Abstract
On Bird Island, South Georgia, a new strain of Chlamydophila abortus was detected in one Brown skua out of 37 specimens from six different seabird species. Phylogenetic analysis of the rnpB and omp1 genes indicated the strain to be more closely related to C. abortus than to 6BC, the type strain of Chlamydophila psittaci.
The family Chlamydiaceae was recently reclassified and now comprises nine separate species (5) that infect a wide variety of animals. Chlamydophila psittaci (previously Chlamydia psittaci) has been detected in at least 130 bird species (1, 14, 19).
In this study we detected Chlamydophila abortus (previously a member of the Chlamydia psittaci group) in a Brown skua (Catharacta antarctica lonnbergi) on Bird Island (54°1′S, 58°3′W), South Georgian archipelago. The avifauna is abundant, with 24 breeding species of seabirds (17). Shedding organisms from the respiratory tract were obtained by fecal swabs (22) from 37 birds of six different species of subantarctic seabirds. Transportation from Bird Island occurred irregularly; swabs were therefore stored for 3 weeks at −20°C in 0.2 M sucrose-phosphate-buffered saline before being transported to the laboratory. Since culture of C. psittaci requires a biosafety lab of class 3, isolation was not attempted.
DNA was extracted from feces by using a QIAamp tissue kit (Qiagen, Hilden, Germany), and the rnpB and omp1 genes were amplified by PCR. The rnpB gene encodes the catalytically active RNA subunit of RNase P and is present in all prokaryotic cells and therefore useful for taxonomic analysis (10). For PCR amplification and subsequent sequencing of the Chlamydophila sp. obtained from specimen R54 (designated strain R54) from a Brown skua, the primer pair JB1 and JB2 was used (10).
The chlamydial outer membrane protein encoded by omp1 shows variation between species and strains (8, 11), and the gene was used for characterization of strain R54. For amplification of a 1,032-bp gene segment, a seminested PCR method was used according to the work of Kaltenboeck et al. (12) with minor modifications. The primers in the first step were 9CTROMP (5′GCTCTGCCTGTGGGGAATCCTGCTGAACC3′) and CHOMP371 [5′TTAGAAIC(GT)GAATTGIGC(AG)TTIA(TC)GTGIGCIGC3′], and in the second step the upstream primer was replaced by 29CTROMP (5′GGAGATCCTTGCGATCCTTG3′). The resulting PCR products were sequenced by using terminator-labeled cycle sequencing chemistry and sequence primers, including 29CTROMP, 191CHOMP (5′GCIYTITGGGARTGYGGITGYGCIAC3′), CTR215 [5′TCTTCGA(C/T)TTT(A/T)GGTTTAGATTGA3′], and CHOMP371. Sequence reactions were analyzed on a 310 Genetic Analyzer (PE Biosystems, Norwalk, Conn.).
Sequence alignment was based on a previous analysis (10) and use of the CLUSTAL W multiple alignment program (21). Phylogenetic analysis of the calculated distance matrix was done by using the neighbor-joining program, as previously described (10), and the obtained tree was displayed by using TREEVIEW (15).
In 37 samples from seabirds, one case of chlamydial infection (R54) was detected. The rnpB nucleotide sequence of the R54 strain showed a similarity of 97.7% (343 of 351 positions, primer sequences excluded) to a sequence found in nine C. psittaci strains, including serovars A to F (10). All eight discrepant nucleotide positions in the 396-bp-long gene product were located in the variable regions of the rnpB gene. Interestingly, the rnpB gene in strain R54 showed highest similarity (99.2%) to the C. abortus rnpB gene when it was compared with all nine species in the Chlamydiacae family. The previously determined rnpB sequences were identical in eight C. abortus strains of bovine, ovine, or caprine origin, but none were derived from birds. Thus, our data from the rnpB gene demonstrate that the R54 strain is more closely related to C. abortus than to C. psittaci.
The striking similarity between R54 and C. abortus was supported by analysis of the partially determined sequence (979 bp) of the omp1 gene in R54. Comparison with previously described nucleotide sequences showed highest similarity (90.7 to 90.9%) to four C. abortus strains inducing ovine (B577T [13], S26/3 [9]) or bovine (BA1 [7]) abortion or enteritis in cattle (LW508 [13]). The sequences were almost identical (90.1%) when R54 was compared to an avian C. psittaci type C strain, which is reported to have greater homology to abortion-inducing strains of C. psittaci (now C. abortus) than to avian C. psittaci group A or E strains (see National Center for Biotechnology Information [www3.ncbi.nlm.nih.gov/9 July 1999] accession no. L25436). Several isolated C. psittaci strains of porcine origin showed even greater similarity (88.3%) to R54 than to other avian strains of serovar A (type strain 6BC, 81.4% similarity [4]) and serovar D (strain 92-1293, 85.8% similarity [23]). Thus, although serotyping was based on the major outer membrane protein, the immunological reactivity pattern was not directly correlated to the sequence similarity of the omp1 gene. Serotyping may be used to differentiate strains, but does not fully reflect taxonomic relationships.
