Abstract
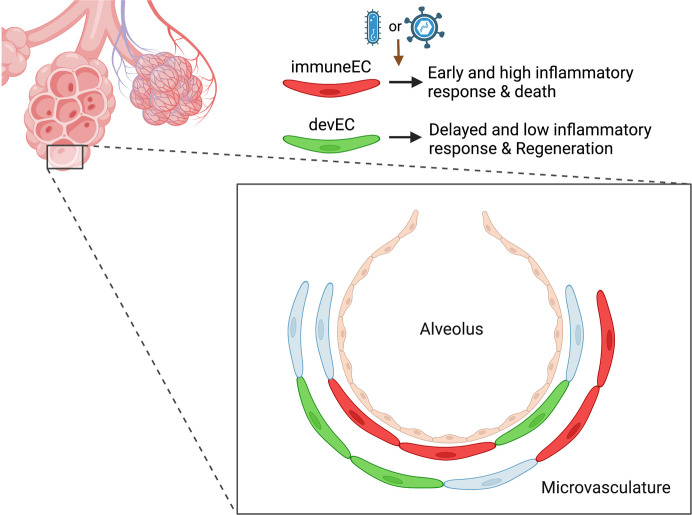
Studies have demonstrated the phenotypic heterogeneity of vascular endothelial cells (ECs) within a vascular bed; however, little is known about how distinct endothelial subpopulations in a particular organ respond to an inflammatory stimulus. We performed single-cell RNA-Seq of 35,973 lung ECs obtained during baseline as well as postinjury time points after inflammatory lung injury induced by LPS. Seurat clustering and gene expression pathway analysis identified 2 major subpopulations in the lung microvascular endothelium, a subpopulation enriched for expression of immune response genes such as MHC genes (immuneEC) and another defined by increased expression of vascular development genes such as Sox17 (devEC). The presence of immuneEC and devEC subpopulations was also observed in nonhuman primate lungs infected with SARS-CoV-2 and murine lungs infected with H1N1 influenza virus. After the peak of inflammatory injury, we observed the emergence of a proliferative lung EC subpopulation. Overexpression of Sox17 prevented inflammatory activation in ECs. Thus, there appeared to be a “division of labor” within the lung microvascular endothelium in which some ECs showed propensity for inflammatory signaling and others for endothelial regeneration. These results provide underpinnings for the development of targeted therapies to limit inflammatory lung injury and promote regeneration.
Keywords: Infectious disease, Pulmonology
Keywords: Bacterial infections, Endothelial cells, Influenza
Introduction
Endothelial cells (ECs) lining all blood vessels exhibit a wide array of functions, such as enabling transmigration of immune cells across the endothelial barrier, controlling hemostasis and thrombosis, regulating the transport of metabolites into tissue, and regenerating or expanding vascular networks after injury and EC death (1). Recent analyses of the vascular endothelium in different organs have identified tissue-specific gene expression signatures in the endothelium, such as expression of genes involved in inflammatory and immune responses in the lung endothelium compared with the endothelium of the heart or brain (2). The critical role of the endothelium in maintaining homeostasis in each tissue as well as mechanisms of dysfunction in organ-specific vascular disease (1) raise fundamental questions regarding the adaptation of ECs in a given vascular bed and how distinct subpopulations of ECs in an organ respond to injury and whether selected ECs can dampen the inflammatory response (3) or promote endothelial regeneration (4).
Single-cell RNA-Seq (scRNA-Seq) provides high-resolution transcriptomic analysis of individual cells and can identify heterogeneity of cell subpopulations serving specialized functions (5). Studies have leveraged scRNA-Seq to define cell subpopulations in the basal state and the responses to inflammation and injury, such as distinct brain macrophage subpopulations during neuroinflammation (6), cardiac macrophage subpopulations after myocardial infarction (7), and lung macrophage subpopulations during fibrosis (8). Studies have also applied scRNA-Seq analysis to identify endothelial subpopulations in the brain vasculature (9), large vessels such as the aorta (10), and the lymphatic endothelium (11) and responses under normal laminar flow (12). Single-cell analysis of lungs in particular has identified a subpopulation of lung ECs expressing carbonic anhydrase 4 (Car4), which depended on VEGFA expression generated by alveolar type I epithelial cells (13). The Car4-high lung ECs have been referred to as “aerocytes” based on the increased expression of genes related to gas exchange, whereas non-aerocytes have been referred to as general capillary ECs (14).
Here, we investigated the responses of distinct lung EC subpopulations to inflammatory injury using an experimental mouse model of acute lung inflammatory injury. Severe injury of the lung endothelium and neutrophilic inflammation is induced by the bacterial endotoxin LPS (15) and is followed by adaptive regeneration of ECs involving upregulation of developmental transcription factors (TFs) such as Sox17 (4). An important question is whether endothelial injury and reparative responses to a stimulus, such as LPS, are shared or distinct across lung EC subpopulations. A key advantage of using LPS-induced injury is that it can be calibrated and allows for temporal quantification due to the defined onset of injury after LPS and the subsequent resolution of injury. Thus, the model is well-suited for studying temporal responses of lung cell subpopulations. A study used LPS to analyze the heterogeneity of alveolar epithelial cells by scRNA-Seq (16). Shortly after LPS challenge, proinflammatory cytokines are released and lead to a systemic inflammatory response syndrome and high mortality, mirroring severe systemic inflammatory responses seen in patients with sepsis (17).
In the present study, we used scRNA-Seq to study distinct lung EC subpopulations during baseline, injury, and resolution of injury. We observed the presence of an EC subpopulation expressing immune response genes (immuneEC) and an EC population expressing developmental or regenerative genes (devEC) during baseline. Surprisingly, the immuneEC population showed an augmented response to the LPS challenge versus the devEC population. Studying the trajectories of these EC subpopulations showed the emergence of a specialized EC subpopulation expressing proliferation and cell cycle genes (i.e., proliferative ECs or proECs) emerging from the developmental EC subpopulation. These findings suggest the importance of EC plasticity and EC phenotype shift in promoting the demands of vascular regeneration. Failure of these adaptation programs may lead to chronicity of inflammatory processes in the lungs.
Results
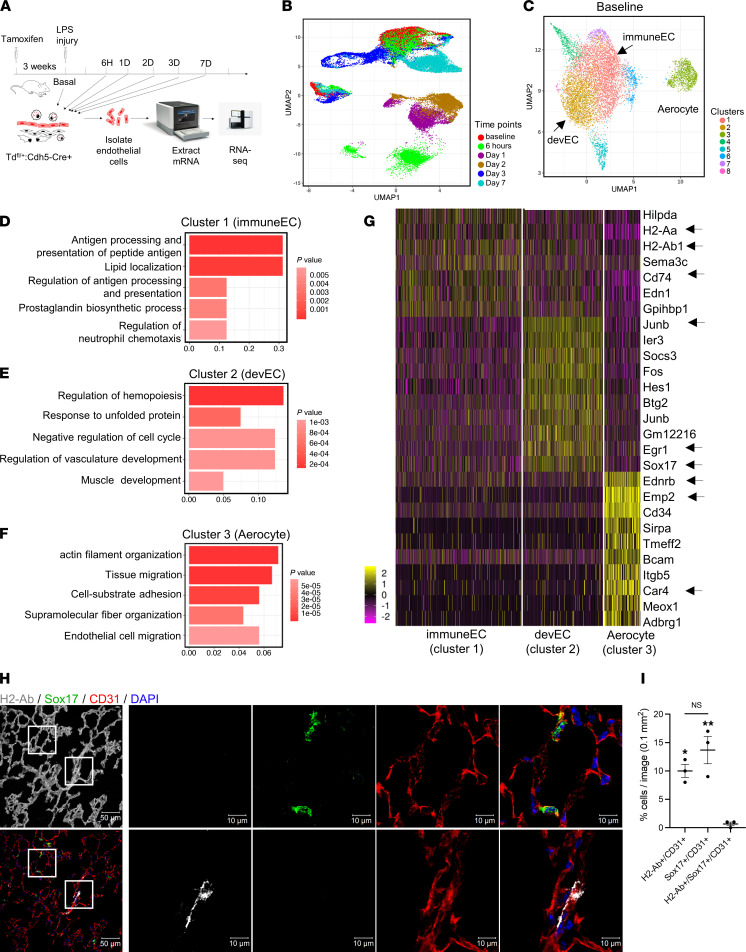
To genetically label the endothelium, we crossed the inducible endothelial-specific Cre (Cdh5-CreERT2) (18) with floxed tdTomato (tdTomatofl/fl) to generate endothelial lineage–specific tdTomatofl/fl Cdh5-CreERT2 mice in which tdTomato protein was expressed in all ECs after induction with tamoxifen. Tamoxifen induction was performed 1 month prior to all experiments to ensure adequate tdTomato labeling of all ECs. We used a model of severe lung vascular injury induced by the bacterial endotoxin LPS (12.5 mg/kg i.p.) with 30% mortality (4). Lung ECs were isolated at baseline and at defined time points after LPS challenge: 6 hours, 1 day, 2 days, 3 days, and 7 days (Figure 1A). DAPI nuclear labeling was used to exclude dead cells; tdTomato+ DAPI– cells were sorted by FACS (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.158079DS1), and subsequently underwent scRNA-Seq. We recovered a total of 35,973 individual ECs with 5000 to 8000 cells at each time point, with a median number of genes expressed ranging from 1100 to 1900 genes per cell (Supplemental Table 1). An overview of gene expression data from all time points are presented in a 2D uniform manifold approximation projection (UMAP) plot (19) (Figure 1B). The specificity of lung ECs was validated by assessing the expression of the endothelial-specific gene Cdh5 (VE-cadherin) and the hematopoietic lineage–specific gene Cd45. More than 99% of the isolated cells expressed the endothelial-specific gene Cdh5 (Supplemental Figure 2, A and B). We also examined the expression of the lymphatic EC markers Prox1 (Prospero homeobox 1) and Nr2f2 (nuclear receptor subfamily 2 group F member 2) and identified a distinct lymphatic endothelial subpopulation (Supplemental Figure 2, C and D), which was filtered out from subsequent data analyses.
Figure 1. Overview of scRNA-Seq of lung ECs and characterization of baseline lung EC subpopulations.
