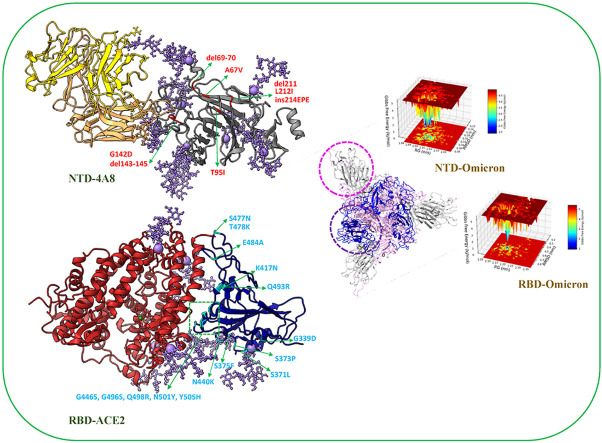
Abstract
Since the new variant of SARS-CoV-2, Omicron (BA.1) has raised serious concerns, it is important to investigate the effects of mutations in the NTD and RBD domains of the spike protein for the development of COVID-19 vaccines. In this study, computational analysis of the Wuhan and Omicron NTDs and RBDs in their unbound and bound states to mAb 4A8 and ACE2 were performed. In addition, the interaction of NTD with antibody and RBD with ACE2 were evaluated in the presence of long glycans. The results show that long glycans at the surface of NTDs can reduce the accessibility of protein epitopes, thereby reducing binding efficiency and neutralizing potency of specific antibodies. Also, our findings indicate that the existence of the long glycans result in increased stability and enhanced affinity of the RBD to ACE2 in the Wuhan and Omicron variant. Key residues that play an important role in increasing the structural stability of the protein were identified using RIN analysis and in the state of interaction with mAb 4A8 and ACE2 through per-residue decomposition analysis. Further, the results of the free energy binding calculation using MM/GBSA method show that the Omicron variant has a higher infectivity than the Wuhan. This study provides a better understanding of the structural changes in the spike protein and can be useful for the development of novel therapeutics.
Keywords: SARS-CoV-2, Omicron (B.1.1.529, BA.1), N-terminal domain (NTD), Receptor binding domain (RBD), Molecular dynamics simulations, COVID-19
Abbreviations: ACE2, Angiotensin Converting Enzyme 2; COVID-19, Coronavirus disease 2019; SARS-CoV-2, severe acute syndrome coronavirus; RBD, receptor-binding domain; NTD, N-terminal domain; BFE, binding free energy; ESURF, surface area; vdW, Van der Waal; GB, generalized born; MMGBSA, the molecular mechanics, the generalized Born model and solvent accessibility; MD, Molecular-dynamics; VOC, Variant of Concern; mAb, monoclonal antibodies
Graphical abstract
1. Introduction
Coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) led to global distortions in everyday life, and millions of people have died as a result of infection by this virus. SARS-CoV-2 is a single-stranded RNA-enveloped virus enclosed with 5′-cap and 3′-poly(A) tail and containing four structural proteins, spike (S), envelope (E), membrane (M), nucleocapsid (N), and 16 non-structural proteins [[1], [2], [3]]. S protein is homotrimeric, consisting of two subunits, S1 and S2. The spike protein is a critical component for binding of the virus to host cells, owing to specific interactions via its receptor binding domain (RBD) with the angiotensin-converting enzyme 2 (ACE2) receptor, thereby mediating virus attachment to host cells. Recent computational results, demonstrated that single-wall carbon nanotube had an affinity to the S1 subunit in the spike glycoprotein [3]. The RBD contains a receptor-binding motif (RBM), which is the most important functional region of the spike protein because of its direct role in the interaction with ACE2 [2]. After two years of global spread of SARS-CoV-2 around the globe, the virus continues to evolve into new variants of concerns (VOCs) with improved transmissibility and resistance to antibodies, especially in vulnerable populations. In November 2021, Botswana, South Africa (SA), and a number of other countries have reported a novel variant of SARS-CoV-2 (lineage B.1.1.529), named Omicron [4]. To date, several Omicron sub-lineages including BA.1, BA.1.1, BA.2, BA.3, BA.4, BA.5 and recombinant forms of BA.1/BA.2 such as XE have been reported [5]. Evidence suggests that Omicron (BA.1) with a large number of mutations, especially in the spike protein (26–32 mutations), containing two deletions and seven substitutions mutations found in the N-terminal domain (NTD) and fifteen substitutions occurred in the RBD, has the feature of escaping the immune system and higher transmissibility [6]. The urgent need for an effective therapeutic approach against this variant has attracted the attention of researchers in the design of novel drugs and the study of databases such as STRING, VARIDT, SYNBIP [[7], [8], [9]]. Computational approaches have a significant role in performing new research studies to recognition and treatment of wide range of diseases like neuroscience diseases and viral diseases including COVID-19 and other diseases [10]. Great efforts by have been paid to disrupt the replication of the virus including drug design techniques, vaccine design, discovering inhibitors of key proteins and design of nAbs and SBPs [8,11,12]. In addition, to facilitate drug discovery efforts, internet resources such as Target Therapy Database (TTD) and DrugBank provide freely accessible and comprehensive information on drug pathways and targets in different development and clinical stages [[13], [14], [15]].
In this study, we used all-atom molecular dynamic simulations to understand the structural dynamics of the N-terminal domain (NTD), the receptor binding domain (RBD), and to characterize peculiarities of the interactions between the ACE2 and RBD from the Wuhan and Omicron variants. In addition, since virus glycosylation is a successful strategy in escaping the immune system, we evaluated the effect of glycosylation on protein structure and protein-protein interactions [16]. Furthermore, the residue interaction networks (RINs) approach was used to investigate the effects of mutations on the RBD and NTD structures. In the RIN model, the network with nodes and edges corresponding to residues and interactions between residue side chains, respectively, is built and studied [17]. The results of this study provide grounds for better understanding of the molecular mechanisms of Omicron infectivity and can be used for the design and development of novel interventions.
2. Computational methods
2.1. Sequence and structure alignment
The sequences of NTD and RBD of Wuhan and Omicron (BA.1) variant of the SARS-CoV-2 spike proteins were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/) entries with the accession numbers YP_009724390.1 and UFO69279.1, respectively. The Wuhan and Omicron NTDs contain 292 (14–305) and 289 (14–302) residues, whereas Wuhan (residues 319–541) and Omicron RBDs (residues 316–538) consist of 223 residues. 3D model of the Wuhan NTD and RBD were made by CHARMM-GUI PDB Reader and (PDB ID: 6VSB) was used as a template. I-TASSER server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) was utilized for prediction of the 3-dimensional structures of corresponding Omicron NTD and RBD. The quality assessment of the predicted structures were done using PROCHECK [2,18], followed by validation using ProSA [19] and SAVES v6.0 (https://saves.mbi.ucla.edu/). Notably, there were no Ramachandran outliers in the NTDs and RBDs (Table S1, Figs. S1 and S2). The structure and sequence alignment were performed using the EMBOSS Needle server [20] and the results were visualized by the ESpript3 software [21].
2.2. System preparation
As the reference structure, the experimental crystal structure of the RBD-ACE2 complexes, PDB ID: 6M0J (residue 333–526 of the spike protein from the Wuhan) were used in our study [21]. Missing protein residues and mutations were then introduced to the Wuhan structure using CHARMM-GUI Solution Builder tool using GalaxyFill [22] and PyMOL program [23,24]. The NTD-4A8 with the PDB code 7C2L was retrieved from the protein data bank [25]. We prepared fourteen different systems that include the spike NTD-4A8 and RBD-ACE2 complexes as well as the NTD and RBD only systems for each of the Wuhan and Omicron variant. CHARMM-GUI was used to attach the N-glycan chains at the glycosylation sites of the six systems including NTD only systems, NTD-4A8 and RBD-ACE2 complexes ( Table S2 ) [26].
2.3. Molecular dynamics simulations
Topologies of all systems were generated by CHARMM-GUI solution builder module using the CHARMM36(m) all-atom force-field with cut off distances 12 Å for van der Waals interactions, and were neutralized with 0.15 M KCl and solvated using the TIP3P water model. The systems were embedded in a rectangular water box with an edge distance of 10 Å [[27], [28], [29]]. Molecular dynamics simulations carried out using the GROMACS 2020.6 with the Charmm36 m force field, WYF parameter [30] and Hydrogen mass repartitioning (HMR). Since cation-π interactions play an important role in biomolecular identification processes and stability of protein structure, WYF parameter was used to improve the MD simulation, which contains free energies, of cation-indole-choline and indole-trimethylated lysines interactions related to protein-protein interfaces [31]. Hydrogen mass repartitioning (HMR) technique permits all-atom MD simulation to employ time steps of up to 4 fs by devoting a heavy atom mass to hydrogen atom(s) linked to the heavy atom [32]. The linear-constraint-solving (LINCS) algorithms was used to limit bonds involving hydrogen atoms [33,34]. The systems were minimized in 5000 steps, and equilibration was performed for 125 ps with constant volume and temperature (NVT). Next, the production step was carried out under a constant pressure and temperature of 310 K (NPT). The single protein systems were run for 100 ns each, and NTD-4A8 and RBD-ACE2 systems production simulations were performed for 50 and 85 ns, respectively. The trajectory analysis was performed using GROMACS tools, the parameters RMSD, RMSF, radius of gyration (Rg), SASA, and the amount of H-bonds were all included, finally trajectories were visualized by PyMol and UCSF Chimera 61 [35].
