Abstract
Fungal keratitis (FK) pathology is driven by both fungal growth and inflammation within the corneal stroma. Standard in vitro infection models–involving co-culture of the pathogen and the corneal cells in tissue culture medium–are sufficient to probe host responses to the fungus; however, they lack the physiological structure and nutrient composition of the stroma to accurately study fungal invasiveness and metabolic processes. We therefore sought to develop a culture model of FK that would allow for both host and fungal cell biology to be evaluated in parallel. Towards this end, we employed a previously described system in which primary human cornea fibroblasts (HCFs) are cultured on transwell membranes, whereupon they secrete a three-dimensional (3D) collagen matrix that resembles the human stroma. We demonstrated that two common mold agents of FK, Fusarium petroliphilum and Aspergillus fumigatus, penetrated into these constructs and caused a disruption of the collagen matrix that is characteristic of infection. HCF morphology appeared altered in the presence of fungus and electron microscopy revealed a clear internalization of fungal spores into these cells. Consistent with this apparent phagocyte-like activity of the HCFs, mRNA and protein levels for several pro-inflammatory cytokines/ chemokines (including TNFα, IL-1β, IL-6, and IL-8) were significantly upregulated compared to uninfected samples. We similarly found an upregulation of several HCF metalloproteases (MMPs), which are enzymes that breakdown collagen during wound healing and may further activate pro-inflammatory signaling molecules. Finally, several fungal collagenase genes were upregulated during growth in the constructs relative to growth in tissue culture media alone, suggesting a fungal metabolic shift towards protein catabolism. Taken together, our results indicate that this 3D-stromal model provides a physiologically relevant system to study host and fungal cell pathobiology during FK.
Keywords: Fungal keratitis, Corneal fibroblasts, Keratocytes, Corneal inflammation, Matrix metalloprotease, Fusarium
1. Introduction
Fungal keratitis (FK) is an infection of the cornea and a significant source of ocular morbidity and unilateral blindness worldwide (Thomas and Kaliamurthy, 2013). Two major risk factors for FK include corneal trauma caused by vegetative debris and contact lens wear (Saha et al., 2009; Cheung et al., 2016). In both contexts, the molds Fusarium and Aspergillus have emerged as predominant agents of infection, likely due their ubiquity in the soil, on vegetation, and even within fresh-water plumbing systems (Rosa et al., 2009; Bharathi et al., 2007; Short et al., 2013). Fusarium species, for example, were the sole agents of FK in a multi-country outbreak (included the United States) associated with a commercial lens cleaning solution (Chang et al., 2016). Regardless of the infectious route or etiology however, medical intervention in the form of topical antifungals fails in up to 50% of FK cases, resulting in the need for corneal transplantation or even enucleation if the infection spreads to intraocular compartments (Mundra et al., 2019). A better understanding of FK pathobiology is therefore required to fuel the development of novel and more efficacious treatment modalities.
FK occurs when a breakdown in corneal epithelial integrity allows the fungus to access and grow within the central stroma. The stroma comprises about 90% of the corneal thickness and accounts for the overall transparency of the tissue due to its avascularity, paucity of resident cells and careful assembly of collagen fibrils (Spadea et al., 2016; Doughty and Jonuscheit, 2019). Fungal growth is inherently damaging to the stoma as the organism secretes copious amounts of hydrolytic enzymes (collagenases) that may directly damage the collagen extracellular matrix (ECM) (Zhu et al., 1990). Just as damaging, however, is the resulting host response. The massive influx of neutrophils from the periphery alters the refractive index of stroma, leading to corneal opacification and acute visual impairment (Leal and Pearlman, 2012). Though ostensibly important for fungal clearance, neutrophils further secrete matrix metalloproteases (MMPs), which are zinc-binding collagenases that can further exacerbate stromal breakdown and promote fungal proliferation, corneal melt and long-term corneal scarring (Taylor et al., 2016). MMPs further cleave and process pro-inflammatory molecules, such as IL-1β, thus propagating the inflammatory cascade (Sivak and Fini, 2002). Both the general induction of neutropenia by cyclophosphamide and the specific blocking of MMPs with neutralizing antibodies mitigate disease development in murine models of fungal or bacterial keratitis, thereby underscoring the significance of the host response in keratitis pathology (Leal et al., 2010; McClellan et al., 2006; Gao et al., 2015). Accordingly, host cell populations and signaling pathways that trigger this inflammatory/fibrotic response may represent ideal targets for FK intervention.
Stromal keratocytes are mesenchymal-derived, non-contractile, dendritic-shaped cells distributed across the stroma. These cells exist in a largely quiescent state, slowly producing crystallins, collagens, proteoglycans, and other products that are critical in maintaining the stromal ECM and corneal transparency. Following corneal injury, or upon exposure to serum in vitro, keratocytes differentiate into fibroblasts, which are characterized by their increased size, spindle shape, increased phagocytic activity, and migratory action (Sato et al., 2019; Fujita et al., 1987; Fukuda et al., 2017; Myrna et al., 2009). Once at the site of stromal damage, fibroblasts remodel the ECM by secreting both MMPs as well as collagen sub-types that may heal the wound but result in scarring due to irregular collagen deposition (Sugioka et al., 2018). In addition, macrophage-like gene expression is induced following the keratocyte-to-fibroblast transition, including an upregulation of mannose receptor type 1, chemokine ligands, CD68 (typically only expressed on monocytes and macrophages), and complement component pathway genes (Chakravarti et al., 2004).
Corneal fibroblasts have been further implicated as modulators of inflammation and corneal damage in the context of microbial keratitis. First, the cells secrete pro-inflammatory cytokines, including the chemokine MCP-1 and IL-8 (aka CXCL8), in the presence of either bacterial lipopolysaccharide (LPS) or peptidoglycan. They also produce various pro-MMPs that are cleaved and activated by bacterial-derived proteases (Fukuda et al., 2005, 2017). Similar results are seen in the presence of fungal antigen, as Nomi et al. demonstrated that cultured corneal fibroblasts secrete IL-6 and IL-8 when challenged with zymosan (purified β-glucan)-coated beads; this response appears to be mediated through the MAP kinase and NF-kB signaling pathways (Nomi et al., 2010). Li and colleagues demonstrated a similar pro-inflammatory response when immortalized human corneal fibroblasts (HCFs) were stimulation with heat-killed A. fumigatus hyphae. Interestingly, the magnitude of the response to hyphal challenge could be attenuated by pre-treating the fibroblasts with lipopolysaccharide and zymosan, which are ligands for TLR 4 and TLR2, respectively (Li et al., 2013). In addition to providing insight into signaling mechanism, the studies suggest that corneal fibroblasts could serve as targets for anti-inflammatory treatment in FK.
Interrogating how specific host cells interact with fungi during infection may be challenging in vivo due to the presence of numerous corneal cell types, particularly after inflammation sets in shortly after fungal inoculation. In vitro models, such as those used in the above- described fibroblast-fungal studies, bypass this problem (Montgomery and Fuller, 2020). However, standard culture models fall short because they lack important features of the stroma, in particular the collagen matrix in which the host-pathogen interaction takes place. Karamichos and colleagues demonstrated that primary HCFs cultured on a polycarbonate transwell membrane secrete a collagen ECM that resembles the composition of a healthy corneal stroma (Karamichos et al., 2007, 2010); Sharif et al., 2018; McKay et al., 2019; Priyadarsini et al., 2016). We reasoned that these three-dimensional (3D) constructs would serve as a valuable in vitro model for FK for several reasons. Not only can HCF activity be probed as they are the only host cell present, but the fungal penetration and damage to the collagen ECM can also be evaluated. Moreover, the presence of ECM components, e.g. collagen, may further influence fungal metabolism such that it more closely mimics that of live infection.
In this study, we challenged the 3D-stromal constructs with two common mold agents of FK, Fusarium petroliphilum and Aspergillus fumigatus, and found they could both penetrate through and damage the stromal ECM. We also found that HCFs phagocytosed fungal conidia and, in agreement with previous studies, expressed both pro- inflammatory and matrix-remodeling genes/proteins. Finally, we demonstrate several A. fumigatus collagenase genes are upregulated when grown in the constructs relative to culture in tissue culture medium. This suggests that the presence of the stomal ECM does influence fungal gene expression, and that this model can be useful for probing both host and fungal biology during FK.
2. Materials and methods
2.1. Culture of primary human corneal fibroblasts
Human corneas were obtained from the National Development and Research Institute (NDRI) and fibroblasts were isolated as described by Guo et al. (2007). Briefly, the corneal epithelium and endothelium were scraped off with a razor blade and the remaining stroma was sectioned into small 2 × 2 mm squares that were placed in T25 flasks and allowed to adhere. These explants were cultured with Eagle’s Minimum Essential Media (EMEM: ATCC; Manassas, VA) that contained 10% fetal bovine serum (FBS: ATCC) and 1% antibiotic and antimycotic 100X (AA: Life Technologies, Grand Island, NY). Cultures were incubated at 37 °C, 5% CO2 for 1–2 weeks or until 100% confluent, and were passaged following trypsin digestion.
2.2. Construct assembly
1.0 × 106 fibroblasts/ml EMEM were seeded onto a polycarbonate transwell membrane insert with 0.4 μm pores for a conventional 6-well tissue culture plate (Corning Costar; Corning Incorporated, Corning, NY, USA). Cells were incubated for 24 h in EMEM. After 24 h, a routine media change was performed every 48 h for 4 weeks with supplemented EMEM that contained 1 μM 2-O-α-D-glucopyranosyl-L-ascorbic acid (American Custom Chemicals Corporation, San Diego, CA), 10% FBS, and 1% AA.
