Abstract
Numerous studies have demonstrated that SARS-CoV-2 can be inactivated by ultraviolet (UV) radiation. However, there are few data available on the relative efficacy of different wavelengths of UV radiation and visible light, which complicates assessments of UV decontamination interventions. The present study evaluated the effects of monochromatic radiation at 16 wavelengths from 222 nm through 488 nm on SARS-CoV-2 in liquid aliquots and dried droplets of water and simulated saliva. The data were used to generate a set of action spectra which quantify the susceptibility of SARS-CoV-2 to genome damage and inactivation across the tested wavelengths. UVC wavelengths (≤280 nm) were most effective for inactivating SARS-CoV-2, although inactivation rates were dependent on sample type. Results from this study suggest that UV radiation can effectively inactivate SARS-CoV-2 in liquids and dried droplets, and provide a foundation for understanding the factors which affect the efficacy of different wavelengths in real-world settings.
Keywords: SARS-CoV-2, COVID-19, Ultraviolet, UV, Inactivation, Disinfection
1. Introduction
Ultraviolet (UV) radiation from both sunlight and artificial sources has been shown to affect the infectivity of viruses, including SARS-CoV-2. Natural sunlight, which includes both UVB (280 nm - 315 nm) and UVA (315 nm - 400 nm) radiation, has been shown to reduce SARS-CoV-2 transmission rates in epidemiological studies (1,2), and laboratory studies have confirmed that simulated sunlight with spectra matching the UVB portion of natural sunlight can rapidly inactivate viruses such as SARS-CoV-2 in both aerosols and on surfaces ([3], [4], [5], [6]). UVC radiation (100 nm - 280 nm), though not present in sunlight at the earth's surface, can be produced by artificial sources and has long been used in various applications for disinfecting water, air, and surfaces. Over the course of the COVID-19 pandemic, multiple studies have demonstrated that UVC radiation sources can effectively inactivate SARS-CoV-2 ([7], [8], [9], [10], [11], [12], [13]). Though significantly lower in energy than UVB and UVC wavelengths, UVA and visible light wavelengths may also be able to reduce the infectivity of SARS-CoV-2 and other viruses if sufficiently high doses can be delivered (8,[14], [15], [16]). Based on these findings, strategies for treating contaminated areas with UV radiation and visible light sources have been suggested as a method for slowing the spread of SARS-CoV-2. However, the implementation of such strategies has been hindered by a lack of directly comparable data on the relative efficacy of different wavelengths for inactivating SARS-CoV-2.
UV radiation has been shown to damage multiple viral structural components, including proteins ([17], [18], [19], [20]). However, the primary mechanism by which UV radiation results in viral inactivation is thought to be by damaging the nucleic acids of the viral genome. These sites of damage, or lesions, may take various forms including strand breaks, protein-RNA crosslinks, and dimers between adjacent pyrimidine residues ([20], [21], [22], [23], [24], [25]). Viruses with lesions in their genomes may still be able to enter a host cell, but the presence of the lesion impairs the function of the polymerase enzymes required for viral replication, and any subsequent progress through the viral replication cycle is thereby blocked. Because of this, wavelengths at which RNA or DNA are highly absorbing tend to be those which are most effective for inactivating viruses (23).
The relative sensitivity of viruses across a range of wavelengths may be quantified in an action spectrum. Action spectra have been reported for many viruses ([26], [27], [28], [29], [30], [31]), and can be useful for evaluating the relative utility of decontamination technologies and modeling expected inactivation in outdoor settings under various sunlight conditions. Commonly observed trends between action spectra for different viruses include a peak in viral susceptibility near 260 nm, the peak absorbance wavelength of nucleic acids, a second peak at wavelengths <240 nm where both nucleic acids and proteins absorb, and an exponential decrease in sensitivity at wavelengths longer than approximately 290 nm. However, despite these common trends, differences in sensitivity at various wavelengths have been observed even for related viruses (32). Additionally, comparisons between studies with the same virus at different wavelengths can be complicated by methodological differences, including the bandwidth of the radiation used for exposure tests, the composition of the viral suspension, and the magnitude of the uncertainties in the UV irradiance delivered to samples. Previous studies on the effects of UV radiation and visible light on SARS-CoV-2, though informative, have used a wide range of radiation sources, experimental designs, and metrics for assessing viral damage (10,12,13,[33], [34], [35], [36], [37]). These methodological differences complicate application of the results to assessments of UV and light-based decontamination technologies.
Therefore, the present study was conducted to evaluate the efficacy of different wavelengths of UV radiation and visible light ranging from 222 nm - 488 nm for inactivating SARS-CoV-2. Inactivation experiments were conducted with monochromatic radiation at 16 discrete wavelengths. Tests were conducted with viral suspensions in simulated saliva, meant to be representative of the composition of human fluids containing SARS-CoV-2, and with viral suspensions in distilled water as a control. At each wavelength, both viral suspensions were evaluated as droplets dried on surfaces and in a liquid state. SARS-CoV-2 inactivation was quantified by microtitration assays for viral infectivity at all tested wavelengths and by quantitative reverse transcriptase polymerase chain reaction (RT-qPCR) at wavelengths from 222 nm - 315 nm to evaluate genomic damage. The results of these tests were used to develop a set of action spectra, which can be used to better define the effects of UV radiation and visible light on SARS-CoV-2.
2. Methods
2.1. Virus Suspensions and Assays
hCoV-19/USA/WA-1/2020 (NR-52281) was obtained from BEI Resources as a passage four sample, and was passaged twice in Vero cells as previously described (4,5). Cells and virus were propagated in cell culture medium with 10% fetal bovine serum (FBS), prepared as described previously ([3], [4], [5],38). For viral stock production, cells were infected at a multiplicity of infection (MOI) of approximately 0.003. After harvest, viral stocks were clarified by centrifugation at 2000 ×g for 10 min, then frozen in 30 mL aliquots at −80 °C until use. Prior to use in exposure tests, frozen aliquots of viral stocks were thawed at 37 °C, underlayed with 8 mL of a solution of 20% sucrose in Tris-NaCl-EDTA (TNE) buffer, and pelleted at 106,000 ×g for 2 h at 4 °C. Viral pellets were re-suspended in 2.65 mL of either UltraPure™ DNase/RNase-free distilled water (Invitrogen PN 10977015) or simulated saliva prepared according to the ASTM E2721–16 standard formulation (39). Absorbance spectra for representative virus suspension batches were measured with a NanoDrop One C spectrophotometer. The absorbance spectrum of a representative sample containing 10.4 log10 genome equivalents (GEQ)/mL of SARS-CoV-2 RNA was also measured; RNA in this sample was extracted from a 1:100 dilution of a sample of SARS-CoV-2 in liquid water.
Simulated saliva prepared with a single batch of porcine mucin (Sigma-Aldrich, PN M1778, batch SLCH5758), was used for the majority of testing in the present study. When a second batch of mucin (Sigma-Aldrich, PN M1778, batch SLCD6129) was used, differences were noted in the absorbance spectrum of the resulting simulated saliva, providing an opportunity to assess the effect of simulated saliva preparations with differing absorbance spectra. Therefore, SARS-CoV-2 inactivation tests at 222, 230, and 253.7 nm were conducted with two different simulated saliva suspensions prepared using the different batches of mucin.
Titers of infectious virus in samples were determined by microtitration assay on confluent monolayers of Vero cells as described previously ([3], [4], [5],40), resulting in concentrations of infectious virus in median tissue culture infectious doses (TCID50) per mL. Microtitration assay plates were read for the presence of cytopathic effect (CPE) on any of days 4–7 post infection (dpi), as preliminary experiments determined that reading plates on any day from 4 to 7 dpi resulted in equivalent titers.
RT-qPCR was performed to quantify SARS-CoV-2 genomic damage following UV exposure for samples exposed to wavelengths ranging from 222 nm - 315 nm. RT-qPCR was performed as described previously (40), with the exception that in the present study the standard curve was prepared using a synthetic RNA fragment matching a conserved region in the viral RNA-Dependent RNA Polymerase (RdRp) gene (BioSynthesis). The primers used for RT-qPCR are based on sequences used previously by Corman et al. (41) and target a similar region in the RdRp, with the forward primer sequence 5′ GTGAAATGGTCATGTGTGGCGG, reverse primer sequence 5′ CAAATGTTAAAAACACTATTAGCATA, and probe sequence of 5′6-FAM/AGGTGGAAC/ZEN/CTCATCAGGAGATGCC/3IABkFQ. The RT-qPCR reaction is expected to produce an amplicon 100 nucleotides long, approximately 0.3% the length of a full SARS-CoV-2 genome. The proportions of potential pyrimidine dimer formation sites to sequence length in the amplified region are similar to the proportions in the full-length genome (Supplemental Table S1). Sample titers of RNA with undamaged amplicon regions were expressed as GEQ per mL.
2.2. Exposure Chamber
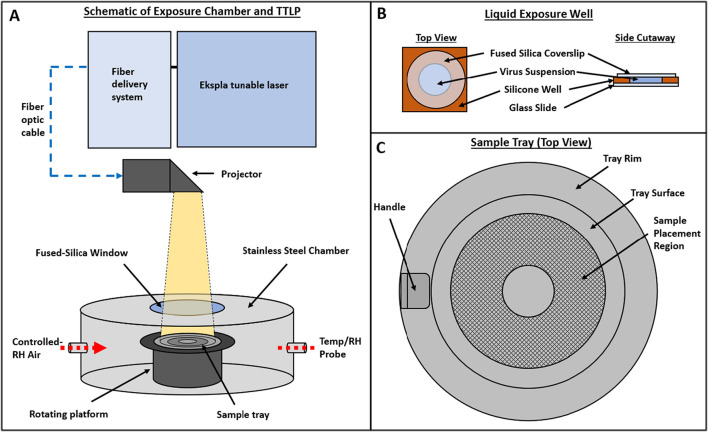
All exposure tests with SARS-CoV-2 were conducted in a cylindrical chamber with a diameter of 25.4 cm and a height of 15.2 cm (Fig. 1A), which was located in a Class II Biological Safety Cabinet (BSC) in a Biosafety Level 3 (BSL-3) laboratory. A 7.6 cm diameter, 0.6 cm thick fused silica window was installed in the center of the chamber lid to allow illumination of the chamber interior. Inside the chamber, a 12.1 cm high platform was rotated at 2 rpm to ensure samples placed on the platform received uniform exposure. The interior surfaces of the chamber were coated with Aeroglaze Z306, a highly absorbent black polyurethane coating, to minimize the potential for reflection inside the chamber. Air, maintained at 20% relative humidity (RH), was supplied at approximately 10 L/min through a port in the side wall of the chamber to maintain constant humidity in the chamber and was exhausted through a HEPA filter to the interior of the BSC. The temperature and RH of the chamber interior were monitored continuously during each test with a Vaisala HMP110 temperature/humidity probe. The mean air temperature and RH values (±SD) in the exposure chamber measured across all tests were 24.5 ± 0.2 °C and 20.8 ± 1.6%, respectively. Transient spikes in RH, typically 1 min or less in duration, occurred in the chamber when the lid was removed to collect samples. The maximum RH recorded in the chamber during a spike was 36.2%.
