Abstract
Native mass spectrometry (MS), the analysis of proteins and protein complexes from solutions that stabilize the native solution structures, is a rapidly expanding area in mass spectrometry. There is strong evidence that supports proteins retain their native fold in the absence of solvent under the experimental timescales of MS experiments. Therefore, instrumentation has been developed to use gas-phase native-like protein ions to exploit the speed, sensitivity, and selectivity of mass spectrometry approaches to solve emerging problems in structural biology. This article reviews some of the recent advances and applications in gas-phase instrumentation for structural proteomics.
Keywords: Native mass spectrometry, ion mobility, tandem mass spectrometry, ion chemistry
Graphical Abstract
1. Ion Mobility and Mass Analyzers
Native MS is a very active area of research, where the ion mobility (IM) and mass spectra (MS) of proteins electrosprayed (ESI) from solutions that stabilize the native state are studied to characterize proteins and protein complexes.1 Several IM and MS technologies have been developed and made commercially available for more informative native MS and IM-MS studies. The mobility of an ion, the quotient of the average drift velocity divided by electric field, is proportional to the rotationally averaged collision cross section (CCS) of the ion and the bath gas and the ion’s effective temperature.2 The first commercially available IM-MS systems used a traveling wave IM (TWIMS) separator, where a short pulse of ions slowly traverses a device filled with an inert buffer gas via the forward motion of a traveling square wave propagating throughout the device, coupled to quadrupole/time-of-flight (qTOF) mass analyzers. Ions having mobilities that result in velocities slower than the velocity of the wave “roll over” the back of the wave, and lower mobilities correspond with more “roll over” events, causing them to travel more slowly through the device, enabling the separation to occur.3 TWIMS instruments allowed for measurements of the topologies of protein complexes.4 An advantage of these instruments includes the comparatively high mass range of TOF MS (20 to 100,000 m/z for the Waters Synapt G2-Si TWIMS TOF, although this can be limited by upfront optics to 32,000)5 to allow for analysis of protein complexes. More recently, other commercial IM/TOF instruments have been introduced, including IM drift tubes (DTIMS) with static electric fields propelling ions slowly through an inert drift gas. 6 In DTIMS, unlike TWIMS, a pulse of ions is dragged along by a weak uniform electric field, with the use of the weak uniform field allowing for direct calculation of an ion/buffer gas collision cross-section (CCS), a measurement of the rotationally averaged overall size of the ion.7 The oscillating field used by TWIMS does not allow for direct measurement of CCS, and so calibration procedures are used.8 Trapped ion mobility (TIMS) has also been recently introduced, where ions are trapped against a trapping field, balanced by forward motion induced by drift gas flow, and released by ramping down the trapping field.9 TIMS offers resolving power over 250 by utilizing slow ramp rates for the trapping field.10 TIMS CCS values are also typically obtained by calibration.11 Recent advances in cyclic TWIMS technology have allowed for very long (tens to hundreds of meter) separation pathlengths, greatly extending resolution for separations.12–13
Although IM devices are traditionally coupled to qTOF MS due to its fast acquisition speed, several examples of Fourier and Hadamard transform DTIMS,14–15 and TIMS,16 coupled to Fourier transform mass spectrometry (FTMS), have been demonstrated to take advantage of the improved m/z resolution offered by FTMS. Recent developments in FTMS technology have increased the m/z range to make these platforms more suitable for native MS, including modifying voltages and voltage profiles to the Orbitrap FTMS platform to increase the m/z range from 350 to 80,000 m/z,17–18 and increasing FT-ion cyclotron resonance (FT-ICR) magnetic field strengths to 21 Tesla,12–13 allowing intact proteins to be measured with resolving powers greater than 2 million. For example, an Orbitrap was used to characterize intact G protein-coupled receptors, revealing differences in sodium binding to the adenosine 2a receptor upon the addition of agonist or antagonist.19 This work illustrated changes in sodium-bound states between the active and inactive forms of the receptor, with the inactivated from (i.e., incubated with antagonist) showing binding of up to seven sodium ions, while the active state (i.e., incubated with agonist) showing only one or two bound sodium ions. A competition assay confirmed that the changes in sodium binding were due to conformational changes upon ligand binding.
Charge detection mass spectrometry (CDMS) has enabled measurements of massive, charged particles, such as the hepatitis B virus capsid,20 and heterogeneous native proteins, including the trimeric SARS-CoV-2 spike protein.21 This exciting new technique detects ions with a conductive cylinder, placed inside an electrostatic linear ion trap. This allows the ions to oscillate through the cylinder, where charge is detected by the image current induced on the cylinder, and m/z is detected by a FT of the ion signal to obtain frequency (and thus, m/z).20 The independent detection of charge and m/z allows for the mass of ions to be directly measured. Orbitrap MS has been also used for charge detection MS; by injecting a very small number of ions such that only one ion is present at each observed m/z. This allows the amplitude of the ion signal to be used to determine the charge state of single ions (since the amplitude of the ion signal is proportional to charge) and the orbiting frequency used to determine m/z.22 As commercial vendors increase their support of native MS experiments through IM/MS and MS development, the application space of native MS methods will likely continue to increase.