2.4. RINs analysis
RING 2.0 (http://old.protein.bio.unipd.it/ring/) [36,37] was used to generate RINs for the free forms of the NTDs and RBDs of Wuhan and Omicron variant. Cytoscape was then used to visualize the RING output. The MCODE plugin in Cytoscape was utilized to find clusters of the resulting networks.
2.4.1. Key residues in the NTD and RBD of Wuhan and Omicron variant
The Network Analyzer program [38] in Cytoscape was used to calculate the local metrics betweenness and stress, and key residues were defined using these two parameters. In order to find key residues, the Z-score of each residue was calculated, and if the Z-score absolute value was more than two, the residue was considered as a key residue.
2.5. Free energy landscape
The free energy landscape of the NTDs and RBDs of the Wuhan and Omicron variant were measured using geo_measures v 0.8 [39]. Geo measures contains a powerful library of g sham and form the MD trajectory against RMSD and Radius of gyration (Rg) energy profile of folding recorded in a 3D plot using the python package matplotlib. The energy is calculated as Gibb's free energy derived from protein enthalpy and entropy.
2.6. Principal component analysis
Principal component analysis (PCA), a statistical method based on covariance analysis, was performed using GROMACS. PCA was used to analyze trajectory data of the MD simulations to recognize the configurational space of a harmonic motion with just a few degrees of freedom [40].
2.7. Free energy binding and ΔΔG (destabilizing)
DynaMut2 server was used to evaluate the effect of some important mutations of the Omicron variant on the folding free energy (ΔΔGDestabilizing) RBD-ACE2 complexes [41]. Overall, if the ΔΔG value is below zero, it indicates that the mutation has caused destabilization of the protein; otherwise, it has induced protein stabilization.
2.8. Binding free energy (BFE) calculation
End-state free energy calculations was carried out by gmx_MMPBSA, which is under MMPBSA.py calculation engine and written with Python 3.8 [42]. gmx_MMPBSA_ana tool, graphical user interface, generates different plots from gmx_MMPBSA output files to visualize and analyze the results [42].
The free binding energy for a complex was calculated by the following equation:
| ΔGbind = ΔGcomplex − ΔGreceptor − ΔGligand |
Per-residue free energy decomposition analysis was performed using MM/GBSA method in order to investigate the key residues involved in protein-protein interaction.
3. Results
3.1. Sequence and structure alignment
The pairwise amino acid sequence alignment revealed that the Wuhan NTD shares 95.24% identity and 97.96% similarity with the Omicron NTD (Fig. 1 A). Between the anti-parallel β-strands, the glycan-free surface in the NTD includes several loops. According to previous studies [43], this region is very electropositive and is targeted by many antibodies. Based on the analysis of the antibody interaction with NTD [44], it's called 5 loops region, as it contains loops N1 (residues 14–26), N2 (residues 67–79), N3 (residues 141–156), N4 (residues 177–186), and N5 (residues 246–260), among which loops 3 and 5 play the most important part in the interaction with antibodies. As it follows from our 3D-structural superimposition (Fig. S3A), Omicron mutations in the NTD have altered the structure of this domains compared to the Wuhan NTD. For example, deletion of residues 143–145 and the G142D mutation led to the N3 loop deformation and in the glycan movement. It is probably one of the viral strategies to escape the antibodies. Moreover, the Wuhan RBD shares 93.3% identity and 95.1% similarity with the Omicron RBD (Fig. 1B and Fig. S3B). The secondary structure of the Wuhan RBD contains five antiparallel β sheets (β1, β2, β3, β4 and β7) with short connecting helices and loops. The receptor-binding motif (RBM), which contains most of the contacting residues of SARS-CoV-2 that bind to ACE2 is an extended region between the β4 and β7 strands, stretching from ARG438 to TYR508 in the Wuhan structure. In comparison with the Wuhan RBD, fifteen mutations are found in the Omicron RBD and ten out of fifteen substitution mutations including N440K, G446S, S477 N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H are located in the RBM which interacts directly with the ACE2. Previous studies indicates that G446S, G496S, and Q498R mutations cooperatively alter the loop residues 443–451 and S371L, S373P, and S375F mutations found in the Omicron RBD not only alter the conformation of loop residues 371–376 that could change the conformation of the N-linked glycosylation at ASN343 which are important for interaction between the RBD and ACE2 [45].
Fig. 1.
Structure and sequence alignment between the (A) NTD-Wuhan and NTD-Omicron. Antibodiy (4A8) intraction pattern with residues is marked by arrows. (B) RBD-Wuhan and RBD-Omicron. ACE2 intraction pattern with residues is marked by arrows. Purple box indicates the RBM region. (α and η: alpha helix with squiggles, β: beta strands with arrows and turns with TT letters).
3.2. Molecular dynamics study
3.2.1. The root-mean-square deviations (RMSDs)
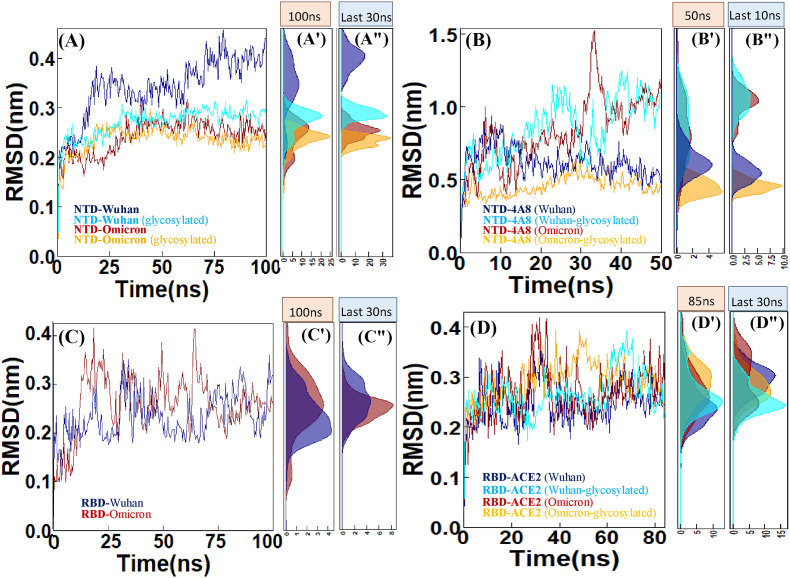
The root-mean-square deviations of the backbone atoms of all systems were analyzed to determine structural stability and convergence. The results of the comparative stability analysis during the 100 ns MD simulation time indicated that the RMSD values of the Wuhan NTD show high fluctuations. It is noticeable that unlike the Wuhan NTD, the RMSD values for the Omicron NTD show less variability during 40–100 ns of simulation. In addition, the analysis of the effect of N-glycosylation with the long glycans on protein dynamics clearly shows a decrease in the flexibility during 40–100 ns of the MD simulation time (Fig. 2 A). RMSDs were also evaluated for the complexes of the Wuhan and Omicron NTDs with mAb 4A8. According to (Fig. 2B), the glycosylated Wuhan NTD-4A8 complex fluctuates more than the glycosylated Omicron NTD-4A8 complex. Presumably one of the reasons for the higher stability of the antibody-Omicron NTD complex is the presence of glycan mass at this position, which restricts the antibody tendency to move and force it to stay in the more fixed position. Because the results of Omicron NTD-4A8 complex clearly show more deviation than Wuhan NTD-4A8.
Fig. 2.
RMSD plots of the (A) NTD-Wuhan and NTD-Omicron and evaluation of these two systems in glycosylated state. (B) NTD-4A8 (Wuhan) and NTD-4A8 (Omicron) and evaluation of these two systems in glycosylated state. (C) RBD-Wuhan and RBD-Omicron. (D) RBD-ACE2 (Wuhan) and RBD-ACE2 (Omicron) and evaluation of these two systems in glycosylated state. (A′, A″, B′, B″,C′, C″, D′ and D″) Density Function of RMSD sampled over the simulations are shown in histograms.
The average values of the RMSDs for the Wuhan RBD and Omicron RBD were found to be ∼0.46 and ∼0.51 nm, respectively. The plot indicates that the highest value of RMSD (∼0.82 nm) was found around 18 ns and 64 ns of simulation. In the last 30 ns of simulation, both systems become stable. Based on the RMSD plot, the Wuhan RBD is more stable than the Omicron RBD (Fig. 2C). As (Fig. 2D) shows, in the last 15 ns of the simulation time both Wuhan and Omicron glycosylated complexes showed less deviations than their counterparts. In addition, based on the RMSD plot both Omicron RBD-ACE2 complexes show stable dynamic and it can be deduced that substitution mutations in the Omicron RBD increase the stability of the RBD-ACE2 complex.
