Abstract
BACKGROUND
Limited information is available on the impact of immunosuppressants on COVID-19 vaccination in patients with immune-mediated inflammatory diseases (IMID).
METHODS
This observational cohort study examined the immunogenicity of SARS-CoV-2 mRNA vaccines in adult patients with inflammatory bowel disease, rheumatoid arthritis, ankylosing spondylitis, or psoriatic disease, with or without maintenance immunosuppressive therapies. Ab and T cell responses to SARS-CoV-2, including neutralization against SARS-CoV-2 variants, were determined before and after 1 and 2 vaccine doses.
RESULTS
We prospectively followed 150 subjects, 26 healthy controls, 9 patients with IMID on no treatment, 44 on anti-TNF, 16 on anti-TNF with methotrexate/azathioprine (MTX/AZA), 10 on anti–IL-23, 28 on anti–IL-12/23, 9 on anti–IL-17, and 8 on MTX/AZA. Ab and T cell responses to SARS-CoV-2 were detected in all participants, increasing from dose 1 to dose 2 and declining 3 months later, with greater attrition in patients with IMID compared with healthy controls. Ab levels and neutralization efficacy against variants of concern were substantially lower in anti-TNF–treated patients than in healthy controls and were undetectable against Omicron by 3 months after dose 2.
CONCLUSIONS
Our findings support the need for a third dose of the mRNA vaccine and for continued monitoring of immunity in these patient groups.
FUNDING
Funded by a donation from Juan and Stefania Speck and by Canadian Institutes of Health (CIHR)/COVID-Immunity Task Force (CITF) grants VR-1 172711 and VS1-175545 (to THW and ACG), CIHR FDN-143250 (to THW), GA2-177716 (to VC, ACG, and THW), and GA1-177703 (to ACG) and the CIHR rapid response network to SARS-CoV-2 variants, CoVaRR-Net (to ACG).
Keywords: COVID-19, Immunology
Keywords: Adaptive immunity, Autoimmune diseases, Immunotherapy
Introduction
The COVID-19 pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), remains a serious health crisis (1, 2). COVID-19 infections vary from asymptomatic or mild through to severe disease, with lethal complications such as progressive pneumonia, acute respiratory distress syndrome, and organ failure driven by hyperinflammation and a cytokine storm syndrome. Patients with immune-mediated inflammatory diseases (IMID), such as inflammatory bowel disease (IBD), psoriatic disease, rheumatoid arthritis (RA), and spondyloarthritis (SpA), are frequently treated with immunosuppressants and biologics and, therefore, may be at increased risk for COVID-19 (3, 4). Age and underlying comorbidities as well as the use of some immunosuppressants have been shown to be risk factors for developing COVID-19 among patients with IMID (3, 5). Glucocorticoids and combination therapy of immunomodulators and biologics have been shown to increase the risk of severe outcomes of COVID-19 (4, 6).
Although many patients with IMID mount adequate serological responses to vaccination after 2 doses of an mRNA vaccine, a proportion of patients with IMID show reduced responses compared with healthy controls (7–14), as confirmed in recent meta-analyses (15, 16). Patients receiving glucocorticoids, methotrexate (MTX), mycophenolate, anti-TNF, and B cell-depleting therapy may have attenuated serological responses to COVID-19 vaccines (7, 11, 13, 15, 17–24). Moreover, 2 recent studies showed that patients on anti-TNF therapy have greater waning of humoral immunity compared with healthy controls (13, 21).
Data regarding the cellular immune responses to vaccination are still relatively scarce and conflicting. Several studies have shown unimpaired T cell responses to SARS-CoV-2 vaccines in patients who are immunocompromised compared with healthy individuals (13, 25–28), although a follow-up study showed that a proportion of patients with IMID on immunosuppression had reduced T cell responses to a second dose of vaccine (29). In another study, MTX limited CD8+ T cell responses to vaccination in a cohort of patients with IMID (30). To gain further insight into immunity to mRNA vaccines in patients with IMID on different maintenance therapies, we investigated serological and T cell responses against SARS-CoV-2 before and after 1 or 2 doses of mRNA vaccine. The results show substantial variation in responses within different treatment groups. Notably, we observed decreased serological responses in anti-TNF–treated patients, including decreased efficacy of neutralization of variants of concern, with no neutralizing capacity against the Omicron variant. T cell cytokine production, including IFN-γ, IL-2, and IL-4, increased from 1 to 2 doses of vaccine and correlated with humoral responses. Importantly, both Ab and T cell responses in the IMID treatment groups showed greater waning by 3 months following the second dose of mRNA vaccine compared with healthy controls. These data highlight the need for third doses of SARS-CoV-2 mRNA vaccines and for continued monitoring of responses in these patients.
Results
Study population and design.
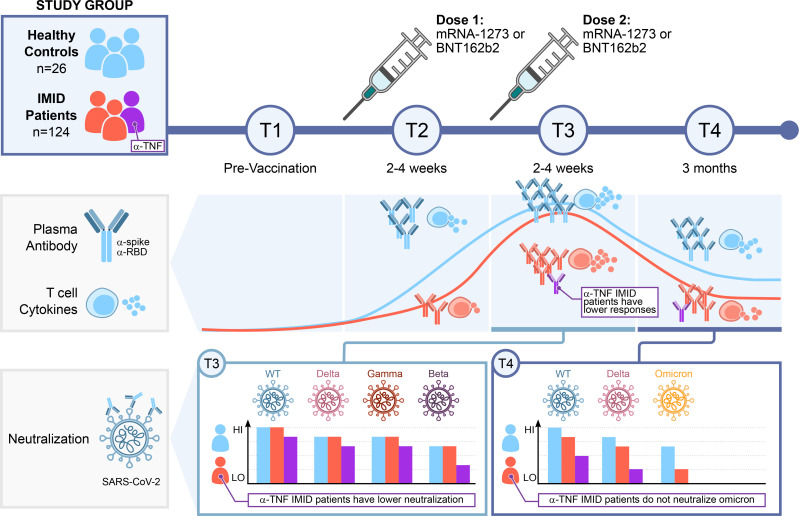
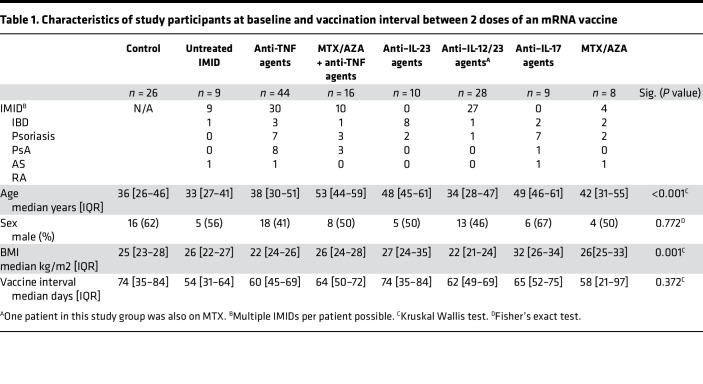
Of 177 initially recruited subjects, 150 met the inclusion criteria for this study (see methods). PBMCs and plasma were collected for T cell and Ab responses at up to 4 time points, before and after vaccination with mRNA vaccines (Figure 1A). This cohort was vaccinated according to Canadian scheduling guidelines at the time, resulting in a median time between dose 1 and dose 2 of the mRNA vaccines of 60.5 days, IQR (45.5–72). Baseline characteristics of the study subjects are shown in Table 1. Of note, age and BMI, but not vaccine interval, were significantly different between groups, and multivariate analysis of the data took these differences into account (Supplemental Tables 1–3; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.159721DS1). The patients who were ultimately analyzed included 26 healthy controls, 9 patients with IMID not on treatment, 44 patients with IMID on anti-TNF, 16 on anti-TNF with MTX/azathioprine (MTX/AZA), 10 on anti–IL-23, 28 on anti–IL-12/23, 9 on anti–IL-17, and 8 on MTX/AZA.