2. Native Tandem MS.
The most straightforward fragmentation method to apply to MS structural determination is collision induced dissociation (CID). In CID, fragmentation is achieved using energetic collisions with a target gas, such as helium, nitrogen, and argon, increasing, stepwise, the internal temperature of the ion.23 However, since CID (at energies commonly used in analytical MS) is a slow heating method, isomerization often occurs prior to fragmentation.24 Therefore, the use of CID to obtain, indirectly, information about the tertiary of proteins in the gas phase is precluded. Additionally, when CID is performed on protein complexes, monomers are first unfolded, then ejected, from the complex, which makes quaternary structural characterization from CID data difficult.25 However, collisional activation has shown broad utility for the unfolding of gas-phase complexes. Collision induced unfolding (CIU) employs energetic collisions prior to IM, using IM arrival time distributions (ATDs) to characterize the extent of unfolding and number of gas-phase unfolding transitions.26 Structural properties of proteins, including their solution conformations, influence gas-phase stabilities, with structural effects such as membrane protein stabilization by lipids,27, mutations,28 and differences in biotherapeutics before and after stress29 yielding unique CIU fingerprints (plots of ATDs versus collision energies). Comparison of CIU fingerprints with solution unfolding can also reveal some of the intrinsic factors in protein stability and unfolding and the role of solvent30. Recently, CIU was applied to differentiate mutational variants of the voltage sensor domain of the KCNQ1 voltage-gated potassium channel.28 CIU fingerprints showed distinct differences for gain and loss of function mutations of the voltage sensor domain, illustrating the ability of CIU to discriminate between various functional forms due to their effects on the overall gas-phase stability of the protein ions. Newly commercially available tandem TIMS31 and cyclic TWIMS32 devices are allowing many users to not only perform traditional pre-mobility CIU experiments, but also to select specific conformer families in CCS space, activate them (or not) and further resolve these activated and/or selected conformers, giving high structural specificity to gas-phase protein characterization. For example, Figure 1 shows CIU fingerprints that detail the evolution of native-like and collisionally activated cytochrome c 7+ ion structures.32 Each of the observed subpopulations was isolated by mobility and activated at various collision energies, allowing for the gas-phase unfolding of each subpopulation to be monitored.
Figure 1.
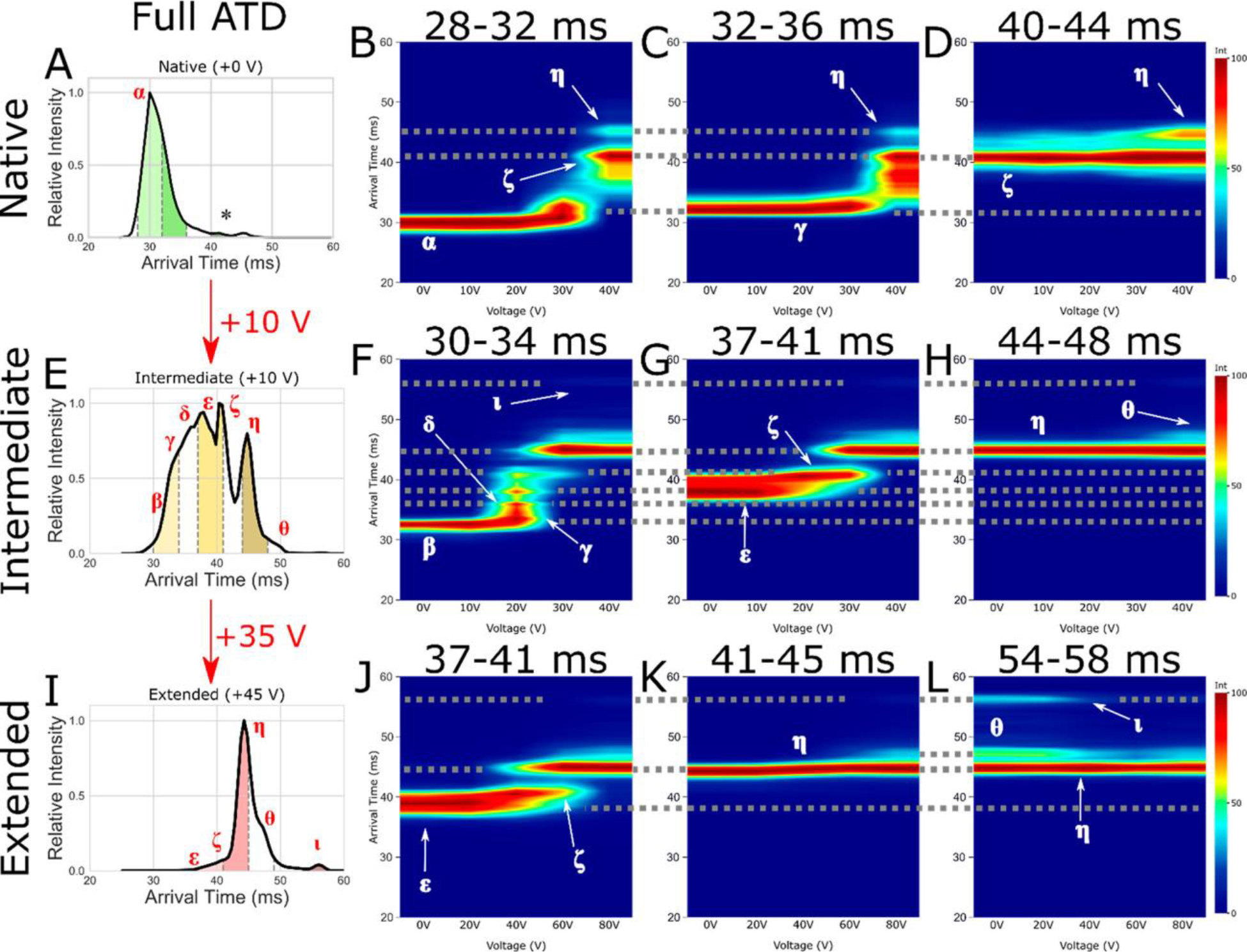
Arrival time distribution for (A) native (+0 V), (E) intermediate (+10 V), and (I) extended (an additional +35 V making +45 V total activation) CytC (+7) with subpopulation slices taken for CA denoted by dashed gray lines, filled with green (native), yellow (intermediate), and red (extended). Also shown are CA fingerprints for subpopulations of interest corresponding to native (B, F, J), intermediate (C, G, K), and extended (D, H, L) states, with interpopulation conformations joined by dotted gray lines. Identified conformations of interest are labeled as α, β, γ, δ, ε, ζ, η, θ, and ι. The * denotes the low intensity species mobility selected in plot (D). Reproduced with permission from ref32 https://pubs.acs.org/doi/10.1021/jasms.1c00018. Copyright 2021 American Chemical Society. Further permissions related to the material excerpted should be directed to the ACS. The identified conformations of interest have the same drift time regardless of the applied voltage.