2.3. Preparation of fungal inoculum and infection of the stromal constructs
The Fusarium petroliphilum strain used in this study is an FK isolate previously described (Lightfoot and Fuller, 2019). To generate the inoculum, F. petroliphilum spores (microconidia) were inoculated into YPD broth and incubated for 2–3 days at 30 °C, shaking at 200 RPM. Microconidia were harvested by passage through Miracloth (EMD Millipore) and pelleted by centrifugation at 3500 RPM. The spore pellets were then washed in PBS twice and ultimately resuspended in PBS and adjusted to a density of 6.74 × 104 conidia/mL. Aspergillus fumigatus was cultured on solid glucose minimal medium (GMM) containing 1% glucose and ammonium tartrate for 48 h at 35 °C. Conidia were collected by washing the surface with PBS and filtering through miracloth (Millipore), followed by two washes in PBS. The conidia were enumerated using a hemocytometer and adjusted to a density of 6.74 × 104 conidia/mL.
72 h preceding the inoculation, the EMEM medium in the upper and lower chambers of the plates were replaced with EMEM containing no antibacterial/antimycotic. On the day of inoculation, media in the top chamber was removed and the constructs were washed twice with 1 ml of PBS. 750 μl of fungal inoculum was overlaid on the constructs, giving a final inoculum 5 × 104 conidia in the top chamber of the transwell system. The bottom chamber was replaced with fresh EMEM containing no FBS and no antibacterial/antimycotic. Samples were incubated undisturbed for their indicated time point at 35 °C and 5% CO2.
2.4. RNA isolation and RNA-seq from stromal constructs
The artificial stroma and fungal inoculum were peeled from the polycarbonate membrane and placed in a 1.5 ml microcentrifuge tube with 0.5 mm diameter glass beads and tissue was homogenized using QIAGEN TissueLyser LT at 50 oscillations/s for two 30 s cycles. RNA extraction was performed following the manufacturer’s protocol for QIAGEN RNeasy Mini Kit (QIAGEN Science, Germantown, Maryland). For RNA-sequencing, 200 ng total RNA from all subjects were used to prepare sequencing libraries with the TaKaRa SMARTer Stranded RNA- Seq kit after ribosomal RNA depletion using the RiboGone - Mammalian kit from TaKaRa (TaKaRa Bio USA, Inc., Mountain View, CA, USA). Briefly, the RiboGone-Mammalian kit removes ribosomal RNA (rRNA) and mitochondrial RNA (mtRNA) sequences from human total RNA samples based on hybridization and RNase H digestion that specifically depletes 5 S, 5.8 S, 18 S, and 28 S nuclear rRNA sequences, as well as 12 S mtRNA sequences. The rRNA-depleted total RNA was reverse- transcribed to synthesize the first-strand complementary (c) DNA followed by PCR amplification using universal forward PCR primer and reverse PCR indexing primer set. Purified RNA-Seq library was validated using the Agilent 2100 Bioanalyzer with Agilent’s High Sensitivity DNA Kit (Agilent, Santa Clara, CA, USA). Pooled RNA-Seq libraries were sequenced with paired-end 75-bp reads using an Illumina NextSeq sequencer with high output v2 (Illumina, Inc., San Diego, CA, USA) at the Georgia Cancer Center Integrated Genomics Core of Augusta University. After quality check and control with all sequencing reads, demultiplexed reads were aligned by TopHat31 using paired-end reading with the approximation of the median library size. Counts of sequencing reads were normalized using Cufflinks in fragments per kilo bases and millions reads (FPKM). After normalization, we annotated the transcripts with a gene transcript file from Ensembl database at the gene isoform level. We performed differential expression analysis using the Cuffdiff package.
2.5. Protein isolation and measurement from stromal constructs
3D constructs were washed with 1 mL 1X PBS and then, using a spatula, were gently scraped off of the membrane and placed in a 2 mL tube containing a 6.35 mm metal ball, T-PER (ThermoScientific, Ref. #78510), and Calbiochem protease inhibitor cocktail set I (Millipore Sigma, Cat. #539131–1VL). Samples were then homogenized using a Qiagen TissueLyser LT at 50 oscillations/second for 30 s (x2). Following 30 min incubation at 4 °C, samples were centrifuged at 12,000 RPM for 15 min at 4 °C. Supernatants were collected and stored at − 80 °C until the time of the experiment. A bead-based multiplex assay for protein detection was used to quantify expression of GM-CSF, IFN-ɣ, IL-10, IL-13, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-12 (p70), and TNF- a (MilliporeSigma, HSTCMAG28SPMX13). Data were graphed in GraphPad Prism 7 and statistics were determined using Welch’s t-test where p ≤ 0.05*, p ≤ 0.01**, p ≤ 0.001***, and p ≤ 0.0001****. Samples with values that were too low to be extrapolated from the standard curve were assigned a value of 0.01.
2.6. Mouse model of Aspergillus keratitis and RNA isolation from infected corneas and EMEM
6–8 wk old, male C57BL/6 J mice (Jackson Laboratory) were immunosuppressed with 100 mg/kg methylprednisolone intraperitoneally (i.p.) on the day preceding fungal inoculation. On the day of inoculation, A. fumigatus (strain Af293) conidia were incubated in YPD broth (yeast extract, peptone, glucose) at a density of 1.0 × 105 conidal/ mL and incubated for 4 h until in shaking culture at 35 °C until conidia were swollen, but not polarized. The swollen conidia were collected via centrifugation, washed with PBS (2X), and resuspended in ~0.5 mL PBS. This served as the inoculum. To infect mice, animals were anesthetized with 100 mg/kg ketamine and 6.6 mg/kg xylazine i.p., and the corneal epithelium over the pupil of the right eye was removed with an alger-brush II. 5 μl of fungal inoculum were topically applied over the abraded cornea and remained in place for 20 min before being removed with a kim wipe. The left eye remained untouched as a control and in accordance with the Association for Research in Vision and Ophthalmology (ARVO) guidelines for the use of animals in vision research. Animals were monitored twice daily until 48 h post-inoculation, at which point animals were photographed, and the eyes were removed following euthanasia under IACUC-approved procedure. The corneas were resected and pooled (4–9 corneas per group), placed in TriZol, and homogenized with 0.5 mm glass beads (same procedure as for the 3D constructs above).
For the EMEM culture samples, conidia (harvested as described above) were inoculated into serum-free, antimycotic-free EMEM (PurMabiologics) to a density of 6.74 × 104/mL, the same density used for the 3D constructs. 3 mls of the inoculated media were added to a 6-well plate (Thermo Scientific). Following 48 h incubation at 35 °C, 5% CO2, the media was aspirated for the well and the fungal biomass was collected with a spatula, dried on filter paper, and added to a microcentrifuge tube containing 100 μl of 0.1 mm glass beads (Biospec Products) and 1 mL Trizol for RNA extraction as describe above.
2.7. qRT-PCR
For quantitative PCR analysis, extracted RNA (described above) was reverse transcribed into cDNA following the manufacturer’s protocol for the SuperScript II First-Strand cDNA Synthesis kit. qPCR was performed using either TaqMan (Applied Biosystems, Foster City) of GAPDH (Hs99999905_m1) or Luna Universal qPCR mastermix (SYBR green; NEB) as indicated. All primers used in the study are listed in Table S2. Fold-expression changes were calculated using the 2ΔΔCt method and analyzed using Graph Pad Prism 9.0.1 and MS-Excel.
2.8. Transmission electron microscopy (TEM)
The constructs were washed twice with PBS and fixed after their indicated time points in 4% paraformaldehyde, 2% glutaraldehyde, and 0.1 M sodium cacodylate fixative for 4 h at 4 °C and processed for TEM following standard procedures that have been previously described (Gipson et al., 1983). Cell constructs were fixed with 4% Paraformaldehyde (EM grade), 2.5% Gluteraldehyde (EM grade), in 0.1 M Sodium Cacodylate buffer for 16 h at 4 °C. Samples were then post fixed for 90 min in 1% Osmium tetroxide (OsO4) in 0.1 M Sodium Cacodylate, and rinsed three times for 5 min each in 0.1 M Sodium Cacodylate buffer. The cell constructs were then dehydrated in a sequential acetone series including 50%, 60%, 75%, 85%, 95%, 100%; the samples were in each a for 15 min shaking at RT. Then the samples were then treated 100% Propylene Oxide for 15 min (x2). Following dehydration, the samples were infiltrated in a graded Epon/Araldite (EMS) resin/Propylene Oxide series (1:3, 1:1, 3:1) for 60 min, overnight, and 120 min the next day respectfully. The cell constructs were further infiltrated with pure resin for 45 min, 90 min, and then overnight. The samples were then embedded in resin plus BDMA (accelerator) and polymerized at 60 °C for 48 h. Semi-thin sections were stained with toluidine blue and were imaged on a Zeiss Axiovert 200 M microscope.
2.9. Histology
Control and infected constructs were washed twice with PBS and fixed with 10% neutral buffered formalin for 24 h. The fixative was then removed and the constructs were placed in 70% ethanol until further processed. For sectioning, fixed constructs were placed on dental wax and cut into 5 μm thick sections while still attached to the polycarbonate membrane. The sections were then stained with toluidine blue as a general stain; Grocott-Gomori’s methenamine silver (GMS) or Periodic acid Schiff-hematoxylin (PASH) stain was used to specifically visualize the fungal cell wall.
3. Results
3.1. Development of a 3D in vitro model of FK to probe the fungal- fibroblast interaction
As described above, we reasoned that the 3D stromal model developed by Karamichos and colleagues could serve as an ideal system to probe host-fungal interactions during FK. Before we could focus on such interactions, however, we first needed to establish that the model would generally mimic features of the disease, namely fungal invasion of and damage to the ECM. We therefore began by characterizing these baseline characteristics with the mold F. petroliphilum, a common agent of FK among contact lens wearers, particularly in the United States.
