Abstract
Background
SARS-CoV-2, the virus that causes COVID-19, is constantly mutating, leading to new variants that culminate in a temporal lineages fluctuation. B.1.1.28 lineage has been evolving in Brazil since February 2020 and originated P.1 (VOC), P.2 (VOI) and other P.Xs proposed as new variants.
Methods and results
In this study, through the Illumina platform, we performed the whole-genome sequencing of 26 positive samples of SARS-CoV-2. Employing variant calling analysis on FASTQ reads and phylogenetic inference, we report a brief dispersion of a potentially new B.1.1.28-derived variant detected between 2021 May and June in individuals crossing the border between Brazil and Argentina, and local spread to inpatients from hospitals at the Rio Grande do Sul state capital (Porto Alegre). Besides, the Rio Grande do Sul State SARS-CoV-2 genomic epidemiological data was analyzed and showed an important B.1.1.28 peak in RS at the same period (May–June), even in the presence of a major Gamma wave.
Conclusions
The emergence of a putative B.1.1.28-derived lineage was identified in travelers crossing Brazil-Argentina border representing an important peak of B.1.1.28 in RS State with a decreased in Gamma variant frequency in the same period of time.
1. Introduction
Since the beginning of the SARS-COV-2 pandemic, in 2019, we have seen a consecutive emergence of Variants of Concern (VOCs), as B.1.1.7 (Alpha) [1], B.1.351 (Beta) [2], P.1 (Gamma) [3] and B.1.617.2 (Delta) [4], which were first identified in UK, South Africa, Brazil, and India, respectively. These VOCs have been responsible for an expressive number of cases and increased transmissibility. In Brazil, more than 30 million COVID-19 confirmed cases were reported, exceeding 600 thousand deaths, being more than 2 million cases and 39 thousand deaths in the Rio Grande do Sul State, according to the authorities. Antibody response, virulence, reinfection potential, and vaccine efficacy against the emergence variants, are not yet fully known, posing a risk for future outbreaks and efficacy in vaccination programs [5].
Rapid sequencing and analysis of SARS-CoV-2 genomes has supporting our ability to monitor the emergence and spread of SARS-CoV-2 variants of concern. Corona-ômica-BR, a Brazilian network of researchers that is dedicated to SARS-CoV-2 genomic surveillance in Brazil, has strongly contributed to keep the monitoring updated and available (http://www.corona-omica.br-mcti.lncc.br/#/). As part of this research, we have been sequencing SARS-CoV-2 positive samples from different regions in the Rio Grande do Sul, the Southernmost Brazilian State, following the emergence and spread of the SARS-CoV-2 lineages and variants.
B.1.1.28 has been evolving in Brazil since February 2020 and originated Gamma (VOC), P.2 (VOI), and other P.Xs proposed as new variants. In this study, we report the finding of a potential newly B.1.1.28-derived lineage. The sequencing of 26 high-quality SARS-CoV-2 whole-genomes were retrieved through the Illumina MiSeq platform. The samples were from passengers crossing the border between Brazil and Argentina and presented a unique additional non-synonymous mutational profile when applying a variant calling approach on FASTQ reads, performed on Geneious Prime software. Analyzing the general Rio Grande do Sul State lineages distribution over the year, a genetic epidemiological study was carried out in order to monitor whether this possible new variant would establish itself in the current scenario. These approaches allow monitoring the emergence of new variants in real time as well as evaluate the temporal distribution of the variants.
2. Methods
2.1. Samples
The genomic SARS-CoV-2 study was conducted at Laboratório de Microbiologia Molecular, Universidade Feevale, Rio Grande do Sul (RS), Brazil. A total of 26 SARS-CoV-2 positive samples were collected between late April and mid-June. There were 20 passengers from Uruguaiana city, crossing the border between Argentina and RS in Southern Brazil; four samples from Secretaria Municipal de Saúde (SMS) and the remaining samples from Garibaldi city, RS, received in our laboratory. SARS-CoV-2 samples were collected from 22 male and 4 female patients, ranging between 22 and 90 years old. From the passengers crossing the Argentina-Brazil border (truck drivers), 13 presented mild to moderate symptoms (including fever, cough, body and back pain, in addition to olfactory loss); five were asymptomatic and from two, no data was reported. The vaccination status from the samples collected by SMS was obtained, and from four patients, only one was reportedly vaccinated (including two doses, Coronavac).