Surface induced dissociation (SID), where ions are accelerated into a surface, a much more massive target than a gas molecule, extensively fragments protein complexes before structural rearrangement can occur.33 Fragmentation by SID at various energies results in the formation of subcomplexes revealing connectivity by assignment of their m/z to specific stoichiometries and identities of subunits.34 Therefore, SID allows for the quaternary structure, including subunit connectivity, to be determined, a strategy that can be combined with cryo electron microscopy (cryo-EM). The work of Seffernick et al. illustrates the combination of SID with cryo-EM and protein docking predictions.35 Measurements and predictions of appearance energies (the energies at which certain protein complex interfaces dissociate) for complex dissociation from SID were combined with low-resolution density maps obtained by cryo-EM and computational docking predictions. The addition of experimental constraints significantly improved the accuracy of structure prediction. As SID devices have recently been simplified,36 SID is amenable to the MS platforms listed above, and is now commercially available on the Waters Select Series Cyclic IM/qTOF platform. The increasing availability of SID, along with its utility in quaternary structure characterization will likely result in its widespread adoption.
While SID has been primarily employed for quaternary structure determination, electron capture dissociation (ECD), electron transfer dissociation (ETD), and ultraviolet photodissocation (UVPD) are fragmentation techniques that can provide primary sequence information for proteoform analysis and ligand binding without extensive isomerization inherent in slow-heating methods such as CID. The two electron dissociation techniques result in fragmentation of the N-Cα bond of the polypeptide after either capture of a low energy electron (ECD)37 or transfer of an electron from a radical anion in an ion/ion reaction (ETD).38 ECD/ETD do not disrupt noncovalent structure or fragment post-translational modification, making them particular useful for characterizing native proteins in the gas phase.39 Recently, ECD performed in an electromagnetostatic cell has been coupled to IM instrumentation, as the ability to perform ECD in transmission mode (versus trapping) is compatible with IM40 and any kind of mass analyzer. Thus, several new applications of ECD have emerged. First, ECD was implemented after a CIU experiment in a commercially-available DTIMS.41 A similar experiment was performed with a commercially-available TWIMS system, where the gas-phase unfolding of holo-hemoglobin was monitored with ECD and ETD.40 This technique holds promise for evaluating which specific regions of the protein are involved in gas-phase protein folding, giving greater specificity to CIU measurements. ECD has also recently been applied to determining metal binding sites to the amyloid β peptide,42 aggregation of human islet amyloid polypeptide (hIAPP),43 to studying the unfolding of apolipoprotein E,44 and to characterizing proteolysis-targeting chimera (PROTAC) complex topology45. ECD of dimers of hIAPP localized the aggregation sites of hIAPP from serine 28/serine 29 to asparagine 35 by the identification of sequence fragments that were linked to an intact monomer.43 This result shows the promise of ECD in detecting protein-protein binding and aggregation regions for pathologically relevant proteins.
UVPD is performed by irradiating ions with UV (typically 193 nm) photons.46 UVPD cleaves all bonds in the polypeptide backbone. By examining the fragments produced by direct bond dissociation (i.e., not peptide amide bond cleavages that can be the result of slower intramolecular vibrational energy relaxation processes), UVPD can be used to characterize native protein structures in the gas phase with high sequence coverage.47 UVPD has also been used to localize gas-phase protonation sites in different protein protomers,48 for example, with alpha synuclein, responsible for Parkinson’s disease, where UVPD fragmentation confirmed that different solution conditions led to different protomers for the protein, depending on the solution conformation and the preferences for different side chains to be protonated.49 Like ECD, UVPD has also been recently used to observe changes in structure upon CIU. CIU-UVPD of the alcohol dehydrogenase tetramer suggested that with increasing CIU energies, the N-terminus unfolds, with charges moving to the unfolded region, followed by the migration of charge away from the N-terminus through salt bridge formation, indicating refolding has occurred, monitored by the isotopic distributions of a and x-type ions produced by direct dissociation of C-C bonds in the polypeptide backbone (Figure 2).46 Recently, several other applications of UVPD to intact protein analysis have been demonstrated, including epitope mapping,50 characterization of ribonucleoprotein complexes and virus-like particles51, and characterization of membrane protein complexes52. For membrane protein complexes, proteins were solubilized in 200 mM ammonium acetate with 0.5% tetraethylene glycol monooctyl ether detergent. Membrane protein complexes were released from the detergent micelles with in-source collisional activation, and UVPD was performed on the intact free complexes. UVPD resulted in sequence fragments across the complexes, giving sequence coverages of 45% and 53% for the membrane proteins aquaporin Z and the mechanosensitive channel of large conductance, respectively. These experiments illustrated the ability of UVPD coupled to native MS to sequence a native, intact, membrane protein complex.
Figure 2.