Fig. 1.
Schematic of exposure chamber, liquid exposure well, and sample tray. A) Exposure chamber configured with the NIST Travelling Tunable UV Laser Projector (TTLP). B) Exposure wells used for liquid samples. C) Tray used for liquid and dried samples. The sample placement region has an interior diameter of 2 cm and an exterior diameter of 6 cm.
In preliminary experiments, the temperature at the surface of the rotating sample platform was measured with a K-type thermocouple during exposure to selected wavelengths spanning the full range of those tested in the present study. Additionally, an ozone monitor (2B Technologies, Model 106-L) was used to measure the concentration of ozone in the exposure chamber. There were no significant increases above baseline in either temperature or ozone concentration in the exposure chamber when the chamber was irradiated (Supplemental Fig. S1).
2.3. Tunable Wavelength Laser
UV radiation and visible light were introduced to the chamber through the fused silica window using the Travelling Tunable UV Laser Projector (TTLP) [80], a tunable wavelength laser source developed by the National Institute of Standards and Technology (NIST). Briefly, the TTLP is comprised of two sub-systems connected by a fiber optic cable. The first sub-system remained outside of the BSL-3 laboratory and was made up of an Ekspla Model NT242-SH/SFG tunable laser and optics for coupling the laser to a fiber optic cable. These optics include an inter-locked safety shutter, timing shutter for controlling sample exposure duration, a Pellin-Broca prism and aperture to reduce the out-of-band radiation, and a turning mirror which directs the beam through a 75 mm focal length UV fused silica, plano-convex focusing lens into the fiber. In the present study, fibers with 1.5 mm diameter cores were used to facilitate acceptance of the radiation without adjustment, despite the chromatic focal shift in the focusing lens. Multiple fibers, with lengths of either 4 m or 5 m, were used over the course of the study due to accumulation of solarization in the fibers with prolonged use.
The second sub-system of the TTLP was positioned inside the Class II Biosafety Cabinet housing the exposure chamber and contained the optics for expanding and projecting the laser beam onto the study samples. This sub-system manipulated the fiber output with an engineered diffuser-based system to produce a uniform beam. The projection system and the output cone were fully enclosed in a 3-D printed enclosure and shroud to fully contain the projector's output and prevent personnel exposure. A window tilted at 45° within the projector provides a low loss pick-off to monitor the system irradiance in real-time.
2.4. Low Pressure Mercury Lamp
In a subset of tests, a low-pressure mercury lamp was used instead of the TTLP for direct comparison between the performance of the TTLP and a type of device more commonly used in UV disinfection studies (i.e., a mercury lamp). For these tests, a 6.5 W ozone-free low-pressure mercury (LP-Hg) lamp (BHK Inc., PN 80-1057-01) in a 3-D printed housing was positioned over the exposure chamber window in place of the TTLP shroud and used to expose SARS-CoV-2 samples to UVC radiation with a primary peak at 253.7 nm.
2.5. Liquid Exposure Wells
Liquid samples were placed in exposure wells that were prepared by affixing silicone isolator wells (13 mm diameter × 2 mm depth; EMS PN 70336–33) to glass microscope slides (Ted Pella PN 260600). For each liquid sample, 240 μL of virus suspension were added to a well, and a 1 mm thick fused silica coverslip (Thorlabs PN WG41010) was placed on top to create a uniform column of liquid with no surface curvature (Fig. 1B). The fraction of incident radiation reflected off the surface (specular and diffuse) of the exposure well bottom was measured at eight degrees off normal with a Perkin-Elmer Lambda 1050 spectrophotometer for each test wavelength. Although the contents of the liquid exposure wells were not stirred during exposures, it is assumed that diffusion and the rotation of the sample platform during the exposures were sufficient to create a well-mixed sample. To recover exposed liquid samples and non-exposed controls, the fused silica coverslip was removed from the exposure well and 150 μL of sample was removed, added to a 2.5 mL aliquot of cell culture medium, and vortexed for 30 s. The use of exposure wells to contain viral suspensions during UV exposure tests was shown to result in dose-response data for MS2 and T1 bacteriophages which were equivalent to those generated using a 10 cm diameter petri dish in a collimated beam exposure system with (Supplemental Methods S1 and Supplemental Fig. S2).
2.6. Dried Droplets
Dried samples were prepared by depositing a 20 μL droplet of viral suspension on a single 7 mm diameter 304 stainless steel coupon and allowing it to dry at room temperature in air maintained at a target RH of ≤20% until the droplets were visually observed to be dry (approximately 75 min after droplet deposition). The diameters of ten representative dried droplet residues of each fluid type were measured with calipers; mean values ± SD were 5.7 ± 0.2 mm and 5.1 ± 0.1 mm for dried residues of virus suspensions in water and simulated saliva, respectively. To recover the exposed dried samples and non-exposed controls, three coupons containing dried material from one fluid type were placed in a single 2.5 mL aliquot of cell culture medium in a 15 mL conical tube and vortexed for 30 s. Each set of three coupons placed in the same aliquot were considered to be a single sample. In a subset of experiments to examine the effect of droplet volume on viral inactivation, droplets with volumes of either 4 μL or 100 μL were also deposited, dried, and recovered in the same manner as the 20 μL droplets. Representative images captured with a 2× objective lens of dried droplets of virus suspensions in water and simulated saliva are shown in Supplemental Fig. S3.
2.7. Dose Determination
Two optical radiation detector types were selected to cover the spectral range of 222 nm −488 nm. A UV sensitive detector (Gigahertz-Optik, model UV-3727-5) was used for 222 nm −305 nm and a visible detector (Gigahertz-Optik, model MD-37-SU100-5 V0) for 310 nm −488 nm. Both detector types were calibrated over a broader spectral range for spectral irradiance responsivity in the automated pulsed laser uniform source (APLUS) laboratory at NIST, and two detectors of each type were used with the TTLP. The “measurement detector” was to quantify the irradiance at the center of the exposure chamber window. However, because the placement of the measurement detector would interfere with sample exposures, a “monitor detector” was positioned in the projector and used to pick off a small fraction of the laser output, providing constant (4 Hz), real-time measurement of the system irradiance during every test. The ratio of responsivities between the monitor detector and the measurement detector was used to calculate the irradiance at the sample plane for each exposure. The ratio between the monitor detector and the measurement detector was confirmed at the beginning and end of each day of testing to ensure the ratio was not changing over time.
At the beginning and end of the study, the radial non-uniformity of the TTLP beam and the transmittance through the exposure chamber window were evaluated at each test wavelength by scanning with the measurement detector at the sample plane across a 5 × 5 grid (25 positions) covering the area of the rotating sample platform. For each wavelength, the irradiance was highest in the center and decreased exponentially to a minimum of approximately 80% of the peak irradiance at the edges of the grid. From these data, a correction factor was determined to account for the transmittance losses through the exposure chamber window and the difference in irradiance between the position of the measurement detector during the daily irradiance checks and the position where the SARS-CoV-2 samples were located. An additional correction factor was determined to account for the transmittance of the fused silica coverslips used for the liquid exposure wells and applied to the doses calculated for the liquid samples. Doses of radiant energy delivered to the samples at each test wavelength were expressed in units of mJ/cm2; the term “dose” used in the present study is equivalent to the terms “fluence” or “radiant exposure” used in other studies.
2.8. Spectral Measurements
A double-monochrometer spectroradiometer (Gooch & Housego Model OL756) was used to measure spectra of the radiation emitted by the LP-Hg lamp and by the TTLP at each tested wavelength. The spectroradiometer was coupled to the radiation source at the exit of the beam shaping optics just above the exposure chamber using a fiber optic cable (Gooch & Housego Model 730-7Q-2.0). The entrance, middle, and exit slits installed in the spectoradiometer were 0.25 mm, 0.5 mm, and 0.25 mm, respectively, a configuration resulting in a half-max bandwidth for the device of 1.2 nm (42). A low-pressure mercury argon lamp (Gooch & Housego Model OL 756–150) was used to verify the wavelength accuracy of the spectroradiometer. Spectra were collected over the wavelength range of 200 nm - 800 nm in 0.5 nm increments with the autogain option enabled and a sampling integration time of 0.1 s.
2.9. Exposure Tests
Suspensions of SARS-CoV-2 (hCoV-19/USA/WA-1/2020) in either distilled water or simulated saliva were exposed to UV radiation and visible light at 222, 230, 240, 253.7, 260, 270, 280, 290, 300, 305, 315, 325, 365, 405, 450, and 488 nm. Two criteria were used for test wavelength selection: first, wavelengths were selected to match the peak output wavelengths of commercially available UV radiation and visible light sources, including excimer and mercury lamps, as well as common LEDs and lasers. Second, wavelengths were selected to maintain approximately regular spacing with intervals of approximately 10 nm through the UVC and UVB regions, and approximately 50 nm through the UVA and visible regions.
Liquid exposure wells or steel coupons were placed on a round tray, (Fig. 1C), to allow them to be placed into and removed from the exposure chamber. As with the chamber interior surfaces, the surface of the sample tray was coated with Aeroglaze Z306 to reduce stray light scattering. After the samples were placed on a tray, the exposure chamber lid was removed and the sample tray was placed on the rotating platform. The chamber lid was replaced and the timing shutter of the TTLP opened to begin the exposure. After the target dose was delivered, the timing shutter was closed and the exposure chamber opened to remove the samples.
Exposure tests were conducted with dried and liquid samples of both simulated saliva and water at the 11 wavelengths from 222 nm - 315 nm with four dose values per wavelength. For these wavelengths, the sample tray was returned to the exposure chamber after a sample was removed up to three times to generate samples exposed to four separate cumulative doses. Three replicate series of exposures were conducted for each combination of wavelength, fluid (virus suspension in water or simulated saliva), and state (liquid or dried). Target doses for each wavelength were chosen based on a combination of the results of preliminary range-finding experiments at a subset of wavelengths, expected shifts in sensitivity based on action spectra for other viruses, and the maximum exposure duration which was logistically feasible to implement (typically ≤ 2 h per replicate of each combination of fluid and state).
Little to no inactivation was observed at 315 nm following exposure to doses >200 mJ/cm2, and further reductions in viral sensitivity were expected for longer wavelengths. Therefore, exposure tests for each combination of fluid and state at the five wavelengths from 325 nm - 488 nm were conducted with a single dose for each wavelength consisting of the highest dose that could be achieved with the irradiance at that wavelength following 2 h of exposure. For each wavelength, three exposed samples and three unexposed samples of each fluid and state were assayed for titers of infectious virus.
