Abstract
We cloned an intact copy of a long terminal repeat retroelement designated marY1 from the ectomycorrhizal basidiomycete Tricholoma matsutake. The reverse transcriptase domain is found in T. matsutake and Tricholoma magnivelare worldwide. This finding suggests that retroelements associate with ectomycorrhizal basidiomycetes and may be useful as genetic markers for identification, phylogenetic analysis, and mutagenesis of this fungal group.
Retroelements are retrovirus-like DNA elements found in eukaryotic genomes (3, 4, 16) which may replicate through an RNA intermediate and are often found at multiple dispersed locations within the genome (2, 3, 4, 13, 15, 16). Retroelements have been recognized in some ascomycetes as genetic markers (1, 6, 7, 14, 17).
Matsutake and American matsutake are economically important edible mushrooms produced by the ectomycorrhizal basidiomycetes Tricholoma matsutake and Tricholoma magnivelare, respectively (12). They form mycorrhizae with some Pinaceae plants (12). We previously identified a 967-bp sequence encoding part of a retroelement polyprotein in the genome of T. matsutake strain Y1 and subsequently found the RNase H domain in T. matsutake and T. magnivelare (18). Our objective in this study was to isolate and characterize a full-length copy of this retroelement from T. matsutake. This retroelement will be used to develop new means of identification, phylogenetic analysis, and mutagenesis of ectomycorrhizal basidiomycetes.
Cloning of a full-length copy of the retroelement.
A genomic library of T. matsutake strain Y1 constructed in λEMBL3 (Stratagene, La Jolla, Calif.) was probed with a 550-bp BamHI-XbaI fragment from the 967-bp multiple-copy sequence previously cloned in plasmid pHHM145 (Fig. 1) (18, 21). The probe DNA was labeled with [α-32P]dCTP by the random primer extension (New England Nuclear, Boston, Mass.). Of approximately 3,000 plaques, 14 hybridized with the probe. DNA was isolated from all the positive λEMBL3 clones, digested with restriction endonucleases, transferred to a nylon membrane, and hybridized with the [α-32P]dCTP-labeled 550-bp BamHI-XbaI fragment (18, 21). Southern hybridizations identified a clone, λA2, which contains the homologue in 2.8-kb BamHI, 9.2-kb EcoRI, and 8.5-kb KpnI fragments whose sizes correspond to those previously identified in genomic Southern hybridizations with strains of T. matsutake (18). Therefore, we hypothesized that λA2 contained a full-length copy of the retroelement.
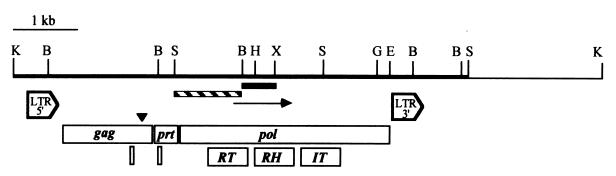
FIG. 1.
Characteristics of marY1 cloned into λA2. Insert DNA is represented by a solid black line. The thin line represents adjacent λEMBL3 vector DNA. Restriction sites of BamHI (B), BglII (G), EcoRI (E), HindIII (H), KpnI (K), SalI (S), and XhoI (X) are given. The arrow denotes the direction of marY1 and the location of the fragment previously cloned in pHHM145 (18). The solid black bar and the hatched bar underneath the insert DNA indicate probes for the plaque hybridizations used to identify λA2 and Southern hybridizations to determine the distribution of the marY1 homologues in fungal strains, respectively. Domains corresponding to 5′ LTR, 3′ LTR, gag, prt, and pol are given. The location of a putative ribosomal frameshifting site is indicated by the arrowhead. A zinc-finger DNA-binding site in the gag gene product, a consensus catalytic site in prt, and domains of reverse transcriptase (RT), RNase H (RH), and integrase (IT) are aligned at the bottom.
The 8.5-kb KpnI fragment from λA2 contains the 2.8-kb BamHI fragment homologous to the 967-bp multiple-copy sequence and is large enough to encode a retroelement of 5 to 8 kb. The KpnI fragment was digested with BglII to generate 5.5-kb and 3.0-kb KpnI-BglII fragments to increase the ligation efficiency and to orient the fragment in the vector. These fragments were ligated into the KpnI-BamHI site of pBluescript SK+ (Stratagene, La Jolla, Calif.), generating plasmids pHHM147 and pHHM148, respectively.
