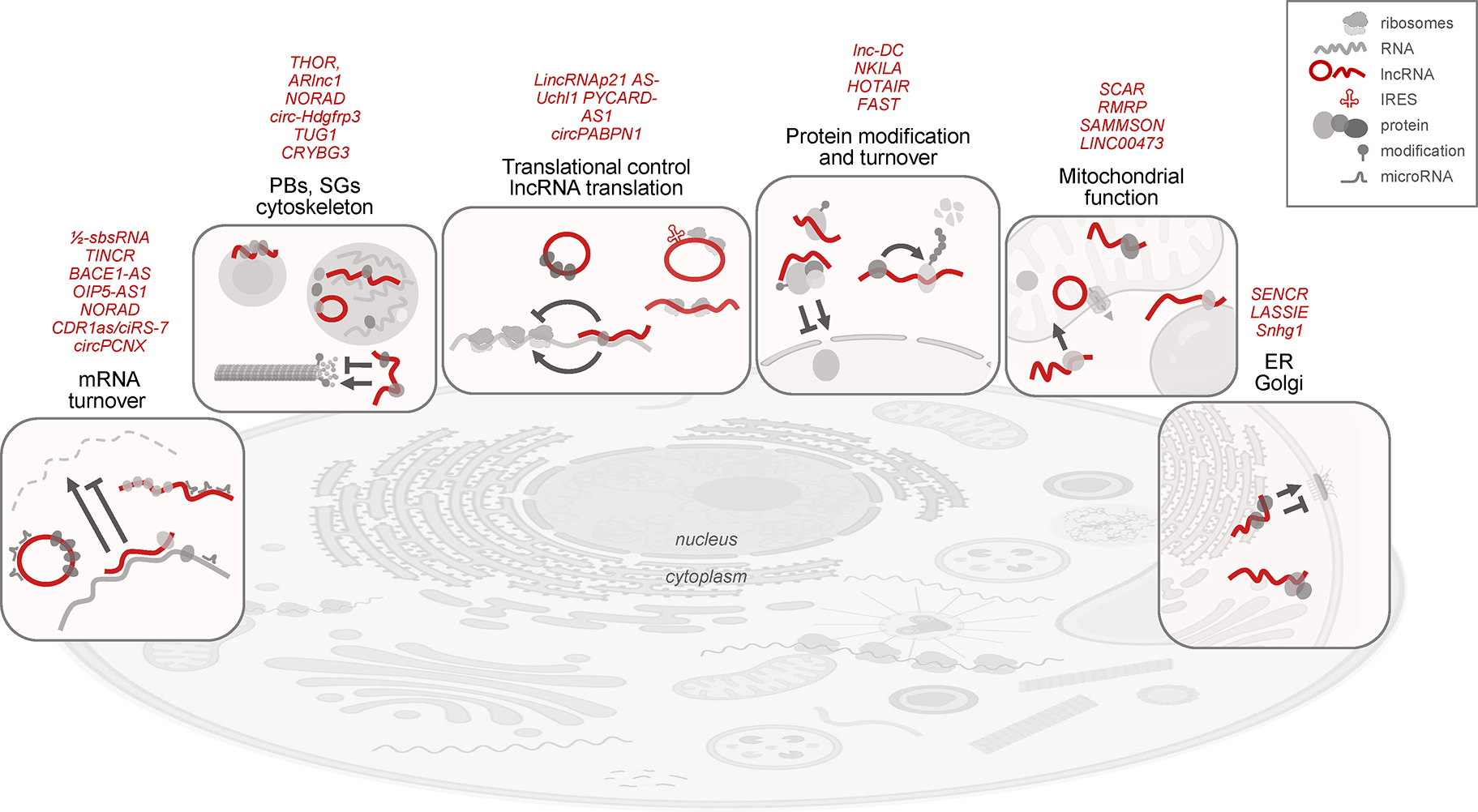
Representative functions of linear and circular lncRNAs in the cytoplasm (red lines), typically elicited by interacting with proteins and altering the stability and localization of mRNAs, as well as the translation and post-translational modification of proteins. From left to right, lncRNAs can promote or repress mRNA decay, either by forming stretches of complementarity with mRNAs or by binding RBPs or microRNAs. LncRNAs associate with RBPs and mRNAs in cytosolic granules (PBs, SGs) to influence mRNA stability and translation, and promote or suppress microtubule polymerization through modification of actin monomers. Some lncRNAs can directly promote or suppress translation of mRNAs through regions of partial complementarity or by sequestering translation regulatory RBPs; moreove, some lncRNAs (linear and circular) may themselves engage with ribosomes and become translated. LncRNAs can affect protein function post-translationally by affecting protein modifications that alter their subcellular distribution or by scaffolding ubiquitin ligases that facilitate protein degradation. In mitochondria, lncRNAs can modulate mitochondrial gene transcription, respiration, and generation of reactive oxygen species; by mediating the interaction with lipid droplets, some lncRNAs can influence mitochondrial fusion and fission. Some lncRNAs associated with ER and Golgi apparatus can help mobilize proteins to the plasma membrane and some ER-associated lncRNAs can promote or reduce adherens junction function. LncRNAs representing these functions are listed above the vignettes; molecules are not drawn at scale. Figure was prepared using BioRender.
LncRNAs affecting mRNA turnover
Cytoplasmic mRNA degradation is driven by deprotecting the 5’ and 3’ ends (5’ decapping and 3’ deadenylation) coupled to exonucleolytic degradation and endonucleolytic cleavage (Schoenberg and Maquat, 2012). These processes are regulated by complex sets of RBPs that recognize labile mRNAs and modulate their recruitment to ribonucleases present in the cytosol or in degradation centers like the exosome. By modulating mRNA stability, RBPs enable adaptive changes in the transcriptome of cells responding to proliferation, differentiation, activation, and stress. The turnover of mRNAs is further governed by microRNAs, a class of small (~22 nt) ncRNAs that can promote the decay of mRNAs with which they share partial complementarity; microRNAs recruit the RNA-induced silencing complex (RISC), a multiprotein complex that includes the endoribonuclease Argonaute that cleaves the mRNA (Pratt and MacRae, 2009). Together, microRNAs and RBPs tightly regulate the steady-state levels of mRNAs.
Several lncRNAs operate upon processes that modify mRNA turnover. In Staufen 1 (STAU1)-mediated mRNA decay (SMD), lncRNAs were found to either stabilize or destabilize specific target mRNAs. For instance, the 3’UTRs of some mRNAs partially complement lncRNAs, and the resulting double-stranded (ds)RNAs can trigger SMD. In the case of lncRNAs containing repetitive elements like Alu [½-sbsRNA (half STAU1-binding site)] or SINEs (short interspersed elements), the resulting dsRNAs trigger mRNA decay through SMD (Gong and Maquat, 2011; Wang et al., 2013). On the other hand, terminal differentiation-induced ncRNA (TINCR), a lncRNA highly abundant during epidermal differentiation and capable of binding mRNAs bearing a 25-nt TINCR box, also interacted with STAU1 but instead stabilized subsets of mRNAs encoding differentiation proteins (Kretz et al., 2013).