3.2.2. Root mean square fluctuations (RMSFs)
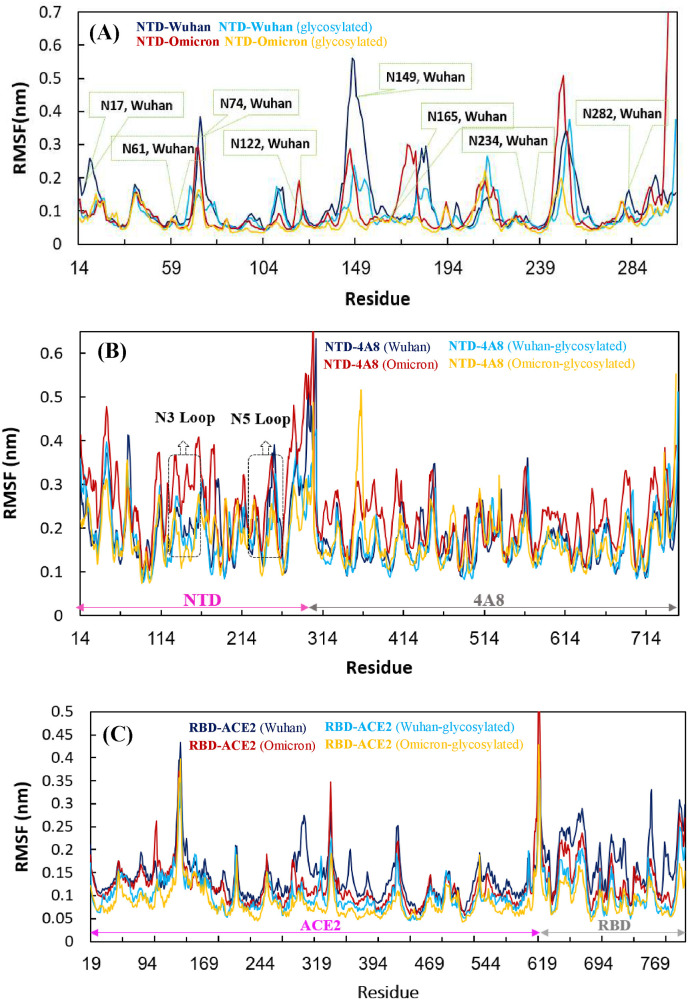
Residue-based backbone fluctuations of the Wuhan and Omicron NTDs in glycosylated and non-glycosylated states were evaluated by plotting their RMSF values (Fig. 3 A). Three major fluctuation peaks were observed in the Wuhan NTD, residues 62–83, 140–158, 177–189, and 239–260 corresponding to the loop regions N2, N3, N4 and N5, respectively. Fluctuations in these regions, except for the N5 loop, were significantly reduced in the glycosylated state. Comparison of the RMSF profiles of the Wuhan and Omicron NTDs revealed that the flexibility of these mentioned loops, except for the residues 240–256, was decreased in the Omicron NTD. By comparing the plots in the glycosylated state, the effect of the mutations, especially in the N5 loop region, can be clearly seen in the stability of the protein relative to Wuhan. The RMSF analysis also revealed higher flexibility in the glycosylated Wuhan NTD-4A8 complex compare to the glycosylated Omicron NTD-4A8 complex. Careful evaluation of the N3 and N5 loop regions, which have the most interaction with the antibody, illustrated less fluctuations in the Omicron structure. This is probably due to the deletion mutation causing removal of the residues 143–145 and the displacement of long glycans, resulting in the contacts of glycans with neighboring residues, as more deviations in the Omicron structure are clearly observed in the non-glycosylated state (Fig. 3, Fig. 5 ).
Fig. 3.
RMSF plots of (A) NTD-Wuhan, NTD-Omicron and evaluation of these two systems in glycosylated state, (B) NTD-4A8 (Wuhan), NTD-4A8 (Omicron) and evaluation of these two systems in glycosylated state. (C) RBD-ACE2 (Wuhan), RBD-ACE2 (Omicron) and evaluation of these two systems in glycosylated state.
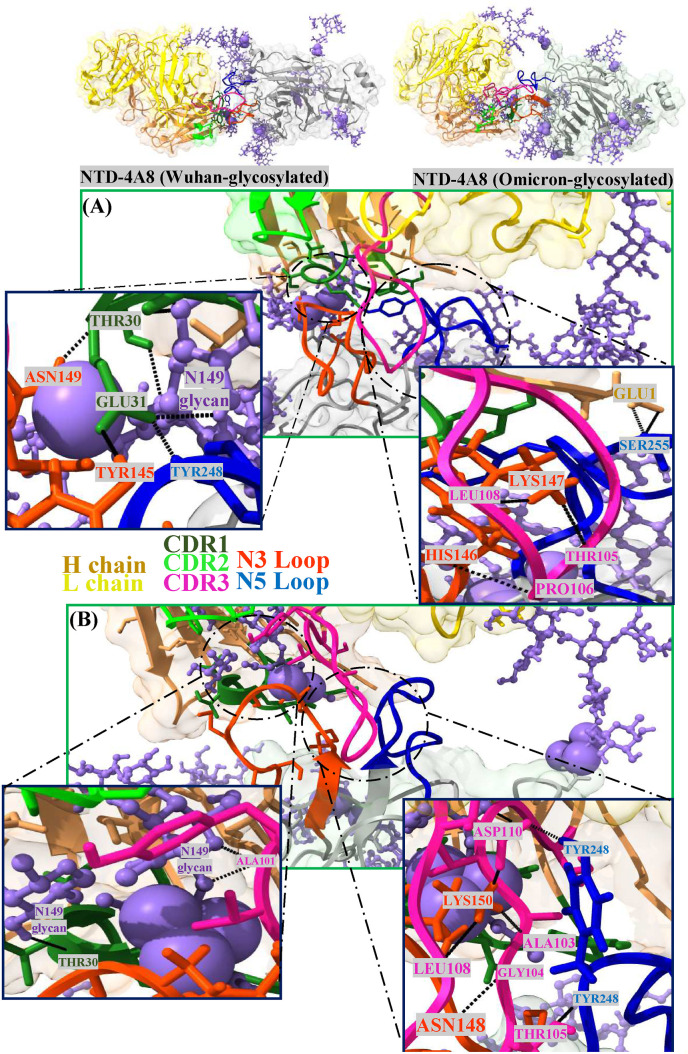
Fig. 5.
(A) NTD-4A8 (Wuhan-glycosylated), (B) NTD-4A8 (Omicron-glycosylated). Long glycans are highlighted in purple. Complement-determining regions (CDRs) in heavy chain 4A8 include CDR1 (residues 25 to 32), CDR2 (residues 51 to 58), and CDR3 (residues 100 to 116), which these regions are mainly mediate the interaction with the NTD. N3 (residues 141 to 156) and N5 (residues 246 to 260) of the NTD mainly participates in binding to the 4A8.
The RMSF for all RBD-ACE2 complexes were also plotted, the average values of RMSF for the Wuhan and Omicron non-glycosylated RBD-ACE2 complexes are ∼0.14 and ∼0.12 nm, respectively, indicating that the Omicron RBD is more rigid than the Wuhan. In addition, the fluctuations of each residues of the glycosylated RBD-ACE2 complexes were evaluated in order to investigate the effect of the glycosylation on protein dynamics. The average values of RMSF for the glycosylated Wuhan and Omicron complexes were 0.10 and 0.08 respectively. Fig. 3C showed that the both non-glycosylated RBD-ACE2 complexes were more dynamic than glycosylated complexes which indicates that dynamic fluctuations of the protein complexes were reduced by the glycosylation. Interestingly, based on the RMSF plots of the both glycosylated RBD-ACE2 complexes, it could be deduced that the decrease of the fluctuations caused by the glycosylation were seen not only near the glycosylated sites but also in the other regions of the glycosylated RBD-ACE2 protein complexes. The Wuhan and Omicron RBD-ACE2 complexes showed similar fluctuations. As Fig. 3C shows, Three loops in the RBDs, (474–485), (488–490), and (494–505), which are critical for binding with ACE2, have slightly less fluctuation in the Omicron RBD than in the Wuhan and Also, in the glycosylated complexes, these regions have less fluctuations. According to Fig. 3C, the RMSF values decreased at the receptor binding motif (RBM, residues 434–508) in the Omicron. This indicates that the Omicron mutations caused variation in conformational dynamics and more stability of the Omicron complex.
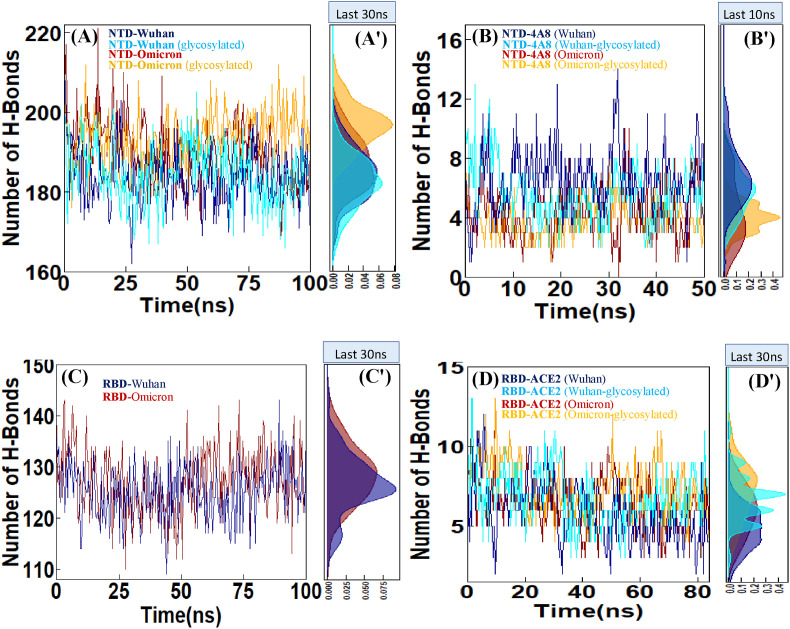
3.2.3. Hydrogen bond analysis
One of the main factors in the stability of the secondary and tertiary structure of proteins is hydrogen bonds. To assess the contribution of this factor to the NTD structure, intramolecular hydrogen bond plots were constructed for the unbound NTD forms during the 100 ns MD simulation time. NTD mutations can affect molecular dynamics, and from the comparative plots, it can be inferred that the Omicron NTD has more hydrogen bonds than the Wuhan NTD. Also, the effects of glycosylation on protein stability are evident in the plots showing an increased number of hydrogen bonds in the glycosylated forms compared with the non-glycosylation states (Fig. 4A). The mean number of hydrogen bonds between Wuhan and Omicron NTDs and mAb 4A8 were 6.56 and 4.31, respectively. On the other hand, hydrogen bonding in the glycosylated state decreased to 5.65 in the Wuhan and to 3.89 in the Omicron (Fig. 4B). As can be seen in (Fig. 5), especially in the Omicron due to mutation and displacement of long glycan in N3 Loop, glycans form the hydrogen bonds with the mAb 4A8.
