Abstract
In order to discover new dual-active agents, a series of novel Biginelli hybrids (tetrahydropyrimidines) and their ruthenium(II) complexes were synthesized. Newly synthesized compounds were characterized by IR, NMR, and X-ray techniques and investigated for their cytotoxic effect on human cancer cell lines HeLa, LS174, A549, A375, K562 and normal fibroblasts (MRC-5). For further examination of the cytotoxic mechanisms of novel complexes, two of them were chosen for analyzing their effects on the distribution of HeLa cells in the cell cycle phases. The results of the flow cytometry analysis suggest that the proportion of cells in G2/M phase was decreased following the increase of subG1 phase in all treatments. These results confirmed that cells treated with 5b and 5c were induced to undergo apoptotic death. The ruthenium complexes 5a-5d show significant inhibitory potency against SARS-CoV-2 Mpro. Therefore, molecule 5b has significance, while 5e possesses the lowest values of ΔGbind and Ki, which are comparable to cinanserin, and hydroxychloroquine. In addition, achieved results will open a new avenue in drug design for more research on the possible therapeutic applications of dual-active Biginelli-based drugs (anticancer-antiviral). Dual-active drugs based on the hybridization concept “one drug curing two diseases” could be a successful tactic in healing patients who have cancer and the virus SARS-CoV-2 at the same time.
Keywords: COVID-19, SARS-CoV-2 Mpro, Tetrahydropyrimidines, Ruthenium, Anticancer
Graphical abstract

1. Introduction
Pietro Biginelli was an Italian chemist who realized the first one-pot multicomponent synthesis. He developed a method that later would be named in his honor as Biginelli synthesis [1,2]. Even though was the first multicomponent reaction, the scientific community does not pay attention to the Biginelli reaction, calling it old-fashioned [3]. However, with the discovery of monastrol [ethyl 1,2,3,4-tetrahydro-4-(3'-hydroxyphenyl)-6-methyl-2-thioxopyrimidine-5-carboxylate] as the kinesin-5 inhibitor, the Biginelli chemistry has become a hot topic [4,5]. To date, thousands of tetrahydropyrimidines-THPMs [former name 3,4-dihydropyrimidine-2-(thi)ones] were produced via the Biginelli reaction. THPMs derivatives possess broad spectra of biological activities such as anti-inflammatory [6], anti-HIV [7], anticancer [8,9], antioxidant [10], and many others [11].
The species severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) [12], the virus that caused COVID-19 the respiratory illness responsible for the COVID-19 pandemic [13]. Coronavirus is 100-nm-wide bubble of genes that caused over 4.5 million deaths, reshaped the world, and put the planet in an uncertain and unpredictable future [14]. The patients exposed to chemotherapy/radiotherapy are more susceptible to broad spectra of infections, and they are denoted as a highly sensitive group during the COVID-19 pandemic [15]. Hence, cancer and COVID-19 are marked as a dangerous mix of diseases whose weaken the immune system. Following mentioned facts, there is reasonable to think that cancer patients undergoing chemotherapy, targeted therapy or immunotherapy could have a more aggressive course if positive for Coronavirus disease [[15], [16], [17]]. At the same time, one of the pharmaceutical industry's mainstream trends is developing dual-active (multitarget) drugs to treat complex associated diseases [18,19]. Besides anticancer activities [[20], [21], [22]], these days the most crucial target of THPMs could be a SARS-CoV-2 Mpro, the protein responsible for COVID-19 disease. A decade ago, Ramajayam et al. synthesized a series of the thiourea-based Biginelli hybrids that are showed significant inhibitory activity (2–10 μM) against SARS-CoV 3CLpro [23].
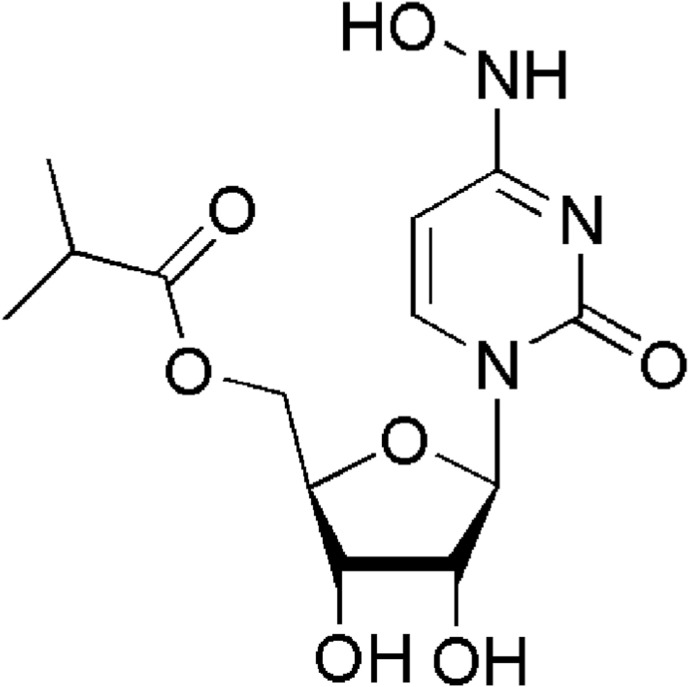
A huge number of already known drugs are employed in the global fight with COVID-19 [24,25]. An excellent example is molnupiravir (Fig. 1 , 2-oxo-pyrimidine) that is an active ingredient in the first approved anti-COVID pill [26,27]. Nevertheless, one ruthenium complex named BOLD-100 [28] has the potential to reduce viral loads in different COVID-19 variants, involving the more virulent B.1.1.7, initially identified in the United Kingdom [29]. Unlike vaccines, which are more effective against certain viral variants, BOLD-100 (Fig. 2 ) with a broad antiviral mechanism of action, appears to remain active on all mutant variants [30]. Very important and promising anticancers are thiolato-bridged dinuclear arene ruthenium complexes family [31].
Fig. 1.
Structure of molnupiravir.
Fig. 2.

Structure of BOLD-100.
There is an burning need to design and synthesis of novel prospective dual-active compounds, especially anticancer-antiviral [[32], [33], [34]]. Our interest is the design, synthesis, and pharmacological evaluation of the heterocyclic compounds, especially those derived from the Biginelli reaction [[35], [36], [37], [38], [39], [40], [41], [42], [43]]. Selected molecules have proven to be excellent ligands for the synthesis of copper and palladium complexes with significant anticancer and antimicrobial activity [[44], [45], [46], [47]]. Considering, primary goal in this study is to synthesise novel THPMs and their iminothiolate-bridged ruthenium complexes, investigation of biological activity, and inhibitory potency towards SARS-CoV-2 main protease Mpro.
2. Material and methods
All solvents and substrates were purchased from Sigma. The melting points (mp) were determined on a Mel-Temp apparatus and are uncorrected. The IR spectra were recorded by a Perkin–Elmer Spectrum One FT-IR spectrometer on a KBr pellet. NMR spectra of ligands and complexes were performed in DMSO‑d 6, with TMS as internal standard on a Varian Gemini 200 MHz NMR spectrometer (1H at 200 and 13C at 50 MHz). NMR spectra of ligands 4a-e and their ruthenium complexes 5a-e are presented in Supplementary Material.
2.1. Synthesis of ligands 4a-e
To 45 mmol of N-methylthiourea, methyl acetoacetate (33 mmol) and aldehyde (30 mmol) was dissolved in boiled ethanol (25 ml), and after that amidosulfonic acid (23 mmol) was added. Twenty-four hours later, solution is diluted with water (10–30 ml) and cooled down. The formed yellow product was filtered, washed with chloroform, cold ethanol and water to achieve the desired product with a high purity grade. Synthesis of 4e was done under different reaction conditions as follow as: 7.5 mmol of N-methylthiourea, 5.5 mmol of methyl acetoacetate, 5 mmol of aldehyde 1e, and 25 mol% MgCl2 (anhydrous) in 15 ml of glacial acetic acid was added at 80 °C. Workup is the same as described above.
2.1.1. Methyl 1,2,3,4-tetrahydro-1,6-dimethyl-2-thioxo-4-phenylpyrimidine-5-carboxylate (4a)
Yield 45%, light yellow powder; 1H NMR (200 MHz, DMSO‑d 6) δ 2.53 (s, 3H, CH3), 3.47 (s, 3H, NCH3), 3.63 (s, 3H, OCH3), 5.19 (d, J = 4.6 Hz, 1H, Bn), 7.22 (dd, J = 8.8, 2.3 Hz, 2H, CHAr), 7.53–7.34 (m, 2H, CHAr), 9.90 (d, J = 4.7 Hz, 1H, NH); 13C NMR (50 MHz, DMSO‑d 6) δ 178.2, 165.8, 148.2, 142.1, 128.7, 127.8, 126.1, 105.3, 52.3, 51.7, 36.4, 16.5 ppm; Calcd. for C14H16N2O2S: C 60.85, H 5.84, N 10.14; Found: C 60.69, H 5.75, N 10.02 (%).
2.1.2. Methyl 4-(4′-chlorophenyl)-1,2,3,4-tetrahydro-1,6-dimethyl-2-thioxopyrimidine-5-carboxylate (4b)
Yield 33%, light yellow powder; 1H NMR (200 MHz, DMSO‑d 6) δ 2.53 (s, 3H, CH3), 3.47 (s, 3H, NCH3), 3.63 (s, 3H, OCH3), 5.19 (d, J = 4.6 Hz, 1H, Bn), 7.22 (dd, J = 8.8, 2.3 Hz, 2H, CHAr), 7.38–7.44 (m, 2H, CHAr), 9.90 (d, J = 4.7 Hz, 1H, NH) ppm; 13C NMR (50 MHz, DMSO‑d 6) δ 178.1, 165.6, 148.7, 140.9, 132.4, 128.7, 128.0, 104.8, 51.7, 36.4, 16.5 ppm; Calcd. for C14H15ClN2O2S: C 54.10; H 4.86; N 9.01; Found: C 53.89, H 4.80, N 8.92 (%).
