Abstract
We examined Cunninghamella elegans to determine its ability to transform amoxapine, a tricyclic antidepressant belonging to the dibenzoxazepine class of drugs. Approximately 57% of the exogenous amoxapine was metabolized to three metabolites that were isolated by high-performance liquid chromatography and were identified by nuclear magnetic resonance and mass spectrometry as 7-hydroxyamoxapine (48%), N-formyl-7-hydroxyamoxapine (31%), and N-formylamoxapine (21%). 7-Hydroxyamoxapine, a mammalian metabolite with biological activity, now can be produced in milligram quantities for toxicological evaluation.
Amoxapine [2-chloro-11-(1-piperazinyl)dibenz-(b,f)(1,4)oxazepine] is a tricyclic antidepressant drug belonging to the dibenzoxazepine class and is the N-demethylated derivative of the neuroleptic compound loxapine (6, 11). Amoxapine, marketed as Asendin, is used to treat depression, as well as anxiety or agitation associated with depression (14). Amoxapine also is considered an atypical antipsychotic agent for the treatment of schizophrenia (12). In mammals, amoxapine is metabolized mainly to two active metabolites, 7-hydroxyamoxapine and 8-hydroxyamoxapine, by hepatic biotransformation (2, 13, 20). Both metabolites inhibit the presynaptic reuptake of norepinephrine, an antidepressant effect, while 7-hydroxyamoxapine blocks postsynaptic dopamine receptors in the central nervous system, an antipsychotic effect (6, 7, 14).
The study of drug metabolism and the toxicity of metabolites are important in drug development. The neurological properties of tricyclic antidepressants have prompted research to evaluate the metabolism of these drugs in various animal models (15). However, identification of metabolites from animal sources and clinical samples can be hindered by insufficient quantities of material. Some microorganisms, especially the zygomycete fungi belonging to the genus Cunninghamella, can metabolize compounds in a manner similar to metabolism by mammals (3–5, 8, 9, 19) and have been used to isolate mammalian drug metabolites (10, 16, 17, 18, 21–24). The advantages of a microbial system as a complementary in vitro model for drug metabolism include its low cost, ease of handling, scale-up capability, and potential to reduce the use of animals. A microbial system also provides an alternative to or complement for organic synthesis and uses milder reaction conditions. In this study, amoxapine was metabolized by Cunninghamella elegans Lendner [Cunninghamella echinulata var. elegans (Lendner) Lunn et Shipton] to produce 7-hydroxyamoxapine and two novel metabolites, which demonstrated the potential of fungal biotransformation to produce compounds of pharmaceutical interest for future toxicological evaluation.
Cultures of C. elegans ATCC 9245 were grown on Sabouraud dextrose agar slants (Remel, Lenexa, Kans.) for 5 days at 24°C and stored at 4°C. The spores and/or mycelia were aseptically transferred to Sabouraud dextrose agar plates and allowed to grow for 5 days at 24°C. The mycelia and agar from two plates were transferred to a sterile blender cup containing 150 ml of a sterile physiological saline solution and homogenized for 5 min. Approximately 5-ml portions of the blended mycelial suspension were used to inoculate 125-ml Erlenmeyer flasks containing 30 ml of Sabouraud dextrose medium. The cultures were incubated at 25°C on a rotary shaker operating at 150 rpm. After 48 h, 10 mg of amoxapine (>99% pure; Research Biochemicals, Inc., Natick, Mass.) dissolved in dimethylformamide (100 mg/ml) was added to each flask. In control experiments we incubated a culture without amoxapine and sterile flasks containing only media and amoxapine. Cultures were extracted after 4, 8, 24, 48, 72, 96, 120, 144, 168, and 192 h. The data below are averages based on three separate experiments performed with replicate batch cultures. The standard deviation was no more than 5% in each case. The incubation time required for maximum metabolite formation was 120 h. Ten flasks were incubated and extracted in each replicate. The organic extracts were dried over sodium sulfate and evaporated to dryness in vacuo at 34°C by using a model Büchi 011 rotary evaporator (Brinkmann Instruments, Westburg, N.Y.). Each residue was dissolved in 5 ml of methanol, transferred to a test tube (13 by 100 mm), and concentrated to a volume of approximately 100 μl with a model SS21 Savant Speed-vac system (Savant Instruments, Holbrook, N.Y.) for analysis by high-performance liquid chromatography (HPLC).