Fig. 4.
Hydrogen bonding analysis of (A) NTD-Wuhan, NTD-Omicron and evaluation of these two systems in the glycosylated state, (B) NTD-4A8 (Wuhan), NTD-4A8 (Omicron) and evaluation of these two systems in the glycosylated state. (C) RBD-Wuhan and RBD-Omicron. (D) RBD-ACE2 (Wuhan), RBD-ACE2 (Omicron) and evaluation of these two systems in glycosylated state. (A′, B′,C'and D′) Density Function of H-bond sampled over the simulations are shown in histograms.
Hydrogen bond profiles of the Wuhan and Omicron RBDs, indicated that the Omicron RBD has slightly more number of hydrogen bond with an average of 127.26 than Wuhan with an average of 125.24 (Fig. 4C). The hydrogen bonds between the RBD and ACE2 in all complexes during the course of the simulations were calculated. The average number of hydrogen bonds were 5.93, 6.46, 6.82, 7.38 for the states Wuhan RBD-ACE2, Omicron RBD-ACE2, glycosylated Wuhan RBD-ACE2, glycosylated Omicron RBD-ACE2 complexes, respectively. The higher number of hydrogen bonds in the Omicron RBD-ACE2 complex (Fig. 4D) suggests higher stability of the interaction between the RBD of Omicron Variant and ACE2 receptor. As Fig. 4D shows, more hydrogen bonds were observed in the glycosylated complexes than in the non-glycosylated counterparts indicating the stronger binding of the RBD to ACE2 in glycosylated forms.
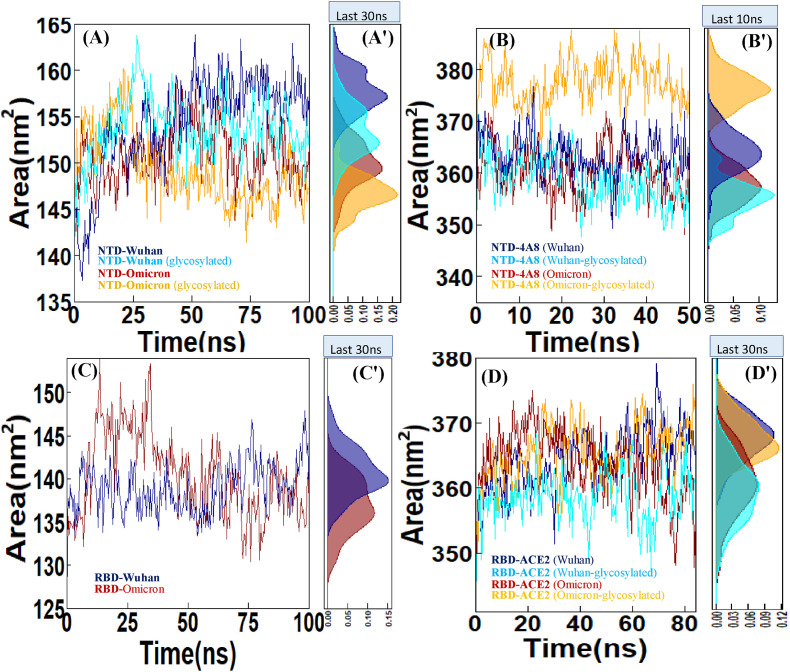
3.2.4. Solvent accessible surface area (SASA)
Solvent accessible surface area (SASA) is one of the key factors in the evaluation of stability and folding of protein structures. The SASA analysis of MD simulation trajectory indicated that Wuhan NTD has a higher SASA with average value ( 154.4 nm2) than the Omicron NTD ( 150.8 nm2), suggesting higher compactness of the Omicron NTD. In addition, by comparison of the plots, it can be inferred that glycosylation reduces SASA indicating more stable and compact structure of the glycosylated form than the non-glycosylated state (Fig. 6 A). Also, by comparing the Wuhan NTD-4A8 with Omicron NTD-4A8, the effect of glycosylation in reducing the interaction and escape of the Omicron NTD from antibody with SASA average value (∼377.55 nm2) can be clearly seen (Fig. 6B). No significant deviation in the SASA was observed between the Wuhan NTD-4A8 (∼363.63 nm2), glycosylated Wuhan NTD-4A8 (∼359.03 nm2) and Omicron NTD-4A8 (∼360.27 nm2) complexes.
Fig. 6.
SASA analysis of (A) NTD-Wuhan, NTD-Omicron and evaluation of these two systems in the glycosylated state, (B) NTD-4A8 (Wuhan), NTD-4A8 (Omicron) and evaluation of these two systems in the glycosylated state. (C) RBD-Wuhan and RBD-Omicron. (D) RBD-ACE2 (Wuhan), RBD-ACE2 (Omicron) and evaluation of these two systems in glycosylated state. (A′, B′,C'and D′) Density Function of SASA sampled over the simulations are shown in histograms.
Conformational changes due to the Omicron RBD mutations caused SASA alterations. The average value of the SASA for the Wuhan and Omicron RBDs are 138.86 nm2 and 140.38 nm2 , respectively. The result indicate that Omicron RBD has higher SASA than the Wuhan (Fig. 6C). Also, the SASA for the RBD-ACE2 complexes was calculated as a function of time, and the results obviously showed that the Wuhan RBD-ACE2 complex has a greater SASA value (∼364.14 nm2) than the Omicron RBD-ACE2 complex (∼363.95 nm2). The SASA analysis for the RBD-ACE2 complexes showed that the non-glycosylated complexes are more exposed in the binding interface therefore less stable than glycosylated forms (Fig. 6D).
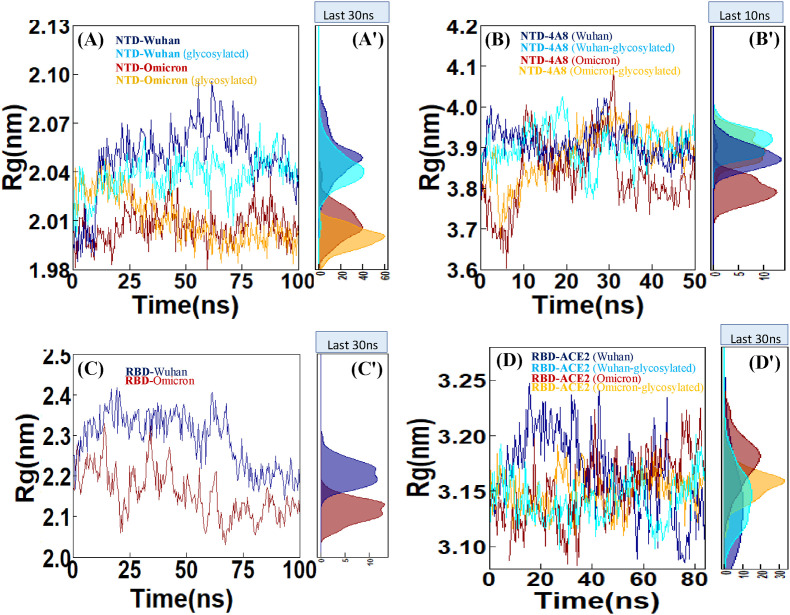
3.2.5. Radius of gyration (Rg)
The radius of gyration (Rg) indicates the system compactness and density, and eventually reflects the folding degree and stability of proteins. The Rg value of the Omicron NTD is smaller than that of the Wuhan NTD. This reflects the effect of mutations on increase in the protein compactness. Furthermore, the largest deviations in the Rg timecourse were detected in the Wuhan NTD, due to the lesser compactness of this protein in comparison with the Omicron NTD. Also in these plots, the glycosylation effect on the efficiency of protein folding, reflected by the increased protein compactness, is clearly seen (Fig. 7 A).
Fig. 7.
Radius of gyration (Rg), (A) NTD-Wuhan, NTD-Omicron and evaluation of these two systems in the glycosylated state, (B) NTD-4A8 (Wuhan), NTD-4A8 (Omicron) and evaluation of these two systems in the glycosylated state. (C) RBD-Wuhan and RBD-Omicron. (D) RBD-ACE2 (Wuhan), RBD-ACE2 (Omicron) and evaluation of these two systems in glycosylated state. (A′, B′,C'and D′) Density Function of Rg sampled over the simulations are shown in histograms.
