Abstract
Coronavirus disease 2019 (COVID-19) initiated global health care challenges such as the necessity for new diagnostic tests. Diagnosis by real-time PCR remains the gold-standard method, yet economical and technical issues prohibit its use in points of care (POC) or for repetitive tests in populations. A lot of effort has been exerted in developing, using, and validating antigen-based tests (ATs). Since individual studies focus on few methodological aspects of ATs, a comparison of different tests is needed. Herein, we perform a systematic review and meta-analysis of data from articles in PubMed, medRxiv and bioRxiv. The bivariate method for meta-analysis of diagnostic tests pooling sensitivities and specificities was used. Most of the AT types for SARS-CoV-2 were lateral flow immunoassays (LFIA), fluorescence immunoassays (FIA), and chemiluminescence enzyme immunoassays (CLEIA). We identified 235 articles containing data from 220,049 individuals. All ATs using nasopharyngeal samples show better performance than those with throat saliva (72% compared to 40%). Moreover, the rapid methods LFIA and FIA show about 10% lower sensitivity compared to the laboratory-based CLEIA method (72% compared to 82%). In addition, rapid ATs show higher sensitivity in symptomatic patients compared to asymptomatic patients, suggesting that viral load is a crucial parameter for ATs performed in POCs. Finally, all methods perform with very high specificity, reaching around 99%. LFIA tests, though with moderate sensitivity, appear as the most attractive method for use in POCs and for performing seroprevalence studies.
Keywords: COVID-19, SARS-CoV-2, antigen test, meta-analysis, diagnostic performance, sensitivity, specificity
1. Introduction
COVID-19, caused by SARS-CoV-2, remains a global public health threat that has already claimed more than six million lives (https://covid19.who.int, accessed on 15 May 2022), with modeling estimates suggesting that this figure is probably much higher [1,2]. Vaccines, however, seem to perform well, especially after the administration of booster doses, providing moderate but short-lived protection from SARS-CoV-2 infection but significantly reducing COVID-19-related morbidity and mortality [3,4,5,6,7,8,9]. Non-pharmaceutical interventions (test-trace-isolate, hand washing, physical distancing, travel restrictions, school closures, closures of businesses, and stay-at-home orders) have also proved their effectiveness in containing the spread of the pandemic virus before the advent of vaccines [10,11,12]. Some of these measures will still be needed in our gradual efforts to return to normalcy. Testing in particular is essential to diagnosis, but also to developing and sustaining a reliable surveillance system for the years to come [13,14].
Real-time reverse transcription polymerase-chain-reaction (rt-PCR) test is the benchmark method for the clinical diagnosis of COVID-19 [15,16,17]. As such, it is designed for use with symptomatic people and has high analytical sensitivity. However, rt-PCR can detect viral genetic material even when the virus does not grow in a cell culture, suggesting that the presence of viral nucleic acid may not always reflect contagiousness. Moreover, it requires advanced laboratory equipment, specialist human resources, and significant infrastructure, often in a centralized setting, which increase costs, though these are less relevant for a single patient who needs a definite answer when he/she is tested. In summary, molecular diagnostic testing (nucleic acid amplification tests) becomes a less appealing method for frequent population screening to detect asymptomatic people with SARS-CoV-2 infection and as a tool to rapidly identify, contact-trace, and isolate highly infectious individuals. Antigen detection tests (AT) are immunoassays performed on pharyngeal, nasopharyngeal, nasal or throat swab specimens that detect the presence of a specific viral protein, which indicates viral activity [18,19]. The currently authorized AT include laboratory-based but also point-of-care (POC tests) and self-tests. AT are less expensive than rt-PCR, and most of them give results in approximately 15–30 min. In terms of weaknesses, AT are generally less sensitive than nucleic acid amplification tests. There are three main categories of AT used for the detection of SARS-CoV-2 infection. Lateral flow immunoassays (LFIA) are small, chromatography-based platforms used in POC. The sample is placed on the slot of the test plastic vector and an optical result (color) is obtained within 5–15 min [20]. Fluorescent immunoassays (FIA) are also small, handy, immunochromatography-based tests. The result is read by a fluorescence immunoassay analyzer within 5–20 min and can be performed in POC [21]. The chemiluminescence enzyme immunoassay (CLEIA) is a quick (about 30 min) and sensitive method to detect SARS-CoV-2 antigens. When the sample antigen reacts with the chemiluminescence substrate (antibody), the reaction product emits a photon of light instead of color development, which is read by an automated chemiluminescence analyzer [20].
Healthcare professionals, laboratory staff, and public health experts should comprehend the performance characteristics of AT, identify determinants of the accuracy of AT, and understand the differences among the three approaches to COVID-19-related testing (diagnostic, screening, and surveillance testing). In this respect, the aim of this meta-analysis is to comprehensively search the literature, to identify all relevant studies, to synthesize individual study estimates, and to determine the overall sensitivity and specificity of antigen-based methods for the detection of SARS-CoV-2, in comparison to quantitative rt-PCR (qPCR), for different types of clinical samples, and among both asymptomatic and symptomatic individuals.
2. Material and Methods
2.1. Literature Search Strategy
We conducted this systematic review and meta-analysis following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [22] along with the advice for best practices [23]. We performed the literature search in Pubmed (https://pubmed.ncbi.nlm.nih.gov accessed on 15 May 2022), medRxiv (https://www.medrxiv.org accessed on 15 May 2022) and BioRxiv (https://www.biorxiv.org, accessed on 15 May 2022) up until 4 July 2021. The search terms were “(SARS-CoV-2 OR “Coronavirus disease 2019” OR COVID-19) AND antigen”. References from the selected studies were also scrutinized. Four independent researchers (AT, MP, HM, GB) evaluated search results; potential disagreements were resolved by discussion with GB and PB and consensus. Articles of all languages were considered to avoid gray literature publication bias [24].
2.2. Study Selection Criteria
Eligible criteria for inclusion in the meta-analysis were: (a) diagnosis of SARS-CoV-2 infection based on detection/quantitation of the viral genome by qPCR, according to World Health Organization (WHO)-, Centers for Disease Control (CDC)-, and European Centre for Disease Prevention and Control (ECDC)-approved methods [16,25,26,27]; (b) detection or measurement of nucleocapsid (N) or spike (S) proteins of SARS-CoV-2 (qualitatively or quantitatively depending on the method used); and (c) providing the necessary data that allow the calculation of sensitivity and specificity. We included studies that reported data on cases (positive samples) and healthy controls (negative samples) as well as studies with data available only for cases (see also Section 2.5).
2.3. Data Extraction
Data extraction was performed in a predetermined Microsoft Excel® sheet. From each study we extracted the following information: first author’s last name, type of antigen used, type of sample, method of detection used, and the qPCR cycle threshold (Ct) values used for the detection of SARS-CoV-2 RNA. Additionally, the method of antigen testing used was recorded along with the brand name and the name of the manufacturer and the existence of data from the virus culture. Symptomatic and asymptomatic cases as well as male/female ratios were also recorded, if given. To obtain sensitivity and specificity measures, a 2 × 2 contingency table was constructed; thus, true positive (TP), false negative (FN), true negative (TN), and false positive (FP) results were recorded. In cases where no controls were used, we used TP and FN values only.
2.4. Study Outcomes
The primary outcome of this meta-analysis was the sensitivity and specificity of AT in relation to qPCR. Secondary outcomes included the performance of AT on different sample types (namely, nasopharyngeal, saliva, and throat samples) and by symptoms (asymptomatic versus symptomatic SARS-CoV-2 infected persons). We also explored the performance of AT across the number of qPCR Ct values (a higher Ct indicated lower viral load).
2.5. Data Analysis
The Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2 tool) was used to assess the quality of the included studies in terms of diagnostic accuracy [28]. The four domains assessed were patient selection, index test, reference standard, and flow and timing. Each domain was evaluated following classifications according to judgment, i.e., low risk, high risk, and unclear risk.
The bivariate meta-analytic method modified for the meta-analysis of diagnostic tests was used [29]. This method has been reported to be equivalent to the so-called hsROC method [30]. It uses logit-transforms of true positive rate (TPR) and false positive rate (FPR) in order to model sensitivity and specificity; it can also be used for the evaluation of between-studies variability (heterogeneity). Studies that include information only for logit (TPR)—that is, only for sensitivity—were included in the bivariate model under the missing at random (MAR) assumption in order to maximize statistical power and allow the modeling of between-studies variability and correlation [31]. Begg’s rank correlation test [32] and Egger’s regression test [33] were used on logit (TPR) to evaluate the presence of publication bias. Stata13 [34] was used to perform the analysis and run the command “mvmeta” with the method of moments for multivariate meta-analyses and meta-regression [35]. Statistical significance was set at p < 0.05; meta-analysis was performed when two or more studies were available, whereas tests for publication bias and meta-regression were performed when five or more studies were available.
3. Results
3.1. Characteristics of Studies
Following the literature search in Pubmed, MedRxiv, and BioRxiv by 4 July 2021, we retrieved 4700 unique articles (Figure 1). After scrutinizing abstracts and full papers and testing for eligibility criteria, we ended up with 235 articles, which included 31,387 SARS-CoV-2 infected individuals and 188,636 individuals without SARS-CoV-2 infection (total: 220,049 individuals). Two hundred and sixteen studies provided data on both cases and controls, while 19 studies reported results only for people with SARS-CoV-2 infection (Figure 1). Table 1 shows the characteristics of the included studies. All studies reported that SARS-CoV-2 infection was confirmed with qPCR of envelope (E), S or N protein according to WHO, CDC and ECDC guidelines. Various methods were used to identify or measure an antigen of SARS-CoV-2. The N antigen was investigated in 225 studies, the S antigen was investigated in eight studies, and in two studies, cumulative estimates were given for N + S or S + E + M (membrane) antigens. Four articles evaluated both N- and S-based assays. Most studies focused on rapid POC tests such as LFIA (181 studies), or FIA (38 studies). Chemiluminescence was used in 21 studies. In total, 83 different kits from 74 manufacturers and 18 in-house tests (LFIA, FIA, CLEIA) from the respective laboratories were used. Thirty-six studies used the same samples to compare different tests from different companies. Twelve studies used twelve unique techniques that are under development (LC-mass spectrometry [36,37], field-effect transistor (FET) based biosensing devices [38], organic electrochemical transistors-OECT [39], voltametric-based immunosensor [40], optical waveguide-based biosensor technology [41], deep learning-based surface-enhanced Raman spectroscopy [42], paper-based impedance sensor [43], high-field asymmetric waveform ion mobility spectrometry (FAIMS)–parallel reaction monitoring (PRM) [44], a colorimetric biosensor [45], an electrochemical glucose sensor [46], and a urine foaming test [47]). Finally, two studies were performed with urine samples [36,47]. Most studies used nasopharyngeal, nasal, pharyngeal, throat, oropharyngeal or saliva samples. We classified the samples into two groups, named “NSP”, containing the first three sample types, and “TS”, containing the last three types. The type of sample was clearly mentioned in 207 studies, while all types of samples were used without distinction in 31 studies. The results from different types of samples were compared with the same method in 11 studies. Finally, data from 60 studies on asymptomatic persons and 73 on symptomatic patients were also used to explore differences in diagnostic accuracy between these two patients’ groups. The results of the quality assessment of the research using the QUADAS tool are provided in Supplementary Table S1 and in Supplementary File S1.
Figure 1.
Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flow diagram.
Table 1.
Characteristics of the 235 studies included in the meta-analysis.
| Author | Country of Study | Ag | Type of Sample | Ag Detection Method/Virus Culture Data |
Kit Name | Kit Company | Ct Values Tested | Signal Detection [Rapid (w/wo Detector)/Quick] |
Total Individuals |
Cases | Controls |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mak et al. [48] | China | N | 1. nsp 2. ts |
1. LFIA 2. LFIA 3. LFIA/virus culture data |
1. COVID-19 Ag Respi-Strip 2. NADAL COVID-19 Ag Test 3. Standard Q COVID-19 Ag |
1. Coris Bioconcept, Belgium 2. Nal Von Minden GmbH, Germany 3. SD Biosensor, Korea |
up to 20/up to 30/up to 40/0–20/20–30/30–40 | Rapid | 35 | 35 | NA |
| Linares et al. [49] | Spain | N | nsp | LFIA | Panbio COVID-19 Ag Rapid Test Device | Abbott Rapid Diagnostic Jena GmbH, Jena, Germany | Up to 40 | Rapid | 255 | 60 | NA |
| Gupta et al. [50] | India | N | nsp | LFIA | Standard Q rapid antigen detection test | SD Biosensor, Inc., Gurugram | Up to 40 | Rapid | 330 | 77 | 253 |
| Fenollar et al. [51] | France | N | nsp | LFIA | PANBIO COVID-19 Ag rapid test device | Abbott, USA | Up to 40 | Rapid | 341 | 204 | 137 |
| Nalumansi et al. [52] | Uganda | N | nsp | LFIA | STANDARD Q COVID-19 Ag Test | SD -Biosensor, Republic of Korea | Up to 30/up to 40/30–40 | Rapid | 262 | 90 | 172 |
| Parada-Ricart et al. [53] |
Spain | N | nsp | FIA | 2019-nCoV Antigen Rapid Test Kit (FIA) | Shenzhen Bioeasy Biotechnology CO LTD, China | Up to 40 | Rapid/detector | 172 | 26 | 146 |
| Lee et al. [54] |
Korea | S | nsp | LFIA/virus culture data | In-house test | Up to 40 | Rapid/detector | 8 | 3 | 5 | |
| Cerutti et al. [55] |
Italy | N | nsp | LFIA | STANDARD Q COVID19 Ag | SD-Biosensor, RELAB, I | Up to 40 | Rapid | 330 | 109 | 221 |
| Diao et al. [56] |
China | N | nsp | FIA | In-house test | Up to 40 | Rapid/detector | 502 | 356 | 146 | |
| Young et al. [57] |
USA | N | nsp | 1. LFIA 2. FIA |
1. BD Veritor™ System 2. Sofia 2 SARS Antigen FIA |
1. Becton-Dickinson and Company, USA 2. Quidel, San Diego, CA |
Up to 40 | 1. Rapid/optional detector 2. Rapid/detector |
612 | 81 | 531 |
| Liotti et al. [58] |
Italy | N | nsp | FIA | STANDARD F COVID19 Ag (FIA) | SD Biosensor, Suwon, Korea | Up to 20/up to 30/up to 40/0–20 | Rapid/detector | 359 | 104 | 255 |
| Ogawa et al. [59] |
Japan | N | Nsp | CLEIA | Lumipulse SARS-CoV-2 Ag | Fujirebio, Tokyo, Japan | Up to 40 | Detector | 325 | 24 | 301 |
| Hirotsu et al. [60] |
Japan | N | nsp | CLEIA | LUMIPULSE SARS-CoV-2 Ag kit | Fujirebio, Inc. (Tokyo, Japan) | Up to 40 | Detector | 313 | 58 | 255 |
| Nagura-Ikeda et al. [61] |
Japan | N | ts | LFIA | Espline SARS-CoV-2 | Fuji Rebio Inc. | Up to 40 | Rapid | 103 | 84 | 19 |
| Mak et al. [62] |
Hong Kong | N | 1. nsp/ts 2. ts |
LFIA | BIOCREDIT COVID-19 Ag kit | RapiGEN Inc. | Up to 20/up to 30/up to 40/0–20/20–30 | Rapid optional detector | 160 | 51 | 109 |
| Mertens et al. [63] |
Belgium | N | nsp | LFIA/virus culture data | COVID-19 Ag RespiStrip | Coris BioConcept | Up to 30/up to 40 | Rapid | 328 | 132 | 196 |
| Blairon et al. [64] |
Belgium | N | nsp | LFIA | COVID-19 Ag Respi-Strip | Coris Bioconcept, Gembloux, Belgium | Up to 40 | Rapid | 774 | 159 | 615 |
| Porte et al. [21] |
Chile | N | nsp/ts | FIA | 2019-nCoV Antigen Rapid Test Kit (FIA) | Bioeasy Biotechnology Co., Shenzhen, China | Up to 30 | Rapid/detector | 127 | 82 | 45 |
| Scohy et al. [65] |
Belgium | N | nsp | LFIA | COVID-19 Ag Respi-Strip | Coris BioConcept, Gembloux, Belgium | Up to 40 | Rapid | 148 | 106 | 62 |
| Lambert-Niclot et al. [66] |
France | N | nsp | LFIA | COVID-19 Ag Respi-Strip | Coris BioConcept, Gembloux, Belgium | Up to 40 | Rapid | 138 | 94 | 44 |
| Diao et al. [67] |
China | N | nsp | FIA | In-house test | Up to 30/up to 40/30–40 | Rapid | 239 | 208 | 31 | |
| Beck et al. [68] |
Milwaukee | N | nsp | FIA | Sofia SARS FIA test (SOFIA) | Quidel, San Diego, CA | Up to 40 | Rapid/detector | 346 | 61 | 285 |
| Krüttgen et al. [69] |
Germany | N | nsp | LFIA | SARS-CoV-2 Rapid Antigen Test | Roche, Switzerland | Up to 20/up to 30/up to 40/0–20/20–30/30–40 | Rapid | 150 | 75 | 75 |
| Albert et al. [70] |
Spain | N | nsp | LFIA/virus culture data | Panbio™ COVID-19 Ag Rapid Test Device | Abbott Diagnostic GmbH, Jena, Germany | Up to 40 | Rapid | 412 | 54 | 358 |
| Chaimayo et al. [71] |
Thailand | N | nsp/ts | LFIA | Standard Q COVID-19 Ag test | SD Biosensor®, Chuncheongbuk-do, Republic of Korea | Up to 40 | Rapid | 454 | 60 | 394 |
| Lanser et al. [72] |
Austria | N | nsp | LFIA | Panbio™ COVID-19 Ag Rapid test | Abbott, Chicago, Illinοis | Up to 30/up to 40/30–40 | Rapid | 53 | 51 | 2 |
| Gremmels et al. [73] |
The Netherlands/Aruba |
N | nsp | LFIA | Panbio COVID-19 Ag rapid test device | Abbott, Lake Country, IL, USA | Up to 40 | Rapid | 2948 | 202 | 2746 |
| Drevinek et al. [74] |
Czech Republic | N | nsp | 1. LFIA 2. FIA |
1. Panbio COVID-19 Ag Rapid Test 2. Standard F COVID-19 Ag FIA |
1. Abbott, Germany 2. SD Biosensor, Republic of Korea |
Up to 20/up to 30/up to 40/0–20/20–30/30–40 | 1. Rapid 2. Rapid/detector |
591 | 223 | 368 |
| Schwob et al. [75] |
Switzerland | N | nsp | 1. LFIA 2. LFIA 3. LFIA |
1. STANDARD Q COVID-19 Ag Test 2. Panbio COVID-19 Ag Test 3. COVID-VIRO |
1. SD -Biosensor, Republik of Korea 2. Abbott, Germany 3. AAZ-LMB |
Up to 40 | Rapid | 928 | 372 | 556 |
| Corman et al. [76] |
Germany | N | nsp | 1. LFIA 2. LFIA 3. LFIA 4. LFIA 5. LFIA 6. LFIA 7. LFIA/virus culture data |
1. Panbio COVID-19 Ag Test 2. BIOCREDIT COVID-19 Ag kit 3. Coronavirus Ag Rapid Test Cassette (swab) 4. COVID-19 Ag Respi-Strip 5. RIDA®QUICK SARS-CoV-2 antigen 6. NADAL COVID19-Ag Test 7. SARS-CoV-2 Rapid Antigen Test |
1. Abbott, Germany 2. RapiGEN Inc. 3. Healgen 4. Coris Bioconcept, Gembloux, Belgium 5. R-Biopharm 6. NAL von minden 7. Roche |
Up to 40 | Rapid | 150 | 115 | 35 |
| Abdulrahman et al. [77] | Bahrain | N | nsp | LFIA | Panbio COVID 19 antigen rapid test device | Abbott Rapid Diagnostic Jena GmbH, Jena, Germany | Up to 30 | Rapid | 4183 | 733 | 3450 |
| Yokota et al. [78] |
Japan | N | Nsp, ts | 1. LFIA 2. CLEIA |
1. Espline SARS-CoV-2 2. Lumipulse SARS-CoV-2 Ag kit |
1. Fujirebio, Tokyo, Japan 2. Fujirebio, Tokyo, Japan |
Up to 30/up to 40/20–30 | 1. Rapid 2. Quick/detector |
34 | 34 | NA |
| Nash et al. [79] |
USA/Brazil | 1. N 2. S |
nsp | LFIA | In-house | Up to 20/up to 30/up to 40/0–20/20–30/30–40 | Rapid | 311 | 172 | 139 | |
| Van der Moeren et al. [80] |
The Netherlands | N | nsp | LFIA | BD Veritor™ System | Becton-Dickinson and Company, USA | Up to 20/up to 30/up to 40/0–20/20–30 | Rapid/optional detector | 351 | 17 | 334 |
| Porte et al. [81] |
Chile | N | nsp/ts | 1. FIA 2. FIA |
1. SOFIA SARS Antigen FIA 2. STANDARD® F COVID-19 Ag FIA |
