Abstract
Streptomyces coelicolor 1A and Pseudomonas citronellolis were able to degrade synthetic high-molecular-weight poly(cis-1,4-isoprene) and vulcanized natural rubber. Growth on the polymers was poor but significantly greater than that of the nondegrading strain Streptomyces lividans 1326 (control). Measurement of the molecular weight distribution of the polymer before and after degradation showed a time-dependent increase in low-molecular-weight polymer molecules for S. coelicolor 1A and P. citronellolis, whereas the molecular weight distribution for the control (S. lividans 1326) remained almost constant. Three degradation products were isolated from the culture fluid of S. coelicolor 1A grown on vulcanized rubber and were identified as (6Z)-2,6-dimethyl-10-oxo-undec-6-enoic acid, (5Z)-6-methyl-undec-5-ene-2,9-dione, and (5Z,9Z)-6,10-dimethyl-pentadec-5,9-diene-2,13-dione. An oxidative pathway from poly(cis-1,4-isoprene) to methyl-branched diketones is proposed. It includes (i) oxidation of an aldehyde intermediate to a carboxylic acid, (ii) one cycle of β-oxidation, (iii) oxidation of the conjugated double bond resulting in a β-keto acid, and (iv) decarboxylation.
Poly(cis-1,4-isoprene) is a natural polymer that is synthesized by many plants and some fungi. This polymer has been commercially produced at a level of several million tons per year since the beginning of the last century by cultivating and tapping the rubber tree (Hevea brasiliensis). The natural polymer has a high molecular weight, about 106, and, after cross-linking of the linear polymer chains by sulfur bridges (vulcanization), has superior physical properties. Despite the development of chemosynthetic poly(cis-1,4-isoprene), natural rubber is still necessary as a basic material for several items, such as tires, latex gloves, condoms, seals and many other things.
As a naturally occurring compound, poly(cis-1,4-isoprene) is subjected to biological mineralization cycles, and many reports on biodegradation of rubbers have been published since the study by Söhngen and Fol in 1914 (9). Even chemically cross-linked (e.g., vulcanized) rubbers have been shown to be biodegradable (2, 10). The most extensive investigation of the molecular basis of biological rubber degradation was done by Tsuchii and coworkers (for a summary see reference 12). They isolated a Nocardia strain which is able to degrade natural and vulcanized rubber and identified low-molecular-weight degradation products (Mw, 103 to 104) as intermediates. Degradation of vulcanized rubber by this isolate is dependent on colonization of the polymer surface, and no soluble rubber-degrading enzymes were detected (10). Previously, a Xanthomonas sp. was the only known gram-negative bacterium that has the ability to utilize poly(cis-1,4-isoprene) as a sole carbon and energy source (11). It produces clearing zones on opaque latex-containing agar, which indicates the extracellular nature of the degrading enzyme(s). It is assumed that enzymatic degradation of poly(cis-1,4-isoprene) is initiated by oxidative cleavage of the isoprene double bond. Low-molecular-weight oligo(cis-1,4-isoprene) molecules with aldehyde and keto groups at their ends, such as 12-oxo-4,8-dimethyltrideca-4,8-diene-1-al (compound 1) (acetonyl diprenyl acetoaldehyde), have been identified as degradation products (12). Recently, another gram-negative bacterium, Pseudomonas aeruginosa AL98, has been isolated. Strain AL98 is able to produce holes in polymer films if the polymer is used as the sole source of carbon and energy on a solid mineral salts medium (4a). Interestingly, this bacterium is not able to produce clearing zones on latex agar.
In an earlier study we isolated and identified several dozen poly(cis-1,4-isoprene)-degrading bacteria (3). All isolates produced clearing zones on opaque rubber-containing solid media. It turned out that the ability to utilize rubber latex is widely distributed among many genera of the Actinomycetes (e. g., Streptomyces, Nocardia, Micromonospora, and Actinoplanes). In this study, we used natural rubber latex, vulcanized natural rubber, and nonvulcanized synthetic rubber as polymers. We were able to show by high-resolution gel permeation chromatography (GPC) and by weight loss measurement that the bacteria used in this study are able to cleave the carbon backbone of synthetic poly(cis,1,4-isoprene) and to utilize the low-molecular-weight degradation products for growth. These bacteria include Pseudomonas citronellolis, which was isolated on the basis of its ability to grow on linear terpenes (8); however, its ability to degrade poly(cis-1,4-isoprene) has not been described previously. In addition, we isolated three degradation products from a latex glove-grown culture of Streptomyces coelicolor 1A and identified them. A biochemical pathway explaining how the bacteria could metabolize the methyl-branched carbon backbone of rubber is proposed.
