Abstract
We investigated the fungus Aspergillus fumigatus PD-18 responses when subjected to the multimetal combination (Total Cr, Cd2+, Cu2+, Ni2+, Pb2+, and Zn2+) in synthetic composite media. To understand how multimetal stress impacts fungal cells at the molecular level, the cellular response of A. fumigatus PD-18 to 30 mg/L multimetal stress (5 mg/L of each heavy metal) was determined by proteomics. The comparative fungal proteomics displayed the remarkable inherent intracellular and extracellular mechanism of metal resistance and tolerance potential of A. fumigatus PD-18. This study reported 2,238 proteins of which 434 proteins were exclusively expressed in multimetal extracts. The most predominant functional class expressed was for cellular processing and signaling. The type of proteins and the number of proteins that were upregulated due to various stress tolerance mechanisms were post-translational modification, protein turnover, and chaperones (42); translation, ribosomal structure, and biogenesis (60); and intracellular trafficking, secretion, and vesicular transport (18). In addition, free radical scavenging antioxidant proteins, such as superoxide dismutase, were upregulated upto 3.45-fold and transporter systems, such as protein transport (SEC31), upto 3.31-fold to combat the oxidative stress caused by the multiple metals. Also, protein–protein interaction network analysis revealed that cytochrome c oxidase and 60S ribosomal protein played key roles to detoxify the multimetal. To the best of our knowledge, this study of A. fumigatus PD-18 provides valuable insights toward the growing research in comprehending the metal microbe interactions in the presence of multimetal. This will facilitate in development of novel molecular markers for contaminant bioremediation.
Keywords: multi-metal, proteomics, mycoremediation, LC-MS/MS, fungi
Graphical Abstract
Highlights
-
-
First proteomic study on filamentous fungus Aspergillus fumigatus for hexametal removal.
-
-
Most upregulated class posttranslational modification, protein turnover, chaperones.
-
-
Major protein player in protein-protein interaction identified as cytochrome oxidase.
-
-
Expressed proteins are environmental biomarkers in heavy metals-fungus interaction.
Introduction
Heavy metals, such as copper (Cu), chromium (Cr), cadmium (Cd), zinc (Zn), lead (Pb), and nickel (Ni), occur in the water bodies, such as river water, and drains, in developing countries that are above the permissible mandates as prescribed by the Food and Agriculture Organization (Bhattacharya et al., 2015). These heavy metals are released from numerous small- and medium-scale enterprises (pesticide, textile, electroplating, fertilizer, batteries, etc.) due to lack of or improper sewage treatment plant systems (Bhattacharya et al., 2015; Bhardwaj et al., 2017; Gola et al., 2020). These hazardous contaminants are discharged into various water bodies via irrigation during agricultural activities. This results in bioaccumulation of these contaminants that enter the food chain and cause detrimental health ailments, such as cancer in human beings (Kou et al., 2018; Li et al., 2019; Abed et al., 2020; Chauhan et al., 2021; Irawati et al., 2021). Thus, these heavy metals are the environmental priority contaminants threatening the environment and therefore must be remediated before discharge into the environment.
The conventional physico-chemical methods, such as chemical precipitation, ion exchange, adsorption, membrane filtration, coagulation-flocculation, and flotation, are usually utilized to remediate these harmful contaminants. These techniques have the disadvantages of being expensive, having low selectivity, production of additional sludge, and further treatment is required for better results (Dey et al., 2021; Zamora-Ledezma et al., 2021).
On the other hand, living and actively growing microbial cells can be a lucrative option for bioremediation (Malik, 2004). Of the bioremediation techniques, mycoremediation has shown to be a promising technology having the potential to ameliorate these hazardous chemicals (Dey et al., 2020; Sabuda et al., 2020). The superiority of fungi over single-celled microbes, such as bacteria, to remediate these recalcitrant heavy metals is well-documented (Deshmukh et al., 2016). In addition, the fungi are omnipresent, multifarious, and have a wider arsenal to acclimatize to environmental limitations, such as immoderations of temperature, extremes of pH, higher metal concentrations, and low nutrient accessibility, due to their morphological diversity (Anand et al., 2006). Besides, fungal mycelia have enhanced enzymatic and mechanical contact with the pollutant due to a greater cell to the surface ratio (Sagar and Singh, 2011).
Further, in microbial cells, such as fungus, heavy metals are key components in the number of catalytic and structural proteins that are integral to biochemical processes. The outcome of these processes differs depending on the type of metal involved and its concentration inside the cell (Gadd, 1994). Particularly, the species of Aspergillus have a high metal uptake capacity for metals, such as Cu (Dusengemungu et al., 2020). Also, filamentous fungi develop signature metabolic pathways which are species-specific to survival in the harsh environment of heavy metals and other contaminants that are utilized as nutrients and energy sources (Kuhn and Käufer, 2003). Accordingly, proteins expressed in cells under such diverse conditions and at different times are dissimilar (Boopathy, 2000; Keller, 2015; Wisecaver and Rokas, 2015).
Also, there is evidence that fungal resistance toward one element does not necessarily infer resistance to another element even though the elements possess similar valency charges (Høiland, 1995). For example, the toxicity of Pb toward the microbe is less compared to other toxic metals, such as Cd, As, and Hg (Jiang et al., 2020). Further, in fungi, the metallothioneins formation is predominantly induced by the heavy metal Cu (Jaeckel et al., 2005). Thus, understanding the altered heavy metal uptake in the fungus in presence of diverse environmental conditions needs to be evaluated.
To gain such mechanistic insight, high throughput techniques, such as proteomic analysis by LC-MS/MS, can be used to identify and characterize proteins involved in the multimetal resistance mechanism in a filamentous fungus (Pandey and Mann, 2000; Kraut et al., 2009; Zhang et al., 2010). Moreover, proteomics facilitates the development of new and important protein biomarkers that specify and monitor metal contamination in the environments as protein type and also their copy numbers are estimated by the translational regulation (Ohno et al., 2014).
Several researchers have studied the change in proteomes under various metal conditions. Selamoglu et al. (2020) have assessed the change in the differential proteome expression of the prokaryotic organism Rhodobacter capsulatus in presence of 5 μM Cu by nano-LC-MS/MS. About 75 proteins were significantly regulated. Most of the proteins present were responsible for maintaining Cu homeostasis. Further, Lotlikar et al. (2020) enumerated the variable expression of proteins in a eukaryotic fungus Penicillium chrysogenum in the presence of 100 mg/L and 500 mg/L Cu. Several key proteins related to genetic information, carbohydrate metabolism, glycan biosynthesis and metabolism, amino acid metabolism, and energy metabolism were expressed.
Cherrad et al. (2012) highlighted the overaccumulation of proteins of the oxidoreductase family when exposed to Cd, Cu, and Ni but not when exposed to Zn. Thus, the secretion of proteins in fungus is extremely dynamic and its production depends on different environmental triggers.
However, despite these growing proteomic studies dealing with the bioremediation of single heavy metal, there is a dearth of information on the modulated proteins triggered by the cumulative toxicity of hexametals, namely, Cd, Cr, Cu, Ni, Pb, and Zn, specifically in a filamentous fungus. Interestingly, Aspergillus fumigatus has high adaptability when subjected to an altered environment (Bakti et al., 2018). Also, as established in our previous works (Dey et al., 2016, 2020; Bhattacharya et al., 2020), A. fumigatus PD-18 is a filamentous fungus capable of removal of 30 mg/L multimetal exceptionally well. Furthermore, there are structural and functional similarities between the numerous genes of lower eukaryotes, such as fungi and mammals (Bae and Chen, 2004).
Thus, this fungus A. fumigatus PD-18 would be a good eukaryotic model for helping us understand how cells adopt various cellular strategies and lay a foundation study to decipher the enzymes produced in the presence of simultaneous effects of multimetal cocktail on interaction with a fungus for scale-up process.
Materials and Methods
Chemicals and Reagents
The stock solutions (10 g/L) of different individual metals were made by dissolving their respective salts, viz. K2Cr2O7, Cd(NO3)2, Ni(NO3)2, Cu(NO3)2, Zn(NO3)2, and Pb(CH3COO)2 in double-distilled water and were diluted to the concentrations that were required for the experiments. For preparing the reagents and calibration standards, deionized ultrapure water (RIONS Ultra 370 series) was used. Rest all other chemicals utilized were of analytical grade and were obtained from Merck, Sigma, and Qualigens.
Microorganism and Culture Media Composition
The fungal strain used was A. fumigatus PD-18 which was isolated from the polluted banks of the river Yamuna, New Delhi, India, and characterized with accession number KX365202 after depositing the sequence to the Genbank (NCBI) (Dey et al., 2016).
The growth media used was the composite medium with following composition (g/L): (NH4NO3, 0.5; K2HPO4, 0.5; MgSO4.7H2O, 0.1; NaCl, 1.0; Yeast extract, 2.5; pH 6.8 ± 0.2). The sterilization of the media was carried out at 121°C for 15 min. Glucose (10.0 g/L) was added to the flasks separately after autoclaving to avoid precipitation.
