Abstract
BACKGROUND
Severe acute respiratory syndrome coronavirus (SARS-CoV-2) omicron variant was first detected in South Africa in November 2021. Since then, the number of cases due to this variant increases enormously every day in different parts of the world. Mutations within omicron genome may impair the molecular detection resulting in false negative results during Coronavirus disease 19 (COVID-19) diagnosis.
OBJECTIVES
To verify if colorimetric reverse transcription loop-mediated isothermal amplification (RT-LAMP) targeting N and E genes would work efficiently to detect omicron SARS-CoV-2 variant and its sub-lineages.
METHODS
SARS-CoV-2 reverse transcription quantitative polymerase chain reaction (RT-qPCR) positive samples were sequenced by next generation DNA sequencing. The consensus sequences generated were submitted to Pangolin tool for SARS-CoV-2 lineage identification. RT-LAMP reactions were performed at 65ºC/30 min targeting N and E.
FINDINGS
SARS-CoV-2 omicron can be detected by RT-LAMP targeting N and E genes despite the genomic mutation of this more transmissible lineage. Omicron SARS-CoV-2 sub-lineages were tested and efficiently detected by RT-LAMP. We demonstrated that this test is very sensitive in detecting omicron variant, with LoD as low as 0.4 copies/µL.
MAIN CONCLUSIONS
Molecular detection of omicron SARS-CoV-2 variant and its sub-lineages can be achieved by RT-LAMP despite the genomic mutations as a very sensitive surveillance tool for COVID-19 molecular diagnosis.
Key words: RT-LAMP, omicron, SARS-CoV-2, mutations, N and E genes
On November 24, 2021 the severe acute respiratory syndrome coronavirus (SARS-CoV-2), PANGO lineage B.1.1.529, was first reported in Botswana and South Africa, pummeling the world by the beginning of December 2021. The World Health Organization (WHO) named B.1.1.529 as omicron and classified as the fifth variant of concern (VOC). 1 Omicron has mutations in genomic coding regions to structural (envelope, nucleocapsid, membrane, spike) and nonstructural proteins that can impair Coronavirus disease (COVID-19) molecular diagnosis based on reverse transcription quantitative polymerase chain reaction (RT-qPCR) tests, leading to misdiagnosis and allowing virus spreading through false negative results. 1 , 2 , 3 As an alternative to RT-qPCR, reverse transcription loop-mediated isothermal amplification (RT-LAMP) has been used with success to detect SARS-CoV-2 RNA from nasopharyngeal swabs and saliva samples. 4 During RT-LAMP reaction, the amplification of genetic material occurs at constant temperature (65ºC) without the need for sophisticated thermal cyclers as in RT-qPCR. 5 Furthermore, the colorimetric reaction allows the result to be interpreted faster than RT-qPCR, since the amplified products can be visually detected due to pH-dependent sensors that changes reaction color from pink to yellow (positive result). 6 Previous results obtained by our group, showed that RT-LAMP reaction targeting SARS-CoV-2 N and E genes, can be equivalent to the gold standard RT-qPCR using nasopharyngeal swabs samples. 4 Given the fact that genomic mutations in SARS-CoV-2 omicron VOC are also present in N and E coding sequences, the main goal of this work was to verify whether RT-LAMP reaction targeting N and E genes (in a multiplex or singleplex manner), would works to detect the virus reinforcing the use of it as an affordable and robust COVID-19 diagnostic tool.
