Abstract
The cellular adaptive response to hypoxia relies on the expression of hypoxia-inducible factors (HIFs), HIF-1 and HIF-2. HIFs regulate global gene expression changes during hypoxia that are necessary for restoring oxygen homeostasis and promoting cell survival. In the early stages of hypoxia, HIF-1 is elevated, whereas at the later stages, HIF-2 becomes the predominant form. What governs the transition between the two HIFs (the HIF switch) and the role of miRNAs in this regulation are not completely clear. Genome-wide expression studies on the miRNA content of RNA-induced silencing complexes (RISC) in HUVECs exposed to hypoxia compared to the global miRNA-Seq analysis revealed very specific differences between these two populations. We analyzed the miRNA and mRNA composition of RISC at 2 hours (mainly HIF-1 driven), 8 hours (HIF-1 and HIF-2 elevated), and 16 hours (mainly HIF-2 driven) in a gene ontology context. This allowed for determining the direct impact of the miRNAs in modulating the cellular signaling pathways involved in the hypoxic adaptive response. Our results indicate that the miRNA-mRNA RISC components control the adaptive responses, and this doesn’t always rely on the miRNA transcriptional elevations during hypoxia. Furthermore, we demonstrate that the hypoxic levels of the vast majority of HIF-1-dependent miRNAs (including miR-210-3p) are also HIF-2 dependent, and that HIF-2 governs the expression of 11 specific miRNAs. In summary, the switch from HIF-1 to HIF-2 during hypoxia provides important level of miRNA-driven control in the adaptive pathways in endothelial cells.
Keywords: HIF1A, EPAS1, HIF-1α, HIF-2α
Introduction
When cells adapt to an insufficient oxygen supply, they rely on the hypoxia-inducible factors, HIF-1 and HIF-2, as the major regulators of the global gene expression reprograming. This regulated process is necessary in order to restore oxygen homeostasis and facilitate cell survival (1). The hypoxic assembly of heterodimeric HIF complexes is facilitated by the stabilization of HIF-α subunits following inactivation of the oxygen-dependent HIF prolyl-hydroxylases (PHDs) (2). The HIF complexes then initiate the adaptive hypoxic responses by inducing the expression levels of genes that contain hypoxia response elements (HRE) in their promoter or enhancer regions and that stimulate angiogenesis and glycolysis as well as other pathways involved in the adaptive response to hypoxia (3-5).
HIF-1 complexes accumulate in all hypoxic cells, whereas HIF-2 is present in endothelial cells (ECs), cardiomyocytes, hepatocytes, bone marrow and renal erythropoietin-producing cells. HIF-1 stimulates glycolysis, maintains pH, and initiates angiogenesis, and HIF-2 also stimulates erythropoiesis and maturation of the vascular networks (5-9). Although expression of some genes is clearly HIF-1 or HIF-2 dependent, the transcription of many others, including vascular endothelial growth factor A (VEGFA) and the glucose transporter 1 (GLUT1) can be regulated by both (5). Furthermore, recent studies have expanded the HIF signaling networks through the identification of HIF-1 and HIF-2 regulation by microRNAs (miRNAs, miRs) (10-15).
HIF-1 activity occurs early during hypoxia and is first supported and then subsequently replaced by HIF-2 signaling that change the cellular gene expression profiles (4, 5, 11, 13). A similar transitional switch between the two HIFs has been reported in cancer cells (5, 16). Although the distinct regulatory functions of HIF-1 and HIF-2 have been intensively studied during the transcriptional consequences of the HIF switch, the repercussions of the transition on the miRNA networks and their mRNA targets remain unknown.
Using genome-wide expression studies on the RNA-induced silencing complexes (RISC) in HUVECs exposed to hypoxia, we examined the HIF switch effects on miRNA profiles and their target mRNAs. This allowed for determining the direct impact of the miRNAs in regulating the cellular signaling pathways involved in the adaptive response to hypoxia. The vast majority of HIF-1-dependent miRNAs including miR-210 were also regulated by HIF-2. We also identified 11 novel HIF-2 regulated miRNAs.
Materials and Methods
Cell culture.
Primary human umbilical vein endothelial cells (HUVEC) that were pooled from ten individual donors were purchased from Cellworks (Caltag Medsystems Ltd, UK) and cultured in EGM-2 Bulletkit Medium (Lonza). All experiments were conducted between passage 2 and 6 at a confluence of 70-80%.
Induction of hypoxia.
Hypoxia was induced in a CO2/O2 incubator/chamber specific for hypoxia research (Invivo2 Baker Ruskin). Briefly, cells were cultured in 35 mm or 60 mm dishes (for RNA isolation and protein isolation, respectively) at 0.9% O2 for the time periods specified (PO2 was 10-12 mm Hg). Control cells were maintained in normoxia in a CO2/O2 incubator (Binder).
siRNA transfections.
siRNA for HIF1A (Ambion assay id s6539) and EPAS1 (Ambion assay id s4698) were purchased from Ambion. HUVECs were transfected using the Lipofectamine RNAiMax (Invitrogen, 13778030) according to manufacturer's protocol. The siRNAs were used at a final concentrations of 40 nM for single knockdowns or 60 nM each for the double knockdowns. The transfected cells were cultured for 2 days prior to further analysis. Ambion siRNA Negative Control 1 (Ambion assay id MC22484) was used as a control. After 24 h, the transfected cells were put into a hypoxia chamber, whereas the control cells remained in an incubator in normoxic conditions.
RNA Immunoprecipitation.
The RIP assay kit for miRNA (#RN1005, MBL) was used following the manufacturer’s protocol. HUVECs (80% confluence) from four 100 mm culture dishes were collected for each condition (17, 18). Cell lysis was conducted in hypoxia or normoxia using a cell scraper and ice-cold PBS and mi-Lysis Buffer (+) was added to the cell pellet. Lysates were snap-frozen and stored at −70°C until use. For immunoprecipitation, 15 μg of anti-EIF2C2/AGO2 (#RN003M, MBL) or mouse IgG2a (isotype control; # M076-3, MBL) and agarose beads (Pierce Protein A/G Agarose, #20421, Thermo Scientific) were used. RNA was isolated using a miRNeasy Mini Kit (Qiagen, Hilden, Germany). 700 μl of QIAzol Lysis Reagent was added to the washed beads and the obtained RNA was eluted in 30 μl of nuclease-free water and used in the next generation RNA sequencing experiments. Prior to NGS, post-RIP and Input samples quality were verified with Western blot and qPCR as described in the protocol (Supplemental Figure 1).
Isolation of RNA.
Total RNA (containing both mRNA and miRNA) was isolated using miRNeasy kit (Qiagen). RNA concentrations were calculated based on the absorbance at 260 nm. RNA samples were stored at −70°C until use.
Measurement of miRNA and mRNA using quantitative Real Time PCR (qRT-PCR).
We used TaqMan One-Step RT-PCR Master Mix Reagents (Applied Biosystems) as described previously (19-23) using the manufacturer’s protocol (retrotranscription: 15 min, 48°). The relative expressions were calculated using the 2−ΔΔCt method (24) with the Ribosomal Protein Lateral Stalk Subunit P0 (RPLP0) and 18S rRNA genes as reference genes for the mRNA, and RNU48 and RNU44 as references for the miRNA. TaqMan probe ids used are given in the Supplemental Table 1.
Western Blots.
Western Blot analysis was performed as previously described (23). Following the normalization of protein concentrations, the lysates were mixed with an equal volume of 6X Laemmli sample buffer (12% SDS, 60% glycerol, 0.06% bromophenol blue, 375 mM Tris-HCl pH = 6.8) and incubated for 5 min at 95°C prior to separation by SDS-PAGE on a 4-15% Criterion TGX Stain-Free Gel (Bio-Rad, Hercules, CA, USA). Following SDS-PAGE, the proteins were transferred to polyvinylidene fluoride membranes (Bio-Rad) using the wet electroblotting method (300 mA, 4°C, 90 min for one gel and 180 min for two gels). The membranes were blocked with BSA dissolved in TBS/Tween-20 (3% BSA, 0.5% Tween-20 for 1 h), followed by immunoblotting with the primary antibodies (overnight, 4°C): mouse anti–HIF-1α (1:2000, ab16066; Abcam), rabbit anti–HIF-2α (1:1000, ab199; Abcam), mouse anti-EIF2C2/AGO2 (1:1000, RN003M; MBL), rabbit anti-Dicer (1:1000, ab227518; Abcam), rabbit anti-Drosha (ab12286; Abcam) and rabbit anti-actin (1:1000, ab1801; Abcam). After the washing steps, the membranes were incubated with goat anti-rabbit IgG (heavy and light chains) or with goat anti-mouse IgG (heavy and light chains) horseradish peroxidase-conjugated secondary antibodies (Bio-Rad) for 1 h at room temperature and detected using SuperSignal West Pico ECL (Thermo Scientific). Densitometry was performed using the Image Lab software v.4.1 (Bio-Rad).
Next generation RNA sequencing.
HUVECs were used for the RNA isolation and analyses. Following total RNA isolation, samples were validated with quantitative real-time PCR for activation of the hypoxic response prior to further analysis. Following rRNA depletion, the remaining RNA fraction was used for the library construction and subjected to 75-bp paired-end sequencing on an Illumina NextSeq 500 instrument (San Diego, CA, USA). Sequencing reads were aligned to the Gencode human reference genome assembly (GRCh38 p13 Release 32) using STAR version 2.7.3a (25). Transcript assembly and estimation of the relative abundance and tests for differential expression were carried out with Cufflinks and Cuffdiff (26). The resulting data were validated with quantitative real-time PCR.
Small RNA sequencing libraries were prepared using QIAseq miRNA library kit (Qiagen) following the manufacturer’s instructions and followed by sequencing on an Illumina NextSeq 500 instrument. Using Qiagen’s Gene Globe Software, sequencing reads were aligned to the human reference genome assembly (hg19) followed by transcript assembly and estimation of the relative abundances. The analysis of the differential expression of small RNAs between control and experimental samples were performed with geNorm (27) in the Gene Globe Software. Low read miRNA changes (below 10) were ignored. For comparing biological replicates, the quantile normalization was performed as recommended in (28). The RIP-Seq data miRNA and mRNA next generation sequencing results were corrected for the signal obtained for normal rabbit IgG which was the negative control.
The heatmap generation and hierarchical clustering were performed with the Morpheus Webserver (https://software.broadinstitute.org/morpheus). The Enrichr Webserver (https://amp.pharm.mssm.edu/Enrichr/) (29) was applied to assign the NGS results into the "Gene Ontology " categories with the selection based on a Q value < 0.05. Furthermore, the analyses were limited to experimentally verified interactions and no extended gene enrichment set analyses were performed. Finally, the miRNA-mRNA target interactions were analyzed with the use of miRTarBase (30).
Hypoxia Response Element (HRE) analysis.