In other examples of lncRNAs forming dsRNAs that affect mRNA outcome, the lncRNA BACE1-AS stabilized BACE1 mRNA, which encodes β-secretase 1 (BACE1), the enzyme that cleaves amyloid precursor protein (APP) to release the neurotoxic Aβ peptide in Alzheimer’s disease (Faghihi et al., 2008). The dsRNA region of complementarity shared by BACE1 mRNA and BACE1-AS blocked a miR-485 site, rendering BACE1 mRNA stable and increasing BACE1 production (Faghihi et al., 2010). In a recent example, lncRNA OIP5-AS1, abundant in human skeletal myoblasts, associated through partial complementarity with MEF2C mRNA and stabilized it by recruiting HuR to the MEF2C 3’UTR, rising MEF2C production and promoting myogenesis (Yang et al., 2020).
Additional lncRNAs have been identified that influence mRNA turnover by sequestering decay-promoting RBPs. The abundant cytoplasmic lncRNA NORAD (noncoding RNA activated by DNA damage) contains many binding sites for Pumilio 1/2 (PUM1/2), an RBP that typically reduces the stability and translation of target mRNAs. The efficient sequestration of Pumilio by NORAD enabled the production of several proteins involved in maintaining genomic stability. In cells, the genomic instability seen after ablating NORAD was rescued by ectopic expression of NORAD containing Pumilio binding sites but not by mutant NORAD lacking Pumilio binding sites (Lee et al., 2016).
Given their intrinsic stability, circRNAs have been proposed to modulate the decay of mRNAs by binding and ‘sponging’ microRNAs that would otherwise repress such mRNA. A prominent example of this function was provided by competing endogenous (ce)RNA CDR1as/ciRS-7, which bears dozens of miR-7 binding sites and can sponge this microRNA (Memczak et al., 2013; Hansen et al., 2013). Some examples of circRNAs sequestering decay-promoting RBPs such as AUF1 (AU-binding factor 1) for circPCNX have also been reported (Tsitsipatis et al., 2021). The low abundance of most circRNAs makes such sponging functions relative rare.
LncRNAs modulating functions of cytoplasmic phase-separation bodies (SGs, PBs)
RBPs and mRNAs assemble into cytoplasmic membrane-less phase-separation bodies, such as stress granules (SGs) and processing bodies (PBs). These particles typically harbor untranslated mRNAs, likely serving as constitutive or stress-induced reservoirs of specific mRNA subsets (Hubstenberger et al., 2017; Kedersha et al., 2005). Although lncRNAs are much less abundant than mRNAs overall in SGs and PBs, they are increasingly recognized as contributing to their assembly and function.
A function was proposed for specific lncRNAs at the interface between PBs and SGs (Pitchiaya et al., 2019). LncRNAs THOR and ARlnc1, interacting with the PB proteins IGF2BP1 and HuR, respectively (Pitchiaya et al., 2019; Zhang et al., 2018), were found in the outer shell of PBs. The lncRNAs influenced the translation and stability of interacting mRNAs by recruiting them to sites of translational repression (PB cores, SGs), degradation or translation (Pitchiaya et al., 2019).
SGs form dynamically in response to stress stimuli and represent sites of aggregation of untranslating mRNAs (Aulas et al., 2017; Kedersha et al., 2005; Protter and Parker, 2016). SGs assemble via protein-protein interaction networks and recruit subsets of mRNAs during times of stress, but the mechanisms that select these mRNAs are unknown. Specialized transcriptomic analysis recently found that NORAD is present in SGs and interacts with other SG RNAs (Khong et al., 2017). Moreover, the RNA helicase eIF4A, which disrupts RNA-RNA associations, prevented the recruitment of RNAs including NORAD to SGs (Tauber et al., 2020), although the association of NORAD with SG proteins like TIAR and TIA-1 may also contribute to its recruitment to SGs (Namkoong et al., 2018).
While some roles for linear lncRNAs upon the assembly and function of phase-separation bodies are emerging, less is known about circRNAs in these spaces. Oxidative stress was recently found to promote the association of circ-Hdgfrp3 with neuronal SGs, although in amyotrophic lateral sclerosis (ALS), circ-Hdgfrp3 associated instead with cytoplasmic aggregates of a mutant form of the RBP FUS linked to ALS pathology (D’Ambra et al., 2021).
The cytoskeleton in lncRNA localization
LncRNAs are beginning to gain recognition in cytoskeletal dynamics. In one study, the lncRNA TUG1 (taurin upregulated gene 1) promoted the interaction of enhancer of zeste homolog 2 (EZH2) with α-actin (ACTA1). This interaction led to the methylation of ACTA1 and to an acceleration of the polymerization of filamentous F-actin in vascular smooth muscle cells (Chen et al., 2017). Another lncRNA capable of influencing the function of actin filaments, CRYBG3 bound instead to globular actin (G-actin), blocked the polymerization of actin filaments, and suppressed cytokinesis (Pei et al., 2018). In addition to suppressing the formation of a functional contractile ring needed to complete cell division, CRYBG3-bound G-actin sequestered the protein MAL in the cytoplasm, preventing the formation of the transcriptionally active MAL-SRF (serum response factor) complex, and blocking the transcription of immediate early genes (Pei et al., 2018).
CircRNAs are also believed to interact with the cytoskeleton, as they can be found at sites distant from the nucleus, such as neuronal synapses. The molecular details of their mobilization to synaptic regions are unknown, although they were postulated to recruit RBPs, microRNAs or other nucleic acids to distal sites (You et al., 2015).
LncRNAs influencing translation
Translation is a complex process whereby mRNA molecules associate with ribosomes to serve as templates for protein synthesis. LncRNAs provide a regulatory overlay that influences protein production in different ways: they can base-pair with mRNAs to promote or repress translation, alter the availability of translation regulatory factors, and associate with ribosomes directly, the latter scenario resulting in protein production and altered lncRNA turnover.
Base pairing of LincRNAp21 with the JUNB and CTNNB1 mRNAs through several regions of complementarity along these two mRNAs led to reduced mRNA association with polysomes and lowered production of JUNB and β-catenin (CTNNB) in cancer cells. This repression was linked to the recruitment of the translational repressor RCK to the LincRNAp21-mRNA complexes (Yoon et al., 2012). In an example of the opposite mode of action, base-pairing between the antisense lncRNA AS-Uchl1 and Uchl1 mRNA (encoding ubiquitin carboxy-terminal hydrolase L1) promoted translation of UCHL1. Although AS-Uchl1 is typically nuclear, stress conditions led to its export to the cytoplasm, where the SINE B2 RNA element in AS-Uchl1 bound to Uchl1 mRNA and enhanced its translation (Carrieri et al., 2012). Antisense lncRNAs can also suppress translation, as documented for the interaction of antisense lncRNA PYCARD-AS1 with PYCARD mRNA, which reduced ribosome assembly and PYCARD translation. Interestingly, PYCARD-AS1 further repressed PYCARD mRNA transcription in the nucleus by recruiting transcriptional repressors DNMT1 and G9a to the PYCARD promoter (Miao et al., 2019).
Some circRNAs may also influence translation. For example, binding of HuR to the PABPN1 3’UTR promoted PABPN1 translation. In this paradigm, high levels of circPABPN1, generated from an exon of PABPN1 pre-mRNA bearing numerous HuR-binding sites, selectively sequestered HuR away from PABPN1 mRNA and lowered PABPN translation (Abdelmohsen et al., 2017).
