Abstract
With the wide use of carbapenems, carbapenem-resistant Enterobacterales have been increasingly reported worldwide. In this study, one blaOXA-181-positive Pseudocitrobacter faecalis strain was isolated from the blood culture of a patient with a bloodstream infection in China, which was its first clinical report outside Pakistan. Species identification of P. faecalis was initially performed using MALDI-TOF/MS and further confirmed by 16S rRNA gene and housekeeping gene sequencing. The antimicrobial susceptibility testing was determined through the broth microdilution method, and their clonal relationship was analyzed by pulsed-field gel electrophoresis. To study the transmission and genetic structure of the blaOXA-181 gene, a transformation test and whole-genome sequencing (WGS) were performed. The results of the antimicrobial susceptibility testing indicated this P. faecalis was resistant to carbapenems, quinolones, and commonly used β-lactam/β-lactamase inhibitor combinations. Through WGS and transformation experiments, blaOXA-181 and qnrS1 genes causing antibiotic resistance were located on a 55,148-bp length IncX3 type plasmid with a truncated ColKp3 replicon gene. As a rare species of Enterobacterales, P. faecalis was clinically reported in China for the first time, and the blaOXA-181 gene it carried was located on a globally disseminated IncX3 plasmid. The spread of such bacteria and antibiotic resistance requires more clinical attention.
Keywords: Pseudocitrobacter faecalis, bla OXA-181 , carbapenem, IncX3
1. Introduction
The emergence of multidrug-resistant Enterobacterales severely threatens public health, and carbapenems have been regarded as its therapeutic choice due to their broad spectrum of activity, stability of extended-spectrum β-lactamase, and proven safety [1,2,3]. However, with rising clinical use, carbapenem-resistant Enterobacterales (CRE) have been increasingly reported worldwide. With limited therapeutic regimens, the infections associated with CRE could even lead to a high mortality rate of above 30% [4]. The detection rate of CRE is undergoing rapid growth worldwide, and that in China has increased over 2.5 times during the past fifteen years, and even reached 10.5% in 2021, based on data from the China Antimicrobial Surveillance Network (CHINET) [5,6,7].
The primary resistance mechanism of CRE is carbapenemase production, mainly including Ambler class A (KPC), Ambler class B (NDM, IMP, VIM), and Ambler class D (OXA-48-like) [8]. Worldwide, KPC-type carbapenemase represented the majority, followed by the NDM- and OXA-type [5]. In China, the proportion of OXA-48-like carbapenemase is rising, especially in children, with OXA-232-type as the primary type (97.1%), while in countries such as Angola, OXA-181-type was dominant [9,10]. Worryingly, such resistance was no longer confined to common bacteria, such as Escherichia coli and Klebsiella pneumoniae, but extensively occurred in other Enterobacterales clinical isolates [11].
Pseudocitrobacter faecalis was a kind of Gram-negative facultatively anaerobic bacteria discovered in 2010 [12]. It is a rare kind of Enterobacterales and has only been clinically reported in Pakistan. Such clinical isolates showed carbapenem resistance due to the production of NDM-1 carbapenemase. Herein, we report the first blaOXA-181-positive Pseudocitrobacter faecalis from a patient with bloodstream infection (BSI) in China.
2. Results
2.1. Case Presentation
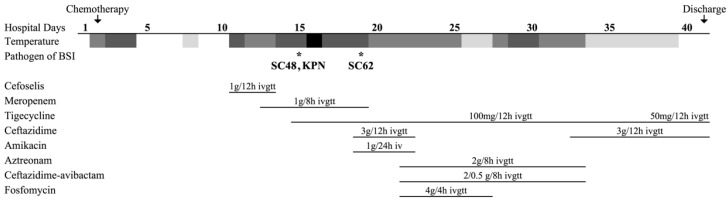
A 19-year-old man was diagnosed with acute myeloid leukemia (AML) type M2 and was first admitted in August 2019. HAA regimen (homoharringtonine, cytarabine, and aclarubicin) was employed as his chemotherapy, while post-chemotherapeutic bone marrow suppression and severe pneumonia caused by bacterial and fungal infection appeared subsequently. On 6 February 2020 (defined as day 1), the patient went through his fifth chemotherapy and soon developed septic shock with positive blood cultures. Meanwhile, lung computed tomography (CT) revealed new ground-glass patchy shadows in the multi-lobar of bilateral lungs. Meropenem (1g q8h) and tigecycline (100g q12h) were used as the empirical therapy based on his previous medical history. Further workup showed that the strains isolated from blood on day 16 were carbapenem-resistant P. faecalis strain SC48 and Klebsiella pneumoniae strain KPN. Strain KPN was sensitive to major commonly used antibiotics, including cephalosporins, carbapenem, quinolones, etc. Thus meropenem was replaced by ceftazidime (3g q12h) and amikacin (1g QD) 3 days later.