Methodology
The methodology adopted in this study was to estimate the differential expression of proteins in A. fumigatus PD18 brought about by 30 mg/L multimetal (MM) viz. Five milligram per liter of each of the individual Cd, Total Cr, Cu, Ni, Pb, and Zn amended in the composite media in addition to 1% of glucose. The concentration of 5 mg/L of each heavy metal was chosen to take into consideration the permissible mandates for heavy metal occurrence in water that can be utilized for irrigation according to Food and Agriculture Organization (FAO). The prescribed limit for each heavy metal is as follows: Cd, 0.01; Total Cr, 0.1; Cu, 0.2; Ni, 0.2; Pb, 5.0; Zn, 2.0. Also, the typical concentration of heavy metals occurring in a mixture in the Yamuna river was considered (Bhattacharya et al., 2015). The biotic control included composite media added with only 1% of glucose.
Studies were conducted in a series of Erlenmeyer flasks (250 mL) comprising 100 mL of composite growth media. One milliliter of spore suspension (having a concentration of 107 spores eluted with sterile distilled water containing 0.01% Tween 80) was inoculated in the flasks and incubated at 30°C and 150 rpm agitation for 72 h to ensure complete uptake of metal ions. The flasks were withdrawn after the late log phase incubation period. Analysis was done in three technical replicates for biotic control and multimetal treated samples separately.
Proteomics Analysis
Cell Lysis and Protein Extraction
The fungal pellets were separated from the media by inversion and followed by centrifugation at 4,000 g for 10 min at 4°C. The mycelia were further snap-freezed in liquid nitrogen and then lyophilized and stored as a dry powder at –20°C before the subsequent steps. The total protein from the dried powdered fungal cells was extracted in a buffer of composition (8M Urea and 2M Thiourea) as per the modified protocol (Rughöft et al., 2020). The supernatant was collected by centrifugation at 14,000 rpm, 4°C for 10 min, and precipitated overnight in five volumes of ice-cold acetone and the pellet was stored at –20°C for further use. The concentration of protein to be measured was estimated according to the Bradford assay of protein quantification (Bradford, 1976). In brief, a 2-mg/mL concentration stock solution of Bovine Serum Albumin (BSA) was prepared in water. BSA standard curve was prepared using the following concentration range (0–2.00 μg/μL) followed by the addition of Coomassie Brilliant Blue dye reagent in 95% ethanol and 100 mL 85% (w/v) phosphoric acid and incubated for 5 min. The absorbance was measured at 595 nm.
Protein Separation by 1D-SDS PAGE and in Gel Tryptic Digestion of the Proteins
The separation of the proteins was carried out by one-dimensional SDS-PAGE and the separated protein bands were visualized by Coomassie brilliant blue staining. The individual gel bands were cut into small pieces and washed with 10 mM ammonium bicarbonate followed by reduction with 10 mM 1,4-Dithiothreitol and 100 mM 2-Iodoacetamide. The proteins lysates were subjected to tryptic digestion and incubated overnight and kept at 37°C. The peptides were extracted from the gel pieces in 5 mM ammonium bicarbonate and further purified by C-18 ZipTip columns according to the protocol (Haange et al., 2012). Thereafter, the peptide lysates were dried in a vacuum centrifuge and stored at –20°C until further analysis.
Mass Spectrometric Analysis
The dried peptides lysates were reconstituted in 0.1% (v/v) formic acid and loaded and separated by reversed-phase chromatography before being measured on mass spectrometer UPLC-coupled (Waters) LTQ Orbitrap Velos MS/MS (Thermo Fisher Scientific).
Data Analysis (Identification and Characterization of the Proteins)
The raw MS-MS ion spectra data from the instrument were processed using the Proteome Discoverer software (v1.0 build 43, Thermo Fisher Scientific) (Orsburn, 2021). The fixed and variable modifications (Carbamidomethylation at cysteines was given as fixed and oxidation of methionines as variable modifications) were taken into consideration. Only ranked one peptide hits with ≥1 high confidence and less than 1% false discovery rate (FDR) were considered as identified and taken up for further analysis. The intensity data were further processed by logarithmic transformation and normalization for the determination of changes in the abundance of proteins (at least 1.5-fold upregulated or downregulated). Only the values of p < 0.05 were considered as statistically significant. Triplicate gels were run for biotic control and MM samples. For functional annotation of the proteins, the generated protein lists were analyzed by prophane.1
The creation of the VENN diagrams was done by the online tool VENNY.2 The heatmaps and volcano plots were constructed using the heatmap and volcano programs from R tool version 3.2.0. The WoLF PSORT tool3 was used for the prediction of the subcellular localization of the proteins. The protein–protein interactions between the differentially expressed proteins were studied by the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database.4
Results
Fungi usually acclimate by making dynamic changes in the cell structure and composition when exposed to heavy metals. In our previous study on the toxicity of individual metals, that is, Cd, Cr, Cu, Ni, Pb, and Zn (500 mg/L) in A. fumigatus PD-18, we determined the highest tolerance index for each metal in solid media. We calculated the cube root growth (k) constant of A. fumigatus PD-18 when subjected to 30 mg/L multimetal in liquid composite media. This fungal strain had exceptional multimetal removal ability after ∼72 h. The growth profile of the fungus was altered when multimetal was added to the composite media and substantial variations were observed when compared with individual metals (Dey et al., 2016).
Further, the morphological changes in this fungus in response to the 30 mg/L multimetal were determined by scanning electron microscope, the localization of the heavy metals inside the fungal cell by transmission electron microscopy, and chelation of the heavy metals with the functional groups occurring in the fungus by Fourier-transform infrared spectroscopy (Dey et al., 2020).
Expression of Proteins in Biotic Control and Multimetal Extracts of Aspergillus fumigatus PD-18
From the six sample preparations (conditions, including one biological control and one multimetal exposure and their three technical replicates), 434 proteins were uniquely present in multimetal extracts and 400 proteins in biotic control of A. fumigatus PD-18.
Figure 1 depicts the expression of the proteins isolated from A. fumigatus PD-18 mycelia as established with SDS-PAGE. This expression of proteins from the biotic control and multimetal (MM) stressed PD-18 were observed after Coomassie Brilliant Blue staining.
FIGURE 1.
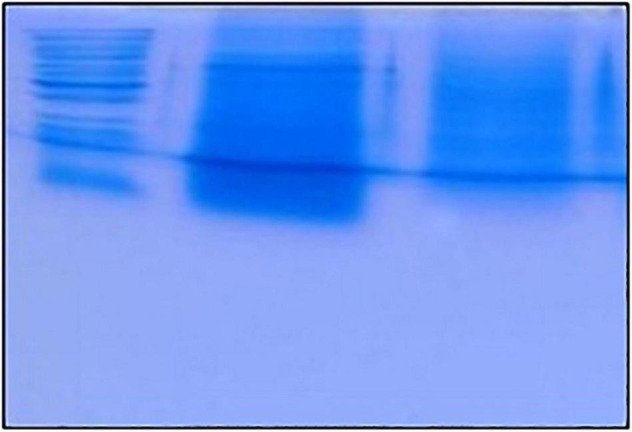
Coomassie brilliant blue stained segment of SDS-PAGE protein expression patterns in heavy metal treated Aspergillus fumigatus in absence of multimetal (Biotic control in 2nd lane); 30 mg/L multimetal (3rd lane). Molecular-weight markers (M) are shown on the left (1st lane).