(A) Schematic diagram of single-cell analysis of lung ECs at different time points using 10x Genomics Chromium platform. (B) Two-dimensional UMAP representation of 35,973 individual lung ECs obtained from all time points. Different time points are labeled as different colors. (C) UMAP of 8191 individual lung ECs at baseline. Different colors indicate distinct clusters. immuneEC, immune EC subpopulation; devEC, developmental EC subpopulation. The third subpopulation was labeled as “aerocyte” (Car4+). (D–F) The GO terms of cluster 1 (D), cluster 2 (E), and cluster 3 (F) during homeostasis (basal state) indicate the enriched biological processes in each cluster. P values are indicated on the left in a color scale. The x axis represents the percentage of differentially expressed genes in each GO term. (G) Heatmap of the most differentially expressed genes of lung ECs in cluster 1 (immuneECs), cluster 2 (devECs), and cluster 3. The color bars indicate gene expression level in log2 scale. (H) Confocal images of frozen lung sections during homeostasis immunostaining for CD31 (red), H2-Ab (gray), and Sox17 (green). The lung structure is shown as the first image by enhancing autofluorescence of lung tissue. The 2 regions in the white squares are shown at higher magnification in the right panels. Nuclei were stained by DAPI. Scale bars: 50 μm on left; 10 μm on right. (I) Quantification of CD31+/H2-Ab+, CD31+/Sox17+, and CD31+/Sox17+/H2-Ab+ cells in lung sections. Data are shown as mean ± SEM from 3 independent mice. **P < 0.01 and *P < 0.05 compared with Td+/Sox17+/Irf7+ cells by 1-way ANOVA.
Lung EC subpopulations at baseline.
We sequenced 8191 ECs during the basal homeostasis condition to perform an unbiased cell clustering analysis. Distinct EC subpopulations were identified using the Seurat algorithm (20). We identified 8 clusters based on their transcriptomic signatures (Figure 1C). We then performed Gene Ontology (GO) analysis of the differentially expressed genes in each cluster to assess potentially distinct functions of each subpopulation. The GO terms describing biological processes and molecular functions for the 3 largest EC clusters are shown in Figure 1, D–F, whereas the analysis for the smaller clusters is shown in Supplemental Figure 3, A–E.
For the 3 predominant EC subpopulations, we found that cluster 1 primarily exhibited immune regulatory signatures, cluster 2 was characterized by vascular development signatures, and cluster 3 showed promigratory gene expression signatures (Figure 1, D–F). Gene expression levels of the most differentially expressed genes in these 3 clusters are shown in a heatmap (Figure 1G). Signature genes for cluster 1 included Cd74, H2-Aa (histocompatibility 2, class II antigen A, alpha), and H2-Ab1 (histocompatibility 2, class II antigen A, beta 1) (Figure 1G and Supplemental Figure 4A), which play critical roles in MHC class II antigen processing and presentation and regulation of innate and adaptive immune response (21, 22). The signature genes most enriched in cluster 2 were Junb (Jun B proto-oncogene), Egr1 (early growth response 1), and Sox17 (sex determining region Y box17) (Figure 1G and Supplemental Figure 4B), which are all key regulators of cell development and differentiation (23). The differentially expressed signature genes for cluster 3 included Ednrb (endothelin receptor type B), Emp2 (epithelial membrane protein 2), and Itgb5 (integrin β 5) (Figure 1G and Supplemental Figure 4C), which are mediators of cell migration, actin filament organization, and cell adhesion (24, 25), as well as Car4, which encodes for Car4 and has resulted in this lung EC cluster being referred to as an aerocyte population (14).
The expression of these signature genes in each of these 3 predominant clusters are shown in UMAP projections (Supplemental Figure 4, A–C). We also validated the expression of these signature genes of EC subpopulations at the protein level by immunofluorescence staining for Sox17, an essential regulator of endothelial regeneration (4) and H2-Ab, a major component of MHC class II antigen complex (21, 22). Confocal microscopy of intact frozen OCT-embedded lung tissue demonstrated that CD31+ lung ECs (red) expressed either H2-Ab (white) or Sox17 (green) (Figure 1, H and I). Importantly, quantification demonstrated that there was only a negligible fraction of ECs double-positive for H2-Ab and Sox17 (Figure 1I), suggesting that ECs exhibited either a predominantly immune regulatory or developmental phenotype such that hybrid phenotypes were extremely rare.
Thus, using scRNA-Seq, Seurat clustering, and differential gene expression analysis, we identified predominant lung EC clusters during baseline: an immune regulatory H2-Ab1hi/Sox17lo EC subpopulation (immuneEC), a developmental Sox17hi/H2-Ab1lo EC subpopulation (devEC), and an EC subpopulation enriched in genes regulating cell migration and cell structure, mirroring the recently described Car4+ EC or aerocyte populations (13, 14). Although greater than 13,000 genes were shared across these 3 lung EC clusters, we found 857, 774, and 313 genes were differentially expressed, respectively, in immuneECs, devECs, and cluster 3 ECs (Supplemental Figure 4D).
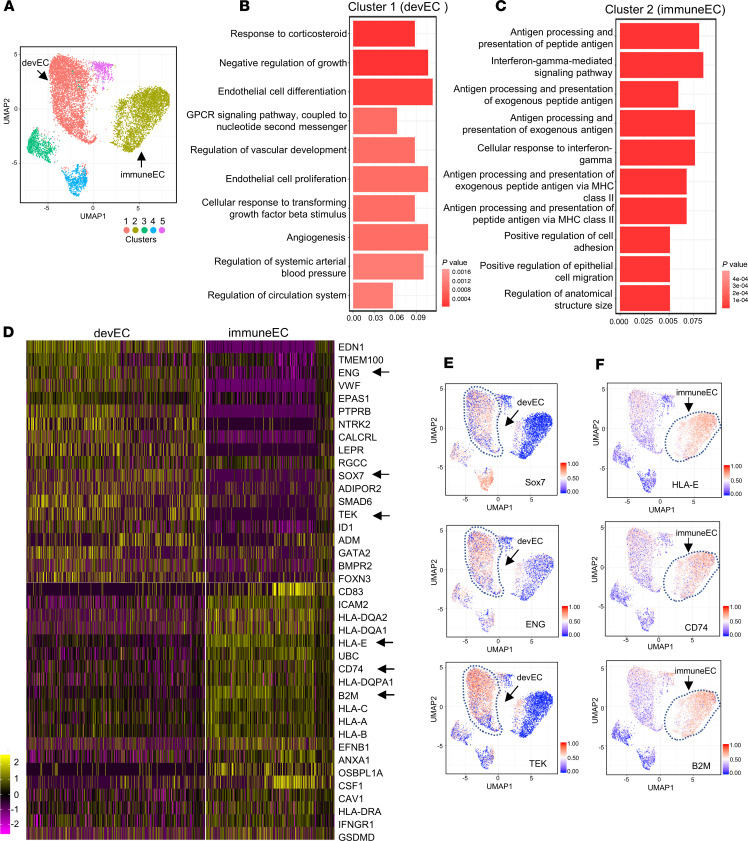
Furthermore, we analyzed scRNA-Seq data of 18,253 lung ECs obtained from human lungs during homeostasis (26). Five cell clusters were identified by unbiased cell clustering analysis (20) based on their differential transcriptomic signatures (Figure 2A). Of these 5 cell clusters, 2 major EC subpopulations were defined as devEC (cluster 1) and immuneEC (cluster 2) based on GO analysis of differentially expressed genes in each cluster; cluster 1 primarily exhibited vascular development signatures (Figure 2B), and cluster 2 was defined by immune regulatory signatures (Figure 2C).
Figure 2. Identification of human lung microvascular EC subpopulations.
(A) UMAP of 18,253 nondiseased human individual lung ECs. Different colors indicate distinct clusters. immuneEC, immune EC subpopulation; devEC, developmental EC subpopulation. (B and C) The GO terms of cluster 1 (B) and cluster 2 (C) indicate the enriched biological processes in developmental signature (B) and inflammatory feature (C). P values are indicated on the left in a color scale. The x axis represents the percentage of differentially expressed genes in each GO term. (D) Heatmap of the most differentially expressed genes of lung ECs in cluster 1 (immuneECs) and cluster 2 (devECs). The color bars indicate gene expression level in log2 scale. Arrows highlight genes shown in E and F. (E and F) UMAP of visualization of expression levels of the representative marker genes in devEC (E) and immuneEC (F). The color bars in UMAP indicate gene expression level in log2 scale.
A heatmap of the most differentially expressed genes in devEC and immuneEC is shown in Figure 2D. UMAPs of representative developmental genes — Sox7, ENG (endoglin), and TEK (tyrosine-protein kinase receptor tie-2) — showed preferential expression in devEC subpopulations (Figure 2E). UMAPs of representative genes encoding antigen processing and presentation molecule HLA-E (MHC class I antigen E), CD74, and B2M (β chain of MHC class I molecules) showed that these were expressed at higher levels in the immuneEC subpopulation (Figure 2F). These findings suggest that the human lung microvascular endothelium also exhibits distinct subpopulations with gene expression signatures during homeostasis that are similar to the immuneEC and devEC populations of the homeostatic mouse lung (Figure 1).
Distinct immune regulatory and developmental EC subpopulations during the early inflammatory phase.