2.10. Calculation of Inactivation Constants
UV dose-response data are often presented as inactivation constants (k values) (22,23,27,[43], [44], [45]), which describe inactivation or damage to the virus as a function of UV dose. A k value is equivalent to a one-phase exponential decay constant for non-log transformed data and the slope of a linear regression fit of natural-log transformed data. In the present study, data from tests at wavelengths from 222 nm - 315 nm were used to calculate separate k values for inactivation (k Inactivation) and genome damage (k Genome) for each combination of wavelength, suspension fluid, and state (liquid or dried). k Genome values were determined using all five points in each data set and were calculated using Microsoft Excel. However, to account for a tailing effect in the infectious virus titers in some data sets, k Inactivation values were determined using only the first three points in each data set for all combinations of wavelength, fluid, and state except for liquid samples at 315 nm. For these samples, no measurable inactivation had occurred by the third dose, and therefore the k values for liquid samples at 315 nm were calculated using all five samples. All titers of infectious virus used for calculating k Inactivation values were above the limit of quantification (LOQ) for the experiments.
k values for the virus suspensions in liquid water represent the effects of each wavelength on SARS-CoV-2 in an idealized controlled circumstance. However, the liquid water suspensions do absorb some radiation, particularly at wavelengths <300 nm. Therefore, in the present study, fluid absorbance adjustment factors were calculated for each fluid type based on the absorbance spectrum of that fluid using the method of Bolton and Linden for calculating dose adjustment “water factor” values (46). Additionally, photons which pass completely through the liquid samples have a chance of reflecting off the glass bottom of the exposure wells and back into the liquid, essentially having a second opportunity to interact with material in the well. The fraction of radiation reflected from the exposure well bottom was used in combination with the fluid absorbance adjustment factors to adjust the k Inactivation and k Genome values for each wavelength. Adjusted k values were calculated using Eq. (1), where F is the fluid absorbance adjustment factor and R is the fraction of radiation reflected from the exposure well bottom.
| (1) |
Surface reflectance values were also measured for stainless steel coupons used for dried droplet exposures; these values increased in a nearly linear manner from 0.095 at 222 nm to 0.390 at 488 nm. However, k values for dried samples were not adjusted for absorbance or reflectance due to an inability to accurately measure the thickness and absorbance of the dried sample residue, values which would be necessary for calculating adjustment factors for those samples. Non-adjusted k values for both liquid and dried samples should be understood as an indication of the sensitivity of SARS-CoV-2 in a particular matrix, rather than the intrinsic sensitivity of the virus itself.
2.11. Statistical Analysis
k values from tests at UVC and UVB wavelengths were compared by two-way ANOVA with post-tests using wavelength and sample type as factors, with separate two-way ANOVA analyses performed for k Inactivation and k Genome values. k values from tests with the TTLP at 253.7 nm and the LP-Hg system were compared by two-way ANOVA with radiation source and sample type as factors, with separate two-way ANOVA analyses performed for k Inactivation and k Genome values. For the experiments assessing the effect of initial droplet volume on the susceptibility of SARS-CoV-2 in dried droplets to inactivation by 253.7 nm radiation, log10TCID50 reductions for each droplet volume were compared by two-way ANOVA analysis with fluid type and droplet volume as factors. For wavelengths from 315 to 488 nm, where only a single dose was used for each wavelength, log10-transformed titers of infectious virus in exposed samples were compared by unpaired t-tests to titers from matched non-exposed samples for each combination of wavelength, fluid, and state to determine if there were any changes in titer following exposure to the delivered dose. All statistical tests were performed using GraphPad Prism v.9.0.0.
3. Results
3.1. Spectra
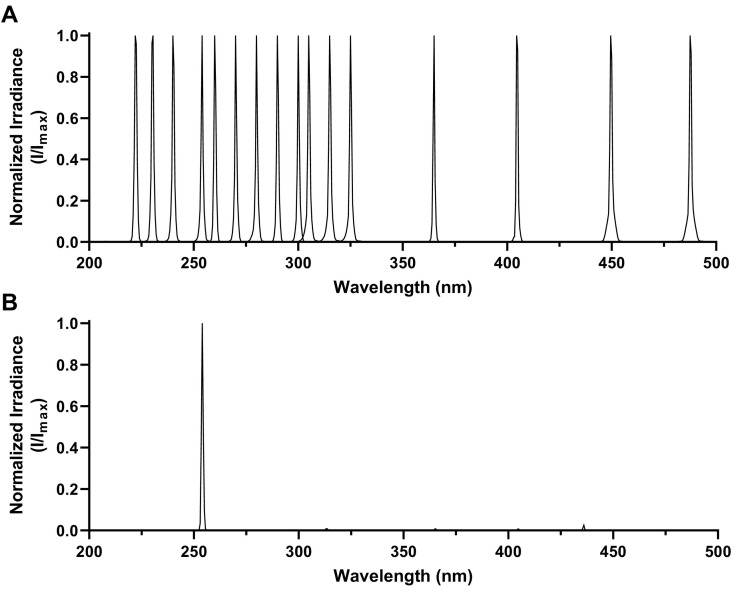
Spectra for the test wavelengths are shown in Fig. 2 . For all the laser wavelengths used in this study, the difference between the nominal wavelength target and the corresponding measured peak irradiance wavelength was ≤0.3 nm. The mean ± SD of the measured full width half-maximum (FWHM) values for the peaks of all laser wavelengths shown in Fig. 2A was 1.09 ± 0.13 nm; for the LP-Hg lamp the measured FWHM value of the primary peak at 253.7 nm in Fig. 2B was 0.98 nm. However, these FWHM values are overestimates due to the operating configuration of the spectroradiometer used in the present study; actual FWHM values for all values are likely <0.2 nm (47,48).
Fig. 2.
Spectra of UV radiation and visible light used for SARS-CoV-2 exposure tests. Spectra are plotted as irradiance values normalized to the peak wavelength for each spectrum, as the total irradiance delivered by the systems varied significantly at different wavelengths. A) Spectra from the TTLP. Sixteen separate spectra are displayed, for target wavelengths of 222, 230, 240, 253.7, 260, 270, 280, 290, 300, 305, 315, 325, 365, 405, 450, and 488 nm. B) Spectrum from the LP-Hg lamp. While minor peaks are present at higher wavelengths, the majority of the irradiance produced by this system is within the primary peak at 253.7 nm.
3.2. UV Dose-Response Data for SARS-CoV-2 in Liquid and Dried Samples
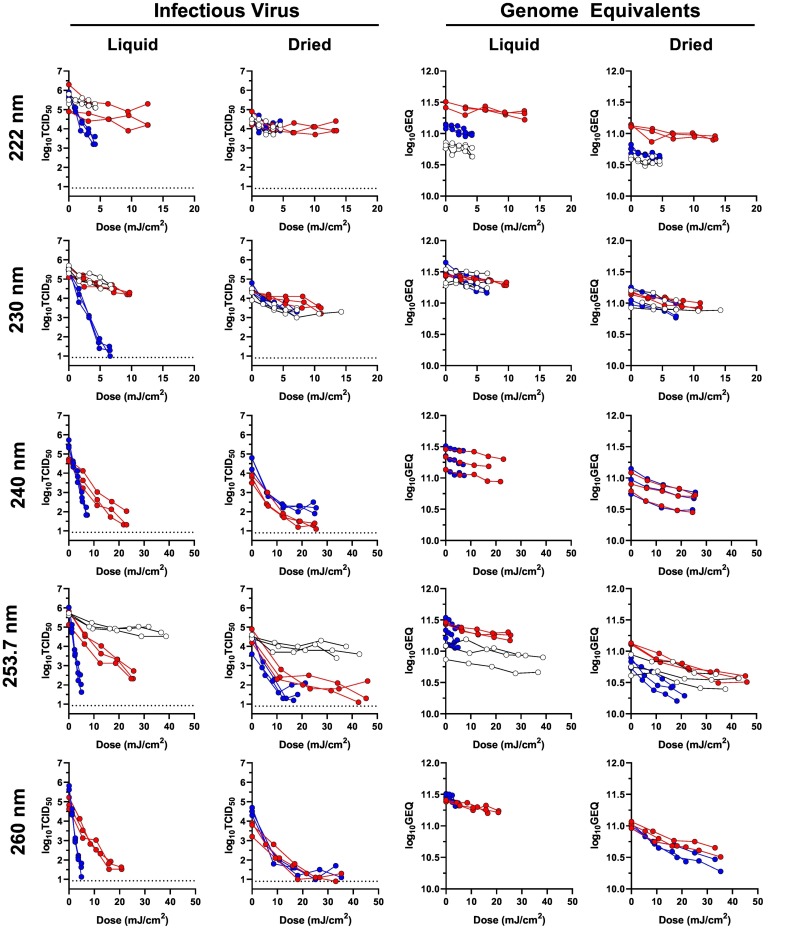
Inactivation and genome damage dose-response data for SARS-CoV-2 exposed to selected wavelengths are shown in Fig. 3 and Supplemental Fig. S4. For several combinations of wavelength, fluid, and state, the log-transformed infectious virus titers were non-linear, particularly near the assay LOQ. This “tailing” phenomenon has been previously described by other investigators (13,27). In the present study, tailing was particularly common for dried samples at wavelengths below 300 nm, and may be a result of the dried residue providing a shielding effect for intact viruses in the interior of the dried residue. Notably, the dose-response data for genome damage, quantified by RT-qPCR, remained log-linear for all combinations of wavelength, fluid, and state, an outcome which may result from the fact that there were approximately six orders of magnitude more GEQ than TCID50 in each sample, and the GEQ titers never approached the LOQ for the assay.
Fig. 3.
SARS-CoV-2 UV Dose-response data for 222, 230, 240, 253.7, and 260 nm. Data from these wavelengths are shown as examples of the dose-response data generated in the present study for SARS-CoV-2 at wavelengths from 222 through 315 nm; the data from the additional tested wavelengths are shown in Supplemental Fig. S1. Data from virus suspensions in water are shown in blue, data from virus suspensions in simulated saliva prepared with porcine mucin batch SLCH5758 are shown in red, and data from virus suspensions in simulated saliva prepared with porcine mucin batch SLCD6129 are shown in white. Dashed lines on the graphs of infectious virus indicate the LOQ of the experiments. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Effect of Different Mucin Batches
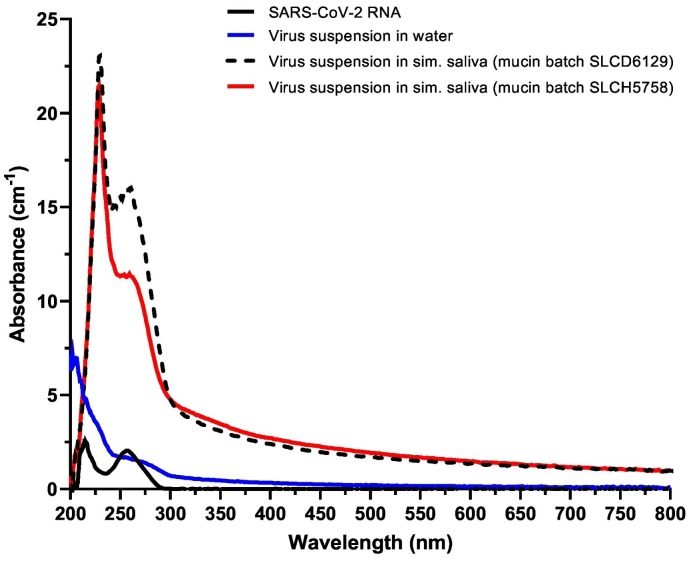
The absorbance spectra of the two simulated saliva virus suspensions (Fig. 4 ) were similar across most of the measured wavelength range, but there was a notable difference in absorbance between approximately 240 nm and 290 nm. At 222 and 230 nm, where there was little difference between the absorbance spectra, UV dose-response results were similar for the two simulated saliva suspensions (Fig. 3). However, at 253.7 nm, where there was a difference of 4.5 absorbance units (cm−1) between the two simulated saliva suspensions, there was a distinct difference between the infectious virus dose-response data for the two fluids. It should be noted that the absorbance values measured in the present study quantify transmission losses resulting from multiple mechanisms, including both molecular absorbance and scattering off of suspended material; references to absorbance values in this manuscript are understood to include these other mechanisms of loss. The reason for the difference in fluid absorbance between 240 nm and 290 nm is not known, and the only quantitative difference between the two mucin batches were different percentages of free and bound sialic acid reported on the manufacturer's certificate of analysis for each batch (0.8% bound and 0.2% free for batch SLCD6129, 1.0% bound and 0.0% free for batch SLCH5758) (49,50). However, further testing would be necessary to determine if these concentration differences are the cause of the differences between the simulated saliva absorbance spectra, or if other factors are involved.