Figure 1. Ab responses after 1 or 2 doses of mRNA vaccine.
(A) Schematic diagram of the sampling schedule. (B) IgG response to vaccination across all participants at each time point (defined in A). Anti-spike (y-axis) and anti-RBD (x-axis) IgG levels at indicated time points. The dark blue line is the median ratio in convalescent patients (340 samples collected 21–115 days after symptom onset). The red line is the seropositivity threshold, set to pass both a 1% FPR and greater than or equal to 3 SDs from the log10 means of the negative controls. See Supplemental Table 5 for percentage of samples that pass these thresholds. (C) IgG responses to vaccination in patients with IMID. Violin plots show the relative ratios of RBD and spike at the indicated time points in patients with IMID under monotherapy and combination therapy (0.0039 μL sample used, see Supplemental Figure 1 for the second dilution and Supplemental Table 4 for conversion to BAU/mL). T1, n = 111; T2, n = 130; T3, n = 130; and T4, n = 87. The dot colors indicate the type of vaccine; Pfizer refers to BNT162b; Moderna to mRNA-1273; and mRNA mix = first dose BNT162b and second dose mRNA-1273. Black and gray lines indicate median and mean ratio values for each violin, respectively. Plots are faceted based on the groups/treatments. Comparisons were made by Dunn’s multiple comparisons test based only on the BNT162b/BNT162b group. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. M/A+TNF inh, MTX/AZA+TNF inh.
Table 1. Characteristics of study participants at baseline and vaccination interval between 2 doses of an mRNA vaccine.
Ab responses are reduced in anti-TNF–treated subjects and wane over time.
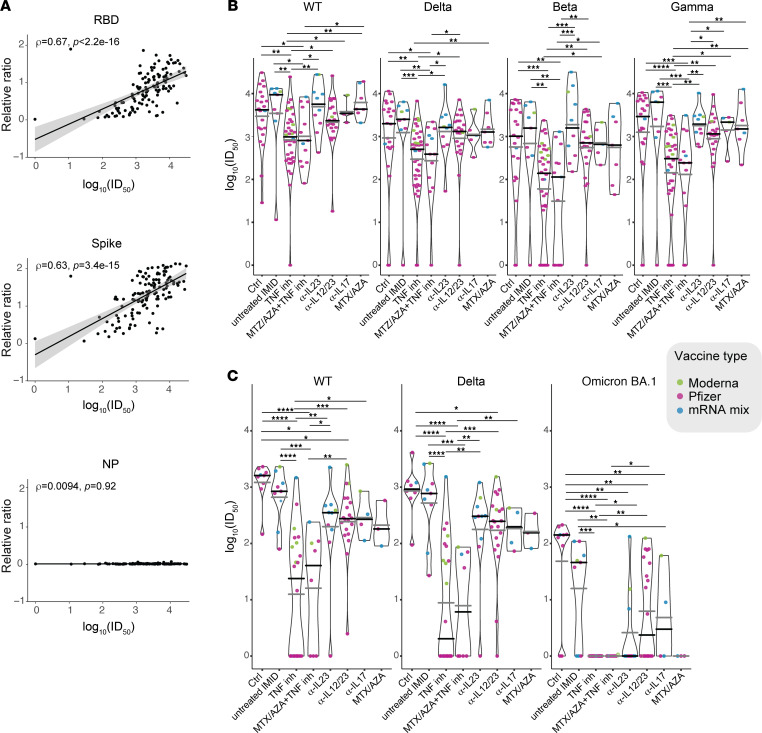
Ab responses were measured by automated ELISA (31; see methods). For the entire cohort, Ab responses increased from T1 to T2 to T3 and then decreased by T4 (Figure 1B; see Supplemental Table 4 for conversion to WHO standards). Responses to nucleocapsid (NP) were used to rule out exposure to SARS-CoV-2 (Supplemental Figure 1). After the first dose (T2), 97.6% and 80% of participants seroconverted for Coronavirus s protein (spike) and receptor binding domain (RBD) IgG, respectively, and the relative ratios were greater than the medians of the convalescents in 44.6% and 13% of the participants, respectively (Figure 1B and Supplemental Table 5). Seroconversion increased to 100% for spike and 99.2% for RBD soon after the second dose (T3) and the anti-spike and anti-RBD IgG levels were greater than the median levels of convalescent patients in 96.1% and 85.3% of participants, with a median relative ratio of 1.91 for spike and 1.55 for RBD (Figure 1B and Supplemental Table 5). Analysis of Ab responses by vaccine type at T3 showed that 2 doses of the mRNA-1273 vaccine elicits a stronger humoral response than BNT162b, with mixed mRNA vaccines inducing significantly higher levels of anti-spike and anti-RBD IgG than 2 doses of BNT162b (Supplemental Figure 2A). Although all data were included in the figures, as most of the cohort was vaccinated twice with BNT162b2, univariate statistical analysis between treatment groups was performed only on samples from the BNT162b/BNT162b participants. Among the BNT162b/BNT162b cohort, males had a slightly lower response to RBD than females, whereas Ab response differences by age were not significant (Supplemental Figure 2, B and C).
Participants undergoing anti-TNF and anti–TNF+MTX/AZA therapies had significantly lower levels of anti-RBD and anti-spike Abs than those in the healthy control, IMID–untreated, and anti–IL-12/23 groups after the first dose of vaccine (Figure 1C and Supplemental Figure 1). Comparison between the groups after the second dose (T3) indicates that for the BNT162b/BNT162b group, participants taking anti-TNF had significantly lower levels of anti-spike IgG than those in the healthy control, untreated patients with IMID, and anti–IL-12/23 groups (Figure 1C; anti-RBD was significant against the untreated IMID but not the healthy controls). Multivariate analyses of treatment groups controlling for age and sex confirmed the deficits in anti-RBD and anti-spike in the anti-TNF group after the second dose, whether the entire cohort or only the BNT162b/BNT162b participants were evaluated (Supplemental Figure 3 and Supplemental Table 1).