LncRNAs with protein-coding potential
In addition to modulating translation of mRNAs through binding the mRNAs directly or by altering the availability of RBPs that modulate translation, many cytoplasmic lncRNAs directly associate with ribosomes (Carlevaro-Fita and Johnson, 2019; Ruiz-Orera and Albà, 2019). Although the consequences of these interactions are not uniform, it is clear that many lncRNAs encode small peptides like myoregulin (MLN), dwarf open reading frame (DWORF), mitoregulin (MTLN), HOXB-AS3 peptide, and many others (Anderson et al., 2015; Huang et al., 2017; Matsumoto et al., 2017; Nelson et al., 2016; Stein et al., 2018). However, not all lncRNAs associated with polysomes are translated (Banfai et al., 2012; Guttman et al., 2013) and many are instead degraded by nonsense-mediated decay (NMD; Carlevaro-Fita et al., 2016). Moreover, short open reading frames in the 5’ segments of lncRNAs led to the ribosomal localization and NMD sensitivity of some lncRNAs (Smith et al., 2014), as shown for lncRNA GAS5, which bears premature stop codons (Smith and Steitz, 1998; Tani et al., 2013).
Although circRNAs lack 5’ cap structures, a recent report identified IRES elements in thousands of circRNAs that facilitated the translation of encoded proteins in a tissue-specific manner (Chen et al., 2021). The authors followed up on circFGFR1, expressing protein circFGFR1p, a protein capable of functioning as a dominant-negative FGF receptor to inhibit proliferation in response to heat stress (Chen et al., 2021). The proteins encoded by linear and circular lncRNAs are intensely studied at present. The apparent paradox embodied by lncRNAs that have coding potential is discussed below (Perspectives section).
LncRNAs influencing post-translational protein modification and stability
With the flow of genetic information typically including post-translational modification, cytoplasmic lncRNAs are increasingly recognized to alter protein functionality after proteins are synthesized. For example, lnc-DC, preferentially expressed in dendritic cells, associated with the C-terminus of STAT3 in the cytoplasm. This interaction increased the levels of STAT3 phosphorylation, as lnc-DC binding to STAT3 suppressed the phosphatase activity of SHP1/PTPN6 (Wang et al., 2014). In another example, the lncRNA NKILA (NF-κB-interacting lncRNA) helped to keep low levels of NF-κB activity by associating with the NF-κB–IκB complex, as NKILA binding blocked IκB phosphorylation by the kinase IKK and prevented the activation of NF-κB; accordingly, low levels of NKILA led to elevated NF-κB activity in cancer cells (Liu et al., 2015).
Protein expression programs are also controlled via complex and precise protein degradation mechanisms. Here too, lncRNAs can offer a key layer of control to coordinate the degradation of existing proteins. For example, the lncRNA HOTAIR promoted ubiquitin-mediated proteolysis in the cytoplasm by binding E3 ubiquitin ligases DZIP3 and MEX3B and their respective ubiquitination substrates, ATXN1 and SNUPN. Through these associations, HOTAIR facilitated the ubiquitination of ATXN1 and SNUPN and accelerated their degradation, as shown in senescent cells (Yoon et al., 2013).
In an example of protein stabilization, the lncRNA FAST (FOXD3-AS1) prevented the degradation of β-catenin. In human embryonic stem cells, FAST associated with the WD40 motif (implicated in protein-protein interactions) of the E3 ubiquitin ligase β-TrCP (β-transducin repeats-containing protein). This interaction prevented the association of β-TrCP with phosphorylated β-catenin and blocked β-catenin degradation, in turn activating WNT signaling (Guo et al., 2020a).
Mitochondrial lncRNAs
Mitochondria are membrane-enclosed organelles essential for energy production in mammalian cells. In humans, the mitochondrial DNA genome (mtDNA) is transcribed into 11 mRNAs that encode 13 proteins, 2 rRNAs (12S and 16S rRNA), and 22 transfer tRNAs (Mercer et al., 2011). In addition, mitochondria-encoded linear RNAs (mtlncRNAs) such as lncND5, lncND6, and lncCyt b were identified by high-throughput RNA-seq analysis (Rackham et al., 2011; Zhang et al., 2021a). The past two decades have begun to identify the supragenomic impact of mitochondrial lncRNAs, including lncRNAs transcribed in the nucleus and imported into mitochondria, on mitochondrial homeostasis and energy metabolism.
In fibroblasts from patients with nonalcoholic steatohepatitis (NASH), three mtcircRNAs were selectively reduced. Ectopic overexpression of one of them, SCAR (steatohepatitis-associated circRNA ATP5B regulator), significantly decreased mitochondrial and cytosolic ROS (Zhao et al., 2020a). SCAR was found mainly in the mitochondrial matrix and matrix-facing inner membrane and associated with ATP5B; through this interaction, SCAR blocked the mitochondrial permeability transition pore (mPTP) and reduced mROS output.
Mitochondria may also harbor lncRNAs transcribed from nuclear DNA (Jeandard et al., 2019). The lncRNA component of the RNA processing endoribonuclease (RMRP) was transcribed in the nucleus, exported to the cytosol by HuR, internalized into mitochondria by PNPase, and retained in mitochondria by the RBP GRSF1; these effects were linked to a role for RMRP in mtDNA replication (Noh et al., 2016). Another prominent lncRNA transcribed in the nucleus, SAMMSON, positively regulated the localization of p32 (C1QBP) in mitochondria, in turn increasing the levels of 16S rRNA, the abundance of mitochondrially encoded proteins COX2 and ATP6, and the activity of the respiratory complexes I and IV (Leucci et al., 2016). In one more example, in response to norepinephrine, LINC00473 was actively transcribed in the nucleus, translocated to the cytoplasm, and resided at the interphase of mitochondria and lipid droplets. Direct interaction of LINC00473 with Perilipin 1 (PLIN1) modulated mitochondrial fusion and fission activity, underscoring a role for LINC00473 as regulator of mitochondrial metabolic signaling (Tran et al., 2020).
Supragenomic impact of lncRNAs in endoplasmic reticulum, Golgi, and plasma membrane
The endoplasmic reticulum (ER) is a complex organelle primarily responsible for the proper folding of proteins. Among the chaperones in the ER, GRP78 facilitates the folding and assembly of nascent polypeptides and senses the accumulation of unfolded proteins, which activate the mammalian unfolded protein response (UPR) and trigger ER stress (Pobre et al., 2019). Many lncRNAs have been implicated in the UPR (Zhao et al., 2020b), but most of them did not directly associate with the ER.
The laminar shear stress-induced SENCR lncRNA interacted with the ER-anchoring domain of cytoskeleton-associated protein 4 (CKAP4), retaining CKAP4 at the rough ER. This complex prevented the association of CKAP4 with Cadherin 5 (CDH5), in turn promoting the localization of CDH5 to the plasma membrane. When SENCR levels declined, CKAP4 bound CDH5 and this led to the internalization and degradation of CDH5, in turn impairing adherens junction (AJ) function (Lyu et al., 2019). Similarly, another shear stress-induced lncRNA, LASSIE, associated with the ER but was required for the barrier function of endothelial cells in the direction of fluid flow. LASSIE helped anchor the cytoskeleton with AJs by binding cytoskeletal proteins such as nestin and AJ proteins PECAM-1 (platelet endothelial cell adhesion molecule-1) and VE-cadherin (Stanicek et al., 2020).