1. Quidel Corporation, San Diego, CA, USA 2. SD Biosensor Inc., Gyeonggi-do, Republic of Korea |
Up to 30/up to 40/30–40 | Rapid/detector | 91 | 59 | 32 |
| Krüger et al. [82] |
Germany/UK | N | nsp/ts | 1. FIA 2. LFIA 3. LFIA/virus culture data |
1. 2019-nCoV Ag Fluorescence Rapid Test Kit 2. COVID-19 Ag Respi-Strip 3. STANDARD Q COVID-19 Ag Test |
1. Shenzhen Bioeasy Biotechnology Co. Ltd., Guangdong Province, China 2. Coris Bioconcept, Gembloux, Belgium 3. SD Biosensor, Inc., Gyeonggi-do, Korea |
Up to 30/up to 40/30–40 | 1. Rapid/detector 2. Rapid 3. Rapid |
2407 | 72 | 2335 |
| Singh et al. [46] |
San Diego | S | nsp | ECGluS | In-house | Up to 40 | Quick * | 24 | 16 | 8 | |
| Ventura et al. [83] |
Italy | S + E + M | Nsp/ts | CBS | In-house | Up to 40 | Detector | 94 | 45 | 49 | |
| Herrera et al. [84] |
Florida | N | nsp | LFIA | NR/AdventHealth Centra Care | Up to 40 | Rapid | 1669 | 486 | 1183 | |
| Renuse et al. [44] |
USA | N | nsp | FAIMS-PRM | In-house | Up to 40 | Detector | 176 | 88 | 88 | |
| Pickering et al. [85] |
UK | N | nsp-ts | LFIA/virus culture data | 1. Innova Rapid SARS-CoV-2 Antigen Test 2. Spring Healthcare SARS-CoV-2 Antigen Rapid Test Cassette 3. E25Bio Rapid Diagnostic Test 4. Encode SARS-CoV-2 Antigen Rapid Test Device 5. SureScreen COVID-19 Rapid Antigen Test Cassette |
1. Xiamen Biotime Biotechnology, Fujian, China 2. Shanghai ZJ Bio-Tech, Shanghai, China 3. E25Bio, Cambridge, MA, USA 4. Zhuhai Encode Medical Engineering, Zhuhai, China 5. SureScreen Diagnostics, Derby, UK |
20–30 | Rapid | 200 | 100 | 100 |
| Harmon et al. [86] |
Washington | N | nsp | FIA | Sofia-2 SARS-CoV-2 Antigen Tests | Quidel, San Diego, CA | Up to 40 | Rapid/detector | 23,462 | 83 | 23,379 |
| Korenkov et al. [87] |
Germany | N | nsp-ts | LFIA/virus culture data | STANDARD Q COVID-19 Ag Test | SD Biosensor, Inc., Gyeonggi-do, Korea | Up to 20/up to 30/up to 40/0–20/20–30/30–40 | Rapid | 2028 | 210 | 1818 |
| Ehsan et al. [43] | Saudi Arabia | S | nsp | Paper-based impedance sensor | In-house | Up to 40 | Detector | 5 | 3 | 2 | |
| Seynaeve et al. [88] | Belgium | N | nsp | LFIA | 1. COVID-19 Ag Respi-Strip 2. coronavirus antigen rapid test cassette |
1. Coris Bioconcept, Belgium 2. Healgen Scientific, LLC, USA |
Up to 30/ Up to 40/30–40 | Rapid | 163 | 98 | 65 |
| Di Domenico et al. [89] | Italy | 1. N 2. S |
1. nsp 2. ts |
1. ELISA based 2. LFIA/virus culture data |
1. Portable COVID-19 Antigen Lab Test 2. Panbio™ COVID-19 Ag Rapid Test Device |
1. Stark 2. Abbott Diagnostic GmbH, Jena, Germany |
Up to 40 | Rapid | 433 | 36 | 397 |
| Kiro et al. [90] | India | N | nsp | FIA | STANDARD® F COVID-19 Ag FIA | SD Biosensor Inc., Gyeonggi-do, Republic of Korea | Up to 40 | Rapid/detector | 354 | 136 | 218 |
| Smith et al. [91] | Illinois | N | 1. nsp-ts 2. nsp |
FIA/virus culture data | SOFIA SARS Antigen FIA | Quidel Corporation, San Diego, CA, USA | Up to 40 | Rapid/detector | 43 | 43 | NA |
| L’Huillier et al. [92] |
Switzerland | N | nsp | LFIA | Panbio™ COVID-19 Ag Rapid Test Device | Abbott Diagnostic GmbH, Jena, German | Up to 40 | Rapid | 822 | 119 | 703 |
| Gupta et al. [93] |
India | S | nsp | ELISA | In-house | Up to 40 | Quick | 232 | 44 | 188 | |
| Wagenhäuser et al. [94] |
Germany | N | ts | LFIA | 1. NADAL COVID-19 Ag Test 2. Panbio COVID-19 Ag rapid test device 3. MEDsan SARS-CoV-2 Antigen Rapid Test |
1. Nal Von Minden GmbH, Germany 2. Abbott Laboratories, Abbott Park IL, USA 3. MEDsan GmbH, Hamburg, Germany |
Up to 40 | Rapid | 5056 | 101 | 4955 |
| Fernandez et al. [95] |
Spain | N | nsp | FIA | LumiraDx™ | LumiraDx™ Limited, Londres, Reino Unido | Up to 40 | Rapid/detector | 46 | 24 | 22 |
| Amer et al. [96] |
Egypt | N | nsp-ts | LFIA | STANDARD Q COVID-19 Ag Test | SD Biosensor, Inc., Gyeonggi-do, Korea | Up to 40 | Rapid | 47 | 45 | 2 |
| Baccani et al. [97] |
Italy | N | nsp | 1. CLEIA 2. FIA 3. FIA |
1. Lumipulse G SARS-CoV-2 Ag 2. STANDARD® F COVID-19 Ag FIA 3. AFIAS COVID-19 Ag |
1. Fujirebio, Tokio, Japan 2. SD Biosensor; Suwon-si, Korea 3. Menarini; Florence, Italy |
Up to 30/Up to 40/30–40 | 1. Quick/detector 2. Rapid/detector 3. Rapid/detector |
375 | 85 | 290 |
| Matsuzaki et al. [98] |
Japan | N | nsp | CLEIA | 1. VITROS® SARS-CoV-2 Antigen Test 2. LUMIPULSE® SASR-CoV-2 Ag Test |
2. Ortho Clinical Diagnostics, Rochester, NY, USA 3. Fujirebio, Tokio, Japan |
Up to 40 | 1. Quick/detector 2. Quick/ detector |
128 | 49 | 79 |
| Jakobsen et al. [99] |
Denmark | N | nsp | LFIA | STANDARD Q COVID-19 Ag Test | SD Biosensor, Inc., Gyeonggi-do, Korea | Up to 40 | Rapid | 4811 | 221 | 4590 |
| Ngo Nsoga et al. [100] |
Switzerland | N | nsp-ts | LFIA/virus culture data | Panbio™ COVID-19 Ag Rapid Test Device | Abbott Diagnostic GmbH, Jena, German | Up to 40 | Rapid | 402 | 168 | 234 |
| Funabashi et al. [41] |
Japan | N | nsp | Optical waveguide-based biosensor technology | In-house | Up to 40 | Detector | 64 | 34 | 30 | |
| Smith et al. [101] |
Maryland | N | nsp | FIA | SOFIA SARS Antigen FIA | Quidel Corporation, San Diego, CA, USA | Up to 40 | Rapid/detector | 2887 | 235 | 2652 |
| Eleftheriou et al. [102] |
Greece | N | nsp | LFIA | Panbio™ COVID-19 Ag Rapid Test Device | Abbott Diagnostic GmbH, Jena, German | Up to 40 | Rapid | 744 | 51 | 693 |
| Huang et al. [42] |
China | S | ts | Deep learning-based surface-enhanced Raman spectroscopy | In-house | Up to 40 | NA/detector | 57 | 30 | 27 | |
| Lindner et al. [103] |
Germany | N | nsp-ts | LFIA | STANDARD Q COVID-19 Ag Test | SD Biosensor, Inc., Gyeonggi-do, Korea | Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | Rapid | 146 | 40 | 106 |
| Ferte et al. [104] |
France | N | nsp | LFIA | Panbio™ COVID-19 Ag Rapid Test Device | Abbott Diagnostic GmbH, Jena, German | Up to 40 | Rapid | 688 | 52 | 636 |
| Fernandez-Montero et al. [105] |
Spain | N | nsp-ts | LFIA | SARS-CoV-2 Rapid Antigen Test | Roche | Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | Rapid | 2543 | 49 | 2494 |
| Hoehl et al. [106] |
Germany | N | nsp | LFIA | RIDA®QUICK SARS-CoV-2 Antigen | R-Biopharm AG | Up to 30 | Rapid | 9 | 9 | NA |
| Lee et al. [107] |
Korea | N | nsp | LFIA | STANDARD Q COVID-19 Ag Test | SD Biosensor, Inc., Gyeonggi-do, Korea | Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | Rapid | 680 | 380 | 300 |
| Mayanskiy et al. [108] |
Russia | N | nsp | ELISA | CoviNAg EIA | XEMA, Russia | Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | Detector | 277 | 182 | 95 |
| Leixner et al. [109] |
Austria | N | nsp | LFIA | AMP Rapid Test SARS-CoV-2 Ag | AMP Diagnostics, AMEDA Labordiagnostik GmbH, Graz, Austria | Up to 30/Up to 40/30–40 | Rapid | 392 | 94 | 298 |
| Hirotsu et al. [110] |
Japan | N | nsp | 1. CLEIA 2. CLEIA |
1. LUMIPULSE® SASR-CoV-2 Ag Test 2. Elecsys1 SARS-CoV-2 Antigen Assay |
1. Fujirebio, Tokio, Japan 2. Roche, Basel, Switzerland |
Up to 40 | Detector | 637 | 487 | 150 |
| Chavan et al. [36] |
USA | N | urine | mass spectrometry | In-house | Up to 40 | Detector | 50 | 39 | 11 | |
| Fiedler et al. [111] |
Germany | N | nsp | CLEIA/virus culture data | LIAISON® SARS-CoV-2 Ag | DiaSorin | Up to 40 | Detector | 182 | 110 | 72 |
| Dierks et al. [112] |
Germany | N | nsp | 1. FIA 2. LFIA |
1. LumiraDx™ 2. NADAL COVID-19 Ag Test |
1. LumiraDx™ Limited, London, United Kingdom 2. Nal Von Minden GmbH, Germany |
Up to 40 | 1. Rapid/detector 2. Rapid |
444 | 11 | 433 |
| Terpos et al. [113] |
Slovenia | N | nsp | LFIA | COVID-19 Antigen Detection Kit (Colloidal Gold) | Zhuhai Lituo Biotechnology Co., Ltd. | Up to 30/Up to 40/30–40 | Rapid | 358 | 114 | 244 |
| Osmanodja et al. [114] |
Germany | N | nsp-ts | LFIA | Dräger Antigen Test SARS-CoV-2 | Dräger Safety AG and Co. KGaA, Lübeck, Germany | Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | Rapid | 379 | 70 | 309 |
| Harris et al. [115] |
USA | N | nsp | FIA | SOFIA SARS Antigen FIA | Quidel Corporation, San Diego, CA, USA | Up to 30/Up to 40/30–40 | Rapid/detector | 2429 | 324 | 2105 |
| Cento et al. [116] |
Italy | N | nsp | FIA | LumiraDx™ | LumiraDx™ Limited, Londres, Reino Unido | Up to 30/Up to 40/30–40 | Rapid/detector | 960 | 347 | 613 |
| Kumar et al. [117] |
India | N | nsp | LFIA | STANDARD Q COVID-19 Ag Test | SD Biosensor, Inc., Gyeonggi-do, Korea | Up to 40 | Rapid | 6 | 6 | NA |
| Orsi et al. [118] |
Italy | N | nsp | FIA | 1. FREND™ COVID-19 Ag 2. STANDARD® F COVID-19 Ag FIA |
1. NanoEntek, Korea 2. SD Biosensor; Suwon-si, Korea |
Up to 30/Up to 40/30–40 | Rapid/detector | 110 | 60 | 50 |
| Blairon et al. [119] |
Belgium | N | nsp | LFIA/virus culture data | 1. Coronavirus Ag Rapid Test Cassette 2. GSD NovaGen SARS-CoV-2 (COVID-19) Antigen Rapid Test 3. Aegle Coronavirus Ag Rapid Test Cassette |
1. BioRad 2. NovaTec Immunodiagnostica GmbH 3. LumiraDx |
Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | Rapid | 199 | 97 | 102 |
| Bornemann et al. [120] |
Germany | N | nsp | FIA | SOFIA SARS Antigen FIA | Quidel Corporation, San Diego, CA, USA | Up to 30/Up to 40/30–40 | Rapid/detector | 1391 | 91 | 1300 |
| Kruger et al. [121] |
Germany | N | 1. nsp 2. ts 3. nsp-ts |
LFIA | Panbio™ COVID-19 Ag Rapid Test Device | Abbott Diagnostic GmbH, Jena, German | Up to 30/Up to 40/30–40 | Rapid | 1108 | 106 | 1002 |
| Eissa et al. [40] |
Saudi Arabia | N | nsp | Voltammetric-based immunosensor | In-house | Up to 30/Up to 40/30–40 | Detector | 6 | 5 | 1 | |
| Shaikh et al. [122] |
USA | N | nsp | LFIA | BinaxNOWTM COVID-19 Ag Card | Abbott Diagnostics Scarborough, Inc., USA | Up to 40 | Rapid | 199 | 39 | 160 |
| Diez Flecha et al. [123] |
Spain | N | nsp | LFIA | Panbio™ COVID-19 Ag Rapid Test Device | Abbott Diagnostic GmbH, Jena, German | Up to 30/Up to 40/30–40 | Rapid | 55 | 49 | 6 |
| Yokota et al. [124] |
Japan | N | ts | CLEIA | In-house | Up to 40 | detector | 2056 | 89 | 1967 | |
| Guo et al. [39] |
Saudi Arabia | N | 1. nsp 2. ts 3. nsp-ts |
OECT | In-house | Up to 40 | detector | 24 | 11 | 13 | |
| Klein et al. [125] |
Germany | N | nsp | LFIA | Panbio™ Ag-RDT | Abbott Diagnostics, Jena, Germany | Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | Rapid | 290 | 39 | 251 |
| Caramello et al. [126] |
Italy | N | nsp | 1. LFIA 2. FIA |
1. SD BIOSENSOR Ag-RDT 2. LUMIRADX Ag-RDT |
1. SD BIOSENSOR Ag-RDT 2. LumiraDx UK Ltd., Dumyat Business Park, Alloa, FK10 2PB, UK) |
Up to 40 | 1. Rapid 2. Rapid/detector |
324 | 210 | 114 |
| Koeleman et al. [127] |
Netherlands | N | nsp-ts | LFIA | 1. Certest SARS-CoV-2 2. Roche SARS-CoV-2 Rapid Antigen Test 3. Romed Coronavirus Ag Rapid Test 4. BD Veritor SARS-CoV-2 point-of-care test 5. Panbio™ COVID-19 Antigen rapid test |
1. Certest Biotec S.L., Spain 2. Roche, Switzerland 3. Romed, The Netherlands 4. Becton, Dickinson and Company, USA 5. Abbott, USA |
Up to 40 | Rapid | 980 | 340 | 640 |
| Šterbenc et al. [128] |
Slovenia | N | nsp | LFIA | SARS-CoV-2 rapid antigen test (Roche) | Roche Diagnostics GmbH, Mannheim, Germany) | Up to 40 | Rapid | 191 | 2 | 189 |
| Kumar et al. [129] |
India | N | nsp-ts | FIA | STANDARD™ Q COVID-19 Ag test kit | SD Biosensor; Suwon-si, Korea | Up to 40 | Rapid/detector | 204 | 12 | 192 |
| Soleimani et al. [130] |
Belgium | N | nsp | FIA | 1. COVID19Speed-antigen test 2. Panbio™ COVID-19 Ag rapid test |
1. BioSpeedia 2. Abbott |
Up to 30/Up to 40/30–40 | Rapid/detector | 401 | 196 | 205 |
| Takeuchi et al. [131] |
Japan | N | nsp | LFIA | QuickNavi-COVID19 Ag | Denka Co., Ltd., Tokyo, Japan | Up to 30 | Rapid | 862 | 51 | 811 |
| Linares et al. [49] |
Spain | N | nsp | 1. LFIA 2. FIA |
1. Panbio COVID-19 Ag Rapid Test Device 2. D-Biosensor STANDARD F COVID-19 Ag |
1. Abbot Rapid Diagnostics GmbH, Jena 2. SD Biosensor, Inc. |
Up to 20/Up to 30/20–30 | 1. Rapid 2. Rapid/detector |
356 | 170 | 186 |
| Homza et al. [132] |
Czech Republic | N | nsp | LFIA | Ecotest COVID-19 Antigen Rapid Test | Assure Tech, Hangzhou, China | Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | Rapid | 491 | 164 | 327 |
| Van der Moeren et al. [133] |
Netherlands | N | nsp-ts | CLEIA | BD veritor system for rapid detection of SARS-CoV-2 (VRD) | Becton-Dickinson and Company, USA | 20–30 | Detector | 978 | 161 | 817 |
| Brihn et al. [134] |
USA | N | nsp | FIA | Quidel Sofia 2 SARS Antigen Fluorescent Immunoassay | Quidel Corporation | Up to 30 | Rapid/detector | 2039 | 149 | 1890 |
| Nordgren et al. [135] |
Sweden | N | nsp | LFIA/virus culture data | 1. Panbio™ COVID-19 Ag Rapid Test 2. Zhejiang Orient Gene |
1. Abbott 2. Healgen Biotech Coronavirus Ag rapid test cassette |
Up to 20/Up to 40/20–30 | Rapid | 462 | 156 | 306 |
| Holzner et al. [136] |
Germany | N | nsp | LFIA | Standard Q COVID-19 Ag | SD Biosensor, Korea | Up to 30 | Rapid | 2280 | 456 | 1824 |
| Kim et al. [137] |
Korea | N | nsp | LFIA | GenBody COVID-19 Ag Test (COVAG025) | GenBody Inc. | Up to 40/20–30 | Rapid | 330 | 130 | 200 |
| Bianco et al. [138] |
Italy | N | nsp | FIA | LumiraDx™ SARS-CoV-2 Antigen Test | LumiraDx | 30–40 | Rapid/detector | 907 | 298 | 609 |
| Peña et al. [139] |
Chile | N | nsp | LFIA | SARS-CoV-2 RAT | SD Biosensor | Up to 30 | Rapid | 842 | 73 | 769 |
| Muhi et al. [140] |
Australia | N | nsp | LFIA/virus culture data | PanBioTM COVID-19 Ag | Abbott | Up to 40 | Rapid | 189 | 26 | 163 |
| Uwamino et al. [141] |
Japan | N | nsp | LFIA/virus culture data | Espline SARS-CoV-2 RAD | FUJIREBIO, Tokyo, Japan | Up to 40 | Rapid | 117 | 25 | 92 |
| Thakur et al. [142] |
India | N | nsp-ts | LFIA | PathoCatch | ACCUCARE | 20–30 | Rapid | 677 | 84 | 593 |
| Homza et al. [143] |
Czech Republic | N | nsp | LFIA/virus culture data | 1. SARS-CoV-2 Antigen Rapid Test Kit 2. Ecotest COVID-19 Antigen Rapid Test 3. Standard Q COVID-19 Ag 4. Immupass VivaDiag™ SARS-CoV-2 Ag Rapid Test 5. ND COVID-19 Ag test |
1. JOYSBIO (Tianjin) Biotechnology Co., Ltd., Tianjin, China 2. Assure Tech, Hangzhou, China 3. SD Biosensor, Korea 4. VivaChek Biotech (Hangzhou) Co., Ltd., Hangzhou, China 5. NDFOS, Eumseong, Korea |
Up to 40 | Rapid | 1141 | 407 | 734 |
| Shah et al. [144] |
USA | N | nsp | LFIA | BinaxNOW COVID-19 Ag | Abbott | 20–30 | Rapid | 2110 | 334 | 1776 |
| McKay [145] |
USA | N | nsp | LFIA/virus culture data | BinaxNOW Rapid Antigen Test | Abbott | Up to 40 | Rapid | 532 | 105 | 427 |
| Yin et al. [146] |
Belgium | N | nsp | LFIA | 1. Panbio™ COVID-19 Ag Rapid Test Device 2. BD Veritor™ SARS-CoV-2 3. COVID-19 Ag Respi-Strip 4. SARS-CoV-2 Rapid Antigen Test |
1. Abbott Rapid Diagnostics, Germany 2. Becton-Dickinson and Company, USA 3. Coris BioConcept, Belgium 4. SD Biosensor, Republic of Korea |
30–40 | Rapid | 760 | 722 | 38 |
| Baro et al. [147] |
Spain | N | nsp | LFIA | 1. PanBioTM COVID-19 Ag Rapid test 2. CLINITEST® Rapid COVID-19 Antigen Test 3. SARS-CoV-2 Rapid Antigen Test 4. SARS-CoV-2 Antigen Rapid Test Kit 5. COVID-19 Coronavirus Rapid Antigen Test Cassette |
1. Abbott 2. Siemens 3. Roche 4. Lepu Medica 5. Surescreen |
Up to 30 | Rapid | 286 | 101 | 185 |
| Caputo et al. [148] |
Italy | N | nsp-ts | CLEIA | Lumipulse G SARS-CoV-2 Ag | Fujirebio, Tokio, Japan | Up to 40 | Quick/detector | 4266 | 503 | 3763 |
| Kenyeres et al. [149] |
Hungary | N | nsp | LFIA | BIOCREDIT COVID-19 Ag | RapiGEN Inc. | Up to 30 | Rapid | 37 | 37 | NA |
| Häuser et al. [150] |
Germany | N | nsp | CLEIA/virus culture data | LIAISON SARS-CoV-2 antigen test | Diasorin | 20–30 | Detector | 196 | 196 | 27 |
| Lefever et al. [151] |
Belgium | N | nsp | LFIA/virus culture data | Liaison antigen test | Diasorin | 20–30 | Rapid | 410 | 200 | 210 |
| Zacharias et al. [152] |
Austria | N | nsp | LFIA | SARS-CoV-2 RAT | Roche | 30–40 | Rapid | 30 | 24 | 6 |
| Oh et al. [153] |
Korea | N | nsp | LFIA | Standard Q COVID-19 Ag | SD Biosensor, Inc. Gyeonggi-do, Korea | Up to 30 | Rapid | 118 | 26 | 92 |
| Asai et al. [154] |
Japan | N | nsp | CLEIA | LUMIPULSE SARS-CoV-2 antigen kit | Fujirebio, Japan | 30–40 | Detector | 305 | 63 | 242 |
| Kweon et al. [155] |
Korea | N | nsp | LFIA | 1. AFIAS COVID-19 Ag 2. ichromaTM COVID-19 Ag |
1. Boditech Med., Chuncheon-si, Gang-won-do, Republic of Korea 2. Boditech Med. |
Up to 30/Up to 40/30–40 | Rapid | 167 | 167 | NA |
| Menchinelli et al. [156] |
Italy | N | nsp | CLEIA/virus culture data | LUMIPULSE SARS-CoV-2 antigen kit | Fujirebio, Japan | Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | Detector | 594 | 194 | 400 |
| Sood et al. [157] |
USA | N | nsp | LFIA | BinaxNOW rapid antigen test | Abbott | 20–30 | Rapid | 774 | 226 | 548 |
| Epstude et al. [158] |
Germany | N | nsp | LFIA | SARS-CoV-2 Rapid Antigen test | Roche® | Up to 40 | Rapid | 30 | 30 | NA |
| Smith et al. [91] |
USA | N | nsp | FIA/virus culture data | SARS Sofia FIA rapid antigen tests | Quidel | Up to 40 | Rapid/detector | 286 | 286 | NA |
| Berger et al. [159] |
Switzerland | N | nsp | LFIA/virus culture data | 1. PanbioTM COVID-19 Ag Rapid Test device 2. Standard Q Ag-RDT |
1. Abbott 2. SD Biosensor, Roche |
20–30 | Rapid | 1064 | 315 | 749 |
| Matsuda et al. [160] |
Brazil | N | nsp | LFIA | 1. COVID-19 Ag ECO Test 2. Panbio COVID-19 Ag Rapid Test |
1. ECO Diagnóstica 2. Abbott, Ludwigshafen, Germany |
Up to 40 | Rapid | 108 | 29 | 80 |
| Van Honacker et al. [161] |
Belgium | N | nsp | LFIA | 1. COVID-19 ag BSS 2. SARS-CoV-2 Ag card 3. Coronavirus AG Rapid test cassette 4. Panbio COVID-19 Ag Rapid Test Device 5. SARS-CoV-2 Rapid Antigen test |
1. Biosynex, Fribourg, Switzerland 2. Biotical health, Madrid, Spain 3. Zhejiang Orient Gene Biotech Co., Zhejiang, China 4. Abbott, Ludwigshafen, Germany 5. SD Biosensor, Gyeonggi-do, Korea |
Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | Rapid | 98 | 58 | 40 |
| Boum et al. [162] |
Cameroon | N | nsp | LFIA | SARS-CoV-2 Rapid Antigen test | SD Biosensor | 20–30 | Rapid | 1090 | 291 | 799 |
| Mboumba Bouassa et al. [163] |
France | N | nsp | LFIA | SIENNA™ COVID-19 Antigen Rapid Test Cassette | Salofa Oy, Salo, Finland; manufactured under license of T&D Diagnostics Canada Pvt. Ltd., Halifax, Canada | Up to 20/Up to 40 | Rapid | 150 | 100 | 50 |
| Stokes et al. [164] |
Canada | N | 1. nsp 2. ts |
LFIA | Panbio COVID-19 antigen Rapid Test Device | Abbott, IL, USA | Up to 40 | Rapid | 1888 | 497 | 1391 |
| Landaas et al. [165] |
Norway | N | nsp-ts | LFIA | Panbio™ COVID-19 Ag Rapid Test Device | Abbott | Up to 30/Up to 40/30–40 | Rapid | 3991 | 250 | 3741 |
| Takeuchi et al. [166] |
Japan | N | nsp | LFIA/virus culture data | QuickNavi™-COVID19 Ag | Denka Co., Ltd., Tokyo, Japan | Up to 40 | Rapid | 1186 | 105 | 1081 |
| Igloi et al. [167] |
Netherlands | N | nsp | LFIA/virus culture data | Roche SD Biosensor SARS-CoV-2 rapid antigen test | Roche Diagnostics | Up to 30/Up to 40/30–40 | Rapid | 970 | 186 | 784 |
| Masiá et al. [168] |
Spain | N | 1. nsp 2. ts |
LFIA | Panbio COVID-19 antigen Rapid Test Device | Abbott Rapid Diagnostic Jena GmbH, Jena, Germany | Up to 40 | Rapid | 2174 | 448 | 1726 |
| Jääskeläinen et al. [169] |
Finland | N | nsp | 1. FIA 2. LFIA/virus culture data |
1. Quidel Sofia SARS FIA 2. Standard Q COVID-19 Ag test 3. Panbio™ |
1. Quidel, San Diego, CA 2. SD Biosensor, Republic of Korea 3. Abbott Diagnostic GmbH, Jena, Germany |
Up to 30/Up to 40/30–40 | 1. Rapid/detector 2. Rapid 3. Rapid |
198 | 185 | 40 |
| Olearo et al. [170] |
Germany | N | nsp | LFIA/virus culture data | 1. SARS-CoV-2 Rapid Antigen Test (Roche) 2. COVID-19 Rapid Test Device (Abbott) 3. MEDsan SARS-CoV-2 Antigen Rapid Test 4. CLINITEST Rapid COVID-19 Antigen Test |
1. Roche Diagnostics SD Biosensor Korea 2. Abbott Rapid Diagnostics Panbio Ltd. Australia 3. MEDsan GmbH Germany 4. Zhejiang Orient Biotech Co. China |
Up to 40 | Rapid | 184 | 84 | 100 |
| Toshiaki Ishii et al. [171] | Japan | N | 1. nsp 2. ts |
1. LFIA 2. CLEIA |
1. Espline® SARS-CoV-2 2. Lumipulse® SARS-CoV-2 |
1. Fujirebio Inc., Tokyo, Japan 2. Fujirebio Inc., Tokyo, Japan |
Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | 1. Rapid 2. Quick/detector |
893 | 44 | 849 |
| Peña-Rodríguez et al. [172] | Mexico | N | nsp | LFIA | STANDARD™ Q COVID-19 Ag Test | SD BIOSENSOR | Up to 40 | Rapid | 369 | 104 | 265 |
| Gili et al. [173] | Italy | N | nsp | CLEIA | Lumipulse® SARS-CoV-2 antigen assay | Fujirebio, Inc., Tokyo, Japan | Up to 40 | Quick/detector | 1964 | 185 | 1779 |