Fig. 4.
Absorbance spectra for representative viral suspensions and SARS-CoV-2 RNA. Viral suspensions in simulated saliva prepared with two different batches of mucin had similar absorbance across most measured wavelengths but differed in the region between approximately 240 nm and 290 nm. The viral suspension in water had lower absorbance than the simulated saliva suspensions for all wavelengths above 213 nm.
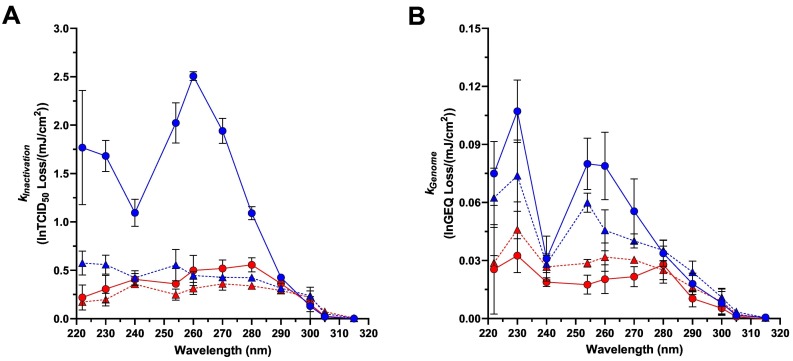
3.4. Action Spectra for SARS-CoV-2 in Liquid and Dried Samples
kInactivation and k Genome values were used to develop separate action spectra for SARS-CoV-2 infectivity and genome damage for each combination of fluid and state (Fig. 5 and Supplemental Tables S2 and S3). All k values for SARS-CoV-2 in suspensions of simulated saliva that are depicted in these action spectra were generated with simulated saliva prepared with mucin batch SLCH5758. By 2-way ANOVA with Tukey's post-test, k Inactivation values in liquid water were significantly higher than k Inactivation values in liquid simulated saliva or in dried droplets of either fluid (Adj. P < 0.0001 for all comparisons) for all wavelengths <290 nm. For these wavelengths, k Inactivation values were similar for dried water droplets and for liquid and dried simulated saliva, though k Inactivation values for dried water diverged from those of simulated saliva in either state for wavelengths <240 nm. There were no significant differences between sample type at all wavelengths from 290 nm through 315 nm (Adj. P ≥ 0.5561 for all comparisons). k Genome values, quantified by RT-qPCR, generally followed the same trends as those for infectivity, with the notable exception that the dried water genome damage spectrum more closely paralleled that of the liquid water spectrum instead of the liquid and dried simulated saliva spectra. By 2-way ANOVA with Tukey's post-test, there were no statistically significant differences between liquid and dried simulated saliva k Genome values at any wavelength (Adj. P ≥ 0.4224 for all comparisons), and there were no statistically significant differences among any of the sample types for all wavelengths ≥280 nm (Adj. P ≥ 0.2625 for all comparisons).
Fig. 5.
SARS-CoV-2 Action Spectra. A) kInactivation values; B) kGenome values. Data from viral suspensions in simulated saliva and water are shown by red and blue symbols, respectively. Data from liquid and dried samples are shown by circles with solid connecting lines and triangles with dashed connecting lines, respectively. Each symbol represents the mean of three replicate values, with error bars indicating SD, except for the dried simulated saliva kGenome values at 315 nm where only two replicates are present. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
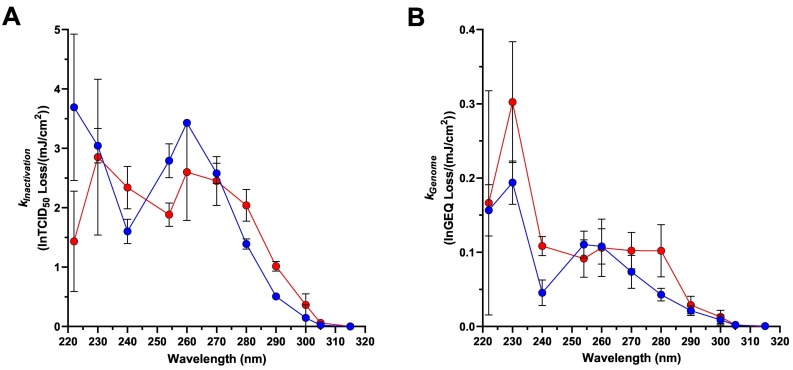
3.5. Action Spectra for SARS-CoV-2 Corrected for Matrix Effects
k values adjusted for absorbance and reflectance (Fig. 6 ) represent the UV dose-response results of the virus itself, independent of interference from the surrounding fluid or artifacts of the experimental test system. These k values were higher than non-adjusted k values for both suspension fluids, although the magnitude of the differences between adjusted and non-adjusted values decreased as wavelength increased. Notably, the absorbance-adjusted k values for simulated saliva samples were similar in magnitude to and followed the same overall pattern as the k values for the liquid water samples. For the adjusted k Inactivation values, there were no statistically significant differences (Adj. P ≥ 0.2789 for all comparisons) between the two suspension fluids for all wavelengths except 222 nm (Adj. P < 0.0001). For the adjusted k Genome values, there were no statistically significant differences (Adj. P ≥ 0.5454 for all comparisons) between the two suspension fluids for all wavelengths except 230 nm (Adj. P = 0.0272).
Fig. 6.
SARS-CoV-2 Action Spectra adjusted for sample absorbance and exposure well reflectance. A) kInactivation values; B) kGenome values. These data represent the intrinsic sensitivity of SARS-CoV-2 to radiation at each of the tested wavelengths. Data from virus suspensions in simulated saliva and water are shown by red and blue symbols, respectively. Each symbol represents the mean of three replicate values, with error bars indicating SDs. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.6. Effects of UVA and Visible Wavelengths
For tests at wavelengths from 325 to 488 nm, the magnitude of the dose received by the samples during the test interval was dependent on the output of the laser system at each wavelength and ranged from average values (±SD) of 82.6 ± 3.2 mJ/cm2 for 365 nm to 956.2 ± 51.3 mJ/cm2 for 450 nm. The results of these tests (Table 1 ) demonstrate that little to no inactivation of SARS-CoV-2 occurred in these tests for any evaluated sample type, with only two combinations of wavelength, state, and fluid having a significant decrease in virus titers following exposure (P > 0.0668 for all other comparisons). At 450 nm, there was a statistically significant decrease (P = 0.0033) following exposure of virus in the liquid simulated saliva samples. At 488 nm, all replicates of exposed dried water samples had the same titer, and all replicates of non-exposed samples had the same titer, resulting in the mean difference between exposed and non-exposed samples of 0.1 log10TCID50 being statistically significant. Additional replicate tests would be needed to better assess the variability associated with these measurements and determine whether the difference remains significant.
Table 1.
Doses of UV radiation or visible light and total quantities of infectious virus in each exposed and unexposed sample for tests at wavelengths from 325 to 488 nm. Virus titers are presented as the mean ± SD of three replicate values. Asterisks indicate titers of exposed samples which are significantly different from matched non-exposed samples.
| Wavelength (nm) | Sample Type | Dose (mJ/cm2) | Total Infectious Virus (Log10TCID50) |
|
|---|---|---|---|---|
| Unexposed | Exposed | |||
| 325 | Liquid Water | 179.9 | 6.46 ± 0.38 | 5.96 ± 0.12 |
| Liquid Sim. Saliva | 205.0 | 4.99 ± 0.06 | 4.86 ± 0.21 | |
| Dried Water | 204.7 | 5.23 ± 0.21 | 5.06 ± 0.68 | |
| Dried Sim. Saliva | 204.7 | 4.43 ± 0.06 | 4.63 ± 0.25 | |
| 365 | Liquid Water | 78.2 | 5.39 ± 0.21 | 5.29 ± 0.12 |
| Liquid Sim. Saliva | 82.2 | 3.92 ± 0.17 | 4.06 ± 0.06 | |
| Dried Water | 85.0 | 4.60 ± 0.10 | 4.46 ± 0.12 | |
| Dried Sim. Saliva | 85.0 | 3.43 ± 0.15 | 3.4 ± 0.35 | |
| 405 | Liquid Water | 219.7 | 6.19 ± 0.29 | 6.46 ± 0.15 |
| Liquid Sim. Saliva | 737.9 | 4.86 ± 0.38 | 4.56 ± 0.29 | |
| Dried Water | 452.7 | 5.13 ± 0.12 | 5.16 ± 0.12 | |
| Dried Sim. Saliva | 452.7 | 4.70 ± 0.26 | 4.83 ± 0.06 | |
| 450 | Liquid Water | 880.3 | 6.39 ± 0.12 | 6.06 ± 0.25 |
| Liquid Sim. Saliva | 969.8 | 4.86 ± 0.12 | *4.39 ± 0.06 | |
| Dried Water | 987.4 | 5.76 ± 0.64 | 5.30 ± 0.10 | |
| Dried Sim. Saliva | 987.4 | 4.40 ± 0.10 | 4.23 ± 0.06 | |
| 488 | Liquid Water | 757.7 | 5.42 ± 0.17 | 5.36 ± 0.21 |
| Liquid Sim. Saliva | 790.4 | 4.32 ± 0.2 | 4.26 ± 0.23 | |
| Dried Water | 808.6 | 4.30 ± 0.00 | *4.20 ± 0.00 | |
| Dried Sim. Saliva | 808.6 | 3.70 ± 0.17 | 3.46 ± 0.06 | |
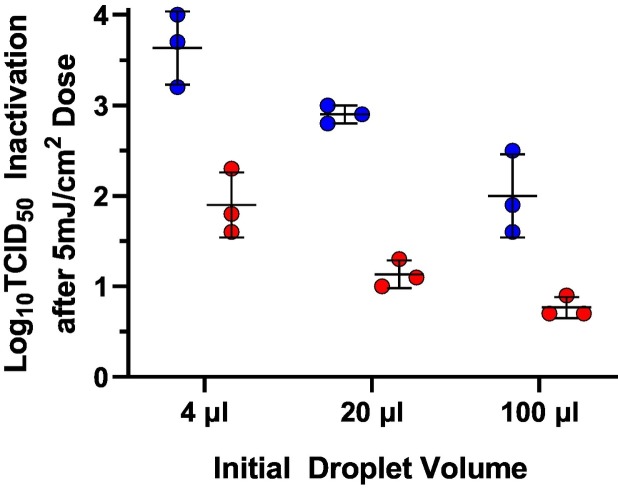
3.7. Evaluation of the Effect of Initial Droplet Volume
Tests to evaluate the effect of initial droplet volume on the susceptibility of SARS-CoV-2 to inactivation by UV radiation in dried droplets of both fluids were conducted with droplets of 4, 20, and 100 μL using the laser system at 253.7 nm. The results of these tests (Fig. 7 ) indicate that the amount of SARS-CoV-2 inactivation following a single 5.3 ± 0.2 mJ/cm2 dose is inversely proportional to the initial droplet volume, presumably due to shielding from the additional mass of residual solids in the larger droplets after drying. The amount of additional shielding provided by each increase in volume was similar for both fluid types, though there was significantly less inactivation in dried simulated saliva samples than in dried water samples. By two-way ANOVA, both the fluid type and the initial droplet volume were significant factors affecting the amount of virus inactivation, but not their interaction (P < 0.0001, P < 0.0001, and P = 0.2706, respectively).
