Abstract
The physiological and regulatory effects of overproduction of five cold shock proteins (CSPs) of Lactococcus lactis were studied. CspB, CspD, and CspE could be overproduced at high levels (up to 19% of the total protein), whereas for CspA and CspC limited overproduction (0.3 to 0.5% of the total protein) was obtained. Northern blot analysis revealed low abundance of the cspC transcript, indicating that the stability of cspC mRNA is low. The limited overproduction of CspA is likely to be caused by low stability of CspA since when there was an Arg-Pro mutation at position 58, the level of CspA production increased. Using two-dimensional gel electrophoresis, it was found that upon overproduction of the CSPs several proteins, including a number of cold-induced proteins of L. lactis, were induced. Strikingly, upon overproduction of CspC induction of CspB, putative CspF, and putative CspG was also observed. Overproduction of CspB and overproduction of CspE result in increased survival when L. lactis is frozen (maximum increases, 10- and 5-fold, respectively, after 4 freeze-thaw cycles). It is concluded that in L. lactis CSPs play a regulatory role in the cascade of events that are initiated by cold shock treatment and that they either have a direct protective effect during freezing (e.g., RNA stabilization) or induce other factors involved in the freeze-adaptive response or both.
In a variety of bacteria, cold shock proteins (CSPs) are the major induced proteins upon exposure to cold shock. Different functions, e.g., as transcriptional activators, RNA chaperones, and anti-freeze proteins, have been attributed to CSPs (for reviews see references 11 and 36). CspA of Escherichia coli and CspB of Bacillus subtilis have been shown to bind single-stranded DNA, and E. coli CspA has been shown to act as a transcriptional activator for the hns and gyrA genes encoding proteins involved in DNA supercoiling (2, 17, 22). E. coli CspA and B. subtilis CspBB have very similar five-stranded β-barrel structures with several outward-facing residues important for single-stranded DNA binding. Furthermore, CSPs contain two highly conserved RNA-binding motifs, named RNP-1 and RNP-2, and indeed, mRNA-binding capacity has been demonstrated for E. coli CspA and B. subtilis CspB (13, 15). It has been proposed that members of the CSP family bind to RNA in a cooperative manner and function as RNA chaperones, thereby facilitating the translation process (13). Disruption of B. subtilis cspB results in a freeze-sensitive phenotype (33) and also affects the level of induction of other cold-induced proteins (CIPs) upon temperature downshock in B. subtilis (12). Deletion of three CSPs in B. subtilis was shown to be lethal (13). Since not all members of the CSP family are cold induced, it has been suggested that CSPs play a role in multiple cellular processes, such as chromosomal condensation and/or cell division (36).
The mesophilic lactic acid bacterium Lactococcus lactis is widely used to start industrial food fermentations. A variety of genes involved in the stress response that probably are important for cell survival under stress conditions have been studied for this organism (7, 26). The L. lactis MG1363 chromosome was found to contain two pairs of tandemly located, cold-inducible csp genes (cspA-cspB and cspC-cspD) and a single, constitutively expressed cspE gene. The CSPs encoded by these genes can be divided in two groups based on isoelectric point (pI) and homology. One group consists of CspA and CspC, which have 80% identical residues, and these CSPs have a pI of 9; the other group includes CspB, CspD, and CspE, which have up to 85% identical residues, and these CSPs have a pI of 5 (35). Upon cold shock of L. lactis IL1403 by transfer from 30 to 15°C, 10-fold induction of cspB-directed β-galactosidase activity is observed (4). Similar cold-induced expression has been reported for E. coli cspA, and the data revealed that the transient induction of E. coli CspA occurs at the level of transcription (14) and at the level of mRNA stabilization (1, 8, 9). Furthermore, it has been reported that mRNAs of CIPs are still translated under cold shock conditions because of the presence of a so-called downstream box, which enhances the ability to form the translation initiation complex with nonadapted ribosomes at low temperatures (23). The presence of CSPs in a cell is also determined by the stability of the proteins. The CSPs of B. subtilis undergo very rapid folding and unfolding transitions, and they exhibit low conformational stability in solution. These CSPs are rapidly degraded by proteases in vitro but are protected against proteolysis by binding to RNA (13).
Overproduction of E. coli CspA leads to increases in the levels of three CIPs (16). Moreover, heterologous expression of B. subtilis CspB in E. coli results in a reduction of cellular growth and in production of several proteins that resembles the cold shock response (10). For B. subtilis strains from which csp genes have been deleted compensatory effects of the remaining CSPs have been reported (13), and a similar response might be expected for L. lactis. Moreover, a comparative analysis of the physiological effects of overproduction of different members of a CSP family has never been presented before. For these reasons, we used the nisin-controlled expression system (21) to overproduce the CSPs of L. lactis and, subsequently, to monitor the physiological and regulatory effects of the CSPs. CspB, CspD, and CspE could be overproduced at high levels, whereas for CspA and CspC only low levels of overproduction were detected, probably due to low protein and mRNA stability at 30°C, respectively. Overproduction of specific CSPs resulted in major induction of other CSPs and CIPs, indicating that these proteins have a regulatory function. L. lactis strains overproducing CspB or CspE did not have a shorter lag time upon cold shock but did show enhanced survival after freezing.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The L. lactis strains used in this study were cultured at 30°C without aeration in M17 medium containing 0.5% glucose. L. lactis was transformed by electroporation as described by Wells et al. (32). E. coli M1601 was used as a host for cloning experiments and was grown in tryptone-yeast extract medium with aeration at 37°C (3). Chloramphenicol was used as a selection marker at a concentration of 10 μg ml−1. Growth of L. lactis was monitored by measurement of the optical density at 600 nm (OD600).
DNA techniques and DNA sequence analysis.