2.1.3. Methyl 4-(4′-nitrophenyl)-1,2,3,4-tetrahydro-1,6-dimethyl-2-thioxopyrimidine-5-carboxylate (4c)
Yield 30%, yellow powder; 1H NMR (200 MHz, DMSO‑d 6) δ 2.55 (s, 3H, CH3), 3.48 (s, 3H, NCH3), 3.64 (s, 3H, OCH3), 5.33 (s, 1H, Bn), 7.45–7.50 (m, 2H, CHAr), 8.17–8.23 (m, 2H, CHAr), 9.99 (s, 1H) ppm; 13C NMR (50 MHz, DMSO‑d 6) δ 178.4, 165.6, 149.4, 149.1, 147.2, 127.6, 124.1, 104.3, 51.9, 51.9, 36.5, 16.6 ppm; Calcd. for C14H15N3O4S: C 52.33, H 4.70, N 13.08; Found: C 52.25, H 4.57, N 12.94 (%).
2.1.4. 4-(5-(methoxycarbonyl)-1,2,3,4-tetrahydro-1,6-dimethyl-2-thioxopyrimidin-4-yl)benzoic acid (4d)
Yield 59%, light yellow powder; 1H NMR (200 MHz, DMSO‑d 6) δ 2.54 (s, 3H, CH3), 3.48 (s, 3H, NCH3), 3.64 (s, 3H, OCH3), 5.27 (d, J = 4.7 Hz, 1H, Bn), 7.32 (d, J = 8.3 Hz, 2H, CHAr), 7.92 (d, J = 8.3 Hz, 2H, CHAr), 9.95 (d, J = 4.8 Hz, 1H, NH), 12.96 (br.s, 1H, COOH) ppm; 13C NMR (50 MHz, DMSO‑d 6) δ 178.3, 167.0, 165.7, 148.8, 146.7, 130.3, 129.9, 126.3, 104.8, 52.2, 51.8, 36.4, 16.5 ppm; Calcd. for C15H16N2O4S: C 56.24, H 5.03, N 8.74; Found: C 55.92, H 5.07, N 8.70 (%).
2.1.5. Methyl 1,2,3,4-tetrahydro-4-(4′-hydroxy-3′-methoxyphenyl)-1,6-dimethyl-2-thioxopyrimidine-5-carboxylate (4e)
Yield 25%, light yellow powder; 1H NMR (200 MHz, DMSO) δ 2.51 (s, 3H, CH3), 3.48 (s, 3H, NCH3), 3.62 (s, 3H, OCH3), 3.71 (s, 3H, OCH3), 5.10 (d, J = 4.6 Hz, 1H, Bn), 6.56 (dd, J = 8.2, 2.1 Hz, 1H, CHAr), 6.68–6.76 (m, 2H, CHAr), 9.01 (s, 1H, OH), 9.77 (d, J = 4.6 Hz, 1H, NH) ppm; 13C NMR (50 MHz, DMSO) δ 178.0, 165.9, 147.8, 147.6, 146.3, 133.1, 118.3, 115.5, 110.6, 105.7, 55.7, 52.0, 51.7, 36.3, 16.5 ppm; Calcd. for C15H18N2O4S: C 55.88, H 5.63, N, 8.69; Found: C 55.74, H 5.51, N 8.64 (%).
2.2. Synthesis of Ru(II) complexes 5a-e
Appropriate ligands 4a-e (0.25 mmol) were dissolved at 50 °C in solvent mixture EtOH: acetone: H2O (15 : 5 : 2 ml) that contains sodium acetate (0.4 mmol), afterwards complexing salt [Ru(p-cymene)Cl2]2 (0.1 mmol) was added. Twelve hours ago reaction mixture was cooled at room temperature. The precipitation of complex salt was induced using an excess of NH4PF6. Immediately after adding of ammonium salt, the orange crystalline product falls out of the solution. The resulting complexes were filtrated and washed with cold water, ethanol, and ether.
2.2.1. Complex 5a [(p-cymene)Ru(4a)]2 (PF6)2
Yield 64%, orange powder; Mp = 264 °C; IR ν 3424, 1715, 1647, 1572, 1362, 1170 cm−1; 1H NMR (200 MHz, DMSO‑d 6) δ 0.82 (d, J = 6.8 Hz, 3H, cym–CH–(CH 3)2), 1.09 (d, J = 6.8 Hz, 3H, cym–CH–(CH 3)2), 2.16 (s, 3H, C CH3), 2.32 (s, 3H, cym-CH3), 2.55–2.62 (m, 1H, cym-CH-(CH3)2), 3.28 (s, 3H, NCH3), 3.54 (s, 3H, OCH3), 3.86 (d, J = 5.6 Hz, 1H, cym-CHAr), 5.22 (s, 1H, Bn), 5.45 (d, J = 6.3 Hz, 1H, cym-CHAr), 5.80 (d, J = 6.3 Hz, 1H, cym-CHAr), 6.22 (d, J = 6.1 Hz, 1H, cym-CHAr), 7.49–7.68 (m, 5H, CHAr) ppm; 13C NMR (50 MHz, DMSO‑d 6) δ 165.0, 163.0, 144.0, 142.8, 129.1, 128.2, 108.8, 103.1, 84.4, 82.7, 82.0, 80.7, 63.9, 51.4, 31.9, 30.7, 23.0, 21.3, 17.6, 15.7 ppm; Calcd. for C48H58F12N4O4P2Ru2S2: C 43.97, H 4.46, N 4.27; Found: C 44.15, H 4.35, N 4.20 (%).
2.2.2. Complex 5b [(p-cymene)Ru(4b)]2 (PF6)2
Yield 56%, orange crystals; Mp = 275 °C; IR ν 3424, 1716, 1647, 1568, 1362, 1169 cm−1; 1H NMR (200 MHz, DMSO‑d 6) δ 0.85 (d, J = 6.9 Hz, 3H, cym–CH–(CH 3)2), 1.11 (t, J = 7.4 Hz, 3H, cym–CH–(CH 3)2), 2.17 (s, 3H, C CH3), 2.30 (s, 3H, cym-CH3), 2.70–2.54 (m, 1H, cym- CH-(CH3)2), 3.25 (s, 3H, NCH3), 3.54 (s, 3H, OCH3), 3.99 (d, J = 5.7 Hz, 1H, cym-CHAr), 5.24 (s, 1H, Bn), 5.52 (d, J = 6.2 Hz, 1H, cym-CHAr), 5.81 (d, J = 6.0 Hz, 1H, cym-CHAr), 6.25 (d, J = 6.4 Hz, 1H, cym-CHAr), 7.54 (d, J = 8.4 Hz, 2H. CHAr), 7.72 (d, J = 8.5 Hz, 2H, CHAr) ppm; 13C NMR (50 MHz, DMSO‑d 6) δ 165.0, 163.5, 144.5, 141.9, 134.1, 130.3, 129.2, 108.9, 103.3, 84.7, 82.9, 82.2, 80.9, 63.1, 51.7, 32.2, 30.8, 23.3, 21.2, 17.8, 15.6 ppm; Calcd. for C48H56Cl2F12N4O4P2Ru2S2: C 41.77, H 4.09, N 4.06; Found: C 41.95, H 4.19, N 4.11 (%).
2.2.3. Complex 5c [(p-cymene)Ru(4c)]2 (PF6)2
Yield 50%, orange crystals; Mp = 278 °C; IR ν 3443, 1639, 1433, 1352, 1169 cm−1; 1H NMR (200 MHz, DMSO‑d 6) δ 0.84 (d, J = 6.9 Hz, 3H, cym–CH–(CH 3)2), 1.11 (d, J = 6.9 Hz, 3H, cym–CH–(CH 3)2), 2.19 (s, 3H, C CH3), 2.32 (s, 3H, cym-CH3), 2.63–2.73 (m, 1H, cym- CH-(CH3)2), 3.29 (s, 3H, NCH3), 3.54 (s, 3H, OCH3), 3.99 (d, J = 5.7 Hz, 1H, cym-CHAr), 5.39 (s, 1H, Bn), 5.57 (d, J = 6.2 Hz, 1H, cym-CHAr), 5.78 (d, J = 6.0 Hz, 1H, cym-CHAr), 6.25 (d, J = 6.4 Hz, 1H, cym-CHAr), 8.01 (d, J = 8.7 Hz, 2H. CHAr), 8.30 (d, J = 8.7 Hz, 2H, CHAr) ppm; 13C NMR (50 MHz, DMSO‑d 6) δ 164.9, 164.2, 149.5, 148.1, 145.3, 129.8, 124.3, 109.2, 103.1, 102.5, 85.2, 83.0, 82.3, 80.6, 63.1, 51.8, 32.5, 31.0, 23.5, 20.8, 17.9, 15.8 ppm; Calcd. for C48H56F12N6O8P2Ru2S2: C 41.14, H 4.03, N 6.00; Found: C 39.85, H 4.15, N 6.10 (%).