Fig. 7.
Effect of initial droplet volume on SARS-CoV-2 inactivation following exposure to a dose of 253.7 nm radiation of 5.3 ± 0.2 mJ/cm2. Data from tests with dried droplets of virus suspensions in simulated saliva and water are shown in red and blue circles, respectively. Both fluid type and initial droplet volume were statistically significant factors affecting the amount of inactivation. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
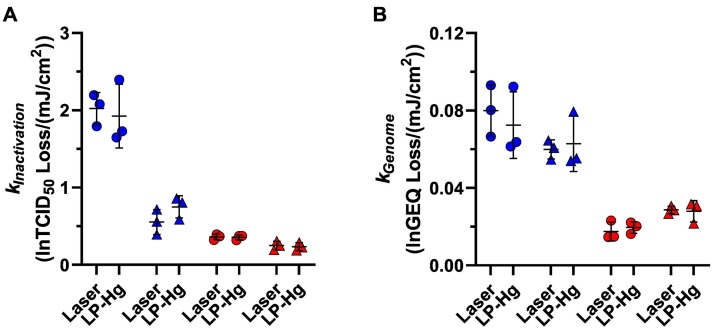
3.8. Comparison of Low-Pressure Mercury Lamp and Laser System
UV dose-response data for SARS-CoV-2 in both suspensions and both states (liquid and dried) generated with a low-pressure mercury lamp system are shown alongside those from the laser system at 253.7 nm in Fig. 8 . There were no significant differences identified by 2-way ANOVA (P > 0.8029 for both data sets) between 253.7 nm radiation sources in SARS-CoV-2 k Inactivation and k Genome values. These data serve as a point of comparison for the SARS-CoV-2 data generated with the laser system across all wavelengths to more common approaches to evaluating viral inactivation by UVC radiation.
Fig. 8.
SARS-CoV-2 kInactivation and kGenome values at 253.7 nm, with radiation supplied by either the TTLP or the LP-Hg lamp. There were no significant differences between radiation source for kInactivation values (A) or kGenome values (B) of any sample type. Data from virus suspensions in simulated saliva and water are shown by red and blue symbols, respectively; data from liquid and dried samples are shown by circles and triangles, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
The present study provides novel data on the effects of sixteen wavelengths of UV radiation and visible light on the infectivity and genome integrity of SARS-CoV-2 in multiple sample types. Rates of inactivation (k Inactivation) and genome damage (k Genome) were highest in the UVC region for all sample types and decreased exponentially through the UVB region. Little to no inactivation of SARS-CoV-2 was observed with any sample type for UVA and visible wavelengths. Taken together, these data suggest that UVC and short-wavelength UVB radiation are far more effective for inactivating SARS-CoV-2 than longer wavelengths. Additionally, the results from liquid samples which are corrected for absorbance and reflectance define the intrinsic sensitivity of SARS-CoV-2 to UV irradiation across the wavelength range of 222–315 nm. These data have broad applicability for the design and implementation of UV-based interventions for SARS-CoV-2 and provide a foundation for understanding the effects of UV radiation and visible light on this virus in real-world scenarios.
The potential for sample matrices to affect UV dose-response relationships has been demonstrated in previous studies with SARS-CoV-2 at single wavelengths (51,52). In the present study, k Inactivation and k Genome values for liquid simulated saliva samples were determined to be lower than equivalent values for liquid water samples for all tested wavelengths below 300 nm, indicating that simulated saliva samples required higher doses of UV radiation to achieve comparable levels of viral inactivation. Across these wavelengths, absorbance values for viral suspensions in simulated saliva were substantially higher than absorbance values for suspensions in water. Measured k values were generally similar between fluid types across the UVC and UVB spectrum when results were adjusted for sample absorbance, suggesting that the optical properties of the fluid alone, and not a secondary phenomenon, are the reason for the protective effect of the simulated saliva on SARS-CoV-2 in liquid samples. This conclusion is further supported by the results of the tests with simulated saliva prepared with different batches of porcine mucin. In these tests, a large difference was observed between the dose-response data of the two simulated saliva preparations at 253.7 nm, where fluid absorbance differed. However, no differences were observed at wavelengths where absorbance values were similar (222 nm and 230 nm).
Although no attempt was made to mathematically account for the absorbance and depth of the dried residue as was done for the liquid samples, the thickness of the dried residues was substantially less than the depth of the liquid samples. However, k Inactivation values for dried samples were lower than values for liquid samples of the same fluid for all tested wavelengths below 300 nm, suggesting that the dried residue served to shield the virus from UV exposure to a greater extent than did the same fluid in liquid form. In tests at 253.7 nm, the degree of protection afforded by dried residues of both fluids was inversely proportional to the initial droplet volume, suggesting that the amount of inactivation achieved with UV sources in real-world surface decontamination applications will likely depend on the size of the contaminated droplets. Very small droplets, including those expelled from the oral cavity and respiratory tract which may contribute to aerosol transmission ([53], [54], [55], [56], [57]), may be small enough that there is little to no matrix shielding effect, and k Inactivation values for these residues may approach those of the absorbance and reflectance-adjusted k Inactivation values for SARS-CoV-2 determined in the present study. However, additional testing would be required to confirm this hypothesis.
While surface effects have been observed in other SARS-CoV-2 UV inactivation studies (36,58,59), the results of the droplet volume tests also serve as an indication that the surface roughness of the stainless steel coupons did not appreciably shield viruses from UV exposure in the present study, as samples with higher ratios of dried residue volume to coupon surface were easier to inactivate than samples with lower ratios – the opposite of what would be expected if virions shielded in the “valleys” of the coupon surface were a significant proportion of the total virus population in each sample.
The residues of the dried simulated saliva droplets contain salts and mucin, whereas the residues of the dried water samples are assumed to be comprised primarily of the virus itself along with any residual cellular and viral components which were not removed during the process of preparing the viral suspensions. While the liquid samples contain the same non-viral components as the dried samples, the components are dissolved or in suspension and therefore do not represent a fixed barrier between the virus and the source of radiation in the manner of the dried residues. In the dried samples, the residual material was enough to substantially reduce k Inactivation values in dried water suspension samples relative to liquid water samples, suggesting that even a minimal amount of shielding may have significant ramifications for the efficacy of UV disinfection platforms in real-world scenarios. However, it is notable that while k Genome values for dried water samples were lower than k Genome values for liquid water samples at most UVC wavelengths, there was less shielding evident in these results than in the k Inactivation values for the same samples. Furthermore, k Genome values for dried simulated saliva samples were not significantly different from k Genome values for liquid simulated saliva samples, suggesting that drying did not confer a protective effect against genome damage for these samples. This outcome may follow from the fact that there were several orders of magnitude more GEQ in each sample than infectious virus, and therefore there was more GEQ available to be inactivated in the regions of the residue accessible to UV radiation near the surface. Additionally, viruses in drying droplets have been shown to partition into discrete regions of the final residue (60). If free RNA and intact infectious virions partitioned into different regions of the dried residues, it is likely that they would receive different amounts of shielding from the incident radiation and therefore have different dose-response relationships. However, more testing would be necessary to explore these hypotheses.
In addition to the slower rates of inactivation observed for dried samples at UVC wavelengths, the k Inactivation values for dried samples reflect only the initial phase of rapid inactivation, and for many wavelengths there appeared to be a second phase for dried samples where inactivation was slower. This second phase began most frequently as viral titers approached the LOQ of the experiment, although there were some combinations of wavelength and suspension fluid (e.g., simulated saliva at 253.7 nm and water at 240 nm) for which the second phase of inactivation began well above the LOQ. The presence of a second phase or “tail” has been observed in previous UV disinfection studies, and has been attributed to various causes including saturation of available targets (22), inherently resistant sub-populations (61,62), recombination of damaged and undamaged viruses (63), and heterogenous protection due to the sample matrix and/or aggregates of the target microorganism ([64], [65], [66], [67]). The fact that tailing in the present study was observed for dried samples but not liquid samples of both fluids suggests that neither the availability of targets nor the physiology of the virus were responsible, and instead suggests that there were variable amounts of shielding in different regions of the dried residue. Unfortunately, in the present study it was not possible to deliver sufficient doses of UV radiation at every wavelength to achieve multiple orders of magnitude of inactivation with each sample type, though such experiments would be useful to evaluate the biphasic inactivation phenomenon observed with some combinations of wavelength and sample type.
Although it remains unclear which anatomical sites of origin and corresponding carrier fluids contribute most significantly to person-to-person transmission of SARS-CoV-2, the viral suspensions in simulated saliva used in the present study were intended to be generally representative of fluid known to be expelled from the oral cavity during speaking and coughing (55). However, bodily fluids vary significantly in composition and optical properties depending on the fluid type and an individual's physiological state (68), and it is likely that the rates of SARS-CoV-2 inactivation in real human fluids will differ from the rates measured in simulated saliva suspensions in the present study to a degree proportional to the differences in the absorbance of the fluids. Despite this, it is likely that the larger principle of high fluid absorbance reducing the efficacy of UV inactivation rates would still hold, and it is important to recognize that achieving sufficient levels of inactivation in real-world applications will require higher doses of UV radiation than might be expected based on data which does not account for matrix effects. Calculating fluid absorbance adjustment factors for other matrices and applying them in a reverse manner to the absorbance and reflectance-adjusted SARS-CoV-2 k values in the present study should provide a means of estimating the sensitivity of this virus in other fluids, including actual human bodily fluids.
