Abstract
The spatial distribution of antibiotic resistance to streptomycin and kanamycin was examined in natural bacterial communities of two streams. The proportion of resistant bacteria was substantially higher (P < 0.05) in the midreaches of an industrially perturbed stream, but no such pattern was apparent in an undisturbed reference stream. The highest relative frequency of resistance was found at the confluence of a tributary draining a nuclear reactor and industrial complex. Antibiotic resistance increased with distance upstream from the confluence and was positively correlated (r2 = 0.54, P = 0.023) with mercury concentrations in the sediments. When the data for two years were compared, this pattern was stable for streptomycin resistance (paired t test, P < 0.05) but not for kanamycin resistance (P > 0.05). Our results imply that heavy metal pollution may contribute to increased antibiotic resistance through indirect selection.
Stream ecosystems are usually connected from headwaters to mouth, and this connectivity provides a means for dissemination and colonization of species. Most streams validate Vannote et al.'s (22) river continuum concept with serial replacement of diverse plants and animals (19, 20, 23). The river continuum concept provides a theoretical basis for the distribution of organisms and biogeochemical transformations along river systems. Interestingly, microbes are mentioned in this concept but no meaningful predictions of their distributions are presented. Since bacteria are important components of all river systems, it is important to know if they further validate the continuum concept.
McArthur et al. (12) demonstrated genetic changes among populations of Burkholderia cepacia and Pseudomonas pickettii along a stream continuum, although both species were persistent at all sampling sites. Wise et al. (25, 26) further validated these results. Since they reported that specific genotypes were repeatedly associated with restricted local stream conditions, selection among genotypes was inferred.
Since most bacteria cannot be cultured, molecular technologies have been used to detect them or their genes in natural environments (3). Using some of these techniques, Leff et al. (8) reported the nptII gene to occur nonrandomly in a stream. nptII abundance was greater in bank sediments than in channel sediments or on submerged leaf surfaces. The results were similar at each sampling site. The nptII gene, from transposon Tn5, encodes the neomycin phosphotransferase responsible for resistance to kanamycin and neomycin; thus, nptII distribution is useful in studying genetic adaptations of bacteria under natural conditions.
Selection for resistance to antibiotics by bacteria in natural and modified stream environments, e.g., below sewage treatment plants (16, 17), below feedlots where feed for livestock has been supplemented with antibiotics (7), and even in streams assumed to be pristine (8), may be important to managing streams for human health. However, it is unclear what the selective advantage of resistance to antibiotics is in unpolluted streams, and there may even be a selective disadvantage (evolutionary cost) in unpolluted streams (1, 10).
Few studies have focused on antibiotic resistance in streams (8), and none have included samples collected systematically to determine spatial patterns in stream systems. Furthermore, antibiotic resistance under field conditions is made more complex by frequent genetic association with metal tolerance and resistance genes (2, 18, 24). Metal resistance and antibiotic resistance may be linked too closely to conclude that they are independent through incidental field sampling. Antibiotic resistance may not prove to be a singular event but may be a complex of events dependent on exposure to metals.
McArthur and Tuckfield (11) presented a concept that predicted the distribution of stream bacteria based on information length or the geographical distance in a stream where various bacteria and/or their genes are adaptive. Two predictions arising from this model were (i) information length for a particular trait would be measurable only when selection was present and (ii) information lengths for antibiotic resistance or carbon utilization could be used as measures of ecosystem health (i.e., there could be measurable and repeatable differences between impacted and unimpacted streams).
Our objectives in this study were to validate aspects of the McArthur-Tuckfield information length hypothesis (ILH). Specifically, we sought to determine whether spatial patterns of antibiotic resistance differ in a chemically and thermally contaminated stream compared with an uncontaminated stream.