2.2. Genomic study
Whole genome library preparation was performed using QIAseq SARS-CoV-2 Primer Panel paired for library enrichment and QIAseq FX DNA Library UDI Kit, according to the manufacturer instructions (Qiagen, Hilden, Germany). The sequencing was implemented on an Illumina MiSeq platform using MiSeq Reagent Kit v3 (600-cycle). The FASTQ reads were imported to Geneious Prime, trimmed (BBDuk 37.25), and mapped against the reference sequence hCoV-19/Wuhan/WIV04/2019 (EPI_ISL_402,124) available in EpiCoV database from GISAID (https://www.gisaid.org/45).
A total of 100 Brazilian SARS-CoV-2 complete genomes and the reference sequence (EPI_ISL_402,124) (>29 kb) were retrieved from the GISAID database and aligned with the sequences generated herein. Sequence alignment was performed using Clustal Omega and the reference sequence from Wuhan was applied as an outgroup. The Maximum Likelihood phylogenetic analysis under General Time Reversible model allowing for a proportion of invariable sites and substitution rates were inferred empirically in IQ-TREE v2.1.2 web server [6] applying 200 replicates and 1000 bootstrap and the analysis corroborated the previous results. Finally, variant calling approach was conducted through Geneious Prime software, between a FASTQ samples dataset and a known reference sequence (hCoV-19/Wuhan/WIV04/2019), analyzing Single Nucleotide Polymorphisms (SNPs) and amino acid changes consequences.
2.3. Epidemiological analysis
To ascertain the SARS-CoV-2 scenario, a total of 1256 complete genomes from Rio Grande do Sul State (including 334 sequences from our laboratory) were retrieved from GISAID database. The total of sequences, including the 26 putative newly B.1.1.28-derived lineage, were collected between 2021 January and September and characterized using Pangolin COVID-19 Lineage Assigner tool ( https://github.com/hCoV-2019/pangolin ). The data were grouped by month and the lineages scenario before and soon after this finding were analyzed to further contextualize the genetic epidemiology.
3. Results
3.1. Genomic description of a putative novel B.1.1.28-derived lineage
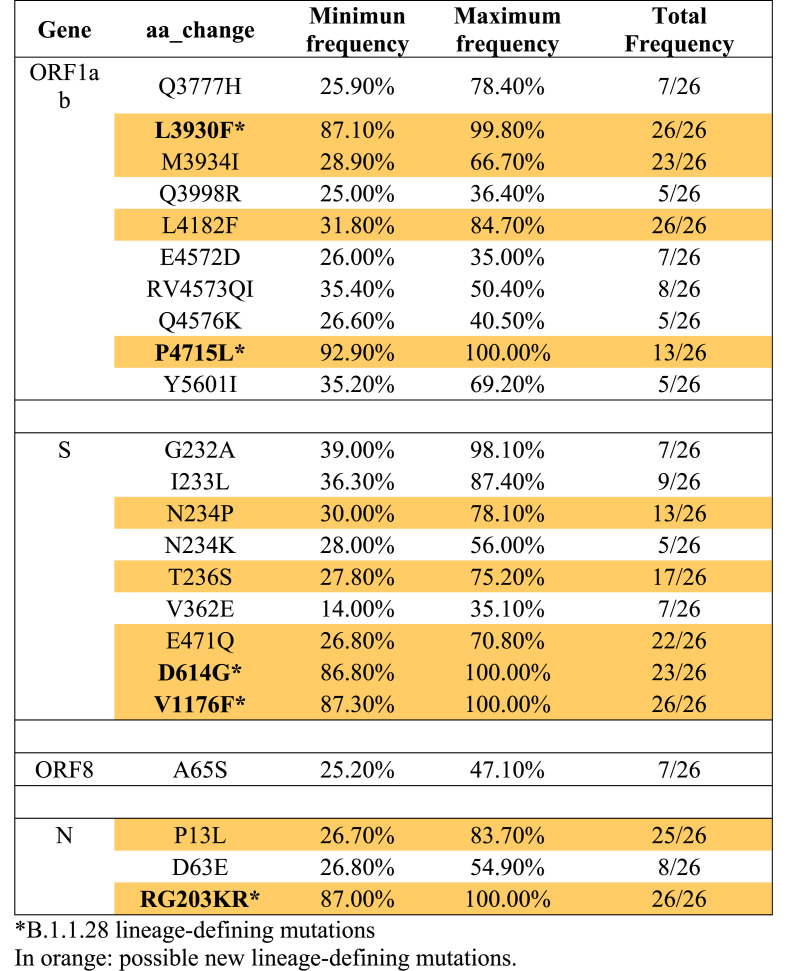
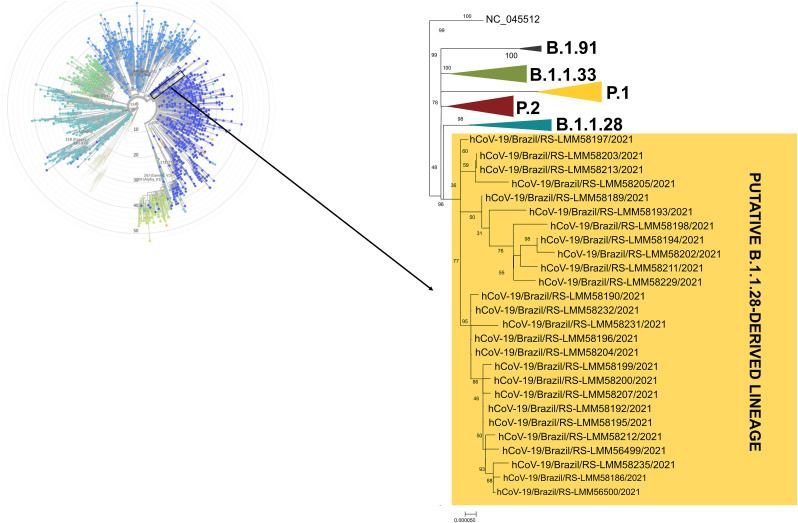