Anti-RBD and anti-spike Ab levels decreased by T4 (median 106 days after dose 2), with a more rapid decline in anti-RBD than anti-spike levels (Figure 1B). At that time point, only 67.8% and 50.5% of the participants showed relative ratios greater than the medians of the convalescents for spike and RBD, respectively (Figure 1B and Supplemental Table 5). When data were analyzed by study group, we again observed that the anti-TNF and anti–TNF+MTX/AZA therapy groups were associated with a statistically significant drop in anti-RBD and anti-spike IgG levels compared with the healthy control, untreated IMID, and anti–IL-12/23 groups (Figure 1C and Supplemental Table 1). Multivariate analyses of treatment groups controlling for age and sex confirmed the deficits in anti-RBD and anti-spike in the anti-TNF and anti–TNF+MTX/AZA groups at T4, whether the entire cohort or only the BNT162b/BNT162b participants were evaluated (Supplemental Figure 3 and Supplemental Table 1).
Patients with IMID undergoing anti-TNF therapy show significantly lower neutralization responses than other groups.
To verify whether the observed deficits in binding Ab detected by ELISA were accompanied by alterations in neutralization potential, we performed spike-pseudotyped lentiviral neutralization assays with serum from T3 and T4 using spike protein from either the WT strain or the B.1.351 (Beta), P.1 (Gamma), B.1.617.2 (Delta), and/or B.1.1.529 (Omicron/BA.1) variants of concern (VOCs). Across all participants at T3, samples neutralized the WT strain more efficiently than any of the VOCs tested (Supplemental Figure 4A). Previous studies have shown that Ab binding to either spike or RBD generally correlates with neutralization activity (32). Consistently, Spearman’s correlations (ρ = 0.59–0.67) were detected between anti-spike or anti-RBD, but not NP IgG levels and neutralization of either the WT lentivirus (Figure 2A) or each of the VOCs (Supplemental Figure 4B). Participants on anti-TNF and anti–TNF+MTX/AZA showed significantly lower neutralization response to the WT and all variants, as compared with controls or untreated IMID groups (Figure 2B), consistent with the ELISA data.
Figure 2. Variant neutralization after 2 doses of vaccine.
(A) Spearman’s correlation at T3 between the indicated Ab levels determined by ELISA (y axis) and the neutralization of the WT spike lentivirus (x axis; see Supplemental Figure 4B for correlations with the VOCs). (B and C) Violin plots of log10 (ID50) — the serum dilution that inhibits 50% of the lentivirus infection — values of samples at B, T3 (2–4 weeks after dose 2), n = 129; and C, T4 (3 months after dose 2), n = 86. Lentiviral particles used: WT (Wuhan Hu-1 sequence with a D614G mutation); B.1.617.2 (Delta); B.1.351 (Beta); P.1 (Gamma); and B.1.1.529 (Omicron, BA.1). The distribution is stratified by study groups/treatments. The dots colors indicate the type of vaccine. Black lines indicate the median and the gray lines the mean ratio value for each violin. Comparisons were made by Dunn’s multiple comparisons test for the entire cohort. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
In accordance with the waning binding Ab levels detected across all samples at T4, median neutralization was reduced in comparison to T3 for both the WT and Delta lentiviruses that were profiled across both time points (Supplemental Figure 4, compare Supplemental Figure 4, A and C). The participants on anti-TNF and anti–TNF+MTX/AZA again showed a significantly lower neutralization response in comparison with the control groups against the WT and Delta lentiviruses (Figure 2C). Reduction in the neutralization ability in other treatment groups was also observed at T4, including the anti–IL-12/IL-23 group, which showed significantly lower neutralization of both the WT and the Delta lentiviruses as compared with the control group (Figure 2C). Consistent with recent reports (33–37), Omicron spike-pseudotyped lentiviral particles were about an order of magnitude more difficult to neutralize than the WT and Delta variants across the T4 samples (Supplemental Figure 4C). Moreover, sera from anti-TNF, anti–TNF+MTX/AZA, or MTX/AZA showed no detectable neutralization of Omicron in our assay, with all treatment groups showing significant defects in neutralization compared with the controls (Figure 2C). Overall, these data demonstrate weaker neutralization responses to mRNA vaccines at all time points and for all VOCs tested for the participants on anti-TNF agents, and a mixed response for the other treatment groups, with an exacerbation of these deficits at T4 and against Omicron.
Patients with IMID show increased T responses to successive vaccine doses, with greater waning after dose 2.
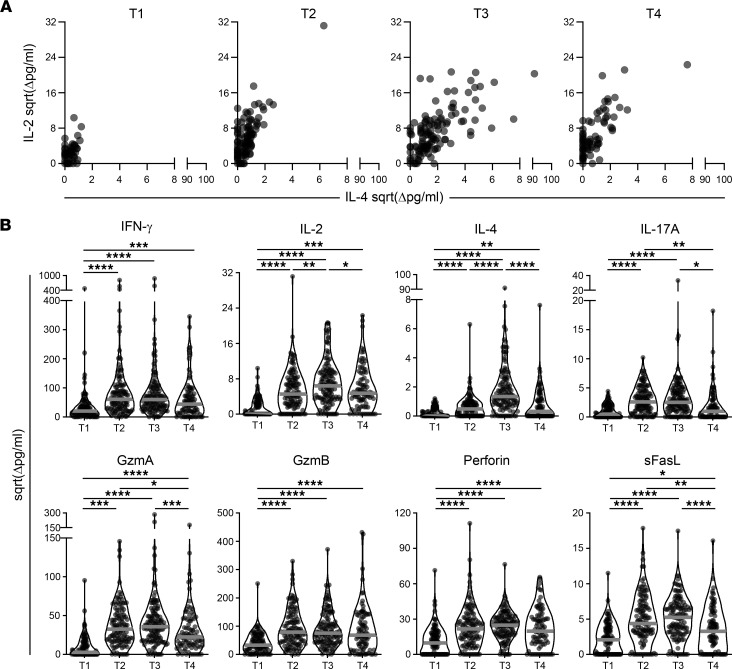
To assess memory T cell responses to SARS-CoV-2, PBMCs were stimulated with spike or NP peptide pools for 48 hours. A quantitative multiplex bead-based immunoassay was used to measure the levels of 9 secreted cytokines and cytotoxic molecules in the supernatants in response to spike peptide stimulation and results are reported after subtracting the values from negative control wells. The response to NP was used as an additional control to detect memory responses to previous virus exposure. NP-specific responses prevaccination were minimal, consistent with study subjects being SARS-CoV-2 naive and suggesting minimal impact of cross-reactive T cells from previous coronavirus infections (Supplemental Figure 5). The cytokines IFN-γ, IL-2, IL-17A, and IL-4 were increased over baseline (T1) after 1 or 2 doses of mRNA vaccine in all patient groups (T2 and T3), with the response predominantly of the Th1 phenotype as characterized by high levels of IFN-γ and IL-2 (Figure 3 and Supplemental Figure 6). Molecules associated with cytotoxicity such as granzyme A (GzmA), GzmB, perforin, and sFasL were also increased over baseline following 1 dose of vaccine and did not consistently increase with the second dose (Figure 4 and Supplemental Figure 6). TNF was not detected over baseline (data not shown). Most study groups showed a wide range of responses to spike peptide pools after first or second vaccine doses (Figures 3 and 4, and Supplemental Figure 6). When multivariate analysis was performed on the BNT162b/BNT162b group only, after controlling for age and sex, we observed deficits in IFN-γ production in the untreated IMID, anti-TNF, MTX/AZA, anti–IL-12/23 and anti–IL-23 treatment groups relative to healthy controls at T2, which had largely recovered by T3 (Supplemental Figure 7 and Supplemental Table 1). However, by T4, IFN-γ and IL-2 responses were lower in most treatment groups as well as in untreated patients with IMID relative to controls (Supplemental Figure 7 and Supplemental Table 1). When results from all subjects were pooled, there was an increase in response from first to second dose for all 8 readouts (Figure 5, A and B). By 3 months after dose 2 (T4), we saw an overall decrease in IL-2, IL-4, IL-17, sFasL, and GzmA (Figure 5, A and B). We also noted higher IL-4 responses following vaccination with mRNA-1273 compared with BNT162b or mixed doses (Supplemental Figure 8A). Although T cell responses overall were similar based on age or sex (Supplemental Figure 8, B and C), multivariate analysis revealed lower IL-4 responses in the over 60 group (Supplemental Table 2).