Proteins Present in High Abundances in Aspergillus fumigatus PD-18
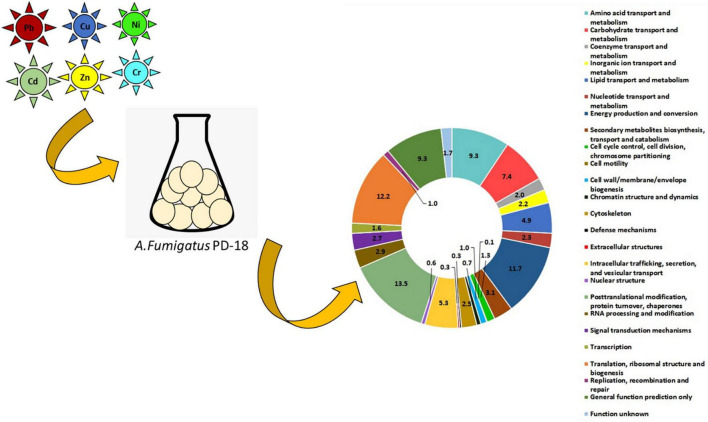
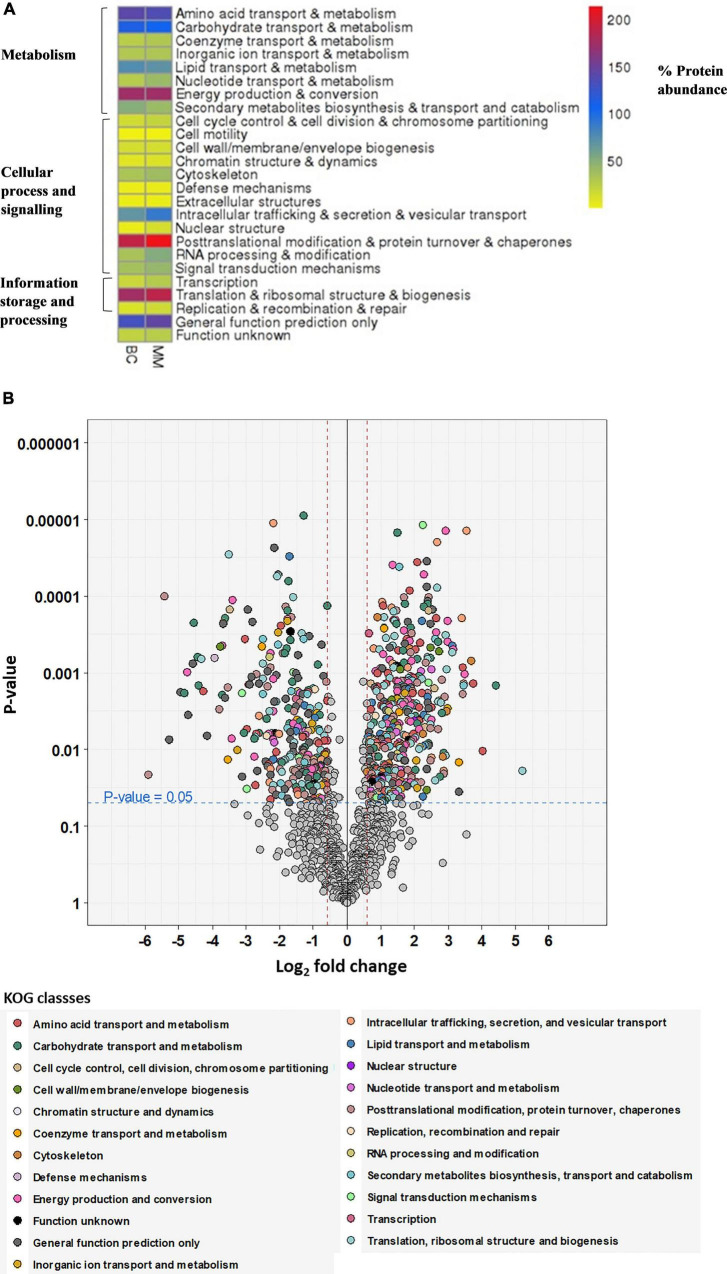
Figure 2A depicts the heatmap of differently regulated proteins of the multimetal exposure when compared to the biotic control without multimetal. The detected proteins were categorized under three functional KOG groups related to metabolism, cellular process and signaling, and information storage and processing. These categories were associated with fungal physiology and most of the identified proteins were further classified on the basis of different 23 KOG classes which are discussed as follows:
FIGURE 2.
(A) Heatmap visualization of the abundance of the proteins as red = higher values, dark blue = mid values, yellow = lower values depicted on (x-axis) for proteome abundance of 30 mg/L multimetal (MM) and proteome abundance without heavy metal exposure (BC). (B) Volcano plot showing the distribution of quantified proteins according to p-value and log2 fold change, indicating significance level at 0.05 (blue dashed line) and fold change at –1.5 and 1.5 (red dashed lines), colored according to their KOG class. Proteins, which were not significant or fold change within –1.5 to 1.5 are colored in gray.
-
•
metabolism [amino acid transport and metabolism (9%), carbohydrate transport and metabolism (7%), coenzyme transport and metabolism (2%), inorganic ion transport and metabolism (2%), lipid transport and metabolism (5%), nucleotide transport and metabolism (3%), energy production and conversion (11%), and secondary metabolites biosynthesis and transport and catabolism (3%)];
-
•
cellular process and signaling [cell cycle control and cell division and chromosome partitioning (1%), cell motility (0.1%), cell wall/membrane/envelope biogenesis (1%), chromatin structure and dynamics (1%), cytoskeleton (3%), defense mechanisms (0.3%), extracellular structures (0.3%), intracellular trafficking, secretion, and vesicular transport (6%), nuclear structure (1%), post-translation modification and protein turnover and chaperones (14%), RNA processing and modification (3%), and signal transduction mechanism (3%)];
-
•
information storage and processing [transcription (2%), translation and ribosomal structure and biogenesis (12%), replication, recombination, and repair (1%), proteins with general function (9%), and function unknown (2%)].
From Figure 2A, the proteins that were expressed in higher abundance in A. fumigatus when treated with 30 mg/L multimetal belonged to the main metabolic functional classes of RNA processing & modification, post-translational modification & protein turnover & chaperones, nuclear structure, intracellular trafficking & secretion & vesicular transport, translation & ribosomal structure & biogenesis, and nucleotide transport & metabolism. Further, hypothetical proteins with general function prediction were also expressed which could play role in tackling the multimetal stress. The afflicted metabolic functions classes where production of proteins decreased belonged to amino acid transport & metabolism, carbohydrate transport & metabolism, lipid transport & metabolism, and secondary metabolites biosynthesis & transport and catabolism. This proteomic study displayed proteins in A. fumigatus PD-18 that showed a significant fold increase and decrease of differentially expressed proteins during growth in 30 mg/L multimetal supplemented composite media as depicted in the volcano plot in Figure 2B. Among the differentially expressed proteins identified, 399 proteins were differentially upregulated while 266 proteins were differentially downregulated. The highly upregulated protein identified hydroxymethylglutaryl-CoA synthase upregulated upto 5.2-fold times. Other important proteins with general function prediction were upregulation of serine/threonine-protein phosphatase upto 3.42-fold times and G-protein beta subunit SfaD upto 2.98-fold. The highly upregulated and downregulated proteins by fold changes and their locations inside the fungal cell are depicted in Table 1. Furthermore, it was found that the majority of the upregulated proteins were located in the cytoplasm (18), mitochondria (12) followed by extracellular (6), nuclear (5), plasma membrane (2), peroxisome (2), nuclear and cytoplasmic (1), and endoplasmic reticulum lumen (1).
TABLE 1.
Differential abundance of highly regulated selected proteins in Aspergillus fumigatus grown in presence 30 mg/L MM.
| Protein accession number | Protein name | Fold change | Cellular location of protein | Protein KOG class | p-value |
| Upregulated proteins | |||||
| A1CQV3 | hydroxymethylglutaryl-CoA synthase | 5.19 | Cytoplasm | Lipid transport and metabolism | 0.019 |
| B0XZB6 | aminotransferase family protein | 4.03 | Mitochondria | Amino acid transport and metabolism | 0.010 |
| Q4WV94 | proteasome regulatory particle subunit Rpt3 | 3.74 | Peroxisome | Post-translational modification, protein turnover, chaperones | 0.001 |
| Q4WXX9 | fibrillarin | 3.68 | Cytoplasm | RNA processing and modification | 0.001 |
| Q4X0E9 | glutamate dehydrogenase | 3.54 | Mitochondria | Amino acid transport and metabolism | 0.001 |
| Q4WLA8 | superoxide dismutase | 3.45 | Mitochondria | Inorganic ion transport and metabolism | 0.001 |
| A0A017SPG0 | calcium/calmodulin-dependent protein kinase | 3.45 | Cytoplasm | Signal transduction mechanisms | 0.001 |
| A0A017SD45 | ATP synthase | 3.44 | Mitochondria | Energy production and conversion | 0.001 |
| Q4WM81 | serine/threonine-protein phosphatase | 3.42 | Nuclear | Signal transduction mechanisms | 0.001 |
| A1C6S1 | histone H4 | 3.40 | Mitochondria | Chromatin structure and dynamics | 0.001 |
| A0A084BYE7 | protein transport protein (SEC31) | 3.31 | Mitochondria | Intracellular trafficking, secretion, and vesicular transport | 0.036 |
| A0A017S654 | glutamate decarboxylase | 3.16 | Peroxisome | Amino acid transport and metabolism | 0.001 |
| A2R0I8 | 26S protease regulatory subunit | 3.12 | Nuclear and cytoplasmic | Post-translational modification, protein turnover, chaperones | 0.001 |
| B0XVU3 | RNA binding effector protein Scp160 | 3.11 | Endoplasmic reticulum lumen | Lipid transport and metabolism | 0.001 |
| O60022 | probable HECT-type ubiquitin ligase | 2.99 | Extracellular | General function prediction only | 0.002 |
| A1CI51 | G protein beta subunit SfaD | 2.98 | Cytoplasm | General function prediction only | 0.001 |
| Q4WLV1 | 26S proteasome regulatory particle subunit Rpn8 | 2.96 | Extracellular | Post-translational modification, protein turnover, chaperones | 0.001 |
| Q4WFD3 | coatomer subunit gamma | 2.94 | Cytoplasm | Intracellular trafficking, secretion, and vesicular transport | 0.012 |
| A0A017S6B4 | phosphoglycerate kinase | 2.91 | Mitochondria | Carbohydrate transport and metabolism | 0.001 |
| Q4WX43 | endosomal cargo receptor (P24) | 2.86 | Nucleus | Intracellular trafficking, secretion, and vesicular transport | 0.020 |