Identification of the LTR retroelement marY1.
The inserts of pHHM147 and pHHM148 were sequenced with an ABI prism 377 autosequencer (Perkin-Elmer Japan, Urayasu, Japan). Data were analyzed using the computer software GENETIX-Mac ver. 9.0 (Software Development Co., Tokyo, Japan), Clustal W (23), and Advanced BLAST Search provided by the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov /BLAST). Unless stated otherwise, the position of a sequence is expressed as the number of base pairs from the 5′ end of marY1 (marY1 = a retroelement of T. matsutake strain Y1 [see below]).
The 6,046-bp element carries identical 425-bp long terminal direct repeats (LTRs) (Fig. 1). The LTRs contain the terminal sequences of 5′-TG…TA-3′ instead of 5′-TG…CA-3′, which are present in the LTRs of many, but not all, retroelements (3, 4). The LTR also has several conserved sequences, including a 5-bp direct repeat of ATGTT immediately outside the 5′ and the 3′ LTRs. This apparent target site duplication is a typical feature generated during the retrotransposon integration process (13, 16). In addition, the AT-rich target site duplication associated with marY1 is consistent with those reported for many retrotransposon insertion sites in other organisms (3, 16).
The 5′ LTR of marY1 has three CT-rich sequences, i.e., a 33-bp stretch starting at bp 77, a 25-bp stretch at bp 231, and a 43-bp stretch at bp 318. The third CT-rich sequence may correspond to the ct box immediately upstream of the transcription initiation site in the Aspergillus nidulans gpdA gene promoter (20). This conclusion is based on the position of the third CT-rich sequence relative to that of the TTCCA sequence at bp 104, which is identical to the enhancer of Ty1 of Saccharomyces cerevisiae (25) and the qa-2 gene of Neurospora crassa (9). Like the ct box of A. nidulans gpdA (20), the third CT-rich sequence of marY1 associates with a stretch of direct repeat. The occurrence of CT-rich sequence in the 5′ LTR was also reported in Grasshopper, an LTR retroelement of Magnaporthe grisea (6). Upstream and downstream of the TTCCA sequence, we identified a sequence similar to a CCAAT-like transcription activation signal and two putative TATA domains (4, 24). A GC-rich sequence upstream of the putative CCAAT-like signal is similar to the GC-rich box required for DNA binding by some transcription factors (24). We did not identify a potential tRNA primer binding site for minus-strand DNA synthesis, which is generally located immediately after the 3′ end of the 5′ LTR, but we did find a 13-bp purine-rich sequence, which could be the primer binding site for plus-strand DNA synthesis, immediately upstream of the 5′ end of the 3′ LTR (3, 24).
Characterization of ORFs in marY1.
Three open reading frames (ORFs) occur on the same strand in the same direction in marY1 (Fig. 1). The first ORF begins at the ATG codon at bp 592 to 594 and continues to a TGA stop codon at bp 1708 to 1710 (Fig. 1). This ORF is predicted to encode a protein with 352 amino acids that is 41 and 39% similar to the amino acid sequences of the putative gag gene products of skippy, which is a retroelement of Fusarium oxysporum, and MAGGY of M. grisea, respectively (1, 7). The putative protein of marY1 also was similar to the gag gene products of a number of other retrotransposons and retroviruses. This protein should have a zinc-finger DNA-binding domain (C-X2-C-X9-C) at amino acid positions 304 to 321 (5). The amino acid sequence of the putative zinc-finger DNA-binding domain encoded in marY1 is 78% similar to that of ovine pulmonary carcinoma virus (10). The molecular mass is predicted to be 42 kDa, which is smaller than most gag gene products, i.e., 60 to 80 kDa (24). However, a putative ribosomal frameshift site occurs at the 3′ end of gag (Fig. 1). If the TGA at bp 1708 to 1710 is bypassed by a ribosomal frameshift at bp 1691 to 1692, then the predicted molecular mass is 63.6 kDa and it would be a gag-prt fusion. Such polyproteins are known in some retroviruses, such as human immunodeficiency virus, mouse mammary tumor virus, and human T-cell leukemia virus (24).
A second ORF extends from an ATG start codon at bp 1962 to 1964 to a TGA stop codon at bp 2292 to 2294 (Fig. 1). This ORF is predicted to encode a peptide of 111 amino acid residues with a molecular mass of 12.2 kDa. It is similar in size to the acid protease of retroviruses (24) and has a conserved catalytic site that is similar to those of other acid proteases (24). The amino acid sequence of a putative acid protease encoded in marY1 is 52 and 49% similar to those of skippy and MAGGY, respectively (1, 7).