MATERIALS AND METHODS
Bacteria, media, and culture conditions.
The bacteria used in this study are shown in Table 1. The bacteria were grown in 100-ml Erlenmeyer flasks containing a solution (pentane)-cast film of synthetic poly(cis-1,4-polyisoprene) (100 mg [dry weight]) as a carbon source and 20 ml of a mineral salts solution as described earlier (7), except that the medium contained 0.05% (wt/vol) yeast extract and was diluted twofold with water. The media were inoculated with washed cells from Luria-Bertani medium seed culture (gram-negative strains) or directly with material from a stock spore suspension (in 0.9% [wt/vol] NaCl) and were incubated without shaking at 30°C. Occasionally (at least three times a week), the flasks were shaken gently. Evaporated water (≈5 ml) was replaced after 4 weeks. All experiments were performed in duplicate. For each experiment two sets of five flasks were inoculated at the same time, and 2 flasks were harvested at intervals of 2 weeks. The bacteria were grown on purified latex agar as described recently (3). For growth on vulcanized rubber, S. coelicolor 1A was incubated in 1-liter Erlenmeyer flasks by using undiluted mineral salts medium (200 ml) without yeast extract that contained 2 g of autoclaved nonpowdered latex gloves as a carbon source with continuous shaking (100 rpm) at 28°C. The cultures were inoculated with 10 ml of a 3-day-old glucose-grown seed culture.
TABLE 1.
Bacteria used in this study
| Organism | Relevant characteristics | Reference(s) |
|---|---|---|
| Streptomyces lividans 1326 | No utilization of synthetic poly(cis,1,4-isoprene), no halo formation on latex agar | 5 |
| Streptomyces coelicolor 1A | Utilization of synthetic poly(cis,1,4-isoprene), halo formation on latex agar | 3 |
| Pseudomonas citronellolis | Utilization of linear terpenes such as citronellol, utilization of synthetic poly(cis,1,4-isoprene), no halo formation on latex agar | 8; this study |
Polymers.
Three poly(cis-1,4-isoprene) polymers were used in this study: (i) purified natural latex of H. brasiliensis, which was a gift from the Rubber Research Institute of Malaysia; (ii) synthetic poly(cis-1,4-isoprene) (molecular weight, about 800,000), which was obtained from Aldrich (the polymer did not contain any low-molecular-weight impurities, as confirmed by GPC analysis and weight loss studies); and (iii) unpowdered vulcanized latex gloves (Romed) that were rinsed with water before use. The gloves were cut into pieces of the desired size, transferred to Erlenmeyer flasks, and autoclaved together with the growth medium. Extraction of vulcanized rubber with solvents (water, methanol, ethanol, or acetone) and subsequent weight loss analysis confirmed the absence of significant amounts of soluble low-molecular-weight impurities.
Harvesting of cultures grown on synthetic poly(cis-1,4-isoprene).
A complete culture was lyophilized, resuspended in distilled water, transferred to 10-ml vials, and lyophilized again. The dried samples were stored sealed at room temperature in the dark until they were used.
Isolation of metabolites from vulcanized latex gloves.
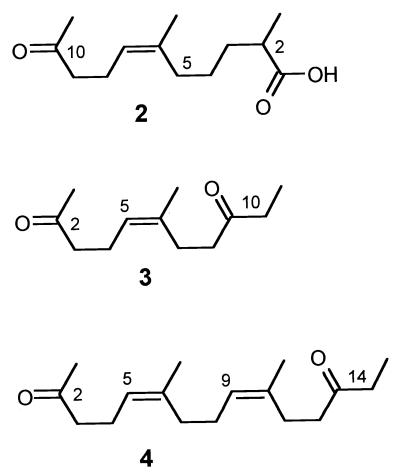
Fermentation broth was filtered through a Celite pad, and the filtrate (∼6 liters) was lyophilized. The brown residue was dissolved in 100 ml of water and extracted four times with ethyl acetate (200 ml each), which yielded a yellow organic phase. The combined organic phases were dried over anhydrous Na2SO4 and concentrated in vacuo, which yielded a brown gum. Column chromatography of the crude material adsorbed on 2 g of silica gel (ICN SiliTech 32-63 [ICN Biomedicals GmbH]; 25 by 3 cm; 500 ml of cyclohexane, 500 ml of CH2Cl2, 500 ml of CH3OH) yielded a mixture of compounds 3 and 4 (Fig. 1) in the most lipophilic fraction and compound 2 in a polar fraction visible as green-colored spots on high-performance thin-layer chromatography (HPTLC) plates (Silica Gel KG 60 F254 [Merck]; CHCl3–methanol [MeOH], 9:1) after staining with anisaldehyde-H2SO4. Further repeated chromatography on silica gel (40 by 1 cm; ethyl acetate-petrol ether, 1:6) yielded 1 mg of compound 4 (90% pure) and 12 mg of pure compound 3. Compound 2 was purified to homogeneity by reversed-phase medium-pressure liquid chromatography (Kronwald Sepapress HPP-100I 50; RP-18; acetone-water, 4:1) and gel permeation chromatography on Sephadex LH-20 (acetone), which yielded 10 mg of compound 2.