2.2.4. Complex 5d [(p-cymene)Ru(4d)]2 (PF6)2
Yield 62%, orange powder; Mp = 258 °C; IR ν 3442, 1712, 1645, 1361, 1171 cm−1; 1H NMR (200 MHz, DMSO‑d 6) δ 0.82 (d, J = 6.9 Hz, 3H, cym–CH–(CH 3)2), 1.11 (d, J = 6.9 Hz, 3H, cym–CH–(CH 3)2), 2.16 (s, 3H, C CH3), 2.32 (s, 3H, cym-CH3), 2.69–2.54 (m, 1H, cym- CH-(CH3)2), 3.28 (s, 3H, NCH3), 3.53 (s, 3H, OCH3), 3.88 (overlapped with water, d, 1H, cym-CHAr), 5.29 (s, 1H, Bn), 5.48 (d, J = 6.0 Hz, 1H, cym-CHAr), 5.79 (d, J = 5.9 Hz, 1H, cym-CHAr), 6.23 (d, J = 6.7 Hz, 1H, cym-CHAr), 7.79 (d, J = 8.3 Hz, 2H. CHAr), 8.02 (d, J = 8.2 Hz, 2H, CHAr) ppm; 13C NMR (50 MHz, DMSO‑d 6) δ 167.4, 165.0, 163.6, 146.9, 144.6, 130.0, 128.6, 109.0, 103.2, 102.8, 84.8, 82.9, 82.3, 80.7, 63.6, 51.7, 40.9, 40.5, 40.1, 39.7, 39.3, 38.9, 38.5, 32.3, 30.8, 23.3, 21.2, 17.9, 15.7 ppm; Calcd. for C50H58F12N4O8P2Ru2S2: C 42.92, H 4.18, N, 4.00; Found: C 42.75, H 3.94, N 4.10 (%).
2.2.5. Complex 5e [(p-cymene)Ru(4e)]2 (PF6)2
Yield 78%, orange crystals; Mp = 197 °C; IR ν 3423, 1702, 1644, 1514, 1360 cm−1; 1H NMR (200 MHz, DMSO‑d 6) δ 0.88 (d, J = 6.8 Hz, 3H, cym–CH–(CH 3)2), 1.14 (d, J = 6.8 Hz, 3H, cym–CH–(CH 3)2), 2.17 (s, 3H, C CH3), 2.29 (s, 3H, cym-CH3), 2.68–2.53 (m, 1H, cym- CH-(CH3)2), 3.25 (s, 3H, NCH3), 3.82 (s, 3H, OCH3), 4.05 (d, J = 5.7 Hz, 1H, cym-CHAr), 5.13 (s, 1H, Bn), 5.48 (d, J = 5.9 Hz, 1H, cym-CHAr), 5.83 (d, J = 5.5 Hz, 1H, cym-CHAr), 6.23 (d, J = 6.1 Hz, 1H, cym-CHAr), 6.84 (d, J = 8.3 Hz, 1H, CHAr), 6.99 (d, J = 8.3 Hz, 1H, CHAr), 7.12 (s, 1H, CHAr) ppm; 13C NMR (50 MHz, DMSO‑d 6) δ 165.3, 162.6, 147.6, 147.5, 143.5, 134.2, 121.1, 116.0, 112.2, 108.8, 103.6, 102.9, 84.8, 82.9, 82.1, 80.9, 63.5, 56.1, 51.5, 32.0, 30.9, 23.2, 21.3, 17.8, 15.4 ppm; Calcd. for C50H62F12N4O8P2Ru2S2: C 42.80, H 4.45, N 3.99; Found: C 42.95, H 4.27, N 4.08 (%).
2.3. Single-crystal X-ray crystallography
A suitable single crystal of 5a was mounted on a glass fiber and crystallographic data were collected using the Rigaku (Oxford Diffraction) Gemini S diffractometer with a CCD area detector and wavelength of graphite monochromated incident radiation λ MoKα = 0.71073 Å at 295 K. CrysAlisPro and CrysAlis RED software packages [48] were employed for data collection and data integration. Final cell parameters were determined by a global refinement of 7733 refiections (2.6 < θ < 28.2°). Numerical absorption correction was performed using Gaussian integration over multifaceted crystal model [49]. Crystal structure was solved using intrisic phasing algorithm SHELXT [50] implemented within OLEX2 [51] graphical user interface. The structure was subsequently refined with SHELXL-2018/3 [52]. Non-hydrogen atoms were refined freely with anisotropic displacement parameters. All hydrogen atoms were placed in geometrically idealized positions and refined as riding on their parent atoms, with their Uiso values approximated by the Ueq values of their carrier atoms. Finally, MERCURY [53] software was employed for molecular graphics.
Several crystals of 5a were non-merohedrally twinned, indicated by splitting of the reflections during crystal screening procedure. Owing to the fact that no untwinned crystal could be found, full data collection was performed for twinned crystal. Successful indexing of observed diffraction peaks was attained on two independently oriented identical lattices. Their mutual orientation was characterized by the twin law that was determined by CrysAlisPRO [48] software. Reflections from both domains were simultaneously integrated and deconvolution of the intensities of partially overlapped reflections yielded twin ratios of 51.64% for domain 1 and 48.36% for domain 2. In total, 31488 composite observations were collected (14709 fully isolated and 16779 overlapped). Reduced data were prepared for the structure solution by selecting 24125 reflections belonging to domain 1, 18258 of which represented reflections with an overlapping factor <0.547 (chosen so that the data completeness was 80%). After successful solution using SHELXT [50], a full dataset (17179 unique reflections), containing both isolated and overlapped reflections of both domains (HKLF5), was used for the structure refinement with SHELXL-2018/3 [52] using the BASF command for twin domain ratio refinement.
2.3.1. Accession code
CCDC 2103331 contain the supplementary crystallographic data for this paper. These data can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif or by emailing data_request@ccdc.cam.ac.uk, or by contacting The Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK; fax: +44 1223 336033.
2.4. Determination of the cytotoxic activity of the compounds
Cytotoxic activity of the compounds was examined against five human cancer cell lines: cervical adenocarcinoma HeLa, lung carcinoma A549, colon adenocarcinoma LS174, malignant melanoma A375, chronic myelogenous leukemia K562 cells, as well as against one normal human cell line, lung fibroblasts MRC-5. Human cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 25 mM HEPES, and penicillin-streptomycin (100 U/mL of penicillin and 0.1 mg/mL of streptomycin) at 37 °C in an atmosphere of 5% CO2 and humidified air. In brief, HeLa (2000 cells per well), A549 (5000 cells per well), LS174 (7000 cells per well), A375 (3000 cells per well), MRC-5 (5000 cells per well) were seeded in 96-well cell culture plates and were allowed to adhere for 20 h before treatment, while K562 cells which grow in a suspension were seeded in plates (5000 cells per well) 2 h before addition of compounds. After treatment which lasted 72 h, MTT cell survival test was performed.
2.5. Cell cycle phase analysis
The cell cycle phase distribution of HeLa cells exposed for 24 h and 48 h to IC50 and 2IC50 concentrations of the compounds 5b and 5c was examined by flow cytometry. HeLa cells (200000 cells per well) were seeded into the 6-well cell culture plates. After 20 h incubation, the cells were treated with the compounds. After 24 h or 48 h incubation, the cell samples were collected, washed, and fixed in 70% cold ethanol. Before performing the flow cytometry analysis, the cells were incubated with RNase A for 30 min at 37 °C. Subsequently, the propidium iodide was added to the cells. HeLa cell cycle phase distribution was analyzed on the BD FACSCaliburTM flow cytometer using BD CellQuest ProTM software (BD Biosciences).
2.6. FITC annexin V/propidium iodide double staining assay
HeLa cells (200000 cells per well) were seeded in 6-well plates. The next day, the cells were treated with IC50 and 2IC50 concentrations of the compounds 5b and 5c for 24 h. After incubation the cells were harvested by trypsinization, washed with PBS and stained with FITC Annexin V and propidium iodide, incubated for 15 min at room temperature in the dark and analyzed on BD FACSCalibur™ flow cytometer using BD CellQuest Pro™ software (BD Biosciences Franklin Lakes, NJ, USA). For each examined sample 10000 cells were collected in a gate and analyzed.
2.7. Determination of in vitro cytotoxicity of complexes against EA.hy926 cell line
The human umbilical vein cell line, EA.hy926 was maintained in Dulbecco's Modified Eagles Medium (DMEM) supplemented with 10% fetal calf serum (FCS) at 37 °C in humidified atmosphere with 5% CO2. EA.hy926 cells were seeded in 96-well flat-bottomed microtiter plates at density of 7000 cells/100 μL in each well. After 24 h of incubation cells were treated with 50 μL of investigated complexes. Treatments were applied at final concentrations in the range from 200 μM to 12.5 μM. Clear medium only was added to the control cell samples, while treatments without cells were used as blank. Experiments were performed in triplicates. Microculture tetrazolium test (MTT) was used for determination of cell viability [54]. After 48 h of treatment, 10 μL of MTT solution was added to each well. Samples were incubated for 4 h and then 100 μL of 10% SDS (100 g/L sodium dodecyl sulfate) was added to dissolve formazan, the product of conversion of MTT dye by viable cells. Twenty four hours later, absorbance was measured at 570 nm using Multiskan EX reader (Thermo Labsystems Beverly, MA, USA). The intensity of the absorbance was proportional to the number of viable cells in each well. IC50 is defined as a concentration of the treatment which inhibits cell survival by 50% compared to the control, and is used for representing the cytotoxic effect of the investigated complexes.
2.8. Endothelial cell tube formation assay
EA.hy926 cell line was used for determining the anti-angiogenic effect of complexes. The volume of 200 μL of Corning® Matrigel® basement membrane matrix was added in 24-well plates. After an hour, 40.000 of EA.hy926 cells in volume of 400 μL were added into coated wells, as well as additional 200 μL of treatments at sub-toxic concentrations (IC20). Control sample was treated with clear nutrient medium only. After incubation of 48 h at 37 °C in an atmosphere of 5% CO2 and humidified air photomicrographs of cells were captured under the inverted phase-contrast microscope.
2.9. Antimigratory effect – in vitro “scratch” assay
EA.hy926 cells were seeded (70.000 cells/600 μL per well) in DMEM with 10% FCS in 24-well plates. After the 48 h cells formed a confluent monolayer. Medium was removed and cell monolayer was scraped in a straight line to create a “scratch” using the yellow tip for the pipette (p200) in each sample. Cells were gently washed with the growth medium, and then treated with investigated complexes at subtoxic concentrations (IC20). The images of cell migration were captured by camera (Olympus® Corporation, Tokyo, Japan) at three time points (0h, 24h and 48h from treatment) and used for analysis of antimigratory effect of complexes in comparison to control sample.