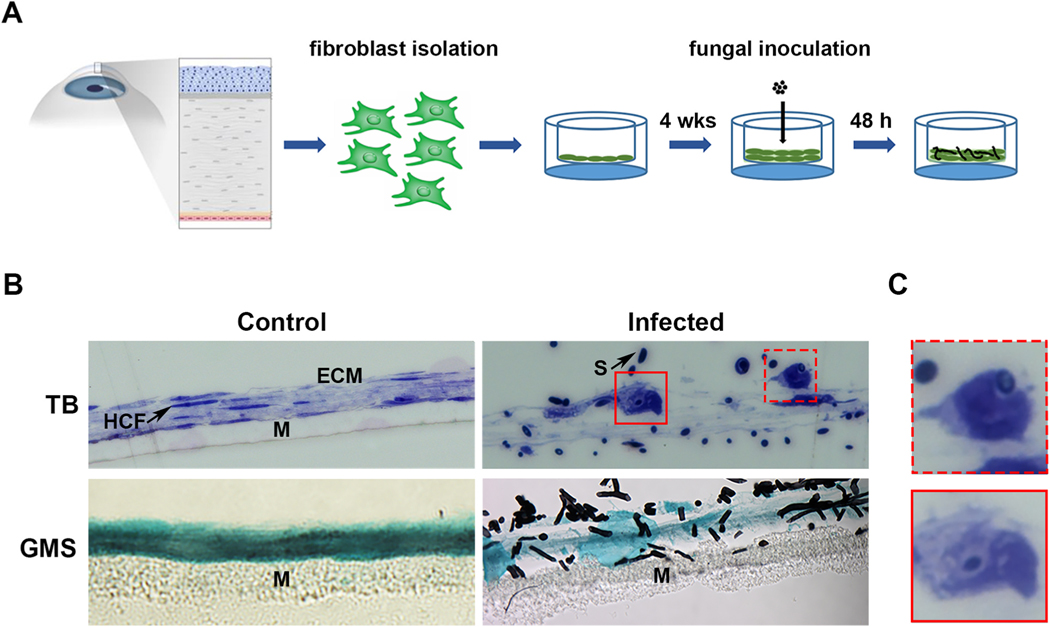
The experimental workflow is summarized in Fig. 1A and began with the generation of stromal constructs as previously described (Karamichos et al., 2010). Briefly, HCFs were isolated from healthy donor corneas, expanded in culture, and seeded onto 6-well polycarbonate inserts at a density of 1.0 × 106 cells/well. The plates were then cultured for four weeks in the presence of ascorbic acid (vitamin C) to promote matrix secretion by the cells. At the end of this incubation period, constructs of approximately 20 μm and 4–5 layers of HCFs thick were observed (Fig. 1B). The constructs were next overlaid with 5.0 × 104 conidia (spores) of F. petroliphilum. In an initial experiment, the fungal inoculum was prepared in tissue culture medium (EMEM), however, we noted that the spores germinated and formed a thick biofilm within the medium, rather than attaching and growing into the stromal constructs as desired (data not shown). Accordingly, we subsequently prepared the inocula in PBS, thus making the construct ECM the primary nutrient source for the fungus; EMEM (without serum or antimycotic) remained in the lower chamber of the transwell post-inoculation as a source of nutrients for the fibroblasts. Using this setup, we performed an initial time course and found that Fusarium conidia had partially germinated at 24 h post-inoculation and the construct ECM was largely intact (Figure S1). At 48 h, by contrast, the conidia had more fully developed into short hyphae and the constructs appeared degraded relative to un-infected controls (Fig. 1B). This suggested the fungus is able to invade the stromal matrix comparable to an in vivo infection. As these were the minimal requirements for pursuing this model, we next turned our attention to the fibroblasts.
Fig. 1.
Development of an in vitro 3D stromal model for fungal keratitis. A) 3D stromal constructs are generated following the isolation of HCFs from healthy human donors and culturing on a transwell membrane for up to 4 weeks. On the day of inoculation, the media in the upper chamber is replaced with the fungal spore inoculum prepared in PBS. B) Brightfield images demonstrating healthy and Fusarium-infected constructs at 48 h post-inoculation. Toluidine Blue (TB) is used to show all features of system, including the human corneal fibroblasts (HCF), the collagen extracellular matrix (ECM), the fungal spores (S), and the nylon membrane (M). Grocott-Gomori’s methanamine silver (GMS) stain is used to stain fungal cell wall black. C) Close up image from panel B, demonstrating fibroblasts that have apparently internalized fungal spores.
3.2. HCFs can engulf and internalize Fusarium conidia
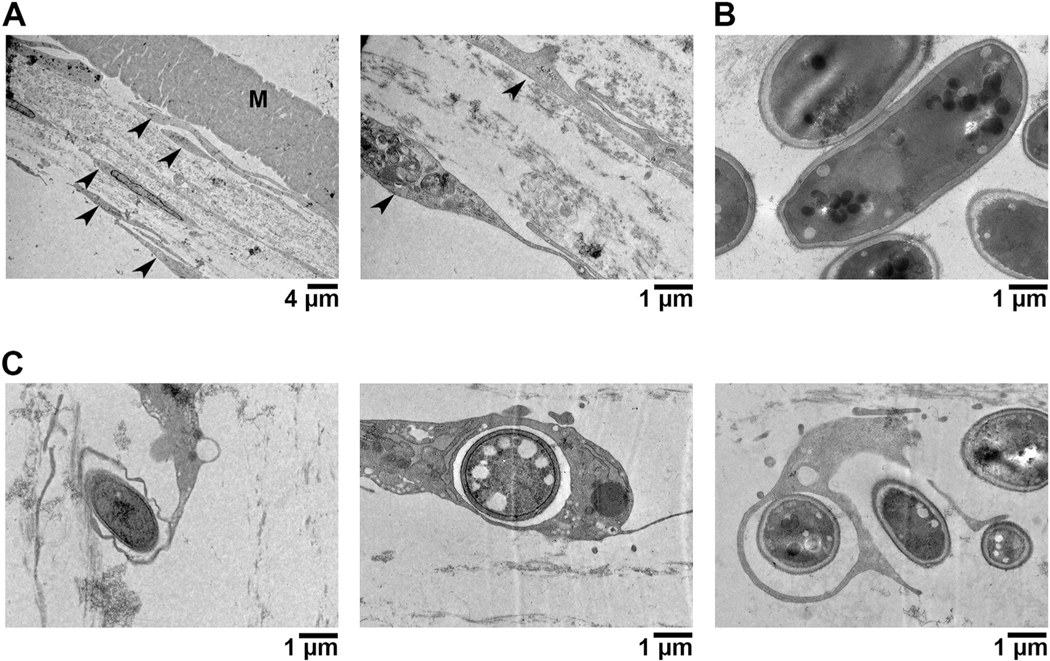
Compared to the flat appearance of the HCFs in the control constructs, the cells often appeared thicker or more rounded in the Fusarium-infected samples (Fig. 1B). Fungal conidia contacted the surface of these fibroblasts and, in some cases, appeared to be internalized within them (Fig. 1C). We then imaged these constructs by transmission electron microscopy (TEM) to observe this interaction more closely. Within such images, the fibroblasts were readily distinguishable from conidia, the latter of which are ovoid/circular in shape and surrounded with an electron-dense cell wall (Figure 2A and 2B). TEM confirmed a direct fungal-fibroblast interaction as conidia were observed within distinct endomembranes. Remarkably, several HCFs appeared to have formed membrane/cytoplasmic projections that engulfed conidia otherwise not in contact with the primary cell body (Fig. 2C). Taken together, these data suggest that corneal fibroblasts have the capacity to detect and endocytose Fusarium conidia.
Fig. 2.
Transmission Electron Microscopy (TEM) of Fusarium infected HCFs. A) Uninfected (control) constructs highlighting the flat apparencu of HCFs (arrowheads) and intact cellular matrix. B) Images of Fusarium microconidia, which are smooth and have an electron-dense cell wall. C) Infected constructs demonstrating an internalization of conidia within HCF endomembranes or cytoplasmic projections.
3.3. HCFs become pro-inflammatory in response to Fusarium infection
To elucidate the HCF response to Fusarium more fully, we performed Illumina sequencing of total RNA isolated from control (un-infected) and Fusarium-infected constructs at 48 h post-inoculation (n = 4/group). A total of 12,198 and 11,314 HCF genes with at least one normalized read were detected in the control and infected groups, respectively; the full dataset containing all detected genes and their RPKM values can be found in Table S1. Differentially expressed genes (DEGs) were considered to be those with at least 10 normalized reads in the control or infected groups and at least a 4-fold expression (q ≤ 0.01) difference between the two groups. Based on these criteria, 156 genes were downregulated in the infected samples relative to the controls. The 30 genes with the strongest down-regulation are shown in Table 1 and include several with a described role in cell-growth or differentiation. Pathway enrichment analysis (KEGG) demonstrated that genes involved in metabolism, proteasome function, glycolysis/gluconeogenesis, and amino acid biosynthesis were statistically enriched among the entire downregulated gene list (Table 2). These results together suggest that growth and metabolism are largely repressed in HCFs following Fusarium infection.
Table 1.