However, the patient had a recurring fever, and progressing pneumonia was shown by CT reexamination. On day 22, blood culture was positive again with an isolate (strain SC62) similar to strain SC48, so the therapeutic regime was switched to aztreonam, ceftazidime-avibactam, and fosfomycin combined with tigecycline. Mercifully, infection was thereafter brought under control gradually evincing decreased inflammatory index, pulmonary foci absorption, and negative blood culture. For economic concerns, tigecycline and ceftazidime were given again on day 33, based on the clinical situation and the dosage was then reduced by degrees. Antifungal drugs, voriconazole and amphotericin B, covered the whole treatment process. The patient was finally discharged 41 days after admission. The other clinical and microbiologic details are summarized in Figure 1.
Figure 1.
Patient treatment course and microbiological characteristics of patients with bloodstream infection. * Pathogen SC48 and SC62 were Pseudocitrobacter faecalis clinical isolates. Pathogen KPN indicated a highly sensitive Klebsiella pneumoniae clinical isolate. The blocks from dark to light indicated high, moderate, low-grade fever, and normal temperature.
2.2. Characteristics of P. faecalis Strains
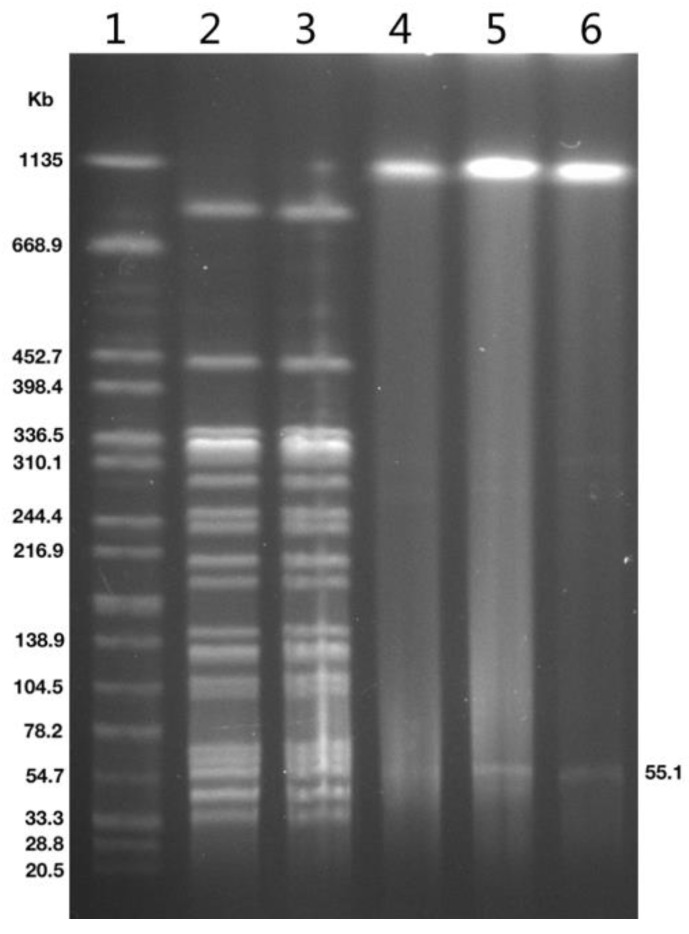
Based on the PFGE fingerprint shown in Figure 2, P. faecalis SC48 and SC62 isolated from blood culture were identical in genomic pulsotype. Both isolates were high-level resistant to carbapenems and quinolones (Table 1).
Figure 2.
PFGE of Pseudocitrobacter faecalis clinical isolates and transformant. Lanes 1, marker Salmonella braenderup H9812; line 2 to 3, PFGE image of P. faecalis SC48, SC62; line 4 to 6, S1-PFGE image of P. faecalis SC48, SC62 and E. coli DH5α-SC48-T.
Table 1.