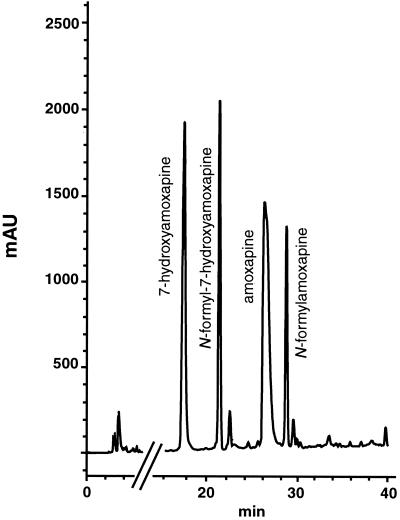
The metabolites were isolated by reversed-phase HPLC by using a semipreparative scale HPLC system consisting of a model 100A pump (Beckman Instruments, Inc., Fullerton, Calif.), a model 486 tunable UV absorbance detector (Waters Corp., Milford, Mass.), and a model CR601 Chromatopac integrator (Shimadzu, Kyoto, Japan). The compounds were eluted (Fig. 1) by using a linear 40 to 95% methanol gradient in water for 40 min, a 5-μm C18 Inertsil ODS-3 column (10 by 250 mm; MetaChem Technologies, Torrance, Calif.), and a flow rate of 5 ml/min. The major fungal metabolites were collected, and their structures were determined by 1H nuclear magnetic resonance (NMR) and mass spectrometry.
FIG. 1.
HPLC chromatogram of amoxapine and its metabolites formed by C. elegans.
In another experiment, the major metabolite, 7-hydroxyamoxapine, and the residual amoxapine were quantified. Cultures of C. elegans were grown in triplicate flasks, 5 mg of amoxapine was added to each flask, and the preparations were incubated and extracted as described above. The residues were concentrated and redissolved in 5 ml of methanol, and 20-μl aliquots were injected into the HPLC column. Amounts of 7-hydroxyamoxapine and residual amoxapine were determined by comparing the peak areas of these compounds with the peak areas of known concentrations of amoxapine.
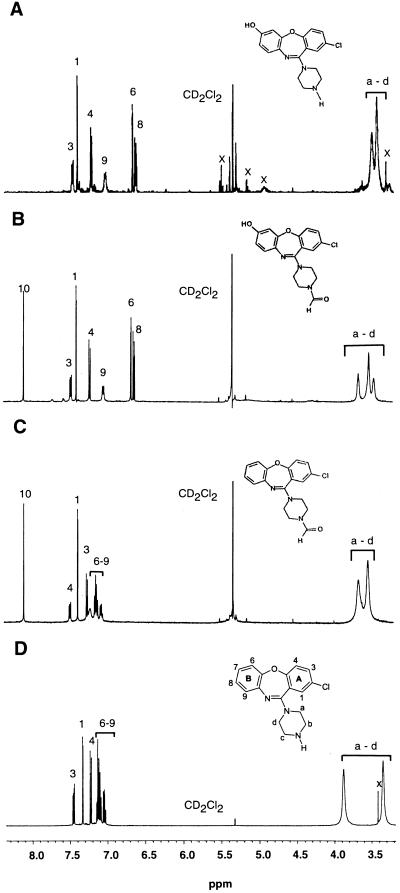
For NMR analysis (Table 1, Fig. 2), each metabolite was dissolved in 0.5 ml of methylene-d2-chloride (99.96 atom% 2H). The NMR measurements were obtained at 500.13 MHz with a model AM500 spectrometer (Bruker Instruments, Billerica, Mass.). Chemical shifts were determined on the δ scale (parts per million) by assigning the residual solvent peak to 5.32 ppm. Typical data acquisition parameters were as follows: data size, 32 K; sweep width, 7,042 Hz; filter width, 8,900 Hz; acquisition time, 2.33 s; flip angle, 90°; relaxation delay, 0 s; and temperature, 301 K.
TABLE 1.