Top 30 downregulated genes in Fusarium-infected HCFs based on fold-change compared to uninfected controls.
| Gene | Description | Fold change | p value |
|---|---|---|---|
|
| |||
| EAPP | E2F-associated Phosphoprotein that promotes proliferation | − 27.9098 | .00005 |
| CDC123 | Cell Division Cycle 123 protein required for entry into S phase | − 18.3722 | .00005 |
| AKR1C3 | Aldo-keto reductase family 1 member C3 | − 15.7936 | .00005 |
| TXNDC9 | Thioredoxin domain-containing protein involved in proliferation | − 15.4993 | .00005 |
| C10orf10 | also called DEPP1 involved in regulating autophagy | − 13.1128 | .00005 |
| AKR1B10 | Aldo-keto reductase family 1 member B10 | − 11.5138 | .00005 |
| AHNAK2 | Nucleoprotein | − 10.5996 | .00005 |
| ADH1B | Alcohol dehydrogenase 1 B | − 9.87931 | .00005 |
| TSEN15 | tRNA splicing endonuclease | − 9.86378 | .00005 |
| PSMB6 | Proteasome subunit beta type-6 | − 9.78533 | .00005 |
| TSNAX | Translin Associated Factor X involved in activation of the RNA-induced silencing complex | − 9.56424 | .00005 |
| ABHD10 | Abhydrolase Domain Containing 10 | − 9.54715 | .00005 |
| EHD3 | EH Domain Containing 3 | − 9.48107 | .00005 |
| TXNIP | Thioredoxin Interacting Protein | − 9.01758 | .00005 |
| FIBIN | Fin Bud Inititation Factor | − 8.98376 | .00005 |
| SLC1A5 | Soluble Carrier Family 1 Member 3 encodes a sodium-dependent neutral amino acid transporter | − 8.91757 | .00005 |
| CBR1 | Carbonyl reductase 1 | − 8.45052 | .00005 |
| GAS1 | Growth Arrest Specific 1 involved in growth suppression | − 8.31952 | .00005 |
| ZFYVE21 | Zinc Finger FYVE-Type Containing 21 involved in cell adhesion | − 8.15068 | .00005 |
| TRAPPC1 | Trafficking Protein Particle Complex 1 involved in ER to Golgi transport | − 8.1408 | .00005 |
| THYN1 | Thymocyte Nuclear Protein 1 potentially involved in the induction of apoptosis | − 8.07952 | .00005 |
| PITX2 | Paired Like Homeodomain 2 encoding a transcription factor that regulates eye development |
− 7.90015 | .00005 |
| TNRC18 | Trinucleotide Repeat Containing 18 | − 7.81345 | .00005 |
| UQCR10 | Ubiquinol-Cytochrome C Reductase involved in respiratory metabolism | − 7.60601 | .00005 |
| PSAT1 | Phosphoserine Aminotransferase 1 | − 7.57891 | .00005 |
| MRPS17 | Mitochondrial Ribosomal Protein S17 | − 7.52802 | .00005 |
| LRIG3 | Leucine Rich Repeats and Immunoglobulin Like Domain | − 7.36012 | .00005 |
| ZFHX4 | Zinc Finger Homeobox 4 | − 7.2105 | .00005 |
| PREB | Prolactin Regulatory Element-Binding Protein | − 7.18451 | .00005 |
| FBXO38 | F-Box Protein 38 | − 7.18292 | .00005 |
Table 2.
KEGG analysis of the 156 downregulated genes in Fusarium-infected HCFs.
| Name | p-value | Genes |
|---|---|---|
|
| ||
| Metabolic Signaling | 2.89e-04 | AKR1C3,AKR1B10,ADH1B,CBR1,UQCR10,PSAT1,ALDH3A2,GCLM,ADH5,IDH1,NDUFV3,CHPF2,NDUFA9, PDE5A,CAT,RDH10,MAT2B,ASNS, MGAT2,UROD, SACM1L,PDHB,PCK2,NDUFA12, EPRS,AGPS,RPE,ADCY7,HIBADH,NQO1 |
| Proteosome | 4.39e-03 | PSMB6,PSMA3,PSMB3,PSMA4,PSMC2 |
| Glycolysis/ Gluconeogenesis | 3.17e-02 | ADH1B,ALDH3A2,ADH5,PDHB,PCK2 |
| Biosynthesis of Amino Acids | 5.00e-02 | PSAT1,IDH1,MAT2B,ASNS,RPE |
A total of 396 genes were upregulated in the infected samples relative to the controls. Among the 30 genes with the greatest fold- induction, 10 were cytokines/chemokines with a well-described role in the inflammatory cell recruitment (CXCL 1,2,3,8) and activation (CSF-1, CSF-2, CSF-3, IL-11, IL-6, IL1α, IL1β, LIF) (Table 3). KEGG analysis demonstrated that IL-17 A, TNF, NFΚB, C-type lectin, and Th17 signaling pathways were statistically enriched among the complete upregulated gene set, further supporting the interpretation that HCFs become pro-inflammatory in response to Fusarium infection (Table 4).
Table 3.
Top 30 upregulated genes in Fusarium-infected HCFs based on fold-change compared to uninfected controls.
| Gene | Description | Fold change | p value |
|---|---|---|---|
|
| |||
| HSPA6 | Heat Shock Protein Family A (Hsp70) Member 6 | 1,794 | .00005 |
| CXCL8 | chemokine involved in neutrophil recruitment and angiogenesis | 1,496 | .00005 |
| CSF3 | cytokine involved in granulocyte production and function | 1,308 | .00005 |
| CXCL2 | chemokine involved inflammation and angiogenesis | 895 | .00005 |
| CXCL3 | chemokine involved in neutrophil recruitment and angiogenesis | 651 | .00005 |
| C11orf96 | 537 | .00005 | |
| TNFAIP3 | TNF-α induced protein that inhibits NF-kappa-B activation and TNF-mediated apoptosis | 420 | .00005 |
| FOSB | Fos proto-oncogene, AP-1 transcription factor subunit implicated in cell proliferation and differentiation | 388 | .00005 |
| HSPA1A | Heat Shock Protein (Hsp70) family member | 263 | .00005 |
| IL11 | Gp130 cytokine family member that play a role in T-cell-dependent development of B-cells | 245 | .00005 |
| HSPA1B | Heat Shock Protein (Hsp70) family member | 213 | .00005 |
| TFPI2 | Kunitz-type serine proteinase inhibitor family | 212 | .00005 |
| G0S2 | G0/G1 switch 2 | 205 | .00005 |
| IL6 | cytokine involved in inflammation and B cell maturation | 199 | .00005 |
| MFSD2A | Sodium-dependent lysophosphatidylcholine transporter | 184 | .00005 |
| DNAJB1 | Heat Shock Family (Hsp40) member | 151 | .00005 |
| BIRC3 | IAP family member that inhibits serum- deprivation-induced apoptosis | 144 | .00005 |
| LIF | Leukemia Inhibitory Factor, IL-6 family cytokine | 123 | .00005 |
| NR4A3 | Nuclear receptor subfamily 4 A member 3 | 91 | .00005 |
| IL1A | cytokine involved in inflammation and hematopoiesis | 81 | .00005 |
| CXCL1 | chemokine involved in neutrophil recruitment | 70 | .00005 |
| NR4A2 | Nuclear receptor subfamily 4 A member 2 | 68 | .00005 |
| CLDN14 | Claudin 14 involved in cell-to-cell adhesion in epithelial and endothelial sheets | 63 | .00005 |
| ATF3 | mammalian Activation Transcription Factor/ CREB protein family of transcription factors | 61 | .00005 |
| NFKBIA | NF-kappa-B inhibitor family | 59 | .00005 |
| CD83 | Immunoglobulin superfamily of receptors involved in the regulation antigen presentation | 58 | .00005 |
| MMP1 | M10 family of matrix metalloproteinases that breaks down interstitial collagens | 57 | .00005 |
| NIPAL4 | NIPA like domain containing membrane receptor | 52 | .00005 |
| IL1B | cytokine involved in inflammation produced by activated macrophages | 51 | .00005 |
| TRIM36 | E3 ubiquitin-protein ligase | 50 | .00005 |
Table 4.
KEGG analysis of the 396 upregulated genes in Fusarium-infected HCFs.