PCR amplifications were performed as previously described by Kuipers et al. (20) with 25 cycles (denaturation at 95°C for 30 s, primer annealing at an appropriate temperature for 1 min, and primer extension at 72°C for 2 min). All manipulations with recombinant DNA were carried out by using standard procedures (25). Plasmid DNA and chromosomal DNA of L. lactis were isolated as described previously (31). DNA sequences were determined on both strands with an ALF DNA sequencer (Pharmacia Biotech, Uppsula, Sweden) and were analyzed by using Clone (version 4.0; Clone Manager) and Lasergene (DNASTAR Inc.) software.
Construction of plasmids for overexpression.
cspA, cspB, cspC, and cspE were amplified by PCR with the primers listed in Table 1, which contain either an NcoI site (forward primers) or an HindIII site (all reverse primers except OECspERev, which contains an XhoI site because of the presence of an HindIII site 6 bp downstream of the structural cspE gene). The PCR products obtained were digested with NcoI and HindIII (or XhoI) and cloned in pNZ8032 (6) digested with the same restriction enzymes. In this way, a translational fusion of the nisA promoter to the start codon of the respective csp gene that replaced the gusA gene that was originally present in pNZ8032 was obtained. The constructs were made in such a way that each csp gene contained its own putative terminator. Each of the resulting plasmids (Table 1) was transformed into L. lactis NZ3900, which contains the nisR and nisK genes of the two-component nisin transduction pathway integrated on the chromosome (5). Overproduction of CspD was obtained by using L. lactis NZ3900 containing pNZOECspD as described previously (34). To obtain overproduction, the appropriate strain was grown to an OD600 of 0.3, and then M17W-nisin, which has a higher induction capacity and a lower growth-inhibitory effect than wild-type nisin (18), was added. For optimal overexpression the strains were incubated at 30°C for 90 min. By inserting of an NcoI site 1 bp was mutated in the second codon of cspA, cspB, and cspC. Consequently, the second amino acid was changed from isoleucine to valine, from threonine to alanine, and from asparagine to aspartic acid in CspA, CspB, and CspC, respectively.
TABLE 1.
Plasmids and oligonucleotides used in this study
| Plasmid or oligonucleotide | Relevant property(ies) or sequencea | Reference |
|---|---|---|
| Plasmids | ||
| pNZ8020 | Cmr | 6 |
| pNZ8032 | Cmr | 6 |
| pNZ8048 | Cmr | 21 |
| pNZOECspA | Cmr, overexpression of cspA | This work |
| pNZOECspB | Cmr, overexpression of cspB | This work |
| pNZOECspC | Cmr, overexpression of cspC | This work |
| pNZOECspD | Cmr, overexpression of cspD | 34 |
| pNZOECspE | Cmr, overexpression of cspE | This work |
| pNZOECspA* | Cmr, overexpression of cspA* | This work |
| Oligonucleotides | ||
| OECspAFor | 5′-GCTGC CATGG TAAAT GGAAC AGTAA AATGG-3′ | |
| OECspARev | 5′-GGTCA AGCTT ATAAA CTGTT AGGAA AGCAA-3′ | |
| OECspBFor | 5′-GCTGC CATGG CAAAA GGAAC TGTAA AATGG-3′ | |
| OECspBRev | 5′-CGACA AGCTT GGAAA GCAAC TAATC TTTCC-3′ | |
| OECspCFor | 5′-GCTGC CATGG ATAAA GGAAC AATAA AATTGG-3′ | |
| OECspCRev | 5′-GCTGA AGCTT AGGGA AGTGT GAGTT TCCTC-3′ | |
| OECspEFor | 5′-GCTGC CATGG CACAA GGAAC TGTTA AATGG-3′ | |
| OECspERev | 5′-GCAGC TCGAG TGTTA AGGCT TTCAT TATAA G-3′ | |
| OECspA*Rev | 5′-GCTGG GTACC CAAAA TTTCT ACTTA ATCTA TATTT GAAGC ATANG GCCCT CGACG-3′ |
Underlined sequences indicate the NcoI sites (forward primers) or the HindIII sites (reverse primers, except for OECspERev, which contains an XhoI site).
A mutant CspA in which the Arg residue at position 58 was replaced by Pro was constructed. Using primers OECspAFor and CspA*Rev (containing a KpnI site [Table 1]), the mutated cspA gene was amplified, digested with NcoI and KpnI, and subsequently cloned in the vector pNZ8048 (Table 1) (21). The plasmid generated was sequenced on both strands, which revealed the presence of the desired mutation leading to the Arg-58-Pro substitution. Moreover, a frame shift occurred in penultimate codon 65, which resulted in a C-terminal deletion of Lys and Val, producing a 64-residue R58P-CspAΔK65V66 mutant protein designated CspA*.
Freeze challenge.
To study the effects of CSPs on the survival of L. lactis after freezing, a freeze-thaw challenge experiment with strains overproducing the CSPs at different levels was performed as described previously (34). In short, 1-ml samples of cultures were withdrawn, spun down, and resuspended in fresh medium, the numbers of CFU were determined, and the samples were immediately frozen at −20°C for 24 h. After this freezing period the samples were thawed at 30°C for 4 min, and the numbers of CFU were determined. Next, each sample was frozen again at −20°C, and the cycle was repeated another three times. Freeze-thaw challenges were performed in duplicate, and the results were expressed as the percentage of cells remaining alive relative to the number of cells before the first freeze period (defined as 100%).
Protein extraction and protein analysis using 1D-EF and 2D-EF.
Proteins were extracted by homogenization with an MSK cell homogenizer (B. Braun Biotech International, Melsungen, Germany) and zirconium beads (diameter, 0.1 mm; Biospec Products, Bartlesville, Okla.). Protein analysis was performed by using one-dimensional Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis (1D-EF) as described by Schägger and Von Jagow (27) or by using two-dimensional gel electrophoresis (2D-EF) as described by Wouters et al. (34). Samples containing equal amounts of protein were loaded on the 1D-EF gels (20 μg) and 2D-EF gels (40 μg). A low-molecular-weight marker (protein bands at 16.9, 14.4, 10.7, 8.2, and 6.2 kDa) and a high-molecular-weight marker (protein bands at 94, 68, 43, 29, 18.4, and 14.4 kDa) were used as size markers (both were purchased from Pharmacia Biotech, Uppsula, Sweden). 1D-EF gels were stained with Coomassie brilliant blue, and 2D-EF gels were stained with silver stain. The 1D-EF and 2D-EF gels were analyzed by using the Chromoscan program (Joyce Loebl, Gateshead, England) and GEMINI software (Applied Imaging, Sunderland, England), respectively. The intensity of a specific band and the intensity of a specific spot were calculated as a percentage of the total intensity of the bands visualized in a lane and as a percentage of the total intensity of the spots visualized on a gel, respectively. The protein gels were electrophoresed at least in duplicate, and a representative gel is shown below.