| A0A084C1K3 | glutamyl-tRNA synthetase | 2.86 | Nucleus | Translation, ribosomal structure and biogenesis | 0.011 |
| A0A017S4K9 | pyridoxine biosynthesis protein | 2.82 | Cytoplasm | Coenzyme transport and metabolism | 0.001 |
| B0XXM2 | CBS and PB1 domain protein | 2.73 | Extracellular | Energy production and conversion | 0.002 |
| A1CPB0 | sulfate adenylyltransferase | 2.71 | Cytoplasm | Inorganic ion transport and metabolism | 0.001 |
| A0A084BD28 | importin beta 4 subunit | 2.69 | Nucleus | Nuclear structure | 0.002 |
| Q4WHV3 | oxidoreductase short chain dehydrogenase | 2.58 | Plasma membrane | Secondary metabolites biosynthesis, transport and catabolism | 0.001 |
| Q4 × 0N5 | T-complex protein 1 subunit gamma | 2.5 | Nucleus | Post-translational modification, protein turnover, chaperones | 0.012 |
| A0A084BP78 | acetolactate synthase | 2.49 | Mitochondria | Amino acid transport and metabolism | 0.005 |
| Q0CSN0 | Mitochondrial inner membrane translocase subunit TIM44 | 2.41 | Mitochondria | Intracellular trafficking, secretion, and vesicular transport | 0.002 |
| Q4WE44 | Cytochrome P450 phenylacetate 2-hydroxylase | 2.39 | Cytoplasm | Secondary metabolites biosynthesis, transport and catabolism | 0.001 |
| I7ZRN7 | glyceraldehyde-3-phosphate dehydrogenase | 2.38 | Cytoplasm | Carbohydrate transport and metabolism | 0.001 |
| A4FSH5 | RAB proteins geranylgeranyltransferase component A | 2.37 | Extracellular | Post-translational modification, protein turnover, chaperones | 0.001 |
| Q5B2V1 | Peptidyl-prolyl cis-trans isomerase D | 2.37 | Cytoplasm | Post-translational modification, protein turnover, chaperones | 0.001 |
| B8NQD2 | mitochondrial enoyl reductase | 2.35 | Cytoplasm | Transcription | 0.033 |
| Q4WGJ9 | Tropomyosin | 2.26 | Mitochondria | Cytoskeleton | 0.041 |
| Q4WIT8 | Ubiquitin DskB | 2.13 | Extracellular | Post-translational modification, protein turnover, chaperones | 0.004 |
| A0A084BHN1 | Importin beta 5 subunit | 2.02 | Cytoskeleton | Intracellular trafficking, secretion, and vesicular transport | 0.018 |
| Upregulated proteins | |||||
| Q4WJB0 | ribonucleoside-diphosphate reductase | 1.93 | Mitochondria | Nucleotide transport and metabolism | 0.001 |
| Q4W9K3 | Mitochondrial outer membrane translocase receptor (TOM70) | 1.84 | Extracellular | Intracellular trafficking, secretion, and vesicular transport | 0.001 |
| G7XPM0 | ATP-dependent bile acid permease | 1.72 | Cytoplasm | Secondary metabolites biosynthesis, transport and catabolism | 0.014 |
| Q4 × 237 | translation release factor eRF3 | 1.71 | Mitochondria | Translation, ribosomal structure and biogenesis | 0.002 |
| Q4WVZ4 | isocitrate dehydrogenase LysB | 1.69 | Nucleus | Amino acid transport and metabolism | 0.013 |
| Q96UX3 | 14-alpha sterol demethylase | 1.69 | Cytoplasmic and nuclear | Secondary metabolites biosynthesis, transport and catabolism | 0.001 |
| A0A084C1V0 | DNA damage inducible v SNARE binding protein Ddi1 | 1.65 | Cytoplasm | Replication, recombination and repair | 0.018 |
| Q5B7M1 | Vesicular-fusion protein sec17 | 1.65 | Cytoplasm | Intracellular trafficking, secretion, and vesicular transport | 0.001 |
| A0A084BWQ6 | Nuclear pore complex subunit (SEC13) | 1.62 | Cytoplasmic and nuclear | Intracellular trafficking, secretion, and vesicular transport | 0.001 |
| Q0CCZ9 | bifunctional purine biosynthetic protein Ade1 | 1.60 | Cytoplasm | Nucleotide transport and metabolism | 0.020 |
| A0A017SJP6 | protein phosphatase 2a 65kd regulatory subunit | 1.54 | Plasma membrane | Signal transduction mechanisms | 0.044 |
| Q4WN17 | septin | 1.54 | Cytoplasm | Cell cycle control, cell division, chromosome partitioning | 0.001 |
| A1CB85 | transcription elongation factor spt4 | 1.49 | Cytoplasm | Post-translational modification, protein turnover, chaperones | 0.001 |
| P20445 | MICOS complex subunit mic60 | 1.39 | Cytoplasm | Cell wall/membrane/envelope biogenesis | 0.003 |
| Q6MYF0 | plasma membrane H +-ATPase Pma1 | 1.28 | Cytoplasm | Inorganic ion transport and metabolism | 0.005 |
| Downregulated proteins | |||||
| Q4WHK1 | Small nuclear ribonucleoprotein Sm D1 | −0.59 | Nucleus | Translation, ribosomal structure, and biogenesis | 0.034 |
| Q4WHE2 | NADH-ubiquinone oxidoreductase | −0.59 | Cytoplasm | Energy production and conversion | 0.002 |
| B0Y5W4 | Pyruvate carboxylase | −0.61 | Peroxisome | Post-translational modification, protein turnover, chaperones | 0.029 |
| G3XYJ1 | GST C-terminal domain-containing protein | −0.63 | Cytoplasm and nucleus | Amino acid transport and metabolism | 0.011 |
| Q4WCQ8 | ATP synthase subunit E | −0.64 | Nucleus | Energy production and conversion | 0.012 |
| Q4 × 0B7 | Arginyl-tRNA synthetase | −0.68 | Cytoplasm | Lipid transport and metabolism | 0.012 |
| A1CIL4 | Histidine–tRNA ligase | −0.75 | Mitochondria | Coenzyme transport and metabolism | 0.029 |
| B0XS44 | Carbamoyl-phosphate synthase | −0.77 | Mitochondria | Signal transduction mechanisms | 0.013 |
| Q6QNB5 | Squalene monooxygenase | −0.78 | Plasma membrane | Translation, ribosomal structure and biogenesis | 0.010 |
| Q4WY15 | Arginase | −0.77 | Mitochondria | Energy production and conversion | 0.016 |
Enriched Protein Network and Pathways in Aspergillus fumigatus PD-18
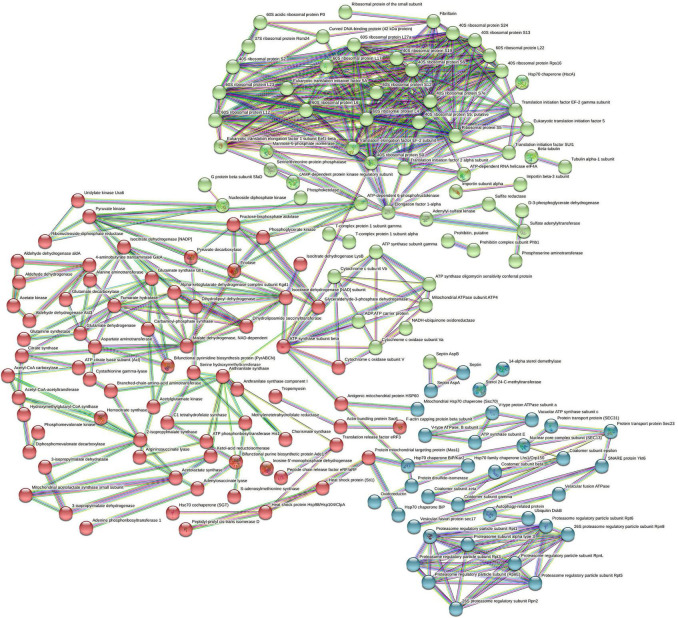
The interaction network as constructed by the STRING database of the highly upregulated proteins in A. fumigatus is depicted in Figure 3. The strength of the association is depicted by the thickness of the line. The number of nodes is 263 that represents the proteins, the number of edges is 467 that represents associations, the average node degree is 3.55, and the average local clustering coefficient is 0.412. The STRING analysis identified three clusters of protein interactions. The first major cluster of proteins (green) was involved in ribosome and preinitiation factors (60S and 40S ribosomal proteins). The predicted partners functionally associated with 60S and 40S ribosomal proteins include translation elongation factor, fibrillarin of class translation, ribosomal structure, and biogenesis. The second cluster proteins (red) have major hub proteins, such as malate dehydrogenase, ATP synthase, pyruvate kinase, fructose biphosphate aldolase, dihydrolipoyl dehydrogenase, glutamate synthase, anthranilate synthase, and isocitrate dehydrogenase. The third cluster proteins (blue) are proteasome regulatory particles, Hsp70 chaperone, protein transport protein (sec31), and V-type proton ATPase.
FIGURE 3.
STRING analysis displaying protein–protein interaction. The colored lines represent the edges that show associations among different proteins. Dark blue: connect proteins that are co-occurring phylogenetically. Dark green: connect proteins that are occurring as gene neighborhood. Red: shows proteins with gene fusions. The colored nodes (red, blue, green) are different clusters of proteins.