Chromatographic, mass spectral, and proton NMR data for amoxapine and its metabolites formed by C. elegans
| Compound | HPLC retention time (min) | Mass spectral ions (m/z)a | NMR chemical shifts (ppm)b |
|---|---|---|---|
| 7-Hydroxyamoxapinec | 17.9 | 329 (24), 299, 285, 273, 261 (100), 244, 209, 85, 69, 56 (7) | 7.37 (H1), 7.44 (H3), 7.19 (H4), 6.64 (H6), 6.60 (H8), 7.00 (H9), 3.40, 3.47 (Ha-d) |
| N-Formyl-7-hydroxyamoxapinec | 21.8 | 357 (100), 328, 313, 299, 285, 273 (57), 260, 245, 209, 113 (7), 98 (4), 85, 70, 56 (5) | 7.38 (H1), 7.45 (H3), 7.20 (H4), 6.65 (H6), 6.61 (H8), 7.02 (H9), 8.08 (H10), 3.43, 3.50, 3.64 (Ha-d) |
| N-Formylamoxapined | 29.1 | 341 (100), 312, 283, 268/270, 257 (84), 244, 228, 229, 193, 113 (7), 98, 85, 69, 56 (6) | 7.37 (H1), 7.47 (H3), 7.25 (H4), 7.06, 7.10–7.15 (H6-9), 8.09 (H10), 3.52, 3.65 (Ha-d) |
| Amoxapined | 26.7 | 313 (10), 283, 263, 257, 245 (100), 228, 216, 193, 56 (4) | 7.33 (H1), 7.45 (H3), 7.22 (H4), 7.30, 7.07–7.15 (H6-9), 3.34, 3.86 (Ha-d) |
Ions above m/z 209 had an isotope pattern consistent with the presence of a single chlorine. Only the most abundant ion is shown. The numbers in parentheses are relative abundance values, expressed as percentages.
Samples were dissolved in methylene-d2-chloride.
The coupling constants were as follows: J1,3 = 2.6 Hz; J3,4 = 8.8 Hz; J6,8 = 2.8 Hz; and J8,9 = 8.6 Hz.
The coupling constants were as follows: J1,3 = 2.6 Hz; and J3,4 = 8.6 Hz.
FIG. 2.
500-MHz 1H NMR spectra of amoxapine and its metabolites formed by C. elegans. (A) 7-Hydroxyamoxapine. (B) N-Formyl-7-hydroxyamoxapine. (C) N-Formylamoxapine. (D) Amoxapine. The peaks labeled x are impurities.
Mass spectral analyses were performed as previously described (23). However, electron ionization (EI) analyses (Table 1) were performed with an electron energy of 25 V to enhance the molecular ion region of the mass spectra. Molecular weights were confirmed by ammonia positive ion chemical ionization analyses (data not shown).
The HPLC chromatogram showed that amoxapine was transformed to one major metabolite, which eluted at 17.9 min and accounted for 48% of the total metabolism. The NMR spectrum of this metabolite produced two resonances, at 3.40 and 3.47 ppm, that were broadened due to chemical exchange (Fig. 2A). These resonances were the protons of the piperazine ring, Ha, Hb, Hc, and Hd. The peaks integrated collectively as 8, indicating that there was no change in this ring. There were no changes in the aromatic resonances of the A ring of the dibenzoxazepine portion of the molecule compared to the resonances of amoxapine, but the B ring had a single substitution. Two aromatic resonances exhibited large (≥0.44-ppm) upfield shifts that were consistent with ring hydroxylation. A nuclear Overhauser enhancement experiment involving saturation of the H4 resonance resulted in enhancement of the meta-coupled doublet at 6.64 ppm, proving that the resonance at 6.64 ppm was H6. The site of substitution was C-7, based on the coupling pattern and the results of homonuclear decoupling experiments. The EI mass spectrum revealed a molecular ion at m/z 329 and a series of ions that included the dibenzoxazepine portion of the molecule with a base peak at m/z 261. The molecular ion at m/z 329 suggested that hydroxylation had occurred, and the base peak at m/z 261 was further evidence that hydroxylation of the dibenzoxazepine portion of the molecule had occurred. Based on the NMR and mass spectral data, the metabolite was identified as 7-hydroxyamoxapine (Table 1).
The NMR experiments used in the analysis of the compound that eluted at 21.8 min revealed that there was a single substitution at the H7 position. The piperazine ring protons at 3.43, 3.50, and 3.64 ppm integrated as 8, as for 7-hydroxyamoxapine. The EI mass spectrum showed that there were a molecular ion at m/z 357, a series of ions similar to those described above for 7-hydroxyamoxapine that indicated that hydroxylation of the dibenzoxazepine portion of the molecule had occurred, and a minor fragment ion at m/z 113. The molecular ion at m/z 357 suggested that both formylation and hydroxylation had occurred. The minor fragment ion at m/z 113 (m/z 85 + C⩵O) was further evidence that formylation of the piperazine ring had occurred (Table 1). The NMR spectrum was consistent with formylation of the piperazine ring in that it showed a singlet at 8.08 ppm that integrated as one (Fig. 2B) and, when selectively saturated, produced a nuclear Overhauser enhancement at 3.50 ppm. The metabolite was identified as N-formyl-7-hydroxyamoxapine.