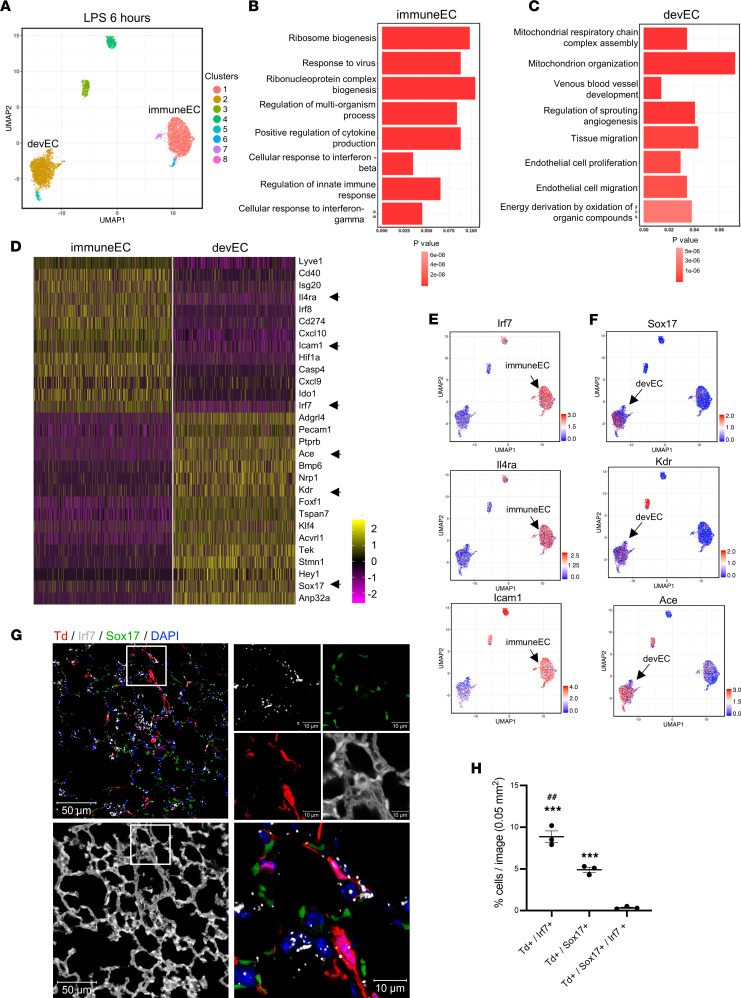
We next investigated how inflammatory injury modulated gene expression profiles of the 2 predominant lung EC clusters, immuneECs and devECs, in a mouse model of LPS injury. At 6 hours after systemic LPS exposure, we found a remarkable shift in the gene expression profiles of the analyzed EC subpopulations. A UMAP plot of ECs at 6 hours after LPS (Figure 3A) showed 2 predominant EC clusters, which were much more divergent from each other than EC clusters we had identified at baseline (Figure 1C). GO analysis allowed inferences regarding the functions of these 2 predominant clusters. We found that cluster 1 was enriched in genes involved in the immune and inflammatory response (Figure 3B), whereas cluster 2 was enriched in vascular development genes (Figure 3C), consistent with the immuneEC and devEC signatures we had seen at baseline (Figure 1), although the differences in expression levels of the individual inflammatory and developmental genes between these 2 populations were far more prominent than during homeostasis.
Figure 3. Divergence of inflammatory and developmental lung EC subpopulations during early immune injury (LPS response at 6 hours).
(A) UMAP of 6527 individual lung ECs isolated at 6 hours after LPS injury. Different colors indicate distinct clusters. The immuneEC (cluster 1) and devEC (cluster 2) are shown. (B and C) The GO terms of immuneEC (B) or devEC (C) indicate enriched biological processes in each cluster. P values are indicated on the left in a color scale. (D) Heatmap of the most differentially expressed genes of immuneEC and devEC. The color bar indicates the gene expression levels using a log2 scale. (E and F) The expression levels of representative genes (arrows labeled in D) of immuneEC (E) and devEC (F) visualized by UMAP. (G and H) Confocal images (G) and quantification (H) of RNA-FISH of mouse lung paraffin-embedded tissues after LPS treatment for 6 hours. The lung structures are shown in G by enhancing autofluorescence of lung tissues. The lung ECs expressed tdTomato (Td) in tamoxifen-treated tdTomatofl/fl Cdh5-CreERT(+) mice. The mRNA levels of Irf7 and Sox17 were probed and labeled in gray and green respectively. Nuclei were stained by DAPI. The small region in a white square is zoomed in as shown on the right. Scale bars: 50 μm on the left; 10 μm on the right. The statistical analysis of numbers of Td+/Irf7+, Td+/Sox17+, and Td+/Sox17+/Irf7+ cells in lung sections of mice are shown in H. Data are shown as mean ± SE from 3 independent mice. ***P < 0.001 compared with Td+/Sox17+/Irf7+ cells, ##P < 0.01 compared with Td+/Sox17+ by 1-way ANOVA.
The expression levels of top differentially expressed signature genes of immuneEC and devEC are shown in a heatmap (Figure 3D). UMAP plots of the representative immuneEC genes Irf7 (IFN regulatory factor 7), Il4ra (IL-4 receptor α), and Icam1 (ICAM 1) are shown (Figure 3E) alongside those of the representative devEC genes Sox17, Kdr (kinase insert domain protein receptor), and Ace (angiotensin converting enzyme 1) (Figure 3F).
To validate the expression of these signature genes in the intact lung tissue, we also used RNA fluorescence in situ hybridization (RNA-FISH) (Figure 3, G and H) and immunofluorescence to detect protein levels (Supplemental Figure 5, A and B) of Sox17, a critical regulator of endothelial regeneration (4), and Irf7, a TF regulating the IFN response during inflammation (27, 28). Confocal microscopy of the intact paraffin-embedded lungs demonstrated that the fraction of Td+/Irf7+ cells (immuneECs) was significantly increased and greater than the fraction of Td+/Sox17+ cells (devECs) (Figure 3H and Supplemental Figure 5B). Furthermore, we found that tdTomato+ (Td+) lung ECs (red) expressed either Irf7 or Sox17 at both mRNA (Figure 3H) and protein levels (Supplemental Figure 5B). Importantly, quantification demonstrated that there was only a negligible fraction of ECs double-positive for Irf7 and Sox17 (Figure 3H and Supplemental Figure 5B), suggesting that lung ECs exhibited either an immune or developmental phenotype and that hybrid phenotypes were extremely rare. Thus, we found that there was prominent gene expression divergence between immuneECs and developmental ECs during the early phase of the inflammatory response.
Immune regulatory and developmental EC subpopulations during the late inflammatory phase.
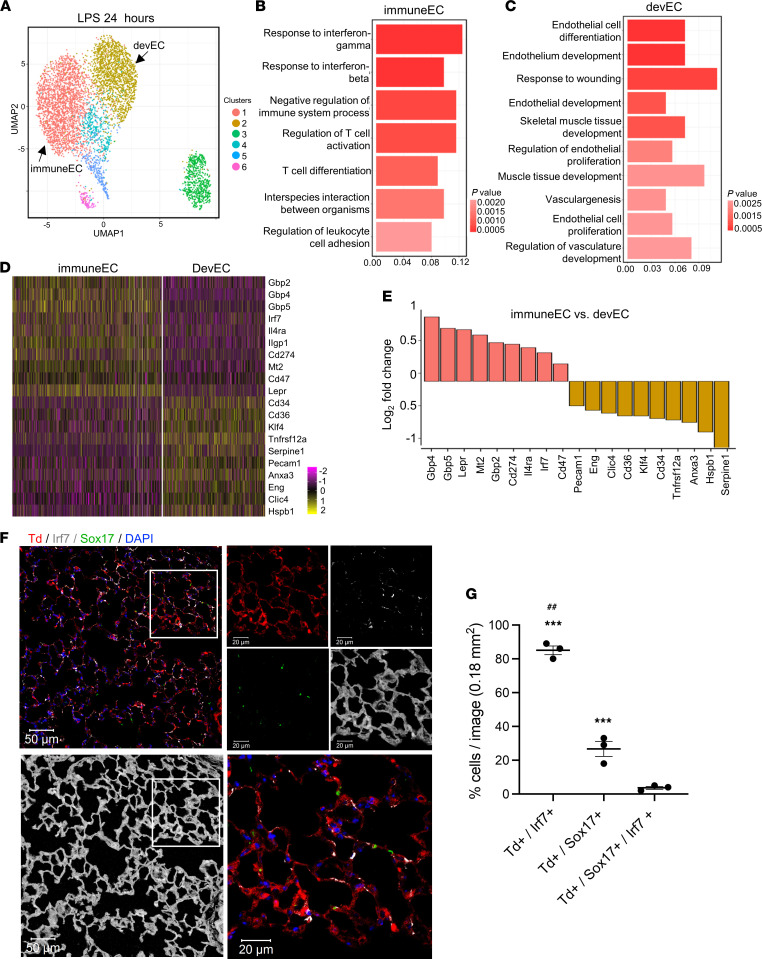
Distinct immuneEC and devEC subpopulations were also present in lungs at 24 hours after induction of inflammatory injury (Figure 4, A–C). The expression of the most differentially expressed genes in immuneECs and devECs are shown in the heatmap and bar graph (Figure 4, D and E). Immunofluorescence (Figure 4, F and G) and RNA-FISH (Supplemental Figure 6, A and B) were used to validate Sox17 and Irf7 as the in situ markers of distinct devEC and immuneEC subpopulations. Quantification demonstrated that the fraction of Td+/Irf7+ cells (immuneECs) was significantly greater than Td+/Sox17+ cells (devECs), and that as we had observed at the 6-hour time point, hybrid cells double-positive for Irf7 and Sox17 were extremely rare (Figure 4G and Supplemental Figure 6, A and B).
Figure 4. Identifying inflammatory and developmental lung EC subpopulations of lung ECs during the late immune phase (24 hours after LPS injury).
(A) UMAP of 5158 individual lung ECs isolated 24 hours after systemic LPS injury with different colors presenting distinct clusters. The immuneEC (cluster 1) and devEC (cluster 2) are indicated. (B and C) The GO terms of immuneEC (B) and devEC (C) indicate the enriched biological processes in each cluster. (D) Heatmap of the most differentially expressed genes from immuneEC and devEC clusters with the color bar showing gene expression levels on a log2 scale. (E) The box plot of top differentially expressed genes in immuneEC compared with devEC. Data are presented as log2 fold changes. (F and G) Confocal images (F) and quantification (G) of protein immunostaining of mouse lung sections for Irf7 (gray) and Sox17 (green) after LPS treatment for 24 hours. The lung structures are shown in G by enhancing autofluorescence of lung tissues. The lung ECs expressed tdTomato (Td) in tamoxifen-treated tdTomatofl/fl Cdh5-CreERT(+) mice. The small region in the white square is shown at higher magnification on the right. Scale bars: 50 μm on the left; 20 μm on the right. Nuclei were stained by DAPI. The quantification of Td+/Irf7+, Td+/Sox17+, and Td+/Sox17+/Irf7+ cells in lung sections of mice are shown in F. Data are shown as mean ± SE from 3 independent mice. ***P < 0.001 compared with Td+/Sox17+/Irf7+ cells, ##P < 0.01 compared with Td+/Sox17+ by 1-way ANOVA.