| Pérez-García et al. [174] | Spain | N | nsp | LFIA | 1. CerTest SARS-CoV-2 Ag One Step Card Test 2. Panbio COVID-19 Ag Rapid Test Device |
1. Certest Biotec S. L., Zaragoza, Spain 2. Abbot Rapid Diagnostics GmbH, Jena, Germany |
Up to 30/Up to 40/30–40 | Rapid | 320 | 170 | 150 |
| Kilic et al. [175] | USA | N | nsp | LFIA | BD Veritor SARS-CoV-2 | Becton, Dickinson, Sparks, MD, USA | Up to 40 | Rapid | 1384 | 116 | 1268 |
| Drain et al. [176] | USA | N | nsp | FIA | LumiraDx SARS-CoV-2 antigen test | LumiraDx UK Ltd., Dumyat Business Park, Alloa, FK10 2PB, UK) | Up to 40 | Rapid/detector | 512 | 123 | 389 |
| Basso et al. [177] | Italy | N | 1. nsp 2. ts |
1. LFIA 2. LFIA 3. CLEIA |
1. ESPLINE rapid test 2. COVID-19 Ag Rapid Test 3. Lumipulse G SARS-CoV-2 Ag |
1. Fujirebio 2. ABBOTT 3. Fujirebio |
Up to 40 | 1. Rapid 2. Rapid 3. Quick/detector |
234 | 87 | 147 |
| Pollock et al. [178] | USA | N | nsp | LFIA | BinaxNOW COVID-19 Ag card | Abbott Diagnostics Scarborough, Inc. | Up to 30/Up to 40/30–40 | Rapid | 2307 | 292 | 2015 |
| Ristić et al. [179] | Serbia | N | nsp | LFIA | STANDARD Q COVID-19 Ag Test | SD Biosensor, Gyeonggi-do, Korea | Up to 40 | Rapid | 120 | 43 | 77 |
| Courtellemont et al. [180] | France | N | nsp | LFIA | COVID-VIRO® | AAZ, Boulogne Billancourt, France | Up to 30/Up to 40/30–40 | Rapid | 248 | 121 | 127 |
| Thommes et al. [181] | Austria | N | nsp | LFIA | 1. Panbio™ COVID-19 Ag Rapid test 2. Novel Coronavirus (2019-nCov) Antigen Detection Kit 3. DIAQUICK COVID-19 Ag Cassette 4. SARS-CoV-2 Rapid Antigen Test |
1. Abbott, Chicago, Illinois 2. CLMSRDL, Sichuan Mass Spectrometry Biotechnology Co., Ltd., Chengdu, Sichuan 3. DIALAB, Wiener Neudorf, Austria 4. Roche Diagnostics Deutschland GmbH, Mannheim, Germany |
Up to 30/Up to 40/30–40 | Rapid | 154 | 154 | NA |
| González-Donapetry et al. [182] | Spain | N | nsp | LFIA | Panbio COVID-19 Ag Rapid Test Device | Abbott Rapid Diagnostics Jena GmbH, Jena, Germany | Up to 40 | Rapid | 440 | 18 | 422 |
| Eshghifar et al. [183] | ? | N | ts | LFIA | 1. BD Veritor™ System for rapid detection of SARS-CoV-2 2. CareStart™ COVID-19 Antigen 3. SG Diagnostics Antigen detection kit 4. Sofia SARS Antigen FIA 5. Rapid Response™ COVID-19 Antigen Rapid Test 6. Shenzhen SARS-CoV-2 Antigen Test kit 7. Genedia W COVID-19 Ag |
1. Becton, Dickinson and Company, MD, USA 2. Accesas Bio, Inc., NJ, USA 3. SG Diagnostics, Singapore 4. Quedel Corporation, Hannover, Germany 5. BNTX, Inc., ON, Canada 6. Shenzhen Ultra-Diagnostics Biotec. Co., Ltd., Shenzhen, PRC 7. Green Cross Medical Sciences Corp., Chungcheongbuk, Republic of Korea |
Up to 40 | Rapid | 5 | 5 | NA |
| Merino et al. [184] | Spain | N | nsp | LFIA | Panbio™ COVID-19 Ag Rapid Test Device | Abbott Diagnostic GmbH, Jena, Germany | Up to 30/Up to 40/30–40 | Rapid | 958 | 359 | 599 |
| Shao et al. [38] | USA | 1. N 2. S |
nsp | FET | In-house | Up to 40 | NA/detector | 38 | 28 | 10 | |
| Bulilete et al. [185] | Spain | N | nsp | LFIA | Panbio™ Ag-RDT | Abbott Diagnostic GmbH, Jena, Germany | Up to 40 | Rapid | 1367 | 140 | 1222 |
| Torres et al. [186] | Spain | N | nsp | LFIA/virus culture data | CLINITEST® Rapid COVID-19 Antigen Test | Siemens, Healthineers, Erlangen, Germany | Up to 40 | Rapid | 270 | 116 | 154 |
| Lindner et al. [187] | Germany | N | nsp | LFIA | STANDARD Q COVID-19 Ag Test | SD Biosensor, Inc., Gyeonggi-do, Korea | Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | Rapid | 179 | 41 | 138 |
| Hirotsu et al. [188] | Japan | N | nsp | CLEIA | LUMIPULSE SARS-CoV-2 antigen test | Fujirebio, Inc., Tokyo, Japan) | Up to 40 | Detector | 1029 | 40 | 989 |
| Salvagno et al. [189] | Italy | N | nsp-ts | LFIA | Roche SARS-CoV-2 Rapid Antigen Test | Roche Diagnostics, Basel, Switzerland | Up to 40 | Rapid | 321 | 149 | 172 |
| Veyrenche et al. [190] | France | N | nsp | LFIA | Coris COVID-19 Ag Respi-Strip | BioConcept | Up to 30/Up to 40/30–40 | Rapid | 65 | 45 | 20 |
| Porte et al. [191] | Chile | N | nsp | FIA | 1. SOFIA SARS Antigen FIA 2. STANDARD F COVID-19 Ag FIA |
1. Quidel Corporation, San Diego, CA, USA 2. SD Biosensor Inc., Gyeonggi-do, Republic of Korea |
Up to 40 | Rapid/detector | 64 | 32 | 32 |
| Domínguez Fernández et al. [192] | Spain | N | nsp | LFIA | Panbio™ rapid antigens test device | Abbott | Up to 40 | Rapid | 30 | 20 | 10 |
| Kobayashi et al. [193] | Japan | N | nsp | 1. CLEIA 2. LFIA |
1. Lumipulse Presto SARS-CoV-2 Ag 2. Espline SARS-CoV-2 |
1. Fujirebio Inc., Tokyo, Japan 2. Fujirebio Inc., Tokyo, Japan |
Up to 40 | 1. Quick/detector 2. Rapid |
300 | 100 | 200 |
| Houston et al. [194] | UK | N | nsp | LFIA | Innova SARS-CoV-2 Antigen Rapid Qualitative Test | Lotus Global Company, London, UK | Up to 40 | Rapid | 728 | 280 | 448 |
| Gremmels et al. [73] | Netherlands/Aruba | N | nsp | LFIA | Panbio™ COVID-19 antigen | Abbott (Lake Country, IL, USA) | Up to 40 | Rapid | 1573 | 202 | 1371 |
| Ciotti et al. [195] | Italy | N | nsp | LFIA | Coris COVID-19 Ag Respi-Strip | Coris BioConcept | Up to 40 | Rapid | 50 | 39 | 11 |
| Okoye et al. [196] | USA | N | nsp | LFIA | Abbott BinaxNOW COVID-19 antigen card | Abbott Diagnostics Scarborough, Inc. | Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | Rapid | 2638 | 45 | 2593 |
| Kurtulmus et al. [47] | Turkey | N | urine | UFT | In-house | Up to 40 | Rapid | 201 | 86 | 115 | |
| Saadi et al. [37] | France | N | nsp | 1. LFIA 2. LFIA 3. LC-MS |
1. NG Test Ag 2. COVID-19 Ag Respi-Strip 3. In-house |
1. NG Biotech, France 2. Coris, Belgium |
Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | 1. Rapid 2. Rapid 3. NA/detector |
19 | 12 | 7 |
| James et al. [197] | USA | N | nsp | LFIA | BinaxNOW COVID-19 Ag Card tests | Abbott Diagnostics, Scarborough | Up to 40 | Rapid | 2339 | 152 | 2187 |
| Villaverde et al. [198] | Spain | N | nsp | LFIA | Panbio COVID-19 Ag Rapid Test | Abbott Rapid Diagnostic | Up to 40 | Rapid | 1620 | 77 | 1543 |
| Pekosz et al. [199] | USA | N | nsp | LFIA/virus culture data | BD Veritor Antigen Test | Becton, Dickinson and Company, BD Life Sciences–, San Diego, California | Up to 40 | Rapid | 38 | 38 | NA |
| Kohmer et al. [200] | Germany | N | nsp | LFIA/virus culture data | 1. RIDA®QUICK SARS-CoV-2 Antigen 2. SARS-CoV-2 Rapid Antigen Test 3. NADAL® COVID-19 Ag Test (test cassette) 4. LumiraDx™ Platform SARS-CoV-2 Ag Test |
1. R-Biopharm AG, Darmstadt, Germany 2. Roche Diagnostics GmbH, Mannheim, Germany 3. Nal von Minden GmbH, Regensburg, Germany 4. LumiraDx GmbH, Cologne, Germany |
Up to 40 | Rapid | 100 | 74 | 26 |
| Prince-Guerra et al. [201] | USA | N | nsp | LFIA/virus culture data | BinaxNOW COVID-19 Ag Card | Abbott Diagnostics Scarborough, Inc. | Up to 40 | Rapid | 3419 | 299 | 3120 |
| Möckel et al. [202] | Germany | N | nsp | LFIA/virus culture data | Roche SARS-CoV-2 rapid antigen test | SD Biosensor | Up to 40 | Rapid | 271 | 89 | 182 |
| Rottenstreich et al. [203] | Israel | N | nsp | LFIA | NowCheck COVID-19 Ag Test | Bionote Inc., Hwaseong-si, Republic of Korea | Up to 30/Up to 40/30–40 | Rapid | 1326 | 9 | 1317 |
| Favresse et al. [204] | Belgium | N | nsp | 1. LFIA 2. LFIA 3. LFIA 4. LFIA 5. CLEIA |
1. Biotical SARS-CoV-2 Ag card 2. Panbio™ COVID-19 Ag Rapid Test Device 3. Coronavirus Ag Rapid Test Cassette 4. Roche SARS-CoV-2 Rapid Antigen Test 5. VITROS Immunodiagnostic Products SARS-CoV-2 Antigen test |
1. Biotical Health, Madrid, Spain 2. Abbott, Chicago, IL, USA 3. Healgen Scientific, Houston, TX, USA 4. Roche Diagnostics, Basel, Switzerland 5. Ortho Clinical Diagnostics, Raritan, NJ, USA |
Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | 1. Rapid 2. Rapid 3. Rapid 4. Rapid 5. Quick/detector |
188 | 96 | 92 |
| Osterman et al. [205] | Germany | N | nsp-ts | 1. LFIA 2. FIA |
1. SARS-CoV-2 Rapid Antigen Test 2. STANDARD™ F COVID-19 Ag |
1. SD Biosensor, Suwon, Korea 2. Roche, Switzerland |
Up to 40 | 1. Rapid 2. Rapid/detector |
1572 | 826 | 746 |
| Pollock et al. [206] | USA | N | nsp | CLEIA/virus culture data | MSD S-PLEX SARS-CoV-2 N assay | MSD Meso Scale Discovery [MSD] | Up to 40 | Quick/detector | 226 | 136 | 90 |
| Aoki et al. [207] | Japan | N | nsp | CLEIA | Lumipulse® SARS-CoV-2 Ag | Fujirebio Inc., Tokyo, Japan | Up to 40 | Quick/detector | 548 | 30 | 518 |
| Torres et al. [208] | Spain | N | nsp | LFIA | Panbio™ COVID-19 Ag | Abbott Diagnostics, Jena, Germany | Up to 40 | Rapid | 634 | 79 | 555 |
| Alemany et al. [209] | Spain | N | nsp | LFIA | Panbio COVID-19 Ag Test | Abbott Rapid Diagnostics, Germany | Up to 30/Up to 40/30–40 | Rapid | 1406 | 951 | 455 |
| Rastawicki et al. [210] | Poland | N | nsp | FIA | PCL COVID-19 Ag | PCL Inc., Korea | Up to 40 | Rapid | 42 | 36 | 6 |
| Yamamoto et al. [211] | Japan | N | nsp | LFIA | ESPLINE SARS-CoV-2 | Fujirebio Inc., Japan | Up to 40 | Rapid | 229 | 128 | 101 |
| Kashiwagi et al. [212] | Japan | N | 1. ts 2. nsp |
LFIA | ESPLINE® SARS-CoV-2 | Fujirebio Inc., Tokyo | Up to 40 | Rapid | 6 | 4 | 2 |
| Pilarowski et al. [213] | USA | N | nsp | LFIA/virus culture data | BinaxNOW rapid antigen test | Abbott Diagnostics Scarborough, Inc. | Up to 30/Up to 40/30–40 | Rapid | 871 | 26 | 845 |
| Aoki et al. [214] | Japan | N | nsp | LFIA | Espline® SARS-CoV-2 | Fujirebio Inc., Japan | Up to 40 | Rapid | 129 | 63 | 66 |
| Pray et al. [215] | Wisconsin | N | nsp | FIA/virus culture data | Sofia SARS Antigen | Quidel Corporation | Up to 40 | Rapid/detector | 1098 | 57 | 1041 |
| Strömer et al. [216] |
Germany | N | nsp | LFIA/virus culture data | 1. NADAL® COVID-19 Ag Test 2. Panbio™ COVID-19 Antigen |
Nal von Minden GmbH, Moers, Germany Abbott Rapid Diagnostics, Germany |
Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | Rapid | 124 | 124 | NA |
| Toptan et al. [217] | Germany | N | nsp-ts | LFIA/virus culture data | novel antigen test | R-Biopharm | Up to 40 | Rapid | 67 | 58 | 9 |
| Turcato et al. [218] | Italy | N | ts | LFIA | STANDARD Q COVID-19 Ag (R-Ag) | SD BIOSENSOR, KR | Up to 40 | Rapid | 3410 | 223 | 3187 |
| Mak et al. [219] | Hong Kong | N | 1. nsp-ts 2. nsp 3. ts |
LFIA/virus culture data | Panbio COVID-19 Ag Rapid Test Device | Abbott Rapid Diagnostics, Germany | Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | Rapid | 35 | 8 | 27 |
| Zhang et al. [220] | China | N | nsp-ts | FIA/virus culture data | SARS-CoV-2 N-protein test strip | Beijing Savant Biotechnology Co., Ltd. | Up to 40 | Rapid/detector | 547 | 247 | 300 |
| Agulló et al. [221] | Spain | N | 1. nsp 2. ts 3. nsp-ts |
LFIA | Panbio COVID-19 Ag-RDT | Abbott Rapid Diagnostic Jena GmbH, Jena, Germany) | Up to 40 | Rapid | 659 | 126 | 527 |
| Tanimoto et al. [222] | Japan | N | nsp | LFIA | ESPLINE SARS-CoV-2® | Fujirebio Inc., Tokyo, Japan | Up to 40 | Rapid | 8 | 2 | 6 |
| Lindner et al. [223] | Germany | N | nsp | LFIA | STANDARD Q COVID-19 Ag Test | SD Biosensor, Inc., Gyeonggi-do, Korea | Up to 20/Up to 30/Up to 40/0–20/20–30/30–40 | Rapid | 39 | 39 | NA |
| Abdelrazik et al. [224] | Egypt | N | nsp | LFIA | BIOCREDIT COVID-19 Ag test | RapiGEN Inc. | Up to 30/Up to 40/30–40 | Rapid | 188 | 188 | NA |
| Weitzel et al. [225] | Chile | N | 1. nsp-ts 2. nsp |
1. LFIA 2. FIA 3. FIA |
1. Biocredit One Step SARS-CoV-2 Antigen Test 2. Huaketai New Coronavirus (SARS-CoV-2) N Protein Detection Kit (FIA) 3. Diagnostic Kit for 2019-Novel Coronavirus (2019-nCoV) |
1. RapiGen Inc., Anyang-si, Gyeonggi-do, Rep. of Korea 2. Savant Biotechnology Co., Beijing, China 3. Bioeasy Biotechnology Co., Shenzhen, China |
Up to 40 | 1. Rapid 2. Rapid/detector 3. Rapid/detector |
111 | 80 | 31 |
| Winkel et al. [226] | Netherlands | N | nsp | LFIA | PanbioTM COVID-19 Ag | Abbott | Up to 40 | Rapid | 2390 | 63 | 2327 |
| Hoehl et al. [227] |
Germany | N | nsp | LFIA | RIDA® QUICK SARS80 CoV-2 Antigen test | R-Biopharm | Up to 20 | Rapid | 602 | 8 | 594 |
| Priya Kannian et al. [228] |
India | N | nsp | LFIA | SARS-CoV2 antigen kit | SD Biosensor | Up to 40 | Rapid | 30 | 20 | 10 |
| Lindner et al. [229] |
Germany | N | nsp | LFIA | STANDARD Q COVID-19 Ag Test | SD Biosensor, Inc., Gyeonggi-do, Korea | Up to 40 | Rapid | 146 | 40 | 106 |
| Filgueiras et al. [230] |
Brazil | N | nsp | LFIA | SARS-CoV-2 rapid antigen test | ECODiagnostica | Up to 40 | Rapid | 139 | 55 | 84 |
| Peto et al. [231] |
UK | N | nsp-ts | LFIA | SARS-CoV-2 Antigen Rapid Qualitative Test | Innova | Up to 30 | Rapid | 834 | 198 | 636 |
| Jakobsen et al. [232] |
Denmark | N | nsp | LFIA | STANDARD Q COVID-19 Ag test | SD BIOSENSOR | Up to 40 | Rapid | 4811 | 221 | 4590 |
| Miyakawa et al. [233] |
Japan | N | nsp | LFIA/virus culture data | 1. SARS-CoV-2 Ag-RDT 2. Panbio COVID-19 Ag Rapid Test 3. SARS-CoV-2 Rapid Antigen Test 4. SD Biosensor Standard Q COVID-19 Ag 5. Espline SARS-CoV-2 |
1. YCU-FF 2. Abbott 3. Roche 4. SD Bio 5. Fujirebio |
Up to 40 | Rapid | 108 | 45 | 63 |
| Torres et al. [186] | Spain | N | nsp | LFIA/virus culture data | CLINITEST® Rapid 29 COVID-19 Antigen Test | Siemens, Healthineers, Erlangen, German | Up to 40 | Rapid | 270 | 33 | 237 |
| Pollock et al. [234] | Massachusetts | N | nsp | LFIA | Access Bio CareStart COVID-19 Antigen test | Up to 30/Up to 40 | Rapid | 1498 | 234 | 1264 | |
| Shidlovskaya et al. [235] |
Russia | N | nsp | LFIA/virus culture data | 1. SGTI-flex COVID-19 Ag 2. Biocredit COVID-19 Ag |
1. SUGENTECH, INC 2. RapiGEN Inc. |
Up to 40 | Rapid | 106 | 14 | 92 |
| Faíco-Filho et al. [236] |
Brazil | N | nsp | LFIA | Panbio™ COVID-19 Ag Rapid Test | Abbott | Up to 30/Up to 40/30–40 | Rapid | 127 | 70 | 57 |
| Schuit et al. [237] |
Netherlands | N | nsp | LFIA/virus culture data | 1. BD VeritorTM System Ag-RDT 2. SD Biosensor Ag-RDT |
1. Becton, Dickinson and Company, Franklin Lakes, NJ, USA 2. Roche |
Up to 40 | Rapid | 4274 | 365 | 4274 |
| Ducrest et al. [238] |
Switzerland | N | nsp | LFIA | COVIDia-Antigen | GaDia SA | Up to 30 | Rapid | 60 | 20 | 40 |
| Vecchio et al. [239] |
Italy | N | nsp | LFIA | Panbio™ COVID-19 Ag test | Abbott | Up to 30 | Rapid | 1441 | 61 | 1380 |
| Bonde et al. [240] |
Denmark | N | ts | LFIA | BD VERITOR Ag Rapid test | Becton-Dickinson and Company, USA | Up to 30 | Rapid | 809 | 65 | 744 |
| Igloi et al. [241] |
Netherlands | N | ts | LFIA/virus culture data | SARS-CoV-2 Rapid Antigen Test | Distributed by Roche (SD Biosensor) | Up to 30 | Rapid | 770 | 30 | 740 |
| Thell et al. [242] | Austria | N | nsp | LFIA | SARS-CoV-2 Rapid Antigen Test | Roche Diagnostics | Up to 30 | Rapid | 541 | 213 | 328 |
| Pollock et al. [243] |
Massachusetts | N | nsp | LFIA | BinaxNOW COVID-19 Ag | Abbott | Up to 30 | Rapid | 98 | 98 | NA |
| Hagbom et al. [244] |
Sweden | N | ts | LFIA/virus culture data | 1. Rapid Response™ COVID-19 Antigen Rapid Test Cassette for oral fluids 2. DIAGNOS™ COVID-19 Antigen Saliva Test |
1. BioServ 2. DIAGNOS |
Up to 30 | Rapid | 34 | 15 | 19 |
| Thirion-Romero et al. [245] |
Mexico | N | nsp | LFIA | Panbio™ | Abbott | Up to 30 | Rapid | 1064 | 474 | 590 |
| Chiu et al. [246] |
Hong Kong | N | nsp | LFIA | INDICAID™ Rapid Test | PHASE Scientific i | Up to 30 | Rapid | 23,343 | 128 | 23,215 |
| Abusrewil et al. [247] | Libya | N | nsp | LFIA | 1. SARS-CoV-2 spike protein test 2. Shenzhen Microprofit Biotech Co 3. ESPLINE SARS-CoV-2 4. RapiGen COVID-19 Ag Detection Kit 5. Panbio™ COVID-19 Ag Rapid Test 6. Flowflex™ SARS-CoV-2 Antigen Rapid Test 7. Europe antigen testing COVID-19 8. Bioperfectus SARSCoV-2 Antigen Rapid Test Kit 9. AMP Rapid Test SARS-CoV-2 Ag 10. Coronavirus ag rapid test cassette |
1. Fluorecare 2. Biotech 3. Fujirebio 4. Biocredit 5. Abbott 6. Acon 7. Assut 8. BIOPERFECTUS 9. AMP 10. Orient GENE |
Up to 30/Up to 40 | Rapid | 231 | 83 | 145 |
| Muthamia et al. [248] |
Kenya | N | nsp | LFIA | BD Veritor antigen test | Becton-Dickinson and Company, USA | Up to 20/Up to 30/0–20/20–30 | Rapid | 272 | 47 | 225 |
| Abdul-Mumin et al. [249] |
Ghana | N | nsp | LFIA | STANDARD Q SARS-CoV-2 Ag Test | SD Biosensor | Up to 40 | Rapid | 193 | 42 | 151 |
| Akashi et al. [250] | Japan | N | nsp | LFIA | QuickNavi™-COVID19 Ag | Otsuka Pharmaceutical Co., Ltd. (Otsuka) and Denka Company | Up to 40 | Rapid | 96 | 96 | NA |
| Lindner et al. [251] |
Germany | N | nsp | LFIA | 1. Espline SARS-CoV-2 2. Sure Status COVID-19 Antigen Card Test 3. Mologic COVID-19 Rapid Test |
1. Fujirebio Inc. 2. Premier Medical Corporation Private Limited 3. Fujirebio Inc |
Up to 40 | Rapid | 329 | 329 | NA |
| Suliman et al. [252] |
Massachusetts | N | nsp | LFIA | Access Bio CareStart™ COVID-19 RDT | CareStart | Up to 30 | Rapid | 631 | 37 | 594 |
| Bruins et al. [253] |
Netherlands | N | nsp | LFIA | Panbio™ COVID-19 Ag Rapid Test | Abbott | Up to 30 | Rapid | 1101 | 84 | 917 |
| Ford et al. [254] |
Wisconsin | N | nsp | LFIA/virus culture data | BinaxNOW SARS-CoV-2 antigen test | Abbott Laboratories, Abbott Park, IL | Up to 40 | Rapid | 2110 | 334 | 1776 |
| Koskinen et al. [255] |
Finland | N | nsp | LFIA/virus culture data | mariPOC SARS-CoV-2 Antigen Test | mariPOC | Up to 30 | Rapid/optional detector | 211 | 13 | 198 |
| Nikolai et al. [256] | Germany | N | nsp | LFIA | STANDARD Q COVID-19 Ag Test | SD Biosensor, Inc. Gyeonggi-do, Korea | Up to 40 | Rapid | 228 | 70 | 188 |
| Stohr et al. [257] |
Netherlands | N | nsp | LFIA/virus culture data | 1. BD Veritor System for Rapid Detection of SARS-CoV-2 2. Roche SARS-CoV-2 antigen detection test |
Becton Dickinson company, USA Roche, Switzerland |
Up to 40 | Rapid | 3239 | 454 | 1528 |
LFIA: Lateral Flow Immunoassay; FIA: Fluorescence Immunoassay; CLEIA: Chemiluminescence Enzyme Immunoassay; FET: Field-Effect Transistors; Ag: Antigen; nsp: nasopharengeal; ts: oropharyngeal/throat/saliva; Rapid: detection time 5–20 min (mainly 15) but never exceeding 30 min; Quick: detection time 30–35 min; Quick *: 60 min; w/wo: with/without; Detector: a detector in needed to read the developed signal; NA: Not applicable; NR: Not reported; Cases: SARS-CoV-2 positive samples according to RT-PCR; Controls: healthy individuals and RT-PCR negative (for SARS-CoV-2); Virus culture data: study that provides any kind of data on the correlation between virus culture [cytopathic effect, tissue culture infective dose 50% (TCID 50), limit of detection (LoD)], and rapid Antigen Test positivity, RNA copies number, Ct values of RT-PCR positive samples.
3.2. Analysis of Diagnostic Performance
A great amount of the available data, for all methods, concerned samples detected with qPCR Ct values of 20, and mostly of 30 and 40. As shown in Table 2, the sensitivity of LFIA tests (using the N antigen) based on NSP samples that were qPCR-positive for Ct < 20 was 0.945 (95% CI: 0.930, 0.961). It declined, however, considerably to 0.329 (95% CI: 0.265, 0.393) for 30 < Ct < 40. LFIA tests using TS samples performed worse in terms of sensitivity, with a highest estimate of 0.805 (95% CI: 0.599, 1.000) in samples positive for Ct < 20 and a lowest of 0.085 (0.000, 0.176) for Ct > 30 (Table 2). The specificity of LFIA on NSP and TS samples (using the N antigen) was very high across all Ct intervals, ranging from 0.959 (95% CI: 0.923, 0.995) to 0.996 (95% CI: 0.993, 0.998). The sensitivity of FIA (using the N antigen) on NSP samples also showed a declining pattern from 0.935 (95% CI: 0.880, 0.990) for Ct < 20 to 0.435 (95% CI: 0.190, 0.680) for 30 < Ct < 40. Specificity was also very high using NSP qPCR positive samples for Ct < 30 (0.992, 95%: 0.979, 1.000). CLEIA (using the N antigen) had high sensitivity based on NSP samples that were PCR-positive for Ct < 30 (0.980, 95% CI: 0.960, 0.999); this estimate, however, was based on a smaller number of studies and dropped considerably at higher Ct (30–40) values (0.515; 95% CI: 0.220, 0.810). The specificity of CLEIA was very high in all comparisons. The evaluation of the performance of other methods (using the N antigen) on NSP and TS samples for the above studied Ct values intervals (0–20, 21–30, and 31–40) was based on a few studies but showed similar patterns. Data on methods using other antigens (i.e., based on S, E or M protein) were too scarce to allow reliable estimations (Table 2).
