Figure 4.
Cellular neighborhood analysis enables functional stratification of tissue microenvironments during viral infection
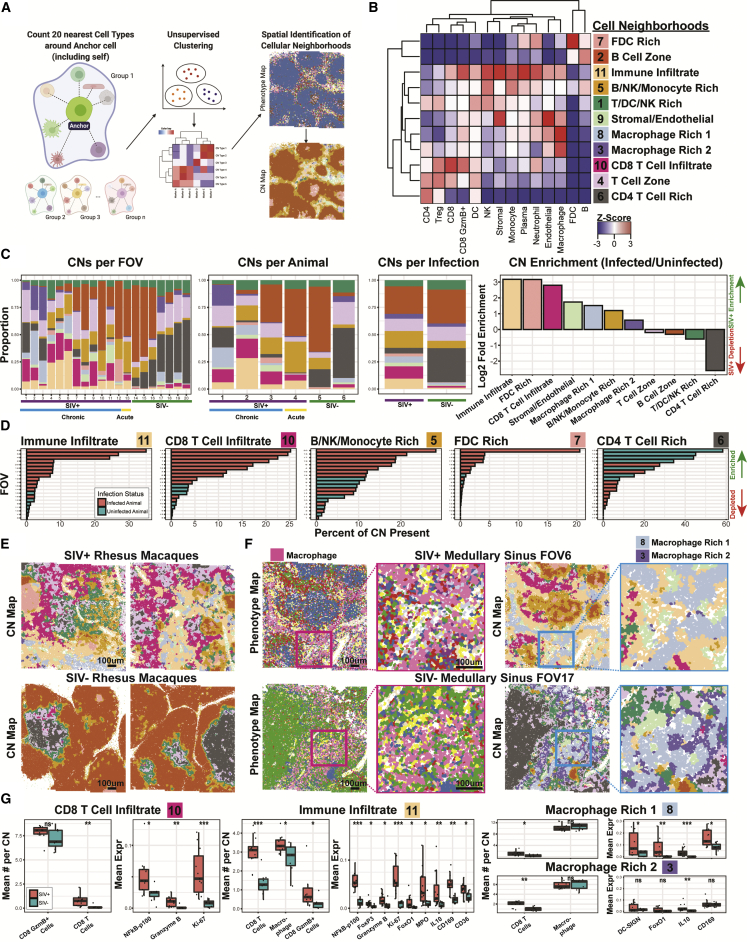
(A) Overview of the method used to define CNs. Twenty nearest neighboring cells (including itself) around each cell were defined, and the cell types were quantified and subjected to unsupervised clustering to define CNs.
(B) A Z score heatmap depicting the 11 CNs identified and cell type enrichment.
(C) From left to right, proportions of each CN aggregated by FOV, animal, and infection status and plot of ranked log2-fold enrichment (infected over uninfected controls) for each CN.
(D) Ranked bar plots showing the percent composition of each CN across the 20 FOVs with bars colored by infection status.
(E) Representative FOVs of infected and control animals (also shown in Figures 2C and 2D) with each individual cell colored by CNs.
(F) Representative FOVs from SIV-infected (top) and SIV-uninfected (bottom) animals containing medullary sinus regions depicted as phenotype maps (left) and CN maps (right). Red and blue boxes indicate regions magnified for zoomed-in views of macrophage-enriched regions. Pink cells in the phenotype map are macrophages. Light blue and purple CNs 8 and 3, respectively, are macrophage rich.
(G) Box plots of mean numbers for cell types within CNs (left) and the mean expression of functional markers within the CN (right). Each dot in the box plot represents data from a single FOV, and the data are divided between infected (orange) and healthy controls (teal). Nonpaired Wilcoxon test: ns, not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
See also Figure S4.