Analysis of the changes in the structural compactness due to the antibody binding with the RMSD plot results shows similar structural stability. A closer evaluation the Rg value at the Omicron NTD-4A8 complex has experienced significant perturbation. In the Omicron NTD-4A8 complex, there was an increase in the Rg values from ∼3.60 nm to ∼4.00 nm during the interval from 6 ns to 10 ns followed by a decrease to ∼3.76 nm in 20 ns, then increase to ∼4.09 nm in 31 ns and then decreased again to ∼3.75 nm in 32 ns and remained consistent until 50 ns. Further, in the glycosylated Omicron NTD-4A8 complex, there was a gradual increase in the Rg values from ∼3.68 nm to ∼3.95 nm during the interval from 5 ns to 30 ns followed by a slight decrease to ∼3.84 nm in 40 ns and then reached a stable state. Indeed, fewer deviations in the Omicron compared to Wuhan indicate more involvement of glycan interactions with mAb 4A8, and escape of protein from antibody binding (Fig. 7B).
Rg was calculated from the 100 ns trajectory to gain insight into the compactness and rigidity of the RBDs. The average Rg value for the Wuhan and Omicron RBDs were ∼2.28 and ∼2.15 nm, respectively. In the last 30 ns of the simulation time both RBD systems became stable. The Rg plot of the RBDs showed the general compactness of Wuhan and Omicron RBDs (Fig. 7C). Also, The Rg for the non-glycosylated Wuhan and Omicron RBD-ACE2 complexes fluctuates between ∼3.07 and ∼3.24 nm, and ∼3.05–∼3.21 nm, respectively. Based on the comparison between the Rg plots of the Wuhan and Omicron RBD-ACE2 complexes, no significant deviation was observed. In addition, the Rg values of the glycosylated RBD-ACE2 complexes have less deviations than that of the glycosylated complexes. This reflects the impact of the glycosylation on the increase in the protein complex compactness (Fig. 7D).
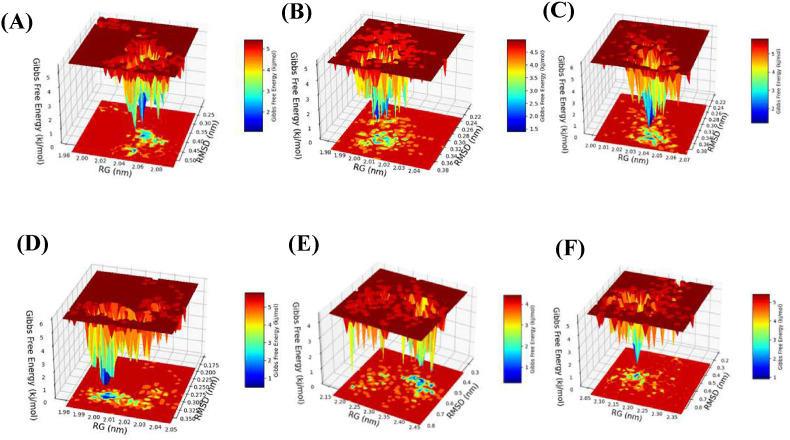
3.2.6. Free energy landscape
The analysis of free energy landscape (FEL) has been used to identify lower-energy basins (minima) during NTDs and RBDs unbound MD simulations. The plots in (Fig. 8 ) indicated a comparative view of the free energy landscape as a function of RMSD and Rg for NTDs and RBDs of the Wuhan and Omicron variant. According to Fig. 8, the energy minima basins were shown from red to blue, and the blue zone denotes more stable conformation with minimal energy. As shown in Fig. 8A and B, Omicron NTD has more blue and dark blue zones than Wuhan NTD and also by comparing the distribution of orange and red zones, it can be deduced that Omicron NTD has higher stability than Wuhan. Moreover, the effect of glycosylation NTD on the reduction of Gibbs free energy compare to non-glycosylated state is clearly observed and it is illustrated that glycosylation results in higher stability of protein structures (Fig. 8C and D). Based on Fig. 8E and F, the more blue and dark blue zones in the Wuhan RBD and the different distribution of orange and red zones, Wuhan RBD has a lower Gibbs free energy level than that of Omicron, which shows higher stability of the Wuhan RBD compared with Omicron RBD.
Fig. 8.
The Free energy landscape (FEL). (A) NTD-Wuhan, (B) NTD-Omicron, (C) NTD-Wuhan (glycosylated), (D) NTD-Omicron (glycosylated), (E) RBD-Wuhan and (F) RBD-Omicron.
3.2.7. Principal component analysis (PCA)
The overall motion of the proteins were analyzed by PCA using construction of the eigenvectors. Figs. S4A and B, exhibited that the Wuhan NTD has more structural motions than the Omicron NTD and is occupying a larger space. The effect of glycosylation can also be inferred from the PCA, and the corresponding analysis revealed that the motions and degrees of freedom within proteins have become more limited and proteins became more stable due to glycosylation (Figs. S4C and D). The comparison of PCA for RBDs has shown that the motion properties described by the first two eigenvectors are different in the Wuhan RBD as compared to the Omicron (Figs. S4E and F).
3.3. Free energy binding and ΔΔG (destabilizing)
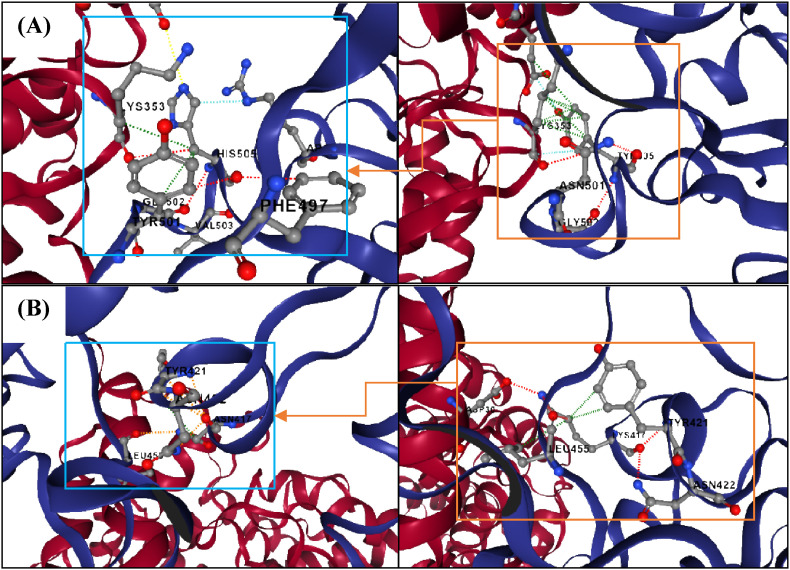
Among the Omicron RBD substitution mutations, residues including K417 N, N440K, G446S, S477 N, T478K, E484A, Q493K, G496S, Q498R, N501Y and Y505H are located at the interface of RBD and ACE2 receptor. Substitution mutations including G339D, T478K, Y505H, S373P, N440K, E484A. Y505H have a value of predicted stability change (ΔΔGStability) above zero and S371L, S375F, Q493R, G496S, Q498R, N501Y have a value of predicted stability change below zero (ΔΔGStability). By the Omicron substitutions, the hydrogen bonding between GLU493 of RBD and ACE2 residue 35 was enhanced and, hydrogen bonds such as interactions HIS505–LYS353 and LYS417-ASP30 are shown in Fig. 9 .
Fig. 9.
Two out of the 15 mutations, namely (A) Y505H, (B) K417 N presented with a ΔΔG value above and below zero respectively. Blue boxes indicate Omicron (left) and orange boxes indicate Wuhan (right). The interface between RBD (blue) and ACE2 (red) is shown (Interaction colors: Hydrogen Bond (Red), Hydrophobic (Green), Polar (Orange), van der Waals (Blue)).
3.4. Binding free energy calculation
The MM/GBSA method was used in order to investigate how the Omicron variant mutations affect on the binding of NTD and RBD with mAb 4A8 and ACE2, respectively. The binding free energy of the NTD to mAb 4A8 within the Wuhan and Omicron NTD complexes were calculated from the last 10 ns of trajectories and also evaluation of these two complexes in glycosylated state. The binding free energy of the Wuhan and Omicron NTDs to mAb 4A8 were −44.90 kcal/mol and −25 kcal/mol, respectively. Furthermore, the binding free energy for the glycosylated Wuhan NTD-4A8 complex was −34.27 kcal/mol and −21.41 kcal/mol for the glycosylated Omicron NTD-4A8 complex (Table 1 ). Overall, these results especially in the glycosylated state show the effect of mutations on the increase of the Omicron variant escape from the host's immune response.
Table 1.
The Binding free energy calculation by using MM/GBSA approach. All units are reported in kcal/mol.
| Energy Terms | NTD-4A8 (Wuhan) | NTD-4A8 (Wuhan-glycosylated) | NTD-4A8 (Omicron) | NTD-4A8 (Omicron-glycosylated) |
|---|---|---|---|---|
| VDWAALS | −70.03 | −58.67 | −40.60 | −30.75 |
| EEL | −889.72 | −566.13 | −680.09 | −147.76 |
| EGB | 925.33 | 599.49 | 702.82 | 162.15 |
| ESURF | −10.48 | −8.95 | −7.13 | −5.03 |
| ΔG gas | −959.75 | −624.81 | −720.70 | −178.52 |
| ΔG solv | 914.85 | 590.53 | 695.69 | 157.11 |
| ΔG TOTAL |
−44.90 |
−34.27 |
−25.00 |
−21.41 |
| Energy Terms | RBD-ACE2 (Wuhan) | RBD-ACE2 (Wuhan-glycosylated) | RBD-ACE2 (Omicron) | RBD-ACE2 (Omicron-glycosylated) |
| VDWAALS | −61.44 | −75.16 | −80.22 | −79.11 |
| EEL | −646.68 | −586.47 | −1366.81 | −1433.12 |
| EGB | 699.60 | 639.77 | 1425.86 | 1482.00 |
| ESURF | −9.73 | −11.42 | −11.46 | −12.67 |
| ΔG gas | −708.13 | −661.64 | −1447.03 | −1512.23 |
| ΔG solv | 689.86 | 628.34 | 1414.39 | 1469.32 |
| ΔG TOTAL | −18.26 | −33.29 | −32.64 | −42.91 |
The binding free energies of the four RBD-ACE2 complexes were calculated from the last 10 ns of the trajectories. The energy values for the Omicron RBD-ACE2 complex were −80.22 kcal/mol (vdW), −1366.81 kcal/mol (electrostatic), and −37.20 kcal/mol (total binding energy). In addition, the binding free energy of the RBD to ACE2 in the Wuhan and Omicron glycosylated complexes were −33.29 kcal/mol and −42.91 kcal/mol, respectively. Importantly, the results indicate that due to existence of the long glycans, the binding free energies are higher for the glycosylated RBD-ACE2 complexes than for the non-glycosylated counterparts. The results demonstrated that the RBD of Omicron variant interacts more effectively with ACE2 than the Wuhan, likely contributing to the faster spread of the Omicron variant (Table 1).