Minimal inhibitory concentrations (MICs) of Pseudocitrobacter faecalis clinical isolate, transformant, and recipient.
| Strains | β-Lactamase Genes | Fluoroquinolone- Resistant Genes |
MIC (mg/L) a | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IMP | MEM | CAZ | FEP | ATM | CZA | SCF | TZP | SXT | CIP | LEV | AMK | TGC | POL | |||
| P. faecalis SC48 | blaDHA-1, blaOXA-1, blaOXA-181 | qnrB4, qnrS1, aac(6’)-Ib-cr | 64 | 32 | 2 | 8 | ≤1 | 1 | 128 | >256 | 0.5 | >8 | 8 | 4 | 1 | 0.5 |
| P. faecalis SC62 | blaDHA-1, blaOXA-1, blaOXA-181 | qnrB4, qnrS1, aac(6’)-Ib-cr | 64 | 64 | 2 | 8 | ≤1 | 1 | 128 | >256 | 0.5 | >8 | 8 | 8 | 1 | 0.5 |
|
E. coli DH5α- SC48-T |
bla OXA-181 | qnrS1 | 1 | 0.25 | 0.5 | 0.125 | ≤1 | 0.25 | 8 | 64 | ≤0.25 | 0.25 | 0.5 | ≤1 | 0.25 | 0.25 |
| E. coli DH5α | - | - | 0.125 | ≤0.03 | 0.5 | ≤0.06 | ≤1 | 0.125 | ≤1 | 4 | ≤0.25 | ≤0.06 | ≤0.125 | ≤1 | 0.125 | 0.25 |
a IPM, imipenem; MEM, meropenem; CAZ, ceftazidime; FEP, cefepime; ATM, aztreonam; CZA, ceftazidime-avibactam; SCF, cefoperazone-sulbactam; TZP, piperacillin-tazobactam; SXT, trimethoprim-sulfamethoxazole; CIP, ciprofloxacin; LEV, levofloxacin; AMK, amikacin; TGC, tigecycline; POL, polymyxin B.
The MICs of the carbapenems ranged from 32 mg/L to 64 mg/L, and that of quinolones ≥ 8 mg/L. Both strains were also resistant to commonly used β-lactam combination agents (cefoperazone-sulbactam and piperacillin-tazobactam) and were intermediate to cefepime. Ceftazidime, aztreonam, ceftazidime-avibactam, trimethoprim-sulfamethoxazole, amikacin, tigecycline, and polymyxin B maintained well in vitro activity for them. The acquisition of blaOXA-181 carrying by plasmid altered the resistance of transformant E. coli DH5α-SC48-T towards β-lactams, increasing the MICs of imipenem, meropenem, cefoperazone-sulbactam, and piperacillin-tazobactam for no less than eight times. Meanwhile, E. coli DH5α-SC48-T also acquired resistance to quinolones with an over 4-fold rise in the MICs of ciprofloxacin and levofloxacin (Table 1).
2.3. Genetic Analysis of Strains and blaOXA-181-Positive Plasmid
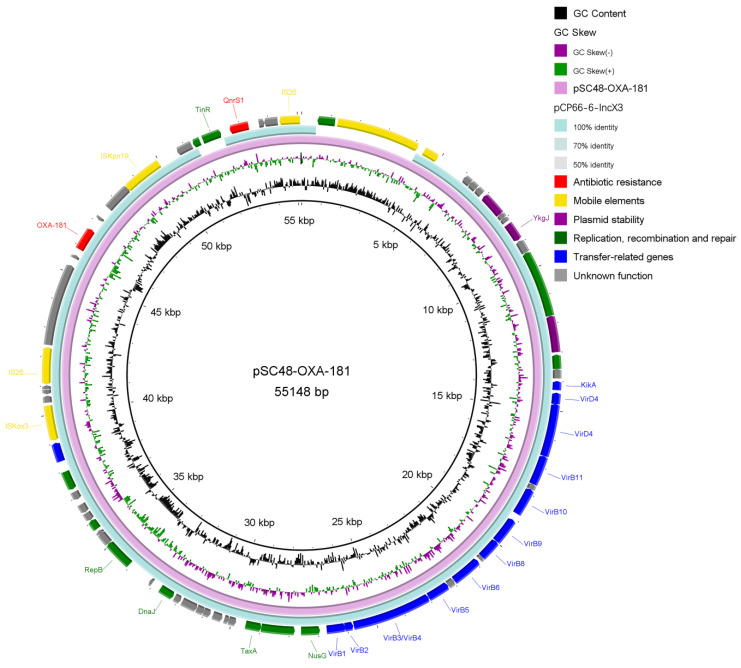
Through the WGS analysis, it was found that both P. faecalis isolates belonged to the same clinical strain, which contained numerous resistance genes, mainly involving blaDHA-1, blaOXA-1, blaOXA-181, aac(6’)-IIc, aac(6’)-Ib-cr, qnrS1, qnrB4, sul1, sul2, tet(A), ere(A), mph(A), catB3, floR, and arr. Each strain bore two plasmids, one of them was a transmissible plasmid harboring blaOXA-181 and qnrS1 that led to carbapenem and quinolone resistance (Figure 2). According to the transformation experiment, the blaOXA-181 gene was located at a 55,148-bp length IncX3 type plasmid with truncated ColKp3 replicon gene and was named pSC48-OXA-181 (Figure 3).