Table 2.
Results of the multivariate meta-analysis for the different types of assays using different samples and stratified according to different cut-off rt-PCR values. Listed information includes the pooled sensitivity and specificity along with the 95% confidence intervals (NSP: pharyngeal, nasopharyngeal, nasal specimens, TS: throat, saliva, N: nucleocapsid protein, S: spike protein, M: membrane E: envelope, NS: nucleocapsid and Spike proteins).
| Sample | Ag | Method | Ct Values |
Studies/Patients/ Controls |
Sensitivity (95% CI) | Specificity (95% CI) | Studies w/o Controls |
|---|---|---|---|---|---|---|---|
| NSP | N | LFIA | 0–20 | 41/7464/3945 | 0.945 (0.930, 0.961) | 0.993 (0.987, 0.998) | 22 |
| NSP | N | LFIA | 0–30 | 99/66,939/47,719 | 0.853 (0.826, 0.879) | 0.991 (0.988, 0.995) | 44 |
| NSP | N | LFIA | 0–40 | 207/88,008/69,415 | 0.702 (0.676, 0.727) | 0.990 (0.987, 0.993) | 30 |
| NSP | N | LFIA | 20–30 | 46/7817/4360 | 0.790 (0.739, 0.841) | 0.987 (0.976, 0.998) | 35 |
| NSP | N | LFIA | 30–40 | 71/5150/911 | 0.329 (0.265, 0.393) | 0.959 (0.923, 0.995) | 51 |
| TS | N | LFIA | 0–20 | 5/90/NA | 0.805 (0.599, 1.000) | - | 5 |
| TS | N | LFIA | 0–30 | 10/2136/1756 | 0.636 (0.477, 0.795) | 0.994 (0.989, 0.998) | 5 |
| TS | N | LFIA | 0–40 | 23/10,249/9232 | 0.354 (0.238, 0.470) | 0.996 (0.993, 0.998) | 12 |
| TS | N | LFIA | 20–30 | 6/160/NA | 0.394 (0.086, 0.702) | - | 6 |
| TS | N | LFIA | 30–40 | 4/44/NA | 0.085 (0.000, 0.176) | - | 4 |
| NSP-TS | N | LFIA | 0–20 | 7/4240/3859 | 0.999 (0.000, 1.000) | 0.999 (0.000, 1.000) | 6 |
| NSP-TS | N | LFIA | 0–30 | 12/9229/8133 | 0.867 (0.792, 0.942) | 0.999 (0.997, 1.000) | 10 |
| NSP-TS | N | LFIA | 0–40 | 30/23,970/21,699 | 0.696 (0.638, 0.754) | 0.992 (0.987, 0.996) | 4 |
| NSP-TS | N | LFIA | 20–30 | 10/1995/1504 | 0.575 (0.279, 0.870) | 0.997 (0.987, 1.000) | 7 |
| NSP-TS | N | LFIA | 30–40 | 10/217/NA | 0.417 (0.242, 0.593) | - | 9 |
| NSP | N | FIA | 0–20 | 3/97/NA | 0.935 (0.880, 0.990) | - | 3 |
| NSP | N | FIA | 0–30 | 10/2221/421 | 0.807 (0.726, 0.889) | 0.992 (0.979, 1.000) | 6 |
| NSP | N | FIA | 0–40 | 29/36,425/33,718 | 0.707 (0.631, 0.783) | 0.984 (0.970, 0.997) | 1 |
| NSP | N | FIA | 20–30 | 3/598/NA | 0.729 (0.544, 0.915) | - | 3 |
| NSP | N | FIA | 30–40 | 12/2283/665 | 0.435 (0.190, 0.680) | 0.983 (0.971, 0.995) | 9 |
| TS | N | FIA | 0–40 | 2/114/31 | 0.162 (0.083, 0.241) | 0.984 (0.941, 1.000) | 1 |
| NSP-TS | N | FIA | 0–30 | 4/195/77 | 0.944 (0.904, 0.985) | 0.975 (0.944, 1.000) | 1 |
| NSP-TS | N | FIA | 0–40 | 11/2779/2018 | 0.691 (0.520, 0.862) | 0.971 (0.953, 0.989) | 2 |
| NSP-TS | N | FIA | 30–40 | 3/72/32 | 0.792 (0.434, 1.000) | 0.969 (0.926, 1.000) | 1 |
| NSP | N | CLEIA | 0–20 | 3/789/152 | 0.955 (0.907, 1.000) | 0.997 (0.000, 1.000) | 2 |
| NSP | N | CLEIA | 0–30 | 3/1268/111 | 0.980 (0.960, 0.999) | 0.995 (0.000, 1.000) | 2 |
| NSP | N | CLEIA | 0–40 | 21/7626/5910 | 0.818 (0.774, 0.862) | 0.978 (0.968, 0.988) | 1 |
| NSP | N | CLEIA | 20–30 | 4/378/68 | 0.900 (0.672, 1.000) | 0.986 (0.960, 1.000) | 2 |
| NSP | N | CLEIA | 30–40 | 4/416/261 | 0.515 (0.220, 0.810) | 0.978 (0.957, 0.999) | 2 |
| TS | N | CLEIA | 0–20 | 1/136/NA | 0.875 (0.550, 1.000) | - | 1 |
| TS | N | CLEIA | 0–30 | 1/136/NA | 0.928 (0.738, 1.000) | - | 1 |
| TS | N | CLEIA | 0–40 | 3/376/179 | 0.709 (0.359, 1.000) | 0.977 (0.950, 1.000) | 1 |
| TS | N | CLEIA | 20–30 | 1/3/NA | 0.875 (0.550, 1.000) | - | 1 |
| TS | N | CLEIA | 30–40 | 1/3/NA | 0.667 (0.000, 1.000) | - | 1 |
| NSP-TS | N | CLEIA | 0–40 | 1/4266/3763 | 0.867 (0.837, 0.896) | 0.973 (0.968, 0.978) | 0 |
| NSP-TS | N | CLEIA | 20–30 | 1/978/817 | 0.795 (0.733, 0.857) | 0.997 (0.000, 1.000) | 0 |
| NSP | N | other | 0–20 | 2/45/7 | 0.973 (0.921, 1.000) | 0.9375 (0.769, 1.000) | 1 |
| NSP | N | other | 0–30 | 4/219/51 | 0.923 (0.807, 1.000) | 0.963 (0.890, 1.000) | 1 |
| NSP | N | other | 0–40 | 8/1228/388 | 0.768 (0.643, 0.894) | 0.915 (0.821, 1.000) | 0 |
| NSP | N | other | 20–30 | 2/110/NA | 0.842 (0.422, 1.000) | - | 2 |
| NSP | N | other | 30–40 | 4/73/NA | 0.540 (0.147, 0.934) | - | 4 |
| NSP | S | LFIA | 0–20 | 1/90/49 | 0.976 (0.928, 1.000) | 0.857 (0.000, 1.000) | 0 |
| NSP | S | LFIA | 0–30 | 2/407/234 | 0.783 (0.627, 0.938) | 0.942 (0.833, 1.000) | 0 |
| NSP | S | LFIA | 0–40 | 2/129/54 | 0.848 (0.768, 0.930) | 0.862 (0.771, 0.954) | 0 |
| NSP | S | LFIA | 20–30 | 1/80/49 | 0.677 (0.513, 0.842) | 0.857 (0.000, 1.000) | 0 |
| NSP | S | other | 0–40 | 4/286/207 | 0.872 (0.780, 0.963) | 0.911 (0.761, 1.000) | 0 |
| TS | S | other | 0–40 | 3/96/42 | 0.817 (0.635, 1.000) | 0.931 (0.856, 1.000) | 0 |
| TS | N, S | other | 0–40 | 1/433/397 | 0.986 (0.949, 1.000) | 0.962 (0.943, 0.981) | 0 |
| NSP-TS | S + E + M | other | 0–40 | 1/94/49 | 0.955 (0.895, 1.000) | 0.959 (0.904, 1.000) | 0 |
| URINE | N, S | other, FIA | 0–40 | 3/271/145 | 0.715 (0.310, 1.000) | 0.869 (0.647, 1.000) | 0 |
Combining all major methods (LFIA, FIA and CLEIA) on NSP and TS samples, measuring both N and S antigens and stratified according to two Ct values (<30 and <40), the maximum sensitivity was estimated at 0.858 (95% CI 0.835, 0.881) for NSP samples positive for Ct < 30 (Table 3). The sensitivity using qPCR positive NSP samples for Ct < 40 is lower at 0.726 (95% CI 0.706, 0.746). Again, antigen testing of NSP samples outperformed that of TS samples for both Ct < 30 and Ct < 40 (0.637 (95% CI: 0.478, 0.795) and 0.438 (95% CI: 0.332, 0.547), respectively). Specificity was very high in all meta-analyses (Table 3).
Table 3.
Results of the multivariate meta-analysis performed cumulatively for methods and/or antigen tested, in <30 and <40 Ct values. Listed information includes the pooled sensitivity and specificity along with the 95% confidence intervals (NSP: pharyngeal, nasopharyngeal, nasal specimens, TS: throat, saliva, oropharyngeal, N: nucleocapsid protein, S: spike protein, M: membrane E: envelope, NS: nucleocapsid and Spike proteins).
| Sample | Ag | Method (LFIA, FIA, CLEIA) | Ct Values | Studies | Sensitivity (95% CI) | Specificity (95% CI) | Studies w/o Controls |
|---|---|---|---|---|---|---|---|
| NSP | NS | LFIA or FIA or CLEIA | 30 | 118 | 0.858 (0.835, 0.881) | 0.991 (0.987, 0.995) | 53 |
| NSP | NS | LFIA or FIA or CLEIA | 40 | 325 | 0.726 (0.706, 0.746) | 0.989 (0.987, 0.992) | 39 |
| TS | NS | LFIA or FIA or CLEIA | 30 | 10 | 0.637 (0.478, 0.795) | 0.994 (0.989, 0.998) | 5 |
| TS | NS | LFIA or FIA or CLEIA | 40 | 36 | 0.438 (0.332, 0.547) | 0.993 (0.987, 0.999) | 14 |
| NSP | NS | LFIA or FIA | 30 | 114 | 0.854 (0.830, 0.878) | 0.991 (0.987, 0.995) | 50 |
| NSP | NS | LFIA or FIA | 40 | 303 | 0.718 (0.697, 0.739) | 0.989 (0.987, 0.992) | 38 |
| TS | NS | LFIA or FIA | 30 | 10 | 0.637 (0.478, 0.795) | 0.994 (0.989, 0.998) | 5 |
| TS | NS | LFIA or FIA | 40 | 32 | 0.395 (0.285, 0.505) | 0.995 (0.993, 0.997) | 13 |
| NSP | NS | LFIA | 30 | 101 | 0.852 (0.825, 0.878) | 0.991 (0.987, 0.995) | 44 |
| NSP | NS | LFIA | 40 | 269 | 0.715 (0.692, 0.738) | 0.990 (0.987, 0.992) | 35 |
| TS | NS | LFIA | 30 | 10 | 0.637 (0.478, 0.795) | 0.994 (0.989, 0.998) | 5 |
| TS | NS | LFIA | 40 | 29 | 0.408 (0.292, 0.523) | 0.995 (0.993, 0.997) | 12 |
| NSP | NS | FIA | 30 | 13 | 0.868 (0.813, 0.924) | 0.991 (0.981, 1.000) | 6 |
| NSP | NS | FIA | 40 | 35 | 0.730 (0.674, 0.785) | 0.986 (0.976, 0.995) | 3 |
| TS | NS | FIA | 30 | - | - | - | - |
| TS | NS | FIA | 40 | 2 | 0.162 (0.083, 0.242) | 0.984 (0.941, 1.000) | 1 |
| NSP | NS | CLEIA | 30 | 4 | 0.977 (0.955, 0.998) | 0.995 (0.000, 1.000) | 3 |
| NSP | NS | CLEIA | 40 | 23 | 0.816 (0.761, 0.870) | 0.979 (0.971, 0.988) | 1 |
| TS | NS | CLEIA | 30 | - | - | - | - |
| TS | NS | CLEIA | 40 | 3 | 0.720 (0.380, 1.000) | 0.957 (0.889, 1.000) | 1 |
To attain a better insight into how each method performs, we compared the meta-analysis results for the sensitivity and specificity of each method (LFIA, FIA, CLEIA) on NSP and TS samples for all antigens cumulatively (N plus S). As shown in Table 3, in terms of sensitivity, the laboratory CLEIA method outperforms the point of care (POC) methods (LFIA and FIA), the NSP samples outperform the TS samples, and the best results are obtained for samples identified positive with PCR for Ct < 30 (0.977 (95% CI: 0.955, 0.998) versus 0.408 (95% CI: 0.292, 0.523) and 0.162 (95% CI: 0.083, 0.242)) (Table 3).
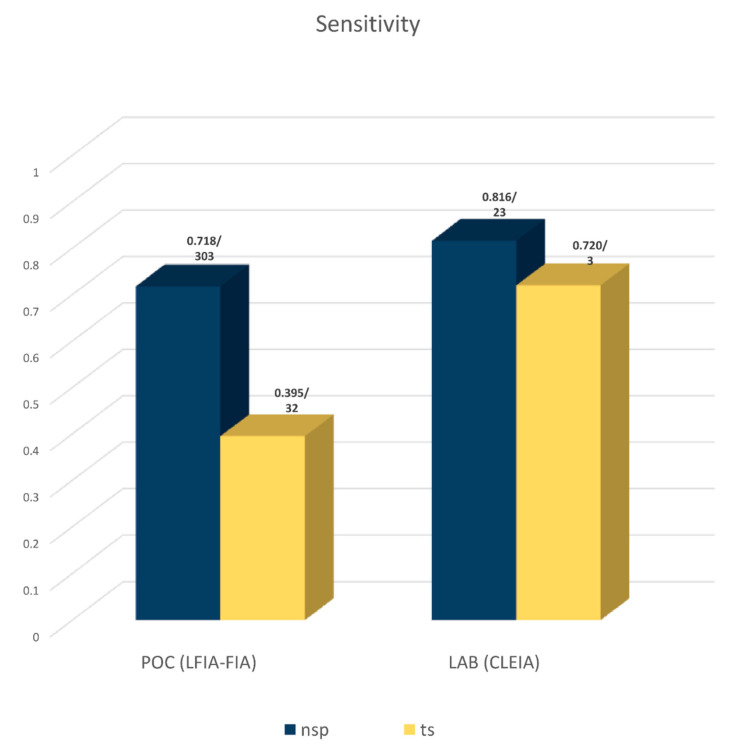
Since the ultimate goal of a diagnostic method for SARS-CoV-2 is to identify an infected person regardless of the low viral load, we compared the overall sensitivity of rapid tests performed in points either of care or where virus surveillance is performed (LFIA or FIA) with laboratory methods (CLEIA) that show the highest sensitivity. As shown in Figure 2 (and Table 3), the overall (for Ct < 40) sensitivity of POC methods is about 10% lower than that of the CLEIA method for NSP samples (0.718 (95% CI: 0.697, 0.739) compared to 0.816 (95% CI: 0.761, 0.870)). Specificity was again high in all cases ranging from 0.957 (95% CI: 0.889, 1.000) to 0.995 (95% CI: 0.993, 0.997), although due to the small number of the included studies in some subgroups, these results may have some uncertainty (Table 3).
Figure 2.
Performance of POC (LFIA and FIA) and laboratory (CLEIA) antigen-based methods in terms of sensitivity. All included assays in the meta-analysis use samples with Ct < 40 and test cumulatively both the nucleocapsid and Spike antigen. Numbers above the bars depict sensitivity values/number of studies included in each meta-analysis.
To investigate the validity of our stratification analysis according to Ct values (<30 and <40), we tried to explore the association between a patient/sample’s infectivity and positivity in POC antigen tests (LFIA and FIA) and PCR tests using data from the included studies. We found 51 studies (Table 1) that used a virus culture to address this issue; however, the results were presented in a plethora of different ways and could not be quantitatively synthesized and analyzed, due to different reported parameters. From them, ten studies used virus cultures to only test the viral load (RNA copies/mL) that a POC test could detect. The remaining 34 studies presented a combination of data such as the limit of detection (LoD) in terms of RNA copies/mL or per swab or in pfus/mL, tissue culture infection dose (TCID), TCID50, TCID95%, sensitivity of POC tests in correlation with virus culture cytopathic effect (CPE) measured in different days and after zero, one or two passages. Nevertheless, sixteen studies [63,85,87,91,101,135,145,151,167,169,199,215,216,217,219,255] determined LoD Ct values ranging from 18.57 [219] to 34 [145], with most of them reporting Ct 30 as an average threshold for a POC test to be positive. Importantly, viral culture positivity (CPE), though measured under various protocols (directly [87,91,101,135,143,145,200,216,241] and indirectly [141,169,201,215,241,254]), has been extensively used as a marker for sample infectivity. Furthermore, twelve studies [54,76,85,143,170,199,213,217,233,235,237,241] presented data providing LoD values for a POC tests ranging from 5.103 (Ct = 27.3 [63]) to 106 RNA copies/swab (Ct = 30) [54,76]. Noteworthily, four studies on the CLEIA method [111,150,156,206] and four studies [41,44,46,47]) on in-house tests also investigated virus infectivity in correlation with either Ct values or positivity of these tests, but these were not analyzed since they were not reporting on POC tests. Taken together, the above observations suggest that if SARS-CoV-2-infected cell culture positivity is an indicator of a patient/sample that is likely to be infectious [202,258,259], this infectivity better correlates with POC test positivity than rt-PCR positivity. As we show herein, POC test positivity corresponds better to PCR positivity for Ct < 30; thus, POC tests are more likely to detect infectious individuals than positive PCR tests.
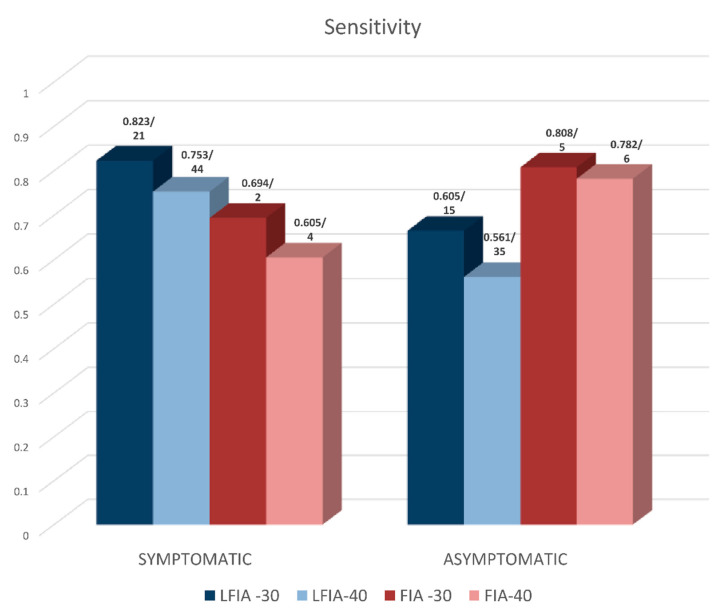
Additional meta-analysis showed that the sensitivity of LFIA (on NSP samples) in symptomatic patients was higher than that in asymptomatic individuals, both for Ct < 30 and Ct < 40 (symptomatic: 0.823 (95% CI: 0.765, 0.882) and 0.753 (95% CI: 0.713, 0.794)—asymptomatic: 0.665 (0.558, 0.772) and 0.561 (95% CI: 0.499, 0.622), respectively) (Table 4 and Figure 3). FIA assays seem to perform worse, but the meta-analysis estimates were based on a smaller number of studies. Specificity was very high for both LFIA and FIA methods (~99%) (Table 4).
Table 4.
Results of the meta-analysis for the different types of assays for symptomatic and asymptomatic patients. Listed information includes the pooled sensitivity and specificity along with the 95% confidence intervals. (NSP: pharyngeal, nasopharyngeal, nasal specimens, TS: throat, saliva, N: nucleocapsid protein, S: spike protein, NS: nucleocapsid and Spike proteins).
| Sample | Ag | Method | Ct | Studies | Sensitivity (95% CI) | Specificity (95% CI) | Studies w/o Controls |
|---|---|---|---|---|---|---|---|
| SYMPTOMATIC INDIVIDUALS | |||||||
| NSP | N | LFIA | 20 | 1 | 0.976 (0.911, 1.000) | - | 1 |
| NSP | N | LFIA | 30 | 21 | 0.823 (0.765, 0.882) | 0.993 (0.989, 0.997) | 7 |
| NSP | N | LFIA | 40 | 44 | 0.753 (0.713, 0.794) | 0.992 (0.987, 0.997) | 7 |
| NSP | N | LFIA | 20–30 | 2 | 0.881 (0.765, 0.996) | - | 2 |
| NSP | N | LFIA | 30–40 | 13 | 0.469 (0.228, 0.709) | 0.947 (0.880, 1.000) | 4 |
| NSP | N | FIA | 30 | 2 | 0.694 (0.509, 0.878) | 0.996 (0.993, 0.998) | 0 |
| NSP | N | FIA | 40 | 4 | 0.605 (0.292, 0.918) | 0.948 (0.827, 1.000) | 1 |
| NSP | N | FIA | 30–40 | 1 | 0.921 (0.868, 0.973) | 0.923 (0.000, 1.000) | 0 |
| TS | N | LFIA | 30 | 2 | 0.669 (0.119, 1.000) | 0.998 (0.994, 1.000) | 0 |
| TS | N | LFIA | 40 | 4 | 0.426 (0.029, 0.823) | 0.986 (0.977, 0.996) | 0 |
| TS | N | LFIA | 30–40 | 1 | 0.025 (0.000, 1.000) | 0.5 (0.000, 1.000) | 0 |
| TS | N | FIA | 40 | 1 | 0.083 (0.000, 1.000) | - | 1 |
| NSP-TS | N | LFIA | 20 | 2 | 0.957 (0.889, 1.000) | - | 2 |
| NSP-TS | N | LFIA | 30 | 4 | 0.873 (0.788, 0.958) | 0.998 (0.993, 1.000) | 3 |
| NSP-TS | N | LFIA | 40 | 11 | 0.767 (0.695, 0.836) | 0.996 (0.992, 0.999) | 3 |
| NSP-TS | N | LFIA | 20–30 | 2 | 0.901 (0.795, 1.000) | - | 2 |
| NSP-TS | N | LFIA | 30–40 | 4 | 0.260 (0.142, 0.378) | 0.500 (0.000, 1.000) | 3 |
| ASYMPTOMATIC INDIVIDUALS | |||||||
| NSP | N | LFIA | 30 | 15 | 0.665 (0.558, 0.772) | 0.992 (0.981, 1.000) | 6 |
| NSP | N | LFIA | 40 | 35 | 0.561 (0.499, 0.622) | 0.995 (0.992, 0.998) | 5 |
| NSP | N | LFIA | 20–30 | 1 | 0.371 (0.270, 0.471) | - | 1 |
| NSP | N | LFIA | 30–40 | 10 | 0.233 (0.061, 0.405) | 0.947 (0.880, 1.000) | 6 |
| NSP | N | FIA | 30 | 5 | 0.808 (0.714, 0.901) | 0.997 (0.989, 1.000) | 3 |
| NSP | N | FIA | 40 | 6 | 0.782 (0.614, 0.949) | 0.949 (0.904, 0.995) | 1 |
| NSP | N | FIA | 30–40 | 2 | 0.734 (0.253, 1.000) | 0.882 (0.774, 0.991) | 1 |
| TS | N | LFIA | 30 | 2 | 0.484 (0.000, 1.000) | 0.995 (0.986, 1.000) | 0 |
| TS | N | LFIA | 40 | 9 | 0.167 (0.034, 0.301) | 0.990 (0.974, 1.000) | 6 |
| TS | N | LFIA | 30–40 | 1 | 0.050 (0.000, 0.185) | 0.5 (0.000, 1.000) | 0 |
| TS | N | FIA | 40 | 1 | 0.166 (0.000, 1.000) | 0.984 (0.941, 1.000) | 0 |
| NSP-TS | N | LFIA | 30 | 1 | 0.300 (0.136, 0.464) | 0.997 (0.000, 1.000) | 0 |
| NSP-TS | N | LFIA | 40 | 5 | 0.481 (0.291, 0.671) | 0.997 (0.995, 0.998) | 1 |
| NSP-TS | N | LFIA | 30–40 | 1 | 0.050 (0.000, 0.185) | 0.997 (0.000, 1.000) | 0 |
| NSP-TS | N | FIA | 40 | 1 | 0.850 (0.772, 0.928) | 0.984 (0.941, 1.000) | 0 |
Figure 3.
Performance of LFIA and FIA methods (N antigen-based) in terms of sensitivity on NSP samples in symptomatic vs. asymptomatic persons. Included assays in the meta-analysis are performed with positive samples for either Ct < 30 or Ct < 40. Numbers above the bars depict sensitivity values/number of studies included in each meta-analysis.
4. Discussion
Test-trace-isolate remains a fundamental strategy to control SARS-CoV-2 transmission. Compared to PCR methods, antigen detection tests do not require specialized laboratory equipment and are less expensive, thus allowing repeated and point-of-care testing on a wide scale [18]. Our meta-analysis, summarizing evidence from thousands of people with and without SARS-CoV-2 infection diagnosed with rt-PCR, and performing various comparisons, shows that the overall performance of AT is comparable to rt-PCR, at least in terms of specificity, with meta-analytic estimates around 99%, irrespective of the method used. Sensitivity is lower and seems to depend on viral concentration being increased if detected at lower PCR cycles (Ct values). AT are also more sensitive when used on NSP samples and in symptomatic individuals. These updated findings are in accordance with previous efforts to summarize the evidence in this field [260,261]. Current best practices in meta-analysis suggest that a frequent update should be performed, and there is active research regarding the identification of the actual time that an update is needed [262,263]. As a matter of fact, previous works include statistical methods and surveillance systems that will identify the need for an update of a published meta-analysis [264,265]. More recently, the concept of a “living” systematic review has emerged, in which the review is continuously updated, incorporating relevant new data as they become available. Such reviews may be particularly important in fields where research evidence is emerging rapidly [266,267], and clearly, the COVID-19 pandemic is a perfect example of a field where new research accumulates in an unprecedented way and an updated meta-analysis is needed.