MATERIALS AND METHODS
Clinical samples obtained from nasopharyngeal swabs were collected in different health units in the municipality of Belo Horizonte and its surrounding metropolitan region, Brazil. Molecular diagnosis using RT-qPCR (Allplex™ SARS-CoV-2 - Seegene, South Korea) - which targets the E, S/RdRp and N genes of SARS-CoV-2 - was performed in the municipal molecular biology laboratory of Belo Horizonte city hall. Twenty positive samples were selected for DNA sequencing and RT-LAMP analysis. For next generation DNA sequencing, the libraries were constructed using Illumina COVIDSeq™ Test (Illumina Inc, San Diego, CA, USA), according to manufacturer’s instructions, and the samples were sequenced through Illumina MiSeq Platform. The trimming tool used in the raw reads was the Trimmomatic 7 with a sliding window of four nucleotides with an average Phred score of 20, and filtered reads smaller than 50 bp were removed. The trimmed reads were aligned to the SARS-CoV-2 reference genome (NC_045512) using BWA 8 with the default parameters. The nucleotide variants were detected using iVar package, 9 with a minimum frequency of 50% and depth of 30 reads. To identify the SARS-CoV-2 lineage and sub-lineages, the consensus sequences generated were submitted to Pangolin tool v3.1.17 (http://pangolin.cog-uk.io/). 10 All sample sequences were deposited at the European Nucleotide Archive, under project number PRJEB49204, and at GISAID Initiative [Supplementary data (181.4KB, pdf) (Table)]. The RT-LAMP test was performed in a singleplex way with the set of primers targeting independently N (primer set N2) and E (primer set E1) genes or multiplexed (combined primer sets N2/E1). Detailed conditions of RT-LAMP reaction, including primer sequences, are described in our previous work. 4 Briefly, reactions were performed at 65ºC, during 30 min, using WarmStart® Colorimetric LAMP 2× Master Mix (New England Biolabs #M1804). To validate the visual colorimetric output, samples resolved in 2% agarose gel stained with DNA intercalator GelRed® (Biotium #41003), confirming the specific amplification when compared to the positive control (RNA of inactivated parental lineage B of SARS-CoV-2 extracted from the supernatant infected Vero E6 cells). The test of the limit of detection (LoD) was performed by RT-LAMP using as input a serial dilution of one of the omicron samples, and the absolute quantification was conducted based on a standard curve prepared using the template SARS-CoV-2 E gene-harboring plasmid (2 × 105 copies/µL; Biogene COVID-19 PCR, Bioclin/Quibasa #K228-1; Lot: 0007). More details of the curve and how the LoD was performed are described in our previous work. 4 Sequencing alignments were performed on SnapGene using embedded multiple alignment Clustal Omega tool. NCBI NC_045512 was included as SARS-CoV-2 reference genomic sequence.
Ethics statement - The studies involving human participants were reviewed and approved by Research Ethics Committee involving human beings at Instituto René Rachou, Fundação Oswaldo Cruz, under license protocol number: 4084902 and CAAE (certificate of presentation for ethical appreciation): 31984720300005091.
RESULTS
Aiming to give experimental evidence that RT-LAMP tests using multiplex strategy (N2/E1) or N2 and E1 primer sets alone are able to detect B.1.1.529 SARS-CoV-2 lineage, we performed the RT-LAMP tests using RT-qPCR positive samples that were confirmed as omicron VOC by whole genome sequencing. Through sequencing, these samples were classified in Omicron sub-lineages (ten BA.1.14, four BA.1.15, two BA.2.10, two BA.2.9, one BA.1.13 and one BA.1.18). The ten BA.1.14 were positive in RT-LAMP when the target was multiplex N/E or the E e N separately (singleplex) turning the original pink color into yellow, as the colorimetric evidence for specific DNA amplification (Fig. 1). Since there was no difference between multiplex or singleplex strategy we performed the multiplex N/E RT-LAMP for the other sub-lineages samples. As demonstrated in Fig. 2, all tubes became yellow after the reaction. The LoD test demonstrated that RT-LAMP using the N/E multiplex is efficient for detection of Omicron samples containing as low as 0.4 copies/µL (Fig. 2B). In summary, either RT-LAMP strategies (multiplex or not) worked to successfully detect omicron RNA. The omicron-derived genomic sequences were aligned against the reference SARS-CoV-2 genome together with RT-LAMP primer sets targeting N and E genes. None of the mutations present in omicron affects N2 primer recognition, since the target region do not overlaps with any of the six nonsynonymous mutations in N gene (Fig. 1). By contrast, the only mutation present in E gene region - a single nucleotide polymorphism (SNP) resulting in a C-to-U transition at position 26270 (C26270U), that yields amino acid change T9I - overlaps with E1 forward inner primer F2 part, apparently without any impact on primer recognition and specific amplification by RT-LAMP (Fig. 1).