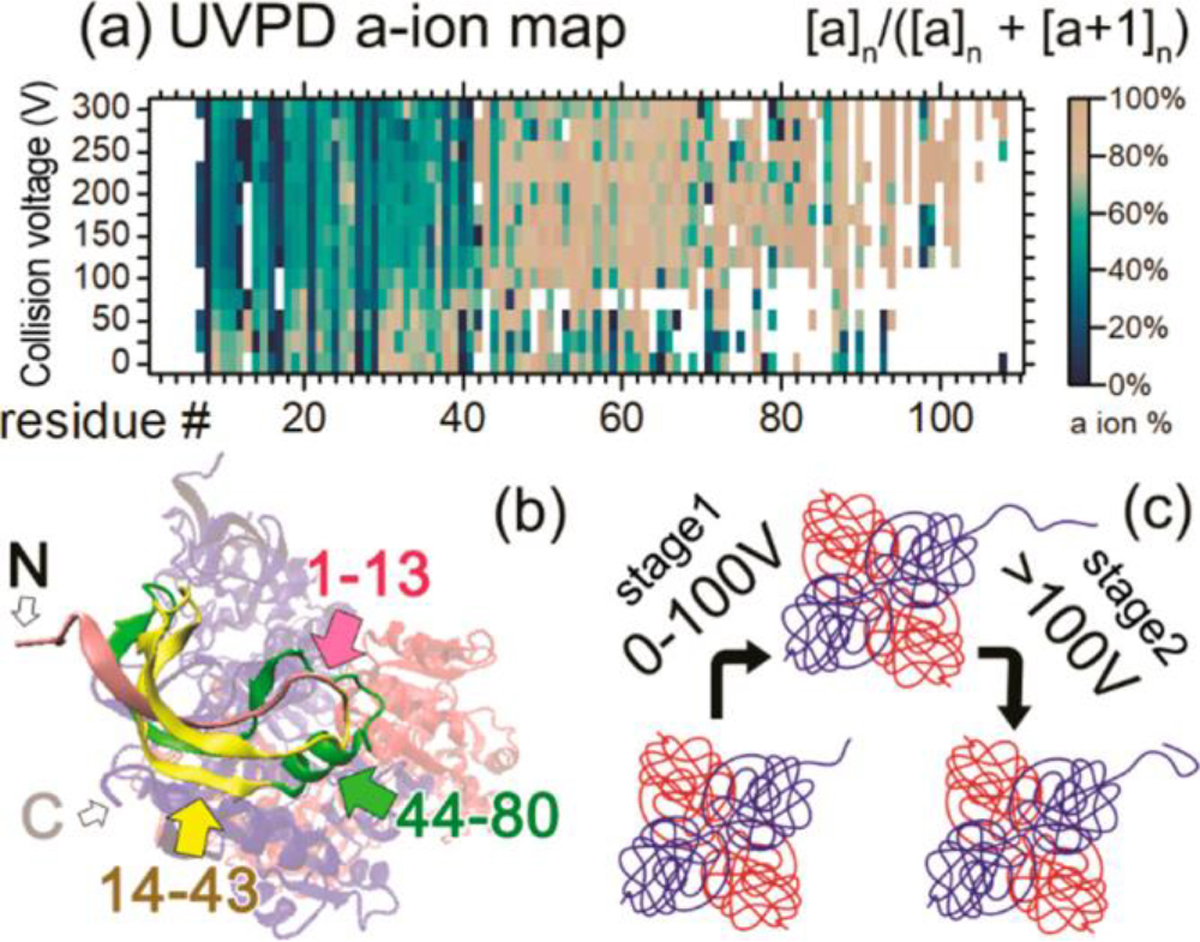
(a) a-ion percentage map for ADH 4mer. Horizontal axis shows the residue number from 1 to 120. Vertical axis shows increasing in-source collision voltage. Color scales are at the right side of the map. (b) Crystal structure of the ADH 4mer. Residues 1–13 of one 1mer subunit are in pink, 14–43 are in yellow, and 44–90 are in green. N/C-termini of the 1mer are noted with “N” and “C”. (c) Proposed ADH unfolding and refolding mechanism based on the native TD data. Reproduced with permission from ref46 https://pubs.acs.org/doi/abs/10.1021/acs.analchem.9b03469. Copyright 2019 American Chemical Society. Further permissions related to the material excerpted should be directed to the ACS.
3. Ion Chemistry.
Ion/ion reactions, reactions between two oppositely charged ions giving a net charged product,53 and ion molecule reactions, between an ion and a neutral, have allowed solution-phase probes of structure to be used in the gas-phase. These reactions are orders of magnitude faster than their solution counterparts and can be applied to mass-to-charge or conformer-selected species, a distinct advantage over solution where measurements can only be made in bulk averages. Hydrogen deuterium exchange (HDX) in the gas phase is an important example of an ion molecule reaction that has its origins in the solution phase. By comparing several conditions, the dynamicity of various regions of the proteins can be determined by localizing which side chains have undergone exchange with deuterons by tandem MS techniques such as ECD.54 Ion/ion chemistry allows for nonvolatile compounds to be used as reagents via their introduction as ions by electrospray, including reagents that can perform electrostatic or covalent labeling.53 Recently, these reactions have been used to gain distance constraints on the small model protein ubiquitin55 (Figure 3) between accessible lysine or other basic residues as well as determine the relative surface accessibilities56 of various nucleophilic sidechains, used as constraints for atomistic molecular models of the gas-phase protein structures. Cheung See Kit and coworkers used the ion/ion reagent sulfobenzoyl−−1-hydroxy-azobenzotriazole (HOAt), (SBH), which is spaced 6.4 Å from the sulfonate oxygen to the carbonyl carbon (Figure 3 A). In the ion/ion reaction between SBH and ubiquitin 6+, electrostatic binding to a protonated side chain on one end of SBH facilitates the covalent reaction with a nitrogen lone pair on the other end. Through ECD, it was determined that, for ubiquitin 6+ electrosprayed from native-like conditions (10 μM ubiquitin dissolved in 10 mM ammonium acetate), SBH linked protonated proline 19 to neutral lysine 29, and protonated lysine 63 to neutral arginine 54, and for ubiquitin 6+ electrosprayed from denaturing conditions (10 μM ubiquitin dissolved in 50/50/0.1 vol/vol/vol water/methanol/formic acid), SBH linked protonated lysine 11 to neutral lysine 48 and protonated lysine 63 to neutral arginine 54. Using ethylene glycol bis(sulfosuccinimidyl succinate), (BS3), (Figure 3 B) under conditions that do not promote covalent bond formation (i.e., minimal collision energy), the 26.9 Å sulfonate oxygen to sulfonate oxygen electrostatic cross-linker linked various charged residues for the native-like and denatured ubiquitin ions by forming a strong electrostatic bond on each side of the linker with a protonated residue. Using ECD, it was determined that sulfo-EGS linked protonated proline 19 to protonated arginine 42 for native-like ubiquitin 6+, and protonated lysine 33 to arginine 72 for denatured ubiquitin 6+. These cross-links agreed with clustered gas-phase trajectories of ubiquitin from molecular dynamics for 6+ ubiquitin from aqueous and denaturing conditions.55 The protonated sites used in assigning the electrostatic linking sites were determined by ECD of unreacted ubiquitin. Since the covalent and electrostatic reaction conditions do not measurably perturb the structures,57 the ion/ion reaction methods are promising for gaining higher structural resolution than is offered by other native mass spectrometry techniques.