Among the few lncRNAs have been found in the Golgi apparatus, the lncRNA Snhg1 (small nucleolar RNA host gene 1) was found to interact with VPS13D (vesicle trafficking protein vacuolar protein sorting 13 homolog D), a protein implicated in trafficking between the Golgi apparatus and the vascular system. The complex Snhg1-VPS13D was required for VPS13D to mobilize IL-7Rα (CD127) to the membrane, in turn affecting the differentiation of memory CD8 T cells (Zhang et al., 2021b). As our understanding of the noncoding transcriptome grows, other prominent lncRNAs with distinct cytoplasmic localization and function will certainly emerge.
Perspectives
The past 25 years have brought into view a legion of lncRNAs that enable critical layers of precision and complexity in gene expression programs. Highly versatile and diverse, lncRNAs create a supragenomic tier of gene regulation that integrates fundamental genomic and epigenomic tiers of gene control. Their rich influence is linked to the wide heterogeneity in lncRNA sequences, structures, interaction partners, and subcellular spaces of residence. In the nucleus, lncRNAs help organize chromatin, direct transcription, and modify nascent RNAs. In the cytoplasm, they modulate mRNA turnover, storage, and translation, as well as orchestrate protein processing events. Given the impact of lncRNAs on these essential cellular processes, their influence in development, physiology, and disease continues to be intensely studied.
A provocative contradiction in lncRNA nomenclature has surfaced in recent years. With the rapid improvement of proteomic methods to detect ever smaller and rarer peptides in the cell, the numbers of lncRNAs found to be translated is also increasing. Thus, the very name ‘long noncoding RNA’ appears inadequate to describe this growing group of lncRNAs. However, given that our knowledge of ‘potentially coding’ lncRNAs (Ruiz-Orera and Albà, 2019) is still evolving, it is not possible to categorically classify them according to translational status. Perhaps we are progressing towards a time in which we will not distinguish lncRNA from mRNA, and instead we consider RNAs on a ‘continuum of engagement in translation’. On one end of this continuum are the RNAs that are largely associated with ribosomes, primarily cytoplasmic, and actively translated; somewhere down the continuum are RNAs that are translated when needed, but may have other structural or catalytic activities; further down the continuum are those RNAs that mainly function outside of translation, perhaps are mostly nuclear, but could occasionally be translated to meet specific needs of the cell; and at the far end of this continuum, we would find RNAs that never appear to leave the nucleus, have no recognizable reading frames, and are not found associated with ribosomes. It is unclear at present if or when such view may be adopted.
Besides the information gained from progress in proteomic detection, current and future advances in lncRNA biology are being fueled by increasingly precise single-cell and spatial multiomics, high-resolution microscopy, single-molecule analysis methods, and advanced computing. Further progress towards elucidating molecular details and functions of lncRNAs will be accelerated as suitable animal models become routinely available, lncRNA nomenclatures are harmonized, and cell-specific and extracellular lncRNAs are comprehensively catalogued.
In sum, we have presented an integrated overview of lncRNA functions that have emerged over the past quarter century. We propose that lncRNAs associate with diverse molecules to build upon genomic processes (e.g., transcription, mRNA turnover, translation) and epigenomic processes (e.g., modifications of DNA, RNA, and histones) a supragenomic level of gene regulation. Beyond nucleic acids and proteins, lncRNAs might associate with molecules like lipids or carbohydrates to further expand their supragenomic influence on cell biology.
We anticipate that future lncRNA research will continue to uncover stunning and unanticipated biology illuminating the broad influence of lncRNAs on protein programs and cell functions. With the landscape of lncRNA biology increasingly coming into view, the diagnostic, prognostic, and therapeutic value of lncRNAs will also become more clearly understood. Judging from the escalation in interest over the past quarter century, the next 25 years of lncRNA research promise to be filled with progress, excitement, and opportunity.
Acknowledgments
The authors are supported entirely by the NIA IRP, NIH. We apologize to many colleagues whose work we were unable to include due to space limitations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, et al. (2017). Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 14, 361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Leonardi T, Han N, Vire E, Gascoigne DK, Arias-Carrasco R, Buscher M, Pandolfini L, Zhang A, Pluchino S, et al. (2018). Genomic positional conservation identifies topological anchor point RNAs linked to developmental loci. Genome Biol 19, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DM, Anderson KM, Chang CL, Makarewich CA, Nelson BR, McAnally JR, Kasaragod P, Shelton JM, Liou J, Bassel-Duby R, et al. (2015). A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160, 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel-Duby R, and Olson EN (2016). Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 539, 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arab K, Karaulanov E, Musheev M, Trnka P, Schafer A, Grummt I, and Niehrs C (2019). GADD45A binds R-loops and recruits TET1 to CpG island promoters. Nat Genet 51, 217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F, Lucero L, Christ A, Mammarella MF, Jegu T, Veluchamy A, Mariappan K, Latrasse D, Blein T, Liu C, et al. (2020). R-Loop Mediated trans Action of the APOLO Long Noncoding RNA. Mol Cell 77, 1055–1065 e1054. [DOI] [PubMed] [Google Scholar]
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, and Kadener S (2014). circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56, 55–66. [DOI] [PubMed] [Google Scholar]
- Aulas A, Fay MM, Lyons SM, Achorn CA, Kedersha N, Anderson P, and Ivanov P (2017). Stress-specific differences in assembly and composition of stress granules and related foci. J Cell Sci 130, 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfai B, Jia H, Khatun J, Wood E, Risk B, Gundling WE Jr., Kundaje A, Gunawardena HP, Yu Y, Xie L, et al. (2012). Long noncoding RNAs are rarely translated in two human cell lines. Genome Res 22, 1646–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardou F, Merchan F, Ariel F, and Crespi M (2011). Dual RNAs in plants. Biochimie 93, 1950–1954. [DOI] [PubMed] [Google Scholar]
- Belak ZR, and Ovsenek N (2007). Assembly of the Yin Yang 1 transcription factor into messenger ribonucleoprotein particles requires direct RNA binding activity. J Biol Chem 282, 37913–37920. [DOI] [PubMed] [Google Scholar]
- Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, and de Herreros AG (2008). A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev 22, 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran M, Yates CM, Skalska L, Dawson M, Reis FP, Viiri K, Fisher CL, Sibley CR, Foster BM, Bartke T, et al. (2016). The interaction of PRC2 with RNA or chromatin is mutually antagonistic. Genome Res 26, 896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi P, Ramirez-Martinez A, Li H, Cannavino J, McAnally JR, Shelton JM, Sanchez-Ortiz E, Bassel-Duby R, and Olson EN (2017). Control of muscle formation by the fusogenic micropeptide myomixer. Science 356, 323–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank-Giwojna A, Postepska-Igielska A, and Grummt I (2019). lncRNA KHPS1 Activates a Poised Enhancer by Triplex-Dependent Recruitment of Epigenomic Regulators. Cell Rep 26, 2904–2915 e2904. [DOI] [PubMed] [Google Scholar]
- Brenner S, Jacob F, and Meselson M (1961). An unstable intermediate carrying information from genes to ribosomes for protein synthesis. Nature 190, 576–581. [DOI] [PubMed] [Google Scholar]