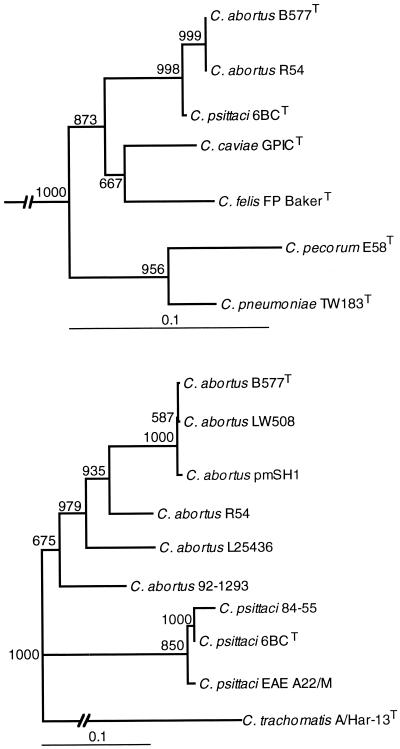
Based on the recent reclassification of members of the Chlamydiaceae (5), R54 may be classified as C. abortus. Phylogenetic analysis by a neighbor-joining program of both the rnpB and the omp1 gene indicated that strain R54 is genetically distinct from C. abortus and is still more separated from C. psittaci (Fig. 1). Further analysis of other genes, such as the 16S RNA (16), the 23S RNA, and the ribosomal intergenic spacer (3) genes might reveal if congruent evolution has occurred and lead to reclassification of some avian Chlamydophila strains.
FIG. 1.
Neighbor-joining trees based on rnpB (upper), showing relationships between the avian strain R54 and type strains of Chlamydophila species, and omp1 (lower), showing relationships among strain R54 and strains of Chlamydophila species. Both trees were outgrouped to Chlamydia trachomatis strain A/Har-13T (indicated with broken line in the upper tree). The denoted bootstrap values were obtained from 1,000 resamplings of each data set. The bar indicates 0.1 substitutions per nucleotide.
Certain avian C. psittaci strains have been reported to show similarity to ovine abortion-inducing strains in restriction enzyme analysis (6) and in the omp1 sequence (20); likewise, an avian serovar B was reported from a case of bovine abortion (2). However, the risk of laboratory contamination between avian and abortion-inducing C. psittaci strains has been suggested as a possible explanation for the exceptional cases of similarity between strains of different host origins (9, 18). Since both the rnpB and the omp1 sequences of our R54 strain are unique, yet similar to sequences in abortion-inducing strains, it is evident that some avian strains are more similar to abortion-inducing strains than to other avian strains. Further investigations are needed to demonstrate if C. abortus-like strains in birds cause clinical manifestations that are different from infections with typical C. psittaci strains.
In conclusion, phylogenetic analysis indicated that avian strain R54 is genetically more closely related to C. abortus than to the type strain of C. psittaci. The impact of Chlamydophila infection on the Antarctic fauna must be one of the aims of further studies.
Nucleotide sequence accession numbers.
Sequences obtained from the R54 specimen were sent to GenBank with the accession numbers AJ243523 for rnpB and AJ243525 for omp1.
Acknowledgments
We gratefully acknowledge the support given to this project by the British Antarctic Survey and the Swedish Polar Research Secretariat. Furthermore, we thank Paul Haemig for valuable comments on the manuscript.
This work was supported financially by the Centre for Environmental Research, the Medical Faculty of Umeå University, the Swedish Council for Agricultural Research (23.0161), and the Swedish Society of Medicine.
ADDENDUM IN PROOF
In an analysis of ompA and rRNA sequences by R. Bush and K. D. E. Everett (personal communication) C. abortus appears to be evolving away from a cluster of R54-like strains, rather than as a sister clade to C. psittaci. Therefore, both ecological data and DNA sequence data must be considered for species identification of a new chlamydial strain. Strain R54 groups ecologically with C. psittaci because it was isolated from birds and because there is no evidence that it targets placenta or causes abortion in mammals.