The similar patterns of the infectivity and genome damage spectra when corrected for fluid absorbance and exposure well reflectance, as well as the similarity of the patterns of both action spectra to the absorbance spectra of SARS-CoV-2 RNA, suggest that genome damage is the primary mechanism by which infectivity was lost in the present study. Notably, the action spectra for SARS-CoV-2 infectivity display the bimodal form characteristic of action spectra for other viruses (26,27,[29], [30], [31]), with a peak at 260 nm corresponding to the peak absorbance of the nucleic acids of the genome, and a second peak or shoulder at wavelengths below 240 nm where both nucleic acids and proteins absorb. However, despite these similarities, the attribution of genome damage as the primary mechanism of infectivity reduction in the present study is tentative, and additional testing would be necessary to evaluate the contributions of other potential inactivation mechanisms. Other such mechanisms may include direct damage to viral structural proteins (19,69) and/or indirect effects resulting from the formation of toxic compounds, including reactive oxygen species, in the matrix surrounding the virus ([70], [71], [72]).
It has been proposed that a single point of damage, or lesion, in a viral genome is sufficient to result in a loss of infectivity (23), as stopping the action of a polymerase enzyme anywhere in its progress across a viral genome should be sufficient to interrupt the viral replication cycle. However, it is notable that there is approximately a 300-fold difference between the length of a SARS-CoV-2 genome and the amplicon produced by the qPCR reaction in the present study. If viral inactivation occurred following a “single hit” to the genome, where a single point of damage anywhere in the genome blocks replication and the infection cycle, it would be expected that the k Inactivation values should be no <300-fold higher than the k Genome values, even if genome damage was responsible for 100% of viral inactivation. However, k Inactivation values measured in the present study were only approximately 20-fold higher than the k Genome values. This difference may suggest that the “single hit” UV inactivation hypothesis is not valid for SARS-CoV-2, possibly because of an ability for the viral RdRp to read through genome lesions or a limited proof-reading ability (73). It is also possible due to the high ratio of GEQ to TCID50 in the samples that rates of genome damage quantified in the present study reflect damage to free RNA in the samples rather than damage to the genomes of intact, infectious virions. The free RNA in the samples likely includes both sub-genomic RNAs and full-length unpackaged genomes (74). These RNAs are not only unshielded by the structural proteins of a complete virion, but also have a degree of redundancy due to their high copy number. While damage to the genomes of infectious virions is likely still occurring, its “signal” would be lost in the “noise” of the damage to free RNA. It should be noted that the RT-qPCR assay used in the present study to quantify damage to viral RNA does not distinguish between the various types of genome lesions which are known to occur. Further testing would be required to quantify the relative contributions of different lesion types to virus inactivation at each tested wavelength.
kInactivation values for SARS-CoV-2 determined in the present study are generally similar in magnitude to those reported in several other studies for comparable samples at equivalent wavelengths, including 222 and 253.7 nm (9,13,35,[75], [76], [77]). Furthermore, there were no differences between SARS-CoV-2 k values in tests with 253.7 nm radiation delivered by either the TTLP or the LP-Hg lamp. Taken together, these results suggest that the novel methods used in the present study, including the combined use of the TTLP and exposure chamber and the use of the liquid exposure wells for liquid samples, can be used to generate data which are directly comparable to UV inactivation data generated in more traditional test systems, including collimated beam exposure systems. However, the approach used in the present study can be used across a wider range of wavelengths, with narrower wavebands, and with lower sample volumes than many other approaches.
One limitation of the test system used in the present study was the inability to deliver doses of UVA and visible radiation that were high enough to cause a measurable effect on SARS-CoV-2 infectivity. While some studies have demonstrated that SARS-CoV-2 and other coronaviruses can be inactivated by UVA and visible radiation (8,[14], [15], [16]), the results of those studies suggest that impractically long test durations would be required to achieve measurable reductions of infectivity with UVA and visible wavelengths using the test system in the present study. For example, Lau et al. report that a dose of 57.5 J/cm2 of 405 nm radiation was required to achieve a single log10 reduction in titer of a bovine coronavirus in liquid (14); achieving a similar dose of 405 nm radiation with the system used in the present study would have required approximately five days of continuous exposure. Therefore, while we can conclude that the UVC and UVB wavelengths tested in the present study were more effective at inactivating SARS-CoV-2 than the tested UVA and visible wavelengths, additional testing with more powerful sources would be required to fully evaluate the potential of UVA and visible wavelengths to inactivate SARS-CoV-2 in the sample types tested in the present study.In addition to informing the implementation of germicidal UV sources to mitigate SARS-CoV-2 contamination, the data from the present study can also be used to better understand the disinfecting potential of natural sunlight. For example, estimates for k Inactivation values for 1 nm increments across the UVB region can be generated by interpolating between wavelengths tested in the present study. By applying these interpolated k Inactivation values to fluence values from sunlight spectra and summing the results, decay rate estimates may be generated for a wide range of natural sunlight conditions. We used this approach with the absorbance and reflectance-adjusted k Inactivation values from liquid water tests in the present study and a model sunlight spectrum from the National Center for Atmospheric Research's tropospheric ultraviolet and visible (TUV) radiation model (78) for mid-day at 40°N latitude on the summer solstice. This resulted in an estimated infectivity exponential decay constant value of 0.21 min−1, which is similar to the value of 0.22 min−1 previously measured by our laboratory for SARS-CoV-2 hCoV-19/USA/WA1/2020 in aerosols at 20 °C, 20 %RH, in simulated sunlight with a spectrum matching that of the summer solstice TUV model spectrum (4). Similar results were also obtained when comparing decay constants estimated from dried simulated saliva k Inactivation values from the present study to decay constants measured for SARS-CoV-2 in 5 μL droplets of simulated saliva dried on steel surfaces under equivalent conditions (3).
Absorbance and reflectance corrections were performed for viral suspensions in both water and simulated saliva. However, these corrections were smaller in magnitude for the water data than they were for the simulated saliva. Therefore, of the two data sets, we believe that the absorbance and reflectance corrected data from the water suspensions provides the best quantification of the intrinsic sensitivity of SARS-CoV-2 to inactivation by UV wavelengths. Although aerosols were not evaluated in the present study, the similarity of the decay constant estimates developed using the absorbance and reflectance-adjusted k Inactivation values from liquid water tests to our previous data from SARS-CoV-2 in aerosols exposed to simulated sunlight suggests that the former data set may provide a reasonable approximation of the UV dose-response relationship of SARS-CoV-2 in small particle aerosols. Repeating this approach with other sunlight spectra (real or modeled) should provide a means of estimating the effect of sunlight on SARS-COV-2 for a range of locations, dates, and atmospheric conditions.
Data from the present study may be useful for informing evaluations of the relative efficacy of different wavelengths of UV radiation and visible light for inactivating SARS-CoV-2 in a variety of circumstances. Experiments with multiple isolates of SARS-CoV-2 indicate that the sensitivity of newer variants of SARS-CoV-2 is similar to that of the earlier hCoV-19/USA/WA-1/2020 used in the present study, suggesting that the results of the present study will likely continue to be applicable as the pandemic progresses (4,9,79). It was beyond the scope of the present study to evaluate the potential for any of the tested wavelengths to cause harm to human health or the integrity of materials in the built environment. Decisions about the use of UV sources for disinfection applications should include consideration of such factors, in addition to evaluations of the virucidal efficacy data presented in both this study and others. However, if used appropriately, approaches to treating SARS-CoV-2 contamination with UV radiation may prove to be a useful component of strategies for mitigating the spread of the COVID-19 pandemic.
Funding
This work was supported by the DHS Science and Technology Directorate (agreement number HSHQDC-15-C-00064 to Battelle National Biodefense Institute for the management and operation of the National Biodefense Analysis and Countermeasures Center, a Federally Funded Research and Development Center).
Additional support was provided by the Department of Commerce.
Disclaimer
The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the Department of Homeland Security (DHS), the Department of Commerce (DoC), or the US Government. The DHS and DoC do not endorse any products or commercial services mentioned in this presentation. In no event shall the DHS, DoC, Battelle National Biodefense Institute, the National Biodefense Analysis and Countermeasures Center, or the National Institute of Standards and Technology have any responsibility or liability for any use, misuse, inability to use, or reliance upon the information contained herein. In addition, no warranty of fitness for a particular purpose, merchantability, accuracy, or adequacy is provided regarding the contents of this document.
Notice
This manuscript has been authored by Battelle National Biodefense Institute, LLC under Contract No. HSHQDC-15-C-00064 with the US Department of Homeland Security. The United States Government retains, and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes.
CRediT authorship contribution statement
Michael A. Schuit: Conceptualization, Methodology, Investigation, Writing – original draft, Writing – review & editing. Thomas C. Larason: Methodology, Investigation, Software, Writing – review & editing. Melissa L. Krause: Methodology, Investigation, Writing – review & editing. Brian M. Green: Methodology, Investigation, Writing – review & editing. Brian P. Holland: Methodology, Investigation, Writing – review & editing. Stewart P. Wood: Methodology, Investigation, Writing – review & editing. Steven Grantham: Methodology, Investigation, Writing – review & editing. Yuqin Zong: Methodology, Investigation, Writing – review & editing. Clarence J. Zarobila: Methodology, Investigation, Writing – review & editing. Denise L. Freeburger: Investigation, Writing – review & editing. David M. Miller: Investigation, Writing – review & editing. Jordan K. Bohannon: Methodology, Investigation, Writing – review & editing. Shanna A. Ratnesar-Shumate: Conceptualization, Writing – review & editing. Ernest R. Blatchley: Conceptualization, Methodology, Writing – review & editing. Xing Li: Methodology, Investigation, Writing – review & editing. Paul A. Dabisch: Conceptualization, Writing – review & editing. C. Cameron Miller: Conceptualization, Methodology, Investigation, Writing – review & editing.
Declaration of Competing Interest
Prof. Blatchley is working with XCMR, a start-up company developing UV-based devices to protect against airborne pathogens. He has been compensated by stock in the company for his efforts. All other authors declare they have no competing interests.
Acknowledgments
The authors thank Dr. Michael Hevey for providing critical review and input on the study plan and this manuscript, Dr. Victoria Wahl for assistance acquiring the initial sample of SARS-CoV-2, Amy Reese for assistance with sample assays, Catherine Cooksey for reflectance and transmittance measurements and Gregory Williams for assistance with viral stock production.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jphotobiol.2022.112503.