Consensus sequences were classified as B.1.1.28 according to the Pangolin COVID-19 Lineage Assigner tool. Nevertheless, analyzing the FASTq reads through variant calling method, additional non-synonymous mutations were identified when compared to those of B.1.1.28. From 26 complete genomes, eight additional non-synonymous mutations were identified in ORF1ab (Q3777H, M3934I, Q3998R, L4182F, E4572D, RV4573QI, Q4576K, Y5601I), seven in S protein (G232A, I233L, N234P, N234K, T236S, V362E, E471Q), two in N protein (P13L, D63E) and one in ORF8 (A65S). Among those, six drew attention due to the total frequency in the samples and appear to be signatures of the putative new B.1.1.28-derived variant, specially N234P and E471Q in S protein, and M3934I and L4182F in ORF1ab (Table 1 ). ML phylogenetic tree showed that the sequences generated herein clearly clustered into a separated group in a highly supported monophyletic clade (Fig. 1 ). The other clades were grouped and represent B.1.91, B.1.133, P.1, P.2 and B.1.1.8 lineages. Of all the aligned sequences, non-clustered with our sequences (Fig. 1).
Table 1.
FAST Q reads variant calling analysis. “Minimum and maximum frequencies” representing the smallest and largest percentage of the referred mutation in a sample and “total frequency” representing the number of samples in which the mutation was found.
Fig. 1.
SARS-CoV-2 complete genome phylogenetic trees. On the left, a global SARS-CoV-2 complete genome phylogeny performed on the Nextclade online platform. On the right, Maximum Likelihood phylogenetic analysis under General Time Reversible allowing for a proportion of invariable sites and with empirical and the substitution rates were inferred empirically in IQ-TREE v2.1.2 web server applying 200 replicates and 1000 bootstrap. GISAID accession numbers: EPI_ISL_2918997, EPI_ISL_2919009, EPI_ISL_2918996, EPI_ISL_2919005, EPI_ISL_2919016, EPI_ISL_2919006, EPI_ISL_2918999, EPI_ISL_2919007, EPI_ISL_2918998, EPI_ISL_2919008, EPI_ISL_2919001, EPI_ISL_2919012, EPI_ISL_2919002, EPI_ISL_2919013, EPI_ISL_2919003, EPI_ISL_2919014, EPI_ISL_2919004, EPI_ISL_2919015, EPI_ISL_2919010, EPI_ISL_2919000, EPI_ISL_2919011, EPI_ISL_2925736, EPI_ISL_2928141, EPI_ISL_2928341, EPI_ISL_2928344, EPI_ISL_2928147, EPI_ISL_2928510, EPI_ISL_2928509, EPI_ISL_2928508, EPI_ISL_2928507.