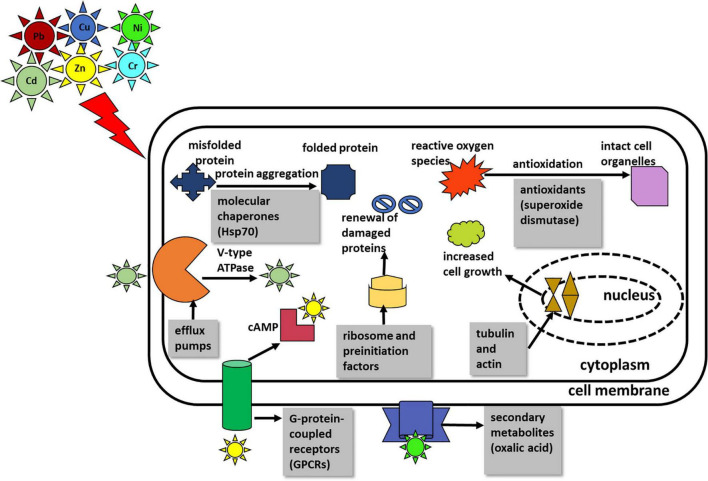
The significant junction proteins in protein–protein interaction identified were cytochrome c, oxidase, and septin suggest that energy production and conversion and cell cycle control, cell division, and chromosome partitioning that play a pivotal role in the resistance toward multimetal exposure. Figure 4 summarizes the salient mechanism of the multimetal detoxification process by A. fumigatus PD-18.
FIGURE 4.
Summary of the salient mechanism of multimetal detoxification process by Aspergillus fumigatus PD-18.
Discussion
In this study, we identified the chief classes of proteins that are crucial for the resistance and tolerance of A. fumigatus PD-18 for multimetals, including intracellular and extracellular mechanisms of metal uptake. These two processes are further elaborated.
Intracellular and Extracellular Mechanism of Metal Uptake by Fungus
The mechanism of the fungal resistance and tolerance could be attributed to its occurrence at the metal-contaminated site (Baker, 1987; Gadd and White, 1993). The fungal detoxification process involves strategies, such as intracellular bioaccumulation, extracellular precipitation, biotransformation, biomineralization, and biosorption, that involve several signaling pathways (Wang et al., 2019). In addition, the reduction of heavy metals from the cell comprises of activation of different metabolic processes in the cell that are stimulated by heavy metals (Goyal et al., 2003).
Fungi largely counter heavy metals in two ways. The first way involves averting the metal uptake and its passage inside the fungal cell. This happens chiefly by a decrease in metal uptake or increased efflux of metals, metal biosorption to the impermeable cell walls by metal binding polysaccharides, peptides, extracellular formation of complexes, and the release of organic acids that chelates heavy metals outside the cell. Thus, the extracellular mechanism operates in the cell to counter heavy metals by circumventing the entry of heavy metals. Particularly, the secondary metabolites, such as citric acid, oxalic acids, succinate, and fumarate, which are low molecular weight compounds (<900 daltons) are secreted by fungal species namely A. niger specifically in response to heavy metals exposure and they bind the heavy metals extracellularly (Kolen, 2013). Also, the secondary metabolite oxalic acid is produced as an intermediate compound in the biochemical tricarboxylic acid cycle (TCA) (Munir et al., 2001). Here, we found the upregulation of the key enzymes that are involved in the TCA cycle viz. phosphoglycerate kinase (2.91-fold), glyceraldehyde-3-phosphate dehydrogenase (2.38-fold), enolase (1.8-fold), and pyruvate kinase (1.86-fold) which mediates the enhanced secretion of oxalic acids.
In the second way, the fungus subsists the high concentration of metals inside the cell by tolerance after the process of detoxification via metal chelation by synthesizing ligands, such as metallothioneins and phytochelatins, that bind heavy metals intracellularly or by compartmentalization of heavy metals within the cell organelles of vacuoles by polyphosphates. The three main classes of intracellular peptides binding metal ions are phytochelatins (PCs), metallothioneins (MTs), and glutathione (GSH). MTs are low molecular weight cysteine-rich metal-binding proteins that have high affinity toward both the essential metal ions, such as Cu and Zn, non-essential metal ions, such as Cd, Hg, and Ag, and also have large metal-binding capacities (Reddy et al., 2014). Further, MTs chelate heavy metals by forming thiolate bonds with the heavy metals. Glutathione S transferases (GSTs) are enzymes that metabolize heavy metals and other contaminants by catalyzing the binding of glutathione to non-polar compounds comprising of electrophilic nitrogen, carbon, and sulfur atom (Morel et al., 2009). Usually, the metals Cd, Cu, Pb, and Zn are removed via glutathione (GSH)-mediated sequestration. However, in this study, there was no evidence of the production of proteins glutathione, metallothioneins, and phytochelatins despite the presence of these heavy metals Cd, Cu, Pb, and Zn in the multimetal mixture. The reason for this phenomenon could be attributed to the dynamics of individual heavy metals when present in a mixture. As the expression of GST is related to the type of heavy metal, its concentration, and the extent of treatment time of the heavy metal (Shen et al., 2015).
Further, it is reported that heavy metals induce oxidative damage to the cell membranes of fungi by the generation of reactive oxygen species (ROS). These ROS are detoxified by the production of antioxidants that are components from the thioredoxin system, such as peroxiredoxins, NADPH dehydrogenases, catalase, superoxide dismutase, and peroxidase, that enables the fungus to confront the reactive-oxygen species that accumulate in the cell on exposure to the metals (Zhang et al., 2015). Thus, in principle, intracellular mechanisms decrease the metal load in the cytosol (Sandau et al., 1996; Gadd, 2000, 2007, 2010). In this study, we found 3.45-fold upregulation of the antioxidant protein Cu-Zn superoxide dismutase. Other important functional groups were detected that expressed amino acid metabolism, lipid metabolism, energy metabolism, and also the proteins involved in signal transduction, transcription, translation, or DNA repair. In general, the upregulated proteins are stimulated to display the fungal resistance against the contaminants’ stress, while the downregulated proteins are suppressed by the action of the pollutants’ toxicity.
The highly upregulated protein hydroxymethylglutaryl-CoA synthase with 5.19-fold upregulation is responsible for the production of secondary metabolite carotenoid from the precursor molecule of acetyl-CoA when stimulated by the heavy metal stress (Bhosale, 2004). The protein serine/threonine phosphatase with 3.42-fold upregulation is responsible for maintaining the conformation of cell organelles and proteasomes (Dias et al., 2019).
The other proteins, such as G-protein beta subunit SfaD, with 2.98-fold upregulation depicted the role of G-protein-coupled receptors (GPCRs) in heavy metal bioremediation. These are the largest transmembrane receptors that aid in communicating the extracellular signals, such as stresses of heavy metals into the intracellular sites. G-protein-coupled receptors (GPCRs) regulate the important effector molecules, such as adenylate cyclase and phospholipase C, and regulate the function of kinase and ion channel by producing secondary messengers, such as cAMP, thereby inducing signaling cascades (El-Defrawy and Hesham, 2020).
From Figures 2A, discussion of the highly regulated selected KOG classes are as follows:
Post-translational Modification, Protein Turnover, Chaperones
Protein homeostasis is crucial for the cell proliferation and viability of all organisms. Further, cellular signaling is greatly affected by the protein homeostasis under different physiological conditions and environmental stresses, such as heavy metals, and therefore they can be suitable biomarkers. Molecular chaperones aid in the delivery of metal ions to the cell organelles and metalloproteins (Lotlikar and Damare, 2018). The two vital regulators of molecular chaperones in the proteostasis network are heat shock transcription factor Hsf1 and heat shock protein Hsp90. Under a stressful environment, heat shock protein enables the folding of newly synthesized proteins and helps in the degradation of damaged/misfolded proteins with the help of the ubiquitin-proteasome system (Hossain et al., 2020). Heat shock proteins bind to the denatured proteins, compelling them to refold into their native conformation and regain their original structure (Feng et al., 2018). In this study, we observed 42 proteins upregulated that belonged to post-translational modification, protein turnover, and chaperones. There was upto 4-fold upregulation of proteasome regulatory particle subunit Rpt3 (KOG0727), upto 2-fold upregulation of Hsp70 chaperone (HscA) (KOG0101), and upto 2.5-fold upregulation of protein geranylgeranyltransferase (KOG1439). The enzyme geranylgeranyltransferase I (GGTase I) aids in the catalysis of the post-translational transfer of lipophilic diterpenoid geranylgeranyl molecule to the cysteine residue of proteins with the termination at CaaX motif (Rho1p and Cdc42p). This alteration helps in the membrane localization of the protein and thereby rendering it biologically active. Rho1p is a regulatory subunit of 1,3-β-D-glucan synthesis and contributes to the cell wall synthesis in fungi which is vital for cell viability under stressful condition of excess metals (Singh et al., 2005).
Translation, Ribosomal Structure, and Biogenesis
Different proteins related to protein translation under multimetal stress were overexpressed. Here, we found 60 proteins of translation, ribosomal structure and biogenesis upregulated. There was upto 3.0-fold increase in glutamyl-tRNA synthetase (KOG1147), upto 2.5-fold increase in eukaryotic translation initiation factor 3 subunit B (KOG2314), upto 2.0-fold increase in 60S ribosomal protein L23 (KOG1751), upto 2.3-fold increase in mitochondrial translation initiation factor IF 2 (KOG1144), and 2.0-fold increase 40S ribosomal protein S10b (KOG3344). Similar elements of protein synthesis, such as translation initiation factor 5A, elongation factor 2, 40S and 60S ribosomal proteins, ATP-dependent RNA helicase, and aspartyl-tRNA synthetase, were overexpressed in Phanerochaete chrysosporium under Cu stress as a result of the need for production of new proteins or renewal of the damaged proteins (Okay et al., 2020).