The promoters of gene transcripts that were affected by hypoxia in the microarrays experiments were analyzed for HIF-1 or HIF-2 binding sites. In each gene promoter sequence that was defined as a 20kb window around the TSS, we examined only the open chromatin regions that were established in the HUVEC cell line by the ENCODE (31) project. We merged both DNase I-seq HUVEC datasets found in Ensemb l (32) (v.79). We focused on two distinct HRE motifs annotated to HIF-1 (M00139, alias HIF1A) and HIF-2 (M00074, alias EPAS1) from the HOCOMOCO v.9 database (33). We used the Nencki Genomics Database(34) (v. 79_1) to obtain genomic coordinates of these motif instances. For each miRNA, we calculated the number of instances found in the open chromatin regions for both HIF-1 and HIF-2. In the current work, we use two distinct HRE motifs annotated to HIF1A and EPAS1 in the Homo sapiens Comprehensive Model Collection (HOCOMOCO) version 9 database (35). We note that from the version 10 onwards, the creators of HOCOMOCO have decided to choose a single generic model for each TF (36). This results in the assignment of nearly identical motifs to HIF1A and EPAS1. Based on previous specific research reports (37-40), however, we chose to continue to use the two distinct HRE motifs annotated to HIF1A and EPAS1.
Statistical analysis.
Results were expressed as a mean ± standard deviation. Statistical significance was determined using the Student’s t-test and ANOVA on ranks with P value <0.05 considered significant. The correlations were accessed via the Pearson product-moment correlation method.
Results
Total cellular miRNA expression during the HIF switch
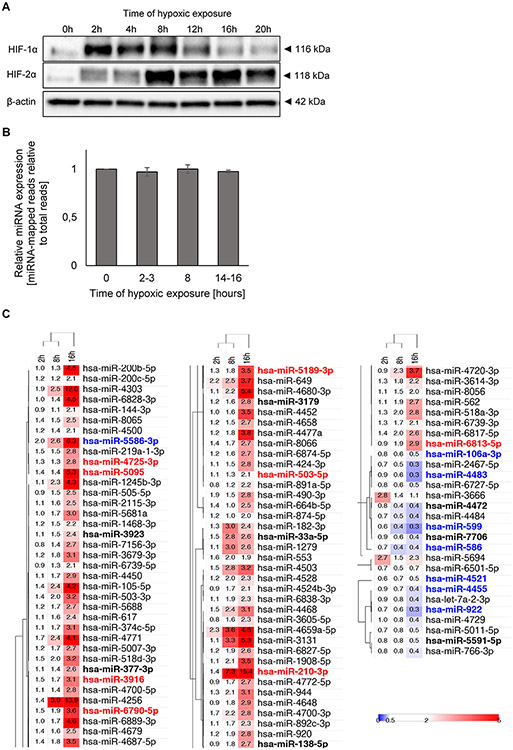
During the development of the adaptive response to hypoxia, HIF-1 is eventually replaced by HIF-2 in human ECs (4, 11, 13). In the present studies, our goal was to explore the consequences of this transition on the miRNA-dependent regulatory networks. To examine this, we followed the HIF-1α and HIF-2α protein levels in human umbilical vein endothelial cells (HUVECs) over a 20-hour time course of hypoxia exposure (0.9% O2) (Figure 1A). Consistent with our previous findings (13, 19, 41), we observed that HIF-1α rapidly accumulated in HUVECs exposed to hypoxia for 2 up to 8 hours and then was dramatically reduced by 12 hours. HIF-2α reached the maximum at 8 hours and remained elevated up to 20 hours. Although both HIF-1α and HIF-2α rapidly accumulate during hypoxia, the reduction of HIF-1 levels leads to the switch to HIF-2 signaling after 8 hours. In order to test for the HIF switch effect on global miRNA expression, we utilized HUVECs as our primary cell model using a 10-donor pool. We isolated total RNA from cells during normoxia and from cells exposed to hypoxia for 2-3 hours (mostly HIF-1 expression), 8 hours (both activities) and 14-16 hours (mostly HIF-2 expression). Next, the samples were subjected to genome-wide next generation sequencing followed by bioinformatics analyses. We only selected mRNAs that were affected significantly (p-value below P = 0.05) by more than a 2-fold change in all three biological replicates. We accessed whether exposure to hypoxia could result in global attenuation of miRNA processing. However, as shown in Figure 1B, global miRNA levels remained constant throughout the entire time of HUVEC exposure to hypoxia. The NGS observed miRNA changes were qualitative rather than quantitative and probably result from the specific activation of the adaptive response to hypoxia signaling networks. As shown in Figure 1C (Supplemental Data Set 1), the acute and more specific HIF-1 response (2 h) to hypoxia resulted in a significant increase of only 4 miRNAs (miR-649; mir-4659a-5p; miR-3666 and miR-5694); whereas after 8 h of exposure to hypoxia, 18 miRNAs were induced (including the well-known miR-210-3p), and 4 were reduced. Finally, 16 hours of exposure to hypoxia resulted in induction of 74 miRNAs and the reduction of 15 others. Furthermore, for the vast majority of hypoxia affected miRNAs we observed a gradual accumulation or reduction with time, and this presumably allowed them to reach maximal or minimal levels after 16 hours of hypoxia. These data suggest that hypoxia-related development of signaling networks is illustrated by the changes in cellular miRNA levels.
Figure 1. Hypoxia results in accumulation of HIF-1α and HIF-2α in HUVECs.
(A) Cells were exposed to hypoxia for the time periods specified and protein lysates were collected. The representative changes in HIF-1α and HIF-2α protein levels were evaluated by western blot normalized to β-actin. (B) Hypoxia does not affect the total miRNA expression in HUVECs. The mapped miRNA reads in NGS analysis were normalized to total mapped RNA reads and expressed as fold change over normoxia control, and 3 biological replicates were used. * P < 0.05 was considered significant, the error bars represent SD. (C) The heat map represents the global distribution of unique miRNAs significantly affected during 2h, 8h and 16h of hypoxia exposure (Supplemental Data Set 1). At each time point the miRNAs levels from 3 biological replicates were expressed as fold change of hypoxia-treated samples compared to the normoxic control samples (P < 0.05 was considered significant). The heatmap generation and hierarchical clustering were performed with the Morpheus Webserver (https://software.broadinstitute.org/morpheus). The miRNAs that were enriched or depleted in the RISC components are denoted with red and blue fonts, respectfully.
Changing mRNA and miRNA content of RISC components during the HIF switch
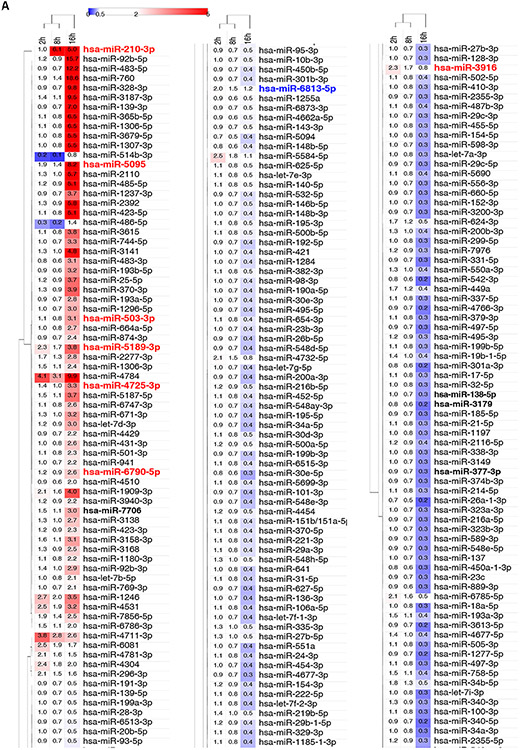
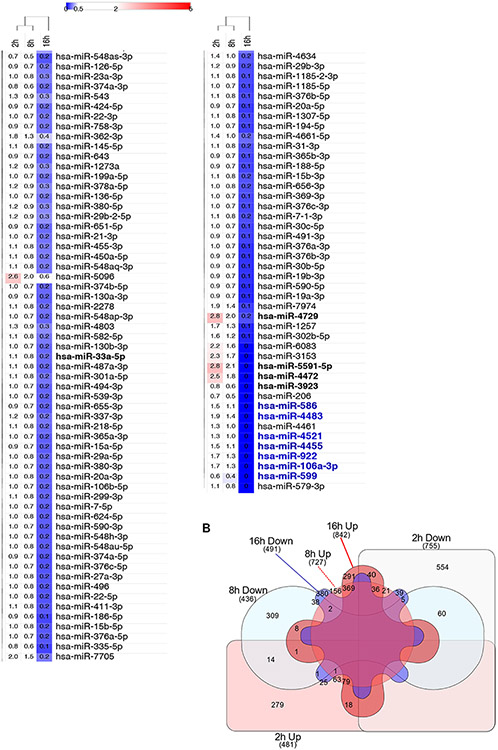
Next, we determined if these hypoxia-related global miRNA changes occurred at a functional level as well. To accomplish this, we isolated miRNAs and mRNA that were incorporated in Ago2-conatining RISC components in HUVECs exposed to hypoxia for 2, 8 and 16 hours by immunoprecipitation with anti-AGO2 (Argonaute 2) antibodies (Supplemental Figure 1ABC). We the analyzed the co-isolated miRNA and mRNA subpopulations with NGS. Although, the global miRNA content in RISC components remained unaffected by hypoxia up to 16 hours (Supplemental Figure 1D), even after 2 hours exposure to hypoxia, we observed at least a 2-fold enrichment of 22 miRNAs in RISC components and depletion of 2 miRNAs when compared to the normoxic RISC complex content (Figure 2A, Supplemental Dataset 2). Whereas in RISC components isolated after 8 hours of exposure to hypoxia, only 6 miRNAs levels were elevated, and 3 miRNAs reduced. The 16 hours exposure to hypoxia, however, resulted in dramatic changes of RISC complex composition that led to enrichment in 61 miRNAs levels, and a reduction of 252 miRNAs. miR-210-3p, one that we consider as a positive control for our study, started to accumulate in RISC components after 8 hours of hypoxia and remained elevated up to 16 hours. Interestingly, the hypoxic distribution of miRNA within RISC components differed drastically from the changes observed at the global miRNA levels (Supplemental Figure 1E). Similar changes in the miRNA expression patterns between these 2 pools (global population versus RISC) were observed for only 15 miRNAs (increasing: hsa-miR-210-3p, hsa-miR-3916, hsa-miR-4725, hsa-miR-503-3p, hsa-miR-5095, hsa-miR-5189-3p, hsa-miR-6790-5p, and hsa-miR-6813-5p; and decreasing: hsa-miR-106a-3p, hsa-miR-4455, hsa-miR-4483, hsa-miR-4521, hsa-miR-586, hsa-miR-599, and hsa-miR-922). In contrast, 6 miRNAs show the opposite pattern of changes between these two pools. The miRNA levels that increased in global population and decreased in RISC components were hsa-miR-138-5p, hsa-miR-3179, hsa-miR-33a-5p, hsa-miR-377-3p, and hsa-miR-3923. Whereas, hsa-miR-7706 was reduced in global population and accumulated in RISC. Furthermore, hsa-miR-4729 was enriched in RISC after 2- and 16-hours of exposure to hypoxia and depleted after 16 hours, whereas this miRNA level was decreased in global population after 16 hours, and similar changes were observed for hsa-miR-4472 and hsa-miR-5591-5p. Although some of the observed differences in miRNA distribution between these two experimental models could be explained by the different assay sensitivity and the related statistical selectivity, it was clear that the hypoxia-related functional changes in miRNA levels in the RISC components differed significantly from global population changes.
Figure 2. RISC miRNA compositional change during the hypoxia time course in HUVECs.