MATERIALS AND METHODS
Four Mile Creek (FMC) is a third-order upper-coastal-plain stream draining a 5,894-ha watershed located on the U.S. Department of Energy's Savannah River Site (SRS). Stream temperatures range from 9.0 to 25°C, and the pH is slightly below neutral (pH range, 5.10 to 8.10; median pH = 6.09) (14). FMC received thermal effluent (>50°C) from reactor operations between 1955 and 1985 at flow rates 10 times higher than the ambient flow rate (from 40 m−3 s−1 [ambient] to more than 400 m−3 s−1 during reactor operation). These flows caused major geomorphological changes within the stream, essentially scrubbing the channel of all organic matter and instream structure. All riparian vegetation was killed. In addition, several chemical seepage basins were established in the headwater reaches of the stream and were used continuously for more than 30 years. These basins received chemical effluent containing tritium, nitrate, and various metals. The amounts of mercury released into these basins ranged from 0.45 to 9.07 kg year−1 (5). The basins were capped in 1992. However, their leachates continue to seep into the stream; e.g., the nitrate concentrations along the seep line range from 4.88 to 5.00 mg of NO3 N liter−1. FMC has been undergoing natural recovery since the cessation of thermal flows in 1987. In 1992 a sewage treatment facility was established alongside FMC, and this facility discharges up to 106 gallons per day. Outfalls from this facility are located approximately midway along the stream.
Meyers Branch (MB) is a historically unimpacted stream on the SRS set-aside for ecological research. MB drains approximately 5,085 ha. It originates in the sand hills of the upper coastal plain and has an extensive riparian floodplain. It has a mean annual temperature of ∼16°C. The pH ranges from 5.8 to 8.3, with a median of 6.9.
Sixty-seven sampling sites on FMC and 62 sites on MB were located from geographical information system (GIS) maps of the SRS. Each site was separated from the next site by 200 m, and its geographic coordinates were determined. These sampling sites spanned lengths of stream reach from near each stream's confluence with a larger stream system (the Savannah River and Steel Creek for FMC and MB, respectively) to near the stream's headwaters. Each sampling site was located with a Trimble global positioning system unit which was accurate to within ±1.5 m.
FMC was sampled during June 1998. At each sampling site one 10-cm-long core (diameter, ∼2.5 cm) was taken from stream bottom sediments adjacent to the bank, placed on ice in a sterile bag, and transported to the laboratory. The integrity of the core was not maintained after placement in the bag and transport. For each sample approximately 5 g of the resulting sediment slurry was removed, gently sonicated (Fisher Sonic water bath) to detach bacteria, serially diluted, and plated on three different plates. Sediments were removed, dried (60°C), and weighed. Control plates consisted of half-strength nutrient broth agar with 100 μg of cycloheximide per ml added to control fungal growth. Previous studies had shown that this medium resulted in the highest densities (9). The remaining plates were identical to the control plates except that 100 μg of either streptomycin, kanamycin, tetracycline, or chloramphenicol per ml was added. Previous studies (8) had determined the effect of 50- and 200-μg ml concentrations of these antibiotics on aquatic bacteria. We chose 100 μg ml−1 as an intermediate concentration. Bacterial colonies on inoculated plates were counted after 6 days of incubation at room temperature (∼20°C). A representative sample of each core was removed, dried at 60°C, weighed, ashed at 600°C for 8 h, rewetted with deionized water, redried, and reweighed to determine the organic matter content.
Each bacterial colony count was divided by the dry weight of the sediment in the corresponding sample. This adjustment was made to reduce sample count bias wherein larger numbers of bacteria are expected in larger amounts of sediment. The relative frequency of antibiotic-resistant bacteria was calculated as the ratio of the adjusted antibiotic-resistant bacterial count to the adjusted control count. The latter value was simply the proportion of the colonies plated that were resistant to a specific antibiotic. Data that are proportions typically are not normally distributed. Therefore, several mathematical data transformations (12) were performed, and the results were subjected to the Shapiro-Wilks W test (13) to validate normality, an assumption important to the subsequent application of parametric statistical models. Three transformations of the relative frequency measurement were performed: (i) square root, (ii) arcsine square root, and (iii) common logarithm. Of these, only the common log transformation showed a nonsignificant departure from normality for both streptomycin (P = 0.84) and kanamycin (P = 0.79). Significance probabilities less than 5% (P < 0.05) were obtained for the Shapiro-Wilks W test results for relative frequency and the other two transformed variables.