3.2. Epidemiological data
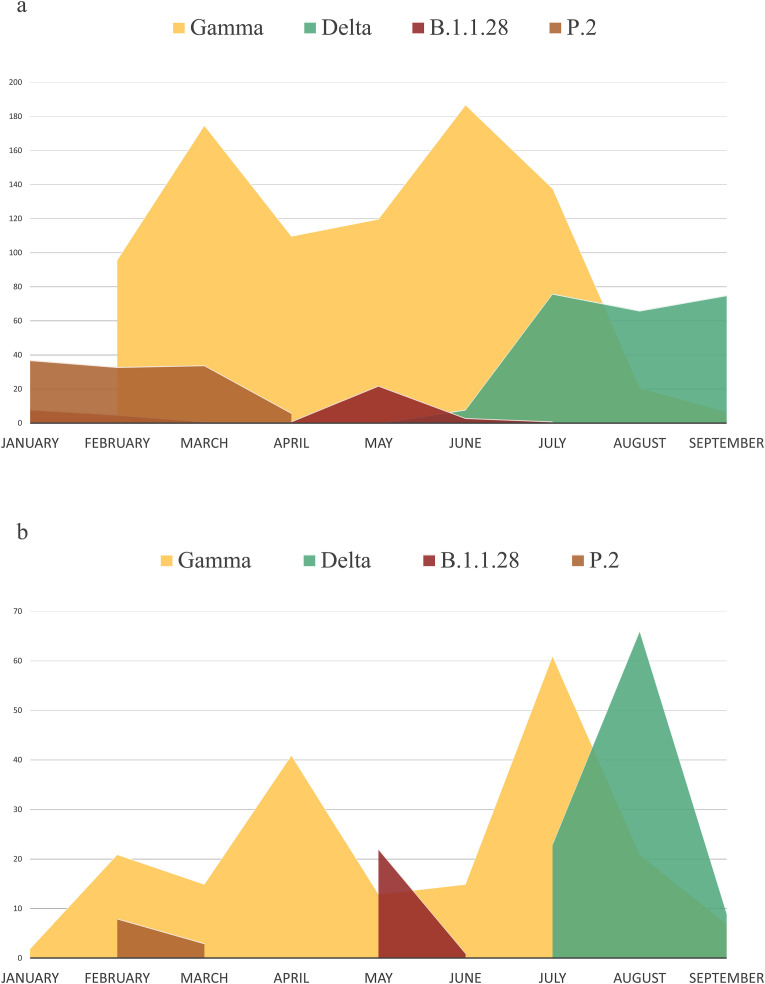
The total of 1256 sequences from RS was analyzed in order to better understand the major variants distribution scenario in 2021 (Fig. 2 ). From January to mid-July, we observe the Gamma lineage predominance, with two biggest waves between January to March, and May to July. Interestingly, it's possible to note that B.1.1.28 lineage presented an important peak that was concomitant with a decrease in the Gamma lineage and occurred in the same period of the putative newly B.1.1.28-derived variant detection. At the end of July, the entrance of Delta lineage was observed and, soon after, its occurrence started to increase, possibly responsible for the decreasing in Gamma and B.1.1.28 lineages frequencies. Complete Rio Grande do Sul State scenario could be observed in Fig. 2a. Reproducing this analysis with the sequences generated by our research group in the same period of time, we could observe the same lineages fluctuation, corroborating the representativeness of our studies when compared to the RS state current scenario. (Fig. 2b).
Fig. 2.
SARS-CoV 2 major lineages distribution. In 2a, the total of 1256 SARS-CoV-2 complete genomes retrieved from GISAID database between January and September in Rio Grande do Sul state. In 2 b, the total of 334 complete SARS-CoV 2 genomes sequenced by our Molecular Microbiology laboratory at the same period, considering only samples taken from individuals passing the Argentina-Brazil border and those living in the most populous region of the state.