2.10. Computational methodology
The compounds investigated in this study are prepared for molecular docking simulations by optimization of their geometries in the gas phase applying density functional theory (DFT). These calculations are performed using Gaussian09 software [55]. Global hybrid Generalized Gradient Approximation (GGA) functional B3LYP with empirical dispersion corrections D3BJ (with Becke and Johnson damping) [56] is employed, in combination with the 6-311G (d,p) basis set for C, N, O, Cl, S and H atoms, and def2-TZVP basis set for Ru atom. The three-dimensional (3D) crystal structure of SARS-CoV-2 main protease Mpro is downloaded from the Protein Data Bank (PDB ID: 6LU7) [57]. The molecular docking simulations are carried out using the AutoDock 4.0 software [58]. The preparation of protein for docking simulations is carried out in Discovery Studio 4.0 [59]. The affinity maps of the target protein are established using AGFR (AutoGridFR) software [59]. These data are further used by AutoDock to predict binding sites by detecting pockets and cavities of the known 3D structure of proteins. Binding site with the lowest expected binding energy, according to AGFR, were used for molecular docking simulations. The grid box center with dimensions −13.642 Å × 13.303 Å × 73.551 Å in -x, -y, and -z directions are used to cover the protein binding site and accommodate ligand to move freely. A grid point spacing of 0.375 Å was used for auto grid runs. The protein structure is released from the co-crystallized ligand, water molecules, and co-factors. The AutoDockTools (ADT) graphical user interface is used for the addition of polar hydrogen atoms and the calculation of Kollman charges. During the preparation for molecular docking simulation, the bonds in the ligands are set to be rotatable and ligand is set to be flexible. The structure of protein remains standing as rigid. The Lamarckian Genetic Algorithm (LGA) is used for protein-ligand flexible molecular docking simulations. All the molecular docking simulations are performed at a temperature of 298.15 K. Analysis of molecular docking simulation results and visualizations are performed using BIOVIA Discovery Studio.
For estimation of the binding affinity, AutoDock uses empirical scoring functions based on the free energy of binding (ΔGbind). The value of ΔGbind depends on the values of Final Intermolecular Energy (FIE), Final Total Internal Energy (FTIE), Torsional Free Energy (TFE) and Unbound System's Energy (USE) (eq. (1)). The value of FIE is a summary of the van der Waals energy, energy of hydrogen bonds, desolvation energy of the system, and electrostatic energy. The value of ΔGbind represents energy that is realized in the formation of interactions between a ligand and protein. This value is calculated by AutoDock program according to the following equation:
| ΔGbind = [(FIE)+(FTIE)+(TFE)-(USE)] | (1) |
There is one more significant parameter in the prediction of binding affinity and that is the constant of inhibition (Ki). AutoDock calculates this value after estimation of free energy of binding, by the following equation:
| Ki = exp(ΔGbind /RT) |
where R is the gas constant (R = 1.99 cal/molK), and T is the room temperature (298.15 K).
3. Result and discussion
3.1. Synthesis and characterization
In the first step, five ligands (THPMs 4a-d) were synthesized via acid-catalyzed one-pot Biginelli reaction using aromatic aldehydes 1a-e, N-methylthiourea 2, and methyl acetoacetate 3 (Scheme 1 ). Amidosulfonic acid in ethanol was used for the synthesis of 4a-d. However, a catalytic system CH3COOH/MgCl2 was employed to synthesise of 4e. Compounds 4c-e are synthesized and characterized for the first time. With the ligands (4a-e) in hands, we have started with optimization the reaction conditions. Dichloro(p-cymene)ruthenium(II) dimer (RD) was used as a complexing agent. THPMs are in thione form in solid-state, and also in solution. Therefore, easy trapping of THPMs by ruthenium ions can be realized by moving thione-thiole equilibrium towards thiol form. Tautomerism in THPM core can be reached exclusively in solution in the presence of a base. Sodium acetate was chosen as the best solution for this synthetic manipulation. The newly-synthesized ruthenium complexes 5a-e were produced heating excess (2.5 eq.) of ligands 4a-e with RD in solvent mixture EtOH: acetone: H2O (15 : 5 : 2 ml). Finally, appropriate complexes fall out from the solution by adding ammonium hexafluorophosphate. The ligands (4a-e) and corresponding ruthenium complexes (5a-e) were assigned using NMR and IR spectroscopy (Figs. S1–S20). One complex (5a) was suitable for crystallographic analysis. The proton NMR spectra of 4a-e showed signal of benzylic proton in the range 5.2–5.4 ppm. Thioamidic proton has appeared around 9.9 ppm as broad singlet or doublet. Aromatic protons are in range from 6.5 to 8.5 ppm. The carbon of thioketo function (C S) was at 178 ppm in all cases.
Scheme 1.
Synthesis of Biginelli hybrids 4a-e and their ruthenium(II) complexes 5a-e. Reaction conditions: A = ethanol, H2NSO3H, reflux (4a-d) or acetic acid, MgCl2, reflux (4e); B = a) [Ru(p-cymene)Cl2]2, CH3COONa, EtOH: acetone: H2O (15 : 5 : 2 ml), reflux, b) NH4PF6.
3.2. Crystal structure analysis
A perspective view of the molecular structure of 5a with adopted atom-numbering scheme is shown in Fig. 3 . Crystal data, selected bond lengths, bond angles, and torsion angles are listed in Tables S1 and S2.
Fig. 3.
MERCURY [53] drawing of the molecular structure of the complex 5a.
The complex has two ruthenium ions, both η6−coordinated to p-cymene molecule and to methyl 1,2,3,4-tetrahydro-1,6-dimethyl-2-thioxo-4-phenylpyrimidine-5-carboxylate (4a) ligand (L) (via N1A,B and S1A,B atoms) leading to the formation of a three-legged piano-stool coordination environment. Conformation around both ruthenium ions are between staggered and eclipsed orientations with regard to the p-cymene ring and the other three coordinated atoms. Distances between ruthenium ions and the p-cymene ring centroids are 1.699 Å for Ru1A and 1.709 Å for Ru1B. The bond angles between ring centroid−metal−ligand atoms (N1A,B and S1A,B) are in the range 125.64° − 136.15°. Complex 5a crystallized as racemic mixture. In Fig. 3 is presented R,R enantiomer.
The p-cymene ring is slightly puckered with Ru–C distances in the range 2.182 (8) − 2.261 (11) Å. The groups Ru1N1S1C1 (A and B) are almost perfectly planar since the largest displacement from the same weighted least-squares plane in both LA and LB ligands is 0.01 Å for C1B atom. Comparing the angles between Ru1N1C1S1 planes and C2C3C4N1C1N2 planes in ligand LA (3.34°) and LB (5.58°), one can observe that the non− phenyl part of ligand B is more disordered from planarity (Table S2).
The crystal packing and intermolecular interactions in the solid 5a complex are presented on Fig. 4 . Additional hexafluorophosphate (PF6 +) anions, inserted between the molecules of Ru(II) complex, creates complicated pattern of C–H⋯F contacts leading to the formation of 1D chain that is aligned approximately along the a−axis. Parameters of intermolecular C–H⋯F as well as intramolecular C–H⋯O contacts are collected in Table S3.
Fig. 4.
MERCURY [53] drawing showing the crystal packing of complex 5a. Hydrogen bonds are shown as black and red dashed lines.
Fingerplot analysis. The Hirshfeld surface surrounds the molecule and defines the volume of space where the promolecule electron density exceeds of all neighboring molecules. The quantitative contribution of different intermolecular contacts to the Hirshfield surface is given by the 2D finger−print plot, thus enabling a clarification of the crystal packing of 5a. Reciprocal contacts are also included while calculating the percentage contribution of interatomic contacts. Fig. 5 shows the finger−print plots of 5a, displaying the normalized distances, de and di to the Hirshfield surface, where de is the distance from the surface to the nearest outside atom and di is the distance from the surface to the nearest atom inside [60,61]. F⋯H and dispersive H⋯H contacts accounted for more than 86% of the Hirshfield surface in this solvate form of the complex.
Fig. 5.
The finger-print plot of 5a decomposed into different interactions.
3.3. Biological activity
3.3.1. Cytotoxicity
The newly synthesized ruthenium compounds based on Biginelli hybrids (4a-e) were investigated for in vitro cytotoxic activity against five human cancer cell lines, cervix adenocarcinoma cell line (HeLa), human colon carcinoma (LS174), chronic myelogenous leukemia (K562), malignant melanoma (A375), non-small cell lung carcinoma (A549) and normal cell line, human fetal lung fibroblast cell line (MRC-5) using a MTT assay (Table 1 ). Based on the obtained IC50 values, it was observed that compounds 5b and 5c showed strong cytotoxic activity against HeLa cell lines. Compounds 5a, 5d, and 5e showed weaker to moderate cytotoxic activity against tested cell lines. IC50 values for 5b and 5c are ranging from 16.39 ± 0.43 to 36.53 ± 0.17 μM indicate that these compounds exhibited significant in vitro cytotoxic activity against HeLa as well as moderate cytotoxicity towards A549 and LS174. The compounds 5a, 5b, and 5c exerted strong cytotoxic effects on melanoma A375 and chronic myelogenous leukemia K562 cell lines with IC50 values ranging from 8.63 μM to 14.14 μM. The K562 cells were more sensitive to the activity of these three compounds than A375 cells. The other seven tested compounds did not exhibit significant cytotoxic activity against K562 cells (IC50 = 95.88–198.09 μM) as well as A375 cells (IC50 = 102.41 μM–199.58 μM). Ruthenium complexes 5a-c displayed higher selectivity index [SI = IC50 (MRC-5)/IC50(K562)] compared to cis-DDP. Most significant selectivity index [SI (K562) = 3] among others has ruthenim complex 5a. Furthermore, complex 5c also demonstrated promising selectivity [SI (K562) = 1.5 that is followed by lowest IC50 value (8.63 ± 0.24 μM). Obtained results suggest that chlorine and nitro group as substituents on the aromatic ring (Scheme 1) increased cytotoxicity.