Fig. 1: colorimetric reverse transcription loop-mediated isothermal amplification (RT-LAMP) targeting N and E genes allows the detection of severe acute respiratory syndrome coronavirus (SARS-CoV-2) variant omicron. Selected nasopharyngeal swab-derived samples, previously detected positive for SARS-CoV-2 by reverse transcription quantitative polymerase chain reaction (RT-qPCR) and identified as the new omicron variant of concern (VOC) by next generation sequencing, were included as input for colorimetric RT-LAMP reaction targeting N and E genes. RT-LAMP reactions were performed combining the primer sets N2/E1 in the same reaction (multiplex) or independently.(4) Both strategies resulted in positive reaction as evidenced by yellow colour. Negative or absence of specific amplification is represented by pink colour and no bands on gel electrophoresis. RT-LAMP reaction was performed at 65ºC during 30 min, using the WarmStart® colorimetric LAMP 2X master mix (NEB #M1804). RT-LAMP amplification products were resolved in 2% agarose gel and stained with GelRed® (Biotium #41003) to confirm DNA amplification. +C: positive control using SARS-CoV-2 RNA extracted from laboratory-cultured inactivated SARS-CoV-2; NTC: non-template control. Upper panel cartoon is showing SARS-CoV-2 genome where N and E gene regions are zoomed in. Polymorphisms are indicated in their corresponding positions. The SNP C26270U, highlighted by the red arrow, is the only polymorphism that overlaps with F2 primer (magnifying glass). F3: forward; B3: backward; LF: loop forward; LB: loop backward; F2 and F1c: parts of FIP - forward internal primer; B1 and B2c: parts of BIP - backward inner primer. P: proline; L: leucine; R: arginine; K: lysine; G: glycine; S: serine; T: threonine; I: isoleucine; E: glutamic acid. BA.2, also known as 21L, is considered a sub-lineage of SARS-CoV-2 omicron. Genomic representation was created using SnapGene. Parts of this figure were created with BioRender.com and are licensed under the agreement number: QC23MCKTFE.
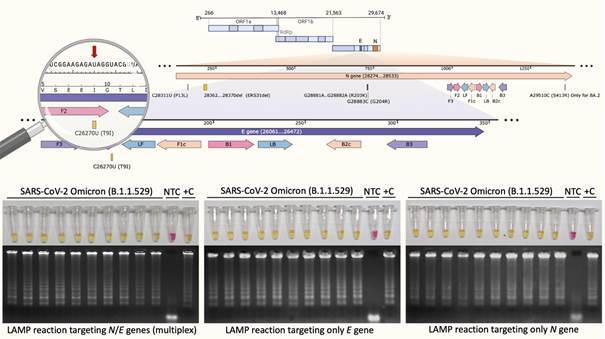
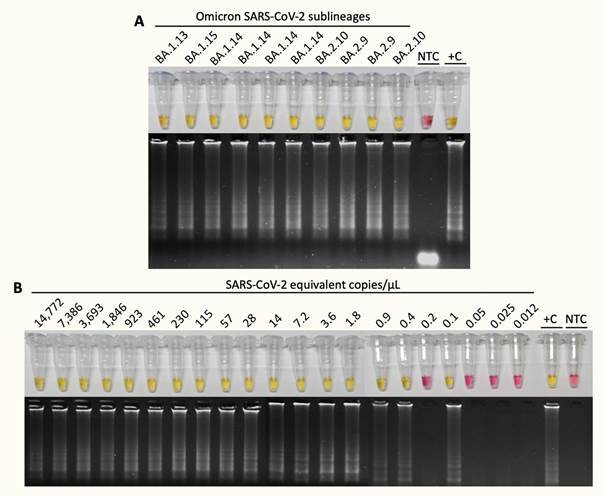
Fig. 2: reverse transcription loop-mediated isothermal amplification (RT-LAMP) of Omicron variant sub-lineages. (A) The test was performed using multiplex N/E strategy. All samples were positive (yellow colour) after 30 min reaction. RT-LAMP amplification products were resolved in 2% agarose gel and stained with GelRed® (Biotium #41003) to confirm DNA amplification. +C: positive control using severe acute respiratory syndrome coronavirus (SARS-CoV-2) RNA extracted from laboratory-cultured inactivated SARS-CoV-2; NTC: non-template control. From the left to the right hand side, clinical samples corresponds to Supplementary data (181.4KB, pdf) (Table samples 11 to 20. (B) Limit of detection (LoD) of Omicron samples. The test was performed using as input a serial dilution of the sample 3 (randomly chosen). The absolute quantification was performed based on a standard curve prepared using the template SARS-CoV-2 E gene-harboring plasmid 2 × 105 