A key feature of the endotoxemia model of inflammatory lung injury is the initiation of endothelial repair and regeneration beginning on day 2 after injury at the LPS dosage used (4). A UMAP plot of scRNA-Seq data obtained at 2 days after LPS injury showed overall convergence of the gene expression profiles in immuneEC and devEC subpopulations (Supplemental Figure 7, A–C). The most differentially expressed genes in the devEC and immuneEC subpopulations at 2 days after LPS are shown in Supplemental Figure 7D. Expression of representative genes in the devEC population included Ace, Acvrl1 (activin A receptor type II-like 1), Calcrl (calcitonin receptor-like), and Sox17 alongside representative genes in the immuneEC population Cd24, Ctla2a (cytotoxic T lymphocyte-associated protein 2 α), Cebpb (CCAAT/enhancer binding protein [C/EBP] β), and Nfkbia (nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor α). These gene expression profiles suggest the beginning of the resolution of inflammation in immuneECs because in particular, Cd24, Ctla2a, and Nfkbia are known to negatively regulate inflammation (29–31).
A volcano plot of immuneECs and devECs underscores their distinct transcriptomic signatures (Supplemental Figure 7E). Confocal imaging of immunofluorescence (Supplemental Figure 7, F and G) for Irf7 and Sox17 of lung tissues showed reversal of the ratio of immuneECs and devECs with a reduction of Irf7+ ECs and peak level of Sox17+ ECs (Supplemental Figure 7, F and G), suggesting a shift away from inflammation toward increased population of endothelial regeneration.
Appearance of proliferating EC subpopulation during recovery.
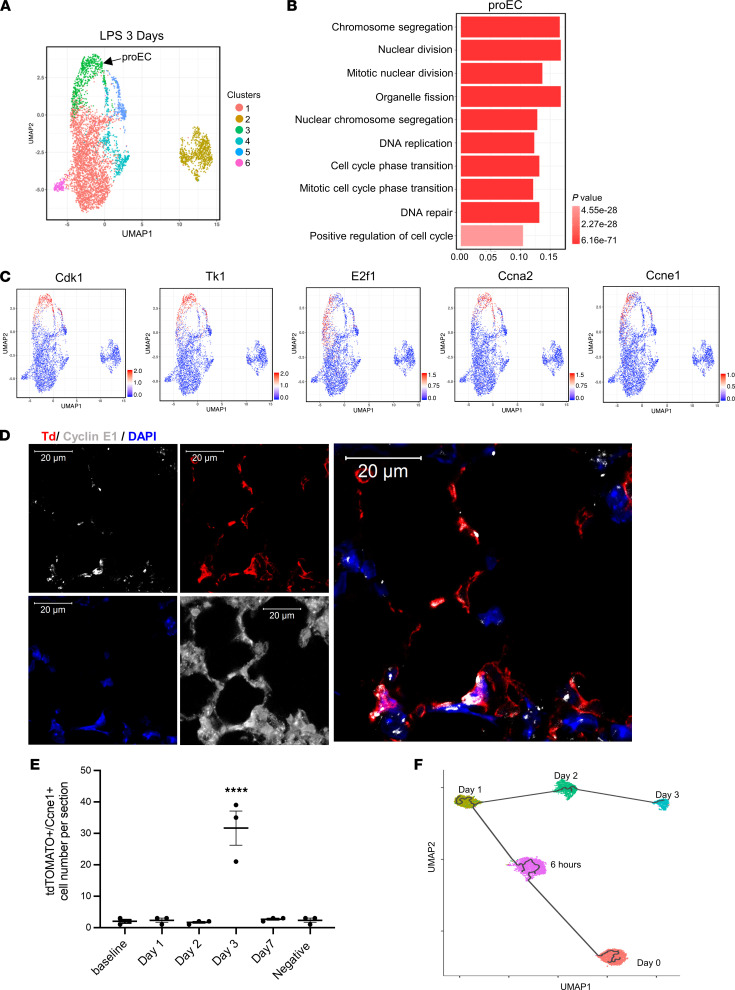
At day 3 after injury, a novel EC subpopulation emerged with highly proliferative transcriptomic signatures. This EC subpopulation was contiguous with the devEC population in the UMAP projection (Figure 5A). GO analysis identified chromosome segregation, nuclear division, DNA replication, and cell cycle phase transition enriched in this subpopulation (Figure 5B). Expression levels of representative cell cycle genes Cdk1 (cyclin-dependent kinase 1), Tk1 (thymidine kinase 1), E2f1 (E2f transcriptional factor 1), Ccna2 (cyclin A2), and Ccne1 (cyclin E1) are shown in UMAP projections (Figure 5C). These results demarcated this newly emerged cluster as a distinct proEC subpopulation.
Figure 5. Identification of the proliferative lung EC subpopulation during the regeneration phase.
(A) UMAP of 5318 individual lung ECs isolated at 3 days after systemic LPS injury. (B) The GO terms indicate the enriched biological processes of proEC. P values are indicated on the left in a color scale. (C) The expression levels of representative marker genes are visualized by UMAP. The color bars in UMAP indicate gene expression level in log2 scale. (D) Confocal images of RNA-FISH of mouse lung sections obtained at 3 days after LPS injury. The lung structures are shown in D by enhancing autofluorescence of lung tissues. The lung ECs express tdTomato (Td). The RNAs of Ccne1 were probed and labeled with gray. Scale bar: 20 μm. Nuclei were stained by DAPI. (E) The quantification of cell number of Td+/Ccne1+ cells in lung sections of mice at baseline, LPS 24 hours, LPS 2 days, and LPS 3 days. Data are shown as mean ± SE from 3 independent mice at different time points. ****P < 0.0001 compared with baseline by 1-way ANOVA. (F) Pseudo-time trajectory of devECs from baseline, LPS 6 hours, LPS 1 day, LPS 2 days, and proEC of LPS 3 days. Colored by time points of LPS treatment.
We verified the expression of the cell cycle regulator Ccne1 in the lung tissue using RNA-FISH. Confocal images of lung sections showed that Ccne1+ ECs were greatly increased at 3 days after LPS (Figure 5D), consistent with prior reports of an EC proliferation burst and upregulation of cyclin E1 at day 3 after induction of inflammatory vascular injury (4). Importantly, RNA-FISH analysis found no significant increase in Ccne1+ ECs at any other time points (Figure 5E), suggesting that the emergence of the proEC subpopulation occurred at a defined time point during the course of the tissue repair after inflammatory injury.
We next sought to identify the origin of the proEC population using Monocle3 transcriptomic trajectory modeling, which identifies the most proximate cell populations by generating a pseudo-time trajectory in which cells are ordered according to dynamic changes in their gene expression profiles (32, 33). The pseudo-time of trajectory of proECs at 3 days after LPS and devECs at baseline, LPS 6 hours, LPS day 1, and LPS day 2 showed that proECs at day 3 gradually traced back to devECs at baseline through devECs of LPS day 2, LPS day 1, and LPS 6 hours as temporal ancestors of previous time points (Figure 5F). These findings suggest that the proEC subpopulation that emerged at 3 days after LPS was most likely derived from devECs at baseline.
We also analyzed the transcriptomic profiles of lung ECs at day 7 after LPS, at which time injury of lung ECs was reversed (4). Consistent with reversal of injury, we found the transcriptomic profiles of lung EC subpopulations at 7 days after LPS were similar to the baseline (Supplemental Figure 8, A and B). The dynamic expression patterns of representative genes for immuneECs and devECs are shown by violin plots across all time points (Supplemental Figure 9, A and B). The violin plots of Irf7, Cd47, and Ifit3 (IFN-induced protein with tetratricopeptide repeats 3) of immuneECs indicated that inflammatory gene expression peaked between LPS 6 hours and 24 hours, then gradually resolved and returned to baseline levels by 7 days after the initial LPS insult. The violin plots of Sox17, Ace1, and Eng suggest that expression of developmental genes was suppressed during the initial phase of inflammatory injury and progressively upregulated during repair concomitant with the resolution of inflammation, consistent with distinct transcriptional programming in the EC subpopulations during the injury and recovery phases.
High expression of Sox17 prevents excessive inflammatory activation in the devEC population.
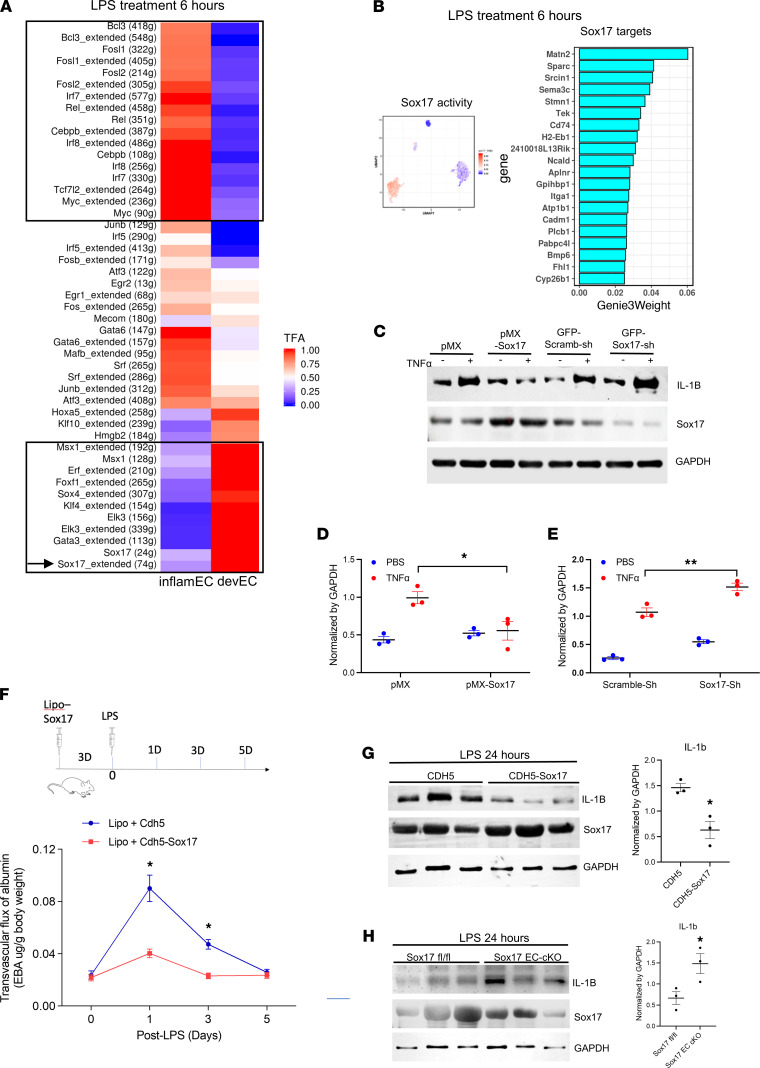
We next inferred the activities of TFs that could regulate the functions of the distinct EC subpopulations described above using the SCENIC (single-cell regulatory network inference and clustering) model, which can inform the activities of TFs and establish gene regulatory networks by combining binding motif and coexpressing modules between TFs and candidate target genes (34). The activities of TFs estimated by SCENIC in immuneEC and devEC subpopulations were compared at LPS 6 hours. As expected, activities of developmental TFs were enhanced in the devEC subpopulation, while the activities of inflammatory TFs were highly upregulated in the immuneEC subpopulation (Figure 6A). The key identified transcriptional regulators in the devEC subpopulation are presented in a UMAP as well as a ranking of the associated target genes of Sox17 (Figure 6B). The transcriptional activity of Sox17 was highly induced in the devEC subpopulation (Figure 6B), suggesting that the observed upregulation of Sox17 mRNA corresponded to upregulation of its inferred transcriptional activity. Considering the attenuated inflammatory response of the devEC subpopulation at LPS 6 hours (Figure 3D), these data suggest that Sox17 itself was crucial in suppressing inflammatory activation in devECs.