The third ORF begins at the ATG codon at bp 2436, ends at the TAG codon at bp 5609, and is predicted to encode a protein product of 1,057 amino acid residues with a molecular mass of 122.2 kDa (Fig. 1). The putative amino acid sequence is similar to those of the pol gene products of a number of retroviruses and retrotransposons and is particularly similar to the putative reverse transcriptase, RNase H, and integrase domains of MAGGY of M. grisea (59% similarity), skippy of F. oxysporum (54% similarity), and CfT-I of Cladosporium fulvum (54% similarity) (1, 7, 17). Of those, the domain V of reverse transcriptase in marY1 is most conserved, and has a perfect match with the YXDD sequence (where X is unknown) of the proposed reverse transcriptase active site (24).
In general, LTR retroelements are classified into two groups (3, 4, 15). Those in the gypsy group have two or three ORFs, the first encoding gag, the second pol, and the third env, and are regarded as a group closely related to retroviruses (3, 13, 15). Although pol generally encodes the protease gene (prt) in retrotransposons (3, 4, 13, 15), prt in some retroviruses occurs as the second ORF prior to the ORF of pol (24). These ORFs are transcribed into a polycistronic mRNA and translated into several polyproteins, which often require ribosomal frameshifting, nonsense suppression, and/or splicing to bypass stop codons located between the ORFs, especially the one associated with the 3′ end of gag (13, 24). The pol gene in gypsy elements generally encodes domains in the following order: prt-reverse transcriptase-RNase H-integrase (2, 3, 15). The other group of LTR retroelements is the copia group, which features a single ORF encoding domains in the following order: gag-prt-integrase-reverse transcriptase-RNase H (2, 3, 15). While a significant number of copia-type retroelements are recognized in many plant species (2, 11, 15), all LTR retroelements identified so far in plant-infecting fungi belong to the gypsy group (1, 6, 7, 17). marY1, the first reported retroelement in an ectomycorrhizal basidiomycete, also belongs to the gypsy group of LTR retroelements.
Conservation of marY1 in the genome of T. matsutake.
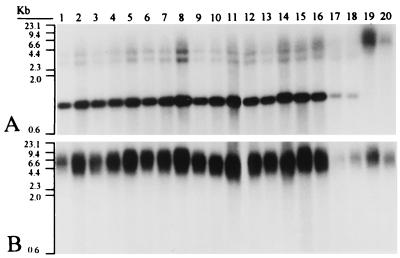
To determine whether marY1 is ubiquitous in T. matsutake and T. magnivelare, we hybridized the [α-32P]dCTP-labeled 1.1-kb SalI-BamHI fragment, which contains the reverse transcriptase domain (Fig. 1), to genomic digests of 18 T. matsutake strains and two T. magnivelare strains from Canada (Fig. 2) (18). Hybridization was detected in all 20 strains (Fig. 2). A reverse transcriptase domain was found on a 1.2-kb BamHI fragment in T. matsutake and on a 5.7-kb BamHI fragment in T. magnivelare (Fig. 2). In addition, two larger BamHI fragments that hybridized weakly with the probe also were present in all samples of T. matsutake (Fig. 2). The length of the EcoRI fragment carrying the homologue of the probe, however, was apparently conserved in both T. matsutake and T. magnivelare (Fig. 2).
FIG. 2.
Southern hybridization analysis of genomic digests of T. matsutake and T. magnivelare probed with the reverse transcriptase domain of marY1. (A) BamHI digests. (B) EcoRI digests. Lanes 1 to 18, T. matsutake (18): lane 1, Y1 (from Ibaraki, eastern Honshu, the main island of Japan); lane 2, Y59 (from Yamaguchi, far western Honshu); lane 3, OK-T4 (from Okayama, western Honshu); lane 4, MR32 (from Hyogo, western Honshu); lane 5, TM-8 (from Kyoto, western Honshu); lane 6, TM15 (from Iwate, northern Honshu); lane 7, Tm-H507 (from Hiroshima, far western Honshu); lane 8, TMT4 (from Tokushima, Shikoku island of Japan); lane 9, Tm31 (from Kyongsang North, Republic of Korea); lane 10, K1 (from the Republic of Korea through a retailer); lane 11, NK1 (from Democratic People's Republic of Korea through a retailer); lane 12, TM-9 (from People's Republic of China through a retailer); lane 13, CHI1 (from People's Republic of China through a retailer); lane 14, BH1 (from Kingdom of Bhutan through a retailer); lane 15, TM-5 (from Kingdom of Morocco through a retailer); lane 16, MC1 (from Kingdom of Morocco through a retailer); lane 17, TM-4 (from Mexico through a retailer); lane 18, MX1 (from Mexico through a retailer). Lanes 19 to 20, T. magnivelare (18): lane 19, CA1 (from Canada through a retailer); lane 20, Tp-C3 (from Canada through a retailer). Molecular markers (kb) are indicated at the left.