copies/µL; Biogene Coronavirus disease 19 (COVID-19) polymerase chain reaction (PCR), Bioclin/Quibasa #K228-1; Lot: 0007. The equation of a straight line is y = -3,476x+38,063, where Y is the Ct for E gene of the sample 3 in reverse transcription quantitative polymerase chain reaction (RT-qPCR) and X is the value of quantification. RT-LAMP amplification products were resolved in 2% agarose gel and stained with GelRed® (Biotium #41003) to confirm DNA amplification. +C: positive control using SARS-CoV-2 RNA extracted from laboratory-cultured inactivated SARS-CoV-2; NTC: non-template control. Figure representative of independent assays and signs of cutted and overlapping images can be seen for this reason. Original raw material can be send upon request.
DISCUSSION
The circulation of new SARS-CoV-2 variants is challenging for diagnostic tests since these strains accumulate mutations that can affect primer or probe recognition, leading to false negative results in standard RT-qPCR diagnostic tests. Indeed, molecular diagnostic test sensitivity is intrinsically related to the target choice. Mismatches could affect reaction’s efficiency, especially if it occurs at the 3’-end of a primer. 11 Studies demonstrated that RT-qPCR can fail in detecting omicron VOC, yielding false negative results. 2 , 12 , 13 Lesbon et al. 2 reported that high viral load samples (Ct < 33) could be detected when targeting E and RdRp genes but failed when considering N gene as target, even the latter being considered the most sensitive region for RT-qPCR. 2 In addition, the mutation C26340U at E gene was associated with a failure on detecting SARS-CoV-2 by RT-qPCR, 13 compromising RT-qPCR-based SARS-CoV-2 detection kits such as GeneFinderTM COVID-19 Plus RealAmp Kit and cobas® SARS-CoV-2 test. 2 , 12 , 13 These evidences lead us to wonder if omicron genomic polymorphisms would affect molecular detection by RT-LAMP. Until now, there is no evidence that mutations in SARS-CoV-2 N or E genes would affect omicron detection by RT-LAMP. Indeed, here we report that the only detected potential mismatch in E gene (mutation C26270U) does not impair specific recognition by E1 primer set, resulting in successful target amplification. Concerning mutations in N gene, Bei et al. 14 reported that the presence of C28311U SNP, that overlaps N1 probe, does not impact omicron detection by CDC 2019-nCoV_N1 primer-probe set from the US Centre for Disease Control and Prevention (CDC) RT-qPCR-based SARS-CoV-2 detection kit. 14 None of the mutations present in N gene perturb RT-LAMP based omicron detection, since N2 primer set is based in a conserved region. It is worth to note that SNPs C28311U, G28881A, G28882A, G28883C in N gene are ancestral and present in alpha (B1.1.7) and Gamma (P.1) SARS-CoV-2 VOCs. 15 , 16 The latter are derive from a trinucleotide substitution GGG > AAC yielding nucleocapsid mutation R203K and G204R, linked to increased: (I) RNA expression 17 and (II) viral load 18 (https://covariants.org/variants/21K.Omicron and https://covariants.org/variants/21L.Omicron).