Table 1.
Cytotoxic activity (IC50 values) of ligands 4a-e and complexes 5a-e against human cancer cell lines. IC50 values were expressed as the mean ± SD determined from the results of MTT assay in three independent experiments. cis-DDP was used as a positive control.
| Compounds | IC50(μM) |
|||||
|---|---|---|---|---|---|---|
| HeLa | A549 | LS174 | A375 | K562 | MRC-5 | |
| 4a | 128.20 ± 15.37 | N/A | 170.46 ± 1.28 | 122.94 ± 7.17 | 162.77 ± 4.62 | 92.97 ± 0.59 |
| 4b | 81.84 ± 9.57 | 122.72 ± 6.05 | 153.13 ± 9.08 | 102.41 ± 6.26 | 95.88 ± 1.85 | 28.17 ± 0.32 |
| 4c | 121.80 ± 12.83 | N/A | 185.11 ± 5.50 | 157.58 ± 7.84 | 193.00 ± 3.75 | 88.92 ± 6.31 |
| 4d | 138.99 ± 7.64 | N/A | >200 | 199.58 ± 0.59 | 194.61 ± 7.62 | 169.13 ± 8.98 |
| 4e | 156.55 ± 6.52 | N/A | 195.55 ± 2.87 | 175.58 ± 2.90 | 197.00 ± 0.54 | 114.33 ± 2.36 |
| 5a | 34.70 ± 1.23 | 61.99 ± 0.36 | 67.43 ± 1.24 | 14.14 ± 1.11 | 11.44 ± 1.19 | 32.46 ± 0.47 |
| 5b | 16.39 ± 0.43 | 24.87 ± 1.14 | 32.78 ± 3.38 | 14.00 ± 0.10 | 11.45 ± 0.15 | 14.81 ± 2.16 |
| 5c | 17.89 ± 0.7 | 33.85 ± 2.74 | 34.00 ± 1.39 | 13.94 ± 0.25 | 8.63 ± 0.24 | 14.28 ± 0.66 |
| 5d | 69.66 ± 4.33 | N/A | 81.79 ± 4.28 | 199.53 ± 0.67 | 198.09 ± 1.58 | 66.49 ± 4.73 |
| 5e | 78.28 ± 3.26 | N/A | 97.77 ± 1.43 | 116.66 ± 5.72 | 130.48 ± 3.13 | 57.17 ± 1.18 |
| cisplatin | 2.36 ± 0.28 | 17.93 ± 0.44 | 20.8 ± 0.44 | 2.56 ± 0.42 | 5.56 ± 0.23 | 4.26 ± 0.46 |
3.3.2. In vitro cytotoxic activity of complexes against EA.hy926 cell lin
The anticancer activity of investigated Ru complexes was examined against human umbilical vein cell line, EA.hy926, by MTT test. The obtained IC50 concentrations are shown in Table 2 . Complexes 5b and 5c showed the highest cytotoxic activities against EA.hy926 cells, with IC50 of 35.24 and 33.85 μM, respectively. Complex 5a showed moderate cytotoxic activity, while 5d and 5e didn't show measurable IC50 values at all. These results were used for further investigation of antimigratory and antiangiogenic potential of investigated complexes.
Table 2.
Concentrations of compounds that induced a 50% decrease in EA.hy926 [expressed as IC50 (μM)].
| Compounds | IC50(μM) EA.hy |
|---|---|
| 5a | 56.96 ± 11.50 |
| 5b | 35.24 ± 1.08 |
| 5c | 33.85 ± 1.68 |
| 5d | N/A |
| 5e | N/A |
The antiangiogenic potential of ruthenium complexes was examined against EA.hy926 cell line using in vitro endothelial cell tube formation assay. Complexes 5a, 5b and 5c at subtoxic IC20 concentrations induced strong morphological changes of EA.hy926 cells. After 48 h of treatment cells became rounded and they lost their connectivity. Anti-angiogenic effects of complexes 5d and 5e were much weaker, but they led to the decreasing the number of cell junctions and inhibiting the tube formation. All treated samples were compared with the control sample of EA.hy926 cells incubated in medium only. As shown in Fig. 6 , cells in the control sample formed three-dimensional capillary-like tubes.
Fig. 6.
Effects of compounds 5a-e on angiogenesis of endothelial EA.hy926 cells. Applied concentrations of compounds are subtoxic (IC20). Representative photomicrographs of one out of two independent experiments are shown.
3.3.3. Antimigratory activity of 5a-e
Antimigratory effect of complexes is determined using in vitro “scratch” assay. All tested complexes showed antimigratory effect after 24 h and 48 h of treatment applied on EA.hy926 cell line when compared to the control sample. The control sample was incubated with a clear medium only, while investigated complexes were applied at their subtoxic concentrations (IC20). Antimigratory effect of the complexes is represented in Fig. 7 . After 48 h of incubation control sample almost formed smooth and confluent monolayer. According to the changes of the wound width, the strongest antimigratory activity had complexes 5a, 5b, and 5c. Cells lost their original morphology, and the wound width retained the same as it was at time point 0h. Even complexes 5d and 5e showed weaker antimigratory activity, as it could be seen in Fig. 7, samples treated with those complexes also retained formed wound but to a lesser extent. A small number of cells grew into the space of the wound.
Fig. 7.
Antimigratory effect of complexes 5a-e after 24 h and 48 h of treatment applied on EA.hy926 cell line.
3.3.4. Cell cycle distribution
The inhibition of cancer cell proliferation, the cessation of cell cycle progression and the induction of apoptosis have all been targeted in chemotherapeutic strategies for cancer treatment. Therefore we, evaluated whether 5b and 5c, as complexes with the strongest cytotoxic activity, altered the cell cycle of HeLa cells and induced apoptosis of HeLa cells using flow cytometry. The cell-cycle phase distribution in HeLa cells was analyzed after exposure to different concentrations of 5b and 5c for 24 and 48 h.
As shown in Fig. 8 , treatment with 5b and 5c led to a marked dose- and time-dependent increase in the proportion of cells in the subG1 phase compared to control. The proportion of cells in G2/M phase was decreased following the increase of subG1 phase in all treatments. This confirmed that cells treated with 5b and 5c were induced to undergo apoptotic death. Furthermore, a population of sub-G1-phase cells, a characteristic of apoptosis, was increased double with increased in the concentrations. For 24 h of treatment the total percentage of cells in the phases subG1 and G2/M together was similar after all treatments. Significant differences in treatments were more visible after 48h.
Fig. 8.
Cell cycle distribution of 5b and 5c.
The alteration of the percentage of cells in the S phase after 24 h and 48 h treatment with different concentrations of complexes showed a slight but not significant increase, while the percentages of cells in G1 phase were decreased.
3.3.5. Determination of HeLa cell death mode
Examination of possible proapoptotic effects of the compounds 5b an 5c on human cervical adenocarcinoma HeLa cells is presented in Fig. 9 . For control and treated HeLa cells, the percentages of Annexin V+ propidium iodide-early apoptotic cells, Annexin V+ propidium iodide+ late apoptotic/secondary necrotic cells, and Annexin V− propidium iodide+ dead cells are shown. Treatment of HeLa with 2IC50 concentrations of the compounds 5b and 5c caused increase in the percentage of early apoptotic and late stage apoptotic/secondary necrotic cells when compared with untreated control cells. The compound 5c applied at lower IC50 concentration also increased the percentage of early apoptotic and late stage apoptotic/secondary necrotic HeLa cells, while the effect of the compound 5b applied at IC50 concentration was weaker. All these changes in treated HeLa cell samples were accompanied with increase in the percentage of dead cells (Annexin V− propidium iodide+) for both tested concentrations of compounds 5b and 5c.
Fig. 9.
Flow cytometric analysis of control (C) and HeLa cells treated with compounds 5b and 5c for 24 h and stained with Annexin V-FITC and propidium iodide. A+P− (FITC Annexin V positive, propidium iodide negative), A+P− (FITC Annexin V positive, propidium iodide positive), and A−P+ (FITC Annexin V negative, propidium iodide positive).
3.4. Anti-SARS-CoV-2 activity
In this study, the protein-ligand interactions between the two drugs used for COVID-19 treatment and synthesized compounds 5a-5e (Fig. 10 ) were investigated toward SARS-CoV-2 main protease Mpro using molecular docking simulations. Drugs cinanserin and hydroxycholoquine are involved in current research as model systems. Cinanserin shows noteworthy antiviral potential against the SARS-CoV-2, since it possesses inhibitory potency of the 3C-like protease toward SARS-coronavirus (SARS) [62]. Hydroxychloroquine has been considered as the drug which can treat COVID-19, but clinical trials found a possible risk of dangerous side effects [63].
Fig. 10.
Structures of cinanserin, hydroxychloroquine, and newly-synthesized compounds 5a-5e.