- Bridges MC, Daulagala AC, and Kourtidis A (2021). LNCcation: lncRNA localization and function. J Cell Biol 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Cao C, Ji L, Ye R, Wang D, Xia C, Wang S, Du Z, Hu N, Yu X, et al. (2020). RIC-seq for global in situ profiling of RNA-RNA spatial interactions. Nature 582, 432–437. [DOI] [PubMed] [Google Scholar]
- Carlevaro-Fita J, and Johnson R (2019). Global Positioning System: Understanding Long Noncoding RNAs through Subcellular Localization. Mol Cell 73, 869–883. [DOI] [PubMed] [Google Scholar]
- Carlevaro-Fita J, Polidori T, Das M, Navarro C, Zoller TI, and Johnson R (2019). Ancient exapted transposable elements promote nuclear enrichment of human long noncoding RNAs. Genome Res 29, 208–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlevaro-Fita J, Rahim A, Guigo R, Vardy LA, and Johnson R (2016). Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA 22, 867–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, et al. (2012). Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 491, 454–457. [DOI] [PubMed] [Google Scholar]
- Chen CK, Cheng R, Demeter J, Chen J, Weingarten-Gabbay S, Jiang L, Snyder MP, Weissman JS, Segal E, Jackson PK, et al. (2021). Structured elements drive extensive circular RNA translation. Mol Cell 81, 4300–4318 e4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LL, and Carmichael GG (2009). Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell 35, 467–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Zhao G, Yan X, Lv Z, Yin H, Zhang S, Song W, Li X, Li L, Du Z, et al. (2018). A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol 19, 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Kong P, Zhang F, Shu YN, Nie X, Dong LH, Lin YL, Xie XL, Zhao LL, Zhang XJ, et al. (2017). EZH2-mediated alpha-actin methylation needs lncRNA TUG1, and promotes the cortex cytoskeleton formation in VSMCs. Gene 616, 52–57. [DOI] [PubMed] [Google Scholar]
- Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, Han K, Chen JW, Judde JG, Deas O, et al. (2019). N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun 10, 4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chillon I, and Pyle AM (2016). Inverted repeat Alu elements in the human lincRNA-p21 adopt a conserved secondary structure that regulates RNA function. Nucleic Acids Res 44, 9462–9471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SW, Kim HW, and Nam JW (2019). The small peptide world in long noncoding RNAs. Brief Bioinform 20, 1853–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Yamazaki T, and Hirose T (2016). Architectural RNAs (arcRNAs): A class of long noncoding RNAs that function as the scaffold of nuclear bodies. Biochim Biophys Acta 1859, 139–146. [DOI] [PubMed] [Google Scholar]
- Cifuentes-Rojas C, Hernandez AJ, Sarma K, and Lee JT (2014). Regulatory interactions between RNA and polycomb repressive complex 2. Mol Cell 55, 171–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, and Lawrence JB (2009). An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33, 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, Cildir G, Jourdain A, Tergaonkar V, Schmid M, Zubieta C, et al. (2017). A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants 3, 17053. [DOI] [PubMed] [Google Scholar]
- Cooper DR, Carter G, Li P, Patel R, Watson JE, and Patel NA (2014). Long Non-Coding RNA NEAT1 Associates with SRp40 to Temporally Regulate PPARgamma2 Splicing during Adipogenesis in 3T3-L1 Cells. Genes (Basel) 5, 1050–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambra E, Santini T, Vitiello E, D’Uva S, Silenzi V, Morlando M, and Bozzoni I (2021). Circ-Hdgfrp3 shuttles along neurites and is trapped in aggregates formed by ALS-associated mutant FUS. iScience 24, 103504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham I and ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson DG, Kelley DR, Tenen D, Bernstein B, and Rinn JL (2016). Widespread RNA binding by chromatin-associated proteins. Genome Biol 17, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Norseen J, Wiedmer A, Riethman H, and Lieberman PM (2009). TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol Cell 35, 403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, et al. (2013). The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz JM, Sirokman K, McDonel P, Shishkin AA, Surka C, Russell P, Grossman SR, Chow AY, Guttman M, and Lander ES (2014). RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell 159, 188–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G 3rd, Kenny PJ, and Wahlestedt C (2008). Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med 14, 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faghihi MA, Zhang M, Huang J, Modarresi F, Van der Brug MP, Nalls MA, Cookson MR, St-Laurent G 3rd, and Wahlestedt C (2010). Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol 11, R56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust T, Frankel A, and D’Orso I (2012). Transcription control by long non-coding RNAs. Transcription 3, 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazal FM, Han S, Parker KR, Kaewsapsak P, Xu J, Boettiger AN, Chang HY, and Ting AY (2019). Atlas of Subcellular RNA Localization Revealed by APEX-Seq. Cell 178, 473–490 e426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Jadaliha M, Harmon TS, Li ITS, Hua B, Hao Q, Holehouse AS, Reyer M, Sun Q, Freier SM, et al. (2017). Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J Cell Sci 130, 4180–4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Nakagawa S, Hirose T, and Bond CS (2018). Paraspeckles: Where Long Noncoding RNA Meets Phase Separation. Trends Biochem Sci 43, 124–135. [DOI] [PubMed] [Google Scholar]
- Gonzalez I, Munita R, Agirre E, Dittmer TA, Gysling K, Misteli T, and Luco RF (2015). A lncRNA regulates alternative splicing via establishment of a splicing-specific chromatin signature. Nat Struct Mol Biol 22, 370–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros F, Hiatt H, Gilbert W, Kurland CG, Risebrough RW, and Watson JD (1961). Unstable ribonucleic acid revealed by pulse labelling of Escherichia coli. Nature 190, 581–585. [DOI] [PubMed] [Google Scholar]
- Guo CJ, Ma XK, Xing YH, Zheng CC, Xu YF, Shan L, Zhang J, Wang S, Wang Y, Carmichael GG, et al. (2020a). Distinct Processing of lncRNAs Contributes to Non-conserved Functions in Stem Cells. Cell 181, 621–636 e622. [DOI] [PubMed] [Google Scholar]
- Guo CJ, Xu G, and Chen LL (2020b). Mechanisms of Long Noncoding RNA Nuclear Retention. Trends Biochem Sci 45, 947–960. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al. (2010). Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Russell P, Ingolia NT, Weissman JS, and Lander ES (2013). Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154, 240–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao-Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR, et al. (2014). Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 21, 198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Shukla CJ, Weiner CL, and Rinn JL (2016). Function and evolution of local repeats in the Firre locus. Nat Commun 7, 11021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Zhang J, Liu Y, Fan X, Ai S, Luo Y, Li X, Jin H, Luo S, Zheng H, et al. (2019). The lncRNA Hand2os1/Uph locus orchestrates heart development through regulation of precise expression of Hand2. Development 146. [DOI] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013). Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388. [DOI] [PubMed] [Google Scholar]
- Hartford CCR, and Lal A (2020). When Long Noncoding Becomes Protein Coding. Mol Cell Biol 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Texari L, Hayes MGB, Urbanowski M, Chang MW, Givarkes N, Rialdi A, White KM, Albrecht RA, Pache L, et al. (2018). Transcription Elongation Can Affect Genome 3D Structure. Cell 174, 1522–1536 e1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Liang D, Tatomer DC, and Wilusz JE (2018). A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev 32, 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JZ, Chen M, Chen, Gao XC, Zhu S, Huang H, Hu M, Zhu H, and Yan GR (2017). A Peptide Encoded by a Putative lncRNA HOXB-AS3 Suppresses Colon Cancer Growth. Mol Cell 68, 171–184 e176. [DOI] [PubMed] [Google Scholar]
- Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. (2010). A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner MR, Eckersley-Maslin MA, and Spector DL (2013). Chromatin organization and transcriptional regulation. Curr Opin Genet Dev 23, 89–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubstenberger A, Courel M, Benard M, Souquere S, Ernoult-Lange M, Chouaib R, Yi Z, Morlot JB, Munier A, Fradet M, et al. (2017). P-Body Purification Reveals the Condensation of Repressed mRNA Regulons. Mol Cell 68, 144–157 e145. [DOI] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P, et al. (2011). Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet 43, 621–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, and Chess A (2007). A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoda T, Moore AJ, He Z, Chandra V, Aida M, Denholtz M, Piet van Hamburg J, Fisch KM, Chang AN, Fahl SP, et al. (2017). Non-coding Transcription Instructs Chromatin Folding and Compartmentalization to Dictate Enhancer-Promoter Communication and T Cell Fate. Cell 171, 103–119 e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeandard D, Smirnova A, Tarassov I, Barrey E, Smirnov A, and Entelis N (2019). Import of Non-Coding RNAs into Human Mitochondria: A Critical Review and Emerging Approaches. Cells 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Z, Song R, Regev A, and Struhl K (2015). Many lncRNAs, 5’UTRs, and pseudogenes are translated and some are likely to express functional proteins. Elife 4, e08890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K, Patel NA, Watson JE, Apostolatos H, Kleiman E, Hanson O, Hagiwara M, and Cooper DR (2009). Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternative splicing of PKCbetaII messenger ribonucleic acid. Endocrinology 150, 2087–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jie M, Wu Y, Gao M, Li X, Liu C, Ouyang Q, Tang Q, Shan C, Lv Y, Zhang K, et al. (2020). CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol Cancer 19, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly C, Usson Y, and Morimoto RI (1999). Rapid and reversible relocalization of heat shock factor 1 within seconds to nuclear stress granules. Proc Natl Acad Sci U S A 96, 6769–6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, and Anderson P (2005). Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol 169, 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khong A, Matheny T, Jain S, Mitchell SF, Wheeler JR, and Parker R (2017). The Stress Granule Transcriptome Reveals Principles of mRNA Accumulation in Stress Granules. Mol Cell 68, 808–820 e805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Siprashvili Z, Chu C, Webster DE, Zehnder A, Qu K, Lee CS, Flockhart RJ, Groff AF, Chow J, et al. (2013). Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang S, and Wang L (2020). Identification and analysis of consensus RNA motifs binding to the genome regulator CTCF. NAR Genom Bioinform 2, lqaa031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CC, Hanzelmann S, Senturk Cetin N, Frank S, Zajzon B, Derks JP, Akhade VS, Ahuja G, Kanduri C, Grummt I, et al. (2019). Detection of RNA-DNA binding sites in long noncoding RNAs. Nucleic Acids Res 47, e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, et al. (2012). Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338, 1469–1472. [DOI] [PubMed] [Google Scholar]
- Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, and Mendell JT (2016). Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 164, 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre P, Witham J, Lacroix CE, Cockerill PN, and Bonifer C (2008). The LPS-induced transcriptional upregulation of the chicken lysozyme locus involves CTCF eviction and noncoding RNA transcription. Mol Cell 32, 129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, Radaelli E, Eyckerman S, Leonelli C, Vanderheyden K, et al. (2016). Melanoma addiction to the long non-coding RNA SAMMSON. Nature 531, 518–522. [DOI] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. (2015). Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22, 256–264. [DOI] [PubMed] [Google Scholar]
- Liang D, Tatomer DC, Luo Z, Wu H, Yang L, Chen LL, Cherry S, and Wilusz JE (2017). The Output of Protein-Coding Genes Shifts to Circular RNAs When the Pre-mRNA Processing Machinery Is Limiting. Mol Cell 68, 940–954 e943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, Lin L, Yao H, Su F, Li D, et al. (2015). A cytoplasmic NF-kappaB interacting long noncoding RNA blocks IkappaB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27, 370–381. [DOI] [PubMed] [Google Scholar]
- Lubelsky Y, and Ulitsky I (2018). Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature 555, 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu Q, Xu S, Lyu Y, Choi M, Christie CK, Slivano OJ, Rahman A, Jin ZG, Long X, Xu Y, et al. (2019). SENCR stabilizes vascular endothelial cell adherens junctions through interaction with CKAP4. Proc Natl Acad Sci U S A 116, 546–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao YS, Zhang B, and Spector DL (2011). Biogenesis and function of nuclear bodies. Trends Genet 27, 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, and Akoulitchev A (2007). Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 445, 666–670. [DOI] [PubMed] [Google Scholar]
- Matera AG, Terns RM, and Terns MP (2007). Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 8, 209–220. [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Pasut A, Matsumoto M, Yamashita R, Fung J, Monteleone E, Saghatelian A, Nakayama KI, Clohessy JG, and Pandolfi PP (2017). mTORC1 and muscle regeneration are regulated by the LINC00961-encoded SPAR polypeptide. Nature 541, 228–232. [DOI] [PubMed] [Google Scholar]
- McHugh CA, Chen CK, Chow A, Surka CF, Tran C, McDonel P, Pandya-Jones A, Blanco M, Burghard C, Moradian A, et al. (2015). The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521, 232–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mele M, and Rinn JL (2016). “Cat’s Cradling” the 3D Genome by the Act of LncRNA Transcription. Mol Cell 62, 657–664. [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. (2013). Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AM, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, et al. (2011). The human mitochondrial transcriptome. Cell 146, 645–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H, Wang L, Zhan H, Dai J, Chang Y, Wu F, Liu T, Liu Z, Gao C, Li L, et al. (2019). A long noncoding RNA distributed in both nucleus and cytoplasm operates in the PYCARD-regulated apoptosis by coordinating the epigenetic and translational regulation. PLoS Genet 15, e1008144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa R, Tano K, Mizuno R, Nakamura Y, Ijiri K, Rakwal R, Shibato J, Masuo Y, Mayeda A, Hirose T, et al. (2012). Identification of cis- and trans-acting factors involved in the localization of MALAT-1 noncoding RNA to nuclear speckles. RNA 18, 738–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero JJ, Lopez-Silanes I, Megias D, M, F.F., Castells-Garcia A, and Blasco MA (2018). TERRA recruitment of polycomb to telomeres is essential for histone trymethylation marks at telomeric heterochromatin. Nat Commun 9, 1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissy AS, Griffith M, and Marra MA (2011). Extensive relationship between antisense transcription and alternative splicing in the human genome. Genome Res 21, 1203–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namkoong S, Ho A, Woo YM, Kwak H, and Lee JH (2018). Systematic Characterization of Stress-Induced RNA Granulation. Mol Cell 70, 175–187 e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, Reese AL, McAnally JR, Chen X, Kavalali ET, et al. (2016). A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TM, Kabotyanski EB, Reineke LC, Shao J, Xiong F, Lee JH, Dubrulle J, Johnson H, Stossi F, Tsoi PS, et al. (2020). The SINEB1 element in the long non-coding RNA Malat1 is necessary for TDP-43 proteostasis. Nucleic Acids Res 48, 2621–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson JA, Krochmalnic G, Wan KM, and Penman S (1989). Chromatin architecture and nuclear RNA. Proc Natl Acad Sci U S A 86, 177–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noh JH, Kim KM, Abdelmohsen K, Yoon JH, Panda AC, Munk R, Kim J, Curtis J, Moad CA, Wohler CM, et al. (2016). HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev 30, 1224–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olins DE, and Olins AL (2003). Chromatin history: our view from the bridge. Nat Rev Mol Cell Biol 4, 809–814. [DOI] [PubMed] [Google Scholar]