Determination of N termini.
Protein (500 μg) was loaded on the 2D-EF gels for determination of the N termini of specific spots by using conditions identical to those used for the analytical gels. The proteins were blotted on an Immobilon-P transfer membrane (Millipore, Bedford, Mass.) by using a Trans Blot unit as recommended by the manufacturer (Bio-Rad, Richmond, Calif.). The proteins were stained with Coomassie brilliant blue, and fragments of the blot were subjected to the Edman procedure and subsequent analysis with the model 476A protein sequencing system (Applied Biosystems, Foster City, Calif.) at the Sequence Center (University of Utrecht, Utrecht, The Netherlands). The N termini derived were screened for sequence similarities by using the BlastP database.
mRNA analysis.
RNA was isolated and Northern blot analysis was performed as described previously (19). Equal amounts of RNA were separated on 1% agarose gels and blotted on a GeneScreen Plus membrane (Dupont, NEN Research Products, Boston, Mass.). The blots were hybridized with [γ-32P]ATP-labelled probes specific for the individual csp genes (35).
RESULTS
Overproduction of CSPs in L. lactis.
To elucidate the role of lactococcal CSPs in cold adaptation, L. lactis strains overproducing these CSPs were constructed (Table 1). Considerable overproduction was achieved for CspB, CspD, and CspE, which were overproduced so that they accounted for 11, 13, and 19% of the total protein, respectively. Strikingly, for CspA and CspC much lower levels of overproduction were obtained (approximately 0.5 and 0.3% of the total protein, respectively) even when a fourfold-higher concentration of nisin was used (Fig. 1A). Using the nisin-controlled expression system, stepwise overproduction of the CSPs could be achieved by adding increasing concentrations of nisin (see below). Upon addition of increasing concentrations of nisin, the growth rate of L. lactis NZ3900 harboring the overexpression constructs decreased significantly (Fig. 1B [only the data for CspD are shown; identical effects were observed for all CSPs), which was possibly explained by occupation of the transcriptional and translational machinery. The growth of the control strain was also reduced (Fig. 1C), probably for the same reasons; however, the extent of growth reduction was lower than those of the CSP-overproducing strains (Fig. 1B). The growth rate of NZ3900 (plasmid free) was not reduced upon incubation with the same concentrations of nisin (Fig. 1D), eliminating the possibility of an antimicrobial effect of nisin at the concentrations used.
FIG. 1.
Protein analysis of cell extracts of L. lactis NZ3900 overproducing CSPs and growth of L. lactis NZ3900 overproducing-sodium CSPs. (A) 1D-EF gels containing cell extracts of L. lactis NZ3900 containing pNZOECspA (lane 2), pNZOECspB (lane 3), pNZOECspC (lane 4), pNZOECspD (lane 5), and pNZOECspE (lane 6) induced with 2.0 ng of nisin/ml for CspA and CspC or with 0.5 ng of nisin/ml for CspB, CspD, and CspE for 90 min at 30°C. Lane 1 contained a low-molecular-weight marker (protein bands at 16.9, 14.4, 10.7, 8.2, and 6.2 kDa). The arrow indicates the position of the CSPs. (B to D) Growth of L. lactis NZ3900 harboring pNZOECspD (B), L. lactis NZ3900 harboring pNZ8020 (C), and L. lactis NZ3900 (D) upon induction with nisin. Growth was measured at 30°C by determining the OD600 without induction (○) or with induction with 0.1 ng of nisin/ml (▵), 0.2 ng of nisin/ml (□), 0.5 ng of nisin/ml (⧫), 1.0 ng of nisin/ml (×), or 2.0 ng of nisin/ml (●). The arrows indicate when nisin was added, and the bulleted arrows indicate when protein was extracted.
Limited overproduction of CspA and CspC is explained by low protein and mRNA stability.
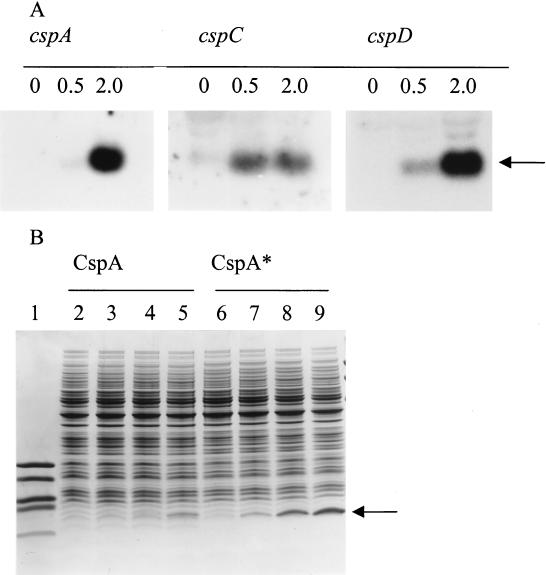
To further investigate the differences in the levels of overproduction obtained for the two groups of lactococcal CSPs, the mRNA levels for lactococcal cspA, cspC, and cspD (positive control) were monitored upon overexpression (Fig. 2A). For cspA and cspD a major increase in the mRNA level was observed upon induction with 0.5 and 2.0 ng of nisin/ml. Thus, despite the similar high mRNA levels for cspA and cspD, a concomitantly high protein level was obtained only for CspD. For cspC a low level of mRNA induction was observed compared to the levels of mRNA induction for cspA and cspD, which provides an explanation for the low amount of CspC obtained. Since the transcription signals for all csp overexpression constructs are identical, the low mRNA level is most likely caused by the low stability of the cspC transcript at 30°C.