Figure 3.
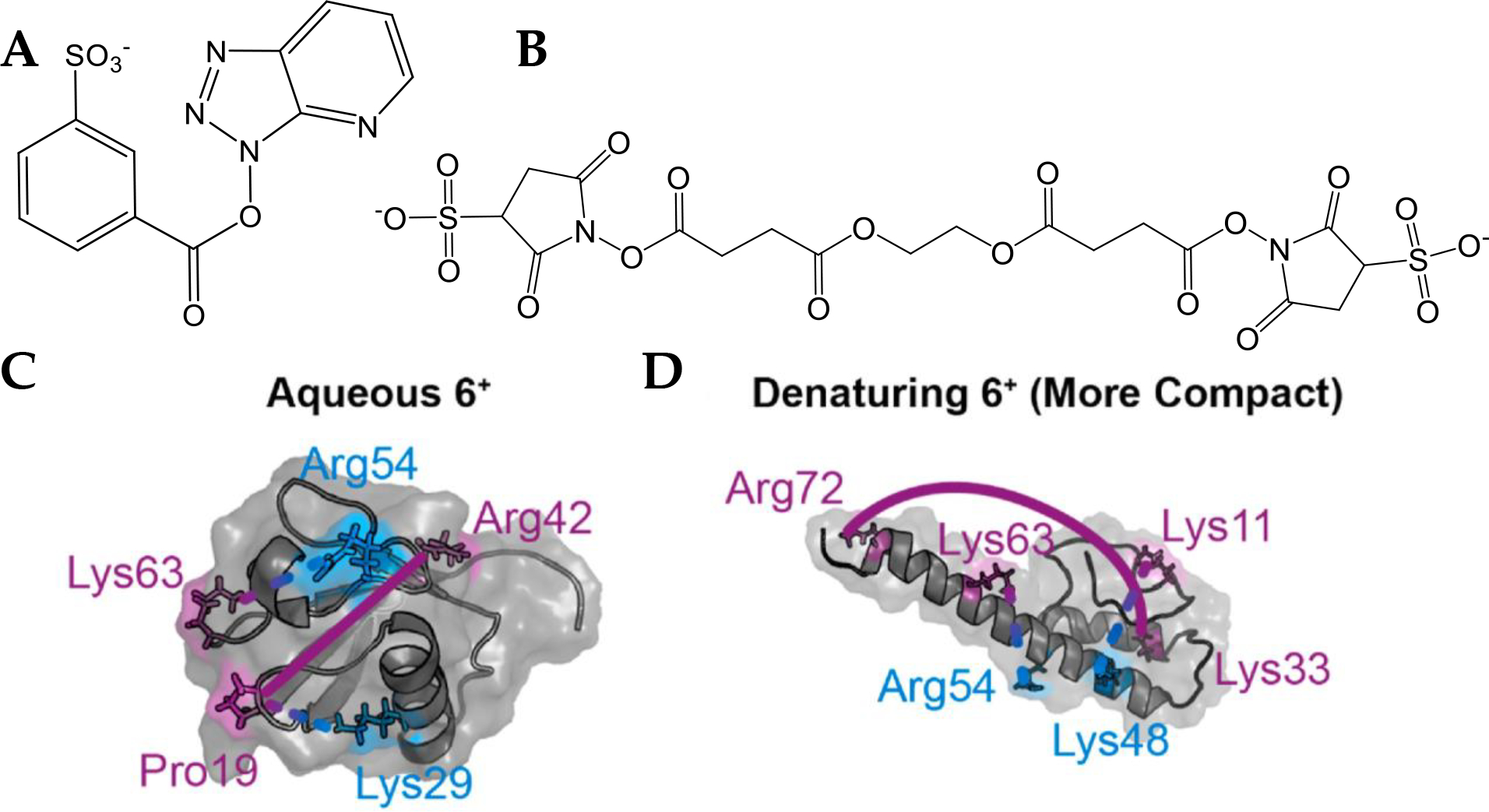
(A) The electrostatic to covalent cross-linker SBH and the (B) electrostatic to electrostatic cross-linker sulfo-EGS. (C, D) Modified residues represented using MD predicted structures of ubiquitin with protonated and covalent sites labeled in purple and blue respectively. (C) Aqueous 6+ is cross-linked by sulfo-EGS at Pro19 – Arg42 (solid line) and by sulfo-benzoyl HOAt at Lys29 – Pro19 and Arg54 – Lys63 (dashed lines). (D) Under denaturing condition, the pair linked by sulfo-EGS is Lys33 – Arg72 (solid line) and the pairs cross-linked by sulfo-benzoyl HOAt are Lys48 – Lys11 and Arg54 – Lys63 (dashed lines). (C) and (D) Reproduced with permission from ref55. f5552 Copyright 2021 Elsevier. Further permissions related to the material excerpted should be directed to Elsevier.