Figure 6. Regulation of central role of Sox17 in the inflammatory response of lung vascular ECs.
(A and B) Activity of TFs and their regulatory targets estimated by SCENIC. Heatmap of estimated mean activities of TFs using SCENIC in each subpopulation at LPS treatment 6 hours (A). Enhanced activities of inflammatory TFs shown in top box and developmental TFs shown in lower box; Sox17 activity marked by an arrow. UMAP of estimated activities and predicted regulatory targets of Sox17 (B). (C–E) Human lung endothelial cells (HLECs) were infected with retroviruses expressing pMX-Sox17 to overexpress Sox17 and lentiviruses expressing GFP-Sox17-shRNA to knock down Sox17. The retroviruses expressing pMX vector and lentiviruses with GFP-scramble-shRNA were applied as control, respectively. Infected HLECs were treated with 20 ng/mL TNF-α for 2 hours. Representative Western blots (C) and quantification (D and E) shown. (F) Time course of lung transvascular permeability after LPS challenge in C57BL/6J mice with liposome delivery of Cdh5 vector or Cdh5-Sox17. Mice were injected with LPS 8 μg/kg 2 days later after liposome delivery of Sox17. Schematic diagram of Sox17 liposome delivery and LPS injection shown on top. n = 4 for each time point. (G) Western blots and quantification of fresh isolated lung ECs from C57BL/6J mice with liposome delivery of CDH5 vector or CDH5-Sox17 for 3 days. (H) Western blots and quantification of fresh isolated lung ECs from Sox17fl/fl mice or Sox17 EC cKO mice. n = 3 for each group. Mice were injected with LPS 8 μg/kg for 24 hours. n = 3 for each group. Data shown as mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001 compared with control by 2-way ANOVA (D and E) and unpaired 2-tailed t test (G and H).
To investigate the role of Sox17 in dampening inflammatory activation, human lung microvascular ECs were activated with 20 ng/mL of the proinflammatory cytokine TNF-α. Sox17 was overexpressed using a pMX-Sox17 lentiviral vector, whereas lentiviral shRNA targeting Sox17 was used to deplete Sox17. We observed that Sox17 overexpression inhibited the induction of the proinflammatory cytokine IL-1β by TNF-α, whereas depletion of Sox17 enhanced TNF-α–induced IL-1β synthesis (Figure 6, C–E). Furthermore, plasmids of Sox17 with the endothelial-specific Cdh5 promoter were delivered to the lung endothelium via i.v. liposomal injection (4) to overexpress Sox17 specifically in lung vascular ECs. We found that overexpression of Sox17 ahead of LPS injury showed decreased LPS-induced vascular injury (Figure 6F). Sox17 overexpression also inhibited IL-1β induction in freshly isolated mouse lung microvascular ECs after LPS activation (Figure 6G). Conversely, EC-specific deletion of Sox17 enhanced IL-1β induction after LPS activation in isolated mouse lung microvascular ECs (Figure 6H). Of note, the EC-specific overexpression or depletion of Sox17 did not influence baseline IL-1β expression (Supplemental Figure 10). Therefore, our results indicate that the expression of the developmental TF Sox17 in devECs not only increased their regenerative potential but also concomitantly served as a “brake” preventing excessive inflammatory activation in devECs.
Transcriptional profiles of devECs and immuneECs after viral infection.
To compare the dynamics of EC subpopulations in a second disease model of inflammatory lung injury, we analyzed the scRNA-Seq profiles of the lung endothelium from nonhuman primates infected with SARS-CoV-2 at day 3 after inoculation (inflammatory injury phase) and day 10 after inoculation (recovery phase). The tissue was obtained from a study that performed scRNA-Seq of whole lung in nonhuman primates (35). We selected lung microvascular ECs based on the expression of classical EC markers VE-cadherin and CD31. UMAPs of lung EC clusters are shown in Supplemental Figure 11A. GO analysis of the differentially expressed genes identified an immuneEC subpopulation enriched for antigen processing and T cell regulation genes (Supplemental Figure 11, B–D). The heatmap of the most differentially expressed genes of immuneECs are shown across different clusters (Supplemental Figure 11C). UMAPs of representative antigen and inflammation genes, such as Chsa-A (MHC class IA antigen), HLA-DRB5 (MHC class II DR β 5 chain), and B2M and IFN signaling — IFIT3 (IFN-induced protein with tetratricopeptide repeats 3), ISG15 (IFN-induced 15 KDa protein), ISG20 (IFN stimulated exonuclease gene 20), IFI16 (IFN-γ inducible protein 16), and ACKR1 (atypical chemokine receptor 1) — highlighted their preferential expression in the immuneEC subpopulation at day 3 after viral infection (Supplemental Figure 11D).
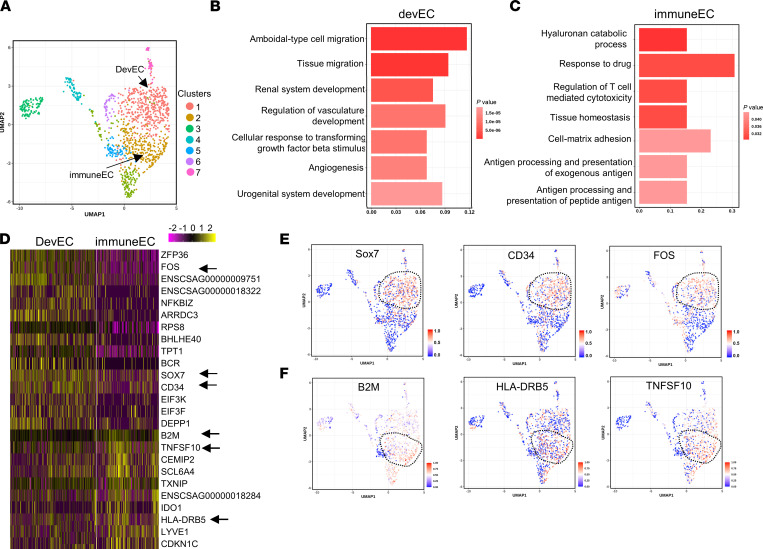
During the recovery phase (day 10 after viral inoculation), devEC and immuneEC subpopulations were present in the nonhuman primate lungs as defined by distinct developmental and immune gene expression signatures (Figure 7, A–C). The heatmap of the most differentially expressed genes in devEC and immuneEC subpopulations is shown in Figure 7D. UMAPs show that developmental TFs and markers — Sox7, CD34 and Fos — were enriched in devECs (Figure 7E), whereas inflammatory genes, such as B2M, HLA-DRB5, and TNFSF10 (TNF superfamily member 10), were enriched in immuneECs (Figure 7F).
Figure 7. Analysis of lung ECs obtained from nonhuman primates infected with SARS-CoV-2.
African green monkeys were inoculated with 2.6 × 106 TCID50 (50% tissue culture infectious dose) live SARS-CoV-2 (nCoV-WA1-2020 MN985325.1). The scRNA-Seq was performed using harvested lung ECs from 4 nonhuman primates at day 10 after inoculation (recovered from SARS-CoV-2 infection). (A) UMAP of 3253 individual lung ECs; different colors indicate distinct clusters (immuneEC, immune EC subpopulation; devEC, developmental EC subpopulation). (B and C) The GO terms of devEC (B) and immuneEC (C) indicate the enriched biological processes in developmental signature (B) and immune signature (C). P values are indicated on the left in a color scale. X axis represents the percentage of differentially expressed genes in each GO term. (D) Heatmap of the most differentially expressed genes in lung ECs from devEC and immuneEC subpopulations. The color bars indicate gene expression level in log2 scale. Arrows highlight genes shown in E and F. UMAP of visualization of expression levels of the representative marker genes in devECs (E) and immuneECs (F). The color bars in UMAP indicate gene expression level using a log2 scale.
We also evaluated the dynamic responses of EC subpopulations in a different mouse model of lung injury, H1N1 influenza–induced lung injury. C57BL/6J mice were inoculated with sublethal dose of H1N1 (50 PFU, influenza A/Puerto Rico/8/34) to induce acute viral lung injury with approximately 30% mortality. Given that the temporal progression of influenza injury is more gradual than LPS, we chose to study lungs at day 7 when we observed peak lung injury after H1N1 infection. Lung ECs were sorted by FACS for live cells that were negative for DAPI and subsequently underwent scRNA-Seq. We recovered 923 individual lung ECs by endothelial-specific markersmCdh5 and Cd31. The UMAP plot of ECs at day 7 after H1N1 (Supplemental Figure 12A) again showed 2 predominant EC clusters. Distinct immuneEC and devEC subpopulations were present based on GO analysis (Supplemental Figure 12, B and C). The expression levels of the most differentially expressed genes in immuneECs and devECs are shown in a heatmap (Supplemental Figure 12D). Therefore, the dynamic responses of distinct immune and developmental subpopulations of lung microvascular ECs were found in multiple lung injury models (SARS-CoV-2, H1N1, and LPS).