Figure 3. Cytokine responses in each group prior to vaccination and after first and second doses of mRNA vaccine.
Cytokine release in cell culture supernatants was analyzed by multiplex bead array following 48 hours stimulation with SARS-CoV-2 spike peptide pools. Violin plots show IFN-γ, IL-2, IL-17A, and IL-4 release at T1 (prevaccination), n = 100; T2, n = 114; T3, n = 123; and T4, n = 85, with time points defined in Figure 1A. Colored dots represent the type of vaccine as indicated in the inset legend. The gray line indicates the median. Values are reported in pg/mL after subtracting background signal from wells containing PBMCs cultured with DMSO alone, as indicated by “Δ”. Ctrl = Healthy controls, inh = inhibitor. Comparisons between groups in entire cohort were made by Dunn’s multiple comparisons test after excluding outliers and subjects with an NP IgG response. *P ≤ 0.05, **P ≤ 0.01.
Figure 4. Cytotoxic responses in each group before or after first and second doses of mRNA vaccine.
The release of cytotoxic molecules in cell culture supernatants was analyzed by multiplex bead array following 48 hours stimulation with SARS-CoV-2 spike peptide pools. Violin plots show release of GzmA, GzmB, perforin, or sFASL release at T1, n = 100; T2, n = 114; T3, n = 123; and T4, n = 85 (with T1–T4 defined in Figure 1A). The dot colors indicate the type of vaccine as indicated in the inset legend. The gray line indicates the median. Values are reported in pg/mL after subtracting background signal from wells containing PBMCs cultured with DMSO alone, as indicated by Δ. Ctrl = Healthy controls, inh = inhibitor. Comparisons on entire cohort were made by Dunn’s multiple comparisons test after excluding outliers and subjects with an NP IgG response. *P ≤ 0.05.
Figure 5. Cytokine and cytotoxic responses in all patient groups in response to spike peptide pools over time.
The release of cytokines in cell culture supernatants were analyzed by multiplex bead array following 48 hours stimulation with SARS-CoV-2 S peptide pools. (A) IL-2 and IL-4 responses across all participants at time points T1–T4 as defined in Figure 1A: T1, n = 100; T2, n = 114; T3, n = 123; and T4, n = 85. (B) Violin plots show release of cytokines and cytotoxic molecules in all study subjects pooled. The median is indicated by the gray line. Pairwise comparisons were made by 1-way mixed-effects ANOVA after excluding outliers and subjects with an NP IgG response. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
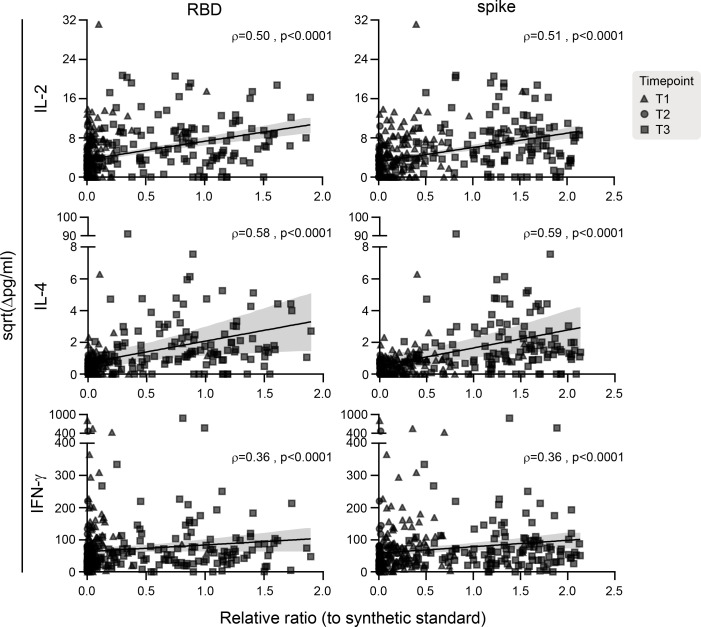
Levels of secreted IL-2 were positively correlated with plasma IgG against RBD (ρ = 0.50) and whole spike trimer (ρ = 0.51). Similarly, there was a positive correlation between IL-4 and plasma IgG against RBD (ρ = 0.58) and whole spike trimer (ρ = 0.59) and between IFN-γ and RBD IgG (ρ = 0.36) and whole spike trimer IgG (ρ = 0.36) (all P values < 0.0001) (Figure 6).
Figure 6. Correlation between IgG levels and T cell cytokine responses at all sampled time points.
The solid black line is the linear regression, and the gray shading indicates the 95% CI. P values and Spearman’s rho coefficients are indicated in each graph, n = 337.
Discussion
Here we studied a cohort of patients with IBD, psoriatic disease, ankylosing spondylitis (AS), or RA treated with biologics (anti-TNF, anti–IL-12/23, anti–IL-23, and anti–IL-17) or antimetabolites to assess their response to COVID-19 mRNA vaccines. There is limited information available on the degree of immunosuppression in this group, raising concern as to how their treatments could impact the response to the vaccines. Although there was considerable variability within groups, 100% of participants seroconverted for spike after 2 doses of vaccine. There was also a clear indication of higher responses to mRNA-1273 vaccine compared with BNT162b vaccine with respect to Ab levels and neutralization titers as well as T cell IL-4 production. Of concern, Ab levels and neutralization activity were lower in the anti-TNF–treated study subjects after dose 2, with neutralization of the Omicron spike-pseudotyped lentivirus undetectable in this group at that time point. We also note that the responses of other treatment groups are markedly reduced against Omicron at T4 compared with healthy controls. Our data are consistent with recent data suggesting reduced vaccine efficacy against Omicron infection in patients who are immunocompromised (14). The observed waning of Ab responses to mRNA vaccines in anti-TNF patients agree with 2 other recently published studies (13, 21). We also observed that patients with IMID overall showed more substantial waning of both Ab and T cell responses compared with healthy controls.