The LoD test demonstrated that RT-LAMP using the N/E multiplex detected at least 0.4 copies/µL of a Omicron sample. Moreover, the RT-LAMP was efficient when six different sub-lineages of Omicron variant were used as input. In this regard, RT-LAMP works as a highly sensitive and specific molecular test to detect SARS-CoV-2 variants. 4 This work highlights the importance of RT-qPCR constant monitoring to detect current circulating SARS-CoV-2 and reinforces RT-LAMP as a robust alternative for massive COVID-19 surveillance.
ACKNOWLEDGEMENTS
To Dr Anna Christina de Matos Salim, Dr Flávio Marcos Gomes de Araújo and Dr Gabriel da Rocha Fernandes - Instituto René Rachou, Fundação Oswaldo Cruz, Belo Horizonte, Brazil, for their help on constructing libraries, performing next generation sequencing and the identification of omicron VOC. We thank the Technological Platform Network from Fundação Oswaldo Cruz, for providing the infrastructure needed for SARS-CoV-2 genomic surveillance. We thank André Luiz de Menezes and Dr Eneida Santos de Oliveira - Molecular Biology Laboratory - Belo Horizonte City Hall, Brazil, for providing the samples used in this study. We are grateful to Cristiane P Gomes and Patrícia PN Miranda for resources management and excellent technical assistance.
Footnotes
Financial support: FAPEMIG (grant number #APQ-00485-20, to RMN); grant number #APQ-00262-20 and Fundação Oswaldo Cruz - Inova Fiocruz Program - Innovative Products (grant number VPPIS-004-FIO-18-51) to RMN; Innovative Products to face COVID-19 pandemics (grant number VPPIS-005-FIO-20-2-45) to PA and from the MCTI - Brazilian Ministry of Science, Technology and Innovation, through the “Rede Virus” initiative to PA (grant number - FINEP 01.20.0005.00). LTA is a post-doctoral fellow from MCTI - CNPq - DTI-A; ABG holds a fellowship from Human Resources Fixation Program (CNPq - C); RLMN research is supported by FAPEMIG, grant PPM-00699-18; RLMN is CNPq research productivity fellow (grant number CNPq 312965/2020-6).
REFERENCES
- 1.Classification of Omicron 2022. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern
- 2.Lesbon JCC, Poleti MD, Oliveira ECM, Patané JSL, Clemente LG, Viala VL. Nucleocapsid (N) gene mutations of SARS-CoV-2 can affect real-time RT-PCR diagnostic and impact false-negative results. Viruses. 2021;13(12):2474–2474. doi: 10.3390/v13122474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxena SK, Kumar S, Ansari S, Paweska JT, Maurya VK, Tripathi AK. Characterization of the novel SARS-CoV-2 Omicron (B 1.1.529) variant of concern and its global perspective. J Med Virol. 2022;94(4):1738–1744. doi: 10.1002/jmv.27524. [DOI] [PubMed] [Google Scholar]
- 4.Alves PA, de Oliveira EG, Franco-Luiz APM, Almeida LT, Gonçalves AB, Borges IA. Optimization and clinical validation of colorimetric reverse transcription loop-mediated isothermal amplification, a fast, highly sensitive and specific COVID-19 molecular diagnostic tool that is robust to detect SARS-CoV-2 variants of concern. Front Microbiol. 2021;12:713713–713713. doi: 10.3389/fmicb.2021.713713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y, Odiwuor N, Xiong J, Sun L, Nyaruaba R, Wei H, et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric. LAMP. 2020 https://europepmc.org/article/ppr/ppr115157 [Google Scholar]