4. Conclusion and Perspective.
Over the past few years, important technological developments in instrumentation, including IM, mass analyzers, fragmentation methods, and gas-phase chemistries, have expanded the utility and applications of gas-phase protein structural methods. Gas-phase methods will continue to be applied to proteins which resist crystallization, are structurally heterogenous, and where small sample consumption is desirable. Future applications will likely continue to target extremely large proteins (>MDa ) and dynamic proteins or proteins with intrinsically disordered regions, pushing the limits of current technology native MS.
Highlights.
New instrument developments provide greater structural detail
Higher resolution and more versatile ion mobility and mass analyzers allow analysis of large, heterogeneous systems
Native mass spectrometric fragmentation provides structural information
Gas-phase ion chemistry for increased structural resolution
Funding.
This work was supported by the National Institute of General Medical Sciences (1R21GM134408).
References
- 1.Leney AC; Heck AJR, Native Mass Spectrometry: What is in the Name? Journal of the American Society for Mass Spectrometry 2017, 28 (1), 5–13. [DOI] [PubMed] [Google Scholar]
- 2.Kinetic Theory of Mobility and Diffusion: Sections 5.1 – 5.2. In Transport Properties of Ions in Gases, 1988; pp 137–193. [Google Scholar]
- 3.Giles K; Williams JP; Campuzano I, Enhancements in travelling wave ion mobility resolution. Rapid Commun Mass Spectrom 2011, 25 (11), 1559–66. [DOI] [PubMed] [Google Scholar]
- 4.Ruotolo BT; Benesch JL; Sandercock AM; Hyung SJ; Robinson CV, Ion mobility-mass spectrometry analysis of large protein complexes. Nat Protoc 2008, 3 (7), 1139–52. [DOI] [PubMed] [Google Scholar]
- 5.Synapt G2Si MS System Instrument Specifications. https://www.waters.com/waters/library.htm?locale=en_US&cid=134740653&lid=134744424 (accessed 10/4/2021).
- 6.May JC; Goodwin CR; Lareau NM; Leaptrot KL; Morris CB; Kurulugama RT; Mordehai A; Klein C; Barry W; Darland E; Overney G; Imatani K; Stafford GC; Fjeldsted JC; McLean JA, Conformational ordering of biomolecules in the gas phase: nitrogen collision cross sections measured on a prototype high resolution drift tube ion mobility-mass spectrometer. Anal Chem 2014, 86 (4), 2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Revercomb HE; Mason EA, Theory of plasma chromatography/gaseous electrophoresis. Review. Analytical Chemistry 2002, 47 (7), 970–983. [Google Scholar]
- 8.Bush MF; Hall Z; Giles K; Hoyes J; Robinson CV; Ruotolo BT, Collision cross sections of proteins and their complexes: a calibration framework and database for gas-phase structural biology. Anal Chem 2010, 82 (22), 9557–65. [DOI] [PubMed] [Google Scholar]
- 9.Michelmann K; Silveira JA; Ridgeway ME; Park MA, Fundamentals of Trapped Ion Mobility Spectrometry. J Am Soc Mass Spectr 2015, 26 (1), 14–24. [DOI] [PubMed] [Google Scholar]
- 10.Silveira JA; Ridgeway ME; Park MA, High resolution trapped ion mobility spectrometery of peptides. Anal Chem 2014, 86 (12), 5624–7. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez DR; Debord JD; Ridgeway ME; Kaplan DA; Park MA; Fernandez-Lima F, Ion dynamics in a trapped ion mobility spectrometer. Analyst 2014, 139 (8), 1913–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw JB; Lin TY; Leach FE 3rd; Tolmachev AV; Tolic N; Robinson EW; Koppenaal DW; Pasa-Tolic L, 21 Tesla Fourier Transform Ion Cyclotron Resonance Mass Spectrometer Greatly Expands Mass Spectrometry Toolbox. J Am Soc Mass Spectrom 2016, 27 (12), 1929–1936. [DOI] [PubMed] [Google Scholar]
- 13.Hendrickson CL; Quinn JP; Kaiser NK; Smith DF; Blakney GT; Chen T; Marshall AG; Weisbrod CR; Beu SC, 21 Tesla Fourier Transform Ion Cyclotron Resonance Mass Spectrometer: A National Resource for Ultrahigh Resolution Mass Analysis. J Am Soc Mass Spectr 2015, 26 (9), 1626–1632. [DOI] [PubMed] [Google Scholar]
- 14.Poltash ML; McCabe JW; Shirzadeh M; Laganowsky A; Clowers BH; Russell DH, Fourier Transform-Ion Mobility-Orbitrap Mass Spectrometer: A Next-Generation Instrument for Native Mass Spectrometry. Anal Chem 2018, 90 (17), 10472–10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ibrahim YM; Garimella SV; Prost SA; Wojcik R; Norheim RV; Baker ES; Rusyn I; Smith RD, Development of an Ion Mobility Spectrometry-Orbitrap Mass Spectrometer Platform. Anal Chem 2016, 88 (24), 12152–12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benigni P; Thompson CJ; Ridgeway ME; Park MA; Fernandez-Lima F, Targeted High-Resolution Ion Mobility Separation Coupled to Ultrahigh-Resolution Mass Spectrometry of Endocrine Disruptors in Complex Mixtures. Anal Chem 2015, 87 (8), 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fort KL; van de Waterbeemd M; Boll D; Reinhardt-Szyba M; Belov ME; Sasaki E; Zschoche R; Hilvert D; Makarov AA; Heck AJR, Expanding the structural analysis capabilities on an Orbitrap-based mass spectrometer for large macromolecular complexes. Analyst 2017, 143 (1), 100–105. [DOI] [PubMed] [Google Scholar]
- 18.Q Exactive™ UHMR Hybrid Quadrupole-Orbitrap™ Mass Spectrometer. https://www.thermofisher.com/order/catalog/product/0726090#/0726090 (accessed 10/4/2021).