The vaccine dose interval used in our study was a median of 60.5 days rather than the standard 21 or 28 days. This was due to the policy in place in Canada at the time, to maximize first doses when vaccines were in limited supply. Subsequent analysis has shown that an interval between vaccine dose 1 and vaccine dose 2 of greater than 8 weeks resulted in enhanced humoral and T cell immunity in healthy subjects (38); therefore, our longer vaccine interval could impact the results relative to studies that used the standard interval. A limitation of our study is that several of our study groups, particularly those on anti–IL-17, anti–IL-23, or MTX/AZA, as well as the untreated patients with IMID, were underpowered, making it difficult to draw conclusions about these specific groups, although they do contribute to the overall analysis of the patients with IMID as a group. Additional limitations are that we grouped patients together by drug class, regardless of disease or the specific drug product, which may contribute heterogeneity to our results. However, we were underpowered to investigate these differences further.
T cell responses, including IL-4, IL-2, and IFN-γ production, showed a significant correlation with RBD and spike-specific Ab responses. There was substantial induction of T cell cytokines and release of cytotoxic molecules following spike peptide pool stimulation of PBMCs collected following 1 dose of vaccine and this increased further for all readouts after 2 doses. Multivariate analysis of the data showed that several groups had decreased IFN-γ after dose 1 of vaccine, but these deficits were largely corrected following the second vaccine dose. When data were pooled for all subjects, it was apparent that cytokine responses including IL-4 and IL-17 were dependent on 2 doses of vaccine. IL-4 is an important mediator of B cell proliferation, which in turn impacts Ab levels and B cell memory (39). This lower IL-4 response after 1 dose as compared with 2 doses of vaccine highlights the need for second doses to maximize B cell responses. Of note, a recent report showed that atopic dermatitis as well as patients with asthma treated with either IL-4 or IL-5 receptor antagonists had reduced Ab responses following 2 doses of mRNA vaccines compared with healthy controls, consistent with the importance of T cell IL-4 in the Ab response to SARS-CoV-2 mRNA vaccines (40).
Taken together, our study shows generally robust T cell responses in most patient groups treated with immunosuppressants or biologics after 2 doses of mRNA vaccine, improving with a second dose but with significantly more attenuation in patients with IMID than healthy controls by 3 months after the second dose. We observed substantial deficits in Ab responses even after 2 doses of vaccine in the anti-TNF–treated patients, with more substantial waning immunity by 3 months after dose 2 and a complete inability to neutralize the Omicron variant. These findings highlight the need for a third vaccine dose, particularly in patients undergoing treatment with anti-TNF agents. As there is limited information available about the duration of immune memory induced by mRNA vaccines, it will also be important to follow these responses for longer time periods and to evaluate the impact of additional vaccine doses in this cohort as well as the possible contribution of natural infection to persistence of the immune response.
Methods
Study design and participants.
Patient recruitment: In this observational multicenter cohort study, we investigated the IMmune resPonse After COVID-19 vaccination during maintenance Therapy in immune-mediated inflammatory diseases (IMPACT). Patients with IMID being treated at Mount Sinai Hospital, University Health Network/Toronto Western Hospital or Women’s College Hospital who were receiving BNT162b (Pfizer-BioNTech) and/or mRNA-1273 (Moderna) SARS-CoV-2 vaccines were recruited between January 8 and October 4, 2021. In Canada, the vaccine schedule between dose 1 and 2 was increased from the standard 21 or 28 days to allow faster rollout of dose 1 and, as a result, in our cohort there was a median of 60.5 days, IQR (45.5–72) between the 2 doses.
Inclusion criteria for this study were adult patients with IMID being treated with anti-TNF therapies (infliximab, adalimumab, golimumab, etanercept, or certolizumab pegol), anti–IL-17 therapy (ixekizumab, secukinumab), MTX or AZA monotherapy, combination therapy of MTX/AZA plus anti-TNF therapies, anti–IL-12/23 (ustekinumab) therapy, anti–IL-23 therapy (guselkumab, risankizumab), or no immunosuppressants. A group of healthy volunteers, without an IMID and without immunosuppression, were also recruited as a control cohort. Excluded were individuals younger than 18 years, those who had a past SARS-CoV-2 infection, patients on vedolizumab or oral steroids, and those receiving COVID-19 vaccines other than mRNA.
Sample and data collection: Patient information and medical history were collected at each visit. Participation was terminated when all the blood samples were collected or when a patient opted out. Clinical data included basic demographics (age, sex, weight, and height), relevant past medical and surgical history, and medication use at inclusion. Questions about prior COVID-19 diagnosis or exposure, vaccination history and side effects, changes in medical history or medication, and disease activity were collected at each study visit. Blood samples were drawn from the participants at up to 4 time points: T1 = prevaccination, T2 = median 26 days after dose 1, T3 = median 16 days after dose 2, and T4 = median 106 days after dose 2. Peripheral blood samples were collected in BD Vacutainer sodium heparin tubes for plasma Ab assessment and PBMC separation. All samples were labelled with unique patient identifiers. Researchers were blinded to the identity and clinical details of the subjects. Plasma samples were stored at –80°C. PBMCs were isolated by density centrifugation using Ficoll-Paque PLUS (GE Healthcare). PBMCs were cryopreserved in 10% DMSO in FBS (Wisent Bioproducts) and stored in liquid nitrogen at a minimum of 2 x 106 mononuclear cells per vial.
Automated ELISAs.
Frozen plasma was thawed and treated with 1% final Triton X-100 for 1 hour. Samples were analyzed by automated ELISA for IgGs to the spike trimer (spike), the spike RBD, and the NP. All antigens and secondary Abs are produced in mammalian cells and were provided by Yves Durocher at the National Research Council of Canada (NRC) Montréal, QC, Canada as previously reported (31). Luminescence values for each sample/assay were normalized to synthetic standards profiled in a 4-fold dilution series on each plate (Human anti-nucleocapsid IgG, clone HC2003, catalog A02039, GenScript; and humanized anti-RBD/spike IgG VHH72hFc1X7, NRC). The synthetic references, as well as a pool of positive samples from convalescent patients with high IgG levels to all 3 antigens and negative controls (pre-COVID era samples, blank and IgG, 1 μg/mL; I4506, Millipore-Sigma) were also added to each plate in a 4-fold dilution series to enable quality controls across the plates and batches of samples. For each assay, log10 raw values and relative ratio of samples were compared with prior runs to confirm that the sample density distribution is within range; and automated scripts, blinded to sample description and metadata, were used to extract relative ratios to the synthetic references. The assay was calibrated to the WHO reference (Code 20/136, National Institute for Biological Standards and Control); a table of conversion of relative ratios for each assay to Binding Antibody Units/mL (BAU/mL) is provided (Supplemental Table 4). Seropositivity was defined based on both receiver operating characteristic (ROC) analysis of negative (pre-COVID era) and positive (PCR-confirmed COVID-19 cases) samples with a less than 1% false positive rate (FPR) threshold and greater than or equal to 3 SDs from the log means of the negative controls. In some of the figures, the median convalescent values for serum samples from 340 PCR-confirmed COVID-19 cases 21–115 days after symptom onset (31) are displayed as reference points. Since the assays saturate in healthy controls after 2 doses of vaccine, all samples were processed both at the dilution used for determination of seroconversion and a 1/16 further dilution for evaluation of the quantitative differences in Ab responses.