Discussion
Our findings demonstrated heterogeneity in the lung vascular endothelium during homeostasis that became more prominent after inflammatory injury. We identified at least 2 predominant EC subpopulations in the lung endothelium with distinct inflammatory and developmental transcriptomic signatures, as well as another lung EC population that arises during the recovery phase. This was accomplished using scRNA-Seq that was performed at defined time points of inflammatory injury and its resolution. In addition, RNA-FISH and immunofluorescence were used to validate the presence of distinct EC subpopulations in the intact lung tissue. Finally, we inferred from these scRNA-Seq data sets the activities of potential regulatory TFs in the EC subpopulations.
Recent studies have used scRNA-Seq to study endothelial heterogeneity within vascular beds. For example, analysis of the brain vasculature revealed distinct transcriptomic profiles of 3 major endothelial clusters: arterial EC, venous EC, and capillary EC, with important clinical relevance in the migration of myeloid cells (9). Another study of lymphatic ECs in human lymph nodes revealed 6 subtypes of lymphatic ECs, which were mapped to particular locations of lymph nodes (11). In our study, we removed the large vessels of the lung prior to disassociating the lung tissue and analyzed solely transcriptomes of ECs from lung microvessels (diameters less than 100 μm). That the microvascular endothelium is the predominant endothelial cell type in lungs (15) coupled to the exclusion of large vessel EC explains why we did not identify dominant arterial or venous EC subpopulations. Instead, we focused on multiple EC subpopulations within the lung microvascular endothelium. In addition to investigating the anatomic-spatial heterogeneity of ECs within an organ, some studies have also explored functional heterogeneity of EC subpopulations. Examples of this analysis include identification of a proliferative capacity or angiogenic ability in the aorta (10) and coronary vasculature (36). scRNA-Seq of high endothelial venule ECs in the basal and inflamed states after antigenic stimulation in lymph nodes defined molecular signatures of the inflammatory responses in lymph nodes (37). In lung microvessels of mice, Car4-high ECs or aerocyte ECs have been characterized (13, 14), as well as a transient subpopulation of proECs responding to influenza-induced lung injury (38). Car4-high ECs showed a gene expression pattern suited for gas exchange, which is why they are referred to as aerocytes, and the majority of microvascular ECs were classified as general capillary ECs (14). In the present study, we also observed the Car4+ aerocyte population (cluster 3) in the lung microvascular endothelium; however, by sequencing over 35,000 microvascular lung ECs, we identified additional diverse EC subpopulations within the general capillary EC population.
Our focus was on studying how distinct lung EC subpopulations change from the basal state during the course of inflammatory injury and resolution. We identified 2 major subpopulations (immuneECs and devECs), which demonstrated dynamic shifts in transcriptional profiles with the greatest divergence in the gene expression profiles seen at 6 hours after systemic endotoxemia, which represents a highly calibrated and reproducible model of inflammatory lung vascular injury (4). In another study, the capillary and venous tumor ECs were reported to express genes involved in immune surveillance (39). These findings comport with our observation that a subpopulation of lung microvascular ECs (immuneECs) may similarly regulate immune responses.
A primary focus of the studies was the identification of the role of TF Sox17, which was previously shown to be required for lung EC regeneration after LPS injury (4). In the present studies, we expanded on these observations by showing that Sox17 is primarily found in a subset of ECs with predominant developmental gene expression signatures. The role of Sox17 in regulating endothelial differentiation and fate has also been previously reported in the hemogenic endothelium (40), engrafting endothelium (41), coronary vessel endothelium (42), and angiogenic endothelium (43). Sox17, as a member of the Sry-related high-mobility group domain family F (SoxF) TFs, regulates development of endothelial and hematopoietic cell lineages (23), tumor angiogenesis (44), differentiation of fibroblasts to ECs (45), and lung endothelial regeneration (4).
In the present studies, we tracked the expression of Sox17 within EC subpopulations over time. Here, we used the SCENIC algorithm to infer activities of TFs that distinguish EC subpopulations. We found that inferred Sox17 activity was among the signature TF activities in the devEC population of lung ECs. The SCENIC algorithm focuses on expression of TF target genes; therefore, the results suggest that Sox17 is not only a marker of the devEC subpopulation described but also transcriptionally active in the population. We observed that Sox17 overexpression suppressed the levels of the proinflammatory cytokine IL-1β, underscoring the potential antiinflammatory role of Sox17+ devECs. Thus, Sox17 is not just a marker of the developmental subpopulation and regulator of EC regeneration but helps maintain distinct EC functions by actively suppressing the release of proinflammatory cytokines. Because lung microvascular ECs with increased Sox17 levels (devECs) showed a delayed and attenuated inflammatory response after LPS injury, therapeutic overexpression of Sox17 in lung ECs may shift ECs away from an inflammatory phenotype toward a regenerative phenotype.
The significance of a devEC subpopulation characterized by increased expression of vascular development regulators, such as Sox17, Kdr, and Klf4, becomes apparent in the context of studying the trajectories of EC subpopulations during the postinjury period. The proEC subpopulation emerged transiently on day 3 of LPS injury and expressed genes regulating chromosome segregation, nuclear division, DNA replication, and cell cycle phase transition. A similar transient proEC subpopulation was also recently observed in an influenza-induced lung injury model (38), thus pointing toward a more general principle of select endothelial subpopulations driving regeneration of the lung endothelium. Using the Monocle3 trajectory modeling approach (32, 33), we inferred the proEC lineage and found a pseudo-time trajectory from devECs at baseline to devECs at LPS day 2 and continuing on to proECs at LPS day 3. Given the transient nature of these shifts in gene expression, traditional genetic lineage tracing approaches require several days of genetic labeling (46) and are therefore unsuitable for labeling cell subpopulations with transiently appearing gene expression markers. However, in the absence of such genetic lineage tracing, the trajectory inference based on our data strongly supports the notion that devECs indeed give rise to proECs and that the early upregulation of developmental TFs at 24 hours after LPS injury represents an early preparatory phase during which an EC subpopulation is in a preparative mode to give rise to a progeny involved in proliferation and regeneration.
We also found that a distinct EC subpopulation (immuneECs) was enriched for the expression of immune regulatory genes during homeostasis such as MHC II related genes — Cd74 and H2-Ab1. Although MHC II antigens mediate innate and adaptive immune responses (21, 22), their potential roles in the endothelium are less well studied. One teleological explanation for a lung EC subpopulation enriched in MHC II–related genes could be the unique role of the lung endothelium due to its continuous exposure to potential pathogens in the air. A lung EC subpopulation expressing higher levels of MHC II–related genes may serve as a vanguard to activate host defense upon sensing of airborne pathogens.
After viral injury, we found that other regulatory genes such as Irf7 predominate in immuneECs. Irf7, as a member of IFN regulatory factors, is a crucial regulator of type I IFNs against pathogenic infections (47). Deficiency of Irf7 can be life-threatening in an influenza infection (27). Irf7 has also recently been identified as a target for nonantibiotic therapy of bacterial infection (28). We observed that the immuneEC subpopulation exhibited faster and more intense inflammatory response than the devEC subpopulation, suggesting that the transcriptomic signatures of devECs actively inhibit or delay their inflammatory activation and thus allow devECs to initiate the repair process.
Interestingly, we found that expression of Irf7 and Sox17 in individual lung ECs was mutually exclusive as assessed by immunofluorescence and RNA-FISH in intact lung tissue after LPS injury, with most lung ECs being either Irf7+ or Sox17+, and only a small number of ECs showing positivity for both. We also observed that the immuneEC subpopulation exhibited a more brisk and intense inflammatory response compared with the devEC subpopulation. Manipulating Sox17 by overexpression or depletion in vivo and in vitro in our studies demonstrated that Sox17 prevented excessive inflammatory activation of ECs, which could explain why EC subpopulations maintain their distinct profiles even during inflammatory injury. Future spatial transcriptomic studies could define the proximity and spatial relationship of devECs and immuneECs.
The scRNA-Seq analysis of lung ECs from nonhuman primates infected with SARS-CoV-2 demonstrated the presence of distinct immuneEC and devEC subpopulations in an inflammatory lung injury model that is highly relevant for the clinical setting. However, we also observed that the specific genes that distinguished immuneEC and devEC populations in the nonhuman primate lungs after SARS-CoV-2 infection were different from the endotoxemic mouse lung. This may reflect either species differences or different injury-specific responses (bacterial endotoxin versus SARS-CoV-2). We additionally observed that after H1N1 influenza infection in murine lungs, 2 distinct EC subpopulations could be identified with either predominantly immune regulatory or developmental gene expression signatures, thus suggesting that the observed heterogeneity in the lung endothelium likely represents a generalized phenomenon present in multiple forms of lung injury.
The identification of immuneEC and devEC subpopulations raises the possibility of a “division of labor” among lung ECs, which engage in distinct yet complementary functions during inflammatory injury and recovery. Such functional heterogeneity allows cells to function efficiently and cooperatively in a synergistic manner. Importantly, these findings could provide a basis for developing targeted therapies to augment EC regeneration and proliferation without compromising the obligatory role of the endothelium in the host defense function.
Methods
Mouse models of inflammatory lung injury.
All animal experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and approved by the IACUC of the University of Illinois. The tdTomatofl/fl mice were crossbred with Cdh5-CreERT2 mice to generate inducible EC lineage tracing mice (tdTomatofl/fl Cdh5-CreERT2; provided by Ralf Adams, Max Planck Institute for Molecular Biomedicine, Münster, Germany). tdTomatofl/fl and Cdh5-CreERT2 transgenic mice were crossbred with C57BL/6J for several generations. To induce the Cre recombinase in tdTomatofl/fl Cdh5-CreERT2 mice, 2- to 4-month-old mice (Cre+) were injected with tamoxifen (0.1 mg/g, i.p., Sigma-Aldrich, T5648) in 100 μL corn oil (Sigma-Aldrich, C8267), once a day for consecutive 3 days. One month later, the mice were subjected to acute lung injury. The C57BL/6J mice were purchased from The Jackson Laboratory (catalog 000664). The mice were injected with a sublethal dose of LPS (12.5 mg/kg, i.p., Sigma-Aldrich, L2030).
Isolation of mouse lung microvessel ECs.