- 19.Agasid MT; Sorensen L; Urner LH; Yan J; Robinson CV, The Effects of Sodium Ions on Ligand Binding and Conformational States of G Protein-Coupled Receptors-Insights from Mass Spectrometry. J Am Chem Soc 2021, 143 (11), 4085–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Todd AR; Barnes LF; Young K; Zlotnick A; Jarrold MF, Higher Resolution Charge Detection Mass Spectrometry. Anal Chem 2020, 92 (16), 11357–11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller LM; Barnes LF; Raab SA; Draper BE; El-Baba TJ; Lutomski CA; Robinson CV; Clemmer DE; Jarrold MF, Heterogeneity of Glycan Processing on Trimeric SARS-CoV-2 Spike Protein Revealed by Charge Detection Mass Spectrometry. J Am Chem Soc 2021, 143 (10), 3959–3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Worner TP; Snijder J; Bennett A; Agbandje-McKenna M; Makarov AA; Heck AJR, Resolving heterogeneous macromolecular assemblies by Orbitrap-based single-particle charge detection mass spectrometry. Nat Methods 2020, 17 (4), 395–398. [DOI] [PubMed] [Google Scholar]
- 23.McLuckey SA, Principles of collisional activation in analytical mass spectrometry. J Am Soc Mass Spectr 1992, 3 (6), 599–614. [DOI] [PubMed] [Google Scholar]
- 24.Polfer NC; Bohrer BC; Plasencia MD; Paizs B; Clemmer DE, On the dynamics of fragment isomerization in collision-induced dissociation of peptides. J Phys Chem A 2008, 112 (6), 1286–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stiving AQ; VanAernum ZL; Busch F; Harvey SR; Sarni SH; Wysocki VH, Surface-Induced Dissociation: An Effective Method for Characterization of Protein Quaternary Structure. Anal Chem 2019, 91 (1), 190–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dixit SM; Polasky DA; Ruotolo BT, Collision induced unfolding of isolated proteins in the gas phase: past, present, and future. Current Opinion in Chemical Biology 2018, 42, 93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laganowsky A; Reading E; Allison TM; Ulmschneider MB; Degiacomi MT; Baldwin AJ; Robinson CV, Membrane proteins bind lipids selectively to modulate their structure and function. Nature 2014, 510 (7503), 172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fantin SM; Huang H; Sanders CR; Ruotolo BT, Collision-Induced Unfolding Differentiates Functional Variants of the KCNQ1 Voltage Sensor Domain. J Am Soc Mass Spectrom 2020, 31 (11), 2348–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang J; Halseth T; Vallejo D; Najafabadi ZI; Sen KI; Ford M; Ruotolo BT; Schwendeman A, Assessment of biosimilarity under native and heat-stressed conditions: rituximab, bevacizumab, and trastuzumab originators and biosimilars. Anal Bioanal Chem 2020, 412 (3), 763–775. [DOI] [PubMed] [Google Scholar]
- 30.Eschweiler JD; Martini RM; Ruotolo BT, Chemical Probes and Engineered Constructs Reveal a Detailed Unfolding Mechanism for a Solvent-Free Multidomain Protein. Journal of the American Chemical Society 2017, 139 (1), 534–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu FC; Ridgeway ME; Park MA; Bleiholder C, Tandem trapped ion mobility spectrometry. Analyst 2018, 143 (10), 2249–2258. [DOI] [PubMed] [Google Scholar]
- 32.Eldrid C; Ben-Younis A; Ujma J; Britt H; Cragnolini T; Kalfas S; Cooper-Shepherd D; Tomczyk N; Giles K; Morris M; Akter R; Raleigh D; Thalassinos K, Cyclic Ion Mobility-Collision Activation Experiments Elucidate Protein Behavior in the Gas Phase. J Am Soc Mass Spectrom 2021, 32 (6), 1545–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Beardsley RL; Jones CM; Galhena AS; Wysocki VH, Noncovalent protein tetramers and pentamers with “n” charges yield monomers with n/4 and n/5 charges. Anal Chem 2009, 81 (4), 1347–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sahasrabuddhe A; Hsia Y; Busch F; Sheffler W; King NP; Baker D; Wysocki VH, Confirmation of intersubunit connectivity and topology of designed protein complexes by native MS. Proceedings of the National Academy of Sciences 2018, 115 (6), 1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seffernick JT; Canfield SM; Harvey SR; Wysocki VH; Lindert S, Prediction of Protein Complex Structure Using Surface-Induced Dissociation and Cryo-Electron Microscopy. Anal Chem 2021, 93 (21), 7596–7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snyder DT; Panczyk EM; Somogyi A; Kaplan DA; Wysocki V, Simple and Minimally Invasive SID Devices for Native Mass Spectrometry. Anal Chem 2020, 92 (16), 11195–11203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zubarev RA; Kelleher NL; McLafferty FW, Electron Capture Dissociation of Multiply Charged Protein Cations. A Nonergodic Process. Journal of the American Chemical Society 1998, 120 (13), 3265–3266. [Google Scholar]