Intracellular Trafficking and Secretion and Vesicular Transport
Proteins such as ion transporters and other solutes are crucial for processes such as detoxification, cell nutrition, cell signaling, cellular homeostasis, and resistance toward metal stress. These polytopic transmembrane proteins are translated altogether and folded in the endoplasmic reticulum (ER) of the eukaryotic cells that are later ultimately arranged to their respective membrane location through vesicular secretion. During any physiological or stressful environment, transporters undertake several regulated turnovers. Thus, in the process, transporters briefly interact dynamically with multiple proteins (Dimou et al., 2021). In this study, we found 18 proteins of intracellular trafficking and secretion and vesicular transport upregulated. The levels of the proteins were expressed in higher amounts (SEC31) (KOG0307) by 3.3-fold, endosomal cargo receptor (P24) (KOG1692) by 2.9-fold, and mitochondrial inner membrane translocase (KOG2580) by 2.4-fold in A. fumigatus under the effect of multimetal stress. The vesicle (Ves) are tissues composed of a lipid bilayer whose size varies ∼nanometers to micrometers. The Ves structures fuse with the plasma membrane of the cell and eject the trapped materials either inside or outside of the cytoplasm. There are three types of intracellular Ves viz. protein complex I (COPI)-coated Ves, protein complex II (COPII)-coated Ves, and BAR-domain protein Ves. These Ves proteins aid in physiological processes, such as the exchange of proteins and RNA intercellularly (Jiang et al., 2020).
Energy Production and Conversion
Heavy metals, such as Cd, Cu, Ni, and Zn, function as cofactors in bacteria and fungi. However, excess amounts of these metals are toxic to these cells and also produce reactive oxygen species (Liu et al., 2017). The need for metabolic energy in the fungal increases during abiotic stress, such as exposure to excess heavy metals. Thus, ATPases are responsible for the biochemical and physiological processes by the production of energy. Heavy metal ATPases (HMAs) or P-type ATPases can be categorized into three groups namely, A, B, and C. Further, P-type ATPases are utilized by numerous organisms to facilitate the transport of cations viz. Na+, K+, and Ca2+. To eliminate these excess metals, fungal HMA Saccharomyces cerevisiae CCC2 (Group A) localizes metals to metal-containing proteins, for example, in the case of copper metal, copper-containing protein FET3 in trans-golgi compartment transports metals to the cell membrane via efflux pumps, such as cadmium efflux pumps, encoded by fungal HMA Saccharomyces cerevisiae PCA1 (Group B and Group C) (Adle et al., 2007). Saitoh et al. (2009) studied the CCC2-type HMA gene that targets copper-containing proteins from the fungus Cochliobolus heterostrophus by cloning. There was upto 0.78-fold upregulation in the production of V-type ATPase and upto 1-fold upregulation in the production of mitochondrial ATPase subunit ATP4. This gene has other multifarious roles, such as in the formation of dark brown colored melanin pigment located in fungal cell walls, that also sequester metals (Chang et al., 2019).
Amino Acid Transport and Metabolism
The nitrogen cycle is essential for nitrogen assimilation and transformation and also for stress tolerance. Heavy metals impact the enzymes that play important role in nitrogen metabolism (Khouja et al., 2014). There was upto 4-fold upregulation in the production of aminotransferase (KOG1549) in response to multimetal. A similar response was observed by Okay et al. (2020), where the production of enzyme aspartate aminotransferase was enhanced in the fungus Phanerochaete chrysosporium to tackle the Cu stress. This enzyme has a possible role in the renewal of the mitochondrial NAD/NADH imbalance.
Analysis of Biological Pathways and Protein–Protein Interactions
In addition to these mechanisms, there are contributions from other regulatory systems, such as cross-talks in various pathways, interconnection amongst these different pathways, and regulation of different genes. These regulatory networks of the microbial proteins are intricate and play crucial roles in resistance to metal contaminants by modifying the series of specific functional proteins/non-proteins and altering the different metabolic enzymes at the cellular level. The metabolic processes related to the detoxification of contaminants are usually regulated by the complete set of proteins and their networks instead of a single enzyme (Zhao and Poh, 2008; Feng et al., 2018).
The STRING analysis displayed protein–protein interaction networks that are related to the resistance and tolerance mechanism A. fumigatus PD-18 for multimetals. Protein interactions in the first cluster (green) are involved in ribosome and preinitiation factors (60S and 40S ribosomal proteins). Ribosomal proteins form the protein part of ribosomes and participate in protein synthesis in cells in conjunction with rRNA. Thus, the increased expression of the large subunit of ribosome renders resistance against abiotic stresses, such as heavy metals, radiation, cold, and salt (Liu et al., 2014).
The important hub proteins expressed were cytochrome-c oxidase that is mitochondrial proteins and catalyst in the electron transport chain and is responsible for the transport of heavy metals particularly copper (Dias et al., 2019). The second cluster (blue) had hub protein ATP synthase. The important interconnecting protein in the network is the septin protein of KOG class cell cycle control, cell division, and chromosome partitioning, and is involved in vesicular trafficking and countering the apoptotic cell death initiated by the toxicity of heavy metals. This energy-intensive process is mediated by ATP synthase to maintain the cellular structure and function under the lethal environment of heavy metals. Excess to or above the permissible limit of heavy metals exposure can substantially delimit the growth of organisms. Here, more growth is initiated in the fungal cell as a result of the enhanced activity of ATP synthase (Yıldırım et al., 2011). This was evident in our study by the upregulation of tubulin by 1.1-fold and actin proteins by 2.2-fold which are responsible for cellular division and growth (Marks et al., 1986). This enhanced growth in the fungus also corroborated with our previous study where there was an increased dry weight of the fungal biomass under 30 mg/L multimetal (Dey et al., 2016). These results showed that the proteins in this network played important functions in cell functioning under heavy metals stress.
The mechanism of hexametal uptake by A. fumigatus PD-18 has been summarized in Figure 4.
Conclusion
In this study, we found the fungus A. fumigatus PD-18 developed stress coping strategies by secreting a suite of proteins that were either unique or upregulated/overexpressed when compared to the control. The proteomics study revealed that maximum proteins that were upregulated belonged to KOG class translation, ribosomal structure, and biogenesis. This study also highlighted the enhanced expression of antioxidants superoxide dismutase, molecular chaperone heat shock proteins, and involvement of proton transporter, such as ATPase. These proteins are involved in the tolerance and detoxification of multimetals by the fungus. A. fumigatus PD-18. Therefore, this investigation on the response of cellular proteomes to multimetal stress enabled us to better understand the cellular mechanism regarding the cumulative effect of the inorganic heavy metals stress on microbes. Further, it will be conducive to screening the key genes coding for enzymes that have higher resistance to these inorganic pollutants with enhanced capability to transform pollutants.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD031741.
Author Contributions
PD, AM, and NJ contributed to conception and design of the study. PD organized the database and wrote the first draft of the manuscript. PD and S-BH performed the statistical analysis. PD, AM, NJ, DS, and MB wrote sections of the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Conflict of Interest
PD, S-BH, MB, and NJ were employed by Helmholtz-Centre for Environmental Research-UFZ GmbH. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
Funding
PD thankfully acknowledges the fellowship from the Ministry of Human Resource and Development (Government of India) and German Academic Exchange Service (DAAD). MB and NJ are grateful for the funding of the UFZ for the ProMetheus platform for proteomics and metabolomics.