Appendix A. Supplementary data
Supplementary material 1
Supplementary material 2
References
- 1.Rendell R., Khazova M., Higlett M., O’Hagan J. Impact of high solar UV radiant exposures in spring 2020 on SARS-CoV-2 viral inactivation in the UK. Photochem. Photobiol. 2021;97(3):542–548. doi: 10.1111/php.13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang L., Liu M., Ren B., Wu Z., Yu X., Peng C., et al. Sunlight ultraviolet radiation dose is negatively correlated with the percent positive of SARS-CoV-2 and four other common human coronaviruses in the US. Sci. Total Environ. 2021;751 doi: 10.1016/j.scitotenv.2020.141816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ratnesar-Shumate S., Williams G., Green B., Krause M., Holland B., Wood S., et al. Simulated sunlight rapidly inactivates SARS-CoV-2 on surfaces. J. Infect. Dis. 2020;222(2):214–222. doi: 10.1093/infdis/jiaa274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schuit M., Biryukov J., Beck K., Yolitz J., Bohannon J., Weaver W., et al. The stability of an isolate of the SARS-CoV-2 B. 1.1. 7 lineage in aerosols is similar to three earlier isolates. J. Infect. Dis. 2021;224(10):1641–1648. doi: 10.1093/infdis/jiab171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schuit M., Ratnesar-Shumate S., Yolitz J., Williams G., Weaver W., Green B., et al. Airborne SARS-CoV-2 is rapidly inactivated by simulated sunlight. J. Infect. Dis. 2020;222(4):564–571. doi: 10.1093/infdis/jiaa334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wondrak G.T., Jandova J., Williams S.J., Schenten D. Solar simulated ultraviolet radiation inactivates HCoV-NL63 and SARS-CoV-2 coronaviruses at environmentally relevant doses. J. Photochem. Photobiol. B Biol. 2021;224 doi: 10.1016/j.jphotobiol.2021.112319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gidari A., Sabbatini S., Bastianelli S., Pierucci S., Busti C., Bartolini D., et al. SARS-CoV-2 survival on surfaces and the effect of UV-C light. Viruses. 2021;13(3):408. doi: 10.3390/v13030408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heilingloh C.S., Aufderhorst U.W., Schipper L., Dittmer U., Witzke O., Yang D., et al. Susceptibility of SARS-CoV-2 to UV irradiation. Am. J. Infect. Control. 2020;48(10):1273–1275. doi: 10.1016/j.ajic.2020.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inagaki H., Saito A., Sugiyama H., Okabayashi T., Fujimoto S. Rapid inactivation of SARS-CoV-2 with deep-UV LED irradiation. Emerg. Microb. Infect. 2020;9(1):1744–1747. doi: 10.1080/22221751.2020.1796529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kitagawa H., Nomura T., Nazmul T., Omori K., Shigemoto N., Sakaguchi T., et al. Effectiveness of 222-nm ultraviolet light on disinfecting SARS-CoV-2 surface contamination. Am. J. Infect. Control. 2021;49(3):299–301. doi: 10.1016/j.ajic.2020.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabino C.P., Sellera F.P., Sales-Medina D.F., Machado R.R.G., Durigon E.L., Freitas-Junior L.H., et al. UV-C (254 nm) lethal doses for SARS-CoV-2. Photodiagn. Photodyn. Ther. 2020;32 doi: 10.1016/j.pdpdt.2020.101995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shimoda H., Matsuda J., Iwasaki T., Hayasaka D. Efficacy of 265-nm ultraviolet light in inactivating infectious SARS-CoV-2. J. Photochem. Photobiol. 2021;100050 doi: 10.1016/j.jpap.2021.100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma B., Gundy P.M., Gerba C.P., Sobsey M.D., Linden K.G. UV inactivation of SARS-CoV-2 across the UVC spectrum: KrCl* excimer, mercury-vapor, and light-emitting-diode (LED) sources. Appl. Environ. Microbiol. 2021;87(22) doi: 10.1128/AEM.01532-21. e01532-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lau B., Becher D., Hessling M. Multidisciplinary Digital Publishing Institute; 2021. High Intensity Violet Light (405 nm) Inactivates Coronaviruses in Phosphate Buffered Saline (PBS) and on Surfaces. Photonics. [Google Scholar]
- 15.De Santis R., Luca V., Näslund J., Ehmann R.K., De Angelis M., Lundmark E., et al. Rapid inactivation of SARS-CoV-2 with LED irradiation of visible spectrum wavelengths. J. Photochem. Photobiol. 2021;100082 doi: 10.1016/j.jpap.2021.100082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stasko N., Kocher J.F., Annas A., Henson I., Seitz T.S., Miller J.M., et al. Visible blue light inhibits infection and replication of SARS-CoV-2 at doses that are well-tolerated by human respiratory tissue. Sci. Rep. 2021;11(1):1–13. doi: 10.1038/s41598-021-99917-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oh C., Sun P.P., Araud E., Nguyen T.H. Mechanism and efficacy of virus inactivation by a microplasma UV lamp generating monochromatic UV irradiation at 222 nm. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116386. [DOI] [PubMed] [Google Scholar]
- 18.Miller R.L., Plagemann P.G. Effect of ultraviolet light on mengovirus: formation of uracil dimers, instability and degradation of capsid, and covalent linkage of protein to viral RNA. J. Virol. 1974;13(3):729–739. doi: 10.1128/jvi.13.3.729-739.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wigginton K.R., Menin L., Sigstam T., Gannon G., Cascella M., Hamidane H.B., et al. UV radiation induces genome-mediated, site-specific cleavage in viral proteins. ChemBioChem. 2012;13(6):837–845. doi: 10.1002/cbic.201100601. [DOI] [PubMed] [Google Scholar]
- 20.Nelson K.L., Boehm A.B., Davies-Colley R.J., Dodd M.C., Kohn T., Linden K.G., et al. Sunlight-mediated inactivation of health-relevant microorganisms in water: a review of mechanisms and modeling approaches. Environ Sci Process Impacts. 2018;20(8):1089–1122. doi: 10.1039/c8em00047f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wurtmann E.J., Wolin S.L. RNA under attack: cellular handling of RNA damage. Crit. Rev. Biochem. Mol. Biol. 2009;44(1):34–49. doi: 10.1080/10409230802594043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beck S.E., Rodriguez R.A., Hawkins M.A., Hargy T.M., Larason T.C., Linden K.G. Comparison of UV-induced inactivation and RNA damage in MS2 phage across the germicidal UV spectrum. Appl. Environ. Microbiol. 2016;82(5):1468–1474. doi: 10.1128/AEM.02773-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cutler T.D., Zimmerman J.J. Ultraviolet irradiation and the mechanisms underlying its inactivation of infectious agents. Anim. Health Res. Rev. 2011;12(1):15–23. doi: 10.1017/S1466252311000016. [DOI] [PubMed] [Google Scholar]
- 24.Besaratinia A., Ji Yoon, Schroeder C., Bradforth S.E., Cockburn M., Pfeifer G.P. Wavelength dependence of ultraviolet radiation-induced DNA damage as determined by laser irradiation suggests that cyclobutane pyrimidine dimers are the principal DNA lesions produced by terrestrial sunlight. FASEB J. 2011;25(9):3079–3091. doi: 10.1096/fj.11-187336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lo C.-W., Matsuura R., Iimura K., Wada S., Shinjo A., Benno Y., et al. UVC disinfects SARS-CoV-2 by induction of viral genome damage without apparent effects on viral morphology and proteins. Sci. Rep. 2021;11(1):1–11. doi: 10.1038/s41598-021-93231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beck S.E., Hull N.M., Poepping C., Linden K.G. Wavelength-dependent damage to adenoviral proteins across the germicidal UV spectrum. Environ. Sci. Technol. 2018;52(1):223–229. doi: 10.1021/acs.est.7b04602. [DOI] [PubMed] [Google Scholar]
- 27.Beck S.E., Rodriguez R.A., Linden K.G., Hargy T.M., Larason T.C., Wright H.B. Wavelength dependent UV inactivation and DNA damage of adenovirus as measured by cell culture infectivity and long range quantitative PCR. Environ. Sci. Technol. 2014;48(1):591–598. doi: 10.1021/es403850b. [DOI] [PubMed] [Google Scholar]
- 28.Kleczkowski A., Govier D. Action spectrum for inactivation of the infectivity of potato virus X by UV radiation. Photochem. Photobiol. 1969;10(1):53–59. doi: 10.1111/j.1751-1097.1969.tb07220.x. [DOI] [PubMed] [Google Scholar]
- 29.Coohill T.P., Moore S.P., Knauer D.J., Fry D.G., Eichenbrenner T.J., Bockstahler L.U. Action spectrum for the in vitro induction of simian virus 40 by ultraviolet radiation. Mutat. Res. 1982;95(2–3):95–103. doi: 10.1016/0027-5107(82)90249-4. [DOI] [PubMed] [Google Scholar]
- 30.Detsch R.M., Bryant F.D., Coohill T.P. The wavelength dependence of herpes simplex virus inactivation by ultraviolet radiation. Photochem. Photobiol. 1980;32(2):173–176. doi: 10.1111/j.1751-1097.1980.tb04005.x. [DOI] [PubMed] [Google Scholar]
- 31.Sime E.H., Bedson H. A comparison of ultraviolet action spectra for vaccinia virus and T2 bacteriophage. J. Gen. Virol. 1973;18(1):55–60. doi: 10.1099/0022-1317-18-1-55. [DOI] [PubMed] [Google Scholar]
- 32.Siegel A., Norman A. Action spectra for two strains of tobacco mosaic virus. Virology. 1958;6(3):725–731. doi: 10.1016/0042-6822(58)90118-1. [DOI] [PubMed] [Google Scholar]
- 33.Jureka A.S., Williams C.G., Basler C.F. Pulsed broad-spectrum UV light effectively inactivates SARS-CoV-2 on multiple surfaces and N95 material. Viruses. 2021;13(3):460. doi: 10.3390/v13030460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kitagawa H., Nomura T., Nazmul T., Kawano R., Omori K., Shigemoto N., et al. Effect of intermittent irradiation and fluence-response of 222 nm ultraviolet light on SARS-CoV-2 contamination. Photodiagn. Photodyn. Ther. 2021;33 doi: 10.1016/j.pdpdt.2021.102184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minamikawa T., Koma T., Suzuki A., Mizuno T., Nagamatsu K., Arimochi H., et al. Quantitative evaluation of SARS-CoV-2 inactivation using a deep ultraviolet light-emitting diode. Sci. Rep. 2021;11(1):1–9. doi: 10.1038/s41598-021-84592-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geldert A., Su A., Roberts A.W., Golovkine G., Grist S.M., Stanley S.A., et al. Mapping of UV-C dose and SARS-CoV-2 viral inactivation across N95 respirators during decontamination. Sci. Rep. 2021;11(1):1–12. doi: 10.1038/s41598-021-98121-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buonanno M., Welch D., Shuryak I., Brenner D.J. Far-UVC light (222 nm) efficiently and safely inactivates airborne human coronaviruses. Sci. Rep. 2020;10(1):1–8. doi: 10.1038/s41598-020-67211-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dabisch P., Schuit M., Herzog A., Beck K., Wood S., Krause M., et al. The influence of temperature, humidity, and simulated sunlight on the infectivity of SARS-CoV-2 in aerosols. Aerosol Sci. Technol. 2021;55(2):142–153. doi: 10.1080/02786826.2020.1829536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.International A, editor . ASTM West Conshohocken, PA; 2016. ASTM E2721-16: Standard Practice for Evaluation of Effectiveness of Decontamination Procedures for Surfaces When Challenged With Droplets Containing Human Pathogenic Viruses. [Google Scholar]