Castor Creek has its confluence with the main channel of FMC in the midreaches of the latter. To determine whether Castor Creek was affecting the antibiotic resistance patterns, we sampled the main channel of FMC and Castor Creek 4 weeks following the initial sampling effort in 1998. This sampling occurred after one major rain or flood that was sufficient to resuspend the sandy bottom sediments, as observed by one of us (JV.M.). In FMC we sampled three locations below and five locations above the confluence at the exact locations sampled in the initial study. In addition, we sampled 10 locations along Castor Creek, including five above and five below a canal that was used to carry thermal water from a nuclear production reactor. The sampling locations along the tributary were not evenly spaced but were ≥200 m apart. The samples obtained were processed by using the 1998 methods. In June 1999 we again sampled five locations above and five locations below the confluence of this tributary with FMC and also collected sediments from 10 additional locations along the tributary stream. These samples were processed by using a replica plate technique described below. The mercury concentrations in these sediment samples were determined by cold vapor atomic fluorescence by using a Brooks Rand model 2 analyzer and methods similar to the methods of Gariboldi et al. (4). Mercury was chosen because of known inputs into FMC. Levels of Hg and antibiotic resistance were regressed against each other by using linear regression.
During May 1999 we sampled MB sediments by using similar procedures. However, the MB samples were processed differently. After sonication and serial dilution only control plates were inoculated. After 3 days of growth at 25°C control plates were counted and used as a source for replica plating (Bel-Art Products, Pequannock, N.J.) onto streptomycin or kanamycin plates. These replica plates were incubated at 25°C and counted 3 to 4 days later.
RESULTS
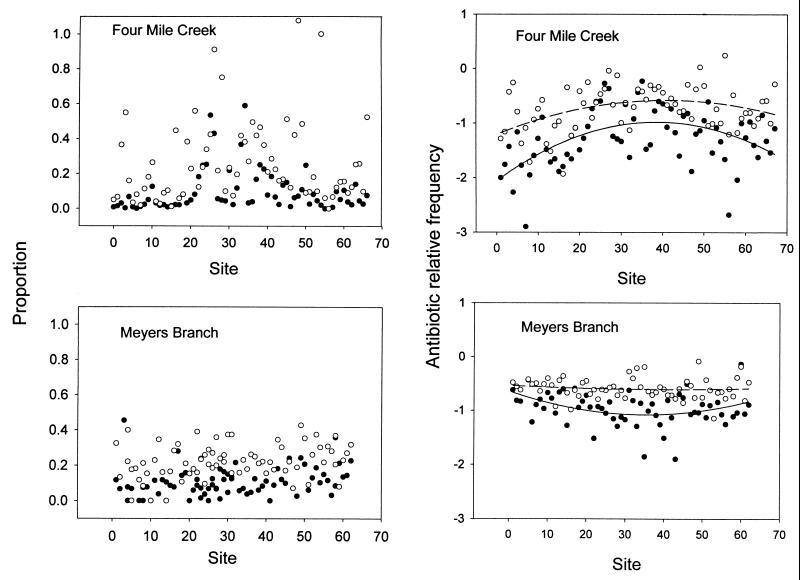
The results for kanamycin, chloramphenicol, and tetracycline were similar. We present the data for kanamycin as representative of the data for these three antibiotics. The patterns for streptomycin were different from those for the other three antibiotics. The spatial distributions of antibiotic resistance along FMC and MB were stream specific for both antibiotics examined (Fig. 1). For both antibiotics in FMC the proportions of antibiotic-resistant bacteria were higher in the midreaches of the stream, and these trends showed a statistically significant and convex quadratic regression relationship with distance (kanamycin, P < 0.0001; streptomycin, P < 0.0005) (Fig. 1). In MB, the distribution of streptomycin-resistant bacteria had no distinct pattern as predicted by the ILH along the stream course (Fig. 1). However, the pattern of kanamycin resistance indicates that there was a slight decrease in the midreaches, as confirmed by a statistically significant (P = 0.017) and concave quadratic regression relationship with distance. Overall, the variability in antibiotic resistance was greater for both antibiotics in the disturbed stream than in the control stream (Fig. 1).