FIG. 1.
Structures of poly(cis1,4-isoprene) degradation products. Compound 2, (6Z)-2,6-dimethyl-10-oxo-undec-6-enoic acid (C13H22O3; molecular weight, 226.32); compound 3, (5Z)-6-methyl-undec-5-ene-2,9-dione (C12H20O2; molecular weight, 196.29); compound 4, (5Z,9Z)-6,10-dimethyl-pentadec-5,9-diene-2,13-dione (C17H28O2; molecular weight, 264.41).
Analysis and identification of the metabolites isolated.
Physical data for the compounds described here were recorded by using the following equipment: an electron ionization (EI) mass spectrometer (Varian 311 A, 70 eV, direct insert, high resolution with fluorokerosine as the standard). 13C and 1H nuclear magnetic resonance spectra were recorded at 295 K with a Bruker AMX 300 (1H, 300 MHz; 13C, 75.5 MHz) and a Varian Inova 500 (1H, 500 MHz; 13C, 125.7 MHz); chemical shifts were expressed as δ values (parts per million) by using the solvent as an internal reference (CDCl3: δH = 7.24, δC = 77.0).
(i) (6Z)-2,6-Dimethyl-10-oxo-undec-6-enoic acid.
The data for (6Z)-2,6-dimethyl-10-oxo-undec-6-enoic acid (C13H22O3; compound 2; molecular weight, 226.32) are as follows: (CHCl3-MeOH, 9:1); colorless oil; m/z (EI) 226 (M+, 1%), 208 ([M-H2O]+, 45), 95 (80), 43 (100); m/z (HREI) found, M+ 226.1568. C13H22O3 requires M 226.1568; δH (300 MHz, CDCl3) 11.10 (br s, -COOH), 5.02 (t, J = 7.0 Hz, 7-H), 2.40 (m, 2-H, 9-H2), 2.20 (m, 8-H2), 2.08 (s, 11-H3), 1.97 (m, 5-H2), 1.61 (m, 3-H2), 1.60 (s, 6-CH3), 1.35 (m, 4-H2), 1.13 (t, J = 6.5 Hz, 2-CH3); δC (75.5 MHz, CDCl3) 209.0 (C-10), 182.9 (C-1), 136.1 (C-6), 123.4 (C-7), 43.8 (C-9), 33.2 (C-3), 31.3 (C-5), 29.8 (C-11), 29.4 (C-2), 25.3 (C-4), 23.2 (6-CH3), 22.1 (C-8), 16.8 (2-CH3).
(ii) (5Z)-6-Methyl-undec-5-ene-2,9-dione.
The data for (5Z)-6-methyl-undec-5-ene-2,9-dione (C12H20O2; compound 3; molecular weight, 196.29) are as follows: Rf 0.65 (CHCl3-MeOH, 9:1); colorless oil; m/z (EI) 196 (M+, 2%), 178 ([M-H2O]+, 10), 57 (60), 43 (100); m/z (high-resolution EI-mass spectroscopy [HREI-MS]) found, M+ 196.1463. C12H20O2 requires M 196.1463; δH (300 MHz, CDCl3) 5.07 (m, 5-H), 2.42 (m, 3-H2, 7-H2, 10-H2), 2.25 (m, 8-H2), 2.20 (m, 4-H2), 2.10 (s, 1-H3), 1.63 (m, 6-CH3), 1.04 (t, J = 7.0 Hz, 11-H3); δC (75.5 MHz, CDCl3) 211.2 (C-8), 208.6 (C-2), 135.0 (C-6), 124.3 (C-5), 43.7 (C-3), 40.5 (C-7), 36.0 (C-10), 29.9 (C-1), 26.0 (C-8), 23.1 (6-CH3), 22.1 (C-4), 7.8 (C-11).
(iii) (5Z,9Z)-6,10-Dimethyl-pentadec-5,9-diene-2,13-dione.