FIG. 2.
Analysis of L. lactis NZ3900 overproduction of CspA and CspC on the mRNA and protein levels. (A) Northern analysis of cspA (left), cspC (center), and cspD (right) in L. lactis NZ3900 containing pNZOECspA, L. lactis NZ3900 containing pNZOECspC, and L. lactis NZ3900 containing pNZOECspD, respectively, upon induction with 0, 0.5, and 2.0 ng of nisin/ml. Equal amounts of total RNA were loaded on the gel, and the arrow indicates the position of csp transcripts. The film used for detection of the cspC transcript was exposed four times longer to the blot than the films used for detection of cspA and cspD. (B) 1D-EF of cell extracts of L. lactis NZ3900 containing pNZOECspA or pNZOECspA* without induction (lanes 2 and 6) and with induction with 0.2 ng of nisin/ml (lanes 3 and 7), 0.5 ng of nisin/ml (lanes 4 and 8), and 2.0 ng of nisin/ml (lanes 5 and 9). Lane 1 contained a low-molecular-weight marker (16.9, 14.4, 10.7, 8.2, and 6.2 kDa). Equal amounts of protein were loaded on the gel, and the arrow indicates the position of CSPs.
CspA and CspC contain an Arg residue at position 58, whereas CspB, CspD, and CspE contain a Pro residue at this position. The absence of the Pro residue might result in lower conformational stability of CspA and CspC because Pro residues are known to reduce the entropy of unfolded proteins (28). The stability of L. lactis CspA was investigated by constructing a mutant CspA (CspA*) in which the Arg residue at position 58 was replaced by Pro. A much higher level of production of CspA* (approximately 20-fold upon induction with 1.0 ng of nisin/ml) was detected compared to the level of production of native CspA, resulting in CspA* that accounted for 9% of the total protein (Fig. 2B). It was calculated that the Arg-Pro substitution results in a change in the calculated pI of CspA from 8.5 to 6.5. A change of Ile to Val in the second codon did not affect the CspA pI, but C-terminal deletion of Lys and Val further reduced the calculated pI of CspA* to 4.8 (see below and Materials and Methods). However, it should be noted that the calculations regarding the CspA* level were based on the 1D-EF gels, in which the more intense band could represent more than one CSP (see below) (Table 2 and Fig. 3).
TABLE 2.
CSP production levels (percentages of total protein visualized on a gel) upon overproduction of the CSPs of L. lactisa
| Plasmid | Production level (% of total protein)
|
||||||
|---|---|---|---|---|---|---|---|
| CspA | CspB | CspC | CspD | CspE | CspF | CspG | |
| pNZOECspA | 0.4 | —b | 0.1 | 0.1 | 0.1 | — | — |
| pNZOECspC | — | 0.1 | 0.4 | 0.1 | 0.1 | 0.1 | 0.1 |
| pNZ8020 (2.0 ng of nisin/ml) | — | — | 0.2 | 0.1 | 0.3 | — | — |
| pNZOECspA* | 0.9 | — | 0.2 | — | 0.4 | — | — |
| pNZOECspB | — | 0.8 | — | — | 0.1 | — | — |
| pNZOECspD | — | — | — | 1.0 | 0.1 | — | — |
| pNZOECspE | — | — | — | — | 0.6 | — | — |
| pNZ8020 (0.1 ng of nisin/ml) | — | — | — | — | 0.2 | — | — |
Values are based on the 2D-EF gels shown in Fig. 3.
—, not detectable.
FIG. 3.
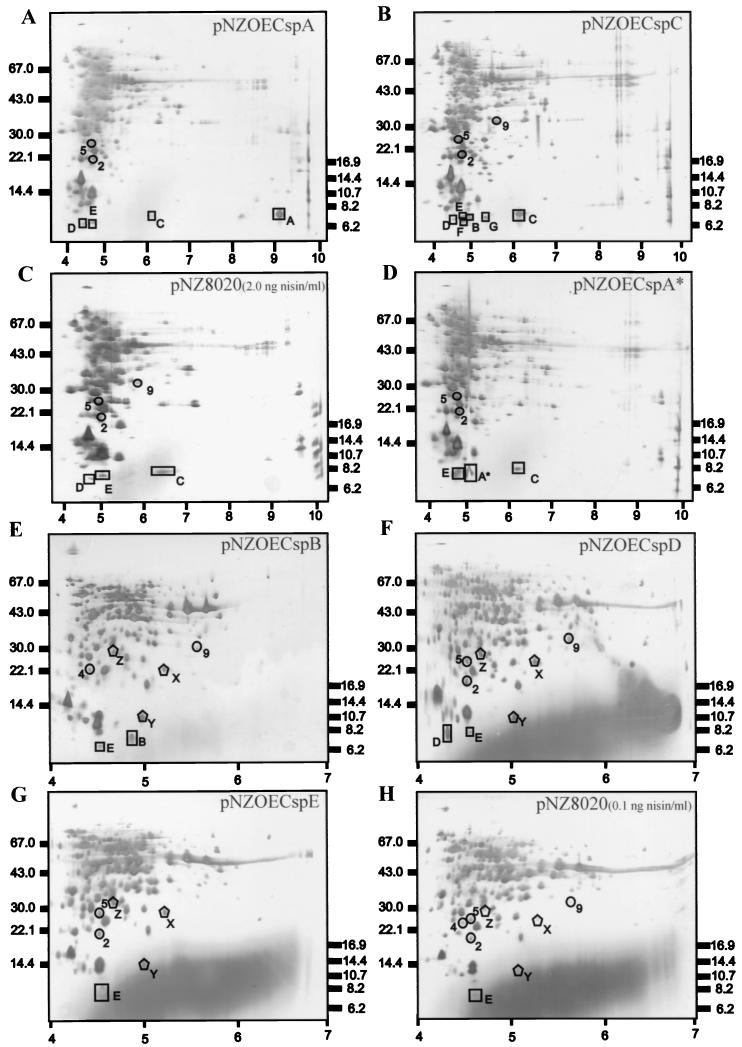
2D-EF analysis of cell extracts of L. lactis NZ3900 overproducing the lactococcal CSPs. Cell extracts were isolated from L. lactis NZ3900 containing pNZOECspA (A), pNZOECspC (B), pNZ8020 (C), or pNZOECspA* (D) upon induction with 2.0 ng of nisin/ml and were separated at pI 3 to 10. Cell extracts were also isolated from L. lactis NZ3900 containing pNZOECspB (E), pNZOECspD (F), pNZOECspE (G), or pNZ8020 (H) induced with 0.1 ng of nisin/ml and were separated at pI 4 to 7. Equal amounts of protein were loaded on the gel and visualized by silver staining. The positions of molecular weight markers are indicated on the left (high-molecular-weight marker) or on the right (low-molecular-weight marker). Spots corresponding to CSPs of L. lactis are enclosed in squares or rectangles, and the letters indicate the CSP families (F = putative CspF, G = putative CspG); the calculated pIs are 8.5, 5.0, 9.1, 4.5, and 4.6 for CspA, CspB, CspC, CspD, and CspE, respectively. Spots induced by overproduction of the CSPs are enclosed in circles and numbered if they belong to previously identified CIPs (Wouters et al., submitted) or are enclosed in pentagons and designated with letters (X, Y, Z) if they are induced during specific overproduction of CSPs. See text for details.