We also attempted to hybridize the probe carrying the 1.1-kb SalI-BamHI fragment to genomic digests of Suillus bovinus strain MR16 (18) and Rhizopogon rubescens strain MR11 (18), fungal species that have been well characterized in terms of ectomycorrhizal symbiosis in Pinus sp. plants (22). However, we could not detect any homologues of a reverse transcriptase domain encoded in marY1.
Retroelements have been recognized in a wide range of eukaryotes and, in some cases, used as genetic markers to investigate phylogeny, to generate mutants and recombinants, and to identify mutated genes (2, 3, 4, 8, 11, 15, 16, 19). In ectomycorrhizal basidiomycetes, however, retroelements had not been previously described. The discovery of a full-length copy of a retroelement in T. matsutake suggests that ectomycorrhizal fungi also carry retroelements, some of which could function as transposons to induce mutants or used as genetic markers for phylogenetic analysis and identification of ectomycorrhizae. The nucleotide and putative amino acid sequences of marY1 will serve as a reference in the search for retroelements in other ectomycorrhizal basidiomycetes.
The genetic manipulation of marY1 could lead to the development of a transposon-mediated analysis system in ectomycorrhizal fungi, since marY1 is apparently intact and might be mobile given the correct stimulus. Transposon-mediated analysis is effective if the organism to be subjected to mutagenesis does not carry any copies of the transposon that is to be introduced (11, 15, 16). In this respect, it is very important that S. bovinus and R. rubescens do not have marY1, and this element might be used for introducing mutations into these ectomycorrhizal symbionts. We are currently investigating whether marY1 functions in heterologous fungal cells and expect to utilize marY1 to further transposon-mediated research with ectomycorrhizal fungi.
Nucleotide sequence accession number.
The nucleotide sequence of the marY1 retroelement has been deposited in the GenBank, EMBL, and DDBJ databases under the accession number AB028236.
Acknowledgments
This work was supported by the Biotechnology in Ectomycorrhizae Program of the Ministry of Agriculture, Forestry and Fisheries of Japan.
The authors thank forest experiment stations located in the following prefectures in Japan for the supply of fungal strains: Chiba, Fukui, Hiroshima, Hyogo, Iwate, Kyoto, Okayama, Shiga, Tokushima, and Yamaguchi.
REFERENCES
- 1.Anaya N, Roncero M I G. skippy, a retrotransposon from the fungal plant pathogen Fusarium oxysporum. Mol Gen Genet. 1995;249:637–647. doi: 10.1007/BF00418033. [DOI] [PubMed] [Google Scholar]
- 2.Bennetzen J L. Transposable element contributions to plant gene and genome evolution. Plant Mol Biol. 2000;42:251–269. [PubMed] [Google Scholar]
- 3.Bingham P M, Zachar Z. Retrotransposons and the FB transposon from Drosophila melanogaster. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 485–502. [Google Scholar]
- 4.Boeke J D. Transposable elements in Saccharomyces cerevisiae. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 335–374. [Google Scholar]
- 5.Covey S N. Amino acid sequence homology in gag region of reverse transcribing elements and the coat protein gene of cauliflower mosaic virus. Nucleic Acids Res. 1986;14:623–633. doi: 10.1093/nar/14.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobinson K F, Harris R E, Hamer J E. Grasshopper, a long terminal repeat (LTR) retroelement in the phytopathogenic fungus Magnaporthe grisea. Mol Plant-Microbe Interact. 1993;6:114–126. doi: 10.1094/mpmi-6-114. [DOI] [PubMed] [Google Scholar]
- 7.Farman M L, Tosa Y, Nitta N, Leong S A. MAGGY, a retrotransposon in the genome of the rice blast fungus Magnaporthe grisea. Mol Gen Genet. 1996;251:665–674. doi: 10.1007/BF02174115. [DOI] [PubMed] [Google Scholar]