The data for (5Z,9Z)-6,10-dimethyl-pentadec-5,9-diene-2,13-dione (C17H28O2; compound 4; molecular weight, 264.41) are as follows: Rf 0.87 (CHCl3-MeOH, 9:1); colorless oil; m/z (EI) 264 (M+, 6%), 246 ([M-H2O]+, 10), 57 (100); m/z (HREI) found, M+ 264.2089. C17H28O2 requires M 264.2089; δH (500 MHz, CDCl3) 5.16 (m, 9-H), 5.10 (m, 5-H), 2.48 (m, 3-H2, 14-H2), 2.45 (m, 12-H2), 2.30 (m, 11-H2), 2.27 (m, 4-H2), 2.16 (s, 1-H3), 2.07 (m, 7-H2, 8-H2), 1.69 (m, 6-CH3, 10-CH3), 1.08 (t, J = 7.0 Hz, 15-H3); δC (125.7 MHz, CDCl3) 211.3 (C-13), 208.8 (C-2), 136.2 (C-10), 134.0 (C-6), 125.8 (C-9), 123.5 (C-5), 43.9 (C-3), 40.8 (C-12), 36.0 (C-14), 32.0 (C-8), 29.9 (C-1), 26.2 (C-7), 26.1 (C-11), 23.4 (10-CH3), 23.2 (6-CH3), 22.3 (C-4), 7.8 (C-15).
Molecular weight measurements.
The polymers were dissolved in toluene at a concentration of 2 g/liter overnight and passed through a 0.45-μm-pore-size filter (Pall Gelman Laboratories, Dreieich, Germany). One hundred microliters of the solution was separated with a GPC system consisting of a PSS SDV 8- by 50-mm precolumn (5 μm; PSS, Mainz, Germany), three 8- by 300-mm PSS SDV columns (column 1, linear XL, 10 μm; column 2, 5 μm, 1,000 Å; column 3, 5 μm, 105 Å), and a refractive index detector (Shodex RI-71) at a flow rate of 1 ml/min. Calibration of the column was performed by using synthetic poly(cis-1,4-isoprene) standard mixtures with narrow molecular weight distributions (PSS). Molecular weight calculations were done with winGPC software obtained from PSS.
Protein determination.
The lyophilized samples or bovine serum albumin standard protein was resuspended in vials in 5 ml of distilled water. After addition of 2 ml of 2 M NaOH, the vials were sealed, incubated in a boiling water bath for 30 min, and allowed to cool to room temperature. A 0.5-ml portion of each solution was used for protein determination by the Lowry procedure (6). If necessary, the solution was diluted with water. The residual polymer was washed extensively with warm water and subsequently dried at 80°C until the weight was constant. The dried polymer was used for GPC measurements after it was dissolved in toluene at a concentration of 2 g/liter.
RESULTS
Growth on natural rubber latex.
Streptomyces lividans 1326, S. coelicolor 1A, and P. citronellolis were grown on opaque latex agar at 30°C for 1 week. Only S. coelicolor 1A showed clearing zone formation which indicated utilization of the polymer. Growth of S. lividans 1326 and growth of P. citronellolis were poor, and the formation of clearing zones could not be observed even after prolonged incubation. Apparently, S. lividans 1326 and P. citronellolis are unable to utilize natural rubber latex as a carbon source.
Growth on synthetic rubber.
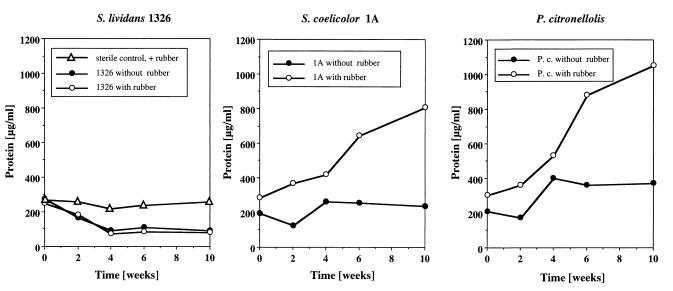
S. lividans 1326, S. coelicolor 1A, and P. citronellolis were grown in 20 ml of growth medium supplemented with 100 mg of a solution-cast film of synthetic rubber as described above. Flasks with polymer that were not inoculated were used as controls. Two cultures of each strain were harvested at 2-week intervals, and the total protein content was determined. A constant low level, 200 to 250 μg of protein/ml, was observed for the noninoculated sterile control throughout the incubation. The protein contents of the S. lividans cultures decreased from 250 μg/ml to less than 100 μg/ml within 4 weeks and remained at constant low values, ≈80 μg/ml, until the end of the experiment (Fig. 2) regardless of whether rubber was present or absent in the culture fluid. The color of the culture did not change significantly. Germination of the S. lividans 1326 spores was visible by microscopical examination. We assume that the traces of yeast extract present induced germination of the spores and were enough to support initiation of cell division, but did not allow significant growth of the bacteria. The residual polymer after cell harvest had the same optical appearance and mechanical properties as the noninoculated control polymer. S. lividans 1326 apparently is not able to utilize synthetic poly(cis-1,4-isoprene) or any soluble or volatile carbon sources that might be present at low concentrations in the air of a microbiology lab within 10 weeks.