Regulatory role of CSPs in cold shock adaptation.
To study the effect of overproduction of CSPs on protein synthesis patterns in L. lactis, 2D-EF was performed. The overproduced CSPs migrated approximately at their calculated pIs except for CspC, as confirmed by N-terminal sequencing (Fig. 3). Overproduction of CspC (Fig. 3B and Table 2) in L. lactis results in induction of CspB, protein F (putative CspF), and previously unidentified protein G (putative CspG) in the 7-kDa region. Upon induction of control cells (L. lactis harboring pNZ8020) (Fig. 3C and Table 2) with 2.0 ng of nisin/ml, increased levels of production of CspC, CspD, and CspE were observed compared to the levels in noninduced cells (data not shown), which is possibly explained by the reduction in growth (Fig. 1C) and the resulting stress conditions. Overproduction of CspA (Fig. 3A and Table 2) did not result in induction of CSPs, whereas overproduction of CspA* (Fig. 3D and Table 2) resulted in a slight increase in the CspE level compared to that in control cells. Furthermore, overproduction of CspB, CspD, or CspE did not affect the level of any of the other CSPs (Fig. 3E through H and Table 2; data not shown for CspA, which was visualized only on gels at pI 3 to 10). Next, upon overproduction of CspA, CspB, CspC, and CspD a slight decrease in the level of CspE was observed, which might indicate possible downregulation of cspE expression by the respective CSPs. This observation is in agreement with data on the disruption of CspA and CspB, which results in derepression of cspE expression at a low temperature (J. A. Wouters, H. Frenkiel, W. M. De Vos, O. P. Kuipers, and T. Abee, submitted for publication).
Since it has been reported that E. coli CspA acts as a transcriptional regulator (2, 17, 22), we analyzed the effects of overproduction of the CSPs of L. lactis on the induction of proteins outside the 7-kDa region by using 2D-EF, and indeed, several induced proteins were observed (Fig. 3). Several CIPs of L. lactis (34) could be identified among the induced proteins. Overproduction of CspA results in induction of CIP2 (hypothetical 50S ribosomal protein L9; ANISKASAHEDTLENFTIE; 68% homology) and CIP5 (N-terminal block), while upon overproduction of CspC CIP9 (N terminus not determined) and several other proteins were also induced. Overproduction of CspA* results in induction of both CIP2 and CIP5. Furthermore, upon overproduction of CspB the levels of CIP4 (N terminus not determined) and CIP9 increased by factors of 2.5 and 3, respectively (compared to values observed in the control gel). Overproduction of CspD resulted in 2.5-, 2-, and 4-fold-higher levels of CIP2, CIP5, and CIP9, respectively, while overproduction of CspE resulted in 2-fold-higher levels of both CIP2 and CIP5. The changes in the levels of expression of these CIPs indicate that CSPs might play a regulatory role in induction of specific proteins involved in the cold adaptation process. On the other hand, increased production is also observed for three non-cold-induced proteins (proteins X, Y [homologous to CelA of B. subtilis; NDKVIALASAAGMSTNLLV; 63% homology], and Z) upon overproduction of CspB, CspD, or CspE. Production of these proteins did not increase upon overproduction of CspA or CspC compared to production in control cells (Fig. 3).
Effect of overproduction of CSPs on cold adaptation and survival of L. lactis after freezing.
The effect of overproduction of CspB or CspD, the major cold-induced CSPs of L. lactis (34, 35), on adaptation to cold shock was tested. These CSPs were overproduced by using relatively low nisin concentrations (0.05 and 0.1 ng of nisin/ml), yielding levels of expression comparable to those observed under cold shock conditions, after which the cultures were exposed to 10°C (without nisin). For all conditions identical adaptation times of 6 to 7 h were observed (data not shown), indicating no beneficial effect on cold adaptation of elevated CspB or CspD expression prior to cold shock. Also, overproduction of CspA and CspC did not result in differences in cold adaptation compared to that of control cells (NZ3900 harboring pNZ8020).