- Palazzo AF, and Lee ES (2018). Sequence Determinants for Nuclear Retention and Cytoplasmic Export of mRNAs and lncRNAs. Front Genet 9, 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, et al. (2017). Translation of CircRNAs. Mol Cell 66, 9–21 e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey PR, Yang JH, Tsitsipatis D, Panda AC, Noh JH, Kim KM, Munk R, Nicholson T, Hanniford D, Argibay D, et al. (2020). circSamd4 represses myogenic transcriptional activity of PUR proteins. Nucleic Acids Res 48, 3789–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paralkar VR, Taborda CC, Huang P, Yao Y, Kossenkov AV, Prasad R, Luan J, Davies JO, Hughes JR, Hardison RC, et al. (2016). Unlinking an lncRNA from Its Associated cis Element. Mol Cell 62, 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei H, Hu W, Guo Z, Chen H, Ma J, Mao W, Li B, Wang A, Wan J, Zhang J, et al. (2018). Long Noncoding RNA CRYBG3 Blocks Cytokinesis by Directly Binding G-Actin. Cancer Res 78, 4563–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitchiaya S, Mourao MDA, Jalihal AP, Xiao L, Jiang X, Chinnaiyan AM, Schnell S, and Walter NG (2019). Dynamic Recruitment of Single RNAs to Processing Bodies Depends on RNA Functionality. Mol Cell 74, 521–533 e526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobre KFR, Poet GJ, and Hendershot LM (2019). The endoplasmic reticulum (ER) chaperone BiP is a master regulator of ER functions: Getting by with a little help from ERdj friends. J Biol Chem 294, 2098–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponjavic J, Oliver PL, Lunter G, and Ponting CP (2009). Genomic and transcriptional co-localization of protein-coding and long non-coding RNA pairs in the developing brain. PLoS Genet 5, e1000617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, and Spector DL (2005). Regulating gene expression through RNA nuclear retention. Cell 123, 249–263. [DOI] [PubMed] [Google Scholar]
- Pratt AJ, and MacRae IJ (2009). The RNA-induced silencing complex: a versatile gene-silencing machine. J Biol Chem 284, 17897–17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protter DSW, and Parker R (2016). Principles and Properties of Stress Granules. Trends Cell Biol 26, 668–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rackham O, Shearwood AM, Mercer TR, Davies SM, Mattick JS, and Filipovska A (2011). Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA 17, 2085–2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. (2007). Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129, 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter N, Ali T, Kopitchinski N, Schuster P, Beisaw A, Hendrix DA, Schulz MH, Muller-McNicoll M, Dimmeler S, and Grote P (2019). The lncRNA Locus Handsdown Regulates Cardiac Gene Programs and Is Essential for Early Mouse Development. Dev Cell 50, 644–657 e648. [DOI] [PubMed] [Google Scholar]
- Ruiz-Orera J and Albà MM (2019). Conserved regions in long non-coding RNAs contain abundant translation and protein-RNA interaction signatures. NAR Genom Bioinform 1, e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki YT, Ideue T, Sano M, Mituyama T, and Hirose T (2009). MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci U S A 106, 2525–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiene-Fischer C (2015). Multidomain Peptidyl Prolyl cis/trans Isomerases. Biochim Biophys Acta 1850, 2005–2016. [DOI] [PubMed] [Google Scholar]
- Schmitz KM, Mayer C, Postepska A, and Grummt I (2010). Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24, 2264–2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenberg DR, and Maquat LE (2012). Regulation of cytoplasmic mRNA decay. Nat Rev Genet 13, 246–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla CJ, McCorkindale AL, Gerhardinger C, Korthauer KD, Cabili MN, Shechner DM, Irizarry RA, Maass PG, and Rinn JL (2018). High-throughput identification of RNA nuclear enrichment sequences. EMBO J 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, and Barlow DP (2002). The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature 415, 810–813. [DOI] [PubMed] [Google Scholar]
- Smith CM, and Steitz JA (1998). Classification of gas5 as a multi-small-nucleolar-RNA (snoRNA) host gene and a member of the 5’-terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol 18, 6897–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JE, Alvarez-Dominguez JR, Kline N, Huynh NJ, Geisler S, Hu W, Coller J, and Baker KE (2014). Translation of small open reading frames within unannotated RNA transcripts in Saccharomyces cerevisiae. Cell Rep 7, 1858–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soibam B, and Zhamangaraeva A (2021). LncRNA:DNA triplex-forming sites are positioned at specific areas of genome organization and are predictors for Topologically Associated Domains. BMC Genomics 22, 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanicek L, Lozano-Vidal N, Bink DI, Hooglugt A, Yao W, Wittig I, van Rijssel J, van Buul JD, van Bergen A, Klems A, et al. (2020). Long non-coding RNA LASSIE regulates shear stress sensing and endothelial barrier function. Commun Biol 3, 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein CS, Jadiya P, Zhang X, McLendon JM, Abouassaly GM, Witmer NH, Anderson EJ, Elrod JW, and Boudreau RL (2018). Mitoregulin: A lncRNA-Encoded Microprotein that Supports Mitochondrial Supercomplexes and Respiratory Efficiency. Cell Rep 23, 3710–3720 e3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojic L, Niemczyk M, Orjalo A, Ito Y, Ruijter AE, Uribe-Lewis S, Joseph N, Weston S, Menon S, Odom DT, et al. (2016). Transcriptional silencing of long noncoding RNA GNG12-AS1 uncouples its transcriptional and product-related functions. Nat Commun 7, 10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork M, Di Lorenzo M, Welch TJ, and Crosa JH (2007). Transcription termination within the iron transport-biosynthesis operon of Vibrio anguillarum requires an antisense RNA. J Bacteriol 189, 3479–3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Del Rosario BC, Szanto A, Ogawa Y, Jeon Y, and Lee JT (2013). Jpx RNA activates Xist by evicting CTCF. Cell 153, 1537–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, and Spector DL (2009). MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 19, 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wong SM, Dhir S, and Proudfoot NJ (2019). R-Loops Promote Antisense Transcription across the Mammalian Genome. Mol Cell 76, 600–616 e606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JY, Smith AAT, Ferreira da Silva M, Matthey-Doret C, Rueedi R, Sonmez R, Ding D, Kutalik Z, Bergmann S, and Marques AC (2017). cis-Acting Complex-Trait-Associated lincRNA Expression Correlates with Modulation of Chromosomal Architecture. Cell Rep 18, 2280–2288. [DOI] [PubMed] [Google Scholar]