FIG. 2.
Growth of selected bacteria on synthetic poly(cis-1,4-isoprene). Twenty Erlenmeyer flasks (100 ml), each containing 20 ml of growth medium with or without rubber, were inoculated with the strain of interest and incubated at 30°C for 10 weeks. Two flasks were harvested at 2-week intervals, and the protein concentrations were determined. Evaporated water was replaced by the addition of sterile water. Mean values of the two determinations are given. A noninoculated sterile culture with rubber served as a control. P.c., P. citronellolis.
The protein content of the S. coelicolor 1A culture increased in the presence of synthetic poly(cis-1,4-isoprene) and was more than 800 μg/ml after 10 weeks. Bacterial growth was visible in the culture fluid and on the surface of the polymer, mainly in polymer areas which floated on top of the culture fluid and were exposed to the air. The protein content of the P. citronellolis culture increased at a comparable rate to 1,000 μg/ml in the presence of rubber after 10 weeks (Fig. 2). The latter result came as a surprise, because P. citronellolis was unable to produce clearing zones on opaque latex agar. After several weeks of incubation of both strains the residual polymer had a dark appearance and differed from the original polymer in its higher degree of stickiness. FTIR analysis confirmed the presence of carbonyl groups in the degraded material (data not shown). Apparently, P. citronellolis and S. coelicolor 1A are able to degrade synthetic poly(cis-1,4-isoprene) and to use the degradation products as carbon sources. Incubation of S. coelicolor and P. citronellolis without rubber resulted in only a small increase in the amount of protein.
Separation efficiency of the GPC system.
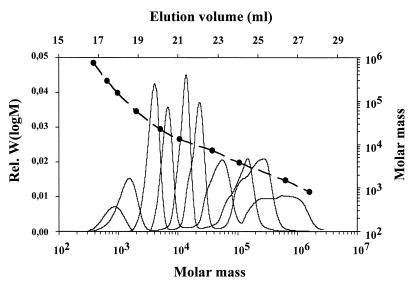
Commercially available GPC columns have a separation range for polymers that differ in molecular weight of about 2 orders of magnitude. However, for analysis of the molecular weight distribution of partially degraded synthetic poly(cis-1,4-isoprene) it was necessary to efficiently separate low-molecular-weight oligomers of isoprene (Mw, <1,000) from high-molecular-weight polymers (Mw, >500,000). Therefore, three GPC columns with different separation ranges were combined. Figure 3 shows separation of poly(cis-1,4-isoprene) standard mixtures by the three columns. Efficient separation occurred in the molecular weight range from ≈500 to about 106. Since poly(cis-1,4-isoprene) standard mixtures with narrow molecular weight ranges were used instead of polystyrol standards, the calibration curve could be used directly for determination of absolute values.
FIG. 3.
Calibration of the GPC system with narrowly distributed poly(cis-1,4-isoprene) standards. The relationship between the elution volumes (in milliliters) at the peak maximum (solid circles) and the molecular masses of the standards and the relationship between the refractive index detector signals [rel. W(logM)] and the molecular masses of the standards are shown. The eluent was toluene and the flow rate was 1 ml/min.
Molecular weight distribution of partially degraded synthetic poly(cis-1,4-isoprene).