Three lungs were harvested at each time point: baseline and LPS treatment at 6 hours, 24 hours, 2 days, 3 days, and 7 days. After perfusing with PBS 20 mL through the left and right ventricular chambers, mouse lungs were minced and digested with 3 mL collagenase A at 1 mg/mL in PBS (Roche, 10103586001) at 37°C in a water bath for 1 hour. Mixtures were titrated with 18 G needles and then pipetted through a 40 μm disposable cell strainer. After centrifuging at 500g for 5 minutes and washing with PBS, the isolated cells were treated with RBC lysis buffer (BioLegend, 420301) for 5 minutes to lyse RBCs. After washing with PBS twice, cells were incubated with DAPI (1 mg/mL) in suspension buffer (Ca2+- and Mg2+-free PBS, 5% BSA, 2 mM EDTA) at 4°C for 30 minutes with gentle tilting. After centrifuging, the lung cells from 3 mice at each time point were mixed and incubated in buffer (Ca2+- and Mg2+-free PBS, 10% BSA, 2 mM EDTA). Six samples from 6 time points were subjected to single-cell FACS of TdTomato+/DAPI– cells (live ECs) in the MoFlow Astrios machine (Supplemental Figure 1).
Single-cell capture and library preparation.
The tdTomato+/DAPI– cells were loaded into a 10x Genomics microfluidics chip and encapsulated with barcoded oligo-dT–containing gel beads using the 10x Genomics Chromium controller according to the manufacturer’s instructions. Cell viability at each time point was more than 80% with a target of sequencing 5000 cells at each time point. Single-cell libraries were constructed using a v2 kit (10x Genomics) according to the manufacturer’s instructions. cDNA quality of each time points was checked (high quality). Libraries from individual samples were multiplexed into 2 lanes for sequencing on NovaSeq 6000 S4 (~100,000 reads per cell and per lane, 2 × 150 bp paired-read) at a depth of approximately 200,000 reads per cell.
Processing of scRNA-Seq data.
For each time point, a raw counts table was generated by Cell Ranger (v3.0.0) with default parameters; 35,973 cells passed the quality control stage (baseline: 8191 cells, 6 hours: 6527 cells, 24 hours: 5158 cells, 2 days: 4608 cells, 3 days: 5318 cells, 7 days: 6171 cells).
Analysis of scRNA-Seq data.
Data analysis and visualization were performed using the Seurat (v2.3) R package (20). The data from each time point were analyzed separately. Data normalization and scaling were performed by NormalizeData() and ScaleData() functions with default parameters. The most variably expressed genes were identified by using FindVariableGenes() function with parameters x.low.cutoff = 0.0125, x.high.cutoff = 3, and y.cutoff = 0.5. The principal component analysis was performed by function RunPCA() on the most variably expressed genes. The subpopulations in each time point were identified by clustering function FindClusters() with the top 20 principal components and the parameter k.param = 10, resolution = 0.4. The UMAP was performed on each time point to visualize the similarity between cells on 2-D space. RunUMAP() was used to generate UMAP of cells (16). Positive markers of each subpopulation were identified by the function FindMarkers() with the parameter only.pos = TRUE. Multiple testing was corrected using the Benjamini-Hochberg method.
GO enrichment analysis.
The marker genes for each subpopulation were tested for enrichment with the DAVID web server on Biological Process level 4 terms. Statistically significant enrichments were defined by a corrected P value (Benjamini-Hochberg method) less than or equal to 0.05.
Analysis of TF activities by SCENIC.
TF activities were identified using SCENIC with default parameters (34). The genes used to identify the TF activities were the most variably expressed genes. The top TF target genes were selected by the top Genie3 weights. Genie3 weight is the weight of a putative regulatory link; higher weights correspond to more likely regulatory links. TFs shown in the heatmap were filtered by maximum activity scores smaller than 0.2. Activity scores were rescaled to [0, 1] across the subpopulations to make them comparable. In the heatmap, the TF activities in each subpopulation were represented by the mean activity scores (SCENIC AUCell) within that subpopulation.
Trajectory building.
The cell trajectory was identified using the Monocle3 R package (32, 33). The variably expressed genes were used to build trajectories. DDRTree was used for dimension reduction required for trajectory building.
Histology and sectioning.
The lungs from PBS-perfused mice (tdTomatofl/fl Cdh5-CreERT2) were harvested and fixed in 4% paraformaldehyde in 4°C for 24 hours. The lung tissues were then embedded in paraffin. The paraffin slides were acquired by sectioning the fixed and embedded lung tissues. For fresh frozen sections, perfused lung tissues were harvested from C57BL/6J mice and were embedded in OCT (Thermo Fisher Scientific, 23-730-571). After 24 hours of being stored in –80°C, the frozen tissues were sectioned, and the slides were fixed in 4% paraformaldehyde for 10 minutes before immunostaining.
Immunofluorescences and confocal microscopy.
For cryosections, the slides were permeabilized with 0.5% Tween 20 for 10 minutes and blocked with 10% goat serum in 2% BSA in PBS/0.5% Tween 20 for 1 hour at room temperature. Then, the slides were incubated with primary antibodies (CD31, 1:50, BD Pharmingen, 550274; H2-Ab, 1:100, BioLegend, 1:16,402; Sox17, 1:100, Aviva Systems Biology, ARP39552_P050) overnight at 4°C. To stain for Irf7 and Sox17, paraffin slides were permeabilized in 0.25% Triton X-100 in PBS for 30 minutes to permeabilize the nuclear membrane and blocked with 1% BSA in PBS for 1 hour. Then, the slides were incubated with primary antibodies (Irf7, 1:100, Biorbyt, orb411601; Sox17, 1:100, OriGene, AM32707PU-N) overnight at 4°C. The next day, the slides were washed and incubated with the fluorescence-conjugated secondary antibody (AF594 goat anti-rat 1:300, Invitrogen, A11007; AF488 goat anti-rabbit, 1:300, Invitrogen, A32731; AF488 goat anti-mouse1:300, Invitrogen, A32723; AF633 goat anti-rabbit 1:300, Invitrogen, A21071; AF633 goat anti-mouse,1:300, Invitrogen, A21052). Images were taken with a confocal microscope LSM880 (Zeiss) and analyzed by Zen software (Zeiss). The structure of the lung is shown by enhancing autofluorescence of lung tissue by Zen software.
RNA-FISH.
RNAscope Multiplex Fluorescent Reagent kit v2 was applied to hybrid RNA in situ (ACD Bio, 323100). The paraffin slides were processed according to the manufacturer’s instructions, and all reagents were obtained from ACD Bio. Slides were washed once in PBS, and then immersed in 1× Target Retrieval reagent at 100°C for 5 minutes. Slides were washed twice in deionized water, immersed in 100% ethanol, air dried, and sections were encircled with a liquid-blocking pen. Sections were incubated with Protease III reagent for 30 minutes at 40°C, and then washed twice with deionized water. Sections were incubated with probes against mouse Sox17 (catalog 493151-C2), Irf7 (catalog 534541-C3), Ccne1(catalog 503801) or a negative control (catalog 320871) and a positive control (320881) and probed for 2 hours at 40°C (all from ACD Bio). Multiplex fluorescence was performed per the manufacturer’s instructions. After RNA-FISH, the slides were permeabilized and blocked with 10% donkey serum, 2% BSA, and 0.05% Tween 20 in PBS for 1 hour at room temperature. The slides were incubated with primary antibodies tdTomato (1:100, Biorbyt, orb182397) at 4°C overnight. The next day, slides were washed and incubated with the fluorescence-conjugated secondary antibody (AF594 donkey anti-goat 1:300, Invitrogen, A32758). Images were taken with a confocal microscope LSM880 and analyzed by Zen software.
Lung EC scRNA-Seq data analysis of H1N1-induced viral lung injury.
C57BL/6J mice were inoculated with a sublethal dose of H1N1 (50 PFU, influenza A/Puerto Rico/8/34) to induce acute viral lung injury. The mice inoculated with H1N1 were harvested at day 7 — the peak of injury as assessed by us in preceding pilot studies. After perfusing PBS (20 mL) through the left and right ventricular chambers, mouse lungs were minced and digested. The isolated cells were treated with RBC lysis buffer (BioLegend, 420301) for 5 minutes to lyse RBCs. After washing with PBS twice, cells were incubated with DAPI (1 mg/mL) in suspension buffer (Ca2+- and Mg2+-free PBS, 5% BSA, 2 mM EDTA) at 4°C for 30 minutes with gentle tilting. After centrifuging, the lung cells from 3 mice at day 7 after inoculation were mixed and incubated in buffer (Ca2+- and Mg2+-free PBS, 10% BSA, 2 mM EDTA). The sample was subjected to single-cell FACS of DAPI– cells (live ECs) in the MoFlow Astrios machine. The DAPI– lung cells were loaded into a 10x Genomics microfluidics chip and encapsulated with barcoded oligo-dT–containing gel beads using the 10x Genomics Chromium controller using the v3 kit according to the manufacturer’s instructions. The library of lung cells was sequenced on NovaSeq 6000 S4 (~100,000 reads per cell and per lane 2× 150 bp paired-read) at a depth of approximately 200,000 reads per cell. Data analysis and visualization were performed using the Seurat (v2.3) R package (20).
Human and nonhuman primate lung EC scRNA-Seq data analysis.
The raw data of human lung ECs were downloaded from the Krasnow Lab Human Lung Cell Atlas at https://cellxgene.cziscience.com, which were published in a human molecular lung atlas (26). The raw data of nonhuman primate (African green monkey) scRNA-Seq data after SARS-CoV-2 infection were obtained from NCBI’s Gene Expression Omnibus (GEO GSE 156755).
Data availability of newly generated mouse scRNA-Seq data.
The scRNA-Seq data generated for this study have been made publicly available via NCBI’s GEO (GSE148499 for the LPS data and GSE201541 for the H1N1 influenza data).
Statistics.
For image analysis, statistical analysis was performed using GraphPad Prism. Quantification is presented as the mean ± SEM from 3 independent biological replicates with each dot presenting the average cell number of 3 sections of 1 mouse. One-way ANOVA was used to determine statistical significance, with a P value threshold less than 0.05 considered as significant. Corrections for multiple comparisons were performed using Tukey’s test. The differential expression analysis of scRNA-Seq data was based on the nonparametric Wilcoxon rank-sum test. The GO enrichment analysis was based on a hypergeometric test. The P values were adjusted using Bonferroni’s correction, and the significance threshold was an adjusted P value less than 0.05.
Study approval.
All aspects of this study were approved by the IACUC and the Office of Environmental Health and Safety at the University of Illinois at Chicago prior to the initiation of this study.