FIG. 1.
Proportion (antibiotic-resistant colony counts/control colony counts) and relative frequency (log10 + 1) of antibiotic resistance among stream bacteria in FMC and MB at the SRS in South Carolina. Symbols: ○, streptomycin-resistant bacteria; ●, kanamycin-resistant bacteria. Solid line, kanamycin; dashed line, streptomycin.
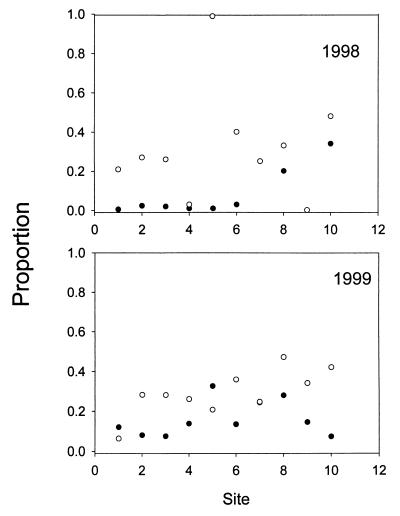
Additional samples were collected 4 weeks after the initial survey of FMC from sites that bracketed the confluence of Castor Creek with FMC (Fig. 2). The same eight sites were sampled again in 1999. The levels of streptomycin resistance in the two years were not significantly different (paired t test, P > 0.05 for transformed and untransformed data). At some sites, the levels of streptomycin resistance were essentially identical in the two years. The kanamycin resistance levels at these eight sites in 1999 were significantly lower than the kanamycin resistance levels in 1998 (paired t test, P < 0.001 for both transformed and untransformed data). In fact, the value for every sample collected in 1999 was lower than the value for the corresponding sample collected in 1998. The differences in the temporal patterns between the two antibiotics indicate that different mechanisms influence the distribution of streptomycin resistance and kanamycin resistance.
FIG. 2.
Proportion (antibiotic-resistant colony counts/control colony counts) of antibiotic-resistant stream bacteria in FMC at selected locations in 1998 and 1999. Symbols: ●, 1998; ○, 1999.
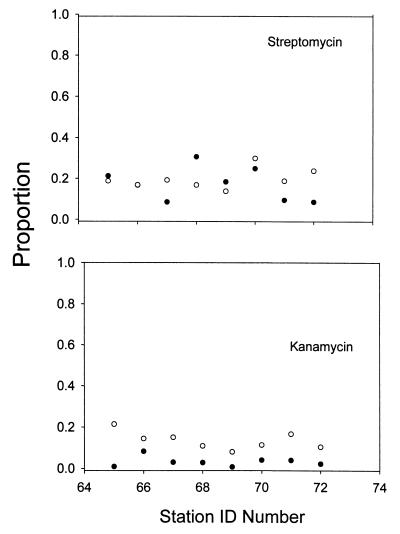
The patterns of antibiotic resistance in Castor Creek in 1998 and 1999 were similar for streptomycin but different for kanamycin (Fig. 3). The level of streptomycin resistance was generally high in Castor Creek. The levels of kanamycin resistance increased upstream in 1998 and were relatively high and constant in 1999.
FIG. 3.
Proportion (antibiotic-resistant colony counts/control colony counts) of antibiotic-resistant stream bacteria in Castor Creek at selected locations in 1998 and 1999. Symbols: ●, kanamycin; ○, streptomycin.
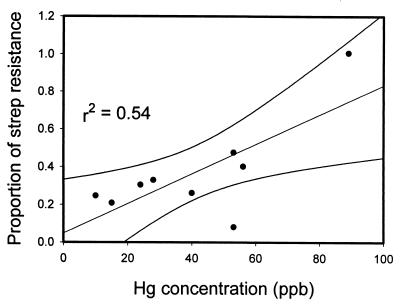
We analyzed the same Castor Creek sediment cores from which the bacteria were isolated to determine the concentration of Hg. The Hg concentrations in the samples ranged from 9 to 127 ppb. The background Hg levels for the SRS range from 5 to 10 ppb. Plotting the proportion of streptomycin-resistant bacteria as a function of Hg concentration (Fig. 4) showed that there was a significant positive correlation (r2 = 0.54, P = 0.023).