Spike-pseudotyped lentivirus neutralization assays.
The lentivirus neutralization assay and the generation of spike-pseudotyped lentivirus particles were performed as described previously (32). Briefly, the lentivirus particles were generated by co-transfection in HEK293TN cells (LV900A-1, System Biosciences) of the Wuhan Hu-1 sequence with a D614G mutation (WT SARS-CoV-2) or the variants B.1.617.2 (Delta), B.1.351 (Beta), P.1 (Gamma), and B.1.1.529 (Omicron) constructs with packaging (psPAX2, catalog 12260, Addgene) and reporter (luciferase-expressing pHAGE-CMV-Luc2-IRES-ZsGreen-W provided by Jesse Bloom and Katharine Crawford, Fred Hutchison Cancer Research Center, Seattle, WA, USA) constructs. Heat-inactivated (30 min at 56°C) plasma was serially diluted and incubated with the lentiviral particles (1 h, 37°C) prior to addition to cells (HEK293T-ACE2/TMPRSS2), previously described (32) for 48 hours; and luminescence signals were detected with the Bright-Glo Luciferase assay system (catalog E2620, Promega) on an EnVision multimode plate reader (Perkin Elmer). GraphPad Prism 9 was used to calculate 50% neutralization titer (ID50) using nonlinear regression. The WHO International Standard (20/136) was evaluated in this assay, and a mean ID50 value of 5744 corresponded with 1000 IU/mL.
T cell cytokine secretion assay.
Cellular immune responses to COVID-19 vaccination were determined by measuring the release of cytokines and cytotoxic molecules in cell culture supernatants following stimulation with peptide arrays using the LEGENDplex multiplex bead assay as previously described (41, 42). Briefly, 1 x 106 PBMCs were seeded per well in 96-well round bottom plates with 1 μg/mL each of SARS-CoV-2 spike or NP peptide pools (JPT Peptide Technologies). PBMCs were cultured with anti-CD28 (clone 9.3, Bio X Cell) and anti-CD3 (clone OKT3, Bio X Cell) as a positive control, or with equimolar DMSO as a negative control. Samples with no response to positive control were not included in the analysis. After 48 hours incubation at 37°C, cell culture supernatants were collected and stored at –80°C. Release of cytokines and cytotoxic molecules (IL-2, IL-4, IL-17, IFN-γ, TNF, GzmA, GzmB, Perforin, and sFASL) in the supernatants were analyzed using LEGENDplex CD8/NK multiplex cytokine bead assay (BioLegend) as per manufacturer’s instructions. Samples were acquired on the BD LSR Fortessa flow cytometer using BD FACSDiva software. Data are reported as square root (sqrt) transformed values in pg/mL after subtracting background signal from wells containing PBMCs cultured with DMSO containing media alone, as indicated by “Δ”.
Statistics.
T cell cytokine secretion data were analyzed using the LEGENDplex Data Analysis Software Suite, pandas data analysis library for Python, and GraphPad Prism v9.3.1 (43). Ab data were analyzed with R (version 4.1.1) using package ggplot2 and custom R scripts. GraphPad Prism v 9.2.0 was used to analyze the neutralization and Ab data. Models controlled for baseline (T1) T cell/Ab data and included an interaction term between the time point and the variable of interest. All multivariate analyses were performed using R (version 4.1.1) and SAS 9.4. Longitudinal multivariate analysis on Ab data and T cell cytokine secretion was performed using linear mixed models and P ≤ 0.05 was considered statistically significant.
Study approval.
This study was approved by the ethics boards of the University of Toronto (REB protocol 27673), Mount Sinai Hospital/Sinai Health System (MSH REB 21-0022-E), University Health Network-Toronto Western Hospital division (REB 21-5096), and Women’s College Hospital (REB approval 2021-0023-E). Written informed consent was obtained from all participants prior to participation.
Author contributions
RMD and JCL shared first authorship, order was decided by mutual agreement. MSS and THW conceived the study and obtained funding. ACG and THW supervised the laboratory assays, analyzed data, acquired funding, and wrote the manuscript. RDI, NH, VP, VC, and MSS contributed to the study design, acquired funding, supervised clinical coordinators, and contributed to data interpretation and manuscript editing. RMD performed neutralization assays, analyzed all Ab data, conducted statistical analysis, prepared figures, and wrote the manuscript. JCL designed and performed T cell assays, analyzed all T cell data, conducted statistical analysis, prepared figures, and wrote the manuscript. RLG contributed to the study conception and design, data analysis, literature review, and wrote the manuscript. GYCC provided overall project management, including patient recruitment and validation of data records. MS conducted statistical analysis. NVB, MG, IL, RL, and MWC assisted with PBMC preparation and/or T cell experiments. BR was responsible for sample intake and ELISA assays. RS and QH generated and titrated the lentiviral stocks and optimized the neutralization assays for VOCs. WRH generated all spike-pseudotyped lentiviral vectors. KTA and JK wrote scripts for data analysis of the Ab data and generated figures. NF contributed to patient recruitment, the literature review, and manuscript preparation. DC contributed to patient recruitment. JMS, DP, LA, SR, RM, KR, and DG contributed to study coordination, and patient recruitment.
Supplementary Material
Acknowledgments
We thank Juan and Stefania Speck for their donation to the University of Toronto for this study. We thank Jesse Bloom and Katharine Crawford for the initial spike lentiviral construct. All antigens and protein reagents for the automated ELISAs were a gift from Yves Durocher at the NRC of Canada generated within the NRC’s Pandemic Response Challenge Program. We thank all members of the serology team at the Network Biology Collaborative Centre for help with ELISA assay development and automated ELISA processing, and in particular Karen Colwill and Adrian Pasculescu for providing the BAU/mL conversion table. We thank Birinder Ghumman for technical assistance and Nathalie Simard and Janine Charron for flow cytometry support. We thank the Royal Bank of Canada and the Krembil Foundation of the Sinai Health System Foundation for donations to fund initial assay development in the Gingras lab. The calibration of the automated assays was supported notably through the COVID-19 Immunity Task Force (CITF). The robotics equipment used is housed in the Network Biology Collaborative Centre at the Lunenfeld-Tanenbaum Research Institute, a facility supported by the Canada Foundation for Innovation, the Ontario Government, and Genome Canada and Ontario Genomics (OGI-139). ACG is the Canada Research Chair in Functional Proteomics and the lead of the functional genomics and structure-function pillar of CoVaRR-Net. THW holds the Canada Research Chair in anti-viral immunity at the University of Toronto. VC is supported by a Pfizer Chair Research Award, Rheumatology, University of Toronto. JCL was supported by an Ontario Graduate Scholarship. KTA and MG were supported by CIHR CGS-D studentships and JK is supported by a NSERC PGS-D studentship.