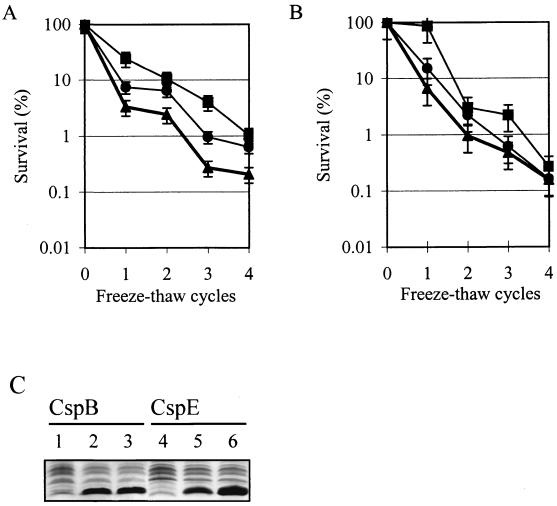
Previously, it has been observed that overproduction of CspD results in at most a 10-fold increase in survival of L. lactis cells after freezing (34). In this study, the impact of overproduction of the other CSPs of L. lactis on survival after freezing was monitored by exposing cultures overproducing one of the CSPs to freeze-thaw challenge. Overproduction of CspB (induced with 0.2 or 0.5 ng of nisin/ml) resulted in an approximately 5- to 10-fold increase in survival compared to that of noninduced cells after four repetitive freeze-thaw cycles (Fig. 4). It should be noted that under these conditions no linear relationship between CspB levels and survival after freezing was observed, which may be explained by an indirect effect of CspB on survival after freezing. Following overproduction of CspE a smaller positive effect (at most a fivefold increase) on survival after freezing was observed (Fig. 4), whereas for control cells (L. lactis harboring pNZ8020) no effect of higher concentrations of nisin was noted (data not shown). For L. lactis cells overproducing CspA, CspC, or CspA* no additional freeze-protective effect was observed compared to control cells (data not shown).
FIG. 4.
Survival of L. lactis NZ3900 after freezing upon overproduction of CspB (A) and CspE (B). Survival is expressed as the percentage of surviving cells compared to the number of cells prior to freezing for L. lactis NZ3900 harboring pNZOECspB or pNZOECspE without induction (▴) or with induction with 0.2 ng of nisin/ml (●) or 0.5 ng of nisin/ml (■). (C) 1D-EF of cell extracts of L. lactis NZ3900 containing pNZOECspB and L. lactis NZ3900 containing pNZOECspE without induction (lanes 1 and 4) and with induction with 0.2 ng of nisin/ml (lanes 2 and 5) and 0.5 ng of nisin/ml (lanes 3 and 6), respectively. Equal amounts of protein were loaded on the gel.
DISCUSSION
In this report, we describe the effects of overproduction of five different CSPs of the lactic acid bacterium L. lactis. The CSPs of L. lactis can be divided into two groups, one consisting of CspA and CspC and the other consisting of CspB, CspD, and CspE, based on amino acid composition and pI (35). Overproduction of CSPs of the latter group resulted in levels of overproduction much higher than those obtained for CspA and CspC (11 to 19% versus 0.3 to 0.5% of the total protein). Most of the CSPs migrated on the 2D-EF gels at their calculated pIs; the only exception was CspC, which had a calculated pI of 8.4 (instead of 9.1) in the overproduction construct because of the Asp-Asn substitution in the second codon. However, the CspC protein was found to migrate at a pI of approximately 6, as confirmed by N-terminal sequencing. No formylation of the N terminus, such as that found for B. subtilis CspB (12), was observed, and it is speculated that the pI shift is caused by posttranslational modifications. Furthermore, two new proteins in the 7-kDa region were identified for L. lactis: one designated putative CspF that was also induced upon cold shock as described previously (34) and the other designated putative CspG that has a pI of 5.5. These putative CSPs might be encoded by genes located on an uncharacterized 3.5-kb HindIII-hybridizing fragment on the L. lactis chromosome (35). The growth rate of L. lactis is reduced upon overproduction of CspB, CspD, and CspE at 30°C. Also, a reduced growth rate was observed for control cells (L. lactis NZ3900 harboring pNZ8020) upon incubation with nisin, but overproduction of CSPs further reduced the growth rate and this might be explained by energy consumption and more intensive occupation of the transcription and translation machinery. Similar growth-inhibitory effects were observed for heterologous expression of B. subtilis CspB in E. coli at 37°C (10).
The observation that CspB, CspD, and CspE of L. lactis can be overproduced to obtain large quantities at 30°C is remarkable. Artificial overexpression of the E. coli cspA gene was very low at 37°C due to the low mRNA stability (9). The low level of overexpression obtained for the lactococcal cspC gene is most likely explained by the low stability of the transcript at 30°C, as shown by Northern blotting. Since high mRNA induction for cspA is observed, the low level of CspA overproduction should be explained by other factors. CspC of B. subtilis contains an Ala residue at position 58 whereas CspB and CspD of B. subtilis contain a Pro residue at this position, and indeed, B. subtilis CspB and CspD were far more stable than B. subtilis CspC (28). At 30°C CspA* can be overproduced to obtain much larger quantities than the quantities of CspA, which is most likely explained by the high stability of the protein as a consequence of the reduced entropy. In addition to the specific Arg-Pro mutation, the decrease in pI may also contribute to CspA* stability. Recently, it was shown that the stabilities of the CSPs of B. subtilis increased significantly in the presence of a nucleic acid ligand. It has been suggested that the stabilities of these CSPs in vivo are mediated by binding to mRNA (28), which might be largely dependent on the overall charge of the protein. Schröder et al. (30) suggested that CSPs may act as RNA chaperones since they possess a positively charged RNA-binding epitope that is backed by a negatively charged surface that would prevent approach of RNA by charge repulsion. Mutation of the surface-exposed Phe residues at positions 15, 17, and 27, which are important for nucleic acid binding, has been found to result in decreased stability of the protein, probably due to a decrease in the nucleic acid-binding capacity that makes the protein more susceptible to protease action (28, 29). This would provide an alternative explanation for the increased stability of CspA* and could also explain why the CSPs of L. lactis that have a low pI can be overproduced to obtain large quantities. Overproduction was also examined at 10°C, and still no overproduction of CspA and CspC was observed, indicating that mRNA and/or proteins were not sufficiently stabilized to yield detectable protein levels (data not shown). For L. lactis NZ3900 harboring pNZOECspD moderate CspD overproduction was observed at 10°C compared to that of control cells (data not shown), which shows that the nisin-controlled expression system is functional at a low temperature but with reduced efficiency.