Author contributions
LZ and JR conceived the project with direct input from ABM. LZ and JR designed the experiments. LZ performed the in vivo studies and single-cell studies. SG designed and performed the analysis of the scRNA-Seq data. LZ performed RNA-FISH studies and confocal imaging. ZW performed immunoblotting and in vitro studies under the supervision of LZ. YD and JR supervised the scRNA-Seq analysis. LZ, SG, and JR wrote the original draft. JR, LZ, and ABM finalized the manuscript. All authors reviewed and revised the manuscript.
Supplementary Material
Acknowledgments
This work was supported in part by NIH grant R01HL157489 to LZ and American Heart Association (AHA) 18CDA34110068 to LZ and NIH grants P01-HL060678, P01-HL151327, R01-HL90152, and R01-HL152515 to JR and ABM. The graphical abstract was generated with the BioRender.com software.
Version 1. 05/05/2022
In-Press Preview
Version 2. 06/08/2022
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2022, Zhang et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2022;7(11):e158079.https://doi.org/10.1172/jci.insight.158079.
Contributor Information
Lianghui Zhang, Email: lhzhang@uic.edu.
Shang Gao, Email: sgao30@uic.edu.
Zachary White, Email: zwhite4@uic.edu.
Yang Dai, Email: yangdai@uic.edu.
Asrar B. Malik, Email: abmalik@uic.edu.
Jalees Rehman, Email: jalees@uic.edu.
References
- 1.Liao JK. Linking endothelial dysfunction with endothelial cell activation. J Clin Invest. 2013;123(2):540–541. doi: 10.1172/JCI66843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jambusaria A, et al. Endothelial heterogeneity across distinct vascular beds during homeostasis and inflammation. Elife. 2020;9:e51413. doi: 10.7554/eLife.51413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langer V, et al. IFN-γ drives inflammatory bowel disease pathogenesis through VE-cadherin-directed vascular barrier disruption. J Clin Invest. 2019;129(11):4691–4707. doi: 10.1172/JCI124884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu M, et al. Sox17 is required for endothelial regeneration following inflammation-induced vascular injury. Nat Commun. 2019;10(1):2126. doi: 10.1038/s41467-019-10134-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kolodziejczyk AA, et al. The technology and biology of single-cell RNA sequencing. Mol Cell. 2015;58(4):610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 6.Jordao MJC, et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science. 2019;363(6425):eaat7554. doi: 10.1126/science.aat7554. [DOI] [PubMed] [Google Scholar]
- 7.Farbehi N, et al. Single-cell expression profiling reveals dynamic flux of cardiac stromal, vascular and immune cells in health and injury. Elife. 2019;8:e43882. doi: 10.7554/eLife.43882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schyns J, et al. Non-classical tissue monocytes and two functionally distinct populations of interstitial macrophages populate the mouse lung. Nat Commun. 2019;10(1):3964. doi: 10.1038/s41467-019-11843-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vanlandewijck M, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554(7693):475–480. doi: 10.1038/nature25739. [DOI] [PubMed] [Google Scholar]
- 10.Lukowski SW, et al. Single-cell transcriptional profiling of aortic endothelium identifies a hierarchy from endovascular progenitors to differentiated cells. Cell Rep. 2019;27(9):2748–2758. doi: 10.1016/j.celrep.2019.04.102. [DOI] [PubMed] [Google Scholar]
- 11.Takeda A, et al. Single-cell survey of human lymphatics unveils marked endothelial cell heterogeneity and mechanisms of homing for neutrophils. Immunity. 2019;51(3):561–572. doi: 10.1016/j.immuni.2019.06.027. [DOI] [PubMed] [Google Scholar]
- 12.Liu Z, et al. Single-cell RNA sequencing reveals endothelial cell transcriptome heterogeneity under homeostatic laminar flow. Arterioscler Thromb Vasc Biol. 2021;41(10):2575–2584. doi: 10.1161/ATVBAHA.121.316797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vila Ellis L, et al. Epithelial Vegfa specifies a distinct endothelial population in the mouse lung. Dev Cell. 2020;52(5):617–630. doi: 10.1016/j.devcel.2020.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillich A, et al. Capillary cell-type specialization in the alveolus. Nature. 2020;586(7831):785–789. doi: 10.1038/s41586-020-2822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matthay MA, et al. The acute respiratory distress syndrome. J Clin Invest. 2012;122(8):2731–2740. doi: 10.1172/JCI60331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Riemondy KA, et al. Single cell RNA sequencing identifies TGFβ as a key regenerative cue following LPS-induced lung injury. JCI Insight. 2019;5:e123637. doi: 10.1172/jci.insight.123637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seemann S, et al. Comprehensive comparison of three different animal models for systemic inflammation. J Biomed Sci. 2017;24(1):60. doi: 10.1186/s12929-017-0370-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorensen I, et al. DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood. 2009;113(22):5680–5688. doi: 10.1182/blood-2008-08-174508. [DOI] [PubMed] [Google Scholar]
- 19.Becht E, et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol. doi: 10.1038/nbt.4314. [published online December 3, 2018]. [DOI] [PubMed] [Google Scholar]
- 20.Butler A, et al. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maizels RM, Withers DR. MHC-II: a mutual support system for ILCs and T cells? Immunity. 2014;41(2):174–176. doi: 10.1016/j.immuni.2014.07.006. [DOI] [PubMed] [Google Scholar]
- 22.Oliphant CJ, et al. MHCII-mediated dialog between group 2 innate lymphoid cells and CD4(+) T cells potentiates type 2 immunity and promotes parasitic helminth expulsion. Immunity. 2014;41(2):283–295. doi: 10.1016/j.immuni.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park C, et al. Transcriptional regulation of endothelial cell and vascular development. Circ Res. 2013;112(10):1380–1400. doi: 10.1161/CIRCRESAHA.113.301078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moreno-Layseca P, et al. Integrin trafficking in cells and tissues. Nat Cell Biol. 2019;21(2):122–132. doi: 10.1038/s41556-018-0223-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishikawa K, et al. Endothelin/endothelin-B receptor signals regulate ventricle-directed interkinetic nuclear migration of cerebral cortical neural progenitors. Neurochem Int. 2011;58(3):261–272. doi: 10.1016/j.neuint.2010.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Travaglini KJ, et al. A molecular cell atlas of the human lung from single-cell RNA sequencing. Nature. 2020;587(7835):619–625. doi: 10.1038/s41586-020-2922-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciancanelli MJ, et al. Infectious disease. Life-threatening influenza and impaired interferon amplification in human IRF7 deficiency. Science. 2015;348(6233):448–453. doi: 10.1126/science.aaa1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Puthia M, et al. IRF7 inhibition prevents destructive innate immunity-A target for nonantibiotic therapy of bacterial infections. Sci Transl Med. 2016;8(336):336ra59. doi: 10.1126/scitranslmed.aaf1156. [DOI] [PubMed] [Google Scholar]
- 29.Chen GY, et al. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science. 2009;323(5922):1722–1725. doi: 10.1126/science.1168988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugita S, et al. Acquisition of T regulatory function in cathepsin L-inhibited T cells by eye-derived CTLA-2alpha during inflammatory conditions. J Immunol. 2009;183(8):5013–5022. doi: 10.4049/jimmunol.0901623. [DOI] [PubMed] [Google Scholar]
- 31.Werner SL, et al. Encoding NF-kappaB temporal control in response to TNF: distinct roles for the negative regulators IkappaBalpha and A20. Genes Dev. 2008;22(15):2093–2101. doi: 10.1101/gad.1680708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao J, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566(7745):496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C, et al. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014;32(4):381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aibar S, et al. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017;14(11):1083–1086. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Speranza E, et al. Single-cell RNA sequencing reveals SARS-CoV-2 infection dynamics in lungs of African green monkeys. Sci Transl Med. 2021;13(578):eabe8146. doi: 10.1126/scitranslmed.abe8146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, et al. Single-cell transcriptome analyses reveal novel targets modulating cardiac neovascularization by resident endothelial cells following myocardial infarction. Eur Heart J. 2019;40(30):2507–2520. doi: 10.1093/eurheartj/ehz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veerman K, et al. Single-cell analysis reveals heterogeneity of high endothelial venules and different regulation of genes controlling lymphocyte entry to lymph nodes. Cell Rep. 2019;26(11):3116–3131. doi: 10.1016/j.celrep.2019.02.042. [DOI] [PubMed] [Google Scholar]
- 38.Niethamer TK, et al. Defining the role of pulmonary endothelial cell heterogeneity in the response to acute lung injury. Elife. 2020;9:e53072. doi: 10.7554/eLife.53072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Su R, et al. Targeting FTO suppresses cancer stem cell maintenance and immune evasion. Cancer Cell. 2020;38(1):79–96. doi: 10.1016/j.ccell.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clarke RL, et al. The expression of Sox17 identifies and regulates haemogenic endothelium. Nat Cell Biol. 2013;15(5):502–510. doi: 10.1038/ncb2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schachterle W, et al. Sox17 drives functional engraftment of endothelium converted from non-vascular cells. Nat Commun. 2017;8:13963. doi: 10.1038/ncomms13963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gonzalez-Hernandez S, et al. Sox17 controls emergence and remodeling of nestin-expressing coronary vessels. Circ Res. 2020;127(11):e252–e270. doi: 10.1161/CIRCRESAHA.120.317121. [DOI] [PubMed] [Google Scholar]
- 43.Lee SH, et al. Notch pathway targets proangiogenic regulator Sox17 to restrict angiogenesis. Circ Res. 2014;115(2):215–226. doi: 10.1161/CIRCRESAHA.115.303142. [DOI] [PubMed] [Google Scholar]
- 44.Yang H, et al. Sox17 promotes tumor angiogenesis and destabilizes tumor vessels in mice. J Clin Invest. 2013;123(1):418–431. doi: 10.1172/JCI64547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L, et al. SOX17 regulates conversion of human fibroblasts into endothelial cells and erythroblasts by dedifferentiation into CD34+ progenitor cells. Circulation. 2017;135(25):2505–2523. doi: 10.1161/CIRCULATIONAHA.116.025722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kretzschmar K, Watt FM. Lineage tracing. Cell. 2012;148(1–2):33–45. doi: 10.1016/j.cell.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 47.Frankiw L, et al. BUD13 promotes a type I interferon response by countering intron retention in Irf7. Mol Cell. 2019;73(4):803–814. doi: 10.1016/j.molcel.2018.11.038. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-Seq data generated for this study have been made publicly available via NCBI’s GEO (GSE148499 for the LPS data and GSE201541 for the H1N1 influenza data).