(A) The heat map represents the distribution of unique miRNAs affected by at least 2-fold during 2h, 8h and 16h of hypoxia exposure (Supplemental Data Set 2). At each time point, the miRNAs levels were corrected for the signal obtained using the normal rabbit IgG control and expressed as fold change relative to normoxic control. The heatmap generation and hierarchical clustering were performed with the Morpheus web server (Morpheus, https://software.broadinstitute.org/morpheus). The miRNAs that were induced or reduced in the RISC component analysis are denoted with red and blue fonts, respectfully. (B) The Venn diagram (80) represents the general distribution of unique mRNAs levels that changed by at least 2 log2 fold in HUVEC RISC samples during 2h, 8h and 16h of hypoxia exposure (Supplemental Data Set 3).
The miRNA composition in the RISC components was affected early during hypoxia, after 2 hours exposure, whereas global population changes in miRNA levels at that time were minimal. This suggested that miRNA-regulated adaptation to early hypoxia occurs through modification of RISC component composition with enrichment in specific miRNAs. Moreover, it also indicated that the functional adaption was not necessarily coupled with the transcriptional induction of these miRNAs. This surprising result was further supported by the observations that after 8 hours of exposure to hypoxia, when we observed significant increases in number of induced miRNAs in the global population that did not correspond to the miRNA changes in the RISC components. Furthermore, after 16 hours of exposure to hypoxia, there were dramatic changes in the miRNA distribution both globally and in the RISC components. There were also a large number of miRNAs that were reduced (Figure 2A). Taken together, the data clearly indicate that during time course of hypoxia, the functional changes in miRNA pool differ significantly from changes in global miRNA expression profiles, particularly during the early stages of hypoxia. This suggests that during these early stages, the changes in RISC miRNA content are based on selection rather than availability. How miRNA selection occurs in the RISC components during hypoxia, however, will require further study.
Since the RIP-Seq procedure permits access to both miRNA and mRNA components in RISC, we next analyzed the NGS for changes in the mRNA distributions. We observed that during the hypoxia time course, the mRNA content in RISC changed in a time-specific manner (Figure 2B, Supplemental Dataset 3). In RISC components from cells exposed to hypoxia for 2 hours, 481 mRNAs levels were increased (by at least 2 log2 fold), interestingly, 279 mRNAs were increased only at this time point, and 755 mRNAs were reduced including 554 mRNAs that were specific for this time point. After 8 hours, 727 mRNAs were increased, whereas only 156 of them were unique for this time point, while 369 were also increased after 16 hours and notably 380 of them were reduced after 16 hours, whereas only 25 mRNAs were induced at the 2-hour time point. However, from 436 mRNAs were reduced at 8 hours, 309 were specific for this time. Similarly, after 16 hours, 842 mRNAs were increased, but only 291 of them were specific for this time point, and the majority was also observed at 8 hours. 16 hours exposure to hypoxia showed a reduction of 491 mRNAs, among which 380 were unique, while the rest were either reduced or induced at 2 and 8 hours (Figure 2B). Taken together, in the RISC we observed a continuous increase in enrichment of mRNAs (from 481 at 2 hours up to 842 at 16 hours) and decreasing numbers of depleted mRNAs (from 755 at 2 hours to 491 at 16 hours). Importantly these changes in RISC mRNA content were unique for each time point, especially for 2 hours, which represented the cellular response to acute hypoxia.
We next asked what the functional role of hypoxia-related changes in mRNA RISC distribution meant. To address this, the mRNA changes at each time point were considered in a gene ontology context and the role of these mRNAs in adaptive response to hypoxia were analyzed using the Erichr database (P value below 0.05) (42). Notably, not all miRNA-targeted mRNAs are ultimately degraded following translational repression imposed by RISC, and furthermore, miRNA target mRNA, which is resistant to miRNA-mediated translational repression, does not undergo degradation despite being bound by RISC (43). Hence, not all RISC-observed miRNA-mRNA complexes will translate to the cellular target mRNA content and the subsequent protein changes. To gain functional insight into hypoxic the RISC composition, however, we assumed the model in which the increased levels of mRNAs in RISC might lead to their reduction in the global population and that the reduced mRNA levels in RISC would support its accumulation in the global population (44).
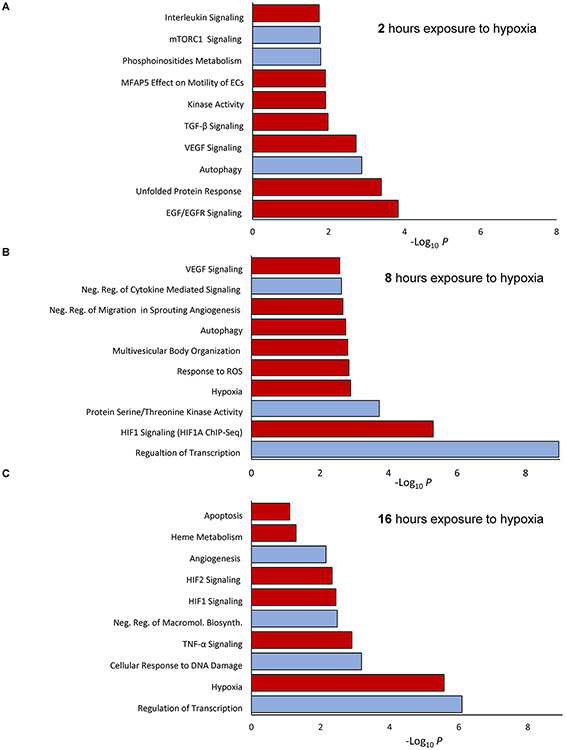
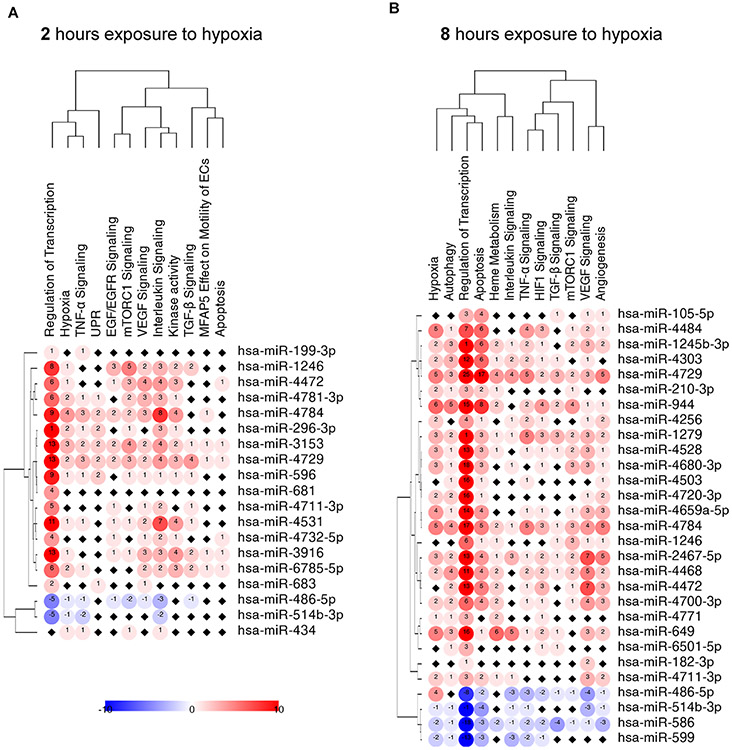
As shown in Figure 3 (Supplemental Dataset 4), the mRNA levels at each time point that increased in RISC composition (red) and those that decreased (blue) were considered separately. mRNAs that enriched RISC components after 2 hours of exposure to hypoxia were related to regulation of epidermal growth factor signaling, the unfolded protein response (UPR), kinase activity and VEGF2 signaling. The mRNAs that were reduced in RISC components at the same time were related to autophagy, mTOR signaling and phosphoinositide metabolism (Figure 3A). Hence, acute hypoxia exposed cells may utilize miRNA networks to prevent cell death and attenuate both VEGF-related signaling and the UPR, and stimulate autophagy.
Figure 3. The predicted cellular responses based on the mRNA composition of RISC samples obtained from HUVECs during the hypoxia time course.
The mRNA levels in RISC components that were increased in by 2 log2 folds (Red color) or decreased 2 log2 folds (Blue color) at (A) 2h, (B) 8h, and (C) 16h of hypoxia were assigned with the Erichr database to cellular signaling pathways (Supplemental Data Set 4) and selected based on P value < 0.05 (only the top 10 assignments are presented for each time point).
After 8 hours exposure to hypoxia, mRNA increases in RISC were assigned to cellular responses to hypoxia, including HIF-1 activity, VEGF signaling, angiogenesis, and responses to ROS (Figure 3B). Notably, many of these mRNAs were identified as HIF-1 transcriptional targets. The mRNAs that were reduced in RISC components were responsible for regulation of genes transcription. Hence, during this time point, miRNAs are used to attenuate the activity of HIF-1 related transcriptional induction of gene expression.
Finally prolonged exposure to hypoxia (16-hour time point) resulted in enriching RISC components in mRNAs involved in the canonical response to hypoxia that includes both HIF-1 and HIF-2 signaling, heme metabolism, and apoptosis (Figure 3C), whereas reduced mRNAs could be assigned as the ones responsible for response to DNA damage and regulation of transcription processes. Taken together, the mRNA changes in RISC components clearly illustrate miRNA’s role in regulating the development and intensity of the adaptive response to hypoxia including the metabolic switch, angiogenesis, prevention of cell death, and the support of HIF-1- and HIF-2-driven global transcriptional reprograming.
To gain a better understanding of the miRNA-mRNA interactions during hypoxic in the RISC components, we used a bioinformatic approach to identify experimentally verified miRNA regulatory effects using the miRTarBase (30). We focused on the miRNA-mRNA interactions that were confirmed with Western blots, qPCR, reporter assays and NGS, to assign RISC-identified miRNA to their potential co-immunoprecipitated mRNAs within the assigned cellular pathways. To visualize the data for each unique cellular pathway miRNA-mRNA interaction, we scored this as 1 for the unique RISC-enriched potential miRNA-mRNA hybrid and −1 for a depleted one. As shown in Figure 4A, the potential miRNA-mRNA pairs that were enriched in RISC components isolated from ECs exposed to hypoxia for 2 hours mainly were genes involved in regulation of transcription, response to hypoxia, and the VEGF and EGF/EGFR signaling pathways. Whereas the miRNA-mRNA pairs that were reduced had a very modest representation in the signaling pathways. Notably, the 8-hour exposure to hypoxia resulted in enrichment of RISC components with potential miRNA-mRNA pairs that mainly could be assigned to HIF-1 and VEGF signaling (Figure 4B), whereas the reduced miRNA-mRNA pairs were associated with mainly with regulation of transcription and apoptosis.
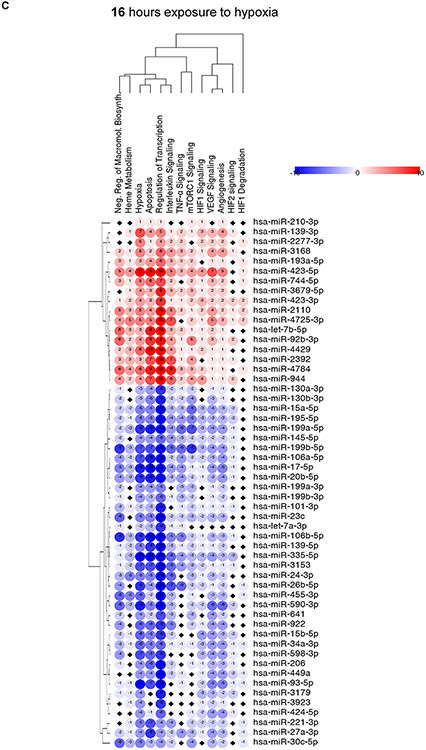
Figure 4. The miRNA-mRNA interactions in RISC samples from HUVECs exposed to hypoxia for (A) 2h, (B) 8h and (C) 16h along with their cellular signaling assignments.