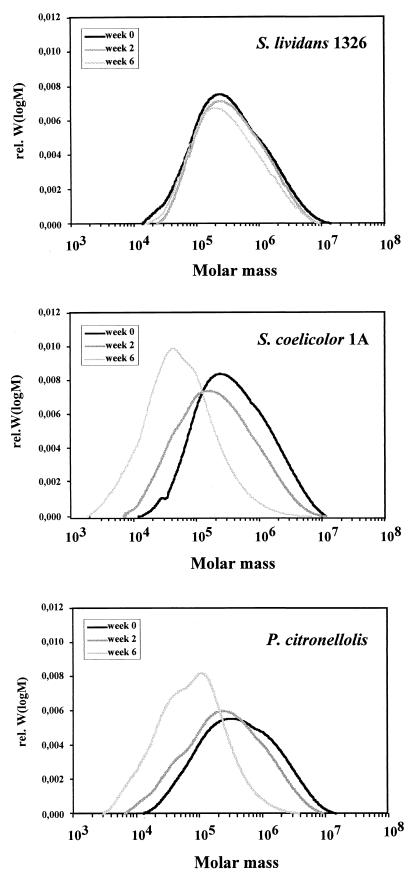
In order to determine the molecular weight distribution of synthetic poly(cis-1,4-isoprene) before and after degradation, GPC analysis of the residual polymer samples at zero time and after 2 and 6 weeks of incubation was performed (Fig. 4). The polymer in the control (zero time) showed an almost Gauss-like distribution of molecular weights between 104 and 107 (Mw, 8 × 105). No signal was obtained for molecular weights below 10,000, confirming the absence of significant amounts of low-molecular-weight impurities in synthetic poly(cis-1,4-isoprene). No significant differences in the molecular weight distributions compared to the zero-time control were found for the sterile control even after several weeks of incubation in the culture fluid. We concluded that the polymer backbone of synthetic poly(cis-1,4-isoprene) is stable and is not cleaved by sterile culture fluid. Similar results were obtained in the analysis of the polymer remaining in the culture inoculated with S. lividans 1326. The polymer recovered had the same optical appearance and indistinguishable mechanical properties. GPC analysis confirmed that no or only very little degradation had taken place (Mw, 5 × 105 to 9 × 105). None of the samples contained material with a molecular weight below ≈10,000. In contrast, the color of the residual polymers in the S. coelicolor 1A and P. citronellolis cultures was brown, and GPC analyses of the recovered polymer revealed a significant shift in the molecular weight distribution to lower values. This shift was moderate after 2 weeks (Mw, 4 × 105 to 5 × 105) and was more evident after 6 weeks (Mw, 1 × 105 to 2 × 105) (Fig. 4). The percentages of residual polymer with Mw less than 10,000 were 0, 0.4, and 7.7% (2 weeks) and 0, 9, and 32% (6 weeks) for S. lividans 1326, S. coelicolor 1A, and P. citronellolis, respectively. Analysis of the relative frequencies of the molecular weight distribution (relative Mw/M) indicated that there was a sharp cutoff in the number of molecules with molecular weights less than ≈5,000 in the polymer recovered (data not shown). These results clearly show that S. coelicolor 1A and P. citronellolis are able to cleave the carbon backbone of synthetic poly(cis-1,4-isoprene).
FIG. 4.
GPC elution profiles for the residual polymers after incubation of S. lividans 1326, S. coelicolor 1A, and P. citronellolis with synthetic poly(cis-1,4-isoprene) for 6 weeks (black line) and 2 weeks (medium grey line) and at zero time (light grey line). rel. W(logM), refractive index detector signal.
Isolation and identification of metabolites from vulcanized rubber degraded by S. coelicolor 1A.
In order to test whether rubber-degrading bacteria might also be able to degrade vulcanized rubber, in which the polymer chains have been cross-linked by sulfur bridges, we performed weight loss experiments with 100-mg pieces of vulcanized latex gloves. In contrast to a noninoculated control culture or a culture containing S. lividans 1326 (weight losses, <3%), weight losses of 10 to 18% were obtained in the presence of degrading bacteria within 6 weeks of inoculation. We were particularly interested in the isolation and identification of degradation products: 34 1-liter Erlenmeyer flasks, each containing 200 ml of mineral salts medium and a 2-g piece of autoclaved powder-free latex glove, were inoculated with a glucose-grown seed culture and were incubated with shaking at 28°C for 10 weeks. The original colorless culture turned yellow-brown, and the rubber pieces visibly disintegrated. S. coelicolor 1A cell growth was observed on the surface of the polymer and in the liquid. A noninoculated control culture showed no growth, and the glove piece remained intact. The S. coelicolor culture was harvested on day 70, and metabolites were extracted as described above. Using the chemical screening approach (1), several metabolites were detected in the crude extract by their intense green colorization reactions with anisaldehyde-H2SO4 on HPTLC plates. Green spots with the same relative mobility on HPTLC plates also occurred after growth of S. coelicolor 1A in liquid culture with films of natural (nonvulcanized) rubber but not when the strain was grown with glucose as the carbon source. Three of the green spots were isolated from the plates (Fig. 1, compounds 2 to 4). Subsequent column chromatography and GPC yielded 10 mg of pure compound 2, 12 mg of compound 3 and 1 mg of compound 4 (90% pure).
The structures of compounds 2 to 4 were elucidated unambiguously by using one-dimensional and two-dimensional nuclear magnetic resonance spectroscopy (1H-1H-correlation spectroscopy [COSY], heteronuclear multiple quantum coherence [HMQC], heteronuclear multiple bond correlation [HMBC]) and HREI-MS. The double bonds revealed the same configuration as poly(cis-1,4-isoprene).