Figure 3.
Ring diagram representation of plasmid pSC48-OXA-181. From the inside to the outside: circle 1, scale; circle 2, GC content; circle 3, GC skew; circle 4, ring diagram of pSC48-OXA-181; circle 5, ring diagram of comparative plasmid pCP66-6-IncX3; circle 6, functional classified genes.
This plasmid was well-matched with another plasmid previously reported in the same province (pCP66-6-IncX3, GenBank accession no. CP053726.1), showing 91% coverage with perfect identity only with the difference of mobile elements. These mobile elements were distributed nearby resistance gene blaOXA-181 and qnrS1 consisting of ISKox3, IS26, and ISKpn19, leading to possible transposon-mediated spread. This plasmid also encoded type IV secretion (T4S) systems mediating transportation, including VirB and VirD proteins.
3. Discussion
Pseudocitrobacter gen. nov. is a novel genus of the Enterobacterales first observed in 2010 with Pseudocitrobacter faecalis sp. nov., Pseudocitrobacter anthropi sp. nov., and Pseudocitrobacter vendiensis sp. nov., among which P. anthropi was a later heterotypic synonym of P. faecalis [12,13]. Unlike P. vendiensis, found only in Denmark and Brazil, P. faecalis was sporadically reported in Asia, America, and Africa, mainly from Pakistan, India, China, and America, based on previous reports and data from the GenBank database (https://www.ncbi.nlm.nih.gov/nuccore/?term=%22Pseudocitrobacter+faecalis%22%5Bporgn%3A__txid1398493%5D, (accessed on 15 February 2022)) [12,13,14,15,16,17]. Though P. faecalis was only clinically reported in Pakistan, it could be found in the environment, animals, and plants, including foods such as egg, cucumber, and mango.
Herein, P. faecalis was first found in China and presumptively associated with bloodstream infection. The prevalence of bloodstream infection increased from 2010 to 2019 in China, among which bacteremia occupied a dominant position (93.1%) [18]. Clinicians attached importance to patients suffering from bloodstream infection, given its high mortality, especially those with high disease severity and inadequate immunologic defenses [19,20]. In this case, neutropenia occurred after high-dose chemotherapy was performed for hematologic malignancy, which significantly impacts the incidence of bloodstream infection in cancer patients [21]. Furthermore, the patient was suggested to have a hospital-acquired bloodstream infection combined with the course of the disease, which was consistent with previous studies showing that the composition ratio of hospital-acquired bloodstream infections was increasing annually and those related to Enterobacterales also dynamically increasing [18,22].
The appearance of a P. faecalis-related hospital-acquired bloodstream infection suggested the possible emergence and prevalence of such rare bacteria, which gave cause for increased vigilance. It has been proved that international travel can transfer resistant bacteria and antimicrobial resistance genes worldwide [23]. These bacteria and resistance genes are likely to invade travelers and further disseminate in the home country before they are lost in the host [24]. Otherwise, P. faecalis has been reported in food and the environment, indicating the possibility that such bacteria and resistance genes have entered the hospital settings via environmental contamination and the food chain [25,26,27].
It is worth noting that all Pseudocitrobacter gen. strains mediating clinical infection are associated with resistance genes encoding carbapenemases, including blaNDM-1, blaKPC-2, and blaIMP-1 [12,13,17]. P. faecalis strains producing OXA-181 carbapenemase were isolated in our study. OXA-48-like carbapenemases such as this are generally found in Enterobacterales worldwide, which even make up the most prevalent carbapenemase type in countries such as the Netherlands (44%) [28]. In China, OXA-48-like carbapenemase-producers accounted for 7.3% of CRE strains ranking behind KPC-2 and NDM [9]. Though OXA-232-type carbapenemase holds an overall majority, OXA-181 type has successively emerged in Enterobacterales since 2014 [29,30].