References
- Abed R. M. M., Shanti M., Muthukrishnan T., Al-Riyami Z., Pracejus B., Moraetis D. (2020). The role of microbial mats in the removal of hexavalent chromium and associated shifts in their bacterial community composition. Front. Microbiol. 11:12. 10.3389/fmicb.2020.00012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adle D. J., Sinani D., Kim H., Lee J. (2007). A cadmium-transporting P1B-type ATPase in yeast Saccharomyces cerevisiae. J. Biol. Chem. 282 947–955. 10.1074/jbc.M609535200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand P., Isar J., Saran S., Saxena R. K. (2006). Bioaccumulation of copper by Trichoderma viride. Bioresour. Technol. 97 1018–1025. 10.1016/j.biortech.2005.04.046 [DOI] [PubMed] [Google Scholar]
- Bae W., Chen X. (2004). Proteomic study for the cellular responses to Cd2+ in Schizosaccharomyces pombe through amino acid-coded mass tagging and liquid chromatography tandem mass spectrometry. Mol. Cell. Proteom. 3 596–607. 10.1074/mcp.M300122-MCP200 [DOI] [PubMed] [Google Scholar]
- Baker A. J. M. (1987). Metal Tolerance. New Phytol. 106 93–111. 10.1111/j.1469-8137.1987.tb04685.x [DOI] [Google Scholar]
- Bakti F., Sasse C., Heinekamp T., Pócsi I., Braus G. H. (2018). Heavy Metal-Induced Expression of PcaA provides cadmium tolerance to Aspergillus fumigatus and supports Its virulence in the Galleria mellonella Model. Front. Microbiol. 9:744. 10.3389/fmicb.2018.00744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj R., Gupta A., Garg J. K. (2017). Evaluation of heavy metal contamination using environmetrics and indexing approach for River Yamuna. Delhi stretch, India. Water Sci. 31 52–66. 10.1016/j.wsj.2017.02.002 [DOI] [Google Scholar]
- Bhattacharya A., Dey P., Gola D., Mishra A., Malik A., Patel N. (2015). Assessment of Yamuna and associated drains used for irrigation in rural and peri-urban settings of Delhi NCR. Environ. Monit. Assess 187:4146. 10.1007/s10661-014-4146-2 [DOI] [PubMed] [Google Scholar]
- Bhattacharya A., Gola D., Dey P., Malik A. (2020). Synergistic and antagonistic effects on metal bioremediation with increasing metal complexity in a hexa-metal environment by Aspergillus fumigatus. Int. J. Environ. Res. 14 761–770. 10.1007/s41742-020-00295-w [DOI] [Google Scholar]
- Bhosale P. (2004). Environmental and cultural stimulants in the production of carotenoids from microorganisms. Appl. Microbiol. Biotechnol. 63 351–361. 10.1007/s00253-003-1441-1 [DOI] [PubMed] [Google Scholar]
- Boopathy R. (2000). Factors limiting bioremediation technologies. Bioresour. Technol. 74 63–67. 10.1016/S0960-8524(99)00144-3 [DOI] [Google Scholar]
- Bradford M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72 248–254. 10.1006/abio.1976.9999 [DOI] [PubMed] [Google Scholar]
- Chang P.-K., Scharfenstein L. L., Mack B., Wei Q., Gilbert M., Lebar M., et al. (2019). Identification of a copper-transporting ATPase involved in biosynthesis of A. flavus conidial pigment. Appl. Microbiol. Biotechnol. 103 4889–4897. 10.1007/s00253-019-09820-0 [DOI] [PubMed] [Google Scholar]
- Chauhan N., Gola S., Surabhi, Sharma S., Khantwal S., Mehrotra R., et al. (2021). “Detrimental effects of industrial wastewater on the environment and health,” in Biological Treatment of Industrial Wastewater, ed. Shah M. P. (London: Royal Society of Chemistry; ), 40–52. 10.1039/9781839165399-00040 [DOI] [Google Scholar]
- Cherrad S., Girard V., Dieryckx C., Gonçalves I. R., Dupuy J.-W., Bonneu M., et al. (2012). Proteomic analysis of proteins secreted by Botrytis cinerea in response to heavy metal toxicity†. Metallomics 4 835–846. 10.1039/c2mt20041d [DOI] [PubMed] [Google Scholar]
- Deshmukh R., Khardenavis A. A., Purohit H. J. (2016). Diverse metabolic capacities of fungi for bioremediation. Indian J. Microbiol. 56 247–264. 10.1007/s12088-016-0584-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey P., Gola D., Chauhan N., Bharti R. K., Malik A. (2021). “Mechanistic insight to bioremediation of hazardous metals and pesticides from water bodies by microbes,” in Removal of Emerging Contaminants Through Microbial Processes, ed. Shah M. P. (Berilin: Springer; ), 467–487. 10.1007/978-981-15-5901-3_23 [DOI] [Google Scholar]
- Dey P., Gola D., Mishra A., Malik A., Kumar P., Singh D. K., et al. (2016). Comparative performance evaluation of multi-metal resistant fungal strains for simultaneous removal of multiple hazardous metals. J. Hazard. Mater. 318 679–685. 10.1016/j.jhazmat.2016.07.025 [DOI] [PubMed] [Google Scholar]
- Dey P., Malik A., Mishra A., Singh D. K., von Bergen M., Jehmlich N. (2020). Mechanistic insight to mycoremediation potential of a metal resistant fungal strain for removal of hazardous metals from multimetal pesticide matrix. Environ. Pollut. 262:114255. 10.1016/j.envpol.2020.114255 [DOI] [PubMed] [Google Scholar]
- Dias M., Gomes de Lacerda J. T. J., Perdigão Cota de Almeida S., de Andrade L. M., Oller do Nascimento C. A., Rozas E. E., et al. (2019). Response mechanism of mine-isolated fungus Aspergillus niger IOC 4687 to copper stress determined by proteomics. Metallomics 11 1558–1566. 10.1039/c9mt00137a [DOI] [PubMed] [Google Scholar]
- Dimou S., Georgiou X., Sarantidi E., Diallinas G., Anagnostopoulos A. K. (2021). Profile of membrane cargo trafficking proteins and transporters expressed under N source derepressing conditions in Aspergillus nidulans. J. Fungi 7:560. 10.3390/jof7070560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusengemungu L., Kasali G., Gwanama C., Ouma K. O. (2020). Recent advances in biosorption of copper and cobalt by filamentous fungi. Front. Microbiol. 11:3285. 10.3389/fmicb.2020.582016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Defrawy M. M. H., Hesham A. E.-L. (2020). G-protein-coupled Receptors in Fungi. Fungal Biotechnol. Bioeng. 37–126. 10.1007/978-3-030-41870-0_3 [DOI] [Google Scholar]
- Feng Y., Zhao Y., Guo Y., Liu S. (2018). Microbial transcript and metabolome analysis uncover discrepant metabolic pathways in autotrophic and mixotrophic anammox consortia. Water Res. 128 402–411. 10.1016/j.watres.2017.10.069 [DOI] [PubMed] [Google Scholar]
- Gadd G. M. (1994). “Interactions of fungi with toxic metals,” in The Genus Aspergillus: From Taxonomy and Genetics to Industrial Application Federation of European Microbiological Societies Symposium Series., eds Powell K. A., Renwick A., Peberdy J. F. Boston, MA: Springer; 361–374. 10.1007/978-1-4899-0981-7_28. [DOI] [Google Scholar]
- Gadd G. M. (2000). Bioremedial potential of microbial mechanisms of metal mobilization and immobilization. Curr. Opin Biotechnol. 11 271–279. 10.1016/s0958-1669(00)00095-1 [DOI] [PubMed] [Google Scholar]
- Gadd G. M. (2007). Geomycology: biogeochemical transformations of rocks, minerals, metals and radionuclides by fungi, bioweathering and bioremediation. Mycolog. Res. 111 3–49. 10.1016/j.mycres.2006.12.001 [DOI] [PubMed] [Google Scholar]
- Gadd G. M. (2010). Metals, minerals and microbes: geomicrobiology and bioremediation. Microbiology 156 609–643. 10.1099/mic.0.037143-0 [DOI] [PubMed] [Google Scholar]
- Gadd G. M., White C. (1993). Microbial treatment of metal pollution — a working biotechnology? Trends Biotechnol. 11 353–359. 10.1016/0167-7799(93)90158-6 [DOI] [PubMed] [Google Scholar]
- Gola D., Bhattacharya A., Dey P., Malik A., Ahammad S. Z. (2020). Assessment of drain water used for irrigation in the delhi region. J. Health Pollut. 10:200610. 10.5696/2156-9614-10.26.200610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal N., Jain S. C., Banerjee U. C. (2003). Comparative studies on the microbial adsorption of heavy metals. Adv. Environ. Res. 7 311–319. 10.1016/S1093-0191(02)00004-7 [DOI] [Google Scholar]
- Haange S.-B., Oberbach A., Schlichting N., Hugenholtz F., Smidt H., von Bergen M., et al. (2012). Metaproteome analysis and molecular genetics of rat intestinal microbiota reveals section and localization resolved species distribution and enzymatic functionalities. J. Proteome Res. 11 5406–5417. 10.1021/pr3006364 [DOI] [PubMed] [Google Scholar]
- Høiland K. (1995). Reaction of some decomposer basidiomycetes to toxic elements. Nordic J. Bot. 15 305–318. 10.1111/J.1756-1051.1995.TB00157.X [DOI] [Google Scholar]
- Hossain S., Veri A. O., Cowen L. E. (2020). The proteasome governs fungal morphogenesis via functional connections with Hsp90 and cAMP-Protein Kinase a Signaling. mBio 11:e00290-20. 10.1128/mBio.00290-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irawati W., Djojo E. S., Kusumawati L., Yuwono T., Pinontoan R. (2021). Optimizing bioremediation: elucidating copper accumulation mechanisms of Acinetobacter sp. IrC2 isolated from an industrial waste treatment center. Front. Microbiol. 12:3049. 10.3389/fmicb.2021.713812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeckel P., Krauss G., Menge S., Schierhorn A., Rücknagel P., Krauss G.-J. (2005). Cadmium induces a novel metallothionein and phytochelatin 2 in an aquatic fungus. Biochem. Biophys. Res. Commun. 333 150–155. 10.1016/j.bbrc.2005.05.083 [DOI] [PubMed] [Google Scholar]