Upon overproduction of CspC the levels of CspB, putative CspF, and putative CspG increased, whereas overproduction of CspA, CspB, CspD, or CspE did not affect the expression of other CSPs. CspC might directly stimulate the expression of the other CSPs by transcriptional activation, as has been reported for genes regulated by E. coli CspA for which Y-box motifs (ATTGG or CCAAT) have been shown to be important (2, 17, 22). Several of these elements are observed in the upstream regions of the lactococcal csp genes (34). The observation that all lactococcal CSPs induce expression of certain proteins, including several CIPs, indicates a regulatory function for this group of proteins. The production of CIP2, CIP4, CIP5, and CIP9 seems to be regulated by several CSPs, indicating that there is overlap in regulatory pathways. The N terminus of CIP2 was identified and shows homology to 50S ribosomal protein L9 of B. subtilis. Cold-induced ribosomal proteins have been reported for both E. coli (S1, S6, L7/L12) (16) and B. subtilis (S6, L7/L12) (12), and it has been suggested that they are essential for correct assembly of rRNA at low temperatures. For overproduction of CspB, CspD, and CspE induction is also observed for non-cold-induced proteins X, Y (CelA), and Z. The observed induction of protein Y, homologous to a cellobiose-specific enzyme II subunit of the phosphotransferase system of B. subtilis (24), might be an indication that CSPs play a role in processes other than cold adaptation (e.g., sugar metabolism).
Overproduction of CSPs did not stimulate adaptation of L. lactis to cold shock conditions. However, similar to overproduction of CspD (34), overproduction of CspB and CspE resulted in 10- and 5-fold-greater survival after freezing compared to that of control cells, respectively. For B. subtilis disruption of B. subtilis cspB resulted in decreased survival after freezing (a 14-fold decrease compared to survival of wild-type cells), indicating an essential role for this gene in protection against freezing (33). CspB, CspD, and CspE may enhance the survival of L. lactis after freezing either directly by protecting RNA or DNA by the moderate, nonspecific binding activities mentioned for CSPs (e.g., RNA stabilization) or indirectly by inducing other factors that provide cryoprotection. The proteins that are induced upon overproduction of the lactococcal CSPs might be involved in cryoprotection of L. lactis.
In this work overproduction of specific CSPs of L. lactis and several physiological effects were studied. Overproduction of different quantities could be obtained for all CSPs depending on mRNA and protein stability. 2D-EF revealed that overproduction of CSPs at 30°C resulted in induction of a specific group of proteins, including CIPs. It is concluded that CSPs of L. lactis play a regulatory role in the cold shock response and that they control the production of both CSPs and CIPs.
REFERENCES
- 1.Brandi A, Pietroni P, Gualerzi C O, Pon C L. Post-transcriptional regulation of CspA expression in Escherichia coli. Mol Microbiol. 1996;19:231–240. doi: 10.1046/j.1365-2958.1996.362897.x. [DOI] [PubMed] [Google Scholar]
- 2.Brandi A, Pon C L, Gualerzi C O. Interaction of the main cold shock protein CS7.4 (CspA) of Escherichia coli with the promoter region of hns. Biochimie. 1994;76:1090–1098. doi: 10.1016/0300-9084(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 3.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 4.Chapot-Chartier M P, Schouler C, Lepeuple A S, Gripon J C, Chopin M C. Characterization of cspB, a cold-shock-inducible gene from Lactococcus lactis, and evidence for a family of genes homologous to the Escherichia coli cspA major cold shock gene. J Bacteriol. 1997;179:5589–5593. doi: 10.1128/jb.179.17.5589-5593.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Ruyter P G G A, Kuipers O P, Beerthuyzen M M, Van Alen-Boerrigter I J, De Vos W M. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J Bacteriol. 1996;178:3434–3439. doi: 10.1128/jb.178.12.3434-3439.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Ruyter P G G A, Kuipers O P, De Vos W M. Controlled gene expression system for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol. 1996;62:3662–3667. doi: 10.1128/aem.62.10.3662-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duwat P, Ehrlich S D, Gruss A. Effects of metabolic flux on stress response pathways in Lactococcus lactis. Mol Microbiol. 1999;31:845–858. doi: 10.1046/j.1365-2958.1999.01222.x. [DOI] [PubMed] [Google Scholar]
- 8.Fang L, Jiang W, Bae W, Inouye M. Promoter-independent cold-shock induction of cspA and its derepression at 37°C by mRNA stabilization. Mol Microbiol. 1997;23:355–364. doi: 10.1046/j.1365-2958.1997.2351592.x. [DOI] [PubMed] [Google Scholar]
- 9.Goldenberg D, Azar I, Oppenheim A. Differential stability of the cspA gene in the cold-shock response of Escherichia coli. Mol Microbiol. 1996;19:241–248. doi: 10.1046/j.1365-2958.1996.363898.x. [DOI] [PubMed] [Google Scholar]
- 10.Graumann P, Marahiel M A. Effects of heterologous expression of CspB, the major cold shock protein of Bacillus subtilis, on protein synthesis in E. coli. Mol Gen Genet. 1997;253:745–752. doi: 10.1007/s004380050379. [DOI] [PubMed] [Google Scholar]
- 11.Graumann P, Marahiel M A. A superfamily of proteins that contain the cold-shock domain. Trends Biochem Sci. 1998;23:286–290. doi: 10.1016/s0968-0004(98)01255-9. [DOI] [PubMed] [Google Scholar]