The aromatic region of the NMR spectrum of the peak eluting at 29.1 min (Fig. 2C) exhibited the same number and multiplicity of resonances as the parent compound, amoxapine. The only difference was a sharp singlet at 8.09 ppm, which was similar to the singlet of N-formyl-7-hydroxyamoxapine. Mass spectral analysis revealed a molecular ion at m/z 341, a series of ions similar to those observed for amoxapine that indicated that the dibenzoxazepine portion of the molecule was unmodified, and a minor fragment ion at m/z 113. The molecular ion at m/z 341 suggested that formylation without hydroxylation had occurred, and the minor fragment ion at m/z 113 again indicated that formylation of the piperazine ring had occurred (Table 1). The metabolite was identified as N-formylamoxapine. Mass spectral and NMR analysis (Fig. 2D) of authentic samples of amoxapine confirmed that the peak eluting at 26.7 min was unmetabolized amoxapine.
The fungus transformed amoxapine to three metabolites. The major type of enzymatic attack was hydroxylation at the C-7 position, and 7-hydroxyamoxapine (48%) was the most common metabolite formed. The other metabolic modification was addition of a formyl group to the piperazine ring to form N-formylamoxapine (21%). The combined modifications at both these locations produced the third metabolite, N-formyl-7-hydroxyamoxapine (31%).
In summary, our experiments demonstrated that C. elegans can be used to biotransform amoxapine to 7-hydroxyamoxapine, the major mammalian metabolite, and two other novel metabolites with a high yield at low cost. These compounds are difficult to chemically synthesize or to isolate from experimental animals in order to obtain clinical samples. Because very little is known about the mechanism of clinical action of amoxapine and its metabolites in patients, the fungal biotransformation system can be used to obtain milligram quantities of amoxapine metabolites for evaluations of their neurotoxicity and biological activity.
Acknowledgments
We thank staff members of the Division of Microbiology for helpful discussions and Kim Cooney, Bob Barringer, Danny Tucker, and Barbara Jacks for illustrations.
This research was supported in part by appointment of Donglu Zhang to the Postgraduate Research Program at the National Center for Toxicological Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration.
REFERENCES
- 1.Ban T A, Fujimori M, Petrie W M, Ragheb M, Wilson W H. Systematic studies with amoxapine, a new antidepressant. Int Pharmacopsychiatry. 1982;17:18–27. doi: 10.1159/000468553. [DOI] [PubMed] [Google Scholar]
- 2.Calvo B, Garcia M J, Pedraz J L, Marino E L, Dominguez-Gil A. Pharmacokinetics of amoxapine and its active metabolites. Int J Clin Pharmacol Ther Toxicol. 1985;23:180–185. [PubMed] [Google Scholar]
- 3.Cerniglia C E. Microbial metabolism of polycyclic aromatic hydrocarbons. Adv Appl Microbiol. 1984;30:31–71. doi: 10.1016/s0065-2164(08)70052-2. [DOI] [PubMed] [Google Scholar]
- 4.Cerniglia C E, Gibson D T. Metabolism of naphthalene by cell-extracts of Cunninghamella elegans. Arch Biochem Biophys. 1978;186:121–127. doi: 10.1016/0003-9861(78)90471-x. [DOI] [PubMed] [Google Scholar]
- 5.Clark A M, Hufford C D. Use of microorganisms for the study of drug metabolism: an update. Med Res Rev. 1991;11:473–501. doi: 10.1002/med.2610110503. [DOI] [PubMed] [Google Scholar]
- 6.Cohen B M, Harries P Q, Altesman R I, Cole J O. Amoxapine: neuroleptic as well as antidepressant? Am J Psychol. 1982;139:1165–1167. doi: 10.1176/ajp.139.9.1165. [DOI] [PubMed] [Google Scholar]
- 7.Coupet J, Rauh C E, Szues-Myers Y A, Yunger L M. 2-Chloro-H-(1-piperazinyl)dibenz(b,f)(1,4) oxazepine (amoxapine), an antidepressant with antipsychotic properties—a possible role for 7-hydroxyamoxapine. Biochem Pharmacol. 1979;28:2514–2515. doi: 10.1016/0006-2952(79)90017-0. [DOI] [PubMed] [Google Scholar]