| Name | p- value | Genes |
|---|---|---|
|
| ||
| IL-17 A Signaling | 3.53e-17 | HSP90AB1,MAPK6,TRAF4,CEBPB, JUND, CXCL6,MMP3,JUN, NFKB1,HSP90AA1,CXCL5,FOS,FOSL1, PTGS2,IL1B,MMP1, NFKBIA, CXCL1,IL6,FOSB, TNFAIP3,CXCL3, CXCL2,CSF3,CXCL8 |
| TNF Signaling | 4.93e-16 | FAS,MAP2K3,CSF1,CEBPB,MAP3K8, ICAM1,CREB5,CXCL6,MMP3,JUNB,IRF1, JUN,NFKB1,TRAF1,CXCL5,FOS,PTGS2, IL1B,NFKBIA, CXCL1,LIF,BIRC3,IL6,TNFAIP3,CXCL3, CXCL2 |
| NFκB Signaling | 5.97e-8 | TICAM1,PLAU, CYLD,ICAM1,NFKB2,RELB, NFKB1,TRAF1,PTGS2, GADD45B,IL1B,NFKBIA, CXCL1,BIRC3, TNFAIP3,CXCL3,CXCL2, CXCL8 |
| MAPK Signaling | 1.45e-12 | HSPA8,FAS,MAP2K3,GADD45A, MAPKAPK2,CSF1,PPP3CC, HSPB1,DUSP10,VEGFA,MAP3K8,JUND, PLA2G4C,NFKB2,RELB, JUN,DUSP1,NFKB1,DUSP5,FOS,GADD45B, NR4A1,DUSP8,EREG, IL1B,IL1A,HSPA1B,HSPA1,HSPA6 |
| C-type Lectin Receptor Signaling | 7.18e-6 | MAPKAPK2,CBLB,PLK3,PPP3CC,CYLD, EGR3,NFKB2,IRF1,RELB, JUN,NFKB1,PTGS2,IL1B,NFKBIA,IL6 |
| Cytokine-cytokine Receptor Interaction | 6.06e-5 | FAS,ACKR3,CSF1,TNFRSF10B,CLCF1, TNFRSF12A,INHBA,IL6R,CXCL6,IL32,IL7, RELT, CXCL5,GDF15,IL1B,CXCL1,IL1A,LIF, IL6,IL11,CXCL3,CXCL2,CSF3,CXCL8 |
| Th17 Cell Differentiation | 2.89e-4 | HSP90AB1,PPP3CC,RORA, NFKBIB,IL6R, JUN,NFKB1,HSP90AA1,NFKBIE,FOS,IL1B, NFKBIA,IL6 |
| NOD-like Receptor Signaling Pathway | 3.35e-4 | HSP90AB1,TICAM1,NFKBIB, NAMPT, RIPK2,JUN,NFKB1, HSP90AA1,IL1B,NFKBIA, CXCL1,BIRC3,IL6, TNFAIP3,CXCL3, CXCL2,CXCL8 |
| Apoptosis | 5.45e-3 | FAS,GADD45A,MCL1,ERN1,TNFRSF10B, JUN,NFKB1,PMAIP1, TRAF1,FOS,GADD45B,NFKBIA, BIRC3 |
| Circadian Rhythm | 1.27e-2 | NR1D1,RORA, BHLHE41,PER1,CRY1, BHLHE40 |
| Cellular Senescence | 2.50e-2 | MAP2K3,GADD45A,MAPKAPK2,PPP3CC, SIRT1,RASSF5,NFKB1,ETS1,SERPINE1, GADD45B,IL1A,IL6,CXCL8 |
| Toll-like Receptor Signaling Pathway | 3.22e-2 | MAP2K3,TICAM1,MAP3K8,JUN,NFKB1, FOS,IL1B,NFKBIA,IL6, CXCL8 |
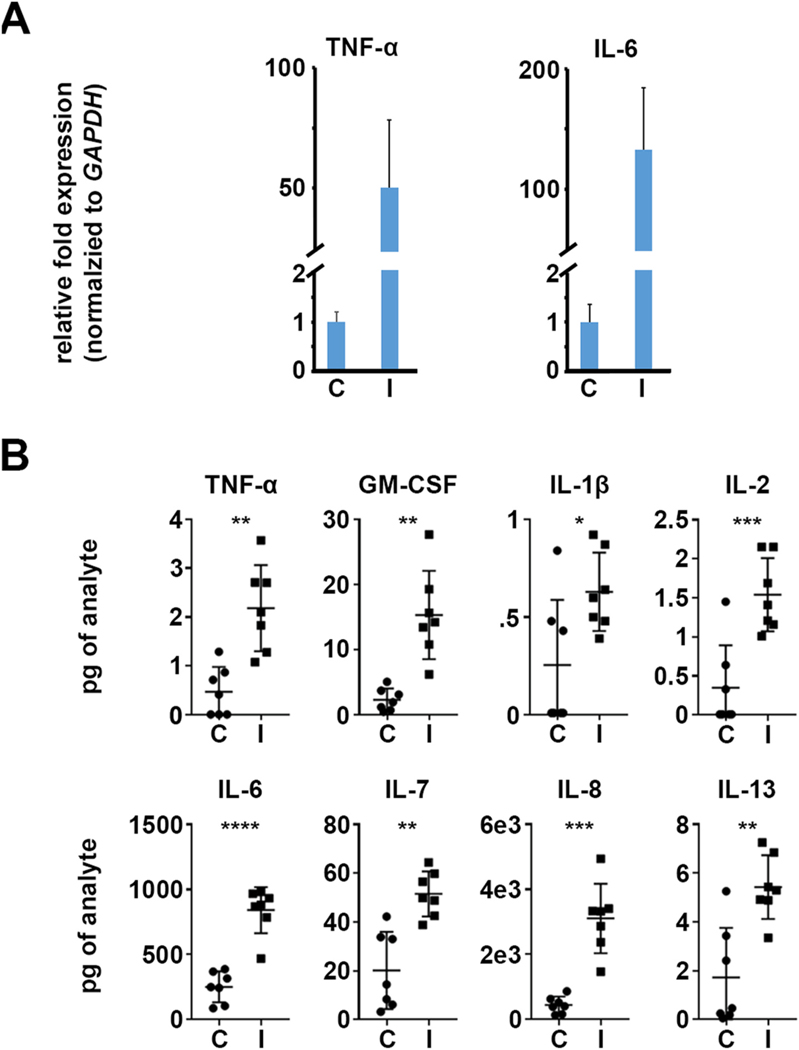
As a means to validate the pro-inflammatory transcriptional response, we next analyzed protein abundance of numerous cytokines and chemokines via a multiplexed immunoassay (Milliplex). Total protein was collected from control and Fusarium-infected constructs at 48 h post-inoculation, in parallel with the constructs harvested for transcriptomics (above). Several genes induced in the RNA-seq data set had corresponding probes in the Milliplex array and were similarly induced, including IL-1β, IL-6, IL-7, and IL-8 (also known as CSCL8) (Fig. 3). Several pro-inflammatory proteins were induced on the protein array that were not detected in RNA-seq data, including TNF-α, IL-2, and IL- 13. We specifically followed up on TNF-α by qRT-PCR since the gene has previously shown to be induced in infected patient or murine FK corneas (Chidambaram et al., 2017; Zhang et al., 2020). In agreement with the protein results, TNF-α did demonstrate an induction in the qRT-PCR assay, perhaps reflecting a greater sensitivity over RNA-seq. IL-6, which is similarly known to be induced in FK corneas and strongly induced in our RNA-seq data, was similarly induced in qRT-PCR and protein assays (Fig. 3A).
Fig. 3.
HCFs become pro-inflammatory during fungal infection. A) qRT-PCR data demonstrating an upregulation of TNF-alpha and IL-6 in Fusarium-infected (I) constructs relative to the uninfected controls (C). GAPDH is used as the internal normalizer for each sample; data plots reflect the ratio of the infected samples relative to the control (set to ‘1’). B) Protein quantitation assay (Milliplex) demonstrating an upregulation of pro-inflammatory cytokines and chemokines.
3.4. Several upstream regulators may contribute to the HCF pro-inflammatory response
We next used Ingenuity Pathway Analysis (IPA, Qiagen) to identify putative regulatory proteins involved in the HCF response to Fusarium infection. This software assesses the overlap between experimental (e.g. RNA-seq) data and an extensively curated database of target genes for each of several hundred known regulatory proteins. It then uses the statistical significance of the overlap and the direction of the differential gene expression to make predictions about activation or repression of these regulatory proteins and associated pathways.
We first focused on HCF cell surface proteins that might interact with the fungus directly to initiate the pro-inflammatory cascade. Of 104 annotated ‘transmembrane’ proteins in the IPA curation, 19 were predicted to be activated. These included well-known pathogen recognition receptors (PRRs), including Dectin-1 (clec7A), TLR2, TLR3, TLR4, TLR5, TLR7, and TLR9. Each of these PRRs, however, share in common several downstream output genes (e.g. pro-inflammatory cytokines regulated by NF-kB) that are upregulated in infected samples. This redundancy creates a bias in the analysis that may lead to false-positives, i.e. proteins considered ‘activated’ by the software even though they are not involved biologically. Indeed, Dectin-1, TLR5 and TLR9 were not detected in either of control or infected RNA-seq datasets or by RT-PCR (data not shown), suggesting that HCFs may not express these proteins. By contrast, TLR3, TLR4, and TLR7 were detected by RNA-seq, but they were not induced in the infected samples relative to the controls. TLR2 was, however, upregulated in the infected samples, suggesting that this protein may detect fungal cell wall moieties (e.g. polysaccharides) to initiate the response. It should be noted, that TLR2 signal remained below 10 FPKM in both the control and infected datasets, indicating a low-level of expression overall.
At the pathway level, IPA demonstrated an enrichment of genes within the IL-17 A, IL-6, TNF-α receptor signaling, and autoinflammatory pathways (Table 5). These cytokine pathways may play a key role in propagating the pro-inflammatory cascade downstream of the initial fungal detection event. Though IL-6 and TNF-α were induced in the infected constructs (Fig. 3), IL-17 A was not detected in the RNA- seq, nor was it induced at the protein level (Table S1 and data not shown).
Table 5.
Upregulated signaling pathways in Fusarium-infected HCFs based on Ingenuity Pathway Analysis.
| Name | p-value | Overlap |
|---|---|---|
|
| ||
| IL-17 A Signaling in Fibroblasts | 1.09E-14 | 37.1% (13/35) |
| Role of IL17-A in Arthritis | 1.84E-14 | 27.3% (15/55) |
| IL-6 Signaling | 5.80E-14 | 15.9% (20/126) |
| Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis | 1.45E-12 | 8.9% (28/313) |
| TNFR2 Signaling | 1.49E-12 | 36.7% (11/30) |
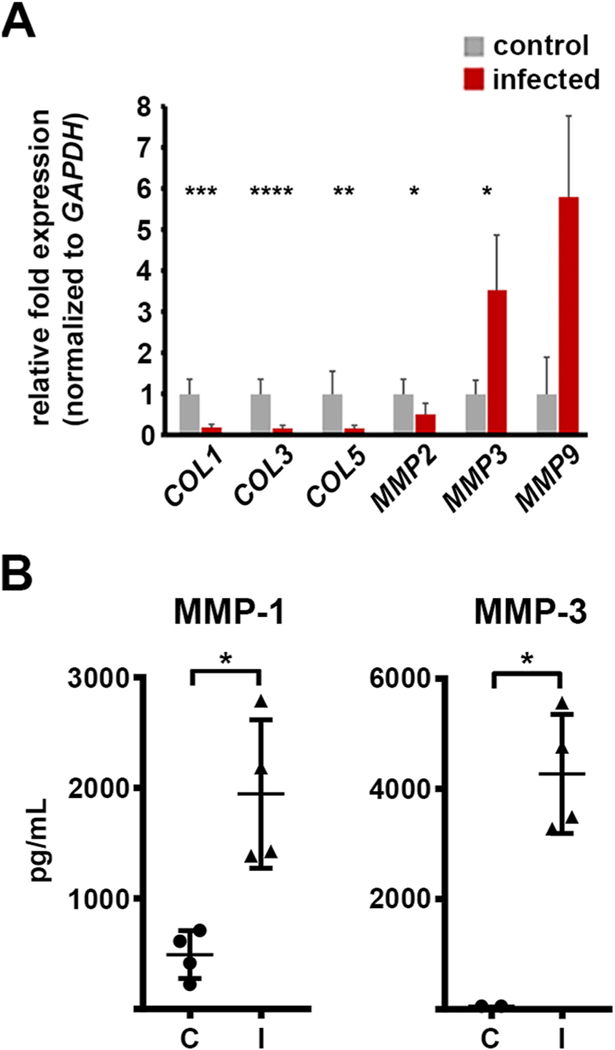
3.5. Fusarium infection regulates HCFs expression of matrix remodeling proteins
As corneal fibroblasts secrete MMPs as part of their normal tissue function as well as in response to bacterial antigen, we predicted they may similarly up-regulate these proteins during fungal infection. Consistent with this prediction, RNA-seq revealed an upregulation of both MMP1 and MMP3 in infected samples relative to controls, and their induction at the protein level was confirmed by the Milliplex assay. MMP9 was not detected in the RNA-seq dataset, but has been described as being highly induced in FK corneas; we, therefore tested its expression by qRT-PCR and found it was detectable and induced in infected constructs (Table S1 and Fig. 4A).