A careful examination of the interactions allows elucidation of the inhibitory potency of the investigated compounds used as ligands in molecular docking simulations toward SARS-CoV-2 main protease Mpro. At the beginning of the research, the pockets and binding sites of the targeted protein were determined using the AGFR software [64]. The bound ligand (N3) was removed from Mpro, and binding pocket analysis was accomplished. After configuring and computing affinity maps for a receptor protein, the one with potentially the lowest expected binding energy was used for AutoDock4 calculations. Further, molecular docking simulations were performed with the two referent drugs and synthesized derivatives 5a-5e in the same docking position. The binding modes of protein-ligand complexes that exhibit the best inhibitory potency are considered and further discussed. The achieved thermodynamic parameters from molecular docking simulations for all examined compounds are given in Table 3 . The interactions in the most stable conformations of complex structures of compounds 5a-5e with Mpro are presented in Fig. 11 . Compounds with the lowest values of ΔGbind and Ki have significant binding affinity to targeted protein and possess good inhibitory potency. Also, the lower values of the Ki indicate the greater binding affinity, and that the smaller concentration of a compound is required to inhibit the activity of the receptor. The results presented in Table 3 point out that the values of ΔGbind and Ki are in correlation. Namely, the lower values of ΔGbind are followed by lower values of Ki. It is observed that values of ΔGbind obtained for M pro -cinanserin and M pro -hydroxychloroquine complexes are very similar. The difference between values of ΔGbind is negligible, minor than 1 μM. On the other hand, the difference between Ki values of M pro -cinanserin and M pro -hydroxychloroquine complexes is significant and based on Ki values it could be said that cinanserin possesses better inhibitory potency against SARS-CoV-2 Mpro.
Table 3.
The important thermodynamical parameters from docking simulations between 5a-5e with SARS-CoV-2 Mpro.
| ΔGbind (kcal/mol) | Ki (uM) | FIE (kcal/mol) | vdW + Hbond + desolv Energy (kcal/mol) | Electrostatic Energy (kcal/mol) | FTIE (kcal/mol) | TFE (kcal/mol) | USE (kcal/mol) | |
|---|---|---|---|---|---|---|---|---|
| Mpro-cinanserin | −7.81 | 1.88 | −9.27 | −9.22 | −0.05 | −1.70 | +2.39 | −0.77 |
| Mpro-hydroxychloroquine | −7.00 | 7.43 | −9.83 | −9.33 | −0.49 | −0.67 | +2.98 | −0.52 |
| Mpro-5a | −6.40 | 20.25 | −8.26 | −8.12 | −0.14 | −4.64 | +2.47 | −4.03 |
| Mpro-5b | −6.24 | 26.84 | −8.28 | −8.15 | −0.13 | −4.66 | +2.47 | −4.23 |
| Mpro-5c | −5.53 | 88.62 | −8.02 | −8.32 | +0.29 | −4.56 | +3.02 | −4.04 |
| Mpro-5d | −5.32 | 124.98 | −8.38 | −8.60 | +0.23 | −4.44 | +3.57 | −3.92 |
| Mpro-5e | −7.34 | 4.18 | −8.77 | −8.85 | +0.08 | −5.88 | +3.57 | −3.75 |
Fig. 11.
The interactions of the most stable conformations of complex structures of compounds 5a-5e with SARS-CoV-2 Mpro.
Further analysis of results presented in Table 3 specifies compound 5e as the compound of interest. Namely, significantly lower values of the ΔGbind and Ki (−7.34 kcal/mol and 4.18 μM, respectively) are obtained for M pro -5e protein-ligand complex than for other examined protein-ligand complexes. Other protein-ligand complexes showed considerably higher values of the ΔGbind and Ki (Table 3). The results achieved in the molecular docking simulations of M pro -5a and M pro -5b are very similar, as well as the results obtained for M pro -5c and M pro -5d complexes. In addition, the compounds 5a-5d show inhibitory potency against SARS-CoV-2 Mpro, but lower than 5e, cinanserin, and hydroxychloroquine. The calculated Ki values for 5a and 5b indicate that these compounds could be considered as potential inhibitors of SARS-CoV-2 Mpro. Additionally, compounds 5c and 5d possesses high values of Ki can not be considered as inhibitors of SARS-CoV-2 Mpro.
To explain the inhibitory potency, first, it will be said about the structure of the SARS-CoV-2 Mpro protein. The SARS-CoV-2 Mpro is a dimer composed of two protomers, and each is formed from three domains: domain I (residues 8–101), domain II (residues 102–184), and domain III (residues 201–303) [57]. Domain III is linked to domain II by a long loop region (residues 185–200). The substrate-binding site is located in a cleft between domain I and domain II. The binding pocket of the SARS-CoV-2 Mpro represents an antiparallel sheet with residues 164–168 of the long strand (residues 155–168) on one side and with residues 189–191 of the loop that links domain II to domain III on the other. The amino acids Phe140, Leu141, Asn142, Glu166, His163, and His172 are involved in the formation of the binding subsite.
A detailed analysis of the interactions achieved in molecular docking simulations shows that all ligands interact with a few amino acids from SARS-CoV-2 Mpro (Phe140, Leu141, Asn142, Met165, Glu 166, Leu167, Pro 168, Ala 191). All of these amino acids are from the binding site of SARS-CoV-2 Mpro [57]. The different types of interactions are established, and that are mainly hydrogen bonds, van der Waals, alkyl, and π-alkyl interactions. Two types of hydrogen bonds are represented in the protein-ligand complex structures. The first type is conventional hydrogen bonds. The second type of hydrogen bond is carbon hydrogen bond, and those are formed with carbon atoms.
The results from Table 3 predict that the binding affinity of 5e is similar as for the referent drugs, and interactions in these protein-ligand complex are carefully analyzed. In Table S4 are collected corresponding amino acid with its position in the structure of the protein, type of interaction during bond formation, and distance between respective active sites of ligand and amino acids from SARS-CoV-2 for used referent drugs and compound 5e. It is noticed that examined referent drugs, cinanserin and hydroxychloroquine formed only few hydrogen bonds and a lot of hydrophobic contacts, such as π-alkyl, alkyl, π –Cation, and π-σ interactions. These interactions are in the range 2.6–5 Å. Cinanserin established one conventional hydrogen bond and one carbon hydrogen bond, with His164 and Asn142, respectively. In the molecular docking simulation between hydroxychloroquine and Mpro two conventional hydrogen bonds are formed (Table S4). From these results, it could be concluded that hydrophobic contacts have a significant impact on the binding affinities of cinanserin and hydroxychloroquine.
Compound 5e possesses a similar binding affinity as referent drugs, and it forms four hydrogen bonds, and two of them are conventional hydrogen bonds established with Phe140 and Asn142. The other two hydrogen bonds are carbon hydrogen bonds formed with Glu166 and Leu167. All these bonds are in the range 2–3 Å. Despite these, alkyl and π-alkyl contacts are established with Pro168 and they are fairly longer (4.3–5.4 Å).
The results collected in Table S5 are obtained from molecular docking simulations between compounds 5a-5e and SARS-CoV-2 Mpro. These results show that a significant number of both types of hydrogen bonds are formed. Since the compounds 5a-5e show lower inhibitory potency than referent drugs, it can be inferred that increase of a number of hydrogen bonds decreases the values of free energy of binding, in this particular case.
4. Conclusion
In the first stage of this article, we have synthesized five (three novel) Biginelli hybrids (2-thioxo-1,2,3,4-tetrahydropyrimidines). Dichloro(p-cymene)ruthenium(II) dimer was used as a complexing salt. Five ruthenium half-sandwich complexes containing Biginelli ligands were synthesized and characterized. X-ray analyses of complex 5a confirmed that two ruthenium atoms and two ligands 4a formatted a three-legged piano-stool coordination environment. Anticancer activities potential on various cell lines (HeLa, LS174, K562, A375 and A549) were checked for ligands (4a-d) and appropriate ruthenium complexes (5a-d). All complexes demonstrated lower (up to ten times) IC50 values on all tested cell lines than ligands. Therefore, most active complexes were included in the investigation of the mechanism of action. The outputs of the flow cytometry analysis imply that the proportion of cells in G2/M phase was decreased following the increase of subG1 phase. Considering these, we verified that cells treated with 5b and 5c were induced apoptosis. Then, we investigate the potential of our complexes for inhibition of SARS-CoV-2 protease at the computational level. Significant inhibitory potency has 5b, therefor 5e possesses the lowest values of ΔGbind and Ki. Experimental and computational results suggest that at least one ruthenium complex (5b) has an affinity to become a possible candidate for dual therapy (anticancer-antiviral) in the future. Among tested novel half-sandwich ruthenium complexes containing Biginelli hybrids, the compounds 5b and 5c showed the most promising anticancer activity due to strong cytotoxicity, proapoptotic effects, antimigratory and antiangiogenic properties. The molecular mechanisms of anticancer effects of these compounds and their in vivo anticancer efficacy need to be investigated in a further robust research. The good results obtained from molecular docking study point out that the inhibitory properties of the synthesized compounds, especially for 5b, against SARS-CoV-2 virus deserve to be studied in a new independent research.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are grateful to the Ministry of Education, Science and Technological Development of the Republic of Serbia for financial support (Agreements No. 451-03-68/2022-14/200378 and 451-03-68/2022-14/200043).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cbi.2022.110025.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bigineli P. Derivati aldeiduredici degli eteri acetil- e dossal-acetico. Gazz. Chim. Ital. 1893;23:360–416. [Google Scholar]