REFERENCES
- 1.Burkhart R L, Page L A. Chlamydiosis (ornithosis-psittacosis) In: Davis J W, Anderson R C, Karstad L, editors. Infectious and parasitic diseases of wild birds. Ames, Iowa: Iowa State University Press; 1971. pp. 118–140. [Google Scholar]
- 2.Cox H U, Hoyt P G, Poston R P, Snider III T G, Lemarchand T X, O'Reilly K L. Isolation of an avian serovar of Chlamydia psittaci from a case of bovine abortion. J Vet Diagn Investig. 1998;10:280–282. doi: 10.1177/104063879801000310. [DOI] [PubMed] [Google Scholar]
- 3.Everett K D, Andersen A A. The ribosomal intergenic spacer and domain I of the 23S rRNA gene are phylogenetic markers for Chlamydia spp. Int J Syst Bacteriol. 1997;47:461–473. doi: 10.1099/00207713-47-2-461. [DOI] [PubMed] [Google Scholar]
- 4.Everett K D, Andersen A A, Plaunt M, Hatch T P. Cloning and sequence analysis of the major outer membrane protein gene of Chlamydia psittaci 6BC. Infect Immun. 1991;59:2853–2855. doi: 10.1128/iai.59.8.2853-2855.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Everett K D E, Bush R M, Andersen A A. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae including a new genus and five new species, and standards for the identification of organisms. Int J Syst Bacteriol. 1999;49:415–440. doi: 10.1099/00207713-49-2-415. [DOI] [PubMed] [Google Scholar]
- 6.Fukushi H, Hirai K. Genetic diversity of avian and mammalian Chlamydia psittaci strains and relation to host origin. J Bacteriol. 1989;171:2850–2855. doi: 10.1128/jb.171.5.2850-2855.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffiths P C, Plater J M, Martin T C, Hughes S L, Hughes K J, Hewinson R G, Dawson M. Epizootic bovine abortion in a dairy herd: characterization of a Chlamydia psittaci isolate and antibody response. Br Vet J. 1995;151:683–693. doi: 10.1016/s0007-1935(95)80149-9. [DOI] [PubMed] [Google Scholar]
- 8.Herring A J. Typing Chlamydia psittaci—a review of methods and recent findings. Br Vet J. 1993;149:455–475. doi: 10.1016/S0007-1935(05)80111-3. [DOI] [PubMed] [Google Scholar]
- 9.Herring A J, Tan T W, Baxter S, Inglis N F, Dunbar S. Sequence analysis of the major outer membrane protein gene of an ovine abortion strain of Chlamydia psittaci. FEMS Microbiol Lett. 1989;53:153–158. doi: 10.1016/0378-1097(89)90383-2. [DOI] [PubMed] [Google Scholar]
- 10.Herrmann B, Pettersson B, Everett K D E, Mikkelsen N E, Kirsebom L A. Characterization of the rnpB gene and the RNase P RNA in the order of Chlamydiales. Int J Syst Evol Microbiol. 2000;50:149–158. doi: 10.1099/00207713-50-1-149. [DOI] [PubMed] [Google Scholar]
- 11.Kaltenböck B, Schmeer N, Schneider R. Evidence for numerous omp1 alleles of porcine Chlamydia trachomatis and novel chlamydial species obtained by PCR. J Clin Microbiol. 1997;35:1835–1841. doi: 10.1128/jcm.35.7.1835-1841.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaltenboeck B, Kousoulas K G, Storz J. Two-step polymerase chain reactions and restriction endonuclease analyses detect and differentiate ompA DNA of Chlamydia spp. J Clin Microbiol. 1992;30:1098–1104. doi: 10.1128/jcm.30.5.1098-1104.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaltenboeck B, Kousoulas K G, Storz J. Structures of and allelic diversity and relationships among the major outer membrane protein (ompA) genes of the four chlamydial species. J Bacteriol. 1993;175:487–502. doi: 10.1128/jb.175.2.487-502.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Page L A. Observations on the involvement of wildlife in an epidemic of chlamydiosis in domestic turkeys. J Am Vet Med Assoc. 1976;169:932–935. [PubMed] [Google Scholar]
- 15.Page R D M. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 16.Pettersson B, Andersson A, Leitner T, Olsvik O, Uhlen M, Storey C, Black C M. Evolutionary relationships among members of the genus Chlamydia based on 16S ribosomal DNA analysis. J Bacteriol. 1997;179:4195–4205. doi: 10.1128/jb.179.13.4195-4205.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prince P A, Croxall J P. Birds of South Georgia: new records and re-evaluations of status. Br Antarct Surv Bull. 1983;59:15–27. [Google Scholar]
- 18.Sayada C, Andersen A A, Storey C, Milon A, Eb F, Hashimoto N, Hirai K, Elion J, Denamur E. Usefulness of omp1 restriction mapping for avian Chlamydia psittaci isolate differentiation. Res Microbiol. 1995;146:155–165. doi: 10.1016/0923-2508(96)80893-x. [DOI] [PubMed] [Google Scholar]
- 19.Simpson V R, Bevan R. Chlamydia psittaci in robins. Vet Rec. 1989;1989(125):537. doi: 10.1136/vr.125.21.537-b. [DOI] [PubMed] [Google Scholar]
- 20.Storey C, Lusher M, Yates P, Richmond S. Use of comparative MOMP gene sequence data for subdivision of Chlamydia psittaci species. In: Mårdh P A, La Placa M, Ward M, editors. Proceedings of the European Society for Chlamydia Research. Uppsala, Sweden: Uppsala University Centre for STD Research; 1992. p. 191. [Google Scholar]
- 21.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanrompay D, Ducatelle R, Haesebrouck F. Chlamydia psittaci infections: a review with emphasis on avian chlamydiosis. Vet Microbiol. 1995;45:93–119. doi: 10.1016/0378-1135(95)00033-7. [DOI] [PubMed] [Google Scholar]
- 23.Vanrompay D, Cox E, Mast J, Goddeeris B, Volckaert G. High-level expression of Chlamydia psittaci major outer membrane protein in COS cells and in skeletal muscles of turkeys. Infect Immun. 1998;66:5494–5500. doi: 10.1128/iai.66.11.5494-5500.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]