3.5. Per-residue free energy decomposition analysis
The key binding residues of NTDs and RBDs were identified with MM/GBSA method. As given in Fig. 10 A and Table S3, there are thirteen key residues (energy contribution < −1.00 kcal/mol) in the NTD for all four systems. The results of the analysis, especially in the residues TYR145, ARG246 and TYR248, emphasize the escape of the Omicron from the antibody. The impact of long glycans on each residue, indicated a decrease in the binding energy of TYR145, ARG246, TYR248, in the Wuhan and ASN148 (→ASN143), ASN149 (→ASN 144), LYS150 (→LYS145) in the Omicron. The per-residue energy decomposition results of the RBD-ACE2 complexes showed that S477 N, Q493R, G496S, Q498R and N501Y with (energy contribution < −1.00 kcal/mol) are identified as key residues (Fig. 10B and Table S4) which have a positive effect on the more binding affinity between the Omicron RBD to ACE2. Also, the impact of long glycans on the key residue, indicated a decrease in the binding energy.
Fig. 10.
Per-residue energetic components of the (A) NTD and (B) RBD. All units are reported in kcal/mol.
3.6. RINs analysis of key residues in the NTDs and RBDs of the Wuhan and Omicron variants
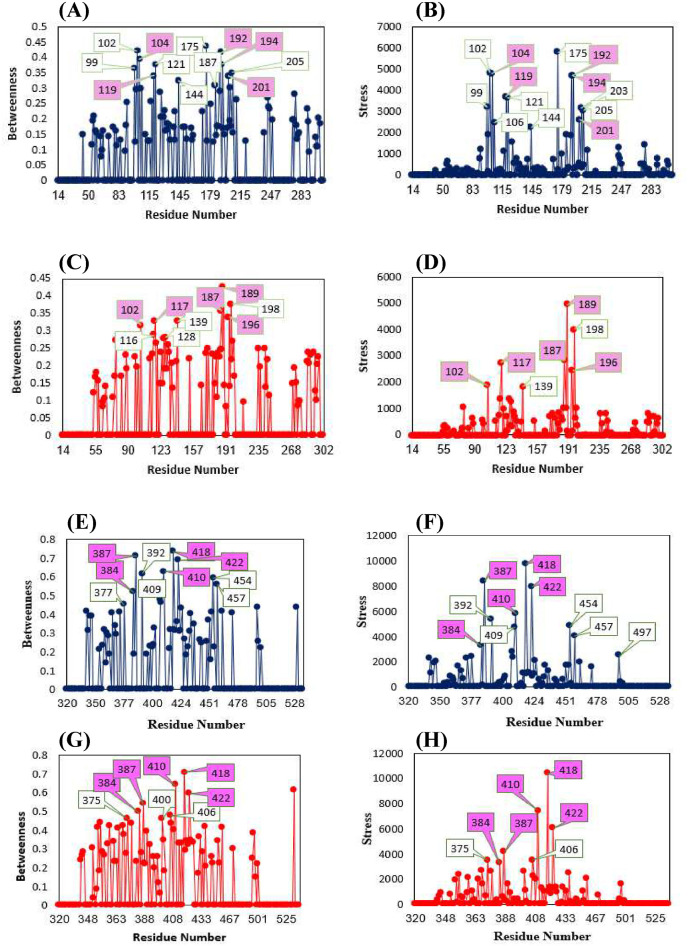
In this study, key residues of the NTD and RBD domains of the Wuhan and Omicron variant were identified by RIN betweenness and stress metrics (Fig. 11 , Tables S5 and S6). Stress is defined as an indicator of the centrality of a node, which indicates the number of shortest paths that pass through a node. Also, to identify the key structural residue as a mediator of interaction with other nodes, betweenness was used, the high values of which indicate the presence of a node in the shortest path between any two other nodes [46]. The key residue agreement between the two structures of Wuhan and Omicron NTDs is TRP104 (→TRP102), ILE119 (→ILE117), PHE192 (→PHE187), PHE194 (→PHE189), and PHE201 (→PHE196) and for RBDs is PRO384, LEU387, ILE410, ILE418, ASN422. The results indicate that the number of clusters and correspondingly MCODE score for Wuhan NTD are higher than that of the Omicron NTD. MCODE scores for clusters with four or more residues and cluster members for NTD structures are presented in Table S5, and cluster members for RBD structures are presented in Table S6. The highest overlap was observed in the cluster members of both types of NTD structures for ILE105 (→ILE103), PHE135 (→PHE133) and TYR160 (→TYR155) and for RBD structures is PRO384, VAL395, VAL524, PHE456, TYR473, TYR489. The small difference in key residues identified in the Wuhan and Omicron NTD and RBD structures is due to the effect of the mutations that have occurred in the structures.
Fig. 11.
Betweenness and stress centralities for NTD of spike glycoprotein for Wuhan (A and B) Omicron variant (C and D). Betweenness and stress centralities for RBD of spike glycoprotein for Wuhan (E and F) Omicron variant (G and H). The data labeled in pink is a key residue agreement NTD and RBD of Spike glycoproteins between Wuhan and Omicron.
4. Discussion
This study has focused on the effects of mutations in the Omicron variant on the structure and function of two important SARS-CoV-2 spike protein domains including the receptor binding domain (RBD) and the N-terminal domain (NTD) by using various computational tools. Our results show the presence of more extensive β-strands in the Omicron NTD compared with the Wuhan NTD structure that can be one of the reasons for the higher stability of the Omicron NTD. Furthermore, our findings indicated a reduction in the protein accessibility, especially in the loop regions of the Omicron NTD compared with that of the Wuhan NTD. Since these mentioned regions are the target of a number of neutralizing antibodies, these changes are indicative of an Omicron strategy to escape the antibodies. In line with these conclusions, the results of analysis of NTD interaction with antibodies show a decrease in the Omicron binding free energies facilitating the virus escape from the immune system. Another key structural feature of the NTD protein is its extensive glycosylation that plays a number of important biological roles by changing the physico-chemical nature of proteins after translation, thereby contributing to the life cycle of the virus and its escape from the immune system [16]. Owing to the importance of glycosylation in the camouflaging of immunogenic protein epitopes, no mutation in the glycosylation sites of SARS-CoV-2 VOCs have been observed during viral evolution [47]. In this study, analysis of the structural role of N-glycans showed an increase in the stability of glycosylated proteins compared to the non-glycosylated state. It is interesting to note that glycans attached to the loop regions are of the complex types for protection of immunogenic epitopes against antibody neutralization. Previous studies have shown the importance of glycans associated with ASN165 and ASN234 for RBD binding to the ACE2 receptor [48], since they are structurally close to the RBD, and our results demonstrate the increased mobility of these glycans and the camouflage of the epitopes. We evaluated the effect of mutations and glycosylation on antibody detection. Similar to previous studies [44,49], structural analysis of the 4A8 epitope indicates the importance of the key residues TYR145, ARG246 and TYR248 of the Wuhan NTD. Molecular explanation for such a wide range of glycosylation and mutations impact on protein structure and consequently the low affinity of TYR145, ARG246 and TYR248 side chains for interaction with antibodies (Fig. S5 and Table S7). Unlike previous studies [50], we included 214 EPE in the Omicron NTD, which can affect the results of the interaction. TYR144 has been mutated in the B.1.1.318, B.1.616, B.1.525, B.1.1.7 and BA.1 variants and, in this work the comparative results of RINs and per-residue decomposition analysis revealed the significant role of TYR144 in the protein structure and interaction with antibody. In addition, the all-atom MD simulations were carried out for the Wuhan and Omicron RBD-ACE2 complexes. Due to the significant role of long glycans in virus-host interactions [51,52], unlike previous studies [50,53] we performed a comparative study between glycosylated and non-glycosylated RBD-ACE2 complexes to evaluate the impact of the existence of glycans on the binding of RBD to ACE2. The RMSD, RMSF, radius of gyration and hydrogen bond analysis confirmed that the Omicron RBD-ACE2 complex is more stable than that of the Wuhan indicating the increased infectivity. Also, at interface region of RBD (residues 434–508), the reduction of RMSF value was demonstrated in the Omicron complexes. This was also confirmed by comparison of the solvent accessible surface area of the Omicron and Wuhan complexes. Furthermore, by comparing the RMSD, radius of gyration, hydrogen bond analysis and the free energy landscape plots of the Wuhan and Omicron RBDs only systems, it is revealed that Omicron substitution mutations affect the structure of the RBD and protein behavior in the course of 100 MD simulation. Overall, the result demonstrated that the Wuhan RBD is more stable than Omicron RBD in unbound form. We further elucidated the effect of different mutations on the interaction of RBD with ACE2. N440K mutation had a significant effect on the increased stability of the Omicron complex. Similar to a previous study [54], Following the results mentioned above, per-residue energy decomposition for the four RBD-ACE2 complexes revealed that positively charged residues including S477 N, G496S, Q498R and N501Y mutations lead to an increase in the binding affinity of the binding of the Omicron RBD to ACE2 [55,56], which are seen in both glycosylated and non-glycosylated forms in our study. The K417 N mutation which is also found in Beta, Gamma, and in some Delta lineage RBDs [57] was the cause of the absence of a salt bridge leading to a decrease in the binding affinity to ACE2 [58]. Importantly the results of the interaction analysis of the Wuhan and Omicron RBD-ACE2 complexes revealed some key interactions found only in the Omicron RBD-ACE2 complex and also, more number of salt-bridge were found in the Omicron complex. The Q493R substitution in the Omicron RBD caused in new formed salt bridge with GLU35 residue of the ACE2. The mutated residues in the RBD indicates strong interactions with some of the residues of the ACE2, including hydrogen bonding that were observed between the ARG493 and ARG498 residues of the Omicron RBD with GLU35 and ASP38 residues of the ACE2 respectively (Table S7). The results of binding free energies calculation of the Wuhan and Omicron RBD-ACE2, indicated the lower total free energy in the Omicron RBD-ACE2 complex than that of the Wuhan. In sum, the results of this study could be useful for the future studies in this field.