The miRNA-mRNA targets interactions were analyzed with the use of miRTarBase (30). For each of signaling pathways, the individual miRNA-mRNA interactions were scored as 1 for the RISC enriched miRNAs or −1 for the RISC depleted miRNAs (Blue). The black square depicts no assignment. The heatmap generation and hierarchical clustering were performed with the Morpheus web server (Morpheus, https://software.broadinstitute.org/morpheus).
Finally in ECs exposed to hypoxia for 16 hours, the potential miRNA-mRNA pairs that were enriched in RISC components were involved in HIF-1 and HIF-2, TNF-α signaling, the VEGF pathway, and the canonical responses to hypoxia (Figure 4C). Furthermore, enriched miRNA-mRNA pairs could be also assigned to regulation of apoptosis. At the same time, RISC components were depleted of potential miRNA-mRNA pairs involved in regulation of transcription, of the negative regulation of macromolecule synthesis, and of the cellular response to hypoxia and apoptosis (Figure 4C).
Taken together, the miRNA-mRNA composition of hypoxic RISC components resembles the time development of all of the main aspects of the adaptive hypoxic response, including the transition from HIF-1 to HIF-2, tuning of angiogenesis, cellular metabolism, and preventing apoptosis and DNA damage. Furthermore, along with the hypoxic time course, the composition of RISC components is modulated and developed in order to fine-tune adaption to hypoxia and to support cellular survival. The changes in the RISC component compositions support the hypothesis that microRNA-mediated gene regulatory networks serve in a regulatory role in buffering the cellular processes by setting threshold settings for their target gene levels and thus conferring the robustness of activated cellular signaling networks (45, 46).
Identification of miRNAs regulated during hypoxia by HIF-1 and HIF-2
Although the both HIF-1 and HIF-2 are the main regulators of the adaptive hypoxic response, the hypoxia-related miRNA changes have been identified as being mainly HIF-1 dependent (11). However, our analysis of the hypoxic changes in miRNAs distributions clearly demarcated the acute (2h) and prolonged (16h) hypoxia effects on both the composition of the RISC components and of the global population miRNA levels. Therefore, next we focused on the impact of ECs transition from HIF-1 to HIF-2 on the miRNA distribution in hypoxia-exposed cells in the global population compared to the RISC components. We analyzed using next generation sequencing the cellular changes in miRNA levels upon HIF-1 and HIF-2 silencing in HUVECs exposed to hypoxia for 8 hours since this was the transitional time point between acute and prolonged hypoxia and a time in which both HIF-1 and HIF-2 were well expressed. We focused on the cellular changes in the miRNA distributions since our goal was to identify the miRNAs that could be transcriptionally regulated by either HIF. As shown in Supplemental Figure 2, HIF1A and EPAS1 silencing in hypoxia-exposed HUVECs was efficient at both protein and mRNA levels, as well as when both HIF1A and EPAS1 were silenced at the same time. Furthermore, although in some experiments we have observed some small increase in HIF1A mRNA levels up on EPAS1 silencing (~ 1.3-fold), this phenomenon was not significant at the HIF-1α protein levels between the three biological replicates. Similarly, silencing HIF1A led to small reduction of EPAS1 mRNA, but again this was not reflected in HIF-2α protein levels (Supplemental Figure 2). We also obtained and analyzed miRNA profiles following efficient HIF1A and EPAS1 silencing in HUVECs exposed to hypoxia for 2 hours, when HIF1A transcript increased upon EPAS1 silencing (Supplemental Figure 2). Given that the silencing related changes in miRNA expression were very limited and similar to what was seen in Figure 2, and we decided to focus on the 8-hour time point.
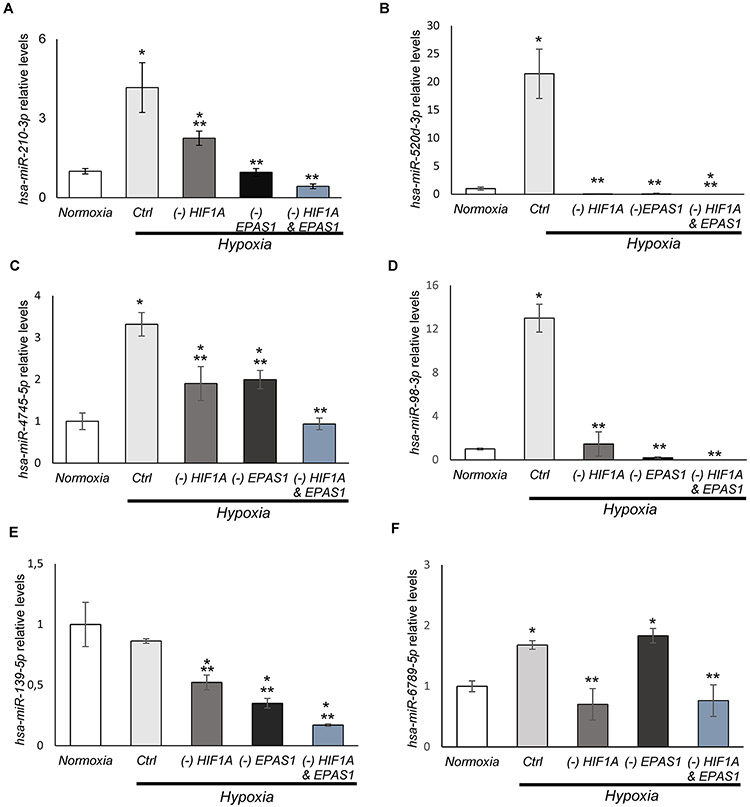
The follow up next generation analysis resulted in the initial selection of 17 miRNAs that were reduced by at least one-fold upon HIF-1α silencing during hypoxia, (Supplemental Data Set 5). Nevertheless, independent validation of next generation sequencing data resulted in identification of 5 potentially both HIF-1 and HIF-2-dependent miRNAs and 1 only HIF-1 dependent (hsa-miR-6789-5p): hsa-miR-210-3p; hsa-miR-520d-3p; hsa-miR-98-3p; hsa-miR-4745-5p; hsa-miR-139-5p and (Figure 5A-F). The majority of these miRNAs were induced in hypoxia exposed cells, except for miR-139-5p, which remained unaffected (Figure 5E). Nevertheless, among them only the hypoxic induction of hsa-miR-210-3p was observed in global population analysis of hypoxic miRNA distribution and in the RISC components content, whereas the changes in miR-98-3p and miR-139-5p were only in RISC components.
Figure 5. The majority of HIF-1 dependent miRNAs are also HIF-2 dependent.
The follow-up qPCR validation of NGS identified miRNA levels that changed by at least log fold upon HIF-1α silencing in HUVECs. This analysis confirmed 6 potentially HIF-1 dependent miRNAs: (A) hsa-miR-210-3p; (B) hsa-miR-520d-3p; (C) hsa-miR-4745-5p; (D) hsa-miR-98-3p; (E) hsa-miR-139-5p; and (F) hsa-miR-6789-5p. The validation was performed upon both HIF-1α (-HIF1A) and HIF-2α (-EPAS1) silencing in HUVECs exposed to hypoxia for 8h. The cells cultured in normoxia and transfected with control siRNA were used as the normoxic control. miRNA levels were normalized to RNU44 and expressed as a fold change over control in normoxia. The data represent mean of 3 biological replicates (3 technical replicates each) ± SEM of three (*P< 0.05 was considered significant).
Next, we analyzed these miRNAs’ genomic locations for the presence of specific HIF-1 and HIF-2 HRE motifs in their target gene promoter regions. For each gene promoter sequence, we looked at the open chromatin regions established in the HUVEC cell line by the ENCODE project and focused on two distinct HRE motifs annotated to HIF-1 and HIF-2 (see Methods section for details). For hsa-miR-210-3p, we identified 12 and 18 HIF-1 and HIF-2 motifs, respectively, 1 HIF-1 and 2 HIF-2 motifs for hsa-miR-139-5p, 2 HIF-1 and 1 HIF-2 motifs for hsa-miR-139-5p and 17 HIF-1 and 14 HIF-2 motifs for hsa-miR-4745-5p. Whereas for some miRNAs like hsa-miR-98-3p, hsa-miR-520d-3p, we could not detect open chromatin regions in our database, so the presence of HIF motifs was unclear in these cases.
Since both the bioinformatic analysis of HRE motifs and the NGS results suggested the possibility that the HIF-1-dependent miRNA could be also regulated by HIF-2, we verified the effects of EPAS1 silencing on their hypoxic expression. Surprisingly, all identified miRNAs except for hsa-miR-6789-5p were also significantly reduced in the absence of HIF-2 (Figure 5). Notably, hsa-miR-210-3p, mainly considered to be HIF-1 dependent miRNA, was more reduced up on HIF-2 silencing than upon HIF-1 silencing (Figure 5A), whereas HIF-2 was mainly responsible for expression of this miRNA after 16 hours exposure to hypoxia (Supplemental Figure 3A). The hsa-miR-520d-3p and miR-98-3p required both HIF-1 and HIF-2 to be expressed during hypoxia (Figure 5BD).