The sensitivity of AT is good but not ideal, and thus rt-PCR remains the gold standard for diagnosis. Given the suboptimal sensitivity of antigen tests, there is a likelihood of false negative results, which should be handled depending on the clinical and epidemiological circumstances. In general, confirmation of an AT result with rt-PCR in a laboratory is necessary when the result is not consistent with clinical and epidemiological information. Given their higher sensitivity among symptomatic people and in those with higher viral load (Ct < 30), ATs are expected to perform better when used for the diagnosis of SARS-CoV-2 infection in people with symptoms, in high-risk contacts of confirmed cases or in high-risk groups as health care workers with known exposure. Moreover, the sole detection of viral RNA with rt-PCR does not seem to overlap with patients’ infectiousness. Rather, POC (rapid) antigen tests that can only detect viral loads detectable with rt-PCR at Ct values <30 seem to more efficiently discriminate infectious SARS-CoV-2 carriers that should stay in isolation [202,255,258,259]. These findings are further supported by CDC recommendations, already posed by the end of 2020, which propose a Ct value of 33 as illustrative of contagiousness [204,268].
Proper interpretation of AT results is important not only for diagnosis but also for screening and surveillance purposes. This meta-analysis did not evaluate screening strategies that used AT. Nevertheless, it seems that AT can be used for regular screening of asymptomatic people in high-risk congregate settings, such as nursing homes, homeless shelters, detention facilities, etc., where the turnaround time of results is critical [269]. The fast identification of highly infected people in these facilities using rapid POC antigen tests will immediately inform infection prevention and control strategies and interventions, and consequently will significantly reduce onward transmission. Due to the lower sensitivity, screening in congregate high-risk settings but also mass screening may suffer from false negative results. Given the presumed direct correlation of rapid ATs’ positivity with patient’s infectivity, and the evidence that the effectiveness of screening depends more on frequency of testing and speed of reporting rather than on very high sensitivity [91,270], it seems that antigen tests can be used for repeated population screening.
In terms of specificity, AT performs extremely well, similarly to rt-PCR, thus minimizing the likelihood of false-positive results. However, false-positive results do occur, especially when the prevalence of SARS-CoV-2 infection in communities is low. This should be considered both in terms of diagnosis and when designing public health interventions or prevalence studies in low-prevalence settings because false positives result in a waste of resources (unnecessary isolation of cases and follow-up actions) and inaccurate estimations.
This meta-analysis is subject to the limitations of the individual studies. Bias and confounding at the study level cannot be easily addressed or corrected at the stage of meta-analysis. There are also issues that could affect the results and are usually not measured, reported, or addressed in studies that evaluate the accuracy of AT: storage and handling, reading of test results (time and interpretation), specimen collection and handling, time from specimen collection to testing, temperature of specimen, and potential cross-contamination, as was shown in the quality assessment of the research performed with the QUADAS tool.
We need to emphasize that the studies included in this meta-analysis were conducted before July 2021. Thus, data collection was completed at a time prior to the emergence of the Omicron variant and thus, the conclusions drawn from this work involve mainly the initial Wuhan strain, Alpha, Beta and Delta (to some extent) variants. A complete treatment of the question regarding the effectiveness of antigen tests against the newly emerged Omicron variant [271] would require a study of its own, but nevertheless we might be able to highlight some of the available evidence. Initially, there were concerns regarding the effectiveness of the tests [272], but the first report with the Abbott BinaxNow SARS-CoV-2 Rapid Antigen Assay provided evidence that it can be used efficiently [273]. Similar results were reported with another approved test (E25Bio, Inc., Cambridge, MA, USA, and Perkin Elmer, Waltham, MA, USA) in a comparison study of the Alpha, Gamma, Delta and Omicron variants [274], and for Panbio™ COVID-19 Ag Rapid Test [275]. Stanley and coworkers examined the analytical sensitivity of the Abbott BinaxNow, the AccessBio CareStart and LumiraDx antigen tests, and found that the level of detection was at least as good for Omicron as for the initial Wuhan strain [276]. Finally, Deerain and coworkers measured the sensitivity of ten different lateral flow devices against the omicron variant and found that the analytical sensitivities of these ten kits were similar for both the Delta and Omicron variants [277]. All in all, even though more studies are needed, the available evidence suggests that the currently used ATs can be used efficiently for detecting the Omicron variant and large discrepancies in sensitivity due to its spread are not expected.
Finally, evaluation of different testing strategies in various settings is also urgently needed [278]. Moreover, the lack of an agreed, universal, standardized protocol starting from specimen collection and handling to performing and reading the test and to the way(s) that its performance is validated (rt-PCR (genes, Ct values) or cytopathic effects of virus cultures (reference virus strain) or RNA copies, etc. [140,279]) has also been revealed through our current systematic review and meta-analysis. Only in such uniform settings can accurate comparisons of methods and individual tests be performed in order to optimally track and manage SARS-CoV-2 infection in the global community.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/diagnostics12061388/s1, Table S1: The QUADAS tool; File S1: QUADAS-2 assessment results for included studies.
Author Contributions
Conceptualization: P.G.B., G.G.B. and P.I.K.; Methodology: P.I.K., G.K.N., P.G.B. and P.G.B.; Validation: G.G.B., G.K.N., P.I.K., A.T., M.P., H.M. and P.G.B.; Formal Analysis: A.T., P.G.B., G.G.B., P.I.K. and G.K.N.; Investigation: A.T., H.M., M.P., P.I.K. and G.G.B.; Resources, A.T. and G.G.B.; Data Curation: A.T., G.G.B. and P.I.K.; Writing—Original Draft Preparation: A.T. and G.G.B.; Writing—Review and Editing, P.I.K., H.M., M.P., G.K.N. and P.G.B.; Visualization: G.G.B. and P.I.K.; Supervision: G.G.B. and P.G.B. All authors have read and agreed to the published version of the manuscript.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research received no external funding.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Estimating excess mortality due to the COVID-19 pandemic: A systematic analysis of COVID-19-related mortality, 2020–2021. Lancet. 2022;399:1513–1536. doi: 10.1016/S0140-6736(21)02796-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.IHME Institute for Health Metrics and Evaluation—COVID-19 Results Briefing. [(accessed on 30 March 2022)]. Available online: https://www.healthdata.org/COVID/updates.
- 3.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heath P.T., Galiza E.P., Baxter D.N., Boffito M., Browne D., Burns F., Chadwick D.R., Clark R., Cosgrove C., Galloway J., et al. Safety and Efficacy of NVX-CoV2373 COVID-19 Vaccine. N. Engl. J. Med. 2021;385:1172–1183. doi: 10.1056/NEJMoa2107659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyams C., Marlow R., Maseko Z., King J., Ward L., Fox K., Heath R., Tuner A., Friedrich Z., Morrison L., et al. Effectiveness of BNT162b2 and ChAdOx1 nCoV-19 COVID-19 vaccination at preventing hospitalisations in people aged at least 80 years: A test-negative, case-control study. Lancet Infect. Dis. 2021;21:1539–1548. doi: 10.1016/S1473-3099(21)00330-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: An interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor C.A., Whitaker M., Anglin O., Milucky J., Patel K., Pham H., Chai S.J., Alden N.B., Yousey-Hindes K., Anderson E.J., et al. COVID-19-Associated Hospitalizations Among Adults During SARS-CoV-2 Delta and Omicron Variant Predominance, by Race/Ethnicity and Vaccination Status—COVID-NET, 14 States, July 2021–January 2022. MMWR Morb. Mortal Wkly. Rep. 2022;71:466–473. doi: 10.15585/mmwr.mm7112e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chemaitelly H., Ayoub H.H., AlMukdad S., Coyle P., Tang P., Yassine H.M., Al-Khatib H.A., Smatti M.K., Hasan M.R., Al-Kanaani Z., et al. Duration of mRNA vaccine protection against SARS-CoV-2 Omicron BA.1 and BA.2 subvariants in Qatar. medRxiv. 2022 doi: 10.1038/s41467-022-30895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flaxman S., Mishra S., Gandy A., Unwin H.J.T., Mellan T.A., Coupland H., Whittaker C., Zhu H., Berah T., Eaton J.W., et al. Estimating the effects of non-pharmaceutical interventions on COVID-19 in Europe. Nature. 2020;584:257–261. doi: 10.1038/s41586-020-2405-7. [DOI] [PubMed] [Google Scholar]
- 11.Fuller J.A., Hakim A., Victory K.R., Date K., Lynch M., Dahl B., Henao O. Mitigation Policies and COVID-19-Associated Mortality—37 European Countries, 23 January–30 June 2020. MMWR Morb. Mortal. Wkly. Rep. 2021;70:58–62. doi: 10.15585/mmwr.mm7002e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piovani D., Christodoulou M.N., Hadjidemetriou A., Pantavou K., Zaza P., Bagos P.G., Bonovas S., Nikolopoulos G.K. Effect of early application of social distancing interventions on COVID-19 mortality over the first pandemic wave: An analysis of longitudinal data from 37 countries. J. Infect. 2021;82:133–142. doi: 10.1016/j.jinf.2020.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ECDC European Centre for Disease Prevention and Control. COVID-19 Testing Strategies and Objectives. [(accessed on 30 March 2022)]. Available online: https://www.ecdc.europa.eu/en/publications-data/COVID-19-testing-strategies-and-objectives.
- 14.TheWhiteHouse National COVID-19 Preparedness Plan. [(accessed on 3 March 2022)]; Available online: https://www.whitehouse.gov/covidplan/
- 15.Chan J.F., Yip C.C., To K.K., Tang T.H., Wong S.C., Leung K.H., Fung A.Y., Ng A.C., Zou Z., Tsoi H.W., et al. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. J. Clin. Microbiol. 2020;58:e00310-20. doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reusken C., Broberg E.K., Haagmans B., Meijer A., Corman V.M., Papa A., Charrel R., Drosten C., Koopmans M., Leitmeyer K., et al. Laboratory readiness and response for novel coronavirus (2019-nCoV) in expert laboratories in 30 EU/EEA countries, January 2020. Euro Surveill. 2020;25:2000082. doi: 10.2807/1560-7917.ES.2020.25.6.2000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mina M.J., Parker R., Larremore D.B. Rethinking COVID-19 Test Sensitivity—A Strategy for Containment. N. Engl. J. Med. 2020;383:e120. doi: 10.1056/NEJMp2025631. [DOI] [PubMed] [Google Scholar]
- 19.Peto T. COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay: A national systematic evaluation of sensitivity and specificity for mass-testing. EClinicalMedicine. 2021;36:100924. doi: 10.1016/j.eclinm.2021.100924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rai P., Kumar B.K., Deekshit V.K., Karunasagar I., Karunasagar I. Detection technologies and recent developments in the diagnosis of COVID-19 infection. Appl. Microbiol. Biotechnol. 2021;105:441–455. doi: 10.1007/s00253-020-11061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porte L., Legarraga P., Vollrath V., Aguilera X., Munita J.M., Araos R., Pizarro G., Vial P., Iruretagoyena M., Dittrich S., et al. Evaluation of a novel antigen-based rapid detection test for the diagnosis of SARS-CoV-2 in respiratory samples. Int. J. Infect. Dis. 2020;99:328–333. doi: 10.1016/j.ijid.2020.05.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moher D., Liberati A., Tetzlaff J., Altman D.G. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forero D.A., Lopez-Leon S., González-Giraldo Y., Bagos P.G. Ten simple rules for carrying out and writing meta-analyses. PLoS Comput. Biol. 2019;15:e1006922. doi: 10.1371/journal.pcbi.1006922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hopewell S., McDonald S., Clarke M., Egger M. Grey literature in meta-analyses of randomized trials of health care interventions. Cochrane Database Syst. Rev. 2007;2007:Mr000010. doi: 10.1002/14651858.MR000010.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.World Health Organization (WHO) Coronavirus Disease (COVID-19) Technical Guidance: Laboratory Testing for 2019-nCoV in Humans. [(accessed on 17 March 2022)]. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/laboratory-guidance/
- 26.Centers for Disease Control and Prevention (CDC) Information for Laboratories about Coronavirus (COVID-19) [(accessed on 13 March 2022)]; Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/index.html.
- 27.ECDC European Centre for Disease Prevention and Control. Novel Coronavirus Disease 2019 (COVID-19) Pandemic: Increased Transmission in the EU/EEA and the UK—Sixth Update—12 March 2020. [(accessed on 12 March 2022)]. Available online: https://www.ecdc.europa.eu/sites/default/files/documents/RRA-sixth-update-Outbreak-of-novel-coronavirus-disease-2019-COVID-19.pdf.
- 28.Whiting P.F., Rutjes A.W., Westwood M.E., Mallett S., Deeks J.J., Reitsma J.B., Leeflang M.M., Sterne J.A., Bossuyt P.M. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 29.Van Houwelingen H.C., Zwinderman K.H., Stijnen T. A bivariate approach to meta-analysis. Stat. Med. 1993;12:2273–2284. doi: 10.1002/sim.4780122405. [DOI] [PubMed] [Google Scholar]
- 30.Harbord R.M., Deeks J.J., Egger M., Whiting P., Sterne J.A. A unification of models for meta-analysis of diagnostic accuracy studies. Biostatistics. 2007;8:239–251. doi: 10.1093/biostatistics/kxl004. [DOI] [PubMed] [Google Scholar]
- 31.Higgins J.P., Whitehead A. Borrowing strength from external trials in a meta-analysis. Stat. Med. 1996;15:2733–2749. doi: 10.1002/(SICI)1097-0258(19961230)15:24<2733::AID-SIM562>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 32.Begg C.B., Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 33.Egger M., Davey Smith G., Schneider M., Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.StataCorp . Stata Statistical Software: Release 13. Stata Press; College Station, TX, USA: 2013. [Google Scholar]
- 35.White I.R. Multivariate random-effects meta-regression: Updates to mvmeta. Stata J. 2011;11:255–270. doi: 10.1177/1536867X1101100206. [DOI] [Google Scholar]
- 36.Chavan S., Mangalaparthi K.K., Singh S., Renuse S., Vanderboom P.M., Madugundu A.K., Budhraja R., McAulay K., Grys T.E., Rule A.D., et al. Mass Spectrometric Analysis of Urine from COVID-19 Patients for Detection of SARS-CoV-2 Viral Antigen and to Study Host Response. J. Proteome Res. 2021;20:3404–3413. doi: 10.1021/acs.jproteome.1c00391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saadi J., Oueslati S., Bellanger L., Gallais F., Dortet L., Roque-Afonso A.M., Junot C., Naas T., Fenaille F., Becher F. Quantitative Assessment of SARS-CoV-2 Virus in Nasopharyngeal Swabs Stored in Transport Medium by a Straightforward LC-MS/MS Assay Targeting Nucleocapsid, Membrane, and Spike Proteins. J. Proteome Res. 2021;20:1434–1443. doi: 10.1021/acs.jproteome.0c00887. [DOI] [PubMed] [Google Scholar]
- 38.Shao W., Shurin M.R., Wheeler S.E., He X., Star A. Rapid Detection of SARS-CoV-2 Antigens Using High-Purity Semiconducting Single-Walled Carbon Nanotube-Based Field-Effect Transistors. ACS Appl. Mater. Interfaces. 2021;13:10321–10327. doi: 10.1021/acsami.0c22589. [DOI] [PubMed] [Google Scholar]
- 39.Guo K., Wustoni S., Koklu A., Díaz-Galicia E., Moser M., Hama A., Alqahtani A.A., Ahmad A.N., Alhamlan F.S., Shuaib M., et al. Rapid single-molecule detection of COVID-19 and MERS antigens via nanobody-functionalized organic electrochemical transistors. Nat. Biomed. Eng. 2021;5:666–677. doi: 10.1038/s41551-021-00734-9. [DOI] [PubMed] [Google Scholar]
- 40.Eissa S., Alhadrami H.A., Al-Mozaini M., Hassan A.M., Zourob M. Voltammetric-based immunosensor for the detection of SARS-CoV-2 nucleocapsid antigen. Mikrochim. Acta. 2021;188:199. doi: 10.1007/s00604-021-04867-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Funabashi R., Miyakawa K., Yamaoka Y., Yoshimura S., Yamane S., Jeremiah S.S., Shimizu K., Ozawa H., Kawakami C., Usuku S., et al. Development of highly sensitive and rapid antigen detection assay for diagnosis of COVID-19 utilizing optical waveguide immunosensor. J. Mol. Cell Biol. 2021;13:763–766. doi: 10.1093/jmcb/mjab037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J., Wen J., Zhou M., Ni S., Le W., Chen G., Wei L., Zeng Y., Qi D., Pan M., et al. On-Site Detection of SARS-CoV-2 Antigen by Deep Learning-Based Surface-Enhanced Raman Spectroscopy and Its Biochemical Foundations. Anal. Chem. 2021;93:9174–9182. doi: 10.1021/acs.analchem.1c01061. [DOI] [PubMed] [Google Scholar]
- 43.Ehsan M.A., Khan S.A., Rehman A. Screen-Printed Graphene/Carbon Electrodes on Paper Substrates as Impedance Sensors for Detection of Coronavirus in Nasopharyngeal Fluid Samples. Diagnostics. 2021;11:1030. doi: 10.3390/diagnostics11061030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Renuse S., Vanderboom P.M., Maus A.D., Kemp J.V., Gurtner K.M., Madugundu A.K., Chavan S., Peterson J.A., Madden B.J., Mangalaparthi K.K., et al. A mass spectrometry-based targeted assay for detection of SARS-CoV-2 antigen from clinical specimens. EBioMedicine. 2021;69:103465. doi: 10.1016/j.ebiom.2021.103465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Della Ventura B., Cennamo M., Minopoli A., Campanile R., Bolletti Censi S., Terracciano D., Portella G., Velotta R. Colorimetric Test for Fast Detection of SARS-CoV-2 in Nasal and Throat Swabs. medRxiv. 2020 doi: 10.1101/2020.08.15.20175489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh N.K., Ray P., Carlin A.F., Magallanes C., Morgan S.C., Laurent L.C., Aronoff-Spencer E.S., Hall D.A. Hitting the diagnostic sweet spot: Point-of-care SARS-CoV-2 salivary antigen testing with an off-the-shelf glucometer. medRxiv. 2020 doi: 10.1016/j.bios.2021.113111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kurtulmus M.S., Kazezoglu C., Cakiroglu B., Yilmaz H., Guner A.E. The urine foaming test in COVID-19 as a useful tool in diagnosis, prognosis and follow-up: Preliminary results. North Clin. Istanb. 2020;7:534–540. doi: 10.14744/nci.2020.42027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mak G.C., Lau S.S., Wong K.K., Chow N.L., Lau C.S., Lam E.T., Chan R.C., Tsang D.N. Analytical sensitivity and clinical sensitivity of the three rapid antigen detection kits for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;133:104684. doi: 10.1016/j.jcv.2020.104684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linares M., Pérez-Tanoira R., Carrero A., Romanyk J., Pérez-García F., Gómez-Herruz P., Arroyo T., Cuadros J. Panbio antigen rapid test is reliable to diagnose SARS-CoV-2 infection in the first 7 days after the onset of symptoms. J. Clin. Virol. 2020;133:104659. doi: 10.1016/j.jcv.2020.104659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta A., Khurana S., Das R., Srigyan D., Singh A., Mittal A., Singh P., Soneja M., Kumar A., Singh A.K., et al. Rapid chromatographic immunoassay-based evaluation of COVID-19: A cross-sectional, diagnostic test accuracy study & its implications for COVID-19 management in India. Indian J. Med. Res. 2021;153:126. doi: 10.4103/ijmr.IJMR_3305_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fenollar F., Bouam A., Ballouche M., Fuster L., Prudent E., Colson P., Tissot-Dupont H., Million M., Drancourt M., Raoult D., et al. Evaluation of the Panbio COVID-19 Rapid Antigen Detection Test Device for the Screening of Patients with COVID-19. J. Clin. Microbiol. 2021;59:e02589-20. doi: 10.1128/JCM.02589-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nalumansi A., Lutalo T., Kayiwa J., Watera C., Balinandi S., Kiconco J., Nakaseegu J., Olara D., Odwilo E., Serwanga J., et al. Field evaluation of the performance of a SARS-CoV-2 antigen rapid diagnostic test in Uganda using nasopharyngeal samples. Int. J. Infect. Dis. 2021;104:282–286. doi: 10.1016/j.ijid.2020.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parada-Ricart E., Gomez-Bertomeu F., Picó-Plana E., Olona-Cabases M. Usefulness of the antigen test for diagnosing SARS-CoV-2 infection in patients with and without symptoms. Enferm. Infecc. Microbiol. Clin. 2021;39:357–358. doi: 10.1016/j.eimc.2020.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee J.H., Choi M., Jung Y., Lee S.K., Lee C.S., Kim J., Kim J., Kim N.H., Kim B.T., Kim H.G. A novel rapid detection for SARS-CoV-2 spike 1 antigens using human angiotensin converting enzyme 2 (ACE2) Biosens. Bioelectron. 2021;171:112715. doi: 10.1016/j.bios.2020.112715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cerutti F., Burdino E., Milia M.G., Allice T., Gregori G., Bruzzone B., Ghisetti V. Urgent need of rapid tests for SARS-CoV-2 antigen detection: Evaluation of the SD-Biosensor antigen test for SARS-CoV-2. J. Clin. Virol. 2020;132:104654. doi: 10.1016/j.jcv.2020.104654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diao B., Wen K., Zhang J., Chen J., Han C., Chen Y., Wang S., Deng G., Zhou H., Wu Y. Accuracy of a nucleocapsid protein antigen rapid test in the diagnosis of SARS-CoV-2 infection. Clin. Microbiol. Infect. 2021;27:289.e281–289.e284. doi: 10.1016/j.cmi.2020.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young S., Taylor S.N., Cammarata C.L., Varnado K.G., Roger-Dalbert C., Montano A., Griego-Fullbright C., Burgard C., Fernandez C., Eckert K., et al. Clinical Evaluation of BD Veritor SARS-CoV-2 Point-of-Care Test Performance Compared to PCR-Based Testing and versus the Sofia 2 SARS Antigen Point-of-Care Test. J. Clin. Microbiol. 2020;59:e02338-20. doi: 10.1128/JCM.02338-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liotti F.M., Menchinelli G., Lalle E., Palucci I., Marchetti S., Colavita F., La Sorda M., Sberna G., Bordi L., Sanguinetti M., et al. Performance of a novel diagnostic assay for rapid SARS-CoV-2 antigen detection in nasopharynx samples. Clin. Microbiol. Infect. 2021;27:487–488. doi: 10.1016/j.cmi.2020.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ogawa T., Fukumori T., Nishihara Y., Sekine T., Okuda N., Nishimura T., Fujikura H., Hirai N., Imakita N., Kasahara K. Another false-positive problem for a SARS-CoV-2 antigen test in Japan. J. Clin. Virol. 2020;131:104612. doi: 10.1016/j.jcv.2020.104612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hirotsu Y., Maejima M., Shibusawa M., Nagakubo Y., Hosaka K., Amemiya K., Sueki H., Hayakawa M., Mochizuki H., Tsutsui T., et al. Comparison of automated SARS-CoV-2 antigen test for COVID-19 infection with quantitative RT-PCR using 313 nasopharyngeal swabs, including from seven serially followed patients. Int. J. Infect. Dis. 2020;99:397–402. doi: 10.1016/j.ijid.2020.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nagura-Ikeda M., Imai K., Tabata S., Miyoshi K., Murahara N., Mizuno T., Horiuchi M., Kato K., Imoto Y., Iwata M., et al. Clinical Evaluation of Self-Collected Saliva by Quantitative Reverse Transcription-PCR (RT-qPCR), Direct RT-qPCR, Reverse Transcription-Loop-Mediated Isothermal Amplification, and a Rapid Antigen Test To Diagnose COVID-19. J. Clin. Microbiol. 2020;58:e01438-20. doi: 10.1128/JCM.01438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mak G.C., Cheng P.K., Lau S.S., Wong K.K., Lau C.S., Lam E.T., Chan R.C., Tsang D.N. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J. Clin. Virol. 2020;129:104500. doi: 10.1016/j.jcv.2020.104500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mertens P., De Vos N., Martiny D., Jassoy C., Mirazimi A., Cuypers L., Van den Wijngaert S., Monteil V., Melin P., Stoffels K., et al. Development and Potential Usefulness of the COVID-19 Ag Respi-Strip Diagnostic Assay in a Pandemic Context. Front. Med. 2020;7:225. doi: 10.3389/fmed.2020.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blairon L., Wilmet A., Beukinga I., Tré-Hardy M. Implementation of rapid SARS-CoV-2 antigenic testing in a laboratory without access to molecular methods: Experiences of a general hospital. J. Clin. Virol. 2020;129:104472. doi: 10.1016/j.jcv.2020.104472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Scohy A., Anantharajah A., Bodéus M., Kabamba-Mukadi B., Verroken A., Rodriguez-Villalobos H. Low performance of rapid antigen detection test as frontline testing for COVID-19 diagnosis. J. Clin. Virol. 2020;129:104455. doi: 10.1016/j.jcv.2020.104455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lambert-Niclot S., Cuffel A., Le Pape S., Vauloup-Fellous C., Morand-Joubert L., Roque-Afonso A.M., Le Goff J., Delaugerre C. Evaluation of a Rapid Diagnostic Assay for Detection of SARS-CoV-2 Antigen in Nasopharyngeal Swabs. J. Clin. Microbiol. 2020;58:e00977-20. doi: 10.1128/JCM.00977-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Diao B., Wen K., Chen J., Liu Y., Yuan Z., Han C., Chen J., Pan Y., Chen L., Dan Y., et al. Diagnosis of Acute Respiratory Syndrome Coronavirus 2 Infection by Detection of Nucleocapsid Protein. medRxiv. 2020 doi: 10.1101/2020.03.07.20032524. [DOI] [Google Scholar]