FIG. 4.
Proportion (antibiotic-resistant colony counts/control colony counts) and 95% confidence intervals of streptomycin-resistant bacteria at selected locations in Castor Creek as a function of Hg concentration in stream sediment samples.
DISCUSSION
Low-level antibiotic resistance in bacteria can be found in pristine habitats (8), suggesting that antibiotic resistance is of minimal importance under natural conditions. Our data demonstrate that there is a substantive relationship between the relative frequency value and distance in a disturbed stream. We predicted, based on the ILH, two peaks of antibiotic resistance in FMC, one near the outfall from the sewage treatment facility and the other near the seep line from the old settling basins; each peak was predicted for different reasons. These predictions were not met. Only one peak was observed, and it was approximately 1 km below the outfall from the sewage treatment facility near the confluence of FMC with a tributary stream draining an industrial area. The predictions arising from the ILH were based on a priori assumptions concerning factors that may affect the distribution of antibiotic resistance traits. We assumed that leachate from the seepage basins and discharge from a sewage treatment plant would be either a source of antibiotic resistance genes (sewage discharge) or a strong selector (seepage leachate). Our data indicate that the strongest predictor of antibiotic resistance is the heavy metal concentration in the sediments, in this case the Hg concentration.
There were significant differences between the chemically and thermally disturbed stream and the reference stream in terms of the patterns of antibiotic resistance. Streptomycin resistance showed a repeatable pattern in FMC and Castor Creek in the two sampling years. Coastal plain streams have shifting-sand bottoms that are easily disturbed with increased flows. The temporal stability of streptomycin resistance at these sites suggests that the gene combinations are maintained despite potential for redistribution and mixing. In contrast, the random distribution of streptomycin resistance in MB is consistent with the expectation of continuous resorting of stream sediments with no selection favoring increased levels of resistance.
Kanamycin resistance was spatially different from streptomycin resistance in the two years in FMC and Castor Creek (Fig. 3). The patterns suggest that different processes and selective factors act on these two antibiotic resistance traits.
No agricultural, medical, or sewage discharge has been recorded for Castor Creek or MB. In fact, the SRS was closed to the public prior to the widespread use of antibiotics either clinically or agriculturally. Therefore, the patterns of antibiotic resistance in Castor Creek are likely to be related to industrial activities in the basin and specifically in the subwatershed draining from the C-reactor and central shops region of the SRS. The selective force may be heavy metal challenge.
Metal tolerance and resistance of bacteria have been shown to increase proportionally along industrial contamination gradients (15, 18). Genes that code for antibiotic resistance traits and genes that code for metal resistance are often carried on the same plasmids or mobile genetic elements (24, 27). While researchers have found relationships between the occurrence of antibiotic resistance and the occurrence of metal tolerance (3), because of insufficient sampling no definitive patterns could be established. The observed correlation between sediment Hg concentration and antibiotic resistance suggests that increased mercury concentration may indirectly select for increased antibiotic resistance in certain stream bacteria. Sundin and Bender (21) state that although the usage of streptomycin in clinical medicine and animal husbandry has diminished, the streptomycin resistance genes persist. This persistence implies that factors other than direct selection are involved in the maintenance of these genes. Wireman et al. (24) showed that bacteria with the mer locus (which codes for Hg resistance) were more likely to be multiresistant than bacteria without the mer locus. Furthermore, the association of antibiotic resistance with mer loci was not random.
The spread of mercury resistance genes is similar to the worldwide spread of antibiotic resistance genes (27) and has resulted in a worldwide population of mercury-resistant species. Indeed, the presence of mer genes in bacteria collected from deep sediment cores indicates that mer is an ancient system (15). Osborn et al. (15) identified three major factors affecting the distribution of mer genes: (i) long ancestry, (ii) coupling of localized selection pressures in the form of mercury compounds, and (iii) spread of mer sequences by a powerful array of broad-host-range plasmids and transposons. Mercury-resistant strains were shown to be resistant to ampicillin, chloramphenicol, kanamycin, and tetracycline (2).