Version 1. 04/26/2022
In-Press Preview
Version 2. 06/08/2022
Electronic publication
Funding Statement
Private donation to the University of Toronto
Footnotes
Conflict of interest: RDI has served as a consultant for AbbVie, Janssen, Eli Lilly, and Novartis and has received research funding support from AbbVie and Novartis. VP has no personal financial ties with any pharmaceutical company, but has received honoraria for speaker and/or advisory board member roles from AbbVie, Almirall, Celgene, Janssen, Kyowa Kirin, LEO Pharma, Novartis, Pfizer, Sanofi, UCB, and Union Therapeutics. In his role as Department Division Director of Dermatology at the University of Toronto, VP has received departmental support in the form of unrestricted educational grants from AbbVie, Bausch Health, Celgene, Janssen, LEO Pharma, Eli Lilly, L’Oréal, NAOS, Novartis, Pfizer, Pierre-Fabre, Sandoz, and Sanofi in the past 36 months. VC has received research grants from AbbVie, Amgen, and Eli Lilly and has received honoraria for advisory board member roles from AbbVie, Amgen, BMS, Eli Lilly, Janssen, Novartis, Pfizer, and UCB. His spouse is an employee of AstraZeneca. MSS has received research support, consulting fees and speaker honoraria from AbbVie, Janssen, Takeda, Pfizer, Gilead, and Amgen.
Copyright: © 2022, Dayam et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2022;7(11):e159721.https://doi.org/10.1172/jci.insight.159721.
Contributor Information
Roya M. Dayam, Email: monicadayam@gmail.com.
Jaclyn C. Law, Email: jaclyn.law@mail.utoronto.ca.
Rogier L. Goetgebuer, Email: Rogier.Goetgebuer@sinaihealth.ca.
Gary Y.C. Chao, Email: gary.chao@utoronto.ca.
Kento T. Abe, Email: kabe@lunenfeld.ca.
Mitchell Sutton, Email: Mitchell.Sutton@uhnresearch.ca.
Naomi Finkelstein, Email: Naomi.Finkelstein@uhnresearch.ca.
Joanne M. Stempak, Email: joanne.stempak@sinaihealth.ca.
Daniel Pereira, Email: Daniel.Pereira@uhnresearch.ca.
David Croitoru, Email: david.croitoru@utoronto.ca.
Lily Acheampong, Email: Lily.Acheampong@wchospital.ca.
Saima Rizwan, Email: Saima.Rizwan@sinaihealth.ca.
Klaudia Rymaszewski, Email: Klaudia.Rymaszewski@sinaihealth.ca.
Raquel Milgrom, Email: Raquel.Milgrom@sinaihealth.ca.
Darshini Ganatra, Email: Darshini.Ganatra@uhnresearch.ca.
Nathalia V. Batista, Email: nathalia.vieirabatista@utoronto.ca.
Melanie Girard, Email: melanie.girard@mail.utoronto.ca.
Irene Lau, Email: ihc.lau@mail.utoronto.ca.
Ryan Law, Email: ryan.law@mail.utoronto.ca.
Michelle W. Cheung, Email: michellew.cheung@mail.utoronto.ca.
Bhavisha Rathod, Email: bhavisha121@gmail.com.
Julia Kitaygorodsky, Email: julia.kitaygorodsky@mail.utoronto.ca.
Reuben Samson, Email: rsamson@lunenfeld.ca.
Queenie Hu, Email: queeniehu@gmail.com.
W. Rod Hardy, Email: whardy@lunenfeld.ca.
Nigil Haroon, Email: Nigil.Haroon@uhn.ca.
Robert D. Inman, Email: robert.inman@uhn.on.ca.
Vincent Piguet, Email: vincent.piguet@utoronto.ca.
Vinod Chandran, Email: vinod.chandran@uhnresearch.ca.
Mark S. Silverberg, Email: msilverberg@mtsinai.on.ca.
Anne-Claude Gingras, Email: gingras@lunenfeld.ca.
Tania H. Watts, Email: tania.watts@utoronto.ca.
References
- 1.Chen N, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner EJ, et al. Corticosteroids, but not TNF antagonists, are associated with adverse COVID-19 outcomes in patients with inflammatory bowel diseases: results from an international registry. Gastroenterology. 2020;159(2):481–491. doi: 10.1053/j.gastro.2020.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akiyama S, et al. Prevalence and clinical outcomes of COVID-19 in patients with autoimmune diseases: a systematic review and meta-analysis. Ann Rheum Dis. doi: 10.1136/annrheumdis-2020-218946. [published online October 13, 2020]. [DOI] [PubMed] [Google Scholar]
- 5.Gianfrancesco M, et al. Characteristics associated with hospitalisation for COVID-19 in people with rheumatic disease: data from the COVID-19 Global Rheumatology Alliance physician-reported registry. Ann Rheum Dis. 2020;79(7):859–866. doi: 10.1136/annrheumdis-2020-217871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brenner EJ, et al. IBD in the COVID-19 era: the value of international collaboration. Lancet Gastroenterol Hepatol. 2020;5(10):887–888. doi: 10.1016/S2468-1253(20)30269-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furer V, et al. Immunogenicity and safety of the BNT162b2 mRNA COVID-19 vaccine in adult patients with autoimmune inflammatory rheumatic diseases and in the general population: a multicentre study. Ann Rheum Dis. 2021;80(10):1330–1338. doi: 10.1136/annrheumdis-2021-220647. [DOI] [PubMed] [Google Scholar]
- 8.Kappelman MD, et al. Humoral immune response to messenger RNA COVID-19 vaccines among patients with inflammatory bowel disease. Gastroenterology. 2021;161(4):1340–1343. doi: 10.1053/j.gastro.2021.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rubbert-Roth A, et al. Anti-SARS-CoV-2 mRNA vaccine in patients with rheumatoid arthritis. Lancet Rheumatol. 2021;3(7):e470–e472. doi: 10.1016/S2665-9913(21)00186-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kearns P, et al. Examining the immunological effects of COVID-19 vaccination in patients with conditions potentially leading to diminished immune response capacity — The OCTAVE Trial [preprint]. Posted on Lancet August 23, 2021. [DOI]
- 11.Kennedy NA, et al. Infliximab is associated with attenuated immunogenicity to BNT162b2 and ChAdOx1 nCoV-19 SARS-CoV-2 vaccines in patients with IBD. Gut. 2021;70(10):1884–1893. doi: 10.1136/gutjnl-2021-324789. [DOI] [PubMed] [Google Scholar]
- 12.Geisen UM, et al. Immunogenicity and safety of anti-SARS-CoV-2 mRNA vaccines in patients with chronic inflammatory conditions and immunosuppressive therapy in a monocentric cohort. Ann Rheum Dis. 2021;80(10):1306–1311. doi: 10.1136/annrheumdis-2021-220272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geisen UM, et al. Humoral protection to SARS-CoV2 declines faster in patients on TNF alpha blocking therapies. RMD Open. 2021;7(3):e002008. doi: 10.1136/rmdopen-2021-002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tseng HF, et al. Effectiveness of mRNA-1273 against SARS-CoV-2 Omicron and Delta variants. Nat Med. doi: 10.1038/s41591.022.01753-y. [published online February 21, 2022]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakuraba A, et al. Serologic response to coronavirus disease 2019 (COVID-19) vaccination in patients with immune-mediated inflammatory diseases: a systematic review and meta-analysis. Gastroenterology. 2022;162(1):88–108. doi: 10.1053/j.gastro.2021.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jena A, et al. Response to SARS-CoV-2 vaccination in immune mediated inflammatory diseases: systematic review and meta-analysis. Autoimmun Rev. 2022;21(1):102927. doi: 10.1016/j.autrev.2021.102927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deepak P, et al. Effect of immunosupperession on the immunogenicity of mRNA vaccines to SARS-CoV-2. Ann Intern Med. 2021;174:1572–1585. doi: 10.7326/M21-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmed S, et al. Postvaccination antibody titres predict protection against COVID-19 in patients with autoimmune diseases: survival analysis in a prospective cohort. Ann Rheum Dis. doi: 10.1136/annrheumdis-2021-221922. [published online February 8, 2022]. https://doi.org/10.1136/annrheumdis–2021–221922 . [DOI] [PubMed] [Google Scholar]