- 68.Beck E.T., Paar W., Fojut L., Serwe J., Jahnke R.R. Comparison of the Quidel Sofia SARS FIA Test to the Hologic Aptima SARS-CoV-2 TMA Test for Diagnosis of COVID-19 in Symptomatic Outpatients. J. Clin. Microbiol. 2021;59:e02727-20. doi: 10.1128/JCM.02727-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Krüttgen A., Cornelissen C.G., Dreher M., Hornef M.W., Imöhl M., Kleines M. Comparison of the SARS-CoV-2 Rapid antigen test to the real star SARS-CoV-2 RT PCR kit. J. Virol. Methods. 2021;288:114024. doi: 10.1016/j.jviromet.2020.114024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Albert E., Torres I., Bueno F., Huntley D., Molla E., Fernández-Fuentes M., Martínez M., Poujois S., Forqué L., Valdivia A., et al. Field evaluation of a rapid antigen test (Panbio™ COVID-19 Ag Rapid Test Device) for COVID-19 diagnosis in primary healthcare centres. Clin. Microbiol. Infect. 2021;27:472.e7–472.e10. doi: 10.1016/j.cmi.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chaimayo C., Kaewnaphan B., Tanlieng N., Athipanyasilp N., Sirijatuphat R., Chayakulkeeree M., Angkasekwinai N., Sutthent R., Puangpunngam N., Tharmviboonsri T., et al. Rapid SARS-CoV-2 antigen detection assay in comparison with real-time RT-PCR assay for laboratory diagnosis of COVID-19 in Thailand. Virol. J. 2020;17:177. doi: 10.1186/s12985-020-01452-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lanser L., Bellmann-Weiler R., Öttl K.W., Huber L., Griesmacher A., Theurl I., Weiss G. Evaluating the clinical utility and sensitivity of SARS-CoV-2 antigen testing in relation to RT-PCR Ct values. Infection. 2021;49:555–557. doi: 10.1007/s15010-020-01542-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gremmels H., Winkel B.M.F., Schuurman R., Rosingh A., Rigter N.A.M., Rodriguez O., Ubijaan J., Wensing A.M.J., Bonten M.J.M., Hofstra L.M. Real-life validation of the Panbio™ COVID-19 antigen rapid test (Abbott) in community-dwelling subjects with symptoms of potential SARS-CoV-2 infection. EClinicalMedicine. 2021;31:100677. doi: 10.1016/j.eclinm.2020.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dřevínek P., Hurych J., Kepka Z., Briksi A., Kulich M., Zajac M., Hubáček P. The sensitivity of SARS-CoV-2 antigen tests in the view of large-scale testing. Epidemiol. Mikrobiol. Imunol. 2021;70:156–160. [PubMed] [Google Scholar]
- 75.Schwob J.M., Miauton A., Petrovic D., Perdrix J., Senn N., Jaton K., Onya O., Maillard A., Minghelli G., Cornuz J., et al. Antigen rapid tests, nasopharyngeal PCR and saliva PCR to detect SARS-CoV-2: A prospective comparative clinical trial. medRxiv. 2020 doi: 10.1101/2020.11.23.20237057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Corman V.M., Haage V.C., Bleicker T., Schmidt M.L., Mühlemann B., Zuchowski M., Jo W.K., Tscheak P., Möncke-Buchner E., Müller M.A., et al. Comparison of seven commercial SARS-CoV-2 rapid point-of-care antigen tests: A single-centre laboratory evaluation study. Lancet Microbe. 2021;2:e311–e319. doi: 10.1016/S2666-5247(21)00056-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abdulrahman A., Mustafa F., AlAwadhi A.I., Alansari Q., AlAlawi B., AlQahtani M. Comparison of SARS-CoV-2 nasal antigen test to nasopharyngeal RT-PCR in mildly symptomatic patients. medRxiv. 2020 doi: 10.1101/2020.11.10.20228973. [DOI] [Google Scholar]
- 78.Yokota I., Sakurazawa T., Sugita J., Iwasaki S., Yasuda K., Yamashita N., Fujisawa S., Nishida M., Konno S., Teshima T. Performance of Qualitative and Quantitative Antigen Tests for SARS-CoV-2 Using Saliva. Infect. Dis. Rep. 2021;13:742–747. doi: 10.3390/idr13030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nash B., Badea A., Reddy A., Bosch M., Salcedo N., Gomez A.R., Versiani A., Dutra Silva G.C., Lopes dos Santos T.M.I., Milhim B.H.G.A., et al. Validating and modeling the impact of high-frequency rapid antigen screening on COVID-19 spread and outcomes. medRxiv. 2021 doi: 10.1101/2020.09.01.20184713. [DOI] [Google Scholar]
- 80.Van der Moeren N., Zwart V.F., Lodder E.B., van den Bijllaardt W., van Esch H.R.J.M., Stohr J.J.J.M., Pot J., Welschen I., van Mechelen P.M.F., Pas S.D., et al. Performance evaluation of a SARS-CoV-2 Rapid antigentest: Test performance in the community in the netherlands. medRxiv. 2020 doi: 10.1101/2020.10.19.20215202. [DOI] [Google Scholar]
- 81.Porte L., Legarraga P., Iruretagoyena M., Vollrath V., Pizarro G., Munita J.M., Araos R., Weitzel T. Rapid SARS-CoV-2 antigen detection by immunofluorescence—A new tool to detect infectivity. medRxiv. 2020 doi: 10.1101/2020.10.04.20206466. [DOI] [Google Scholar]
- 82.Krüger L.J., Gaeddert M., Köppel L., Brümmer L.E., Gottschalk C., Miranda I.B., Schnitzler P., Kräusslich H.G., Lindner A.K., Nikolai O., et al. Evaluation of the accuracy, ease of use and limit of detection of novel, rapid, antigen-detecting point-of-care diagnostics for SARS-CoV-2. medRxiv. 2020 doi: 10.1101/2020.10.01.20203836. [DOI] [Google Scholar]
- 83.Ventura B.D., Cennamo M., Minopoli A., Campanile R., Censi S.B., Terracciano D., Portella G., Velotta R. Colorimetric Test for Fast Detection of SARS-CoV-2 in Nasal and Throat Swabs. ACS Sens. 2020;5:3043–3048. doi: 10.1021/acssensors.0c01742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Herrera V., Hsu V., Adewale A., Johnson L., Hendrix T., Kuhlman J., Finkler N. Testing Healthcare Workers Exposed to COVID19 using Rapid Antigen Detection. medRxiv. 2020 doi: 10.1101/2020.08.12.20172726. [DOI] [Google Scholar]
- 85.Pickering S., Batra R., Merrick B., Snell L.B., Nebbia G., Douthwaite S., Reid F., Patel A., Kia Ik M.T., Patel B., et al. Comparative performance of SARS-CoV-2 lateral flow antigen tests and association with detection of infectious virus in clinical specimens: A single-centre laboratory evaluation study. Lancet Microbe. 2021;2:e461–e471. doi: 10.1016/S2666-5247(21)00143-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harmon K., de St Maurice A.M., Brady A.C., Swaminathan S., Aukerman D.F., Rueda M.A., Terrell K., Cohen R.P., Gamradt S.C., Henry S.D., et al. Surveillance testing for SARS-CoV-2 infection in an asymptomatic athlete population: A prospective cohort study with 123 362 tests and 23 463 paired RT-PCR/antigen samples. BMJ Open Sport Exerc. Med. 2021;7:e001137. doi: 10.1136/bmjsem-2021-001137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Korenkov M., Poopalasingam N., Madler M., Vanshylla K., Eggeling R., Wirtz M., Fish I., Dewald F., Gieselmann L., Lehmann C., et al. Evaluation of a Rapid Antigen Test to Detect SARS-CoV-2 Infection and Identify Potentially Infectious Individuals. J. Clin. Microbiol. 2021;59:e0089621. doi: 10.1128/JCM.00896-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Seynaeve Y., Heylen J., Fontaine C., Maclot F., Meex C., Diep A.N., Donneau A.F., Hayette M.P., Descy J. Evaluation of Two Rapid Antigenic Tests for the Detection of SARS-CoV-2 in Nasopharyngeal Swabs. J. Clin. Med. 2021;10:2774. doi: 10.3390/jcm10132774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Di Domenico M., De Rosa A., Di Gaudio F., Internicola P., Bettini C., Salzano N., Castrianni D., Marotta A., Boccellino M. Diagnostic Accuracy of a New Antigen Test for SARS-CoV-2 Detection. Int. J. Environ. Res. Public Health. 2021;18:6310. doi: 10.3390/ijerph18126310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kiro V.V., Gupta A., Singh P., Sharad N., Khurana S., Prakash S., Dar L., Malhotra R., Wig N., Kumar A., et al. Evaluation of COVID-19 Antigen Fluorescence Immunoassay Test for Rapid Detection of SARS-CoV-2. J. Glob. Infect. Dis. 2021;13:91–93. doi: 10.4103/jgid.jgid_316_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Smith R.L., Gibson L.L., Martinez P.P., Ke R., Mirza A., Conte M., Gallagher N., Conte A., Wang L., Fredrickson R., et al. Longitudinal Assessment of Diagnostic Test Performance Over the Course of Acute SARS-CoV-2 Infection. J. Infect. Dis. 2021;224:976–982. doi: 10.1093/infdis/jiab337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.L’Huillier A.G., Lacour M., Sadiku D., Gadiri M.A., De Siebenthal L., Schibler M., Eckerle I., Pinösch S., Kaiser L., Gervaix A., et al. Diagnostic Accuracy of SARS-CoV-2 Rapid Antigen Detection Testing in Symptomatic and Asymptomatic Children in the Clinical Setting. J. Clin. Microbiol. 2021;59:e0099121. doi: 10.1128/JCM.00991-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gupta A., Anand A., Jain N., Goswami S., Anantharaj A., Patil S., Singh R., Kumar A., Shrivastava T., Bhatnagar S., et al. A novel G-quadruplex aptamer-based spike trimeric antigen test for the detection of SARS-CoV-2. Mol. Nucleic Acids. 2021;26:321–332. doi: 10.1016/j.omtn.2021.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wagenhäuser I., Knies K., Rauschenberger V., Eisenmann M., McDonogh M., Petri N., Andres O., Flemming S., Gawlik M., Papsdorf M., et al. Clinical performance evaluation of SARS-CoV-2 rapid antigen testing in point of care usage in comparison to RT-qPCR. EBioMedicine. 2021;69:103455. doi: 10.1016/j.ebiom.2021.103455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fernández M.D., Estévez A.S., Alfonsín F.L., Arevalo G.B. Usefulness of the Lumiradx ™ SARS-CoV-2 antigen test in nursing home. Enferm. Infecc. Microbiol. Clin. 2021 doi: 10.1016/j.eimc.2021.06.006. [DOI] [Google Scholar]
- 96.Amer R.M., Samir M., Gaber O.A., El-Deeb N.A., Abdelmoaty A.A., Ahmed A.A., Samy W., Atta A.H., Walaa M., Anis R.H. Diagnostic performance of rapid antigen test for COVID-19 and the effect of viral load, sampling time, subject’s clinical and laboratory parameters on test accuracy. J. Infect. Public Health. 2021;14:1446–1453. doi: 10.1016/j.jiph.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baccani I., Morecchiato F., Chilleri C., Cervini C., Gori E., Matarrese D., Bassetti A., Bonizzoli M., Mencarini J., Antonelli A., et al. Evaluation of Three Immunoassays for the Rapid Detection of SARS-CoV-2 antigens. Diagn. Microbiol. Infect. Dis. 2021;101:115434. doi: 10.1016/j.diagmicrobio.2021.115434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Matsuzaki N., Orihara Y., Kodana M., Kitagawa Y., Matsuoka M., Kawamura R., Takeuchi S., Imai K., Tarumoto N., Maesaki S., et al. Evaluation of a chemiluminescent enzyme immunoassay-based high-throughput SARS-CoV-2 antigen assay for the diagnosis of COVID-19: The VITROS® SARS-CoV-2 Antigen Test. J. Med. Virol. 2021;93:6778–6781. doi: 10.1002/jmv.27153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jakobsen K.K., Jensen J.S., Todsen T., Tolsgaard M.G., Kirkby N., Lippert F., Vangsted A.M., Martel C.J., Klokker M., von Buchwald C. Accuracy and cost description of rapid antigen test compared with reverse transcriptase-polymerase chain reaction for SARS-CoV-2 detection. Dan. Med. J. 2021;68:A03210217. [PubMed] [Google Scholar]
- 100.Ngo Nsoga M.T., Kronig I., Perez Rodriguez F.J., Sattonnet-Roche P., Da Silva D., Helbling J., Sacks J.A., de Vos M., Boehm E., Gayet-Ageron A., et al. Diagnostic accuracy of Panbio rapid antigen tests on oropharyngeal swabs for detection of SARS-CoV-2. PLoS ONE. 2021;16:e0253321. doi: 10.1371/journal.pone.0253321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Smith R.D., Johnson J.K., Clay C., Girio-Herrera L., Stevens D., Abraham M., Zimand P., Ahlman M., Gimigliano S., Zhao R., et al. Clinical evaluation of Sofia Rapid Antigen Assay for detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) among emergency department to hospital admissions. Infect. Control Hosp. Epidemiol. 2021:1–6. doi: 10.1017/ice.2021.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Eleftheriou I., Dasoula F., Dimopoulou D., Lebessi E., Serafi E., Spyridis N., Tsolia M. Real-life evaluation of a COVID-19 rapid antigen detection test in hospitalized children. J. Med. Virol. 2021;93:6040–6044. doi: 10.1002/jmv.27149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lindner A.K., Nikolai O., Rohardt C., Kausch F., Wintel M., Gertler M., Burock S., Hörig M., Bernhard J., Tobian F., et al. Diagnostic accuracy and feasibility of patient self-testing with a SARS-CoV-2 antigen-detecting rapid test. J. Clin. Virol. 2021;141:104874. doi: 10.1016/j.jcv.2021.104874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ferté T., Ramel V., Cazanave C., Lafon M.E., Bébéar C., Malvy D., Georges-Walryck A., Dehail P. Accuracy of COVID-19 rapid antigenic tests compared to RT-PCR in a student population: The StudyCov study. J. Clin. Virol. 2021;141:104878. doi: 10.1016/j.jcv.2021.104878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fernandez-Montero A., Argemi J., Rodríguez J.A., Ariño A.H., Moreno-Galarraga L. Validation of a rapid antigen test as a screening tool for SARS-CoV-2 infection in asymptomatic populations. Sensitivity, specificity and predictive values. EClinicalMedicine. 2021;37:100954. doi: 10.1016/j.eclinm.2021.100954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hoehl S., Schenk B., Rudych O., Göttig S., Foppa I., Kohmer N., Karaca O., Toptan T., Ciesek S. High-Frequency Self-Testing by Schoolteachers for SARS-CoV-2 Using a Rapid Antigen Test–Results of the Safe School Hesse study. Dtsch. Arztebl. Int. 2021;118:252–253. doi: 10.3238/arztebl.m2021.0187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lee J., Kim S.Y., Huh H.J., Kim N., Sung H., Lee H., Roh K.H., Kim T.S., Hong K.H. Clinical Performance of the Standard Q COVID-19 Rapid Antigen Test and Simulation of its Real-World Application in Korea. Ann. Lab. Med. 2021;41:588–592. doi: 10.3343/alm.2021.41.6.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mayanskiy N., Brzhozovskaya E., Fedorova N., Lebedin Y. Parallel detection of SARS-CoV-2 RNA and nucleocapsid antigen in nasopharyngeal specimens from a COVID-19 patient screening cohort. Int. J. Infect. Dis. 2021;108:330–332. doi: 10.1016/j.ijid.2021.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Leixner G., Voill-Glaninger A., Bonner E., Kreil A., Zadnikar R., Viveiros A. Evaluation of the AMP SARS-CoV-2 rapid antigen test in a hospital setting. Int. J. Infect. Dis. 2021;108:353–356. doi: 10.1016/j.ijid.2021.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hirotsu Y., Sugiura H., Maejima M., Hayakawa M., Mochizuki H., Tsutsui T., Kakizaki Y., Miyashita Y., Omata M. Comparison of Roche and Lumipulse quantitative SARS-CoV-2 antigen test performance using automated systems for the diagnosis of COVID-19. Int. J. Infect. Dis. 2021;108:263–269. doi: 10.1016/j.ijid.2021.05.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fiedler M., Holtkamp C., Dittmer U., Anastasiou O.E. Performance of the LIAISON(®) SARS-CoV-2 Antigen Assay vs. SARS-CoV-2-RT-PCR. Pathogens. 2021;10:658. doi: 10.3390/pathogens10060658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Dierks S., Bader O., Schwanbeck J., Groß U., Weig M.S., Mese K., Lugert R., Bohne W., Hahn A., Feltgen N., et al. Diagnosing SARS-CoV-2 with Antigen Testing, Transcription-Mediated Amplification and Real-Time PCR. J. Clin. Med. 2021;10:2404. doi: 10.3390/jcm10112404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Terpos E., Ntanasis-Stathopoulos I., Skvarč M. Clinical Application of a New SARS-CoV-2 Antigen Detection Kit (Colloidal Gold) in the Detection of COVID-19. Diagnostics. 2021;11:995. doi: 10.3390/diagnostics11060995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Osmanodja B., Budde K., Zickler D., Naik M.G., Hofmann J., Gertler M., Hülso C., Rössig H., Horn P., Seybold J., et al. Accuracy of a Novel SARS-CoV-2 Antigen-Detecting Rapid Diagnostic Test from Standardized Self-Collected Anterior Nasal Swabs. J. Clin. Med. 2021;10:2099. doi: 10.3390/jcm10102099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Harris D.T., Badowski M., Jernigan B., Sprissler R., Edwards T., Cohen R., Paul S., Merchant N., Weinkauf C.C., Bime C., et al. SARS-CoV-2 Rapid Antigen Testing of Symptomatic and Asymptomatic Individuals on the University of Arizona Campus. Biomedicines. 2021;9:539. doi: 10.3390/biomedicines9050539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cento V., Renica S., Matarazzo E., Antonello M., Colagrossi L., Di Ruscio F., Pani A., Fanti D., Vismara C., Puoti M., et al. Frontline Screening for SARS-CoV-2 Infection at Emergency Department Admission by Third Generation Rapid Antigen Test: Can We Spare RT-qPCR? Viruses. 2021;13:818. doi: 10.3390/v13050818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kumar A., Kunjukutty R., Thaha A., Srikumar S., Madhusoodanan H., David S., Biswas L., Sathyapalan D. Universal screening for SARS-CoV-2 in pregnant women using a combination of antigen and RT-PCR testing. Infez. Med. 2021;29:294–296. [PubMed] [Google Scholar]
- 118.Orsi A., Pennati B.M., Bruzzone B., Ricucci V., Ferone D., Barbera P., Arboscello E., Dentone C., Icardi G. On-field evaluation of a ultra-rapid fluorescence immunoassay as a frontline test for SARS-CoV-2 diagnostic. J. Virol. Methods. 2021;295:114201. doi: 10.1016/j.jviromet.2021.114201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Blairon L., Cupaiolo R., Thomas I., Piteüs S., Wilmet A., Beukinga I., Tré-Hardy M. Efficacy comparison of three rapid antigen tests for SARS-CoV-2 and how viral load impact their performance. J. Med. Virol. 2021;93:5783–5788. doi: 10.1002/jmv.27108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bornemann L., Kaup O., Kleideiter J., Panning M., Ruprecht B., Wehmeier M. Real-life evaluation of the Sofia SARS-CoV-2 antigen assay in a large tertiary care hospital. J. Clin. Virol. 2021;140:104854. doi: 10.1016/j.jcv.2021.104854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Krüger L.J., Gaeddert M., Tobian F., Lainati F., Gottschalk C., Klein J.A.F., Schnitzler P., Kräusslich H.G., Nikolai O., Lindner A.K., et al. The Abbott PanBio WHO emergency use listed, rapid, antigen-detecting point-of-care diagnostic test for SARS-CoV-2-Evaluation of the accuracy and ease-of-use. PLoS ONE. 2021;16:e0247918. doi: 10.1371/journal.pone.0247918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shaikh N., Friedlander E.J., Tate P.J., Liu H., Chang C.H., Wells A., Hoberman A. Performance of a Rapid SARS-CoV-2 Antigen Detection Assay in Symptomatic Children. Pediatrics. 2021;148 doi: 10.1542/peds.2021-050832. [DOI] [PubMed] [Google Scholar]
- 123.Diez Flecha C., Rivero Rodríguez A.M., Fernández-Villa T., Fernández García P., Ferreira de Jesús J.L., Sánchez Antolín G. Internal validity of a rapid test for COVID-19 antigens in a nursing home. Semergen. 2021;47:332–336. doi: 10.1016/j.semerg.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yokota I., Shane P.Y., Okada K., Unoki Y., Yang Y., Iwasaki S., Fujisawa S., Nishida M., Teshima T. A novel strategy for SARS-CoV-2 mass screening with quantitative antigen testing of saliva: A diagnostic accuracy study. Lancet Microbe. 2021;2:e397–e404. doi: 10.1016/S2666-5247(21)00092-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Klein J.A.F., Krüger L.J., Tobian F., Gaeddert M., Lainati F., Schnitzler P., Lindner A.K., Nikolai O., Knorr B., Welker A., et al. Head-to-head performance comparison of self-collected nasal versus professional-collected nasopharyngeal swab for a WHO-listed SARS-CoV-2 antigen-detecting rapid diagnostic test. Med. Microbiol. Immunol. 2021;210:181–186. doi: 10.1007/s00430-021-00710-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Caramello V., Boccuzzi A., Basile V., Ferraro A., Macciotta A., Catalano A., Costa G., Vineis P., Sacerdote C., Ricceri F. Are antigenic tests useful for detecting SARS-CoV-2 infections in patients accessing to emergency departments? Results from a North-West Italy hospital. J. Infect. 2021;83:237–279. doi: 10.1016/j.jinf.2021.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Koeleman J.G.M., Brand H., de Man S.J., Ong D.S.Y. Clinical evaluation of rapid point-of-care antigen tests for diagnosis of SARS-CoV-2 infection. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:1975–1981. doi: 10.1007/s10096-021-04274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Šterbenc A., Tomič V., Bidovec Stojković U., Vrankar K., Rozman A., Zidarn M. Usefulness of rapid antigen testing for SARS-CoV-2 screening of healthcare workers: A pilot study. Clin. Exp. Med. 2022;22:157–160. doi: 10.1007/s10238-021-00722-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kumar K.K., Sampritha U.C., Maganty V., Prakash A.A., Basumatary J., Adappa K., Chandraprabha S., Neeraja T.G., Guru Prasad N.S., Preethi B., et al. Pre-Operative SARS-CoV-2 Rapid Antigen Test and Reverse Transcription Polymerase Chain Reaction: A conundrum in surgical decision making. Indian J. Ophthalmol. 2021;69:1560–1562. doi: 10.4103/ijo.IJO_430_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Soleimani R., Deckers C., Huang T.D., Bogaerts P., Evrard S., Wallemme I., Habib B., Rouzé P., Denis O. Rapid COVID-19 antigenic tests: Usefulness of a modified method for diagnosis. J. Med. Virol. 2021;93:5655–5659. doi: 10.1002/jmv.27094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Takeuchi Y., Akashi Y., Kato D., Kuwahara M., Muramatsu S., Ueda A., Notake S., Nakamura K., Ishikawa H., Suzuki H. Diagnostic performance and characteristics of anterior nasal collection for the SARS-CoV-2 antigen test: A prospective study. Sci. Rep. 2021;11:10519. doi: 10.1038/s41598-021-90026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Homza M., Zelena H., Janosek J., Tomaskova H., Jezo E., Kloudova A., Mrazek J., Svagera Z., Prymula R. COVID-19 antigen testing: Better than we know? A test accuracy study. Infect. Dis. 2021;53:661–668. doi: 10.1080/23744235.2021.1914857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Van der Moeren N., Zwart V.F., Lodder E.B., Van den Bijllaardt W., Van Esch H., Stohr J., Pot J., Welschen I., Van Mechelen P.M.F., Pas S.D., et al. Evaluation of the test accuracy of a SARS-CoV-2 rapid antigen test in symptomatic community dwelling individuals in the Netherlands. PLoS ONE. 2021;16:e0250886. doi: 10.1371/journal.pone.0250886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Brihn A., Chang J., Yong K.O., Balter S., Terashita D., Rubin Z., Yeganeh N. Diagnostic Performance of an Antigen Test with RT-PCR for the Detection of SARS-CoV-2 in a Hospital Setting—Los Angeles County, California, June–August 2020. MMWR Morb. Mortal. Wkly. Rep. 2021;70:702–706. doi: 10.15585/mmwr.mm7019a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nordgren J., Sharma S., Olsson H., Jämtberg M., Falkeborn T., Svensson L., Hagbom M. SARS-CoV-2 rapid antigen test: High sensitivity to detect infectious virus. J. Clin. Virol. 2021;140:104846. doi: 10.1016/j.jcv.2021.104846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Holzner C., Pabst D., Anastasiou O.E., Dittmer U., Manegold R.K., Risse J., Fistera D., Kill C., Falk M. SARS-CoV-2 rapid antigen test: Fast-safe or dangerous? An analysis in the emergency department of an university hospital. J. Med. Virol. 