- Jiang Z., Wang T., Sun Y., Nong Y., Tang L., Gu T., et al. (2020). Application of Pb(II) to probe the physiological responses of fungal intracellular vesicles. Ecotoxicol. Environ. Saf. 194:110441. 10.1016/j.ecoenv.2020.110441 [DOI] [PubMed] [Google Scholar]
- Keller N. P. (2015). Translating biosynthetic gene clusters into fungal armor and weaponry. Nat. Chem. Biol. 11 671–677. 10.1038/nchembio.1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khouja H. R., Daghino S., Abbà S., Boutaraa F., Chalot M., Blaudez D., et al. (2014). OmGOGAT-disruption in the ericoid mycorrhizal fungus Oidiodendron maius induces reorganization of the N pathway and reduces tolerance to heavy-metals. Fungal Genet. Biol. 71 1–8. 10.1016/j.fgb.2014.08.003 [DOI] [PubMed] [Google Scholar]
- Kolen M. (2013). Leaching of zinc, cadmium, lead and copper from electronic scrap using organic acids and the Aspergillus niger strain. Fresenius Environ. Bull. 22:8. [Google Scholar]
- Kou S., Vincent G., Gonzalez E., Pitre F. E., Labrecque M., Brereton N. J. B. (2018). The response of a 16S Ribosomal RNA gene fragment amplified community to lead. zinc, and copper pollution in a shanghai field trial. Front. Microbiol. 9:366. 10.3389/fmicb.2018.00366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut A., Marcellin M., Adrait A., Kuhn L., Louwagie M., Kieffer-Jaquinod S., et al. (2009). Peptide storage: are you getting the best return on your investment? Defining optimal storage conditions for proteomics samples. J. Proteome Res 8 3778–3785. 10.1021/pr900095u [DOI] [PubMed] [Google Scholar]
- Kuhn A. N., Käufer N. F. (2003). Pre-mRNA splicing in Schizosaccharomyces pombe. Curr. Genet 42 241–251. 10.1007/s00294-002-0355-2 [DOI] [PubMed] [Google Scholar]
- Li H., Dong W., Liu Y., Zhang H., Wang G. (2019). Enhanced biosorption of nickel ions on immobilized surface-engineered yeast using nickel-binding peptides. Front. Microbiol. 10:1254. 10.3389/fmicb.2019.01254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.-H., Zeng G.-M., Niu Q.-Y., Liu Y., Zhou L., Jiang L.-H., et al. (2017). Bioremediation mechanisms of combined pollution of PAHs and heavy metals by bacteria and fungi: a mini review. Bioresour. Technol. 224 25–33. 10.1016/j.biortech.2016.11.095 [DOI] [PubMed] [Google Scholar]
- Liu X.-D., Xie L., Wei Y., Zhou X., Jia B., Liu J., et al. (2014). Abiotic stress Resistance, a novel moonlighting function of ribosomal protein RPL44 in the halophilic fungus Aspergillus glaucus. Appl. Environ. Microbiol. 80 4294–4300. 10.1128/AEM.00292-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotlikar N., Damare S., Meena R. M., Jayachandran S. (2020). Variable protein expression in marine-derived filamentous fungus Penicillium chrysogenum in response to varying copper concentrations and salinity. Metallomics. 12 1083–1093. 10.1039/C9MT00316A [DOI] [PubMed] [Google Scholar]
- Lotlikar N. P., Damare S. R. (2018). Variability in protein expression in marine-derived Purpureocillium lilacinum subjected to salt and chromium stresses. Indian J. Microbiol. 58 360–371. 10.1007/s12088-018-0733-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik A. (2004). Metal bioremediation through growing cells. Environ. Int. 30 261–278. 10.1016/j.envint.2003.08.001 [DOI] [PubMed] [Google Scholar]
- Marks J., Hagan I. M., Hyams J. S. (1986). Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton. J. Cell Sci. 1986 229–241. 10.1242/jcs.1986.Supplement_5.15 [DOI] [PubMed] [Google Scholar]
- Morel M., Ngadin A. A., Droux M., Jacquot J.-P., Gelhaye E. (2009). The fungal glutathione S-transferase system. Evidence of new classes in the wood-degrading basidiomycete Phanerochaete chrysosporium. Cell Mol. Life Sci. 66 3711–3725. 10.1007/s00018-009-0104-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munir E., Yoon J. J., Tokimatsu T., Hattori T., Shimada M. (2001). A physiological role for oxalic acid biosynthesis in the wood-rotting basidiomycete Fomitopsis palustris. PNAS 98 11126–11130. 10.1073/pnas.191389598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno M., Karagiannis P., Taniguchi Y. (2014). Protein expression analyses at the single cell level. Molecules 19 13932–13947. 10.3390/molecules190913932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okay S., Yildirim V., Büttner K., Becher D., Özcengiz G. (2020). Dynamic proteomic analysis of Phanerochaete chrysosporium under copper stress. Ecotoxicol. Environ. Saf. 198:110694. 10.1016/j.ecoenv.2020.110694 [DOI] [PubMed] [Google Scholar]
- Orsburn B. C. (2021). Proteome Discoverer—A Community Enhanced Data Processing Suite for Protein Informatics. Proteomes 9:15. 10.3390/proteomes9010015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A., Mann M. (2000). Proteomics to study genes and genomes. Nature 405 837–846. 10.1038/35015709 [DOI] [PubMed] [Google Scholar]
- Reddy M. S., Prasanna L., Marmeisse R., Fraissinet-Tachet L. Y. (2014). Differential expression of metallothioneins in response to heavy metals and their involvement in metal tolerance in the symbiotic basidiomycete Laccaria bicolor. Microbiology 160 2235–2242. 10.1099/mic.0.080218-0 [DOI] [PubMed] [Google Scholar]
- Rughöft S., Jehmlich N., Gutierrez T., Kleindienst S. (2020). Comparative Proteomics of Marinobacter sp. TT1 reveals corexit impacts on hydrocarbon metabolism, chemotactic motility, and biofilm formation. Microorganisms 9:3. 10.3390/microorganisms9010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabuda M. C., Rosenfeld C. E., DeJournett T. D., Schroeder K., Wuolo-Journey K., Santelli C. M. (2020). Fungal bioremediation of selenium-contaminated industrial and municipal wastewaters. Front. Microbiol. 11:2105. 10.3389/fmicb.2020.02105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagar V., Singh D. P. (2011). Biodegradation of lindane pesticide by non white- rots soil fungus Fusarium sp. World J. Microbiol. Biotechnol. 27 1747–1754. 10.1007/s11274-010-0628-8 [DOI] [Google Scholar]
- Saitoh Y., Izumitsu K., Tanaka C. (2009). Phylogenetic analysis of heavy-metal ATPases in fungi and characterization of the copper-transporting ATPase of Cochliobolus heterostrophus. Mycol. Res. 113 737–745. 10.1016/j.mycres.2009.02.009 [DOI] [PubMed] [Google Scholar]
- Sandau E., Sandau P., Pulz O. (1996). Heavy metal sorption by microalgae. Acta Biotechnologica. 16 227–235. 10.1002/abio.370160402 [DOI] [Google Scholar]
- Selamoglu N., Önder Ö, Öztürk Y., Khalfaoui-Hassani B., Blaby-Haas C. E., Garcia B. A., et al. (2020). Comparative differential cuproproteomes of Rhodobacter capsulatus reveal novel copper homeostasis related proteins. Metallomics 12 572–591. 10.1039/C9MT00314B [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M., Zhao D.-K., Qiao Q., Liu L., Wang J.-L., Cao G.-H., et al. (2015). Identification of glutathione S-transferase (GST) genes from a dark septate endophytic fungus (Exophiala pisciphila) and their expression patterns under varied metals stress. PLoS One 10:e0123418. 10.1371/journal.pone.0123418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S. B., Kelly R., Guan Z., Polishook J. D., Dombrowski A. W., Collado J., et al. (2005). New fungal metabolite geranylgeranyltransferase inhibitors with antifungal activity. Nat. Prod. Res. 19 739–747. 10.1080/1478641042000334715 [DOI] [PubMed] [Google Scholar]
- Wang Y., Yi B., Sun X., Yu L., Wu L., Liu W., et al. (2019). Removal and tolerance mechanism of Pb by a filamentous fungus: a case study. Chemosphere 225 200–208. 10.1016/j.chemosphere.2019.03.027 [DOI] [PubMed] [Google Scholar]
- Wisecaver J. H., Rokas A. (2015). Fungal metabolic gene clusters—caravans traveling across genomes and environments. Front. Microbiol. 6:161. 10.3389/fmicb.2015.00161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yıldırım V., Özcan S., Becher D., Büttner K., Hecker M., Özcengiz G. (2011). Characterization of proteome alterations in Phanerochaete chrysosporium in response to lead exposure. Proteome Sci. 9:12. 10.1186/1477-5956-9-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora-Ledezma C., Negrete-Bolagay D., Figueroa F., Zamora-Ledezma E., Ni M., Alexis F., et al. (2021). Heavy metal water pollution: a fresh look about hazards, novel and conventional remediation methods. Environ. Technol. Innov. 22:101504. 10.1016/j.eti.2021.101504 [DOI] [Google Scholar]
- Zhang Q., Zeng G., Chen G., Yan M., Chen A., Du J., et al. (2015). The effect of heavy metal-induced oxidative stress on the enzymes in white rot fungus Phanerochaete chrysosporium. Appl. Biochem. Biotechnol. 175 1281–1293. 10.1007/s12010-014-1298-z [DOI] [PubMed] [Google Scholar]
- Zhang X., Fang A., Riley C. P., Wang M., Regnier F. E., Buck C. (2010). Multi-dimensional Liquid Chromatography in Proteomics. Anal. Chim. Acta 664 101–113. 10.1016/j.aca.2010.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Poh C. L. (2008). Insights into environmental bioremediation by microorganisms through functional genomics and proteomics. Proteomics 8 874–881. 10.1002/pmic.200701005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD031741.