- 19.Alexander JL, et al. COVID-19 vaccine-induced antibody responses in immunosuppressed patients with inflammatory bowel disease (VIP): a multicentre, prospective, case-control study. Lancet Gastroenterol Hepatol. 2022;7(4):342–352. doi: 10.1016/S2468-1253(22)00005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Edelman-Klapper H, et al. Lower serologic response to COVID-19 mRNA vaccine in patients with inflammatory bowel diseases treated with anti-TNFα. Gastroenterology. 2022;162(2):454–467. doi: 10.1053/j.gastro.2021.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin S, et al. Ab decay, T cell immunity and breakthrough infections following two SARS-CoV-2 vaccine doses in inflammatory bowel disease patients treated with infliximab and vedolizumab. Nat Commun. 2022;13(1):1379. doi: 10.1038/s41467-022-28517-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mandl P, et al. Response to SARS-CoV-2 vaccination in systemic autoimmune rheumatic disease depends on immunosuppressive regimen: a matched, prospective cohort study. Ann Rheum Dis. doi: 10.1136/annrheumdis-2021-221788. [published online March 18, 2022]. https://doi.org/10.1136/annrheumdis–2021–221788 . [DOI] [PubMed] [Google Scholar]
- 23.Melmed GY, et al. Antibody responses after SARS-CoV-2 mRNA vaccination in adults with inflammatory bowel disease. Ann Intern Med. 2021;174(12):1768–1770. doi: 10.7326/M21-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wieske L, et al. Humoral responses after second and third SARS-CoV-2 vaccination in patients with immune-mediated inflammatory disorders on immunosuppressants: a cohort study. Lancet Rheumatol. 2022;4(5):e338–e350. doi: 10.1016/S2665-9913(22)00034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mahil SK, et al. The effect of methotrexate and targeted immunosuppression on humoral and cellular immune responses to the COVID-19 vaccine BNT162b2: a cohort study. Lancet Rheumatol. 2021;3(9):e627–e637. doi: 10.1016/S2665-9913(21)00212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reuken PA, et al. T cell response after SARS-CoV-2 vaccination in immunocompromised patients with inflammatory bowel disease. J Crohns Colitis. 2022;16(2):251–258. doi: 10.1093/ecco-jcc/jjab147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prendecki M, et al. Humoral and T-cell responses to SARS-CoV-2 vaccination in patients receiving immunosuppression. Ann Rheum Dis. 2021;80(10):1322–1329. doi: 10.1136/annrheumdis-2021-220626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smetanova J, et al. Humoral and cellular immune responses to mRNA COVID-19 vaccines in patients with axial spondyloarthritis treated with adalimumab or secukinumab. Lancet Rheumatol. 2022;4(3):e163–e166. doi: 10.1016/S2665-9913(21)00393-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mahil SK, et al. Humoral and cellular immunogenicity to a second dose of COVID-19 vaccine BNT162b2 in people receiving methotrexate or targeted immunosuppression: a longitudinal cohort study. Lancet Rheumatol. 2022;4(1):e42–e52. doi: 10.1016/S2665-9913(21)00333-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haberman RH, et al. Methotrexate hampers immunogenicity to BNT162b2 mRNA COVID-19 vaccine in immune-mediated inflammatory disease. Ann Rheum Dis. 2021;80(10):1339–1344. doi: 10.1136/annrheumdis-2021-220597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colwill K, et al. A scalable serology solution for profiling humoral immune responses to SARS-CoV-2 infection and vaccination. Clin Transl Immunology. 2022;11(3):e1380. doi: 10.1002/cti2.1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abe KT, et al. A simple protein-based surrogate neutralization assay for SARS-CoV-2. JCI Insight. 2020;5(19):e142362. doi: 10.1172/jci.insight.142362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng SMS, et al. Neutralizing antibodies against the SARS-CoV-2 Omicron variant following homologous and heterologous CoronaVac or BNT162b2 vaccination. Nat Med. 2022;28(3):486–489. doi: 10.1038/s41591-022-01704-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dejnirattisai W, et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185(3):467–484. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans JP, et al. Neutralizing antibody responses elicited by SARS-CoV-2 mRNA vaccination wane over time and are boosted by breakthrough infection. Sci Transl Med. 2022;14(637):eabn8057. doi: 10.1126/scitranslmed.abn8057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Planas D, et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2021;602(7898):671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- 37.Sievers BL, et al. Antibodies elicited by SARS-CoV-2 infection or mRNA vaccines have reduced neutralizing activity against Beta and Omicron pseudoviruses. Sci Transl Med. 2022;14(634):eabn7842. doi: 10.1126/scitranslmed.abn7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall VG, et al. Delayed-interval BNT162b2 mRNA COVID-19 vaccination enhances humoral immunity and induces robust T cell responses. Nat Immunol. 2022;23(3):380–385. doi: 10.1038/s41590-021-01126-6. [DOI] [PubMed] [Google Scholar]
- 39.Cyster JG, Allen CDC. B cell responses: cell interaction dynamics and decisions. Cell. 2019;177(3):524–540. doi: 10.1016/j.cell.2019.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Runnstrom MC, et al. Reduced COVID-19 vaccine response in patients treated with biologic therapies for asthma. Am J Respir Crit Care Med. doi: 10.1164/rccm. [published online February 18, 2022]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Law JC, et al. Persistence of T cell and antibody responses to SARS-CoV-2 up to 9 months after symptom onset. J Immunol. 2022;208(2):429–443. doi: 10.4049/jimmunol.2100727. [DOI] [PubMed] [Google Scholar]
- 42.Law JC, et al. Systematic examination of antigen-specific recall T cell responses to SARS-CoV-2 versus influenza virus reveals a distinct inflammatory profile. J Immunol. 2021;206(1):37–50. doi: 10.4049/jimmunol.2001067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McKinney W. Data structures for statistical computing in Python. Paper presented at: Proceedings of the 9th Python in Science Conference (SciPy 2010); June 28–July 3, 2010; Austin, Texas, USA. http://conference.scipy.org/proceedings/scipy2010/mckinney.htmL Accessed May 10, 2022.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.