Globally, blaOXA-181 was mainly carried by highly conserved plasmids instead of chromosomally localizing. Such plasmids featured the qnrS1 allele and ColKP3 and IncX3 replicons, which was consistent with our study [31,32,33]. Early research revealed that the blaOXA-181 was inserted into the IncX3 plasmid through the IS3000-mediated co-integration of the ColKP3-type plasmid [34]. This kind of plasmid was also characterized by Tn3 family transposases and T4S systems mediating DNA transportation, different from those with other blaOXA-48-like genes [32,35]. These resistance genes located on plasmids were often colocalized with mobile genetic elements leading to the spread of resistance to carbapenem antibiotics between distinct plasmids and bacteria, which increases the need for vigilance in the clinic [24].
4. Materials and Methods
4.1. Clinical Isolates and Patient Data
A total of three clinical strains were isolated from blood samples of a patient in a tertiary hospital. Among them, P. faecalis SC48 and P. faecalis SC62 were carbapenem-resistant, while one K. pneumoniae was carbapenem-susceptible. Strain identification was performed by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS, bioMérieux, Marcy-l’Étoile, France), and further confirmed by PCR of 16S rRNA gene and housekeeping gene sequences [12]. Clinical features of the patient were then systematically obtained through electronic medical records, mainly including age, gender, disease diagnosis and prognosis, specimen origin and date, antibiotics usage, etc.
4.2. Antimicrobial Susceptibility Testing
According to the Clinical and Laboratory Standards Institute (CLSI), the minimal inhibition concentration (MIC) was determined through the broth microdilution method [36]. Antimicrobial agents, including imipenem, meropenem, ceftazidime, cefepime, aztreonam, ceftazidime-avibactam, cefoperazone-sulbactam, piperacillin-tazobactam, trimethoprim-sulfamethoxazole, ciprofloxacin, levofloxacin, amikacin, tigecycline, and polymyxin B, were tested and results were interpreted by breakpoints of 2021 CLSI, FDA (for tigecycline only) and EUCAST (for polymyxin B only) [36]. E. coli ATCC 25,922 was used as quality control for the antimicrobial susceptibility testing.
4.3. Plasmid Transformation Experiments
Plasmid DNAs were extracted from donor P. faecalis SC48 by phenol-chloroform method and then electroporated into recipient E. coli DH5α. These transformants were selected on Luria–Bertani agar plates containing ampicillin (50 mg/L) and subjected to PCR for detection of the blaOXA-181 gene using primers OXA-F (5′-GCGTGGTTAAGGATGAACAC-3′) and OXA-R (5′-CATCAAGTTCAACCCAACCG-3′) [37]. All PCR positive products were sequenced and comparatively analyzed for homology using BLASTn algorithms (http://blast.ncbi.nlm.nih.gov/Blast.cgi, (accessed on 15 February 2022)).
4.4. Pulsed-Field Gel Electrophoresis (PFGE)
With Salmonella braenderup H9812 as the reference marker, the clonality and plasmids of strains were confirmed by PFGE and S1-PFGE [38]. Briefly, bacterial DNA of P. faecalis was digested with the restriction endonuclease XbaI and that of the donor strain and transformant with S1-nuclease (TaKaRa, Beijing, China). PFGE was carried out at 14 °C for 20 h using a CHEF Mapper system (Bio-Rad Laboratories, Hercules, CA, USA).
4.5. Whole-Genome Sequencing and Analysis
Total DNA of P. faecalis strain SC48, SC62, and transformant E. coli DH5α-SC48-T was extracted and subjected to whole genome sequencing (WGS) via Illumina paired-end sequencing (Illumina, San Diego, CA, USA), and then de novo assembled by SPAdes 3.12.0 [39]. Antimicrobial resistance genes were analyzed by ResFinder 4.1 (https://cge.food.dtu.dk/services/ResFinder/, (accessed on 15 February 2022)) with a 90% threshold for gene identification and a 60% minimum length coverage.
5. Conclusions
As a rare species of Enterobacterales, a P. faecalis-mediating fatal bloodstream infection was found in China, the first to be clinically reported outside Pakistan since its discovery. The blaOXA-181 gene carried by P. faecalis was located on a globally disseminated IncX3 plasmid. The P. faecalis-related nosocomial infection indicated the potential worldwide spread of such bacteria, which was considered to be rare. Population mobility and environmental pollution might be the reason, so more clinical attention is required.