4. Discussion
The high levels of SARS-CoV-2 globally transmission favor the emergence of new variants, which have potential impacts on COVID-19 severity, transmission, diagnostics, therapeutics, and natural and vaccine-induced immunity [7]. Since the beginning of 2021, Gamma was the predominant lineage in Rio Grande do Sul State, responsible for the major COVID-19 epidemic wave [8]. During the P.1 predominance, we observed an important B.1.1.28 peak in the State, concomitant with the sequencing of a putative new B 1.1.28-derived variant in 26 samples. The most samples were from passengers crossing the border between Brazil and Argentina, in Uruguaiana city, an important center of commerce and tourism. Brazil and Argentina's border remained partially closed during the pandemic, but it will be reopened in October 2021. Travelers and border openings represent a possible risk for increasing COVID-19 cases number, as well as entry and/or the emergence of new variants [3,9].
The sequences reported herein presented a unique constellation of specific newly mutations. Newly arising mutations are a natural by-product of viral replication and in the most cases, are determined by natural selection [10]. Among the likely B.1.1.28-derived lineage S protein signatures, N234P was found in 13 genomes and E471Q in 23. Observing the presence of these mutations, the minimum and maximum frequency per sample was around 26–30% and 70–78%, respectively (Table 1). E471Q has already been detected in some countries, such as India, in a local circulating variants study [11], nevertheless, it has never been reported in Brazil, nor has it been associated with B.1.1.28 lineage. A systematic study based on all possible future S protein mutations tested E471Q and the results showed an increased binding affinity on the receptor-binding domain (RBD) [12], which can increase viral fitness. The number of the putative B.1.1.28-derived lineage could be a study limitation, nevertheless, at the same period, 92 SARS-CoV-2 complete genomes were sequenced by our laboratory, so the sequences described herein correspond to 28% of the total.
Mutations that confer a competitive advantage with respect to viral fitness will probably increase in frequency, and those that reduce viral replication, and transmission, do not escape from immunity or lose variants competitions, tend to be culled from the population of circulating lineages [10]. In our genomic surveillance study, we could have observed this competition lineage context; once the pressure caused especially due to the second Gamma wave (June to August) and the Delta lineage emergence in RS (July) could have prevented B.1.1.28 establishment.
Although, B.1.1.28-derived lineage cases had an important representativeness in the RS lineages fluctuation scenario. The documented peak noticed in the State sequencing was concomitant to a decrease in Gamma frequency, showing the possibility of some pressure caused by the lineage. Furthermore, in that moment, represented a real threat, and it can still resume its peak in the course of the time and/or with the reopening of the country's borders. The fluctuation in the presence and spread of different lineages has already been observed in several countries, however, it is important to be careful about the introduction of new lineages, as sporadic detections do not always remain [13]. Besides, SARS-CoV-2 increased sequencing around the world marks a unique opportunity to analyze virus spreading and evolution in a worldwide context. Genomic surveillance studies are indeed a great contribution to COVID-19 pandemic control, especially in highly interconnected borders.
5. Conclusions
In this study, we identified the emergence of a putative B.1.1.28-derived lineage circulating in travelers crossing the Brazil-Argentina border. The sequences presented a unique additional non-synonymous mutational profile. In addition, an epidemiological genomic study was performed with all Rio Grande do Sul State complete SARS-CoV-2 genomes retrieved from GISAID database. This analysis revealed an important peak of B.1.1.28 with a decrease in Gamma variant frequency in the same period of time. SARS-CoV-2 genomic surveillance should be continued to monitor the emergence and spread of new variant strains, enabling evidence-based decision-making.
Funding
This work is an initiative of Rede Corona-ômica BR MCTI/FINEP affiliated to RedeVírus/MCTI (FINEP = 01.20.0029.000462/20, CNPq = 404,096/2020–4), and FAPERGS (grant 21/2551–0000081-3). FRS is a CNPq research fellow.