- 12.Graumann P, Schröder K, Schmid R, Marahiel M A. Cold shock stress-induced proteins in Bacillus subtilis. J Bacteriol. 1996;178:4611–4619. doi: 10.1128/jb.178.15.4611-4619.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graumann P, Wendrich T M, Weber M H W, Schröder K, Marahiel M A. A family of cold shock proteins in Bacillus subtilis is essential for cellular growth and for efficient protein synthesis at optimal and low temperatures. Mol Microbiol. 1997;25:741–756. doi: 10.1046/j.1365-2958.1997.5121878.x. [DOI] [PubMed] [Google Scholar]
- 14.Jiang W, Fang L, Inouye M. Chloramphenicol induces the transcription of the major cold shock gene of Escherichia coli, cspA. J Bacteriol. 1993;175:5824–5828. doi: 10.1128/jb.175.18.5824-5828.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272:196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- 16.Jones P G, Cashel M, Glaser G, Neidhardt F C. Function of a relaxed-like state following temperature downshifts in Escherichia coli. J Bacteriol. 1992;174:3903–3914. doi: 10.1128/jb.174.12.3903-3914.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones P G, Krah R, Tafuri S R, Wolffe A P. DNA gyrase, CS7.4, and the cold shock response in Escherichia coli. J Bacteriol. 1992;174:5798–5802. doi: 10.1128/jb.174.18.5798-5802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuipers O P, Beerthuyzen M M, De Ruyter P G G A, Luesink E J, De Vos W M. Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem. 1995;270:27299–27304. doi: 10.1074/jbc.270.45.27299. [DOI] [PubMed] [Google Scholar]
- 19.Kuipers O P, Beerthuyzen M M, Siezen R J, De Vos W M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis: requirement of the nisA and nisI genes for development of immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 20.Kuipers O P, Boot H J, De Vos W M. Improved site-directed mutagenesis method using PCR. Nucleic Acids Res. 1991;19:4558. doi: 10.1093/nar/19.16.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuipers O P, De Ruyter P G G A, Kleerebezem M, De Vos W M. Quorum sensing-controlled gene expression in lactic acid bacteria. J Biotechnol. 1998;64:15–21. [Google Scholar]
- 22.LaTeana A, Brandi A, Falconi M, Spurio R, Pon C L, Gualerzi C O. Identification of a cold shock transcriptional enhancer of the Escherichia coli major cold shock gene encoding nucleoid protein H-NS. Proc Natl Acad Sci USA. 1991;88:10907–10911. doi: 10.1073/pnas.88.23.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitta M, Fang L, Inouye M. Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding for its cold shock induction. Mol Microbiol. 1997;26:321–335. doi: 10.1046/j.1365-2958.1997.5771943.x. [DOI] [PubMed] [Google Scholar]
- 24.Sadaie Y, Yata K, Fujita M, Sagai H, Itaya M, Kasahara Y, Ogasawara N. Nucleotide sequence and analysis of the phoB-rrnE-groESL region of the Bacillus subtilis chromosome. Microbiology. 1997;143:1861–1866. doi: 10.1099/00221287-143-6-1861. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 26.Sanders J-W, Venema G, Kok J. Environmental stress responses in Lactococcus lactis. FEMS Microbiol Rev. 1999;23:483–501. [Google Scholar]
- 27.Schägger H, Von Jagow G. Tricine-sodium dodecyl sulfate polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 28.Schindler T, Graumann P L, Perl D, Ma S, Schmid F X, Marahiel M A. The family of cold shock proteins of Bacillus subtilis. Stability and dynamics in vitro and in vivo. J Biochem. 1999;274:3407–3413. doi: 10.1074/jbc.274.6.3407. [DOI] [PubMed] [Google Scholar]
- 29.Schindler T, Perl D, Graumann P, Sieber V, Marahiel M A, Schmidt F X. Surface-exposed phenylalanines in the RNP1/RNP2 motif stabilize the cold-shock protein CspB from B. subtilis. Proteins Struct Funct Genet. 1998;30:401–406. doi: 10.1002/(sici)1097-0134(19980301)30:4<401::aid-prot7>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 30.Schröder K, Graumann P, Schnuchel A, Holak T A, Marahiel M A. Mutational analysis of the putative nucleic-acid binding surface of the cold-shock domain, CspB, revealed an essential role of aromatic and basic residues in binding of single stranded DNA containing the Y-box motif. Mol Microbiol. 1995;16:699–708. doi: 10.1111/j.1365-2958.1995.tb02431.x. [DOI] [PubMed] [Google Scholar]
- 31.Vos P, Van Asseldonk M, Van Jeveren F, Siezen R J, Simons G, De Vos W M. A maturation protein is essential for the production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J Bacteriol. 1989;171:2795–2802. doi: 10.1128/jb.171.5.2795-2802.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wells J M, Wilson P W, Le Page R W F. Improved cloning vectors and transformation procedure for Lactococcus lactis. J Appl Bacteriol. 1993;74:629–636. doi: 10.1111/j.1365-2672.1993.tb05195.x. [DOI] [PubMed] [Google Scholar]
- 33.Willimsky G, Bang H, Fischer G, Marahiel M A. Characterization of cspB, a Bacillus subtilis inducible cold shock gene affecting cell viability at low temperatures. J Bacteriol. 1992;174:6326–6335. doi: 10.1128/jb.174.20.6326-6335.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wouters J A, Jeynov B, Rombouts F M, De Vos W M, Kuipers O P, Abee T. Analysis of the role of 7 kDa cold-shock proteins of Lactococcus lactis MG1363 in cryoprotection. Microbiology. 1999;145:3185–3194. doi: 10.1099/00221287-145-11-3185. [DOI] [PubMed] [Google Scholar]
- 35.Wouters J A, Sanders J-W, Kok J, De Vos W M, Kuipers O P, Abee T. Clustered organization and transcriptional analysis of a family of five csp genes of Lactococcus lactis MG1363. Microbiology. 1998;144:2885–2893. doi: 10.1099/00221287-144-10-2885. [DOI] [PubMed] [Google Scholar]
- 36.Yamanaka K, Fang L, Inouye M. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol Microbiol. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]