Identification of miRNAs regulated during hypoxia by HIF-2
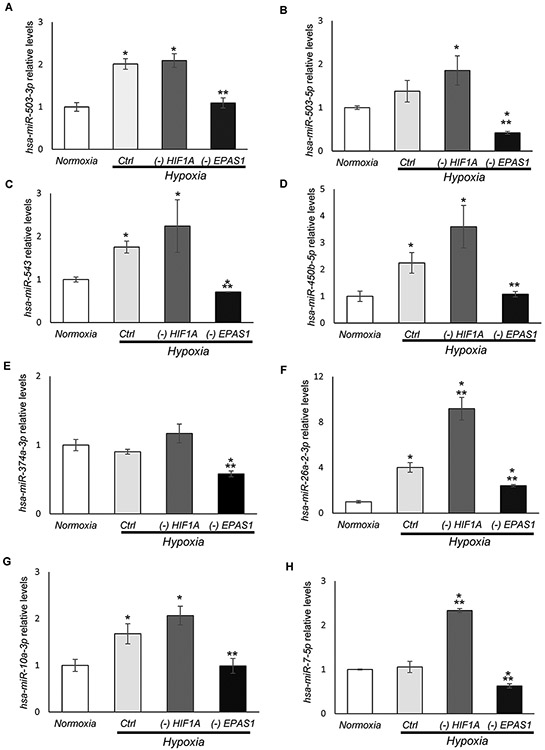
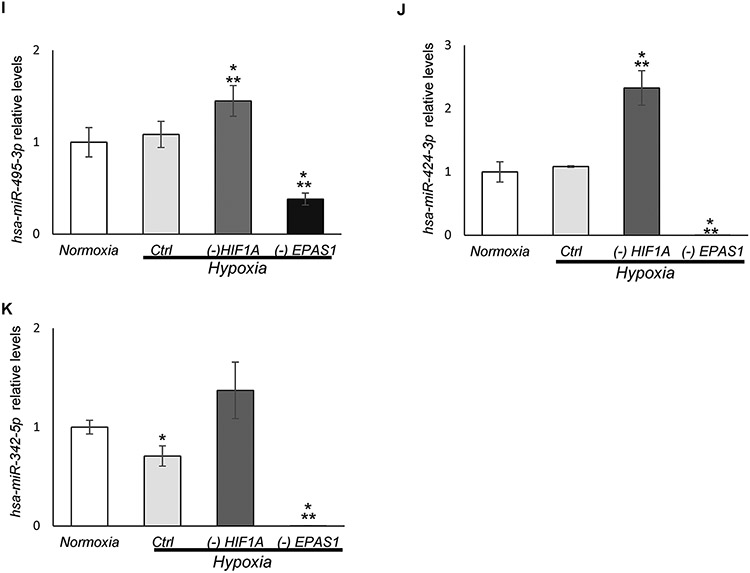
A similar analysis was performed in hypoxia exposed cells during HIF-2α silencing that resulted in initial selection of 8 miRNAs that were reduced by at least one-fold upon HIF-2α silencing during hypoxia (Figure 6, Supplemental Data Set 5). Independent validation of next generation sequencing data resulted in identification of 11 potentially HIF-2 dependent miRNAs: hsa-miR-7-5p; hsa-miR-503-3p; hsa-miR-503-5p. hsa-miR-543; hsa-miR-450b-5p; hsa-miR-374a-3p, hsa-miR-10a-3p, hsa-miR-424-3p, hsa-miR-495-3p and hsa-miR-342-5p; hsa-miR-26a-2-3p (Figure 6A-K). Surprisingly only 5 of these miRNAs were induced in hypoxia exposed cells: hsa-miR-503-3p; hsa-miR-503-5p, hsa-miR-543; hsa-miR-10a-3p and hsa-miR-26a-2-3p (Figure 6), whereas levels of the others remained constant during hypoxia. The hypoxic induction of hsa-miR-503-3p, hsa-miR-503-5p, and hsa-miR-424-3p was observed in the global population analysis, whereas the majority of these miRNA changes were reflected in RISC component content that included hsa-miR-424-3p, hsa-miR-495-3p, hsa-miR-7-5p, hsa-miR-450b-5p; hsa-miR-543, hsa-miR-503-3p and hsa-miR-503-5p. Interestingly, the analysis of HRE motifs revealed that hsa-miR-7-5p has 1 HIF-1 motif, hsa-miR-374a-3p has 1 HIF-1 and 1 HIF-2 motif. Whereas, hsa-miR-503-3p and hsa-miR-10a-3p have 4 HIF-1 and 10 HIF-2 motifs, whereas hsa-miR-26a, hsa-miR-424-3p,hsa-miR-503-5p, hsa-miR-503-3p hsa-miR-543, hsa-miR-450b-5p, hsa-miR-495-3p were expressed from chromatin regions that are not open during normoxia. Furthermore, in case of hsa-miR-342-5p, we did not identify any HIF-1 or HIF-2 motifs. We have also tested the effects of HIF-1α silencing on this miRNA levels in cells exposed to hypoxia (Figure 6), and noted that expression of all of them were HIF-2-specific, and HIF-1 silencing in some cases (hsa-miR-26a-2-3p; hsa-miR-7-5p; hsa-miR-495-3p; and hsa-miR-424-3p) resulted in a significant increase in the expression of these miRNAs. Importantly, none of the identified HIF-2-dependent miRNAs were also HIF-1 dependent (reduced upon HIF-1α silencing). Taken together, we identified 3 different groups of HIF-dependent miRNAs: one HIF-1 specific (hsa-miR-6789-5p); five both HIF-1 and HIF-2 specific, and eleven HIF-2 specific miRNAs. The identified miRNAs were also confirmed as components of the RISC components. These findings support the importance of separating the transcriptional signaling by the HIF switch and its impact on the miRNA profiles.
Figure 6. HIF-2 modulates expression of 11 miRNAs in hypoxia-exposed endothelial cells.
The follow-up qPCR validation of NGS identified miRNA levels that changed by at least log fold upon HIF-1α silencing in HUVECs exposed to hypoxia (Supplemental Data Set 6). This confirmed 11 potentially HIF-2 dependent miRNAs: (A) hsa-miR-503-3p, (B) hsa-miR-503-5p, (C) hsa-miR-543, (D) hsa-miR-450b-5p, (E) hsa-miR-374a-3p, (F) hsa-miR-26a-2-3p, (G) hsa-miR-10a-3p, (H) hsa-miR-7-5p, (I) hsa-miR-495-3p, (J) hsa-miR-424-3p, and (K) hsa-miR-342-5p. The validation was performed upon both HIF-1α (-HIF1A) and HIF-2α (-EPAS1) silencing in HUVECs exposed to hypoxia for 8h, and the cells that were cultured in normoxia and transfected with control siRNA were used as a normoxic control. miRNA levels were normalized to RNU44 and expressed as a fold change over control in normoxia. The data represent mean of 3 biological replicates (3 technical replicates each) ± SEM of three (*P< 0.05 was considered significant).
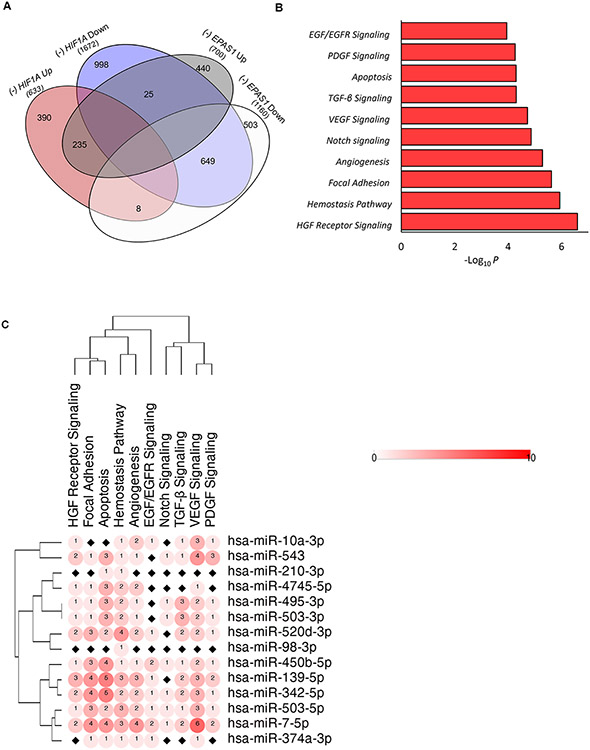
In final step, we used a similar approach as for the RISC components to evaluate the functional role of these HIF-dependent miRNAs in the adaptive response to hypoxia by using the data of the global mRNA changes upon HIF silencing (Supplemental Data Set 6). We focused on the data obtained from ECs exposed for 8 hours to hypoxia (Figure 7A). We observed that silencing of HIF1A reduced (by 2-fold) the expression of 1672 transcripts, among which 649 were also reduced after EPAS1 silencing. Whereas, EPAS1 silencing resulted in reduction of 1160 genes. Furthermore, HIF1A silencing resulted in the upregulation of 633 genes among which 235 were also induced in after EPAS1 silencing. EPAS1 silencing alone resulted in upregulation of total 700 genes. Although these data provided only a brief insight into the HIF transition, they clearly show that despite the common signaling pathways, both of these factors have important distinct functions during hypoxic in ECs.
Figure 7. The Venn diagram represents the NGS-based general distribution of unique mRNAs levels that are changed by at least 2 log2 fold upon HIF-1α and HIF-2α silencing in HUVECs exposed to hypoxia.
(A) The mRNAs that were upregulated in hypoxia exposed HUVECs upon HIF-1α and HIF-2α silencing were assigned with the Erichr database to cellular signaling pathways and selected based on P value < 0.05. (B) The miRNA-mRNA targets interactions were analyzed with the use of miRTarBase (30). For each of the signaling pathways, the individual miRNA-mRNA interactions were scored as 1. (C) The heatmap generation and hierarchical clustering were performed with the Morpheus web server (Morpheus, https://software.broadinstitute.org/morpheus).
Given that our focus was on HIF-related miRNAs, we narrowed our analysis to the transcripts that were upregulated in the absence of HIFs since this upregulation could potentially result from increased mRNA stability caused by the depletion of specific miRNAs. We assigned these upregulated genes into signaling pathways (Figure 7B), and noted that absence of HIFs leads to accumulation of mRNAs similar to that observed in the RISC component analysis. The increased pathways involved included hepatocyte growth factor signaling, hemostasis, focal adhesion, Notch and VEGF, EGF and TGF signaling, and the regulation of apoptosis and angiogenesis. Given that only some of these expression changes could be considered as a direct consequence of HIF-dependent miRNAs on these mRNAs, we performed a similar identification of the miRNA-mRNA pairs as we did for RISC components and assigned these data to the identified pathways as shown in Figure 7C. For the majority of the identified HIF-dependent miRNAs, we found assignments in pathways for hsa-miR-139-5p, hsa-miR-210-3p, miR-7-5p, miR-503-3p, miR-503-5p and miR-342-5p. Furthermore, all of these miRNAs were induced either by HIF-1 and HIF-2 or by HIF-2 alone. Taken together, these data illustrate that the transition from HIF-1 to HIF-2 allows the cells to enhance expression levels of miRNAs initially induced by HIF-1 as well as to induce expression of HIF-2 dependent ones. Thus HIF-2 thorough its effect on miRNAs, modulates mRNA levels in order to both sustain and fine-tune cellular adaptation to prolonged hypoxia and angiogenesis.
Discussion
The molecular mechanisms that determine cell survival during hypoxia that is termed the hypoxic adaptive response (1) offer a number of therapeutic opportunities for many of the deadliest human diseases that include coronary artery disease, stroke, chronic obstructive pulmonary disease, and cancer. In order to take advantage of controling this hypoxia adaptive response, it is important to understand molecular basis of this complex process including the role of miRNAs and their function in mediating the transition from HIF-1 to HIF-2 signaling (11). Although numerous studies have tried to indentify these hypoxia-related changes in the global population miRNA profiles, the majority of these studies examined the steady state analysis of only a single time point after prolonged of exposure to hypoxia (16 or more hours). Furthermore, most of these studies were performed in cancer cells, and did not consider the consequences of the HIF-1 to HIF-2 switch for miRNA-mediated regulation of the adaptive response to hypoxia.
To determine how the miRNA changes influence the transition form HIF-1 to HIF-2, we performed a dedicated genome-wide miRNA time-course study in ECs exposed to hypoxia. Although hypoxia results in changes in the global population miRNA levels for the vast majority of affected miRNAs, these changes are gradual and require at least 8 to 16 hours exposure to reach maximal or minimal levels. Furthermore, only a small number of miRNAs are affected during accute hypoxia (2 hours) and thus there was no clear relationship to the HIF-1 levels since at 8 hours both HIF-1 and HIF-2 are present. Whereas at 16 hours, HIF-2 becomes the main driver of hypoxic signaling in ECs.
Given that hypoxia-related genome-wide changes in gene expression profiles are extremely complex and affect hundreds of transcripts (47-49), we decided to examine the hypoxia-related changes in miRNA and mRNA compositions of the RNA-induced silencing complexes (RISC). Only a small proportion of the total cellular miRNA is contained within the RISC (44, 50), and the level of RISC association of a given endogenous miRNA is regulated by the available mRNA targetome (51). We hypothesized that the level of the miRNAs in RISC might be a better predictor the activity of a given miRNA than the total global miRNA content and it would also allow for a better correlation of the mRNA with miRNA activity during the development of adaptive response to hypoxia.