5. Conclusions
This computational study analyzed structural peculiarities of the Wuhan and Omicron RBDs and revealed structural changes caused by the mutations found in the Omicron variant. Analysis of the effect of mutations through MD simulation shows the stability of the NTD structure in the Omicron variant relative to the Wuhan, and probably provides a basis for the accumulation of dangerous mutations in the RBD, as the results of the RBD plots show a decrease in its stability relative to the Wuhan. Subsequently, the study of the interaction of the NTD with antibody and the RBD with the ACE2 through MD simulation and MMGBSA method showed an increase in the Omicron infectivity compared to the Wuhan. Furthermore, this study evaluated the effect of glycosylation, as an important post-translational modification, on proteins stability and antigenicity. Taken together, these data could be useful for better understanding of the molecular basis of the infectivity of the SARS-CoV-2 Omicron (BA.1) variant and can be utilized to accelerate the development of novel therapeutics.
Author contributions
Conceptualization, Y.S; methodology, Y.S; software, F.B, N.S; validation, Y.S, V.U; investigation, F.B, N.S, V.U, Y.S and S.M; resources, Y.S.; writing—original draft preparation, F.B, N.S, V.U, Y.S and S.M; writing—review and editing, V.U and Y.S; visualization, F.B, N.S, Y.S and V.U; Formal analysis, F.B, N.S, Y.S supervision, Y.S and V.U; project administration, Y.S All authors have read and agreed to the published version of the manuscript.
Declaration of competing interest
There is no conflict of interest regarding this manuscript.
Acknowledgements
The support and resources from the Center for High Performance Computing at the Shahid Beheshti University (SARMAD) of Iran are gratefully acknowledged.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2022.105735.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Rezaei S., Sefidbakht Y., Uskoković V. Tracking the pipeline: immunoinformatics and the COVID-19 vaccine design. Briefings Bioinf. 2021;22 doi: 10.1093/bib/bbab241. [DOI] [PubMed] [Google Scholar]
- 2.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 3.Jomhori M., Mosaddeghi H., Farzin H. Tracking the interaction between single-wall carbon nanotube and SARS-Cov-2 spike glycoprotein: a molecular dynamics simulations study. Comput. Biol. Med. 2021;136 doi: 10.1016/j.compbiomed.2021.104692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma V., Rai H., Gautam D.N.S., Prajapati P.K., Sharma R. Emerging evidence on omicron (B.1.1.529) SARS‐CoV‐2 variant. J. Med. Virol. 2022 doi: 10.1002/jmv.27626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar S., Karuppanan K., Subramaniam G. Omicron (BA.1) and sub-variants (BA.1, BA.2 and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: a comparative sequence and structural-based computational assessment. bioRxiv. 2022 doi: 10.1101/2022.02.11.480029. 2022.02.11.480029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L., Cheng G. Sequence analysis of the emerging SARS‐CoV‐2 variant Omicron in South Africa. J. Med. Virol. 2022;94:1728–1733. doi: 10.1002/jmv.27516. [DOI] [PubMed] [Google Scholar]
- 7.Fu T., Li F., Zhang Y., Yin J., Qiu W., Li X., Liu X., Xin W., Wang C., Yu L., Gao J.-Q., Zheng Q., Zeng S., Zhu F. VARIDT 2.0: structural variability of drug transporter. Nucleic Acids Res. 2022;50:D1417–D1431. doi: 10.1093/nar/gkab1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X., Li F., Qiu W., Xu B., Li Y., Lian X., Yu H., Zhang Z., Wang J., Li Z., Xue W., Zhu F. SYNBIP: synthetic binding proteins for research, diagnosis and therapy. Nucleic Acids Res. 2022;50:D560–D570. doi: 10.1093/nar/gkab926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., Jensen L.J., von Mering C. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue W., Wang P., Tu G., Yang F., Zheng G., Li X., Li X., Chen Y., Yao X., Zhu F. Computational identification of the binding mechanism of a triple reuptake inhibitor amitifadine for the treatment of major depressive disorder. Phys. Chem. Chem. Phys. 2018;20:6606–6616. doi: 10.1039/C7CP07869B. [DOI] [PubMed] [Google Scholar]
- 11.Yang J., Lin X., Xing N., Zhang Z., Zhang H., Wu H., Xue W. Structure-based discovery of novel nonpeptide inhibitors targeting SARS-CoV-2 M pro. J. Chem. Inf. Model. 2021;61:3917–3926. doi: 10.1021/acs.jcim.1c00355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang J., Zhang Z., Yang F., Zhang H., Wu H., Zhu F., Xue W. Computational design and modeling of nanobodies toward SARS‐CoV‐2 receptor binding domain. Chem. Biol. Drug Des. 2021;98:1–18. doi: 10.1111/cbdd.13847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu F., Shi Z., Qin C., Tao L., Liu X., Xu F., Zhang L., Song Y., Liu X., Zhang J., Han B., Zhang P., Chen Y. Therapeutic target database update 2012: a resource for facilitating target-oriented drug discovery. Nucleic Acids Res. 2012;40:D1128–D1136. doi: 10.1093/nar/gkr797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang H., Qin C., Li Y.H., Tao L., Zhou J., Yu C.Y., Xu F., Chen Z., Zhu F., Chen Y.Z. Therapeutic target database update 2016: enriched resource for bench to clinical drug target and targeted pathway information. Nucleic Acids Res. 2016;44:D1069–D1074. doi: 10.1093/nar/gkv1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Y.H., Li X.X., Hong J.J., Wang Y.X., Fu J.B., Yang H., Yu C.Y., Li F.C., Hu J., Xue W.W., Jiang Y.Y., Chen Y.Z., Zhu F. Clinical trials, progression-speed differentiating features and swiftness rule of the innovative targets of first-in-class drugs, Brief. Bioinform. 2020;21:649–662. doi: 10.1093/bib/bby130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao X., Chen H., Wang H. Glycans of SARS-CoV-2 spike protein in virus infection and antibody production. Front. Mol. Biosci. 2021;8 doi: 10.3389/fmolb.2021.629873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rezaei S., Sefidbakht Y., Uskoković V. Comparative molecular dynamics study of the receptor-binding domains in SARS-CoV-2 and SARS- CoV and the effects of mutations on the binding affinity. J. Biomol. Struct. Dyn. 2020:1–20. doi: 10.1080/07391102.2020.1860829. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 19.Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Needleman S.B., Wunsch C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 21.Robert X., Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42:W320–W324. doi: 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coutsias E.A., Seok C., Jacobson M.P., Dill K.A. A kinematic view of loop closure. J. Comput. Chem. 2004;25:510–528. doi: 10.1002/jcc.10416. [DOI] [PubMed] [Google Scholar]
- 23.Schrodinger L.L.C. 2010. The PyMOL Molecular Graphics System. 0. [Google Scholar]
- 24.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinforma. 2016;54:5–6. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., Zepeda S., di Iulio J., Bowen J.E., Montiel-Ruiz M., Zhou J., Rosen L.E., Bianchi S., Guarino B., Fregni C.S., Abdelnabi R., Foo S.-Y.C., Rothlauf P.W., Bloyet L.-M., Benigni F., Cameroni E., Neyts J., Riva A., Snell G., Telenti A., Whelan S.P.J., Virgin H.W., Corti D., Pizzuto M.S., Veesler D. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347. doi: 10.1016/j.cell.2021.03.028. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woo H., Park S.-J., Choi Y.K., Park T., Tanveer M., Cao Y., Kern N.R., Lee J., Yeom M.S., Croll T.I., Seok C., Im W. Developing a fully glycosylated full-length SARS-CoV-2 spike protein model in a viral membrane. J. Phys. Chem. B. 2020;124:7128–7137. doi: 10.1021/acs.jpcb.0c04553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 28.Huang J., Rauscher S., Nawrocki G., Ran T., Feig M., de Groot B.L., Grubmüller H., MacKerell A.D. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hatcher E., Guvench O., MacKerell A.D. CHARMM additive all-atom force field for aldopentofuranoses, methyl-aldopentofuranosides, and fructofuranose. J. Phys. Chem. B. 2009;113:12466–12476. doi: 10.1021/jp905496e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan H.M., MacKerell A.D., Reuter N. Cation-π interactions between methylated ammonium groups and tryptophan in the CHARMM36 additive force field. J. Chem. Theor. Comput. 