Author Contributions
Conceptualization, F.H. and Q.S.; methodology, F.H. and Q.S.; investigation, R.H. and L.D.; data curation, F.H. and Q.S.; writing—original draft preparation, Q.S. and F.H.; writing—review and editing, all authors; project administration, Y.Y. and S.W.; supervision, F.H.; funding acquisition, F.H., Y.G. and D.Y. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study protocol was approved by the Institutional Review Board of Huashan Hospital, Fudan University (No. 2018-408).
Informed Consent Statement
Not applicable.
Data Availability Statement
The nucleotide sequence of pSC48-OXA-181 containing blaOXA-181 and qnrS1 was deposited in the GenBank under accession number OK558605.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the National Natural Science Foundation of China (grant numbers 81871690, 81902101, and 81861138052), Three-year Action Plan for the Construction of Shanghai Public Health System (GWV-10.2-XD02), and China Antimicrobial Surveillance Network (grant number 2020QD049).
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Peleg A.Y., Hooper D.C. Hospital-acquired infections due to gram-negative bacteria. N. Engl. J. Med. 2010;362:1804–1813. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris P.N.A., Tambyah P.A., Lye D.C., Mo Y., Lee T.H., Yilmaz M., Alenazi T.H., Arabi Y., Falcone M., Bassetti M., et al. Effect of Piperacillin-Tazobactam vs Meropenem on 30-Day Mortality for Patients with E. coli or Klebsiella pneumoniae Bloodstream Infection and Ceftriaxone Resistance: A Randomized Clinical Trial. JAMA. 2018;320:984–994. doi: 10.1001/jama.2018.12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhanel G.G., Wiebe R., Dilay L., Thomson K., Rubinstein E., Hoban D.J., Noreddin A.M., Karlowsky J.A. Comparative review of the carbapenems. Drugs. 2007;67:1027–1052. doi: 10.2165/00003495-200767070-00006. [DOI] [PubMed] [Google Scholar]
- 4.Tian L., Tan R., Chen Y., Sun J., Liu J., Qu H., Wang X. Epidemiology of Klebsiella pneumoniae bloodstream infections in a teaching hospital: Factors related to the carbapenem resistance and patient mortality. Antimicrob. Resist. Infect. Control. 2016;5:48. doi: 10.1186/s13756-016-0145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Duin D., Arias C.A., Komarow L., Chen L., Hanson B.M., Weston G., Cober E., Garner O.B., Jacob J.T., Satlin M.J., et al. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): A prospective cohort study. Lancet Infect. Dis. 2020;20:731–741. doi: 10.1016/S1473-3099(19)30755-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu F., Wang M., Zhu D., Wang F. CHINET efforts to control antimicrobial resistance in China. J. Glob. Antimicrob. Resist. 2020;21:76–77. doi: 10.1016/j.jgar.2020.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Hu F., Zhu D., Wang F., Wang M. Current Status and Trends of Antibacterial Resistance in China. Clin. Infect. Dis. 2018;67:S128–S134. doi: 10.1093/cid/ciy657. [DOI] [PubMed] [Google Scholar]
- 8.Tilahun M., Kassa Y., Gedefie A., Ashagire M. Emerging Carbapenem-Resistant Enterobacteriaceae Infection, Its Epidemiology and Novel Treatment Options: A Review. Infect. Drug Resist. 2021;14:4363–4374. doi: 10.2147/IDR.S337611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han R., Shi Q., Wu S., Yin D., Peng M., Dong D., Zheng Y., Guo Y., Zhang R., Hu F. Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) Among Carbapenem-Resistant Enterobacteriaceae Isolated from Adult and Children Patients in China. Front. Cell Infect. Microbiol. 2020;10:314. doi: 10.3389/fcimb.2020.00314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kieffer N., Nordmann P., Aires-de-Sousa M., Poirel L. High Prevalence of Carbapenemase-Producing Enterobacteriaceae among Hospitalized Children in Luanda, Angola. Antimicrob. Agents Chemother. 2016;60:6189–6192. doi: 10.1128/AAC.01201-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang X., Shen S., Shi Q., Ding L., Wu S., Han R., Zhou X., Yu H., Hu F. First Report of blaIMP-4 and blaSRT-2 Coproducing Serratia marcescens Clinical Isolate in China. Front. Microbiol. 