- 38.Syka JE; Coon JJ; Schroeder MJ; Shabanowitz J; Hunt DF, Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A 2004, 101 (26), 9528–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lermyte F; Valkenborg D; Loo JA; Sobott F, Radical solutions: Principles and application of electron-based dissociation in mass spectrometry-based analysis of protein structure. Mass Spectrometry Reviews 2018, 37 (6), 750–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams JP; Morrison LJ; Brown JM; Beckman JS; Voinov VG; Lermyte F, Top-Down Characterization of Denatured Proteins and Native Protein Complexes Using Electron Capture Dissociation Implemented within a Modified Ion Mobility-Mass Spectrometer. Anal Chem 2020, 92 (5), 3674–3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gadkari VV; Ramírez CR; Vallejo DD; Kurulugama RT; Fjeldsted JC; Ruotolo BT, Enhanced Collision Induced Unfolding and Electron Capture Dissociation of Native-like Protein Ions. Analytical Chemistry 2020, 92 (23), 15489–15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lermyte F; Everett J; Lam YPY; Wootton CA; Brooks J; Barrow MP; Telling ND; Sadler PJ; O’Connor PB; Collingwood JF, Metal Ion Binding to the Amyloid beta Monomer Studied by Native Top-Down FTICR Mass Spectrometry. J Am Soc Mass Spectrom 2019, 30 (10), 2123–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lam YPY; Wootton CA; Hands-Portman I; Wei J; Chiu CKC; Romero-Canelon I; Lermyte F; Barrow MP; O’Connor PB, Determination of the Aggregate Binding Site of Amyloid Protofibrils Using Electron Capture Dissociation Tandem Mass Spectrometry. J Am Soc Mass Spectrom 2020, 31 (2), 267–276. [DOI] [PubMed] [Google Scholar]
- 44.Wang H; Eschweiler J; Cui W; Zhang H; Frieden C; Ruotolo BT; Gross ML, Native Mass Spectrometry, Ion Mobility, Electron-Capture Dissociation, and Modeling Provide Structural Information for Gas-Phase Apolipoprotein E Oligomers. J Am Soc Mass Spectr 2019, 30 (5), 876–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song JH; Wagner ND; Yan J; Li J; Huang RY; Balog AJ; Newitt JA; Chen G; Gross ML, Native mass spectrometry and gas-phase fragmentation provide rapid and in-depth topological characterization of a PROTAC ternary complex. Cell Chem Biol 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou M; Liu W; Shaw JB, Charge Movement and Structural Changes in the Gas-Phase Unfolding of Multimeric Protein Complexes Captured by Native Top-Down Mass Spectrometry. Anal Chem 2020, 92 (2), 1788–1795. [DOI] [PubMed] [Google Scholar]
- 47.Brodbelt JS; Morrison LJ; Santos I, Ultraviolet Photodissociation Mass Spectrometry for Analysis of Biological Molecules. Chem Rev 2020, 120 (7), 3328–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Morrison LJ; Brodbelt JS, Charge site assignment in native proteins by ultraviolet photodissociation (UVPD) mass spectrometry. Analyst 2016, 141 (1), 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lermyte F; Theisen A; O’Connor PB, Solution Condition-Dependent Formation of Gas-Phase Protomers of Alpha-Synuclein in Electrospray Ionization. J Am Soc Mass Spectrom 2021, 32 (1), 364–372. [DOI] [PubMed] [Google Scholar]
- 50.Mehaffey MR; Lee J; Jung J; Lanzillotti MB; Escobar EE; Morgenstern KR; Georgiou G; Brodbelt JS, Mapping a Conformational Epitope of Hemagglutinin A Using Native Mass Spectrometry and Ultraviolet Photodissociation. Anal Chem 2020, 92 (17), 11869–11878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greisch JF; Tamara S; Scheltema RA; Maxwell HWR; Fagerlund RD; Fineran PC; Tetter S; Hilvert D; Heck AJR, Expanding the mass range for UVPD-based native top-down mass spectrometry. Chem Sci 2019, 10 (30), 7163–7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sipe SN; Patrick JW; Laganowsky A; Brodbelt JS, Enhanced Characterization of Membrane Protein Complexes by Ultraviolet Photodissociation Mass Spectrometry. Anal Chem 2020, 92 (1), 899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foreman DJ; McLuckey SA, Recent Developments in Gas-Phase Ion/Ion Reactions for Analytical Mass Spectrometry. Anal Chem 2020, 92 (1), 252–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sanguantrakun N; Chanthamontri C; Gross ML, Top-Down Analysis of In-Source HDX of Native Protein Ions. J Am Soc Mass Spectrom 2020, 31 (5), 1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheung See Kit M; Carvalho VV; Vilseck JZ; Webb IK, Gas-Phase Ion/Ion Chemistry for Structurally Sensitive Probes of Gaseous Protein Ion Structure: Electrostatic and Electrostatic to Covalent Cross-Linking. Int J Mass Spectrom 2021, 463, 116549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carvalho VV; See Kit MC; Webb IK, Ion Mobility and Gas-Phase Covalent Labeling Study of the Structure and Reactivity of Gaseous Ubiquitin Ions Electrosprayed from Aqueous and Denaturing Solutions. J Am Soc Mass Spectr 2020, 31 (5), 1037–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheung See Kit M; Shepherd SO; Prell JS; Webb IK, Experimental Determination of Activation Energies for Covalent Bond Formation via Ion/Ion Reactions and Competing Processes. J Am Soc Mass Spectr 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]