- 6.Tanner NA, Zhang Y, Evans TC. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. Biotechniques. 2015;58(2):59–68. doi: 10.2144/000114253. [DOI] [PubMed] [Google Scholar]
- 7.Bolger AM, Lohse M, Usadel B. Trimmomatic a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grubaugh ND, Gangavarapu K, Quick J, Matteson NL, De Jesus JG, Main BJ. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 2019;20(1):8–8. doi: 10.1186/s13059-018-1618-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Toole Á.Scher E.Underwood A.Jackson B.Hill V.McCrone JT Assignment of epidemiological lineages in an emerging pandemic using the pangolin tool. Virus Evol. 2021;7(2):veab064–veab064. doi: 10.1093/ve/veab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allawi HT, SantaLucia J. Thermodynamics and NMR of internal G·T mismatches in DNA. Biochemistry. 1997;36(34):10581–10594. doi: 10.1021/bi962590c. [DOI] [PubMed] [Google Scholar]
- 12.Metzger CMJA, Lienhard R, Seth-Smith HMB, Roloff T, Wegner F, Sieber J. PCR performance in the SARS-CoV-2 omicron variant of concern. Swiss Med Wkly. 2021;151:w30120–w30120. doi: 10.4414/smw.2021.w30120. [DOI] [PubMed] [Google Scholar]
- 13.Artesi M, Bontems S, Göbbels P, Franckh M, Maes P, Boreux R, et al. A recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the E gene quantitative reverse transcription-PCR utilized in a commercial dual-target diagnostic assay. J Clin Microbiol. 2020 doi: 10.1128/JCM.01598-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bei Y, Vrtis KB, Borgaro JG, Langhorst BW, Nichols NM. The omicron variant mutation at position 28,311 in the SARS-CoV-2 N gene does not perturb CDC N1 target detection. medRxiv. 2021 doi: 10.1101/2021.12.16.21267734. [DOI] [Google Scholar]
- 15.Moreira FRR, Bonfim DM, Geddes VEV, Zauli DAG, Silva JP, Lima AB, et al. Increasing frequency of SARS-CoV-2 lineages B.1.1.7, P.1 and P.2 and identification of a novel lineage harboring E484Q and N501T spike mutations in Minas Gerais, Southeast Brazil - SARS-CoV-2 coronavirus [Internet]. Virological. 2021 https://virological.org/t/increasing-frequency-of-sars-cov-2-lineages-b-1-1-7-p-1-and-p-2-and-identification-of-a-novel-lineage-harboring-e484q-and-n501t-spike-mutations-in-minas-gerais-southeast-brazil/676 [Google Scholar]
- 16.Gallaher WR. Omicron is a multiply recombinant set of variants that Have evolved over many months - SARS-CoV-2 coronavirus / SARS-CoV-2 molecular evolution [Internet] Virological. 2021 https://virological.org/t/omicron-is-a-multiply-recombinant-set-of-variants-that-have-evolved-over-many-months/775 [Google Scholar]
- 17.Leary S, Gaudieri S, Parker MD, Chopra A, James I, Pakala S, et al. Generation of a novel SARS-CoV-2 sub-genomic RNA due to the R203K/G204R variant in nucleocapsid: homologous recombination has potential to change SARS-CoV-2 at both protein and RNA level. bioRxiv. 2021 doi: 10.20411/pai.v6i2.460. https://www.biorxiv.org/content/10.1101/2020.04.10.029454v4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mourier T, Shuaib M, Hala S, Mfarrej S, Alofi F, Naeem R, et al. Saudi Arabian SARS-CoV-2 genomes implicate a mutant Nucleocapsid protein in modulating host interactions and increased viral load in COVID-19 patients. medRxiv. 2021 https://www.medrxiv.org/content/10.1101/2021.05.06.21256706v2 [Google Scholar]