2021;12:743312. doi: 10.3389/fmicb.2021.743312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kämpfer P., Glaeser S.P., Raza M.W., Abbasi S.A., Perry J.D. Pseudocitrobacter gen. nov., a novel genus of the Enterobacteriaceae with two new species Pseudocitrobacter faecalis sp. nov., and Pseudocitrobacter anthropi sp. nov, isolated from fecal samples from hospitalized patients in Pakistan. Syst. Appl. Microbiol. 2014;37:17–22. doi: 10.1016/j.syapm.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 13.Kaempfer P., Fuglsang-Damgaard D., Overballe-Petersen S., Hasman H., Hammerum A.M., Fuursted K., Blom J., Glaeser S.P., Hansen F. Taxonomic reassessment of the genus Pseudocitrobacter using whole genome sequencing: Pseudocitrobacter anthropi is a later heterotypic synonym of Pseudocitrobacter faecalis and description of Pseudocitrobacter vendiensis sp. nov. Int. J. Syst. Evol. Microbiol. 2020;70:1315–1320. doi: 10.1099/ijsem.0.003918. [DOI] [PubMed] [Google Scholar]
- 14.Lu Z., Pérez-Díaz I.M., Hayes J.S., Breidt F. Bacteriophages Infecting Gram-Negative Bacteria in a Commercial Cucumber Fermentation. Front. Microbiol. 2020;11:1306. doi: 10.3389/fmicb.2020.01306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abarca J.G., Whitfield S.M., Zuniga-Chaves I., Alvarado G., Kerby J., Murillo-Cruz C., Pinto-Tomás A.A. Genotyping and differential bacterial inhibition of Batrachochytrium dendrobatidis in threatened amphibians in Costa Rica. Microbiology. 2021;167:001017. doi: 10.1099/mic.0.001017. [DOI] [PubMed] [Google Scholar]
- 16.Al-Kharousi Z.S., Guizani N., Al-Sadi A.M., Al-Bulushi I.M., Shaharoona B. Hiding in Fresh Fruits and Vegetables: Opportunistic Pathogens May Cross Geographical Barriers. Int. J. Microbiol. 2016;2016:4292417. doi: 10.1155/2016/4292417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Andrade L.K., Levican A., Cerdeira L., De Morais A.B.Z., Braz M.M., Martins E.R., Casella T., Moura Q., Fuga B., Lincopan N., et al. Carbapenem-resistant IMP-1-producing Pseudocitrobacter vendiensis emerging in a hemodialysis unit. Braz. J. Microbiol. 2022;53:251–254. doi: 10.1007/s42770-021-00638-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cui J., Li M., Cui J., Wang J., Qiang X., Liang Z. The proportion, species distribution and dynamic trends of bloodstream infection cases in a tertiary hospital in China, 2010–2019. Infection. 2021;50:121–130. doi: 10.1007/s15010-021-01649-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhu S., Kang Y., Wang W., Cai L., Sun X., Zong Z. The clinical impacts and risk factors for non-central line-associated bloodstream infection in 5046 intensive care unit patients: An observational study based on electronic medical records. Crit. Care. 2019;23:52. doi: 10.1186/s13054-019-2353-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timsit J.F., Ruppé E., Barbier F., Tabah A., Bassetti M. Bloodstream infections in critically ill patients: An expert statement. Intensive Care Med. 2020;46:266–284. doi: 10.1007/s00134-020-05950-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Widmer A.F., Kern W.V., Roth J.A., Dettenkofer M., Goetting T., Bertz H., Theilacker C. Early versus late onset bloodstream infection during neutropenia after high-dose chemotherapy for hematologic malignancy. Infection. 2019;47:837–845. doi: 10.1007/s15010-019-01327-0. [DOI] [PubMed] [Google Scholar]
- 22.Boev C., Kiss E. Hospital-Acquired Infections: Current Trends and Prevention. Crit. Care Nurs. Clin. 2017;29:51–65. doi: 10.1016/j.cnc.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 23.Frost I., Van Boeckel T.P., Pires J., Craig J., Laxminarayan R. Global geographic trends in antimicrobial resistance: The role of international travel. J. Travel Med. 2019;26:taz036. doi: 10.1093/jtm/taz036. [DOI] [PubMed] [Google Scholar]