- 40.Ratnesar-Shumate S., Bohannon K., Williams G., Holland B., Krause M., Green B., et al. Comparison of the performance of aerosol sampling devices for measuring infectious SARS-CoV-2 aerosols. Aerosol Sci. Technol. 2021:1–15. doi: 10.1080/02786826.2021.1910137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25(3):2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Housego G. OL 756 portable high-accuracy UV-visible spectroradiometer user manual #M000280. Rev. J. 2015 Orlando, FL: Gooch & Housego. [Google Scholar]
- 43.Lytle C.D., Sagripanti J.-L. Predicted inactivation of viruses of relevance to biodefense by solar radiation. J. Virol. 2005;79(22):14244–14252. doi: 10.1128/JVI.79.22.14244-14252.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mbonimpa E.G., Blatchley E., Applegate B., Harper W. Ultraviolet A and B wavelength-dependent inactivation of viruses and bacteria in the water. J. Water Health. 2018;16(5):796–806. doi: 10.2166/wh.2018.071. [DOI] [PubMed] [Google Scholar]
- 45.Park G., Linden K., Sobsey M. Inactivation of murine norovirus, feline calicivirus and echovirus 12 as surrogates for human norovirus (NoV) and coliphage (F+) MS2 by ultraviolet light (254 nm) and the effect of cell association on UV inactivation. Lett. Appl. Microbiol. 2011;52(2):162–167. doi: 10.1111/j.1472-765X.2010.02982.x. [DOI] [PubMed] [Google Scholar]
- 46.Bolton J.R., Linden K.G. Standardization of methods for fluence (UV dose) determination in bench-scale UV experiments. J. Environ. Eng. 2003;129(3):209–215. [Google Scholar]
- 47.Sansonetti C.J., Salit M.L., Reader J. Wavelengths of spectral lines in mercury pencil lamps. Appl. Opt. 1996;35(1):74–77. doi: 10.1364/AO.35.000074. [DOI] [PubMed] [Google Scholar]
- 48.Zong Y., Brown S.W., Eppeldauer G.P., Lykke K.R., Ohno Y. A new method for spectral irradiance and radiance responsivity calibrations using kilohertz pulsed tunable optical parametric oscillators. Metrologia. 2012;49(2):S124. [Google Scholar]
- 49.Sigma-Aldrich. Batch SLCD6129. 2019. Certificate of analysis for mucin from porcine stomach - type III, bound sialic acid 0.5–1.5%, partially purified powder, product number M1778. [Google Scholar]
- 50.Sigma-Aldrich. Batch SLCH5758. 2020. Certificate of analysis for mucin from porcine stomach - type III, bound sialic acid 0.5–1.5%, partially purified powder, product number M1778. [Google Scholar]
- 51.Doughty D.C., Hill S.C., Mackowski D.W. Viruses such as SARS-CoV-2 can be partially shielded from UV radiation when in particles generated by sneezing or coughing: numerical simulations. J. Quant. Spectrosc. Radiat. Transf. 2021;262 doi: 10.1016/j.jqsrt.2020.107489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bispo-dos-Santos K., Barbosa P.P., Granja F., Martini M.C., Oliveira C.F.S., Schuck D.C., et al. Ultraviolet germicidal irradiation is effective against SARS-CoV-2 in contaminated makeup powder and lipstick. J. Photochem. Photobiol. 2021;8 doi: 10.1016/j.jpap.2021.100072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asadi S., Wexler A.S., Cappa C.D., Barreda S., Bouvier N.M., Ristenpart W.D. Effect of voicing and articulation manner on aerosol particle emission during human speech. PLoS One. 2020;15(1) doi: 10.1371/journal.pone.0227699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fabian P., Brain J., Houseman E.A., Gern J., Milton D.K. Origin of exhaled breath particles from healthy and human rhinovirus-infected subjects. J. Aerosol Med. Pulm. Drug Deliv. 2011;24(3):137–147. doi: 10.1089/jamp.2010.0815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson G., Morawska L., Ristovski Z., Hargreaves M., Mengersen K., Chao C.Y.H., et al. Modality of human expired aerosol size distributions. J. Aerosol Sci. 2011;42(12):839–851. doi: 10.1016/j.jaerosci.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson G.R., Morawska L. The mechanism of breath aerosol formation. J. Aerosol Med. Pulm. Drug Deliv. 2009;22(3):229–237. doi: 10.1089/jamp.2008.0720. [DOI] [PubMed] [Google Scholar]
- 57.Leung N.H., Chu D.K., Shiu E.Y., Chan K.-H., McDevitt J.J., Hau B.J., et al. Respiratory virus shedding in exhaled breath and efficacy of face masks. Nat. Med. 2020;26(5):676–680. doi: 10.1038/s41591-020-0843-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ludwig-Begall L.F., Wielick C., Dams L., Nauwynck H., Demeuldre P.-F., Napp A., et al. The use of germicidal ultraviolet light, vaporized hydrogen peroxide and dry heat to decontaminate face masks and filtering respirators contaminated with a SARS-CoV-2 surrogate virus. J. Hosp. Infect. 2020;106(3):577–584. doi: 10.1016/j.jhin.2020.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Criscuolo E., Diotti R.A., Ferrarese R., Alippi C., Viscardi G., Signorelli C., et al. Fast inactivation of SARS-CoV-2 by UV-C and ozone exposure on different materials. Emerg. Microb. Infect. 2021;10(1):206–210. doi: 10.1080/22221751.2021.1872354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vejerano E.P., Marr L.C. Physico-chemical characteristics of evaporating respiratory fluid droplets. J. R. Soc. Interface. 2018;15(139):20170939. doi: 10.1098/rsif.2017.0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pennell K.G., Aronson A., Blatchley E.R., III Phenotypic persistence and external shielding ultraviolet radiation inactivation kinetic model. J. Appl. Microbiol. 2008;104(4):1192–1202. doi: 10.1111/j.1365-2672.2007.03645.x. [DOI] [PubMed] [Google Scholar]
- 62.Zhang J., Leser G.P., Pekosz A., Lamb R.A. The cytoplasmic tails of the influenza virus spike glycoproteins are required for normal genome packaging. Virology. 2000;269(2):325–334. doi: 10.1006/viro.2000.0228. [DOI] [PubMed] [Google Scholar]
- 63.Mattle M.J., Kohn T. Inactivation and tailing during UV254 disinfection of viruses: contributions of viral aggregation, light shielding within viral aggregates, and recombination. Environ. Sci. Technol. 2012;46(18):10022–10030. doi: 10.1021/es302058v. [DOI] [PubMed] [Google Scholar]
- 64.Blatchley E.R., III, Meeusen A., Aronson A.I., Brewster L. Inactivation of Bacillus spores by ultraviolet or gamma radiation. J. Environ. Eng. 2005;131(9):1245–1252. [Google Scholar]
- 65.Kowalski W., Bahnfleth W., Raguse M., Moeller R. The cluster model of ultraviolet disinfection explains tailing kinetics. J. Appl. Microbiol. 2020;128(4):1003–1014. doi: 10.1111/jam.14527. [DOI] [PubMed] [Google Scholar]
- 66.Mamane-Gravetz H., Linden K.G. Relationship between physiochemical properties, aggregation and uv inactivation of isolated indigenous spores in water. J. Appl. Microbiol. 2005;98(2):351–363. doi: 10.1111/j.1365-2672.2004.02455.x. [DOI] [PubMed] [Google Scholar]
- 67.Azimi Y., Allen D., Farnood R. Kinetics of UV inactivation of wastewater bioflocs. Water Res. 2012;46(12):3827–3836. doi: 10.1016/j.watres.2012.04.019. [DOI] [PubMed] [Google Scholar]
- 68.Virkler K., Lednev I.K. Analysis of body fluids for forensic purposes: from laboratory testing to non-destructive rapid confirmatory identification at a crime scene. Forensic Sci. Int. 2009;188(1–3):1–17. doi: 10.1016/j.forsciint.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 69.Eischeid A.C., Linden K.G. Molecular indications of protein damage in adenoviruses after UV disinfection. Appl. Environ. Microbiol. 2011;77(3):1145–1147. doi: 10.1128/AEM.00403-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hotze E.M., Badireddy A.R., Chellam S., Wiesner M.R. Mechanisms of bacteriophage inactivation via singlet oxygen generation in UV illuminated fullerol suspensions. Environ. Sci. Technol. 2009;43(17):6639–6645. doi: 10.1021/es901110m. [DOI] [PubMed] [Google Scholar]
- 71.Costa L., Faustino M.A., Tomé J.P., Neves M.G., Tomé A.C., Cavaleiro J.A., et al. Involvement of type I and type II mechanisms on the photoinactivation of non-enveloped DNA and RNA bacteriophages. J. Photochem. Photobiol. B Biol. 2013;120:10–16. doi: 10.1016/j.jphotobiol.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 72.Rosado-Lausell S.L., Wang H., Gutiérrez L., Romero-Maraccini O.C., Niu X.-Z., Gin K.Y., et al. Roles of singlet oxygen and triplet excited state of dissolved organic matter formed by different organic matters in bacteriophage MS2 inactivation. Water Res. 2013;47(14):4869–4879. doi: 10.1016/j.watres.2013.05.018. [DOI] [PubMed] [Google Scholar]
- 73.Denison M.R., Graham R.L., Donaldson E.F., Eckerle L.D., Baric R.S. Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 2011;8(2):270–279. doi: 10.4161/rna.8.2.15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat. Rev. Microbiol. 2021;19(3):155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Saito Y., Wada S., Nagata K., Makino H., Boyama S., Miwa H., et al. Efficiency improvement of AlGaN-based deep-ultraviolet light-emitting diodes and their virus inactivation application. Jpn. J. Appl. Phys. 2021;60(8) [Google Scholar]
- 76.Biasin M., Bianco A., Pareschi G., Cavalleri A., Cavatorta C., Fenizia C., et al. UV-C irradiation is highly effective in inactivating SARS-CoV-2 replication. Sci. Rep. 2021;11(1):1–7. doi: 10.1038/s41598-021-85425-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Storm N., McKay L.G., Downs S.N., Johnson R.I., Birru D., de Samber M., et al. Rapid and complete inactivation of SARS-CoV-2 by ultraviolet-C irradiation. Sci. Rep. 2020;10(1):1–5. doi: 10.1038/s41598-020-79600-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.National Center for Atmospheric Research Tropospheric Ultraviolet and Visible (TUV) Radiation Model. https://www2.acom.ucar.edu/modeling/tropospheric-ultraviolet-and-visible-tuv-radiation-model (Accessed 5 April 2021)
- 79.Ulloa S., Bravo C., Ramirez E., Fasce R., Fernandez J. Inactivation of SARS-CoV-2 isolates from lineages B. 1.1. 7 (Alpha), P. 1 (Gamma) and B. 1.110 by heating and UV irradiation. J. Virol. Methods. 2021;114216 doi: 10.1016/j.jviromet.2021.114216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Larason Thomas, Grantham Steven, Zarobila Clarence, Zong Yuqin, Schuit Michael, Holland Brian, Wood Stewart, Krause Melissa, Miller C. Cameron. Traveling tunable laser projector for UV-blue disinfection dose determinations. Appl. Optics. 2022;61(19):5559–5566. doi: 10.1364/AO.460317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
Supplementary material 2