CRediT authorship contribution statement
Mariana Soares da Silva: Conceptualization, Investigation, Writing – review & editing. Juliana Schons Gularte: Investigation, Writing – review & editing. Meriane Demoliner: Investigation, Writing – review & editing. Alana Witt Hansen: Investigation, Writing – review & editing. Fágner Henrique Heldt: Investigation, Writing – review & editing. Micheli Filippi: Investigation, Writing – review & editing. Cristiane Borba Luckmann: Investigation, Writing – review & editing. Vyctoria Malayhka de Abreu Góes Pereira: Investigation, Writing – review & editing. Rodrigo de Almeida Vaucher: Methodology, Investigation. Victor dos Santos Barboza: Methodology, Investigation. Janice Luehring Giongo: Methodology, Investigation. Raquel Borba Rosa: Methodology, Investigation. Evelise Tarouco da Rocha: Methodology, Investigation. Bruno Kilpp Goulart: Methodology, Investigation. Fernanda dos Santos Fernandes: Methodology, Investigation. Juliana Maciel Pinto: Methodology, Investigation. Leandro Pergher Bolzan: Methodology, Investigation. Marta Maria Medeiros Frescura Duarte: Methodology, Investigation. Matheus Nunes Weber: Methodology, Investigation, Writing – review & editing. Paula Rodrigues de Almeida: Writing – review & editing. Juliane Deise Fleck: Writing – review & editing. Fernando Rosado Spilki: Writing – review & editing, Supervision.
Acknowledgements
We thank the Brazilian Coordination for the Improvement of Higher-Level Personnel (CAPES), Foundation for Research Support of the State of Rio Grande do Sul (FAPERGS), and Brazilian National Council for Scientific Development (CNPq) for scholarships. See Supplementary Acknowledgments.
References
- 1.Rambaut A., Loman N., Pybus O., et al. Virological.org; 2020. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. [Google Scholar]
- 2.Tegally H., Wilkinson E., Giovanetti M., et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020 doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- 3.Fujino T., Nomoto H., Kutsuna S., et al. Novel SARS-CoV-2 variant in travelers from Brazil to Japan. Emerg Infect Dis. 2021;27(4):1243–1245. doi: 10.3201/eid2704.210138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherian S., Potdar V., Jadhav S., et al. SARS-CoV-2 spike mutations, L452R, T478K, E484Q and P681R, in the second wave of COVID-19 in Maharashtra, India. Microorganisms. 2021;9(7):1542. doi: 10.3390/microorganisms9071542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harvey W.T., Carabelli A.M., Jackson B., et al. SARS-CoV-2 variants, spike mutations and immune escape. Nature. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen L.-T., Schmidt H.A., von Haeseler A., et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.CDC . US Department of Health and Human Services, CDC; Atlanta, GA: 2021. SARS-CoV-2 variant classifications and definitions.https://www.cdc.gov/coronavirus/2019-ncov/variants/variant-info.html [Google Scholar]
- 8.da Silva S.J.R., Pena L. Collapse of the public health system and the emergence of new variants during the second wave of the COVID-19 pandemic in Brazil. One Health. 2021 doi: 10.1016/j.onehlt.2021.100287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yadav P.D., Gupta N., Nyayanit D.A., et al. Imported SARS-CoV-2 V501Y.V2 variant (B.1.351) detected in travelers from South Africa and Tanzania to India. Trav Med Infect Dis. 2021;41 doi: 10.1016/j.tmaid.2021.102023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lauring A.S., Hodcroft E.B. Genetic variants of SARS-CoV-2—what do they mean? JAMA. 2021;325(6):529–531. doi: 10.1001/jama.2020.27124. [DOI] [PubMed] [Google Scholar]
- 11.Chand G.B., Banerjee A., Azad G.K. Identification of twenty-five mutations in surface glycoprotein (Spike) of SARS-CoV-2 among Indian isolates and their impact on protein dynamics. Gene reports. 2020;21 doi: 10.1016/j.genrep.2020.100891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen J., Wang R., Wang M. Mutations strengthened SARS-CoV-2 infectivity. J Mol Biol. 2020;432(19):5212–5226. doi: 10.1016/j.jmb.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arantes I.G., Salvato R.S., Gregianini T.S., et al. Multiple introduction of SARS-CoV-2 C.37 lambda lineage in the southern Brazilian region. Travel Med. 2021;27:153. doi: 10.1093/jtm/taab153. [DOI] [PMC free article] [PubMed] [Google Scholar]