Fig. 4.
Altered expression of matrix remodeling genes during Fusarium infection. A) qRT-PCR data demonstrating an differential regulation of various collagen (COL) and matrix metalloproteases (MMP)-encoding genes in infected (I) constructs relative to the un-infected controls (C). GAPDH is used as the internal normalizer for each sample; data plots reflect the ratio of the infected samples relative to the control (set to ‘1’). B) Protein quantitation assay (Milliplex) demonstrating an upregulation of MMP-1 and MMP-3 in Fusarium- infected HCF constructs.
In contrast to the induction of MMPs, the RNA-seq data suggested a down-regulation of several genes encoding collagen components, including COL1, COL3, and COL5. The reduced expression of these genes was confirmed by qRT-PCR (Fig. 4A). Taken together, these data indicate that Fusarium infection broadly regulates the expression of matrix remodeling genes, including those encoding MMPs and collagen, ultimately favoring a state of collagen degradation.
3.6. A. fumigatus similarly induces a pro-inflammatory and pro-fibrotic in HCFs
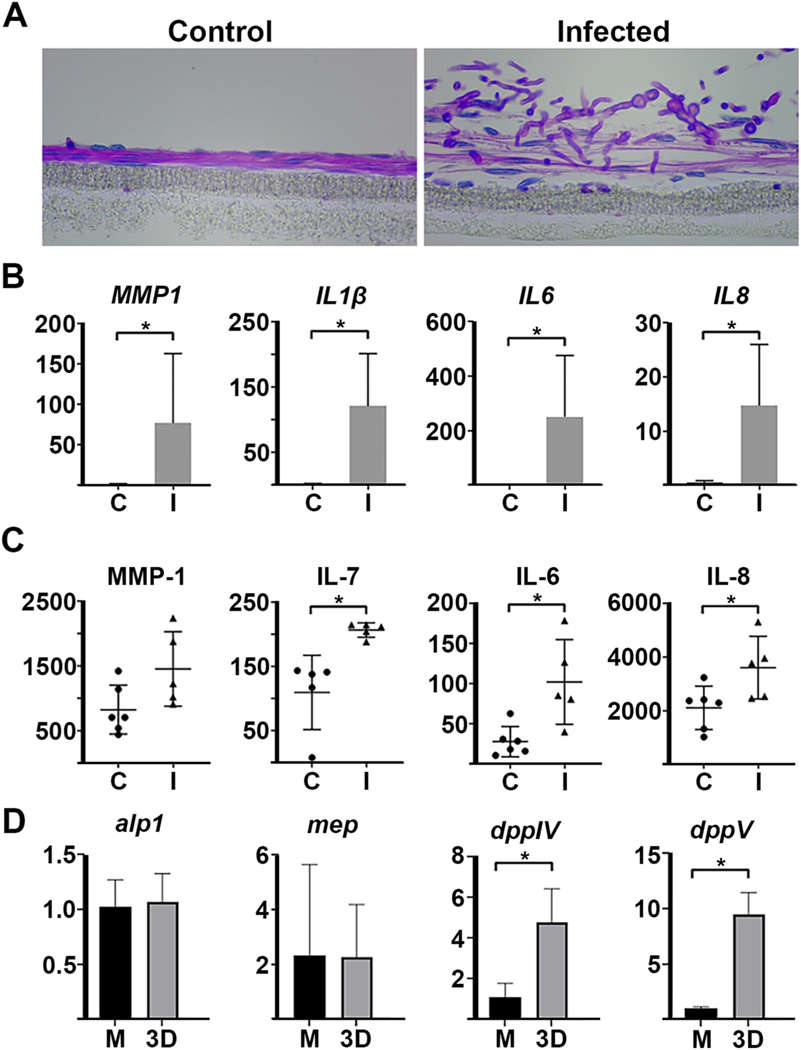
To determine if the model is suitable for other fungal pathogens, we performed a similar experimental workflow for another common FK agent, Aspergillus fumigatus. Using the same inoculation parameters as above, we found that A. fumigatus infected constructs faster than F. petroliphilum, with hyphae present by 12 h and matrix damage appreciable by 24 h post-inoculation (Figure S1). We performed qRT- PCR and Milliplex assays at 48 h, however, to keep the time points consistent between the two organisms. As with Fusarium-infected constructs, Aspergillus similarly induced the upregulation of selected pro- inflammatory cytokines and chemokines, including IL1B, IL6, IL7 and IL8. MMP1 was statistically induced at the level of mRNA and showed the same trend, although not significant, for the protein (Figure 5B and 5C). Taken together, these data suggest that the fibroblast response may be similar across fungi, and that the 3D model can be used to compare growth/infection kinetics of different pathogens.
Fig. 5.
Altered fibroblast and Aspergillus gene expression within the 3D model. A) Periodic Acid Schiff-hematoxylin (PASH) staining of control and A. fumigatus-infected constructs. Fibroblast nuclei stain blue and fungal cell walls stain pink. B) qRT-PCR data demonstrating the upregulation of pro- inflammatory cytokines, chemokines, and matrix metalloprotease genes in Aspergillus- infected (I) constructs relative to uninfected controls (C). C) Milliplex data demonstrating the upregulation of the corresponding proteins. D) qRT-PCR demonstrating the expression of several A. fumigatus collagenase genes grown in the 3D construct model relative to tissue culture media, EMEM media (M). Data for all panels and conditions taken at 48 h post-inoculation.
3.7. A. fumigatus upregulates the expression of secreted collagenase genes during infection of stromal constructs
Standard host-pathogen co-culture models take place in tissue culture medium, which contains free glucose and amino acids for nutrition. Compared to such a system, we hypothesized that the ECM of the 3D constructs would more closely mimic the nutritional environment of an in vivo cornea, as we have previously shown with other corneal conditions including keratoconus and diabetic keratopathy (Priyadarsini et al., 2016). To provide some insight into this, we tested the expression of four A. fumigatus protease/collagenases (mep, alp1, dppIV, dppV) from fungus isolated from three conditions: (1) serum-free EMEM, which reflects the media used in standard culture models, (2) infected mouse corneas, and (3) the 3D stromal constructs. For the corneal infections, 6 week-old C57BL/6 mice were immunosuppressed with steroids on the day preceding infection. To initiate disease, the corneas were abraded and germinated (swollen) conidia were topically applied. Corneas of the infected animals were then isolated at 48 h post-inoculation, which corresponded to the same time point as the two in vitro conditions. Three separate corneal pools (4–9 corneas each) were assessed in the experiment. Relative to the EMEM condition, we found an upregulation infected corneas, indicating the cornea does represent a protein-rich environment to which the fungus must adapt (of alp1 (avg. 233-fold), mep (avg. 539 fold), and Figure S2dppIV (avg. 9-fold) in ). In the 3D constructs, we found a significant induction of dppIV (avg. 5-fold) and dppV (avg 10-fold), but not of alp1 and mep (Fig. 5D). This suggests that the 3D constructs do promote a similar, although not an identical, induction of proteases to that observed in the infected in vivo cornea.
4. Discussion
In vivo models of FK generally capture all of the salient features of the disease, including the pathogen and a myriad of host cell populations interacting within and damaging the corneal tissue. In vitro (co-culture) models, by contrast, allow for the study of specific host cell responses to the fungus, but lack the appropriate physiological environment to explore much else. A 3D fibroblast model developed by Karamichos et al. contains a fibroblast-secreted ECM and has been used to study a variety of physiologic and disease states of the stroma, including corneal fibrosis, corneal diabetes, keratoconus, UV-crosslinking, and cell-cell communication (Priyadarsini et al., 2016; Sharif et al., 2019; McKay et al., 2019; Karamichos et al., 2010). We reasoned that the system could be co-opted as a model for FK that would allow both fibroblast biology to be explored in parallel with fungal growth and penetration kinetics, which would make it the first of its kind. We tested these features of the model with two common mold agents of FK, F. petroliphilum and A. fumigatus.