- 2.Biginelli P. Ueber Aldehyduramide des Acetessigaethers. Ber. Dtsch. Chem. Ges. 1891;24:1317–1319. [Google Scholar]
- 3.Tron G.C., Minassi A., Appendino G., Biginelli Pietro. The man behind the reaction. Eur. J. Org. Chem. 2011;28:5541–5550. [Google Scholar]
- 4.Mayer T.U., Kapoor T.M., Haggarty S.J., King R.W., Schreiber S.L., Mitchison T.J. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science. 1999;286:971–974. doi: 10.1126/science.286.5441.971. [DOI] [PubMed] [Google Scholar]
- 5.Maliga Z., Kapoor T.M., Mitchison T.J. Evidence that monastrol is an allosteric inhibitor of the mitotic kinesin Eg5. Chem. Biol. 2002;9:989–996. doi: 10.1016/s1074-5521(02)00212-0. [DOI] [PubMed] [Google Scholar]
- 6.Bahekar S.S., Shinde D.B. Synthesis and anti-inflammatory activity of some [4,6-(4-substituted aryl)-2-thioxo-1, 2, 3, 4-tetrahydro-pyrimidin-5-yl]-acetic acid derivatives. Bioorg. Med. Chem. Lett. 2004;14:1733–1736. doi: 10.1016/j.bmcl.2004.01.039. [DOI] [PubMed] [Google Scholar]
- 7.Narasimhulu Naidu B., Sorenson M.E., Patel M., Ueda Y., Banville J., Beaulieu F., Bollini S., Dicker I.B., Higley H., Lin Z., Pajor L., Parker D.D., Terry B.J., Zheng M., Martel A., Meanwell N.A., Krystal M., Walker M.A. Synthesis and evaluation of C2-carbon-linked heterocyclic-5-hydroxy-6-oxodihydropyrimidine-4-carboxamides as HIV-1 integrase inhibitors, Bioorg. Med. Chem. Lett. 2015;25:717–720. doi: 10.1016/j.bmcl.2014.11.060. [DOI] [PubMed] [Google Scholar]
- 8.Agbaje O.C., Fadeyi O.O., Fadeyi S.A., Myles L.E., Okoro C.O. Synthesis and in vitro cytotoxicity evaluation of some fluorinated hexahydropyrimidine derivatives. Bioorg. Med. Chem. Lett. 2011;21 doi: 10.1016/j.bmcl.2010.12.022. [DOI] [PubMed] [Google Scholar]
- 9.Cherukupalli S., Chandrasekaran B., Krystof V., Aleti R.R., Sayyad N., Merugu S.R., Kushwaha N.D., Karpoormath R. Synthesis, anticancer evaluation, and molecular docking studies of some novel 4,6-disubstituted pyrazolo[3,4-d]pyrimidines as cyclin-dependent kinase 2 (CDK2) inhibitors. Bioorg. Chem. 2018;79:46–59. doi: 10.1016/j.bioorg.2018.02.030. [DOI] [PubMed] [Google Scholar]
- 10.Attri P., Bhatia R., Gaur J., Arora B., Gupta A., Kumar N., Choi E.H. Triethylammonium acetate ionic liquid assisted one-pot synthesis of dihydropyrimidinones and evaluation of their antioxidant and antibacterial activities. Arab. J. Chem. 2017;10:206–214. [Google Scholar]
- 11.Kaur R., Chaudhary S., Kumar K., Gupta M.K., Rawal R.K. Recent synthetic and medicinal perspectives of dihydropyrimidinones: a review. Eur. J. Med. Chem. 2017;132:108–134. doi: 10.1016/j.ejmech.2017.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nature Microbiology. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdelrahman Z., Li M., Wang X. Comparative review of SARS-CoV-2, SARS-CoV, MERS-CoV, and influenza A respiratory viruses. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.552909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.https://www.who.int/emergencies/diseases/novel-coronavirus-2019
- 15.Di Lorenzo G., Di Trolio R., Kozlakidis Z., Busto G., Ingenito C., Buonerba L., Ferrara C., Libroia A., Ragone G., Dello Ioio C., Savastano B., Polverino M., De Falco F., Iaccarino S., Leo E. COVID 19 therapies and anti-cancer drugs: a systematic review of recent literature. Crit. Rev. Oncol. Hematol. 2020;152 doi: 10.1016/j.critrevonc.2020.102991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saini K.S., Tagliamento M., Lambertini M., McNally R., Romano M., Leone M., Curigliano G., de Azambuja E. Mortality in patients with cancer and coronavirus disease 2019: a systematic review and pooled analysis of 52 studies. Eur. J. Cancer. 2020;139:43–50. doi: 10.1016/j.ejca.2020.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakouny Z., Hawley J.E., Choueiri T.K., Peters S., Rini B.I., Warner J.L., Painter C.A. COVID-19 and cancer: current challenges and perspectives. Cancer Cell. 2020;38:629–646. doi: 10.1016/j.ccell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hegazy G.E., Abu-Serie M.M., Abo-Elela G.M., Ghozlan H., Sabry S.A., Soliman N.A., Abdel-Fattah Y.R. In vitro dual (anticancer and antiviral) activity of the carotenoids produced by haloalkaliphilic archaeon Natrialba sp. M6. Sci. Rep. 2020;10:5986. doi: 10.1038/s41598-020-62663-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanković T., Dinić J., Podolski-Renić A., Musso L., Stojković Burić S., Dallavalle S., Pešić M. Dual inhibitors as a new challenge for cancer multidrug resistance treatment. Curr. Med. Chem. 2019;26:2019. doi: 10.2174/0929867325666180607094856. [DOI] [PubMed] [Google Scholar]
- 20.Đorđić Crnogorac M., Matić I.Z., Damjanović A., Janković N., Krivokuća A., Stanojković T. 3D HeLa spheroids as a model for investigating the anticancer activity of Biginelli-hybrids. Chem. Biol. Interact. 2021;345 doi: 10.1016/j.cbi.2021.109565. [DOI] [PubMed] [Google Scholar]
- 21.Ristovski Trifunović J., Žižak Ž., Marković S., Janković N., Ignjatović N. Chitosan nanobeads loaded with Biginelli hybrids as cell-selective toxicity systems with a homogeneous distribution of the cell cycle in cancer treatment. RSC Adv. 2020;10:41542–41550. doi: 10.1039/d0ra08085c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janković N., Trifunović J., Vraneš M., Tot A., Petronijević J., Joksimović N., Stanojković T., Đorđić Crnogorac M., Petrović N., Boljević I., Matić I.Z., Bogdanović G.A., Mikov M., Bugarčić Z. Discovery of the Biginelli hybrids as novel caspase-9 activators in apoptotic machines: lipophilicity, molecular docking study, influence on angiogenesis gene and miR-21 expression levels. Bioorg. Chem. 2019;86:569–582. doi: 10.1016/j.bioorg.2019.02.026. [DOI] [PubMed] [Google Scholar]
- 23.Ramajayam R., Tan K.-P., Liu H.-G., Liang P.-H. Synthesis, docking studies, and evaluation of pyrimidines as inhibitors of SARS-CoV 3CL protease. Bioorg. Med. Chem. Lett. 2010;20:3569–3572. doi: 10.1016/j.bmcl.2010.04.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pawełczyk A., Zaprutko L. Anti-COVID drugs: repurposing existing drugs or search for new complex entities, strategies and perspectives. Future Med. Chem. 2020;12:1743–1757. doi: 10.4155/fmc-2020-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhou Y.-W., Xie Y., Tang L.-S., Pu D., Zhu Y.-J., Liu J.-Y., Ma X.-L. Therapeutic targets and interventional strategies in COVID-19: mechanisms and clinical studies. Signal Transduct. Targeted Ther. 2021;6:317. doi: 10.1038/s41392-021-00733-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabinger F., Stiller C., Schmitzová J., Dienemann C., Kokic G., Hillen H.S., Höbartner C., Cramer P. Mechanism of molnupiravir-induced SARS-CoV-2 mutagenesis. Nat. Struct. Mol. Biol. 2021;28:740–746. doi: 10.1038/s41594-021-00651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toots M., Yoon J.-J., Cox R.M., Hart M., Sticher Z.M., Makhsous N., Plesker R., Barrena A.H., Reddy P.G., Mitchell D.G., Shean R.C., Bluemling G.R., Kolykhalov A.A., Greninger A.L., Natchus M.G., Painter G.R., Plemper R.K. Characterization of orally efficacious influenza drug with high resistance barrier in ferrets and human airway epithelia. Sci. Transl. Med. 2019;11 doi: 10.1126/scitranslmed.aax5866. eaax5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neuditschko B., Legin A.A., Baier D., Schintlmeister A., Reipert S., Wagner M., Keppler B.K., Berger W., Meier-Menches S.M., Gerner C. Interaction with ribosomal proteins accompanies stress induction of the anticancer metallodrug BOLD-100/KP1339 in the endoplasmic reticulum. Angew. Chem. Int. Ed. Engl. 2021;60:5063–5068. doi: 10.1002/anie.202015962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munteanu A.-C., Uivarosi V. Ruthenium complexes in the fight against pathogenic microorganisms. An Extensive Review, Pharmaceutics. 2021;13:874. doi: 10.3390/pharmaceutics13060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bold therapeutics potential to fight COVID-19. 2021. https://www.bold-therapeutics.com/COVID-19 Available online:
- 31.Furrer J., Süss-Fink G. Thiolato-bridged dinuclear arene ruthenium complexes and their potential as anticancer drugs. Coord. Chem. Rev. 2016;309:36–50. [Google Scholar]