To test this, we performed Ago2-RIP-seq to detect both RISC-associated miRNA and mRNA in ECs exposed to hypoxia for 2 hour, 8 hours and 16 hours (Figure 2). The miRNA changes observed in RISC from cells exposed to hypoxia poorly correlated with their respective total profiles. We also observed that the hypoxic changes in miRNA composition became more pronounced at the later stages of the time course. The number of miRNA affected by 2 hours of exposure to hypoxia was drastically higher in RISC, and the complexes obtained after 16 hours of exposure to hypoxia were depleted of large number of miRNAs when compared to ones obtained from normoxic cells. Given that the miRNA content of the complexes remained unaffected by hypoxia (Supplemental Figure 1D), similar to the global miRNA content (Figure 1B), this suggested that the miRNA depletion of RISC observed after 16 hours of exposure to hypoxia is a part of a specific regulation rather than the general consequences of hypoxia-related impairing of miRNA biogenesis (52-54). Supporting this model, the RISC formation, especially Argonaute 4 (AGO4) levels, were shown to be increased during hypoxia (52). This data is also in good agreement with previous reports of the analyses of pri-miRNAs and mature miRNAs levels during hypoxia that suggested that additional mechanisms are involved in maintaining hypoxic miRNA expression (52).
The 2 hours of exposure to hypoxia indicated that there were significant changes in RISC and suggested that the miRNA-mediated adaptation to low oxygen pressure relies on the efficient utilization of global miRNAs without the necessity of increasing their cellular content through transcription. The depletion of RISC of numerous miRNAs at 16 hours may also suggest that the cells are minimizing miRNAs to prevent reduction of expression of proteins important for adaptive response to hypoxia caused by chronic hypoxia-related translational repression (55, 56).
Given that the miRNA presence in RISC results from availability of their mRNA targets (51), we next analyzed their respective mRNA content using RNA-seq analyses. Notably, the mRNA profiles of RISC clearly indicated that at each time of exposure to hypoxia a large number of specific transcripts were uniquely affected. Interestingly, during the time course, the number of mRNAs that are enriched in RISC increased (481 mRNAs at 2 hours, 727 at 8 hours and 842 at 16 hours), whereas a number of depleted transcripts decreased at 8 hours (755 mRNAs at 2 hours, 436 at 8 hours and 491 at 16 hours). We tested that the RISC enrichment in mRNA could be predictor of reduced protein levels, whereas depletion would have the opposite effect. This would suggest that during acute hypoxia, the RISC composition supports the development of adaptive response allowing for the accumulation of proteins crucial for initial cell survival, whereas at later time points, the RISC composition is modified to govern/adjust oxygen homeostasis and metabolism.
To test this hypothesis, we performed a functional assignment analysis for the RISC-observed mRNA changes at each time point and found that at 2 hours the RISC were depleted of mRNAs involved in initiation of transcription as well as autophagy, whereas enriched transcripts were involved in the unfolded protein response (UPR), apoptosis, VEGFA signaling and apoptosis. This suggests that miRNA composition of RISC during acute hypoxia support HIF-1-related activation and changes in mitochondrial metabolism as well as survival through autophagy. At the same time, they prevent activation of apoptosis, UPR and modulate the extend of VEGF signaling (57, 58). After 8 hours, the RISC composition supports RNA binding and mRNA processing as well as chromatin reorganization, and controls the extend of HIF-1 signaling and angiogenesis. At 16 hours, mRNA processing, transcription and chromatin remodeling are promoted, along with the cellular responses to DNA damage, cell cycle progression, and the regulation of both HIF-1 and HIF-2 signaling and apoptosis. Furthermore, these observations are in good agreement with the numerous reports that miRNA modulate their targetome through both positive and negative feedback loops (13, 59). This was further supported by our analysis of involvement of potential miRNA-mRNA complexes that clearly indicate that both mRNA and miRNA changes observed in RISC complement each other and serve to regulate the proposed signaling pathways involved in cellular adaptation to hypoxia.
Furthermore, the composition of RISC components suggest that the miRNAs are modulating the HIF activated cellular responses to hypoxia (45, 46) that could also protect the cells from damage if the oxygen levels are rapidly restored and lead to reoxygenation injury (60). Support for this theory is also provided by the comparisons of RISC mRNA content from the results of our previous studies using microarrays to follow the global transcriptomic changes in HUVECs exposed to hypoxia during the same time periods (61). In this analysis, we found that after 8- and 16-hours of hypoxia about one third of induced transcripts were enriched in RISC (Supplemental Figure 3BC) and these mRNAs coded for proteins involved in HIF-1 signaling, apoptosis as well as glycolysis. That being said, further extended molecular and functional studies will be necessary to confirm the extend of each of these miRNA functions. Given that miRNA-dependent regulation of expression is often tissue and cell specific (62), similar types of studies will be necessary to obtain the composition of RISC components during hypoxia in other experimental models such as hepatocytes, epithelial cells and immune cells.
The changes in miRNA-mRNA composition of RISC may also be an important regulator and controller of both adaptation and the transition from acute to chronic to hypoxia. Interestingly, miRNA comprise only about 0.01% of total RNA (63), whereas mRNA account for up to 4% of cellular RNA (64). Hence, the changes in miRNA-mRNA complexes observed in RISC are an important mechanism for fine tuning the hypoxia adaptive response to prevent its uncontrolled expanse rather than relying on an abrupt shutdown mechanism. Despite the fact that our RISC analysis suggested some level independence from the transcriptional regulation of miRNA levels, the impact of HIF-1 and HIF-2 on miRNA expression should not be overlooked. For example, previous studies reported that hypoxia and ischemia change the expression profiles of many miRNAs that are HIF-dependent (65, 66). A large number of these studies, however, have focused on cancer cell lines only and HIF-1’s role and did not consider the consequences of HIF-2 activity (11). Furthermore, most of the previous studies focused on miRNA and mRNA changes during chronic hypoxia, without relating the observed miRNA changes to the specific HIF-1 and HIF-2 levels. In spite of the fact that HIF-1 and HIF-2 are the main regulators of the adaptive hypoxic response, many of the hypoxia-related miRNAs changes have been identified as being mainly HIF-1 dependent (11). Our results of hypoxic miRNA profiles in both the global population and in the RISC fractions clearly demarcated between acute and chronic hypoxia and illustrated the regulated transition from HIF-1 to HIF-2.
Our focus was on the 8-hour time point when both HIFs were well represented. We analyzed the cellular miRNA and mRNA fractions since our goal was to identify miRNAs that could be transcriptionally regulated by HIF-1 and HIF-2. These resulted in the identification of 6 potentially HIF-1-dependent miRNAs: hsa-miR-210-3p; hsa-miR-520d-3p; hsa-miR-98-3p; hsa-miR-4745-5p; hsa-miR-139-5p and hsa-miR-6789-5p, among which only the well-known hsa-miR-210-3p was induced in both in the global and RISC fractions. Whereas, the induction of miR-98-3p and miR-139-5p was only observed in RISC fraction. The analysis of the miRNAs’ genomic locations for the presence of specific HIF-1 and HIF-2 motifs indicated that all of these that promotors regions were located in open chromatin regions for both HIF-1 and HIF-2. This suggested that these HIF-1-dependent miRNAs could be also regulated by HIF-2, which was further verified with EPAS1 silencing. Notably, all identified miRNAs except for hsa-miR-6789-5p were also significantly reduced in the absence of HIF-2. This observation is in good agreement with our previous study showing that the majority of genes responsible for HIF-dependent cellular responses to hypoxia can be regulated by both HIF-1 and HIF-2 (48) as well as previous report of miR-210 being a target of both HIF-1 and HIF-2 in renal cancer (67).
A parallel analysis that we performed to identify HIF-2α-related miRNAs resulted in identification of 11 miRNAs. The hypoxic induction of hsa-miR-503-3p, hsa-miR-503-5p and hsa-miR-424-3p was observed in global analysis of hypoxic miRNA distribution, whereas the majority of the miRNA changes were indicated in RISC contents that included hsa-miR-424-3p, hsa-miR-495-3p, hsa-miR-7-5p, hsa-miR-450b-5p; hsa-miR-543, hsa-miR-503-3p and hsa-miR-503-5p. Furthermore, 6 of these miRNAs were not induced in hypoxia exposed cells, suggesting that the HIF-2α transcriptional activity is either substituted by some other transcription factors or that HIF-2α driven expression of this miRNA is equalizing their functional utilization in RISC that relies on degradation of these miRNA-mRNA hybrids rather than on translational suppression. Nevertheless, this hypothesis will require further study. The expression of all of these miRNAs were HIF-2-specific since HIF-1α silencing did not decrease their expression. Taken together, we identified 3 different groups of HIF-dependent miRNAs: one HIF-1 specific (hsa-miR-6789-5p); five both HIF-1 and HIF-2 specific, and eleven HIF-2 specific miRNAs. The identified miRNAs were also confirmed as components of the RISC components. Previous reports have indicated that HIF-1 governs the expression of several miRNAs during hypoxia including miR-210 (68), miR-146a (69), miR-145 (70), miR-382 (71), miR-191 (72), miR-363 (73), miR-421 (74), miR-204 (75), miR-30a, miR-21 (76, 77), miR-687 (78), miR-155 (15), and miR-429 (13) and miR-19a (79), whereas to the best to our knowledge this is the first study identifying HIF-1 and HIF-2 specific, and HIF-2 specific miRNAs.
Importantly, we correlated the identified the HIF-dependent miRNAs with the corresponding mRNA changes and observed that the identified HIF-dependent miRNAs are likely involved in governing adaptive response to hypoxia signaling pathways that include angiogenesis-related signaling and the hypothesis that the transition from HIF-1 to HIF-2 modulates miRNA levels in order to both sustain and tune ECs cellular adaptation to prolonged hypoxia. Furthermore, although some of the miRNAs that were identified in this work were already well established as HIF-dependent master regulators of hypoxic response, the role of the majority of them is unknown and will require further study. Notably, deregulation of these miRNAs has been reported in cancer and cardiovascular diseases (Supplemental Table 2), suggesting their potential for developing novel therapeutic and diagnostic approaches.
In summary, our study demonstrates that hypoxia-related changes in miRNA-mRNA RISC composition serve both the development and control of adaptive responses to hypoxia and do not completely rely on transcriptionaly driven changes in miRNA experssion levels. Furthermore, for the first time we show that the hypoxic levels of the vast majority of HIF-1 dependent miRNAs are also HIF-2 dependent, whereas HIF-2 additionaly specifically governs the expression of a number of miRNAs, and thus the switch from HIF-1 to HIF-2 in ECs provides important level of miRNA-driven control in the hypoxia adaptive pathways in endothelial cells.
Supplementary Material
Funding:
This research was funded by National Science Center "SONATA BIS" Program under contract UMO-2015/18/E/NZ3/00687 (R.B.). J.F.C. was funded by an NIH P30 DK072482 Grant and a Research Development Program (RDP) Grant from the Cystic Fibrosis Foundation.