2021;93:5323–5327. doi: 10.1002/jmv.27033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kim D., Lee J., Bal J., Seo S.K., Chong C.K., Lee J.H., Park H. Development and Clinical Evaluation of an Immunochromatography-Based Rapid Antigen Test (GenBody™ COVAG025) for COVID-19 Diagnosis. Viruses. 2021;13:796. doi: 10.3390/v13050796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Bianco G., Boattini M., Barbui A.M., Scozzari G., Riccardini F., Coggiola M., Lupia E., Cavallo R., Costa C. Evaluation of an antigen-based test for hospital point-of-care diagnosis of SARS-CoV-2 infection. J. Clin. Virol. 2021;139:104838. doi: 10.1016/j.jcv.2021.104838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Peña M., Ampuero M., Garcés C., Gaggero A., García P., Velasquez M.S., Luza R., Alvarez P., Paredes F., Acevedo J., et al. Performance of SARS-CoV-2 rapid antigen test compared with real-time RT-PCR in asymptomatic individuals. Int. J. Infect. Dis. 2021;107:201–204. doi: 10.1016/j.ijid.2021.04.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Muhi S., Tayler N., Hoang T., Ballard S.A., Graham M., Rojek A., Kwong J.C., Trubiano J.A., Smibert O., Drewett G., et al. Multi-site assessment of rapid, point-of-care antigen testing for the diagnosis of SARS-CoV-2 infection in a low-prevalence setting: A validation and implementation study. Lancet Reg. Health West Pac. 2021;9:100115. doi: 10.1016/j.lanwpc.2021.100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Uwamino Y., Nagata M., Aoki W., Nakagawa T., Inose R., Yokota H., Furusawa Y., Sakai-Tagawa Y., Iwatsuki-Horimoto K., Kawaoka Y., et al. Accuracy of rapid antigen detection test for nasopharyngeal swab specimens and saliva samples in comparison with RT-PCR and viral culture for SARS-CoV-2 detection. J. Infect. Chemother. 2021;27:1058–1062. doi: 10.1016/j.jiac.2021.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Thakur P., Saxena S., Manchanda V., Rana N., Goel R., Arora R. Utility of Antigen-Based Rapid Diagnostic Test for Detection of SARS-CoV-2 Virus in Routine Hospital Settings. Lab. Med. 2021;52:e154–e158. doi: 10.1093/labmed/lmab033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Homza M., Zelena H., Janosek J., Tomaskova H., Jezo E., Kloudova A., Mrazek J., Svagera Z., Prymula R. Five Antigen Tests for SARS-CoV-2: Virus Viability Matters. Viruses. 2021;13:684. doi: 10.3390/v13040684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Shah M.M., Salvatore P.P., Ford L., Kamitani E., Whaley M.J., Mitchell K., Currie D.W., Morgan C.N., Segaloff H.E., Lecher S., et al. Performance of Repeat BinaxNOW Severe Acute Respiratory Syndrome Coronavirus 2 Antigen Testing in a Community Setting, Wisconsin, November 2020–December 2020. Clin. Infect. Dis. 2021;73:S54–S57. doi: 10.1093/cid/ciab309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.McKay S.L., Tobolowsky F.A., Moritz E.D., Hatfield K.M., Bhatnagar A., LaVoie S.P., Jackson D.A., Lecy K.D., Bryant-Genevier J., Campbell D., et al. Performance Evaluation of Serial SARS-CoV-2 Rapid Antigen Testing During a Nursing Home Outbreak. Ann. Intern. Med. 2021;174:945–951. doi: 10.7326/M21-0422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Yin N., Debuysschere C., Decroly M., Bouazza F.Z., Collot V., Martin C., Ponthieux F., Dahma H., Gilbert M., Wautier M., et al. SARS-CoV-2 Diagnostic Tests: Algorithm and Field Evaluation From the Near Patient Testing to the Automated Diagnostic Platform. Front. Med. 2021;8:650581. doi: 10.3389/fmed.2021.650581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Baro B., Rodo P., Ouchi D., Bordoy A.E., Saya Amaro E.N., Salsench S.V., Molinos S., Alemany A., Ubals M., Corbacho-Monné M., et al. Performance characteristics of five antigen-detecting rapid diagnostic test (Ag-RDT) for SARS-CoV-2 asymptomatic infection: A head-to-head benchmark comparison. J. Infect. 2021;82:269–275. doi: 10.1016/j.jinf.2021.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Caputo V., Bax C., Colantoni L., Peconi C., Termine A., Fabrizio C., Calvino G., Luzzi L., Panunzi G.G., Fusco C., et al. Comparative analysis of antigen and molecular tests for the detection of SARS-CoV-2 and related variants: A study on 4266 samples. Int. J. Infect. Dis. 2021;108:187–189. doi: 10.1016/j.ijid.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kenyeres B., Ánosi N., Bányai K., Mátyus M., Orosz L., Kiss A., Kele B., Burián K., Lengyel G. Comparison of four PCR and two point of care assays used in the laboratory detection of SARS-CoV-2. J. Virol. Methods. 2021;293:114165. doi: 10.1016/j.jviromet.2021.114165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Häuser F., Sprinzl M.F., Dreis K.J., Renzaho A., Youhanen S., Kremer W.M., Podlech J., Galle P.R., Lackner K.J., Rossmann H., et al. Evaluation of a laboratory-based high-throughput SARS-CoV-2 antigen assay for non-COVID-19 patient screening at hospital admission. Med. Microbiol. Immunol. 2021;210:165–171. doi: 10.1007/s00430-021-00706-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lefever S., Indevuyst C., Cuypers L., Dewaele K., Yin N., Cotton F., Padalko E., Oyaert M., Descy J., Cavalier E., et al. Comparison of the Quantitative DiaSorin Liaison Antigen Test to Reverse Transcription-PCR for the Diagnosis of COVID-19 in Symptomatic and Asymptomatic Outpatients. J. Clin. Microbiol. 2021;59:e0037421. doi: 10.1128/JCM.00374-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zacharias M., Stangl V., Thüringer A., Loibner M., Wurm P., Wolfgruber S., Zatloukal K., Kashofer K., Gorkiewicz G. Rapid Antigen Test for Postmortem Evaluation of SARS-CoV-2 Carriage. Emerg. Infect. Dis. 2021;27:1734–1737. doi: 10.3201/eid2706.210226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Oh S.M., Jeong H., Chang E., Choe P.G., Kang C.K., Park W.B., Kim T.S., Kwon W.Y., Oh M.D., Kim N.J. Clinical Application of the Standard Q COVID-19 Ag Test for the Detection of SARS-CoV-2 Infection. J. Korean Med. Sci. 2021;36:e101. doi: 10.3346/jkms.2021.36.e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Asai N., Sakanashi D., Ohashi W., Nakamura A., Kawamoto Y., Miyazaki N., Ohno T., Yamada A., Chida S., Shibata Y., et al. Efficacy and validity of automated quantitative chemiluminescent enzyme immunoassay for SARS-CoV-2 antigen test from saliva specimen in the diagnosis of COVID-19. J. Infect. Chemother. 2021;27:1039–1042. doi: 10.1016/j.jiac.2021.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kweon O.J., Lim Y.K., Kim H.R., Choi Y., Kim M.C., Choi S.H., Chung J.W., Lee M.K. Evaluation of rapid SARS-CoV-2 antigen tests, AFIAS COVID-19 Ag and ichroma COVID-19 Ag, with serial nasopharyngeal specimens from COVID-19 patients. PLoS ONE. 2021;16:e0249972. doi: 10.1371/journal.pone.0249972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Menchinelli G., Bordi L., Liotti F.M., Palucci I., Capobianchi M.R., Sberna G., Lalle E., Romano L., De Angelis G., Marchetti S., et al. Lumipulse G SARS-CoV-2 Ag assay evaluation using clinical samples from different testing groups. Clin. Chem. Lab. Med. 2021;59:1468–1476. doi: 10.1515/cclm-2021-0182. [DOI] [PubMed] [Google Scholar]
- 157.Sood N., Shetgiri R., Rodriguez A., Jimenez D., Treminino S., Daflos A., Simon P. Evaluation of the Abbott BinaxNOW rapid antigen test for SARS-CoV-2 infection in children: Implications for screening in a school setting. PLoS ONE. 2021;16:e0249710. doi: 10.1371/journal.pone.0249710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Epstude J., Skiba M., Harsch I.A. Antibody titers and rapid antigen testing in elderly patients with SARS-CoV-2 pneumonia vs. staff of ICU and “COVID-19” wards. GMS Hyg. Infect. Control. 2021;16:Doc11. doi: 10.3205/dgkh000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Berger A., Nsoga M.T.N., Perez-Rodriguez F.J., Aad Y.A., Sattonnet-Roche P., Gayet-Ageron A., Jaksic C., Torriani G., Boehm E., Kronig I., et al. Diagnostic accuracy of two commercial SARS-CoV-2 antigen-detecting rapid tests at the point of care in community-based testing centers. PLoS ONE. 2021;16:e0248921. doi: 10.1371/journal.pone.0248921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Matsuda E.M., de Campos I.B., de Oliveira I.P., Colpas D.R., Carmo A., Brígido L.F.M. Field evaluation of COVID-19 antigen tests versus RNA based detection: Potential lower sensitivity compensated by immediate results, technical simplicity, and low cost. J. Med. Virol. 2021;93:4405–4410. doi: 10.1002/jmv.26985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Van Honacker E., Van Vaerenbergh K., Boel A., De Beenhouwer H., Leroux-Roels I., Cattoir L. Comparison of five SARS-CoV-2 rapid antigen detection tests in a hospital setting and performance of one antigen assay in routine practice: A useful tool to guide isolation precautions? J. Hosp. Infect. 2021;114:144–152. doi: 10.1016/j.jhin.2021.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boum Y., Fai K.N., Nikolay B., Mboringong A.B., Bebell L.M., Ndifon M., Abbah A., Essaka R., Eteki L., Luquero F., et al. Performance and operational feasibility of antigen and antibody rapid diagnostic tests for COVID-19 in symptomatic and asymptomatic patients in Cameroon: A clinical, prospective, diagnostic accuracy study. Lancet Infect. Dis. 2021;21:1089–1096. doi: 10.1016/S1473-3099(21)00132-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mboumba Bouassa R.S., Veyer D., Péré H., Bélec L. Analytical performances of the point-of-care SIENNA™ COVID-19 Antigen Rapid Test for the detection of SARS-CoV-2 nucleocapsid protein in nasopharyngeal swabs: A prospective evaluation during the COVID-19 second wave in France. Int. J. Infect. Dis. 2021;106:8–12. doi: 10.1016/j.ijid.2021.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Stokes W., Berenger B.M., Portnoy D., Scott B., Szelewicki J., Singh T., Venner A.A., Turnbull L., Pabbaraju K., Shokoples S., et al. Clinical performance of the Abbott Panbio with nasopharyngeal, throat, and saliva swabs among symptomatic individuals with COVID-19. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:1721–1726. doi: 10.1007/s10096-021-04202-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Landaas E.T., Storm M.L., Tollånes M.C., Barlinn R., Kran A.B., Bragstad K., Christensen A., Andreassen T. Diagnostic performance of a SARS-CoV-2 rapid antigen test in a large, Norwegian cohort. J. Clin. Virol. 2021;137:104789. doi: 10.1016/j.jcv.2021.104789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Takeuchi Y., Akashi Y., Kato D., Kuwahara M., Muramatsu S., Ueda A., Notake S., Nakamura K., Ishikawa H., Suzuki H. The evaluation of a newly developed antigen test (QuickNavi™-COVID19 Ag) for SARS-CoV-2: A prospective observational study in Japan. J. Infect. Chemother. 2021;27:890–894. doi: 10.1016/j.jiac.2021.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Igloi Z., Velzing J., van Beek J., van de Vijver D., Aron G., Ensing R., Benschop K., Han W., Boelsums T., Koopmans M., et al. Clinical Evaluation of Roche SD Biosensor Rapid Antigen Test for SARS-CoV-2 in Municipal Health Service Testing Site, the Netherlands. Emerg. Infect. Dis. 2021;27:1323–1329. doi: 10.3201/eid2705.204688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Masiá M., Fernández-González M., Sánchez M., Carvajal M., García J.A., Gonzalo-Jiménez N., Ortiz de la Tabla V., Agulló V., Candela I., Guijarro J., et al. Nasopharyngeal Panbio COVID-19 Antigen Performed at Point-of-Care Has a High Sensitivity in Symptomatic and Asymptomatic Patients With Higher Risk for Transmission and Older Age. Open Forum Infect. Dis. 2021;8:ofab059. doi: 10.1093/ofid/ofab059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Jääskeläinen A.E., Ahava M.J., Jokela P., Szirovicza L., Pohjala S., Vapalahti O., Lappalainen M., Hepojoki J., Kurkela S. Evaluation of three rapid lateral flow antigen detection tests for the diagnosis of SARS-CoV-2 infection. J. Clin. Virol. 2021;137:104785. doi: 10.1016/j.jcv.2021.104785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Olearo F., Nörz D., Heinrich F., Sutter J.P., Roedl K., Schultze A., Wiesch J.S.Z., Braun P., Oestereich L., Kreuels B., et al. Handling and accuracy of four rapid antigen tests for the diagnosis of SARS-CoV-2 compared to RT-qPCR. J. Clin. Virol. 2021;137:104782. doi: 10.1016/j.jcv.2021.104782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ishii T., Sasaki M., Yamada K., Kato D., Osuka H., Aoki K., Morita T., Ishii Y., Tateda K. Immunochromatography and chemiluminescent enzyme immunoassay for COVID-19 diagnosis. J. Infect. Chemother. 2021;27:915–918. doi: 10.1016/j.jiac.2021.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Peña-Rodríguez M., Viera-Segura O., García-Chagollán M., Zepeda-Nuño J.S., Muñoz-Valle J.F., Mora-Mora J., Espinoza-De León G., Bustillo-Armendáriz G., García-Cedillo F., Vega-Magaña N. Performance evaluation of a lateral flow assay for nasopharyngeal antigen detection for SARS-CoV-2 diagnosis. J. Clin. Lab. Anal. 2021;35:e23745. doi: 10.1002/jcla.23745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gili A., Paggi R., Russo C., Cenci E., Pietrella D., Graziani A., Stracci F., Mencacci A. Evaluation of Lumipulse® G SARS-CoV-2 antigen assay automated test for detecting SARS-CoV-2 nucleocapsid protein (NP) in nasopharyngeal swabs for community and population screening. Int. J. Infect. Dis. 2021;105:391–396. doi: 10.1016/j.ijid.2021.02.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Pérez-García F., Romanyk J., Gómez-Herruz P., Arroyo T., Pérez-Tanoira R., Linares M., Pérez Ranz I., Labrador Ballestero A., Moya Gutiérrez H., Ruiz-Álvarez M.J., et al. Diagnostic performance of CerTest and Panbio antigen rapid diagnostic tests to diagnose SARS-CoV-2 infection. J. Clin. Virol. 2021;137:104781. doi: 10.1016/j.jcv.2021.104781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Kilic A., Hiestand B., Palavecino E. Evaluation of Performance of the BD Veritor SARS-CoV-2 Chromatographic Immunoassay Test in Patients with Symptoms of COVID-19. J. Clin. Microbiol. 2021;59:e00260-21. doi: 10.1128/JCM.00260-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Drain P.K., Ampajwala M., Chappel C., Gvozden A.B., Hoppers M., Wang M., Rosen R., Young S., Zissman E., Montano M. A Rapid, High-Sensitivity SARS-CoV-2 Nucleocapsid Immunoassay to Aid Diagnosis of Acute COVID-19 at the Point of Care: A Clinical Performance Study. Infect. Dis. Ther. 2021;10:753–761. doi: 10.1007/s40121-021-00413-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Basso D., Aita A., Padoan A., Cosma C., Navaglia F., Moz S., Contran N., Zambon C.F., Maria Cattelan A., Plebani M. Salivary SARS-CoV-2 antigen rapid detection: A prospective cohort study. Clin. Chim. Acta. 2021;517:54–59. doi: 10.1016/j.cca.2021.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pollock N.R., Jacobs J.R., Tran K., Cranston A.E., Smith S., O’Kane C.Y., Roady T.J., Moran A., Scarry A., Carroll M., et al. Performance and Implementation Evaluation of the Abbott BinaxNOW Rapid Antigen Test in a High-Throughput Drive-Through Community Testing Site in Massachusetts. J. Clin. Microbiol. 2021;59:e2021050832. doi: 10.1128/JCM.00083-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ristić M., Nikolić N., Čabarkapa V., Turkulov V., Petrović V. Validation of the Standard Q COVID-19 antigen test in Vojvodina, Serbia. PLoS ONE. 2021;16:e0247606. doi: 10.1371/journal.pone.0247606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Courtellemont L., Guinard J., Guillaume C., Giaché S., Rzepecki V., Seve A., Gubavu C., Baud K., Le Helloco C., Cassuto G.N., et al. High performance of a novel antigen detection test on nasopharyngeal specimens for diagnosing SARS-CoV-2 infection. J. Med. Virol. 2021;93:3152–3157. doi: 10.1002/jmv.26896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Thommes L., Burkert F.R., Öttl K.W., Goldin D., Loacker L., Lanser L., Griesmacher A., Theurl I., Weiss G., Bellmann-Weiler R. Comparative evaluation of four SARS-CoV-2 antigen tests in hospitalized patients. Int. J. Infect. Dis. 2021;105:144–146. doi: 10.1016/j.ijid.2021.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.González-Donapetry P., García-Clemente P., Bloise I., García-Sánchez C., Sánchez Castellano M., Romero M.P., Gutiérrez Arroyo A., Mingorance J., de Ceano-Vivas La Calle M., García-Rodriguez J. Think of the Children: Evaluation of SARS-CoV-2 Rapid Antigen Test in Pediatric Population. Pediatr. Infect. Dis. J. 2021;40:385–388. doi: 10.1097/INF.0000000000003101. [DOI] [PubMed] [Google Scholar]
- 183.Eshghifar N., Busheri A., Shrestha R., Beqaj S. Evaluation of Analytical Performance of Seven Rapid Antigen Detection Kits for Detection of SARS-CoV-2 Virus. Int. J. Gen. Med. 2021;14:435–440. doi: 10.2147/IJGM.S297762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Merino P., Guinea J., Muñoz-Gallego I., González-Donapetry P., Galán J.C., Antona N., Cilla G., Hernáez-Crespo S., Díaz-de Tuesta J.L., Gual-de Torrella A., et al. Multicenter evaluation of the Panbio™ COVID-19 rapid antigen-detection test for the diagnosis of SARS-CoV-2 infection. Clin. Microbiol. Infect. 2021;27:758–761. doi: 10.1016/j.cmi.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bulilete O., Lorente P., Leiva A., Carandell E., Oliver A., Rojo E., Pericas P., Llobera J. Panbio™ rapid antigen test for SARS-CoV-2 has acceptable accuracy in symptomatic patients in primary health care. J. Infect. 2021;82:391–398. doi: 10.1016/j.jinf.2021.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Torres I., Poujois S., Albert E., Álvarez G., Colomina J., Navarro D. Point-of-care evaluation of a rapid antigen test (CLINITEST(®) Rapid COVID-19 Antigen Test) for diagnosis of SARS-CoV-2 infection in symptomatic and asymptomatic individuals. J. Infect. 2021;82:e11–e12. doi: 10.1016/j.jinf.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lindner A.K., Nikolai O., Rohardt C., Burock S., Hülso C., Bölke A., Gertler M., Krüger L.J., Gaeddert M., Tobian F., et al. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with professional-collected nasal versus nasopharyngeal swab. Eur. Respir J. 2021;57:2004430. doi: 10.1183/13993003.04430-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Hirotsu Y., Maejima M., Shibusawa M., Amemiya K., Nagakubo Y., Hosaka K., Sueki H., Hayakawa M., Mochizuki H., Tsutsui T., et al. Prospective study of 1308 nasopharyngeal swabs from 1033 patients using the LUMIPULSE SARS-CoV-2 antigen test: Comparison with RT-qPCR. Int. J. Infect. Dis. 2021;105:7–14. doi: 10.1016/j.ijid.2021.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Salvagno G.L., Gianfilippi G., Bragantini D., Henry B.M., Lippi G. Clinical assessment of the Roche SARS-CoV-2 rapid antigen test. Diagnosis. 2021;8:322–326. doi: 10.1515/dx-2020-0154. [DOI] [PubMed] [Google Scholar]
- 190.Veyrenche N., Bolloré K., Pisoni A., Bedin A.S., Mondain A.M., Ducos J., Segondy M., Montes B., Pastor P., Morquin D., et al. Diagnosis value of SARS-CoV-2 antigen/antibody combined testing using rapid diagnostic tests at hospital admission. J. Med. Virol. 2021;93:3069–3076. doi: 10.1002/jmv.26855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Porte L., Legarraga P., Iruretagoyena M., Vollrath V., Pizarro G., Munita J., Araos R., Weitzel T. Evaluation of two fluorescence immunoassays for the rapid detection of SARS-CoV-2 antigen-new tool to detect infective COVID-19 patients. PeerJ. 2021;9:e10801. doi: 10.7717/peerj.10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Domínguez Fernández M., Peña Rodríguez M.F., Lamelo Alfonsín F., Bou Arévalo G. Experience with Panbio™ rapid antigens test device for the detection of SARS-CoV-2 in nursing homes. Enferm. Infecc. Microbiol. Clin. 2021;40:42–43. doi: 10.1016/j.eimc.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Kobayashi R., Murai R., Asanuma K., Fujiya Y., Takahashi S. Evaluating a novel, highly sensitive, and quantitative reagent for detecting SARS-CoV-2 antigen. J. Infect. Chemother. 2021;27:800–807. doi: 10.1016/j.jiac.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Houston H., Gupta-Wright A., Toke-Bjolgerud E., Biggin-Lamming J., John L. Diagnostic accuracy and utility of SARS-CoV-2 antigen lateral flow assays in medical admissions with possible COVID-19. J. Hosp. Infect. 2021;110:203–205. doi: 10.1016/j.jhin.2021.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ciotti M., Maurici M., Pieri M., Andreoni M., Bernardini S. Performance of a rapid antigen test in the diagnosis of SARS-CoV-2 infection. J. Med. Virol. 2021;93:2988–2991. doi: 10.1002/jmv.26830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Okoye N.C., Barker A.P., Curtis K., Orlandi R.R., Snavely E.A., Wright C., Hanson K.E., Pearson L.N. Performance Characteristics of BinaxNOW COVID-19 Antigen Card for Screening Asymptomatic Individuals in a University Setting. J. Clin. Microbiol. 2021;59:e03282-20. doi: 10.1128/JCM.03282-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.James A.E., Gulley T., Kothari A., Holder K., Garner K., Patil N. Performance of the BinaxNOW coronavirus disease 2019 (COVID-19) Antigen Card test relative to the severe acute respiratory coronavirus virus 2 (SARS-CoV-2) real-time reverse transcriptase polymerase chain reaction (rRT-PCR) assay among symptomatic and asymptomatic healthcare employees. Infect. Control Hosp. Epidemiol. 2022;43:99–101. doi: 10.1017/ice.2021.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Villaverde S., Domínguez-Rodríguez S., Sabrido G., Pérez-Jorge C., Plata M., Romero M.P., Grasa C.D., Jiménez A.B., Heras E., Broncano A., et al. Diagnostic Accuracy of the Panbio Severe Acute Respiratory Syndrome Coronavirus 2 Antigen Rapid Test Compared with Reverse-Transcriptase Polymerase Chain Reaction Testing of Nasopharyngeal Samples in the Pediatric Population. J. Pediatr. 2021;232:287–289.e284. doi: 10.1016/j.jpeds.2021.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Pekosz A., Parvu V., Li M., Andrews J.C., Manabe Y.C., Kodsi S., Gary D.S., Roger-Dalbert C., Leitch J., Cooper C.K. Antigen-Based Testing but Not Real-Time Polymerase Chain Reaction Correlates with Severe Acute Respiratory Syndrome Coronavirus 2 Viral Culture. Clin. Infect. Dis. 2021;73:e2861–e2866. doi: 10.1093/cid/ciaa1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Kohmer N., Toptan T., Pallas C., Karaca O., Pfeiffer A., Westhaus S., Widera M., Berger A., Hoehl S., Kammel M., et al. The Comparative Clinical Performance of Four SARS-CoV-2 Rapid Antigen Tests and Their Correlation to Infectivity In Vitro. J. Clin. Med. 2021;10:328. doi: 10.3390/jcm10020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Prince-Guerra J.L., Almendares O., Nolen L.D., Gunn J.K.L., Dale A.P., Buono S.A., Deutsch-Feldman M., Suppiah S., Hao L., Zeng Y., et al. Evaluation of Abbott BinaxNOW Rapid Antigen Test for SARS-CoV-2 Infection at Two Community-Based Testing Sites—Pima County, Arizona, 3–17 November 2020. MMWR Morb. Mortal. Wkly. Rep. 2021;70:100–105. doi: 10.15585/mmwr.mm7003e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Möckel M., Corman V.M., Stegemann M.S., Hofmann J., Stein A., Jones T.C., Gastmeier P., Seybold J., Offermann R., Bachmann U., et al. SARS-CoV-2 antigen rapid immunoassay for diagnosis of COVID-19 in the emergency department. Biomarkers. 2021;26:213–220. doi: 10.1080/1354750X.2021.1876769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Rottenstreich A., Zarbiv G., Kabiri D., Porat S., Sompolinsky Y., Reubinoff B., Benenson S., Oster Y. Rapid antigen detection testing for universal screening for severe acute respiratory syndrome coronavirus 2 in women admitted for delivery. Am. J. Obstet. Gynecol. 2021;224:539–540. doi: 10.1016/j.ajog.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Favresse J., Gillot C., Oliveira M., Cadrobbi J., Elsen M., Eucher C., Laffineur K., Rosseels C., Van Eeckhoudt S., Nicolas J.B., et al. Head-to-Head Comparison of Rapid and Automated Antigen Detection Tests for the Diagnosis of SARS-CoV-2 Infection. J. Clin. Med. 2021;10:265. doi: 10.3390/jcm10020265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Osterman A., Baldauf H.M., Eletreby M., Wettengel J.M., Afridi S.Q., Fuchs T., Holzmann E., Maier A., Döring J., Grzimek-Koschewa N., et al. Evaluation of two rapid antigen tests to detect SARS-CoV-2 in a hospital setting. Med. Microbiol. Immunol. 2021;210:65–72. doi: 10.1007/s00430-020-00698-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Pollock N.R., Savage T.J., Wardell H., Lee R.A., Mathew A., Stengelin M., Sigal G.B. Correlation of SARS-CoV-2 Nucleocapsid Antigen and RNA Concentrations in Nasopharyngeal Samples from Children and Adults Using an Ultrasensitive and Quantitative Antigen Assay. J. Clin. Microbiol. 2021;59:e03077-20. doi: 10.1128/JCM.03077-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Aoki K., Nagasawa T., Ishii Y., Yagi S., Okuma S., Kashiwagi K., Maeda T., Miyazaki T., Yoshizawa S., Tateda K. Clinical validation of quantitative SARS-CoV-2 antigen assays to estimate SARS-CoV-2 viral loads in nasopharyngeal swabs. J. Infect. Chemother. 2021;27:613–616. doi: 10.1016/j.jiac.2020.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Torres I., Poujois S., Albert E., Colomina J., Navarro D. Evaluation of a rapid antigen test (Panbio™ COVID-19 Ag rapid test device) for SARS-CoV-2 detection in asymptomatic close contacts of COVID-19 patients. Clin. Microbiol. Infect. 2021;27:636.e631–636.e634. doi: 10.1016/j.cmi.2020.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Alemany A., Baró B., Ouchi D., Rodó P., Ubals M., Corbacho-Monné M., Vergara-Alert J., Rodon J., Segalés J., Esteban C., et al. Analytical and clinical performance of the panbio COVID-19 antigen-detecting rapid diagnostic test. J. Infect. 2021;82:186–230. doi: 10.1016/j.jinf.2020.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rastawicki W., Gierczyński R., Juszczyk G., Mitura K., Henry B.M. Evaluation of PCL rapid point of care antigen test for detection of SARS-CoV-2 in nasopharyngeal swabs. J. Med. Virol. 2021;93:1920–1922. doi: 10.1002/jmv.26765. [DOI] [PubMed] [Google Scholar]