- 24.D’Souza A.W., Boolchandani M., Patel S., Galazzo G., Van Hattem J.M., Arcilla M.S., Melles D.C., De Jong M.D., Schultsz C., Dantas G., et al. Destination shapes antibiotic resistance gene acquisitions, abundance increases, and diversity changes in Dutch travelers. Genome Med. 2021;13:79. doi: 10.1186/s13073-021-00893-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J., Bi Z., Ma S., Chen B., Cai C., He J., Schwarz S., Sun C., Zhou Y., Yin J., et al. Inter-host Transmission of Carbapenemase-Producing Escherichia coli among Humans and Backyard Animals. Environ. Health Perspect. 2019;127:107009. doi: 10.1289/EHP5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi X., Li Y., Yang Y., Shen Z., Cai C., Wang Y., Walsh T.R., Shen J., Wu Y., Wang S. High prevalence and persistence of carbapenem and colistin resistance in livestock farm environments in China. J. Hazard Mater. 2021;406:124298. doi: 10.1016/j.jhazmat.2020.124298. [DOI] [PubMed] [Google Scholar]
- 27.Wang Y., Zhang R., Li J., Wu Z., Yin W., Schwarz S., Tyrrell J.M., Zheng Y., Wang S., Shen Z., et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat. Microbiol. 2017;2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 28.Van Der Zwaluw K., Witteveen S., Wielders L., Van Santen M., Landman F., De Haan A., Schouls L.M., Bosch T. Molecular characteristics of carbapenemase-producing Enterobacterales in the Netherlands; Results of the 2014–2018 national laboratory surveillance. Clin. Microbiol. Infect. 2020;26:1412.e7–1412.e12. doi: 10.1016/j.cmi.2020.01.027. [DOI] [PubMed] [Google Scholar]
- 29.Liu C., Fang Y., Zeng Y., Lu J., Sun Q., Zhou H., Shen Z., Chen G. First Report of OXA-181-Producing Klebsiella pneumoniae in China. Infect. Drug Resist. 2020;13:995–998. doi: 10.2147/IDR.S237793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Y., Feng Y., Wu W., Xie Y., Wang X., Zhang X., Chen X., Zong Z. First Report of OXA-181-Producing Escherichia coli in China and Characterization of the Isolate Using Whole-Genome Sequencing. Antimicrob. Agents Chemother. 2015;59:5022–5025. doi: 10.1128/AAC.00442-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pitout J.D.D., Peirano G., Kock M.M., Strydom K.A., Matsumura Y. The Global Ascendency of OXA-48-Type Carbapenemases. Clin. Microbiol. Rev. 2019;33:e00102–e00119. doi: 10.1128/CMR.00102-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hendrickx A.P.A., Landman F., De Haan A., Witteveen S., Van Santen-Verheuvel M.G., Schouls L.M., Dutch CPE Surveillance Study Group Bla OXA-48-like genome architecture among carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the Netherlands. Microb. Genom. 2021;7:000512. doi: 10.1099/mgen.0.000512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mouftah S.F., Pál T., Darwish D., Ghazawi A., Villa L., Carattoli A., Sonnevend Á. Epidemic IncX3 plasmids spreading carbapenemase genes in the United Arab Emirates and worldwide. Infect. Drug Resist. 2019;12:1729–1742. doi: 10.2147/IDR.S210554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Y., Fang L., Yang Y., Yan R., Fu Y., Shen P., Zhao D., Chen Y., Hua X., Jiang Y., et al. Emergence of carbapenem-resistant Klebsiella pneumoniae harbouring blaOXA-48-like genes in China. J. Med. Microbiol. 2021;70:001306. doi: 10.1099/jmm.0.001306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Redzej A., Ukleja M., Connery S., Trokter M., Felisberto-Rodrigues C., Cryar A., Thalassinos K., Hayward R.D., Orlova E.V., Waksman G. Structure of a VirD4 coupling protein bound to a VirB type IV secretion machinery. EMBO J. 2017;36:3080–3095. doi: 10.15252/embj.201796629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wayne P.A. Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. 31st ed. Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2021. [Google Scholar]
- 37.Poirel L., Walsh T.R., Cuvillier V., Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011;70:119–123. doi: 10.1016/j.diagmicrobio.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 38.Barton B.M., Harding G.P., Zuccarelli A.J. A general method for detecting and sizing large plasmids. Anal. Biochem. 1995;226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 39.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The nucleotide sequence of pSC48-OXA-181 containing blaOXA-181 and qnrS1 was deposited in the GenBank under accession number OK558605.