- Tani H, Torimura M, and Akimitsu N (2013). The RNA degradation pathway regulates the function of GAS5 a non-coding RNA in mammalian cells. PLoS One 8, e55684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber D, Tauber G, Khong A, Van Treeck B, Pelletier J, and Parker R (2020). Modulation of RNA Condensation by the DEAD-Box Protein eIF4A. Cell 180, 411–426 e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran KV, Brown EL, DeSouza T, Jespersen NZ, Nandrup-Bus C, Yang Q, Yang Z, Desai A, Min SY, Rojas-Rodriguez R, et al. (2020). Human thermogenic adipocyte regulation by the long noncoding RNA LINC00473. Nat Metab 2, 397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Song DY, Zong X, Shevtsov SP, Hearn S, Fu XD, Dundr M, and Prasanth KV (2012). SRSF1 regulates the assembly of pre-mRNA processing factors in nuclear speckles. Mol Biol Cell 23, 3694–3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, and Chang HY (2010). Long noncoding RNA as modular scaffold of histone modification complexes. Science 329, 689–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YY, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell TC, et al. (2014). PVT1 dependence in cancer with MYC copy-number increase. Nature 512, 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsitsipatis D, Grammatikakis I, Driscoll RK, Yang X, Abdelmohsen K, Harris SC, Yang JH, Herman AB, Chang MW, Munk R, et al. (2021). AUF1 ligand circPCNX reduces cell proliferation by competing with p21 mRNA to increase p21 production. Nucleic Acids Res. 49, 1631–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachsmuth M, Caudron-Herger M, and Rippe K (2008). Genome organization: balancing stability and plasticity. Biochim Biophys Acta 1783, 2061–2079. [DOI] [PubMed] [Google Scholar]
- Wang CY, Jegu T, Chu HP, Oh HJ, and Lee JT (2018). SMCHD1 Merges Chromosome Compartments and Assists Formation of Super-Structures on the Inactive X. Cell 174, 406–421 e425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Long H, Zheng Q, Bo X, Xiao X, and Li B (2019). Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer 18, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Xue Y, Han Y, Lin L, Wu C, Xu S, Jiang Z, Xu J, Liu Q, and Cao X (2014). The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344, 310–313. [DOI] [PubMed] [Google Scholar]
- Wang XH, and Li J (2020). CircAGFG1 aggravates the progression of cervical cancer by downregulating p53. Eur Rev Med Pharmacol Sci 24, 1704–1711. [DOI] [PubMed] [Google Scholar]
- Wilson RC, and Doudna JA (2013) Molecular mechanisms of RNA interference. Annu Rev Biophys 42, 217–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, JnBaptiste CK, Lu LY, Kuhn CD, Joshua-Tor L, and Sharp PA (2012). A triple helix stabilizes the 3’ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev 26, 2392–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Yin QF, Luo Z, Yao RW, Zheng CC, Zhang J, Xiang JF, Yang L, and Chen LL (2016). Unusual Processing Generates SPA LncRNAs that Sequester Multiple RNA Binding Proteins. Mol Cell 64, 534–548. [DOI] [PubMed] [Google Scholar]
- Xu X, Zhang J, Tian Y, Gao Y, Dong X, Chen W, Yuan X, Yin W, Xu J, Chen K, et al. (2020). CircRNA inhibits DNA damage repair by interacting with host gene. Mol Cancer 19, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Yang L, Chen LL (2017). The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet 33, 540–552. [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Souquere S, Chujo T, Kobelke S, Chong YS, Fox AH, Bond CS, Nakagawa S, Pierron G, and Hirose T (2018). Functional Domains of NEAT1 Architectural lncRNA Induce Paraspeckle Assembly through Phase Separation. Mol Cell 70, 1038–1053 e1037. [DOI] [PubMed] [Google Scholar]
- Yang F, Hu A, Li D, Wang J, Guo Y, Liu Y, Li H, Chen Y, Wang X, Huang K, et al. (2019). Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol Cancer 18, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JH, Chang MW, Pandey PR, Tsitsipatis D, Yang X, Martindale JL, Munk R, De S, Abdelmohsen K, and Gorospe M (2020). Interaction of OIP5-AS1 with MEF2C mRNA promotes myogenic gene expression. Nucleic Acids Res 48, 12943–12956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, and Rosenfeld MG (2011). ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 147, 773–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Du WW, Wu N, Yang W, Awan FM, Fang L, Ma J, Li X, Zeng Y, Yang Z, et al. (2017a). A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ 24, 1609–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen LL, Wang Y, et al. (2017b). Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res 27, 626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZG, Awan FM, Du WW, Zeng Y, Lyu J, Wu, Gupta S, Yang W, and Yang BB (2017c). The Circular RNA Interacts with STAT3, Increasing Its Nuclear Translocation and Wound Repair by Modulating Dnmt3a and miR-17 Function. Mol Ther 25, 2062–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap K, Mukhina S, Zhang G, Tan JSC, Ong HS, and Makeyev EV (2018). A Short Tandem Repeat-Enriched RNA Assembles a Nuclear Compartment to Control Alternative Splicing and Promote Cell Survival. Mol Cell 72, 525–540 e513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap KL, Li S, Munoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, and Zhou MM (2010). Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38, 662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, and Chen LL (2012). Long noncoding RNAs with snoRNA ends. Mol Cell 48, 219–230. [DOI] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, White EJ, Orjalo AV, Rinn JL, Kreft SG, et al. (2013). Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination. Nat Commun 4, 2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, and Gorospe M (2012). LincRNA-p21 suppresses target mRNA translation. Mol Cell 47, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. (2015). Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18, 603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Gunawardane L, Niazi F, Jahanbani F, Chen X, and Valadkhan S (2014). A novel RNA motif mediates the strict nuclear localization of a long noncoding RNA. Mol Cell Biol 34, 2318–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Hou L, Zuo Z, Ji P, Zhang X, Xue Y, and Zhao F (2021a). Comprehensive profiling of circular RNAs with nanopore sequencing and CIRI-long. Nat Biotechnol 39, 836–845. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Li B, Bai Q, Wang P, Wei G, Li Z, Hu L, Tian Q, Zhou J, Huang Q, et al. (2021b). The lncRNA Snhg1-Vps13D vesicle trafficking system promotes memory CD8 T cell establishment via regulating the dual effects of IL-7 signaling. Signal Transduct Target Ther 6, 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pitchiaya S, Cieslik M, Niknafs YS, Tien JC, Hosono Y, Iyer MK, Yazdani S, Subramaniam S, Shukla SK, et al. (2018). Analysis of the androgen receptor-regulated lncRNA landscape identifies a role for ARLNC1 in prostate cancer progression. Nat Genet 50, 814–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, and Chen LL (2013). Circular intronic long noncoding RNAs. Mol Cell 51, 792–806. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, and Lee JT (2008). Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322, 750–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Liu J, Deng H, Ma R, Liao JY, Liang H, Hu J, Li J, Guo Z, Cai J, et al. (2020a). Targeting Mitochondria-Located circRNA SCAR Alleviates NASH via Reducing mROS Output. Cell 183, 76–93 e22. [DOI] [PubMed] [Google Scholar]
- Zhao T, Du J, and Zeng H (2020b). Interplay between endoplasmic reticulum stress and non-coding RNAs in cancer. J Hematol Oncol 13, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Qian Z, Jiang F, Ge D, Tang J, Chen H, Yang J, Yao Y, Yan J, Zhao L, et al. (2019). CircRNA LRP6 promotes the development of osteosarcoma via negatively regulating KLF2 and APC levels. Am J Transl Res 11, 4126–4138. [PMC free article] [PubMed] [Google Scholar]
- Zuckerman B, Ron M, Mikl M, Segal E, and Ulitsky I (2020). Gene Architecture and Sequence Composition Underpin Selective Dependency of Nuclear Export of Long RNAs on NXF1 and the TREX Complex. Mol Cell 79, 251–267 e256. [DOI] [PubMed] [Google Scholar]
- Zuckerman B, and Ulitsky I (2019). Predictive models of subcellular localization of long RNAs. RNA 25, 557–572. [DOI] [PMC free article] [PubMed] [Google Scholar]