- 211.Yamamoto K., Suzuki M., Yamada G., Sudo T., Nomoto H., Kinoshita N., Nakamura K., Tsujimoto Y., Kusaba Y., Morita C., et al. Utility of the antigen test for coronavirus disease 2019: Factors influencing the prediction of the possibility of disease transmission. Int. J. Infect. Dis. 2021;104:65–72. doi: 10.1016/j.ijid.2020.12.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kashiwagi K., Ishii Y., Aoki K., Yagi S., Maeda T., Miyazaki T., Yoshizawa S., Aoyagi K., Tateda K. Immunochromatographic test for the detection of SARS-CoV-2 in saliva. J. Infect. Chemother. 2021;27:384–386. doi: 10.1016/j.jiac.2020.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Pilarowski G., Lebel P., Sunshine S., Liu J., Crawford E., Marquez C., Rubio L., Chamie G., Martinez J., Peng J., et al. Performance Characteristics of a Rapid Severe Acute Respiratory Syndrome Coronavirus 2 Antigen Detection Assay at a Public Plaza Testing Site in San Francisco. J. Infect. Dis. 2021;223:1139–1144. doi: 10.1093/infdis/jiaa802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Aoki K., Nagasawa T., Ishii Y., Yagi S., Kashiwagi K., Miyazaki T., Tateda K. Evaluation of clinical utility of novel coronavirus antigen detection reagent, Espline® SARS-CoV-2. J. Infect. Chemother. 2021;27:319–322. doi: 10.1016/j.jiac.2020.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Pray I.W., Ford L., Cole D., Lee C., Bigouette J.P., Abedi G.R., Bushman D., Delahoy M.J., Currie D., Cherney B., et al. Performance of an Antigen-Based Test for Asymptomatic and Symptomatic SARS-CoV-2 Testing at Two University Campuses—Wisconsin, September–October 2020. MMWR Morb. Mortal. Wkly. Rep. 2021;69:1642–1647. doi: 10.15585/mmwr.mm695152a3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Strömer A., Rose R., Schäfer M., Schön F., Vollersen A., Lorentz T., Fickenscher H., Krumbholz A. Performance of a Point-of-Care Test for the Rapid Detection of SARS-CoV-2 Antigen. Microorganisms. 2020;9:58. doi: 10.3390/microorganisms9010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Toptan T., Eckermann L., Pfeiffer A.E., Hoehl S., Ciesek S., Drosten C., Corman V.M. Evaluation of a SARS-CoV-2 rapid antigen test: Potential to help reduce community spread? J. Clin. Virol. 2021;135:104713. doi: 10.1016/j.jcv.2020.104713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Turcato G., Zaboli A., Pfeifer N., Ciccariello L., Sibilio S., Tezza G., Ausserhofer D. Clinical application of a rapid antigen test for the detection of SARS-CoV-2 infection in symptomatic and asymptomatic patients evaluated in the emergency department: A preliminary report. J. Infect. 2021;82:e14–e16. doi: 10.1016/j.jinf.2020.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Mak G.C.K., Lau S.S.Y., Wong K.K.Y., Chow N.L.S., Lau C.S., Lam E.T.K., Chan R.C.W., Tsang D.N.C. Evaluation of rapid antigen detection kit from the WHO Emergency Use List for detecting SARS-CoV-2. J. Clin. Virol. 2021;134:104712. doi: 10.1016/j.jcv.2020.104712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Zhang C., Zhou L., Du K., Zhang Y., Wang J., Chen L., Lyu Y., Li J., Liu H., Huo J., et al. Foundation and Clinical Evaluation of a New Method for Detecting SARS-CoV-2 Antigen by Fluorescent Microsphere Immunochromatography. Front. Cell Infect. Microbiol. 2020;10:553837. doi: 10.3389/fcimb.2020.553837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Agulló V., Fernández-González M., Ortiz de la Tabla V., Gonzalo-Jiménez N., García J.A., Masiá M., Gutiérrez F. Evaluation of the rapid antigen test Panbio COVID-19 in saliva and nasal swabs in a population-based point-of-care study. J. Infect. 2021;82:186–230. doi: 10.1016/j.jinf.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Tanimoto T., Matsumura M., Tada S., Fujita S., Ueno S., Hamai K., Omoto T., Maeda H., Nishisaka T., Ishikawa N. Need for a high-specificity test for confirming weakly positive result in an immunochromatographic SARS-CoV-2-specific antigen test: A case report. J. Microbiol. Immunol. Infect. 2021;54:534–535. doi: 10.1016/j.jmii.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lindner A.K., Nikolai O., Kausch F., Wintel M., Hommes F., Gertler M., Krüger L.J., Gaeddert M., Tobian F., Lainati F., et al. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with self-collected nasal swab versus professional-collected nasopharyngeal swab. Eur. Respir. J. 2021;57:2003961. doi: 10.1183/13993003.03961-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Abdelrazik A.M., Elshafie S.M., Abdelaziz H.M. Potential Use of Antigen-Based Rapid Test for SARS-CoV-2 in Respiratory Specimens in Low-Resource Settings in Egypt for Symptomatic Patients and High-Risk Contacts. Lab. Med. 2021;52:e46–e49. doi: 10.1093/labmed/lmaa104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Weitzel T., Legarraga P., Iruretagoyena M., Pizarro G., Vollrath V., Araos R., Munita J.M., Porte L. Comparative evaluation of four rapid SARS-CoV-2 antigen detection tests using universal transport medium. Travel Med. Infect. Dis. 2021;39:101942. doi: 10.1016/j.tmaid.2020.101942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Winkel B., Schram E., Gremmels H., Debast S., Schuurman R., Wensing A., Bonten M., Goedhart E., Hofstra M. Screening for SARS-CoV-2 infection in asymptomatic individuals using the Panbio COVID-19 antigen rapid test (Abbott) compared with RT-PCR: A prospective cohort study. BMJ Open. 2021;11:e048206. doi: 10.1136/bmjopen-2020-048206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Hoehl S., Schenk B., Rudych O., Göttig S., Foppa I., Kohmer N., Karaca O., Toptan T., Ciesek S. At-home self-testing of teachers with a SARS-CoV-2 rapid antigen test to reduce potential transmissions in schools. medRxiv. 2020 doi: 10.1101/2020.12.04.20243410. [DOI] [Google Scholar]
- 228.Kannian P., Lavanya C., Ravichandran K., Jayaraman B.G., Mahanathi P., Ashwini V., Kumarasamy N., Rajan G., Ranganathan K., Challacombe S.J., et al. Detection of SARS-CoV2 antigen in human saliva may be a reliable tool for large scale screening. medRxiv. 2020 doi: 10.1101/2020.12.17.20248437. [DOI] [Google Scholar]
- 229.Lindner A.K., Nikolai O., Rohardt C., Kausch F., Wintel M., Gertler M., Burock S., Hörig M., Bernhard J., Tobian F., et al. SARS-CoV-2 patient self-testing with an antigen-detecting rapid test: A head-to-head comparison with professional testing. medRxiv. 2021 doi: 10.1101/2021.01.06.20249009. [DOI] [Google Scholar]
- 230.Filgueiras P.S., Corsini C.A., Almeida N.B.F., Assis J.V., Pedrosa M.L.C., de Oliveira A.K., Amorim R.N.H., de Miranda D.A.P., Coutinho L.A., Gomes S.V.C., et al. COVID-19 Rapid Antigen Test at hospital admission associated to the knowledge of individual risk factors allow overcoming the difficulty of managing suspected patients in hospitals COVID-19 Rapid Antigen Test facilitates the management of suspected patients on hospital admission. medRxiv. 2021 doi: 10.1101/2021.01.06.21249282. [DOI] [Google Scholar]
- 231.Peto T., Team U.C.-L.F.O. COVID-19: Rapid Antigen detection for SARS-CoV-2 by lateral flow assay: A national systematic evaluation for mass-testing. medRxiv. 2021 doi: 10.1101/2021.01.13.21249563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Jakobsen K.K., Jensen J.S., Todsen T., Lippert F., Martel C.J.-M., Klokker M., von Buchwald C. Detection of SARS-CoV-2 infection by rapid antigen test in comparison with RT-PCR in a public setting. medRxiv. 2021 doi: 10.1101/2021.01.22.21250042. [DOI] [Google Scholar]
- 233.Miyakawa K., Funabashi R., Yamaoka Y., Jeremiah S.S., Katada J., Wada A., Takei T., Shimizu K., Ozawa H., Kawakami C., et al. SARS-CoV-2 antigen rapid diagnostic test enhanced with silver amplification technology. medRxiv. 2021 doi: 10.1101/2021.01.27.21250659. [DOI] [Google Scholar]
- 234.Pollock N.R., Tran K., Jacobs J.R., Cranston A.E., Smith S., O’Kane C.Y., Roady T.J., Moran A., Scarry A., Carroll M., et al. Performance and Operational Evaluation of the Access Bio CareStart Rapid Antigen Test in a High-Throughput Drive-Through Community Testing Site in Massachusetts. Open Forum Infect. Dis. 2021;8:ofab243. doi: 10.1093/ofid/ofab243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Shidlovskaya E.V., Kuznetsova N.A., Divisenko E.V., Nikiforova M.A., Siniavin A.E., Ogarkova D.A., Shagaev A.V., Semashko M.A., Tkachuk A.P., Burgasova O.A., et al. The Value of Rapid Antigen Tests to Identify Carriers of Viable SARS-CoV-2. medRxiv. 2021 doi: 10.1101/2021.03.10.21252667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Faíco-Filho K.S., Finamor Júnior F.E., Vinícius Leão Moreira L., Lins P.R.G., Justo A.F.O., Bellei N. Evaluation of the Panbio™ COVID-19 Ag Rapid Test at an Emergency Room in a Hospital in São Paulo, Brazil. medRxiv. 2021 doi: 10.1016/j.bjid.2022.102349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Schuit E., Veldhuijzen I., Venekamp R., van den Bijllaardt W., Pas S., Lodder E., Molenkamp R., GeurtsvanKessel C., Velzing J., Huisman R., et al. Diagnostic accuracy of rapid antigen tests in pre-/asymptomatic close contacts of individuals with a confirmed SARS-CoV-2 infection. medRxiv. 2021 doi: 10.1101/2021.03.18.21253874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Ducrest P.J. Development and Evaluation of a new Swiss Made SARS-CoV-2 antigen-detecting rapid test. medRxiv. 2021 doi: 10.1101/2021.03.25.21252280. [DOI] [Google Scholar]
- 239.Del Vecchio C., Brancaccio G., Brazzale A.R., Lavezzo E., Onelia F., Franchin E., Manuto L., Bianca F., Cianci V., Cattelan A., et al. Emergence of N antigen SARS-CoV-2 genetic variants escaping detection of antigenic tests. medRxiv. 2021 doi: 10.1101/2021.03.25.21253802. [DOI] [Google Scholar]
- 240.Bonde J., Ejegod D., Pedersen H., Smith B., Cortes D., Leding C., Thomsen T., Benfield T., Schnieder U.V., Tingleff J., et al. Clinical validation of point-of-care SARS-CoV-2 BD Veritor antigen test by a single throat swab for rapid COVID-19 status on hospital patients predominantly without overt COVID symptoms. medRxiv. 2021 doi: 10.1101/2021.04.12.21255299. [DOI] [Google Scholar]
- 241.Igloi Z., Velzing J., Huisman R., Geurtsvankessel C., Comvalius A., van Beek J., Ensing R., Boelsums T., Koopmans M., Molenkamp R. Clinical evaluation of the SD Biosensor saliva antigen rapid test with symptomatic and asymptomatic, non-hospitalized patients. medRxiv. 2021 doi: 10.1371/journal.pone.0260894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Thell R., Kallab V., Weinhappel W., Mueckstein W., Heschl L., Heschl M., Korsatko S., Toedling F., Blaschke A., Herzog T., et al. Evaluation of a novel, rapid antigen detection test for the diagnosis of SARS-CoV-2. medRxiv. 2021 doi: 10.1371/journal.pone.0259527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Pollock N.R., Berlin D., Smole S.C., Madoff L.C., Brown C., Henderson K., Larsen E., Hay J., Gabriel S., Gawande A.A., et al. Implementation of SARS-CoV2 Screening in K-12 Schools Using In-School Pooled Molecular Testing and Deconvolution by Rapid Antigen Test. J. Clin. Microbiol. 2021;59:e0112321. doi: 10.1128/JCM.01123-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hagbom M., Carmona-Vicente N., Sharma S., Olsson H., Jämtberg M., Nilsdotter-Augustinsson Å., Sjöwall J., Nordgren J. Evaluation of SARS-CoV-2 rapid antigen diagnostic tests for saliva samples. medRxiv. 2021 doi: 10.1016/j.heliyon.2022.e08998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Thirion-Romero I., Guerrero-Zúñiga S., Arias-Mendoza A., Cornejo-Tjuárez D.P., Meza-Meneses P., Torres-Erazo D.S., Hernández T., Galindo-Fraga A., Villegas-Mota I., Sepúlveda-Delgado J., et al. Evaluation of a rapid antigen test for SARS-CoV-2 in symptomatic patients and their contacts: A multicenter study. medRxiv. 2021 doi: 10.1016/j.ijid.2021.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Chiu R.Y.T., Kojima N., Mosley G.L., Cheng K.K., Pereira D.Y., Brobeck M., Chan T.L., Zee J.S., Kittur H., Chung C.Y.T., et al. Evaluation of the INDICAID COVID-19 Rapid Antigen Test in Symptomatic Populations and Asymptomatic Community Testing. Microbiol. Spectr. 2021;9:e0034221. doi: 10.1128/Spectrum.00342-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Abusrewil Z., Alhudiri I.M., Kaal H.H., El Meshri S.E., Ebrahim F.O., Dalyoum T., Efrefer A.A., Ibrahim K., Elfghi M.B., Abusrewil S., et al. Time scale performance of rapid antigen testing for SARS-CoV-2: Evaluation of 10 rapid antigen assays. J. Med. Virol. 2021;93:6512–6518. doi: 10.1002/jmv.27186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Muthamia E., Mungai S., Mungai M., Bandawe G., Qadri F., Kawser Z., Lockman S., Ivers L.C., Walt D., Suliman S., et al. Assessment of performance and implementation characteristics of rapid point of care SARS-CoV-2 antigen testing. medRxiv. 2021 doi: 10.12688/aasopenres.13323.1. [DOI] [Google Scholar]
- 249.Abdul-Mumin A., Abubakari A., Agbozo F., Abdul-Karim A., Nuertey B.D., Mumuni K., Heuschen A.-K., Hennig L., Denkinger C.M., Müller O., et al. Field evaluation of specificity and sensitivity of a standard SARS-CoV-2 antigen rapid diagnostic test: A prospective study at a teaching hospital in Northern Ghana. medRxiv. 2021 doi: 10.1371/journal.pgph.0000040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Akashi Y., Kiyasu Y., Takeuchi Y., Kato D., Kuwahara M., Muramatsu S., Ueda A., Notake S., Nakamura K., Ishikawa H., et al. Evaluation and clinical implications of the time to a positive results of antigen testing for SARS-CoV-2. J. Infect. Chemother. 2022;28:248–251. doi: 10.1016/j.jiac.2021.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lindner A.K., Krüger L.J., Nikolai O., Klein J.A.F., Rössig H., Schnitzler P., Corman V.M., Jones T.C., Tobian F., Gaeddert M., et al. SARS-CoV-2 variant of concern B.1.1.7: Diagnostic accuracy of three antigen-detecting rapid tests. medRxiv. 2021 doi: 10.1101/2021.06.15.21258502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Suliman S., Matias W.R., Fulcher I.R., Molano F.J., Collins S., Uceta E., Zhu J., Paxton R.M., Gonsalves S.F., Harden M.V., et al. Evaluation of the Access Bio CareStart™ rapid SARS-CoV-2 antigen test in asymptomatic individuals tested at a community mass-testing program in Western Massachusetts. medRxiv. 2021 doi: 10.1101/2021.06.17.21259109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Bruins M.J., dos Santos C.O., Spoelman-Lunsche M., van den Bos-Kromhout M.I., Debast S.B. Evaluation of the Panbio™ rapid antigen test for COVID-19 diagnosis in symptomatic health care workers. medRxiv. 2021 doi: 10.1101/2021.06.21.21259234. [DOI] [Google Scholar]
- 254.Ford L., Whaley M.J., Shah M.M., Salvatore P.P., Segaloff H.E., Delaney A., Currie D.W., Boyle-Estheimer L., O’Hegarty M., Morgan C.N., et al. Antigen Test Performance Among Children and Adults at a SARS-CoV-2 Community Testing Site. J. Pediatric Infect. Dis. Soc. 2021;10:1052–1061. doi: 10.1093/jpids/piab081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Koskinen J.M., Antikainen P., Hotakainen K., Haveri A., Ikonen N., Savolainen-Kopra C., Sundström K., Koskinen J.O. Clinical validation of automated and rapid mariPOC SARS-CoV-2 antigen test. Sci. Rep. 2021;11:20363. doi: 10.1038/s41598-021-99886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Nikolai O., Rohardt C., Tobian F., Junge A., Corman V.M., Jones T.C., Gaeddert M., Lainati F., Sacks J.A., Seybold J., et al. Anterior nasal versus nasal mid-turbinate sampling for a SARS-CoV-2 antigen-detecting rapid test: Does localisation or professional collection matter? Infect. Dis. 2021;53:947–952. doi: 10.1080/23744235.2021.1969426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Stohr J.J.J.M., Zwart V.F., Goderski G., Meijer A., Nagel-Imming C.R.S., Kluytmans-van den Bergh M.F.Q., Pas S.D., van den Oetelaar F., Hellwich M., Gan K.H., et al. Self-testing for the detection of SARS-CoV-2 infection with rapid antigen tests. medRxiv. 2021 doi: 10.1101/2021.02.21.21252153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L., Boodman C., Bello A., Hedley A., Schiffman Z., et al. Predicting Infectious Severe Acute Respiratory Syndrome Coronavirus 2 From Diagnostic Samples. Clin. Infect. Dis. 2020;71:2663–2666. doi: 10.1093/cid/ciaa638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: A systematic review and meta-analysis. Lancet Microbe. 2021;2:e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Dinnes J., Deeks J.J., Berhane S., Taylor M., Adriano A., Davenport C., Dittrich S., Emperador D., Takwoingi Y., Cunningham J., et al. Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst. Rev. 2021;3:Cd013705. doi: 10.1002/14651858.CD013705.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Khandker S.S., Nik Hashim N.H., Deris Z.Z., Shueb R.H., Islam M.A. Diagnostic Accuracy of Rapid Antigen Test Kits for Detecting SARS-CoV-2: A Systematic Review and Meta-Analysis of 17,171 Suspected COVID-19 Patients. J. Clin. Med. 2021;10:3493. doi: 10.3390/jcm10163493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.French S.D., McDonald S., McKenzie J.E., Green S.E. Investing in updating: How do conclusions change when Cochrane systematic reviews are updated? BMC Med. Res. Methodol. 2005;5:33. doi: 10.1186/1471-2288-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Moher D., Tsertsvadze A., Tricco A.C., Eccles M., Grimshaw J., Sampson M., Barrowman N. When and how to update systematic reviews. Cochrane Database Syst. Rev. 2008;2008:Mr000023. doi: 10.1002/14651858.MR000023.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ahmadzai N., Newberry S.J., Maglione M.A., Tsertsvadze A., Ansari M.T., Hempel S., Motala A., Tsouros S., Schneider Chafen J.J., Shanman R., et al. A surveillance system to assess the need for updating systematic reviews. Syst. Rev. 2013;2:104. doi: 10.1186/2046-4053-2-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Moher D., Tsertsvadze A., Tricco A.C., Eccles M., Grimshaw J., Sampson M., Barrowman N. A systematic review identified few methods and strategies describing when and how to update systematic reviews. J. Clin. Epidemiol. 2007;60:1095–1104. doi: 10.1016/j.jclinepi.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 266.Elliott J.H., Synnot A., Turner T., Simmonds M., Akl E.A., McDonald S., Salanti G., Meerpohl J., MacLehose H., Hilton J., et al. Living systematic review: 1. Introduction-the why, what, when, and how. J. Clin. Epidemiol. 2017;91:23–30. doi: 10.1016/j.jclinepi.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 267.Elliott J.H., Turner T., Clavisi O., Thomas J., Higgins J.P.T., Mavergames C., Gruen R.L. Living Systematic Reviews: An Emerging Opportunity to Narrow the Evidence-Practice Gap. PLoS Med. 2014;11:e1001603. doi: 10.1371/journal.pmed.1001603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Centers for Disease Control and Prevention Common Investigation Protocol for Investigating Suspected SARS-CoV-2 Reinfection. [(accessed on 18 May 2022)]; Available online: https://www.cdc.gov/coronavirus/2019-ncov/php/reinfection.html.
- 269.Centers for Disease Control and Prevention (CDC) Guidance for Antigen Testing for SARS-CoV-2 for Healthcare Providers Testing Individuals in the Community. [(accessed on 4 April 2022)]; Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html.
- 270.Larremore D.B., Wilder B., Lester E., Shehata S., Burke J.M., Hay J.A., Tambe M., Mina M.J., Parker R. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci. Adv. 2021;7:eabd5393. doi: 10.1126/sciadv.abd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Arora S., Grover V., Saluja P., Algarni Y.A., Saquib S.A., Asif S.M., Batra K., Alshahrani M.Y., Das G., Jain R., et al. Literature Review of Omicron: A Grim Reality Amidst COVID-19. Microorganisms. 2022;10:451. doi: 10.3390/microorganisms10020451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Food and Drug Administration Omicron variant: Impact on Antigen Diagnostic Tests. [(accessed on 28 December 2021)]; Available online: https://www.fda.gov/medical-devices/coronavirus-COVID-19-and-medical-devices/SARS-CoV-2-viral-mutations-impact-COVID-19-tests#omicronvariantimpact.
- 273.Regan J., Flynn J.P., Choudhary M.C., Uddin R., Lemieux J., Boucau J., Bhattacharyya R.P., Barczak A.K., Li J.Z., Siedner M.J. Detection of the Omicron Variant Virus with the Abbott BinaxNow SARS-CoV-2 Rapid Antigen Assay. Open Forum Infect. Dis. 2022;9:ofac022. doi: 10.1093/ofid/ofac022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Salcedo N., Nandu N., Boucau J., Herrera B.B. Detection of SARS-CoV-2 Omicron, Delta, Alpha and Gamma variants using a rapid antigen test. medRxiv. 2022 doi: 10.1101/2022.01.27.22269299. [DOI] [Google Scholar]
- 275.de Michelena P., Torres I., Ramos-García Á., Gozalbes V., Ruiz N., Sanmartín A., Botija P., Poujois S., Huntley D., Albert E., et al. Real-life performance of a COVID-19 rapid antigen detection test targeting the SARS-CoV-2 nucleoprotein for diagnosis of COVID-19 due to the Omicron variant. J. Infect. 2022;84:e64–e66. doi: 10.1016/j.jinf.2022.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Stanley S., Hamel D.J., Wolf I.D., Riedel S., Dutta S., Cheng A., Kirby J.E., Kanki P.J. Limit of Detection for Rapid Antigen Testing of the SARS-CoV-2 Omicron Variant. medRxiv. 2022 doi: 10.1101/2022.01.28.22269968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Deerain J., Druce J., Tran T., Batty M., Yoga Y., Fennell M., Dwyer D.E., Kok J., Williamson D.A. Assessment of the Analytical Sensitivity of 10 Lateral Flow Devices against the SARS-CoV-2 Omicron Variant. J. Clin. Microbiol. 2022;60:e0247921. doi: 10.1128/jcm.02479-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Diederichs M., Glawion R., Kremsner P.G., Mitze T., Müller G.J., Papies D., Schulz F., Wälde K. Is large-scale rapid CoV-2 testing a substitute for lockdowns? PLoS ONE. 2022;17:e0265207. doi: 10.1371/journal.pone.0265207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Mathuria J.P., Yadav R. Laboratory diagnosis of SARS-CoV-2—A review of current methods. J. Infect. Public Health. 2020;13:901–905. doi: 10.1016/j.jiph.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.