Considering the host cells first, our data agree with previous studies that demonstrate cultured corneal fibroblasts become pro-inflammatory upon challenge with fungal antigen. Li et al. for example, challenged immortalized HCFs (2D culture) with heat-killed A. fumigatus hyphae and found induction of IL6 and IL8 mRNA (Li et al., 2013). We observed the same upregulation of these markers at both the gene and protein level when our constructs were challenged with live A. fumigatus or F. petroliphilum. Indeed, pro-inflammatory cytokines and chemokines were among the most strongly upregulated genes in Fusarium-infected constructs based on RNA-seq, and pro-inflammatory pathways (IL-17, TNF, NFkB, MAPK, TLR) were all statistically enriched within the induced gene category. In line with this analysis, Nomi and colleagues directly demonstrated NFkB and MAPK pathways are activated in HCFs by zymosan. Zymosan is purified fungal β,1–3 glucan and is a known agonist for both Dectin-1 (c-type lectin) TLR2. These two receptors co-localize and synergize within macrophage phagosomes to drive the NF-kB mediated pro-inflammatory response and several lines of evidence have implicated their importance in FK (Choi et al., 2011; Li et al., 2019; Zhong et al., 2016). First, both the mRNA and protein for Dectin-1 and TLR2 are upregulated in fungal-infected corneal epithelial cells in vitro as well as whole mouse corneas in vivo (Jin et al., 2007; Kolar et al., 2017; Guo et al., 2012; Wu et al., 2015; Zhao et al., 2009; Karthikeyan et al., 2011). Second, inhibition of either gene results in attenuated corneal opacity and inflammatory signaling in one or more models of FK (Jin et al., 2007; Leal et al., 2010). Consistent with these observations, IPA analysis of our RNA-seq data indicated that both TLR2 and Dectin-1 (CLEC7A) were activated in Fusarium-infected HCFs; this was based on the upregulation of known downstream genes within our data set, including the NFkB signaling complex. The TLR2 gene itself did indeed demonstrate an induction in Fusarium-infected constructs. By contrast, however, CLEC7A was not detected in either infected or control samples, nor were genes involved in the proximal Dectin-1 signaling module, including Src kinase, Syk kinase, or Card9. Taken together, these data suggest that fungal β-glucan may activate corneal fibroblasts exclusively through the TLR2-NFkB signaling module.
A previously unreported activity of HCFs in our study is the direct internalization of fungal conidia. Although fibroblasts are not “professional” phagocytes, the uptake and turnover of damaged ECM particulate is an important function of these cells at the site of corneal injury. Indeed, the ability of cultured HCFs to take up beads has been described for decades and, most recently, Sato et al. demonstrated this rate of phagocytosis increases following stimulation with plasminogen (Lande et al., 1981; Sato et al., 2019). Whether the uptake of Fusarium conidia we captured reflects stochastic phagocytic activity of the cells or a stimulated response (e.g. through direct interaction with the fungus or damaged ECM) is currently unclear. It is also unclear if endocytosed spores are killed by the HCF, or vice versa. Whereas the endosomes of macrophages become acidified and accumulate microbicidal compounds (e.g. ROS and hydrolases), to our knowledge, such killing capacity has not been demonstrated in corneal fibroblasts. Histological examination of resected corneas from FK patients suggests the fibroblasts are apoptotic at late stages of infection (Vemuganti et al., 2004); but their fate at early stages, i.e. prior to neutrophil infiltration, is unknown. The outcome of the fungal-fibroblast interaction is the topic of ongoing investigation in our group. In any case, our current data cumulatively support a model in which corneal fibroblasts serve as immune surveillance cells within the corneal stroma, similar to macrophages or dendritic cells, and can promote the recruitment of peripheral neutrophils through the expression of pro-inflammatory cytokines and chemokines. Pro-inflammatory signaling is likely exacerbated during early infection by fibroblast-derived MMPs, that can cleave IL-1β into the active form. Moreover, the MMPs may directly breakdown the ECM and promote both fungal invasion and leukocyte migration through the stroma.
Our results concerning fibroblast activity in the presence of microbial antigen is in agreement with studies performed in standard (2D) culture models. While this serves as an important validation of the 3D model, it raises an important question: what new information can the 3D model provide? We propose the answer is manifold, but ultimately relates to the presence of the fibroblast-derived collagen matrix. First, we were able to visualize hyphal penetration through, and degradation of, the ECM, which mimics the pathogenesis within FK corneas. Moreover, we observed that A. fumigatus collagenase genes were differentially upregulated in both the 3D construct and infected murine corneas, relative to tissue culture media (EMEM). Each of the tested protease genes were previously shown to be induced upon culture in proteinaceous media (Bergmann et al., 2009). We hypothesize that collagen and proteoglycans present in the 3D model and in vivo stroma represents the primary nitrogen source for the fungus and thereby drives a metabolic shift towards protein catabolism. EMEM, by contrast, contains free amino acids and glucose that likely suppress the expression of collagenase genes. Indeed, we noticed that F. petroliphilum invaded the stromal construct when the inoculum was prepared in non-nutritive PBS, but not EMEM. Since standard culture models take place in tissue culture media, we suggest that the 3D constructs represent a more physiologically relevant environment to probe fungal metabolic activities. Taken all the above observations together, the 3D construct model may represent an ideal platform to compare the stromal invasiveness (virulence) of various fungal species, or mutants of a particular species, e.g. with metabolic or protease defects. The antigenicity of a particular fungal strain could be assessed in parallel using fibroblast cytokine expression as a readout.
In summary, 3D stromal constructs provide the benefits of a typical in vitro system, e.g. more experimental control, but within a more physiologically relevant context. Beyond the experiments we have described, the model could easily be adapted to screen antifungal and anti- inflammatory characteristics of novel drugs or treatment modalities such as corneal collagen cross-linking (Sharif et al., 2019). The major disadvantages to consider with the model is the initial time needed (1 month) to cultivate the stromal constructs and the lack of additional corneal cells that are important in FK, such as epithelial cells (Jin et al., 2007; Kolar et al., 2017). Karamichos et al. have demonstrated that corneal epithelial and/or nerves can be added to this culture system, suggesting that the model can be fine-tuned based on the research question (McKay et al., 2019; Sharif et al., 2018).
Supplementary Material
Acknowledgements
We would like to thank Linda Boone (DMEI Imaging Core), Mark Dittmar and staff (DMEI Animal Research Facility), and the staff at the Oklahoma Medical Research Foundation Imaging Core Facility for excellent technical assistance. This work was supported by a Research to Prevent Blindness (RPB) Career Development Award to KKF, a RPB Unrestricted Grant to Dean McGee Eye Institute, and a 5P30EY021725– 10 from the National Eye Institute to the OUHSC Department of Ophthalmology. None of the funding sources were involved in any aspect of the study design, the collection or interpretation of the data, writing of the manuscript or the decision to publish.
Footnotes
Declaration of competing interest
None.
Appendix A. Supplementary data
Supplementary data related to this article can be found at https://doi.org/10.1016/j.exer.2021.108581.
References
- Bergmann A, Hartmann T, Cairns T, Bignell E, Krappmann S, 2009. A regulator of Aspergillus fumigatus extracellular proteolytic activity is dispensable for virulence. Infect. Immun. 77, 4041–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathi MJ, Ramakrishnan R, Meenakshi R, Padmavathy S, Shivakumar C, Srinivasan M, 2007. Microbial keratitis in South India: influence of risk factors, climate, and geographical variation. Ophthalmic Epidemiol. 14, 61–69. [DOI] [PubMed] [Google Scholar]
- Chakravarti S, Wu F, Vij N, Roberts L, Joyce S, 2004. Microarray studies reveal macrophage-like function of stromal keratocytes in the cornea. Invest. Ophthalmol. Vis. Sci. 45, 3475–3484. 10.1167/iovs.04-0343. [DOI] [PubMed] [Google Scholar]
- Chang DC, Grant GB, O’Donnell K, Wannemuehler KA, Noble-Wang J, Rao CY, Jacobson LM, Crowell CS, Sneed RS, Lewis FM, Schaffzin JK, Kainer MA, Genese CA, Alfonso, Cheung N, Nagra P, Hammersmith K, 2016. Emerging trends in contact lens-related infections. Curr. Opin. Ophthalmol. 27, 327–332. 10.1097/ICU.0000000000000280. [DOI] [PubMed] [Google Scholar]
- Chidambaram JD, Kannambath S, Srikanthi P, Shah M, Lalitha P, Elakkiya S, Bauer J, Prajna NV, Holland MJ, Burton MJ, 2017. Persistence of innate immune pathways in late stage human bacterial and fungal keratitis: results from a comparative transcriptome analysis. Front. Cell Infect. Microbiol. 7, 193. 10.3389/fcimb.2017.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HJ, Kim MK, Ko JH, Lee HJ, Jeong HJ, Wee WR, Seong SY, Akira S, 2011. Effect of Toll-like receptor 2 and 4 of corneal fibroblasts on cytokine expression with co-cultured antigen presenting cells. Cytokine 56, 265–271. 10.1016/j.cyto.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Doughty MJ, Jonuscheit S, 2019. Corneal structure, transparency, thickness and optical density (densitometry), especially as relevant to contact lens wear-a review. Contact Lens Anterior Eye 42, 238–245. 10.1016/j.clae.2018.11.014. [DOI] [PubMed] [Google Scholar]
- Fujita H, Ueda A, Nishida T, Otori T, 1987. Uptake of India ink particles and latex beads by corneal fibroblasts. Cell Tissue Res. 250, 251–255. 10.1007/BF00219069. [DOI] [PubMed] [Google Scholar]