2019;15:7–12. doi: 10.1021/acs.jctc.8b00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khan H.M., MacKerell A.D., Reuter N. Cation-π interactions between methylated ammonium groups and tryptophan in the CHARMM36 additive force field. J. Chem. Theor. Comput. 2019;15:7–12. doi: 10.1021/acs.jctc.8b00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao Y., Lee J., Smith I.P.S., Lee H., Kim S., Qi Y., Klauda J.B., Widmalm G., Khalid S., Im W. CHARMM-GUI supports hydrogen mass repartitioning and different protonation states of phosphates in lipopolysaccharides. J. Chem. Inf. Model. 2021;61:831–839. doi: 10.1021/acs.jcim.0c01360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 34.Dehury B., Raina V., Misra N., Suar M. Effect of mutation on structure, function and dynamics of receptor binding domain of human SARS-CoV-2 with host cell receptor ACE2: a molecular dynamics simulations study. J. Biomol. Struct. Dyn. 2021;39:7231–7245. doi: 10.1080/07391102.2020.1802348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Martin A.J.M., Vidotto M., Boscariol F., Di Domenico T., Walsh I., Tosatto S.C.E. RING: networking interacting residues, evolutionary information and energetics in protein structures. Bioinformatics. 2011;27:2003–2005. doi: 10.1093/bioinformatics/btr191. [DOI] [PubMed] [Google Scholar]
- 37.Piovesan D., Minervini G., Tosatto S.C.E. The RING 2.0 web server for high quality residue interaction networks. Nucleic Acids Res. 2016;44:W367–W374. doi: 10.1093/nar/gkw315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emerson I.A., Louis P.T. Detection of active site residues in bovine rhodopsin using network analysis. Trends Bioinforma. 2015;8:63–74. doi: 10.3923/tb.2015.63.74. [DOI] [Google Scholar]
- 39.Kagami L.P., das Neves G.M., Timmers L.F.S.M., Caceres R.A., Eifler-Lima V.L. Geo-Measures: a PyMOL plugin for protein structure ensembles analysis. Comput. Biol. Chem. 2020;87 doi: 10.1016/j.compbiolchem.2020.107322. [DOI] [PubMed] [Google Scholar]
- 40.Amadei A., Linssen A.B.M., Berendsen H.J.C. Essential dynamics of proteins. Proteins: Struct., Funct., Bioinf. 1993;17:412–425. doi: 10.1002/prot.340170408. [DOI] [PubMed] [Google Scholar]
- 41.Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46:W350–W355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Valdés-Tresanco M.S., Valdés-Tresanco M.E., Valiente P.A., Moreno E. gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theor. Comput. 2021;17:6281–6291. doi: 10.1021/acs.jctc.1c00645. [DOI] [PubMed] [Google Scholar]
- 43.Cerutti G., Guo Y., Zhou T., Gorman J., Lee M., Rapp M., Reddem E.R., Yu J., Bahna F., Bimela J., Huang Y., Katsamba P.S., Liu L., Nair M.S., Rawi R., Olia A.S., Wang P., Zhang B., Chuang G.-Y., Ho D.D., Sheng Z., Kwong P.D., Shapiro L. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021;29:819–833. doi: 10.1016/j.chom.2021.03.005. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y., Chen Z., Guo Y., Zhang J., Li Y., Song X., Chen Y., Xia L., Fu L., Hou L., Xu J., Yu C., Li J., Zhou Q., Chen W. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science (80- 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cerutti G., Guo Y., Liu L., Liu L., Zhang Z., Luo Y., Huang Y., Wang H.H., Ho D.D., Sheng Z., Shapiro L. Cryo-EM structure of the SARS-CoV-2 Omicron spike. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2022.110428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu G., Yan W., Zhou J., Shen B. Residue interaction network analysis of Dronpa and a DNA clamp. J. Theor. Biol. 2014;348:55–64. doi: 10.1016/j.jtbi.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 47.Huang H.-Y., Liao H.-Y., Chen X., Wang S.-W., Cheng C.-W., Shahed-Al-Mahmud M., Liu Y.-M., Mohapatra A., Chen T.-H., Lo J.M., Wu Y.-M., Ma H.-H., Chang Y.-H., Tsai H.-Y., Chou Y.-C., Hsueh Y.-P., Tsai C.-Y., Huang P.-Y., Chang S.-Y., Chao T.-L., Kao H.-C., Tsai Y.-M., Chen Y.-H., Wu C.-Y., Jan J.-T., Cheng T.-J.R., Lin K.-I., Ma C., Wong C.-H. Vaccination with SARS-CoV-2 spike protein lacking glycan shields elicits enhanced protective responses in animal models. Sci. Transl. Med. 2022 doi: 10.1126/scitranslmed.abm0899. 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casalino L., Gaieb Z., Goldsmith J.A., Hjorth C.K., Dommer A.C., Harbison A.M., Fogarty C.A., Barros E.P., Taylor B.C., McLellan J.S., Fadda E., Amaro R.E. Beyond shielding: the roles of glycans in the SARS-CoV-2 spike protein. ACS Cent. Sci. 2020;6:1722–1734. doi: 10.1021/acscentsci.0c01056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jaafar R., Boschi C., Aherfi S., Bancod A., Le Bideau M., Edouard S., Colson P., Chahinian H., Raoult D., Yahi N., Fantini J., La Scola B. High individual heterogeneity of neutralizing activities against the original strain and nine different variants of SARS-CoV-2. Viruses. 2021;13:2177. doi: 10.3390/v13112177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan A., Waris H., Rafique M., Suleman M., Mohammad A., Ali S.S., Khan T., Waheed Y., Liao C., Wei D.-Q. The Omicron (B.1.1.529) variant of SARS-CoV-2 binds to the hACE2 receptor more strongly and escapes the antibody response: insights from structural and simulation data. Int. J. Biol. Macromol. 2022;200:438–448. doi: 10.1016/j.ijbiomac.2022.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang Y., Harris B.S., Minami S.A., Jung S., Shah P.S., Nandi S., McDonald K.A., Faller R. SARS-CoV-2 spike binding to ACE2 is stronger and longer ranged due to glycan interaction. Biophys. J. 2022;121:79–90. doi: 10.1016/j.bpj.2021.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang C., Tan Z., Zhao K., Zou W., Wang H., Gao H., Sun S., Bu D., Chai W., Li Y. The effect of N-glycosylation of SARS-CoV-2 spike protein on the virus interaction with the host cell ACE2 receptor. iScience. 2021;24 doi: 10.1016/j.isci.2021.103272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim S., Liu Y., Ziarnik M., Cao Y., Zhang X.F., Im W. Binding of human ACE2 and RBD of omicron enhanced by unique interaction patterns among SARS-CoV-2 variants of concern. BioRxiv Prepr. Serv. Biol. 2022 doi: 10.1101/2022.01.24.477633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lan J., He X., Ren Y., Wang Z., Zhou H., Fan S., Zhu C., Liu D., Shao B., Liu T.-Y., Wang Q., Zhang L., Ge J., Wang T., Wang X. Structural insights into the SARS-CoV-2 Omicron RBD-ACE2 interaction. Cell Res. 2022 doi: 10.1038/s41422-022-00644-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang K., Zhang Y., Hui X., Zhao Y., Gong W., Wang T., Zhang S., Yang Y., Deng F., Zhang Q., Chen X., Yang Y., Sun X., Chen H., Tao Y.J., Zou Z., Jin M. Q493K and Q498H substitutions in Spike promote adaptation of SARS-CoV-2 in mice. EBioMedicine. 2021;67 doi: 10.1016/j.ebiom.2021.103381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Niu S., Wang J., Bai B., Wu L., Zheng A., Chen Q., Du P., Han P., Zhang Y., Jia Y., Qiao C., Qi J., Tian W.-X., Wang H.-W., Wang Q., Gao G.F. Molecular basis of cross-species ACE2 interactions with SARS-CoV-2-like viruses of pangolin origin. EMBO J. 2021;40 doi: 10.15252/embj.2021107786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Han P., Li L., Liu S., Wang Q., Zhang D., Xu Z., Han P., Li X., Peng Q., Su C., Huang B., Li D., Zhang R., Tian M., Fu L., Gao Y., Zhao X., Liu K., Qi J., Gao G.F., Wang P. Receptor binding and complex structures of human ACE2 to spike RBD from omicron and delta SARS-CoV-2. Cell. 2022;185:630–640. doi: 10.1016/j.cell.2022.01.001. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han P., Su C., Zhang Y., Bai C., Zheng A., Qiao C., Wang Q., Niu S., Chen Q., Zhang Y., Li W., Liao H., Li J., Zhang Z., Cho H., Yang M., Rong X., Hu Y., Huang N., Yan J., Wang Q., Zhao X., Gao G.F., Qi J. Molecular insights into receptor binding of recent emerging SARS-CoV-2 variants. Nat. Commun. 2021;12:6103. doi: 10.1038/s41467-021-26401-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.