We have demonstrated that there is a repeatable pattern of antibiotic resistance in bacteria collected from a stream not exposed to sewage or agricultural contamination. The potential impact of increased antibiotic resistance due to metal contamination is particularly great considering the very large number of metal-contaminated locations. Microbes may create their own dispersal agents from water and soil through various mechanisms (6) and thus enter the atmosphere. Once in the atmosphere, bacteria can be distributed over large geographical areas and subsequently return to earth through rain, snow, hail, or dryfall (27), thus aiding in the worldwide distribution of various bacteria or their genes. We suggest that the potential impact of chemically polluted, more specifically metal-polluted, locations on human life may be much greater than the direct effect of the pollution.
ACKNOWLEDGMENTS
We thank A. Lindell, C. King, C. Draney, N. Wiedrich, L. Tuckfield, and G. Novak for various aspects of data collection and technical assistance. C. D. Jorgensen and M. H. Smith provided significant comments and recommendations on earlier drafts of the manuscript.
This research was supported by Financial Assistance Award DE-FC09-96SR18546 from the U. S. Department of Energy to the University of Georgia Research Foundation.
REFERENCES
- 1.Bouma J E, Lenski R E. Evolution of bacteria/plasmid association. Nature. 1988;335:351–352. doi: 10.1038/335351a0. [DOI] [PubMed] [Google Scholar]
- 2.Dhakephalkar P K, Chopade B A. High levels of multiple metal resistance and its correlation to antibiotic resistance in environmental isolates of Acinetobacter. Biometals. 1994;7:67–74. doi: 10.1007/BF00205197. [DOI] [PubMed] [Google Scholar]
- 3.Diels L, Springael D, van der Lelie N, Top G, Mergeay M. Use of DNA probes and plasmid capture in a search for new interesting environmental genes. Sci Total Environ. 1993;139/140:471–478. doi: 10.1016/0048-9697(93)90044-7. [DOI] [PubMed] [Google Scholar]
- 4.Gariboldi J C, Jagoe C H, Bryan A L. Dietary exposure to mercury in nestling wood storks (Mycteria americana) in Georgia. Arch Environ Contam Toxicol. 1998;34:398–405. doi: 10.1007/s002449900336. [DOI] [PubMed] [Google Scholar]
- 5.Gladden J B, Lower M W, Mackey H E, Specht W L, Wilde E W. Radionuclide and heavy metal transport, Savannah River Plant. Publication DP-1697-4. IV. Aiken, S.C: E. I. Du Pont de Nemours & Co.; 1985. Comprehensive cooling water study annual report. [Google Scholar]
- 6.Hamilton W D, Lenton T M. Spora and Gaia: how microbes fly with their clouds. Ethol Ecol Evol. 1998;10:1–16. [Google Scholar]
- 7.Holmberg S D, Osterholm M T, Senger K A, Cohen M L. Drug-resistant Salmonella from animals fed antimicrobials. N Engl J Med. 1984;311:617–622. doi: 10.1056/NEJM198409063111001. [DOI] [PubMed] [Google Scholar]
- 8.Leff L G, Dana J R, McArthur J V, Shimkets L G. Detection of Tn5-like sequences in kanamycin-resistant stream bacteria and environmental DNA. Appl Environ Microbiol. 1993;59:417–421. doi: 10.1128/aem.59.2.417-421.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leff L G, McArthur J V, Shimkets L G. Spatial and temporal variability of antibiotic resistance in freshwater bacterial assemblages. FEMS Microbiol Ecol. 1993;13:135–144. [Google Scholar]
- 10.Levin B R, Lenski R E. Coevolution in bacteria and their viruses and plasmids. In: Futuyma D J, Slatkin M, editors. Coevolution. Sunderland, Mass: Sinauer; 1983. pp. 99–127. [Google Scholar]