- 8.Flavell A J, Smith D B. A Ty1-copia group retrotransposon sequence in a vertebrate. Mol Gen Genet. 1992;233:322–326. doi: 10.1007/BF00587596. [DOI] [PubMed] [Google Scholar]
- 9.Geever R F, Case M E, Tyler B M, Buxton F, Giles N H. Point mutations and DNA rearrangements 5′ to the inducible qa-2 gene of Neurospora allow activator protein-independent transcription. Proc Natl Acad Sci USA. 1983;80:7298–7302. doi: 10.1073/pnas.80.23.7298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hecht S J, Carlson J O, DeMartin J C. Analysis of a type D retroviral capsid gene expressed in ovine pulmonary carcinoma and present in both affected and unaffected sheep genomes. Virology. 1994;202:480–484. doi: 10.1006/viro.1994.1366. [DOI] [PubMed] [Google Scholar]
- 11.Hirochika H. Retrotransposons of rice: their regulation and use for genome analysis. Plant Mol Biol. 1997;35:231–240. [PubMed] [Google Scholar]
- 12.Hosford D, Pilz D, Molina R, Amaranthus M. U.S. Department of Agriculture Forest Service Pacific Northwest Research Station General Technical Report PNW-GTR-412. Washington, D.C.: U.S. Department of Agriculture; 1997. Ecology and management of the commercially harvested American matsutake mushroom; pp. 1–68. [Google Scholar]
- 13.Hutchison C A, III, Hardies S C, Loeb D D, Ronald Shehee W, Edgell M H. LINEs and related retroposons: long interspersed repeated sequences in the eukaryotic genome. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 593–617. [Google Scholar]
- 14.Kinsey J A, Helber J. Isolation of a transposable element from Neurospora crassa. Proc Natl Acad Sci USA. 1989;86:1929–1933. doi: 10.1073/pnas.86.6.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar A. The adventures of the Ty1-copia group of retrotransposons in plants. Trends Genet. 1996;12:41–43. doi: 10.1016/0168-9525(96)81393-x. [DOI] [PubMed] [Google Scholar]
- 16.Lucas H, Feuerbach F, Kunert K, Grandbastien M-A, Caboche M. RNA-mediated transposition of the tobacco retrotransposon Tnt1 in Arabidopsis thaliana. EMBO J. 1995;14:2364–2373. doi: 10.1002/j.1460-2075.1995.tb07231.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McHale M T, Roberts I N, Noble S M, Beaumont C, Whitehead M P, Seth D, Oliver R P. CfT-I: an LTR-retrotransposon in Cladosporium fulvum, a fungal pathogen of tomato. Mol Gen Genet. 1992;233:337–347. doi: 10.1007/BF00265429. [DOI] [PubMed] [Google Scholar]
- 18.Murata H, Yamada A, Babasaki K. Identification of repetitive sequences containing motifs of retrotransposons in the ectomycorrhizal basidiomycete Tricholoma matsutake. Mycologia. 1999;91:766–775. [Google Scholar]
- 19.Peterson-Burch B D, Wright D A, Laten H M, Voytas D F. Retroviruses in plants? Trends Genet. 2000;16:151–152. doi: 10.1016/s0168-9525(00)01981-8. [DOI] [PubMed] [Google Scholar]
- 20.Punt P J, Dingemanse M A, Kuyvenhoven A, Soede R D M, Pouwel P H, van den Hondel C A M J J. Functional elements in the promoter region of the Aspergillus nidulans gpdA gene encoding glyceraldehyde-3-phosphate dehydrogenase. Gene. 1990;93:101–109. doi: 10.1016/0378-1119(90)90142-e. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 22.Smith S E, Read D J. Mycorrhizal symbiosis. London, England: Academic Press; 1997. pp. 1–605. [Google Scholar]
- 23.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varmus H, Brown P. Retroviruses. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C.: American Society for Microbiology; 1989. pp. 53–108. [Google Scholar]
- 25.Xu H, Boeke J D. Localization of sequences required in cis for yeast Ty1 element transposition near the long terminal repeats: analysis of mini-Ty1 elements. Mol Cell Biol. 1990;10:2695–2702. doi: 10.1128/mcb.10.6.2695. [DOI] [PMC free article] [PubMed] [Google Scholar]