DISCUSSION
In this study degradation of synthetic poly(cis-1,4-isoprene) was unambiguously shown for a gram-positive bacterium (S. coelicolor 1A) and a gram-negative bacterium (P. citronellolis) by increases in the protein concentration and decreases in the molecular weight of the residual polymer. The ability of P. citronellolis to degrade synthetic rubber was astonishing because this bacterium, in contrast to S. coelicolor 1A, did not produce clearing zones on opaque latex agar. Apparently, bacteria differ in the mechanisms by which the non-water-soluble polymer is degraded. This is in agreement with observations of Tsuchii and coworkers (10–12), who found that degradation of vulcanized rubber by a Nocardia isolate required colonization of the polymer, while a Xanthomonas sp. was able to degrade rubber without significant surface colonization by secreting water-soluble factors into the culture medium. One additional observation concerning the primary attack of rubber has been described for a Gordonia isolate recently (4). This strain is able to solubilize rubber to water-soluble products in a mineral salts solution with solid poly(cis-1,4-isoprene) as the sole carbon source.
On the molecular level, two basic mechanisms can be involved in polymer degradation. The exo type of degradation requires that each polymer chain is degraded from one end to the other before the next molecule is attacked. In this case the molecular weight of the residual polymer remains very high until all polymer molecules are degraded. This has not been observed for any of the rubber-degrading bacteria. In contrast, the polymer chains can be cleaved somewhere within the chain (endo type of degradation), yielding products with significantly lower molecular weights. Analysis of rubber partially degraded by S. coelicolor 1A and P. citronellolis clearly revealed a time-dependent decrease in the molecular weight to low-molecular-weight products for both strains (Fig. 4). Similar results have been found for rubbers partially degraded by all other bacteria that have been analyzed so far (data not shown). Therefore, we conclude that an endo type of mechanism is mainly involved in the primary attack of poly(cis-1,4-isoprene); however, we cannot exclude additional exo-type degradation. Analysis of the relative frequencies of the partially degraded polymer molecules showed that there was a sharp cutoff for compounds with molecular weights less than 5,000. The small degradation products might have been preferentially used by the bacteria and therefore could not be recovered from the remaining polymer.
The biochemical reactions which cleave the polymer and the subsequent reactions leading to central metabolism are unknown. Dioxygenase-mediated cleavage of the polymer has been suggested by Tsuchii and Takeda (11) for rubber degradation by a Xanthomonas sp. This suggestion was based on identification of acetonyl diprenyl acetoaldehyde (compound 1) in the culture fluid of a rubber-grown Xanthomonas sp. culture. Spectroscopical analysis of the remaining synthetic poly(cis-1,4-isoprene) clearly confirmed the presence of carbonyl groups in the surface of the partially degraded material in this and other studies (10; A. Linos, personal communication). Therefore, we assume that oxidative cleavage of the double bond of poly(cis-1,4-isoprene) is a general feature of rubber-degrading microorganisms.
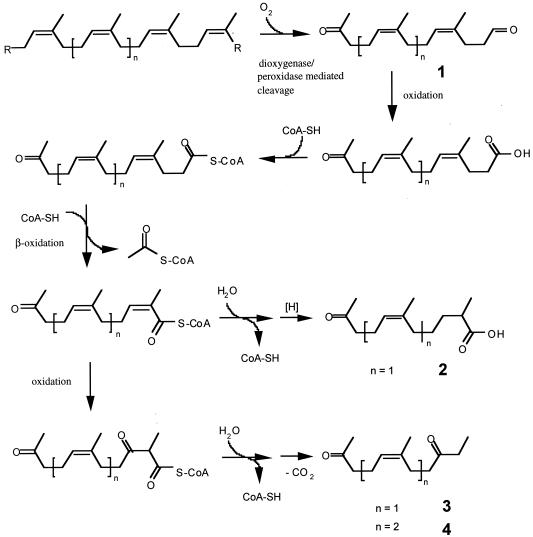
In this study we isolated three intermediates, compounds 2 to 4 (Fig. 1), from a 70-day-old culture of S. coelicolor 1A grown on vulcanized latex gloves. At the time of harvest the polymer had significantly disintegrated, and the surface was colonized with the bacteria. The long incubation time, 70 days, was chosen in order to promote the formation of stable intermediates of rubber degradation rather than accumulation of the primary degradation products. Figure 5 shows a potential biochemical route leading from intact polymer to the metabolites identified. The first step is oxidative cleavage of the double bonds of the polymer, resulting in the formation of cis-1,4-isoprene oligomers such as compound 1 (Fig. 5) with terminal keto and aldehyde groups, as has been proposed for a Nocardia sp. and a Xanthomonas sp. (10, 11). Oligomers with sufficiently low molecular weights are taken up by the bacteria, and the aldehyde group is oxidized to the corresponding carboxylic acid. The acid is activated as a coenzyme A (CoA) ester, followed by one cycle of β-oxidation. An additional complete cycle of β-oxidation might be difficult due to the presence of the α-methyl group, resulting in accumulation of this intermediate. After ester hydrolysis, the unstable β-keto acid spontaneously decarboxylates to compound 3 or 4; compounds 3 and 4 have been isolated from culture broth (Fig. 5). Alternatively, the intermediate after the first cycle of β-oxidation can be reduced to the saturated CoA ester, resulting in the free acid compound 2 after hydrolysis.