Abbreviations
- AGO2
Argonaute protein 2
- AGO4
Argonaute protein 4
- BSA
bovine serum albumin
- DMOG
Dimethyloxalylglycine
- ECs
Endothelial cells
- EGF
Epidermal growth factor
- EGFR
Epidermal growth factor receptor
- EPAS1
hypoxia - inducible factor 2 alpha
- GLUT1
Glucose transporter 1
- HIF
Hypoxia-inducible factor
- HIF1A
Hypoxia-inducible factor 1 alpha
- HIF-1
Hypoxia-inducible factor 1
- HIF-2
Hypoxia-inducible factor 2
- HUVEC
Human umbilical vein endothelial cells
- HRE
Hypoxia response element
- MAPK
Mitogen-activated protein kinase
- miRNA
microRNA
- mTOR
Mammalian target of rapamycin
- NGS
Next generation sequencing
- PHD
Prolyl hydroxylase
- RIP-Seq
RNA Immunoprecipitation Sequencing
- RISC
RNA-induced silencing complex
- RPLP0
Ribosomal Protein Lateral Stalk Subunit P0
- ROS
Reactive oxygen species
- siRNA
Small interfering RNA
- TGF
Transforming growth factor
- TNF-α
Tumor necrosis factor α
- UPR
Unfolded protein response
- VEGFA
Vascular endothelial growth factor A
Footnotes
Conflicts of Interest: The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Data Availability:
Deep sequencing data were deposited in Gene Expression Omnibus (GEO) at accession numbers: GSE116909 (10), GSE190240 and GSE190242.
References
- 1.Semenza GL (1999) Perspectives on oxygen sensing. Cell 98, 281–284 [DOI] [PubMed] [Google Scholar]
- 2.Wang GL, and Semenza GL (1993) General involvement of hypoxia-inducible factor 1 in transcriptional response to hypoxia. Proceedings of the National Academy of Sciences of the United States of America 90, 4304–4308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Semenza GL (1999) Regulation of mammalian O2 homeostasis by hypoxia-inducible factor 1. Annual review of cell and developmental biology 15, 551–578 [DOI] [PubMed] [Google Scholar]
- 4.Skulski M, Bartoszewski R, Majkowski M, Machnicka B, Kuliczkowski K, Sikorski AF, and Boguslawska DM (2019) Efficient method for isolation of reticulocyte RNA from healthy individuals and hemolytic anaemia patients. J Cell Mol Med 23, 487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koh MY, and Powis G (2012) Passing the baton: the HIF switch. Trends in biochemical sciences 37, 364–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keith B, Johnson RS, and Simon MC (2011) HIF1alpha and HIF2alpha: sibling rivalry in hypoxic tumour growth and progression. Nature reviews. Cancer 12, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landau D, London L, Bandach I, and Segev Y (2018) The hypoxia inducible factor/erythropoietin (EPO)/EPO receptor pathway is disturbed in a rat model of chronic kidney disease related anemia. PloS one 13, e0196684–e0196684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su J, Li Z, Cui S, Ji L, Geng H, Chai K, Ma X, Bai Z, Yang Y, Wuren T, Ge R-L, and Rondina MT (2015) The Local HIF-2α/EPO Pathway in the Bone Marrow is Associated with Excessive Erythrocytosis and the Increase in Bone Marrow Microvessel Density in Chronic Mountain Sickness. High Alt Med Biol 16, 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J, Du F, Shen G, Zheng F, and Xu B (2015) The role of hypoxia-inducible factor-2 in digestive system cancers. Cell Death & Disease 6, e1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kochan-Jamrozy K, Kroliczewski J, Moszynska A, Collawn JF, and Bartoszewski R (2019) miRNA networks modulate human endothelial cell adaptation to cyclic hypoxia. Cell Signal 54, 150–160 [DOI] [PubMed] [Google Scholar]
- 11.Serocki M, Bartoszewska S, Janaszak-Jasiecka A, Ochocka RJ, Collawn JF, and Bartoszewski R (2018) miRNAs regulate the HIF switch during hypoxia: a novel therapeutic target. Angiogenesis 21, 183–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janaszak-Jasiecka A, Bartoszewska S, Kochan K, Piotrowski A, Kalinowski L, Kamysz W, Ochocka RJ, Bartoszewski R, and Collawn JF (2016) miR-429 regulates the transition between Hypoxia-Inducible Factor (HIF)1A and HIF3A expression in human endothelial cells. Scientific reports 6, 22775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bartoszewska S, Kochan K, Piotrowski A, Kamysz W, Ochocka RJ, Collawn JF, and Bartoszewski R (2015) The hypoxia-inducible miR-429 regulates hypoxia-inducible factor-1alpha expression in human endothelial cells through a negative feedback loop. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 29, 1467–1479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Madanecki P, Kapoor N, Bebok Z, Ochocka R, Collawn JF, and Bartoszewski R (2013) Regulation of angiogenesis by hypoxia: the role of microRNA. Cellular & Molecular Biology Letters 18, 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruning U, Cerone L, Neufeld Z, Fitzpatrick SF, Cheong A, Scholz CC, Simpson DA, Leonard MO, Tambuwala MM, Cummins EP, and Taylor CT (2011) MicroRNA-155 Promotes Resolution of Hypoxia-Inducible Factor 1 alpha Activity during Prolonged Hypoxia. Molecular and cellular biology 31, 4087–4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seton-Rogers S (2011) HIF switch. Nature Reviews Cancer 11, 391–391 [DOI] [PubMed] [Google Scholar]
- 17.Degrauwe N, Schlumpf TB, Janiszewska M, Martin P, Cauderay A, Provero P, Riggi N, Suva ML, Paro R, and Stamenkovic I (2016) The RNA Binding Protein IMP2 Preserves Glioblastoma Stem Cells by Preventing let-7 Target Gene Silencing. Cell Rep 15, 1634–1647 [DOI] [PubMed] [Google Scholar]
- 18.Bach DH, Luu TT, Kim D, An YJ, Park S, Park HJ, and Lee SK (2018) BMP4 Upregulation Is Associated with Acquired Drug Resistance and Fatty Acid Metabolism in EGFR-Mutant Non-Small-Cell Lung Cancer Cells. Molecular therapy. Nucleic acids 12, 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartoszewska S, Kamysz W, Jakiela B, Sanak M, Kroliczewski J, Bebok Z, Bartoszewski R, and Collawn JF (2017) miR-200b downregulates CFTR during hypoxia in human lung epithelial cells. Cellular & molecular biology letters 22, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gebert M, Sobolewska A, Bartoszewska S, Cabaj A, Crossman DK, Kroliczewski J, Madanecki P, Dabrowski M, Collawn JF, and Bartoszewski R (2021) Genome-wide mRNA profiling identifies X-box-binding protein 1 (XBP1) as an IRE1 and PUMA repressor. Cell Mol Life Sci 78, 7061–7080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartoszewska S, Kroliczewski J, Crossman DK, Pogorzelska A, Baginski M, Collawn JF, and Bartoszewski R (2021) Triazoloacridone C-1305 impairs XBP1 splicing by acting as a potential IRE1 alpha endoribonuclease inhibitor. Cellular & Molecular Biology Letters 26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartoszewski R, Gebert M, Janaszak-Jasiecka A, Cabaj A, Kroliczewski J, Bartoszewska S, Sobolewska A, Crossman DK, Ochocka R, Kamysz W, Kalinowski L, Dabrowski M, and Collawn JF (2020) Genome-wide mRNA profiling identifies RCAN1 and GADD45A as regulators of the transitional switch from survival to apoptosis during ER stress. Febs J 287, 2923–2947 [DOI] [PubMed] [Google Scholar]
- 23.Bartoszewska S, Cabaj A, Dabrowski M, Collawn JF, and Bartoszewski R (2019) miR-34c-5p modulates X-box-binding protein 1 (XBP1) expression during the adaptive phase of the unfolded protein response. Faseb Journal 33, 11541–11554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Livak KJ, and Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif.) 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 25.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29, 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, and Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, and Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome biology 3, RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garmire LX, and Subramaniam S (2012) Evaluation of normalization methods in mammalian microRNA-Seq data. RNA 18, 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, and Ma'ayan A (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic acids research 44, W90–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang H-Y, Lin Y-C-D, Li J, Huang K-Y, Shrestha S, Hong H-C, Tang Y, Chen Y-G, Jin C-N, Yu Y, Xu J-T, Li Y-M, Cai X-X, Zhou Z-Y, Chen X-H, Pei Y-Y, Hu L, Su J-J, Cui S-D, Wang F, Xie Y-Y, Ding S-Y, Luo M-F, Chou C-H, Chang N-W, Chen K-W, Cheng Y-H, Wan X-H, Hsu W-L, Lee T-Y, Wei F-X, and Huang H-D (2020) miRTarBase 2020: updates to the experimentally validated microRNA–target interaction database. Nucleic acids research 48, D148–D154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.(2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Giron CG, Gil L, Gordon L, Haggerty L, Haskell E, Hourlier T, Izuogu OG, Janacek SH, Juettemann T, To JK, Laird MR, Lavidas I, Liu Z, Loveland JE, Maurel T, McLaren W, Moore B, Mudge J, Murphy DN, Newman V, Nuhn M, Ogeh D, Ong CK, Parker A, Patricio M, Riat HS, Schuilenburg H, Sheppard D, Sparrow H, Taylor K, Thormann A, Vullo A, Walts B, Zadissa A, Frankish A, Hunt SE, Kostadima M, Langridge N, Martin FJ, Muffato M, Perry E, Ruffier M, Staines DM, Trevanion SJ, Aken BL, Cunningham F, Yates A, and Flicek P (2018) Ensembl 2018. Nucleic acids research 46, D754–D761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kulakovskiy IV, Vorontsov IE, Yevshin IS, Sharipov RN, Fedorova AD, Rumynskiy EI, Medvedeva YA, Magana-Mora A, Bajic VB, Papatsenko DA, Kolpakov FA, and Makeev VJ (2018) HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic acids research 46, D252–D259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krystkowiak I, Lenart J, Debski K, Kuterba P, Petas M, Kaminska B, and Dabrowski M (2013) Nencki Genomics Database--Ensembl funcgen enhanced with intersections, user data and genome-wide TFBS motifs. Database : the journal of biological databases and curation 2013, bat069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kulakovskiy IV, Medvedeva YA, Schaefer U, Kasianov AS, Vorontsov IE, Bajic VB, and Makeev VJ (2013) HOCOMOCO: a comprehensive collection of human transcription factor binding sites models. Nucleic acids research 41, D195–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kulakovskiy IV, Vorontsov IE, Yevshin IS, Soboleva AV, Kasianov AS, Ashoor H, Ba-Alawi W, Bajic VB, Medvedeva YA, Kolpakov FA, and Makeev VJ (2016) HOCOMOCO: expansion and enhancement of the collection of transcription factor binding sites models. Nucleic acids research 44, D116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schödel J, Oikonomopoulos S, Ragoussis J, Pugh CW, Ratcliffe PJ, and Mole DR (2011) High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood 117, e207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tausendschön M, Rehli M, Dehne N, Schmidl C, Döring C, Hansmann ML, and Brüne B (2015) Genome-wide identification of hypoxia-inducible factor-1 and −2 binding sites in hypoxic human macrophages alternatively activated by IL-10. Biochimica et biophysica acta 1849, 10–22 [DOI] [PubMed] [Google Scholar]