- 8.Davis P J. Microbial models of mammalian drug metabolism. Dev Ind Microbiol. 1988;29:197–219. [Google Scholar]
- 9.Ferris J P, MacDonald L H, Patrie M A, Martin M A. Aryl hydrocarbon hydroxylase activity in the fungus Cunninghamella bainieri: evidence for the presence of cytochrome P450. Arch Biochem Biophys. 1976;175:443–452. doi: 10.1016/0003-9861(76)90532-4. [DOI] [PubMed] [Google Scholar]
- 10.Hansen E B, Jr, Cho B P, Korfmacher W A, Cerniglia C E. Fungal transformations of antihistamines: metabolism of brompheniramine, chlorpheniramine, and pheniramine to N-oxide and N-demethylated metabolites by the fungus Cunninghamella elegans. Xenobiotica. 1995;25:1081–1092. doi: 10.3109/00498259509061908. [DOI] [PubMed] [Google Scholar]
- 11.Hue B, Palomba B, Giacardy-Paty M, Bottai T, Alric R, Petit P. Concurrent high-performance liquid chromatographic measurement of loxapine and amoxapine and of their hydroxylated metabolites in plasma. Ther Drug Monit. 1998;20:335–339. doi: 10.1097/00007691-199806000-00016. [DOI] [PubMed] [Google Scholar]
- 12.Kapur S, Cho R, Jones C, McKay G, Zipursky R B. Is amoxapine an atypical antipsychotic? Positron-emission tomography investigation of its dopamine and serotonin occupancy. Biol Psychol. 1999;45:1217–1220. doi: 10.1016/s0006-3223(98)00204-2. [DOI] [PubMed] [Google Scholar]
- 13.Kobayashi A, Nakazawa K. Determination of amoxapine and its metabolites in human serum by high-performance liquid chromatography. Neuropharmacology. 1985;24:1253–1256. doi: 10.1016/0028-3908(85)90162-5. [DOI] [PubMed] [Google Scholar]
- 14.Medical Economics Press. Physicians's desk reference. Montvale, N.J: Medical Economics Press; 1997. pp. 1419–1421. [Google Scholar]
- 15.Midha K K, Hubbard J W, McKay G, Rawson M J, Hsia D. The role of metabolites in a bioequivalence study. II. Amoxapine, 7-hydroxyamoxapine, and 8-hydroxyamoxapine. Int J Clin Pharmacol Ther Toxicol. 1999;37:428–438. [PubMed] [Google Scholar]
- 16.Moody J D, Freeman J P, Cerniglia C E. Biotransformation of doxepin by Cunninghamella elegans. Drug Metab Dispos. 1999;27:1157–1164. [PubMed] [Google Scholar]
- 17.Rao G P, Davis P J. Microbial models of mammalian metabolism: biotransformations of HP 749 (Besipiridine) using Cunninghamella elegans. Drug Metab Dispos. 1997;25:709–715. [PubMed] [Google Scholar]
- 18.Reighard J B, Knapp J E. Microbial models of mammalian metabolism. Pharm Int. 1986;7:92–94. [Google Scholar]
- 19.Smith R V, Rosazza J P. Microbial transformation as a means of preparing mammalian drug metabolites. In: Rosazza J P, editor. Microbial transformation of bioactive compounds. II. Boca Raton, Fla: CRC Press; 1982. pp. 1–42. [Google Scholar]
- 20.Takeuchi H, Yokota S, Shimada S, Ohtani Y, Miura S, Kubo H. Pharmacokinetics of amoxapine and its active metabolites in healthy subjects. Curr Ther Res Clin Exp. 1993;53:427–434. [Google Scholar]
- 21.Zhang D, Evans F E, Freeman J P, Duhart B T, Jr, Cerniglia C E. Biotransformation of amitriptyline by Cunninghamella elegans. Drug Metab Dispos. 1995;23:1417–1425. [PubMed] [Google Scholar]
- 22.Zhang D, Evans F E, Freeman J P, Yang Y, Deck J, Cerniglia C E. Formation of mammalian metabolites of cyclobenzaprine by the fungus, Cunninghamella elegans. Chem-Biol Interact. 1996;102:79–92. doi: 10.1016/s0009-2797(96)03736-2. [DOI] [PubMed] [Google Scholar]
- 23.Zhang D, Freeman J P, Sutherland J B, Walker A E, Yang Y, Cerniglia C E. Biotransformation of chlorpromazine and methdilazine by Cunninghamella elegans. Appl Environ Microbiol. 1996;62:798–803. doi: 10.1128/aem.62.3.798-803.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang D, Hansen E B, Jr, Deck J, Heinze T M, Sutherland J B, Cerniglia C E. Fungal biotransformation of the antihistamine azatadine by Cunninghamella elegans. Appl Environ Microbiol. 1996;62:3477–3479. doi: 10.1128/aem.62.9.3477-3479.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]