- 11.McArthur J V, Tuckfield R C. Information length: spatial and temporal parameters among stream bacterial assemblages. J North Am Benthol Soc. 1997;16:347–357. [Google Scholar]
- 12.McArthur J V, Leff L G, Smith M H. Genetic diversity of bacteria along a stream continuum. J North Am Benthol Soc. 1992;11:269–277. [Google Scholar]
- 13.Neter J, Wasserman W. Applied linear statistical models. Homewood, Ill: Richard D. Irwin, Inc.; 1974. [Google Scholar]
- 14.Newman M C. Water quality. Report SREL-28. Vol. 2. Springfield, Va: National Technical Information Service; 1986. Comprehensive cooling water report. [Google Scholar]
- 15.Osborn A M, Bruce K D, Strike P, Ritchie D A. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol Rev. 1997;19:239–262. doi: 10.1111/j.1574-6976.1997.tb00300.x. [DOI] [PubMed] [Google Scholar]
- 16.Parveen S, Murphree R L, Edmiston L, Kaspar C W, Portier K M, Tauplin M L. Association of multiple-antibiotic-resistance profiles with point and nonpoint sources of Escherichia coli in Apalachicola Bay. Appl Environ Microbiol. 1997;63:2607–2612. doi: 10.1128/aem.63.7.2607-2612.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pickup R W, Mallinson H E H, Rhodes G, Chatfield L K. A novel nickel resistance determinant found in sewage-associated bacteria. Microb Ecol. 1997;33:230–239. doi: 10.1007/s002489900026. [DOI] [PubMed] [Google Scholar]
- 18.Roane T M, Kellogg S T. Characterization of bacterial communities in heavy metal contaminated soils. Can J Microbiol. 1996;42:593–603. doi: 10.1139/m96-080. [DOI] [PubMed] [Google Scholar]
- 19.Rundle S D, Hildrew A G. The distribution of micro-arthropods in some southern English streams: the influence of physicochemistry. Freshwater Biol. 1990;23:411–432. [Google Scholar]
- 20.Sheldon A L. Species diversity and longitudinal succession in stream fishes. Ecology. 1968;49:193–198. [Google Scholar]
- 21.Sundin G W, Bender C L. Dissemination of the strA-strB streptomycin resistance genes among commensal and pathogenic bacteria from humans, animals, and plants. Mol Ecol. 1996;5:133–143. doi: 10.1111/j.1365-294x.1996.tb00299.x. [DOI] [PubMed] [Google Scholar]
- 22.Vannote R L, Minshall G W, Cummins K W, Sedell J R, Cushing C E. The river continuum concept. Can J Fish Aquat Sci. 1980;37:130–137. [Google Scholar]
- 23.Ward J V. Altitudinal zonation in a Rocky Mountain stream. Arch Hydrobiol Suppl. 1986;74:133–199. [Google Scholar]
- 24.Wireman J, Liebert C A, Smith T, Summers A O. Association of mercury resistance with antibiotic resistance in gram-negative fecal bacteria of primates. Appl Environ Microbiol. 1997;63:4494–4503. doi: 10.1128/aem.63.11.4494-4503.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wise M G, Shimkets L J, McArthur J V. Genetic structure of a lotic population of Burkholderia (Pseudomonas) cepacia. Appl Environ Microbiol. 1995;61:1791–1798. doi: 10.1128/aem.61.5.1791-1798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wise M G, Shimkets L G, Wheat C, McArthur J V. Temporal variation in genetic diversity and structure of a lotic population of Burkholderia (Pseudomonas) cepacia. Appl Environ Microbiol. 1996;62:1558–1562. doi: 10.1128/aem.62.5.1558-1562.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yurieva O, Kholodii G, Minakhin L, Gorlenko Z, Kalyaeva E, Mindlin S, Nikiforov V. Intercontinental spread of promiscuous mercury resistance transposons in environmental bacteria. Mol Microbiol. 1997;24:321–329. doi: 10.1046/j.1365-2958.1997.3261688.x. [DOI] [PubMed] [Google Scholar]