FIG. 5.
Hypothetical biochemical route for degradation of poly(cis-1,4-isoprene) to compounds 2 to 4 by S. coelicolor 1A.
It should be mentioned that compounds 2 to 4 are stable products which accumulated in the culture broth during incubation for 10 weeks but which do not necessarily represent intermediates of poly(cis-1,4-isoprene) utilization. For example, if we assume that β-oxidation can proceed slowly in S. coelicolor 1A despite the presence of α-methyl groups, propionyl-CoA would be generated. S. coelicolor 1A and P. citronellolis grew well on propionate as the sole source of carbon. Analysis of the role of the enzymatic reactions involving the primary degradation products of rubber in the central metabolism is needed.
ACKNOWLEDGMENTS
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (to K.P. and D.J.) and by a grant from the Graduiertenkolleg (GRK 227, “Chemische Aktivitäten von Mikroorganismen”) (to H.B.B. and A.Z.).
REFERENCES
- 1.Grabley S, Thiericke R, Zeeck A. The chemical screening approach. In: Grabley S, Thiericke R, editors. Drug discovery from nature. Berlin, Germany: Springer; 1999. pp. 124–148. [Google Scholar]
- 2.Heisey R M, Papadatos S. Isolation of microorganisms able to metabolize purified rubber. Appl Environ Microbiol. 1995;61:3092–3097. doi: 10.1128/aem.61.8.3092-3097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jendrossek D, Tomasi G, Kroppenstedt R M. Bacterial degradation of natural rubber: a priviledge of actinomycetes? FEMS Microbiol Lett. 1997;150:179–188. doi: 10.1016/s0378-1097(97)00072-4. [DOI] [PubMed] [Google Scholar]
- 4.Linos A, Steinbüchel A, Sproer C, Kroppenstedt R M. Gordonia polyisoprenivorans sp. nov., a rubber-degrading actinomycete isolated from an automobile tyre. Int J Syst Bacteriol. 1999;49:1785–1791. doi: 10.1099/00207713-49-4-1785. [DOI] [PubMed] [Google Scholar]
- 4a.Linos A, Reichelt R, Keller U, Steinbüchel A. Gram-negative bacterium, identified as Pseudomonas aeruginosa AL98, is a potent degrader of natural rubber and synthetic cis-1,4-polyisoprene. FEMS Microbiol Lett. 2000;182:155–161. doi: 10.1111/j.1574-6968.2000.tb08890.x. [DOI] [PubMed] [Google Scholar]
- 5.Lomovskaya N D, Mkrtumian N M, Gostimskaya N L, Danilenka V N. Characterization of the temperate actinophage φC31 isolated from Streptomyces coelicolor A2(2) J Virol. 1972;9:258–262. doi: 10.1128/jvi.9.2.258-262.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 7.Schlegel H G, Kaltwasser H, Gottschalk G. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch Mikrobiol. 1961;38:209–222. [PubMed] [Google Scholar]
- 8.Seubert W. Degradation of isoprenoid compounds by microorganisms. J Bacteriol. 1959;79:426–434. doi: 10.1128/jb.79.3.426-434.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Söhngen N L, Fol J G. Die Zersetzung des Kautschuks durch Mikroben. Zentralbl Bakteriol Parasitenkd Infektionskr Hyg Abt. 1914;40:87–98. [Google Scholar]
- 10.Tsuchii A, Suzuki T, Takeda K. Microbial degradation of natural rubber vulcanisates. Appl Environ Microbiol. 1985;50:965–970. doi: 10.1128/aem.50.4.965-970.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuchii A, Takeda K. Rubber-degrading enzyme from a bacterial culture. Appl Environ Microbiol. 1990;56:269–274. doi: 10.1128/aem.56.1.269-274.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuchii A, Tokiwa Y. Microbial degradation of natural rubber. In: Steinbüchel A, editor. Biochemical principles and mechanisms of biosynthesis and biodegradation of polymers. WeinheimNew York, N.Y.: Wiley-VCH; 1998. pp. 258–264. [Google Scholar]