- 32.Girgis A.S., Panda S.S., Srour A.M., Abdelnaser A., Nasr S., Moatasim Y., Kutkat O., El Taweel A., Kandeil A., Mostafa A., Ali M.A., Fawzy N.G., Bekheit M.S., Shalaby E.M., Gigli L., Fayad W., Soliman A.A.F. 3-Alkenyl-2-oxindoles: synthesis, antiproliferative and antiviral properties against SARS-CoV-2. Bioorg. Chem. 2021;114 doi: 10.1016/j.bioorg.2021.105131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Srour A.M., Panda S.S., Mostafa A., Fayad W., El-Manawaty M.A., Soliman A.A.F., Moatasim Y., El Taweel A., Abdelhameed M.F., Bekheit M.S., Ali M.A., Girgis A.S. Synthesis of aspirin-curcumin mimic conjugates of potential antitumor and anti-SARS-CoV-2 properties. Bioorg. Chem. 2021;117 doi: 10.1016/j.bioorg.2021.105466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Díaz-Carballo D., Acikelli A.H., Klein J., Jastrow H., Dammann P., Wyganowski T., Guemues C., Gustmann S., Bardenheuer W., Malak S., Tefett N.S., Khosrawipour V., Giger-Pabst U., Tannapfel A., Strumberg D. J. Exp. Clin. Cancer Res. 2015;34:81. doi: 10.1186/s13046-015-0199-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Đorđić Crnogorac M., Matić I.Z., Damjanović A., Janković N., Krivokuća A., Stanojković T. 3D HeLa spheroids as a model for investigating the anticancer activity of Biginelli-hybrids. Chem. Biol. Interact. 2021;345 doi: 10.1016/j.cbi.2021.109565. [DOI] [PubMed] [Google Scholar]
- 36.Milović E., Janković N., Bogdanović G., Petronijević J., Joksimović N. On water synthesis of the novel 2-oxo-1,2,3,4-tetrahydropyrimidines. Tetrahedron. 2021;78 [Google Scholar]
- 37.Milović E., Janković N., Vraneš M., Stefanović S., Petronijević J., Joksimović N., Muškinja J., Ratković Z. Green one-pot synthesis of pyrido-dipyrimidine DNA-base hybrids in water. Environ. Chem. Lett. 2021;19:729–736. [Google Scholar]
- 38.Ristovski Trifunović J., Žižak Ž., Marković S., Janković N., Ignjatović N. Chitosan nanobeads loaded with Biginelli hybrids as cell-selective toxicity systems with a homogeneous distribution of the cell cycle in cancer treatment. RSC Adv. 2020;10:41542–41550. doi: 10.1039/d0ra08085c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janković N., Trifunović J., Vraneš M., Tot A., Petronijević J., Joksimović N., Stanojković T., Đorđić Crnogorac M., Petrović N., Boljević I., Matić I.Z., Bogdanović G.A., Mikov M., Bugarčić Z. Discovery of the Biginelli hybrids as novel caspase-9 activators in apoptotic machines: lipophilicity, molecular docking study, influence on angiogenesis gene and miR-21 expression levels. Bioorg. Chem. 2019;86:569–582. doi: 10.1016/j.bioorg.2019.02.026. [DOI] [PubMed] [Google Scholar]
- 40.Janković N., Stefanović S., Petronijević J., Joksimović N., Novaković S.B., Bogdanović G.A., Muškinja J., Vraneš M., Ratković Z., Bugarčić Z. Water-tuned tautomer-selective tandem synthesis of the 5,6-Dihydropyrimidin-4(3H)-ones, driven under the umbrella of sustainable chemistry. ACS Sustain. Chem. Eng. 2018;6:13358–13366. [Google Scholar]
- 41.Gavrilović M., Janković N., Joksović Lj, Petronijević J., Joksimović N., Bugarčić Z. Water ultrasound-assisted oxidation of 2-oxo-1,2,3,4-tetrahydropyrimidines and benzylic acid salts. Environ. Chem. Lett. 2018;16:1501–1506. [Google Scholar]
- 42.Muškinja J., Janković N., Ratković Z., Bogdanović G., Bugarčić Z. Vanillic aldehydes for the one-pot synthesis of novel 2-oxo-1,2,3,4-tetrahydropyrimidines. Mol. Divers. 2016;20:591–604. doi: 10.1007/s11030-016-9658-y. [DOI] [PubMed] [Google Scholar]
- 43.Janković N., Bugarčić Z., Marković S. Double catalytic effect of (PhNH3)2CuCl4 in a novel, highly efficient synthesis of 2-oxo and thioxo-1,2,3,4-tetrahydropyrimidines. J. Serb. Chem. Soc. 2015;80:595–604. [Google Scholar]
- 44.Petronijević J., Joksimović N., Milović E., Đorđić Crnogorac M., Petrović N., Stanojković T., Milivojević D., Janković N. Antitumor activity, DNA and BSA interactions of novel copper(II) complexes with 3,4-dihydro-2(1H)-quinoxalinones. Chem. Biol. Interact. 2021;348 doi: 10.1016/j.cbi.2021.109647. [DOI] [PubMed] [Google Scholar]
- 45.Joksimović N., Janković N., Petronijević J., Baskić D., Popovic S., Todorović D., Zarić M., Klisurić O., Vraneš M., Tot A., Bugarčić Z. Synthesis, аnticancer еvaluation and synergistic еffects with cisplatin of novel palladium complexes: DNA, BSA interactions and molecular docking study. Med. Chem. 2020;16:78–92. doi: 10.2174/1573406415666190128095732. [DOI] [PubMed] [Google Scholar]
- 46.Joksimović N., Petronijević J., Janković N., Kosanić M., Milivojević D., Vraneš M., Tot A., Bugarčić Z. Synthesis, characterization, antioxidant activity of β-diketonates, and effects of coordination to copper(II) ion on their activity: DNA, BSA interactions and molecular docking study. Med. Chem. 2019 doi: 10.2174/1573406415666191024102520. [DOI] [PubMed] [Google Scholar]
- 47.Joksimović N., Baskić D., Popović S., Zarić M., Kosanić M., Ranković B., Stanojković T., Novaković S.B., Davidović G., Bugarčić Z., Janković N. Synthesis, characterization, biological activity, DNA and BSA binding study: novel copper(II) complexes with 2-hydroxy-4-aryl4-oxo-2-butenoate. Dalton Trans. 2016;45:15067–15077. doi: 10.1039/c6dt02257j. [DOI] [PubMed] [Google Scholar]
- 48.CrysAlis P.R.O. Oxford Diffraction Ltd. Yarnton; Engl: 2009. [Google Scholar]
- 49.Coppens P. In: Crystallographic Computing. FR Ahmed, Huber SR.Hall & CP., editors. Munksgaard; Copenhagen: 1970. pp. 255–270. [Google Scholar]
- 50.Sheldrick G.M. Shelxt - integrated space-group and crystal-structure determination. Acta Crystallogr. A. 2015;71:3–8. doi: 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dolomanov O.V., Bourhis L.J., Gildea R.J., Howard J.A.K., Puschmann H. OLEX2: a complete structure solution, refinement and analysis program. J. Appl. Crystallogr. 2009;42:339–341. [Google Scholar]
- 52.Sheldrick G.M. Crystal structure refinement with SHELXL. Acta Crystallogr. C. 2015;71:3–8. doi: 10.1107/S2053229614024218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bruno I.J., Cole J.C., Edgington P.R., Kessler M., Macrae C.F., McCabe P., Pearson J., Taylor R. New software for searching the Cambridge Structural Database and visualizing crystal structures. Acta Crystallogr. Sect. B Struct. Sci. 2002;58:389–397. doi: 10.1107/s0108768102003324. [DOI] [PubMed] [Google Scholar]
- 54.Matić I.Z., Aljančić I., Žižak Ž., Vajs V., Jadranin M., Milosavljević S., Juranić Z.D. In vitro antitumor actions of extracts from endemic plant Helichrysum zivojinii, BMC Complement. Altern. Med. 2013;13:36. doi: 10.1186/1472-6882-13-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frisch M.J., Trucks G.W., Schlegel H.B., Scuseria G.E., Robb M.A., Cheeseman J.R., Scalmani G., Barone V., Mennucci B., Petersson G.A., Nakatsuji H., Caricato M., Li X., Hratchian H.P., Izmaylov A.F., Bloino J., Zheng G., Sonnenberg J.L., Hada M., Ehara M., Toyota K., Fukuda R., Hasegawa J., Ishida M., Nakajima T., Honda Y., Kitao O., Nakai H., Vreven T., Montgomery J.A., Jr., Peralta J.E., Ogliaro F., Bearpark M., Heyd J.J., Brothers E., Kudin K.N., Staroverov V.N., Kobayashi R., Normand J., Raghavachari K., Rendell A., Burant J.C., Iyengar S.S., Tomasi J., Cossi M., Rega N., Millam J.M., Klene M., Knox J.E., Cross J.B., Bakken V., Adamo C., Jaramillo J., Gomperts R., Stratmann R.E., Yazyev O., Austin A.J., Cammi R., Pomelli C., Ochterski J.W., Martin R.L., Morokuma K., Zakrzewski V.G., Voth G.A., Salvador P., Dannenberg J.J., Dapprich S., Daniels A.D., Farkas Ö., Foresman J.B., Ortiz J.V., Cioslowski J., Fox D.J. Revision D.1, Inc.; Wallingford, CT: 2013. Gaussian 09. [Google Scholar]
- 56.Becke A.D. Phys. Rev. A. 1988;38:3098–3100. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 57.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 58.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Biovia D.S. 2017. Discovery Studio Modeling Environment. [Google Scholar]
- 60.Spackman M.A., Jayatilaka D. Hirshfeld surface analysis. CrystEngComm. 2009;11:19–32. [Google Scholar]
- 61.Nefisath P., Prasad Dasappa J., Haripriya B., Chopra D., Venugopala K.N., Deb P.K., Gleiser R.M., Mohanlall V., Maharaj R., Vishwanatha Poojary S.S. Synthesis, characterization and larvicidal activity of novel benzylidene derivatives of fenobam and its thio analogues with crystal insight. J. Mol. Struct. 2021;1226 [Google Scholar]
- 62.Chen L., Gui C., Luo X., Yang Q., Günther S., Scandella E., Drosten C., Bai D., He X., Ludewig B., Chen J., Luo H., Yang Y., Yang Y., Zou J., Thiel V., Chen K., Shen J., Shen X., Jiang H. Cinanserin is an inhibitor of the 3C-like proteinase of severe acute respiratory syndrome coronavirus and strongly reduces virus replication in vitro. J. Virol. 2005;79:7095–7103. doi: 10.1128/JVI.79.11.7095-7103.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chloroquine or Hydroxychloroquine". COVID-19 Treatment Guidelines. National Institutes of Health. Retrieved 14 February 2021.
- 64.Zhang Y., Forli S., Omelchenko A., Sanner M.F. AutoGridFR: improvements on AutoDock affinity maps and associated software tools. J. Comput. Chem. 2019;40:2882–2886. doi: 10.1002/jcc.26054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.