- Fukuda K, Kumagai N, Yamamoto K, Fujitsu Y, Chikamoto N, Nishida T, 2005. Potentiation of lipopolysaccharide-induced chemokine and adhesion molecule expression in corneal fibroblasts by soluble CD14 or LPS-binding protein. Invest. Ophthalmol. Vis. Sci. 46, 3095–3101. 10.1167/iovs.04-1365. [DOI] [PubMed] [Google Scholar]
- Fukuda K, Ishida W, Fukushima A, Nishida T, 2017. Corneal fibroblasts as sentinel cells and local immune modulators in infectious keratitis. Int. J. Mol. Sci. 18, 1831. 10.3390/ijms18091831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao N, Kumar A, Yu FS, 2015. Matrix Metalloproteinase-13 as a target for suppressing corneal ulceration caused by Pseudomonas aeruginosa infection. J. Infect. Dis. 212, 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Hutcheon AE, Melotti SA, Zieske JD, Trinkaus-Randall V, Ruberti JW, 2007. Morphologic characterization of organized extracellular matrix deposition by ascorbic acid-stimulated human corneal fibroblasts. Invest. Ophthalmol. Vis. Sci. 48, 4050–4060. 10.1167/iovs.06-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Gao J, Wu X, 2012. Toll-like receptor 2 siRNA suppresses corneal inflammation and attenuates Aspergillus fumigatus keratitis in rats. Immunol. Cell Biol. 90, 352–357. 10.1038/icb.2011.49. [DOI] [PubMed] [Google Scholar]
- Jin X, Qin Q, Tu L, Zhou X, Lin Y, Qu J, 2007. Toll-like receptors (TLRs) expression and function in response to inactivate hyphae of Fusarium solani in immortalized human corneal epithelial cells. Mol. Vis. 13, 1953–1961. [PMC free article] [PubMed] [Google Scholar]
- Karamichos D, Guo XQ, Hutcheon AE, Zieske JD, 2010. Human corneal fibrosis: an in vitro model. Invest. Ophthalmol. Vis. Sci. 51, 1382–1388. 10.1167/iovs.09-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamichos D, Lakshman N, Petroll WM, 2007. Regulation of corneal fibroblast morphology and collagen reorganization by extracellular matrix mechanical properties. Invest. Ophthalmol. Vis. Sci. 48, 5030–5037. 10.1167/iovs.07-0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan RS, Leal SM Jr., Prajna NV, Dharmalingam K, Geiser DM, Pearlman E, Lalitha P, 2011. Expression of innate and adaptive immune mediators in human corneal tissue infected with Aspergillus or fusarium. J. Infect. Dis. 204, 942–950. 10.1093/infdis/jir426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolar SS, Baidouri H, McDermott AM, 2017. Role of pattern recognition receptors in the modulation of antimicrobial peptide expression in the corneal epithelial innate response to F. solani. Invest. Ophthalmol. Vis. Sci. 58, 2463–2472. 10.1167/iovs.16-20658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande MA, Birk DE, Nagpal ML, Rader RL, 1981. Phagocytic properties of human keratocyte cultures. Invest. Ophthalmol. Vis. Sci. 20, 481–489. [PubMed] [Google Scholar]
- Leal SM Jr., Cowden S, Hsia YC, Ghannoum MA, Momany M, Pearlman E, 2010. Distinct roles for Dectin-1 and TLR4 in the pathogenesis of Aspergillus fumigatus keratitis. PLoS Pathog. 6, e1000976 10.1371/journal.ppat.1000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal SM Jr., Pearlman E, 2012. The role of cytokines and pathogen recognition molecules in fungal keratitis - insights from human disease and animal models. Cytokine 58, 107–111. 10.1016/j.cyto.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Yan J, Yu Y, 2019. Geometrical reorganization of Dectin-1 and TLR2 on single phagosomes alters their synergistic immune signaling. Proc. Natl. Sci. USA 116, 25106–25114. 10.1073/pnas.1909870116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yang H, Wu X, 2013. Pretreatment with TLR2 and TLR4 ligand modulates innate immunity in corneal fibroblasts challenged with Aspergillus fumigatus. Invest. Ophthalmol. Vis. Sci. 54, 4261–4270. 10.1167/iovs.12-11504.PMID:23633661. [DOI] [PubMed] [Google Scholar]
- Lightfoot JD, Fuller KK, 2019. CRISPR/Cas9-Mediated gene replacement in the fungal keratitis pathogen Fusarium solani var. petroliphilum. Microorganisms 7, 457. 10.3390/microorganisms7100457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay TB, Karamichos D, Hutcheon AEK, Guo X, Zieske JD, 2019. Corneal epithelial-stromal fibroblast constructs to study cell-cell communication in vitro. Bioengineering 6, 110. 10.3390/bioengineering6040110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan SA, Huang X, Barrett RP, Lighvani S, Zhang Y, Richiert D, Hazlett LD, 2006. Matrix metalloproteinase-9 amplifies the immune response to Pseudomonas aeruginosa corneal infection. Invest. Ophthalmol. Vis. Sci. 47, 256–264. [DOI] [PubMed] [Google Scholar]
- Mundra J, Dhakal R, Mohamed A, Jha G, Joseph J, Chaurasia S, Murthy S, 2019. Outcomes of therapeutic penetrating keratoplasty in 198 eyes with fungal keratitis. Indian J. Ophthalmol. 67, 1599–1605. 10.4103/ijo.IJO_1952_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery ML, Fuller KK, 2020. Experimental models for fungal keratitis: an overview of principles and protocols. Cells 9, 1713. 10.3390/cells9071713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myrna KE, Pot SA, Murphy CJ, 2009. Meet the corneal myofibroblast: the role of myofibroblast transformation in corneal wound healing and pathology. Vet. Ophthalmol. 12, 25–27. 10.1111/j.1463-5224.2009.00742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomi N, Kimura K, Nishida T, 2010. Release of interleukins 6 and 8 induced by zymosan and mediated by MAP kinase and NF-kappaB signaling pathways in human corneal fibroblasts. Invest. Ophthalmol. Vis. Sci. 51, 2955–2959. 10.1167/iovs.09-4823. [DOI] [PubMed] [Google Scholar]
- Priyadarsini S, Sarker-Nag A, Rowsey TG, Ma JX, Karamichos D, 2016. Establishment of a 3D in vitro model to accelerate the development of human therapies against corneal diabetes. PloS One 11, e0168845. 10.1371/journal.pone.0168845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa RH, Miller D, Alfonso EC, 2009. The changing spectrum of fungal keratitis in south Florida. Ophthalmology 101, 1005–1013. 10.1016/s0161-6420(94)31225-5. [DOI] [PubMed] [Google Scholar]
- Saha S, Banerjee D, Khetan A, Sengupta J, 2009. Epidemiological profile of fungal keratitis in urban population of West Bengal, India. Oman J. Ophthalmol. 2, 114–118. 10.4103/0974-620X.57310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Sugioka K, Kodama-Takahashi A, Murakami J, Saito A, Mishima H, Nishida T, Kusaka S, 2019. Stimulation of phagocytic activity in cultured human corneal fibroblasts by plasminogen. Invest. Ophthalmol. Vis. Sci. 60, 4205–4214. 10.1167/iovs.19-27736. [DOI] [PubMed] [Google Scholar]
- Sivak JM, Fini ME, 2002. MMPs in the eye: emerging roles for matrix metalloproteinases in ocular physiology. Prog. Retin. Eye Res. 21, 1–14. [DOI] [PubMed] [Google Scholar]
- Sharif R, Priyadarsini S, Rowsey TG, Ma JX, Karamichos D, 2018. Corneal tissue engineering: an in vitro model of the stromal-nerve interactions of the human cornea. J. Vis. Exp. 24, 56308. 10.3791/56308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif R, Fowler B, Karamichos D, 2019. Collagen cross-linking impact on keratoconus extracellular matrix. PloS One 13, e0200704. 10.1371/journal.pone.0200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Short DP, O’Donnell K, Thrane U, Nielsen KF, Zhang N, Juba JH, Geiser DM, 2013. Phylogenetic relationships among members of the Fusarium solani species complex in human infections and the descriptions of F. keratoplasticum sp. nov. and F. petroliphilum stat. nov. Fungal Genet. Biol. 53, 59–70. 10.1016/j.fgb.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Spadea L, Maraone G, Verboschi F, Vingolo EM, Tognetto D, 2016. Effect of corneal light scatter on vision: a review of the literature. Int. J. Ophthalmol. 9, 459–464. 10.18240/ijo.2016.03.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugioka K, Kodama-Takahshi A, Sato T, Okada K, Murakami J, Park AM, Mishima H, Shimomura Y, Kusaka S, Nishida T, 2018. Plasminogen-dependent collagenolytic properties of staphylococcus aureus in collagen gel cultures of human corneal fibroblasts. Invest. Ophthalmol. Vis. Sci. 59, 5098–5107. 10.1167/iovs.18-24925. [DOI] [PubMed] [Google Scholar]
- Taylor PR, Roy S, Meszaros EC, Sun Y, Howell SJ, Malemud CJ, Pearlman E, 2016. JAK/STAT regulation of Aspergillus fumigatus corneal infections and IL-6/23- stimulated neutrophil, IL-17, elastase, and MMP9 activity. J. Leukoc. Biol. 100, 213–222. 10.1189/jlb.4A1015-483R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PA, Kaliamurthy J, 2013. Mycotic keratitis: epidemiology, diagnosis and management. Clin. Microbiol. Infect. 19, 210–220. 10.1111/1469-0691.12126. [DOI] [PubMed] [Google Scholar]
- Vemuganti GK, Reddy K, Iftekhar G, Garg P, Sharma S, 2004. Keratocyte loss in corneal infection through apoptosis: a histologic study of 59 cases. BMC Ophthalmol. 4, 16. 10.1186/1471-2415-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Zhang Y, Xin Z, Wu X, 2015. The crosstalk between TLR2 and NOD2 in Aspergillus fumigatus keratitis. Mol. Immunol. 64, 235–243. 10.1016/j.molimm.2014.11.021. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Zhang J, Gong M, Pan R, Liu Y, Tao L, He K, 2020. Transcriptome analysis of the gene expression profiles associated with fungal keratitis in mice based on RNA-Seq. Invest. Ophthalmol. Vis. Sci. 61, 32. 10.1167/iovs.61.6.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Wu X, Yu FS, 2009. Activation of Toll-like receptors 2 and 4 in Aspergillus fumigatus keratitis. Innate Immun. 15, 155–168. 10.1177/1753425908101521. [DOI] [PubMed] [Google Scholar]
- Zhong J, Huang W, Deng Q, Wu M, Jiang H, Lin X, Sun Y, Huang X, Yuan J, 2016. Inhibition of TREM-1 and Dectin-1 alleviates the severity of fungal keratitis by modulating innate immune responses. PloS One 11, eo050114. 10.1371/journal.pone.0150114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W-S, Wojdyla K, Donlon K, Thomas PA, Eberle HI, 1990. Extracellular proteases of Aspergillus flavus: fungal keratitis, proteases, and pathogenesis. Diagn. Microbiol. Infect. Dis. 13, 491–497. 10.1016/0732-8893(90)90081-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.