- 39.Lee MC, Huang HJ, Chang TH, Huang HC, Hsieh SY, Chen YS, Chou WY, Chiang CH, Lai CH, and Shiau CY (2016) Genome-wide analysis of HIF-2α chromatin binding sites under normoxia in human bronchial epithelial cells (BEAS-2B) suggests its diverse functions. Sci Rep 6, 29311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smythies JA, Sun M, Masson N, Salama R, Simpson PD, Murray E, Neumann V, Cockman ME, Choudhry H, Ratcliffe PJ, and Mole DR (2019) Inherent DNA-binding specificities of the HIF-1α and HIF-2α transcription factors in chromatin. EMBO Rep 20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Janaszak-Jasiecka A, Siekierzycka A, Bartoszewska S, Serocki M, Dobrucki LW, Collawn JF, Kalinowski L, and Bartoszewski R (2018) eNOS expression and NO release during hypoxia is inhibited by miR-200b in human endothelial cells. Angiogenesis 21, 711–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, Jeon M, and Ma'ayan A (2021) Gene Set Knowledge Discovery with Enrichr. Current Protocols 1, e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilczynska A, and Bushell M (2015) The complexity of miRNA-mediated repression. Cell Death & Differentiation 22, 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flores O, Kennedy EM, Skalsky RL, and Cullen BR (2014) Differential RISC association of endogenous human microRNAs predicts their inhibitory potential. Nucleic acids research 42, 4629–4639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai X, Wolkenhauer O, and Vera J (2016) Understanding microRNA-mediated gene regulatory networks through mathematical modelling. Nucleic acids research 44, 6019–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ebert MS, and Sharp PA (2012) Roles for microRNAs in conferring robustness to biological processes. Cell 149, 515–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mole DR, Blancher C, Copley RR, Pollard PJ, Gleadle JM, Ragoussis J, and Ratcliffe PJ (2009) Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. The Journal of biological chemistry 284, 16767–16775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartoszewski R, Moszynska A, Serocki M, Cabaj A, Polten A, Ochocka R, Dell'Italia L, Bartoszewska S, Kroliczewski J, Dabrowski M, and Collawn JF (2019) Primary endothelial cell-specific regulation of hypoxia-inducible factor (HIF)-1 and HIF-2 and their target gene expression profiles during hypoxia. FASEB J 33, 7929–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chi J-T, Wang Z, Nuyten DSA, Rodriguez EH, Schaner ME, Salim A, Wang Y, Kristensen GB, Helland Å, Børresen-Dale A-L, Giaccia A, Longaker MT, Hastie T, Yang GP, van de Vijver MJ, and Brown PO (2006) Gene Expression Programs in Response to Hypoxia: Cell Type Specificity and Prognostic Significance in Human Cancers. PLOS Medicine 3, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ayoubian H, Ludwig N, Fehlmann T, Menegatti J, Groger L, Anastasiadou E, Trivedi P, Keller A, Meese E, and Grasser FA (2019) Epstein-Barr Virus Infection of Cell Lines Derived from Diffuse Large B-Cell Lymphomas Alters MicroRNA Loading of the Ago2 Complex. J Virol 93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Janas MM, Wang B, Harris AS, Aguiar M, Shaffer JM, Subrahmanyam YV, Behlke MA, Wucherpfennig KW, Gygi SP, Gagnon E, and Novina CD (2012) Alternative RISC assembly: binding and repression of microRNA-mRNA duplexes by human Ago proteins. RNA 18, 2041–2055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Camps C, Saini HK, Mole DR, Choudhry H, Reczko M, Guerra-Assuncao JA, Tian YM, Buffa FM, Harris AL, Hatzigeorgiou AG, Enright AJ, and Ragoussis J (2014) Integrated analysis of microRNA and mRNA expression and association with HIF binding reveals the complexity of microRNA expression regulation under hypoxia. Molecular cancer 13, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rupaimoole R, Wu SY, Pradeep S, Ivan C, Pecot CV, Gharpure KM, Nagaraja AS, Armaiz-Pena GN, McGuire M, Zand B, Dalton HJ, Filant J, Miller JB, Lu CH, Sadaoui NC, Mangala LS, Taylor M, van den Beucken T, Koch E, Rodriguez-Aguayo C, Huang L, Bar-Eli M, Wouters BG, Radovich M, Ivan M, Calin GA, Zhang W, Lopez-Berestein G, and Sood AK (2014) Hypoxia-mediated downregulation of miRNA biogenesis promotes tumour progression. Nat Commun 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ho JJ, Metcalf JL, Yan MS, Turgeon PJ, Wang JJ, Chalsev M, Petruzziello-Pellegrini TN, Tsui AK, He JZ, Dhamko H, Man HS, Robb GB, Teh BT, Ohh M, and Marsden PA (2012) Functional importance of Dicer protein in the adaptive cellular response to hypoxia. The Journal of biological chemistry 287, 29003–29020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas JD, Dias LM, and Johannes GJ (2008) Translational repression during chronic hypoxia is dependent on glucose levels. RNA 14, 771–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bartoszewska S, and Collawn JF (2020) Unfolded protein response (UPR) integrated signaling networks determine cell fate during hypoxia. Cell Mol Biol Lett 25, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karali E, Bellou S, Stellas D, Klinakis A, Murphy C, and Fotsis T (2014) VEGF Signals through ATF6 and PERK to Promote Endothelial Cell Survival and Angiogenesis in the Absence of ER Stress. Molecular Cell 54, 559–572 [DOI] [PubMed] [Google Scholar]
- 58.Abhinand CS, Raju R, Soumya SJ, Arya PS, and Sudhakaran PR (2016) VEGF-A/VEGFR2 signaling network in endothelial cells relevant to angiogenesis. J Cell Commun Signal 10, 347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsang J, Zhu J, and van Oudenaarden A (2007) MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Molecular cell 26, 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li C, and Jackson RM (2002) Reactive species mechanisms of cellular hypoxia-reoxygenation injury. Am J Physiol Cell Physiol 282, C227–241 [DOI] [PubMed] [Google Scholar]
- 61.Bartoszewski R, Moszynska A, Serocki M, Cabaj A, Polten A, Ochocka R, Dell'Italia L, Bartoszewska S, Kroliczewski J, Dabrowski MD, and Collawn JF (2019) Primary endothelial cell-specific regulation of hypoxia-inducible factor (HIF)-1 and HIF-2 and their target gene expression profiles during hypoxia. Faseb Journal 33, 7929–7941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Juzenas S, Venkatesh G, Hübenthal M, Hoeppner MP, Du ZG, Paulsen M, Rosenstiel P, Senger P, Hofmann-Apitius M, Keller A, Kupcinskas L, Franke A, and Hemmrich-Stanisak G (2017) A comprehensive, cell specific microRNA catalogue of human peripheral blood. Nucleic acids research 45, 9290–9301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dong H, Lei J, Ding L, Wen Y, Ju H, and Zhang X (2013) MicroRNA: function, detection, and bioanalysis. Chemical reviews 113, 6207–6233 [DOI] [PubMed] [Google Scholar]
- 64.Wu J, Xiao J, Zhang Z, Wang X, Hu S, and Yu J (2014) Ribogenomics: the Science and Knowledge of RNA. Genomics, Proteomics & Bioinformatics 12, 57–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, and Ivan M (2007) A microRNA signature of hypoxia. Molecular and cellular biology 27, 1859–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greco S, Gaetano C, and Martelli F (2014) HypoxamiR regulation and function in ischemic cardiovascular diseases. Antioxidants & redox signaling 21, 1202–1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McCormick RI, Blick C, Ragoussis J, Schoedel J, Mole DR, Young AC, Selby PJ, Banks RE, and Harris AL (2013) miR-210 is a target of hypoxia-inducible factors 1 and 2 in renal cancer, regulates ISCU and correlates with good prognosis. British journal of cancer 108, 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, Story M, Le QT, and Giaccia AJ (2009) Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell 35, 856–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Spinello I, Quaranta MT, Paolillo R, Pelosi E, Cerio AM, Saulle E, Lo Coco F, Testa U, and Labbaye C (2015) Differential hypoxic regulation of the microRNA-146a/CXCR4 pathway in normal and leukemic monocytic cells: impact on response to chemotherapy. Haematologica 100, 1160–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blick C, Ramachandran A, McCormick R, Wigfield S, Cranston D, Catto J, and Harris AL (2015) Identification of a hypoxia-regulated miRNA signature in bladder cancer and a role for miR-145 in hypoxia-dependent apoptosis. British journal of cancer 113, 634–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Seok JK, Lee SH, Kim MJ, and Lee YM (2014) MicroRNA-382 induced by HIF-1alpha is an angiogenic miR targeting the tumor suppressor phosphatase and tensin homolog. Nucleic acids research 42, 8062–8072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nagpal N, Ahmad HM, Chameettachal S, Sundar D, Ghosh S, and Kulshreshtha R (2015) HIF-inducible miR-191 promotes migration in breast cancer through complex regulation of TGFbeta-signaling in hypoxic microenvironment. Sci Rep 5, 9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xie Y, Li W, Feng J, Wu T, and Li J (2016) MicroRNA-363 and GATA-1 are regulated by HIF-1alpha in K562 cells under hypoxia. Molecular medicine reports 14, 2503–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ge X, Liu X, Lin F, Li P, Liu K, Geng R, Dai C, Lin Y, Tang W, Wu Z, Chang J, Lu J, and Li J (2016) MicroRNA-421 regulated by HIF-1alpha promotes metastasis, inhibits apoptosis, and induces cisplatin resistance by targeting E-cadherin and caspase-3 in gastric cancer. Oncotarget 7, 24466–24482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang X, Li J, Wu D, Bu X, and Qiao Y (2016) Hypoxia promotes apoptosis of neuronal cells through hypoxia-inducible factor-1alpha-microRNA-204-B-cell lymphoma-2 pathway. Exp Biol Med (Maywood) 241, 177–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang Y, Li Y, Chen X, Cheng X, Liao Y, and Yu X (2016) Exosomal transfer of miR-30a between cardiomyocytes regulates autophagy after hypoxia. J Mol Med (Berl) 94, 711–724 [DOI] [PubMed] [Google Scholar]
- 77.Liu Y, Nie HG, Zhang KK, Ma D, Yang G, Zheng ZL, Liu K, Yu B, Zhai CL, and Yang S (2014) A feedback regulatory loop between HIF-1 alpha and miR-21 in response to hypoxia in cardiomyocytes. Febs Lett 588, 3137–3146 [DOI] [PubMed] [Google Scholar]
- 78.Bhatt K, Wei Q, Pabla N, Dong G, Mi QS, Liang M, Mei C, and Dong Z (2015) MicroRNA-687 Induced by Hypoxia-Inducible Factor-1 Targets Phosphatase and Tensin Homolog in Renal Ischemia-Reperfusion Injury. Journal of the American Society of Nephrology : JASN 26, 1588–1596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Akhtar S, Hartmann P, Karshovska E, Rinderknecht FA, Subramanian P, Gremse F, Grommes J, Jacobs M, Kiessling F, Weber C, Steffens S, and Schober A (2015) Endothelial Hypoxia-Inducible Factor-1alpha Promotes Atherosclerosis and Monocyte Recruitment by Upregulating MicroRNA-19a. Hypertension 66, 1220–1226 [DOI] [PubMed] [Google Scholar]
- 80.Heberle H, Meirelles GV, da Silva FR, Telles GP, and Minghim R (2015) InteractiVenn: a web-based tool for the analysis of sets through Venn diagrams. BMC Bioinformatics 16, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deep sequencing data were deposited in Gene Expression Omnibus (GEO) at accession numbers: GSE116909 (10), GSE190240 and GSE190242.