Abstract
More than 60 years ago, Richard Feynman gave a lecture titled “There’s Plenty of Room at the Bottom: An Invitation to Enter a New Field of Physics”, where he called on others to join the then-nascent field of nanotechnology. In a similar spirit, we wish to invite chemists, biologists, physicists, bioengineers, educators, high school students, and inventors of all backgrounds to join us in the emerging field of frugal science. In this Review, we define frugal science and use six case studies to describe the broad applications of frugal science, from synthetic biology to disease diagnostics. We conclude by establishing an argument for curiosity-driven research through frugal science to enable broader access in chemical and bioengineering research and drive innovation.
Graphical Abstract
MOTIVATION AND BACKGROUND
Scientific Tools Are Both an Enabler and an Impediment.
Scientific hardware and tools form the basis of many scientific disciplines and engineering applications. More than 2000 years ago, Eratosthenes, using only a simple stick, measured the diameter of the Earth to reasonable accuracy.1 Today, however, scientists require much more complex, intricate, and expensive tools to perform measurements and experiments. Thus, the accessibility and affordability to these tools determines who gets to participate in the exciting voyages of scientific discoveries. Even in the United States, there is an ever-growing gap between the “haves and the haves not”, as talented young scientists starting their laboratories realize the sobering astronomical cost of laboratory equipment.2,3 In low- and middle-income countries around the world, with much lower funding rates, conducting state-of-the-art science is a luxury not many can afford. Thus, modern tools that can simultaneously enable marvelous feats of human achievement, such as the first and fastest mRNA vaccine rollout against SARS-CoV-2, can also be a critical impediment toward global manufacturing and last-mile delivery (cold chain).4
DIY and FOSH Movements.
The do-it-yourself (DIY) and free and open-source hardware (FOSH) movements have made encouraging headway to address some of these challenges by reducing cost, offering local manufacturing, and enabling easier access to scientific tools with notable successes.5–10 However, these movements are primarily characterized by utilizing less-expensive components or using three-dimensional (3D) printers for manufacturing, mostly relying on the same underlying operational principles and complexity of their commercial equivalents. As a result, they help improve accessibility, but still succumb to the other challenges faced by traditional tools and occasionally sacrifice on quality. For example, a 3D-printed micropipette still requires access to a 3D printer and a supply of disposable plastic pipettes.11 Are these open source and DIY tools affordable? Affordable is a relative term. While these alternatives cost much less than their commercial closed-box counterparts, they are still out of reach for many millions of promising young scientists and engineers in under-resourced parts of the world.
What if We Could "Reinvent the Wheel"?
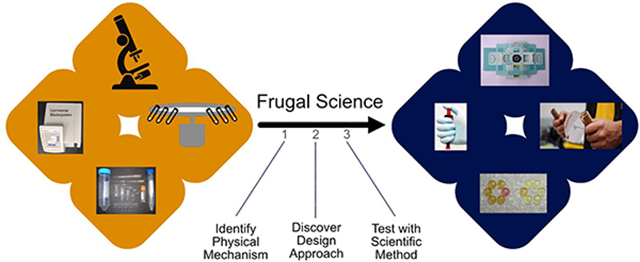
Bioengineering technologies often rely on fundamental physical mechanisms for their design, which, in turn, gives them their functionality. For example, a centrifuge relies on centrifugal force to separate materials based on their density, vortex mixers (or vortexers) use a rapid oscillatory motion to create turbulent flow (a vortex) to mix samples, etc. These technologies also rely on a design approach to provide this functionality. For example, centrifuges rely on complex electronic circuits to power an electrical motor that rotates at a specified speed, vortexers use a similar approach with circuits to power a motor that moves a platform in an oscillatory motion, etc. To “reinvent the wheel”, we seek to identify different design approaches that use similar underlying physical mechanisms to achieve the same final functionality in a scientific instrument, while radically increasing its affordability, accessibility, and scalability.
What is Frugal Science?
How do we arrive at these new design approaches? Here, we define a term representing the process of “utilizing curiosity-driven thinking to leverage common everyday items and repurpose them to solve complex engineering problems for bioengineering”: frugal science (Figure 1). The concept of repurposing existing materials or products for uses other than that they were originally intended is not new. Others have proposed ideas of “jugaad innovation”12 or “adaptive use”,13 where a bicycle may be repurposed into a washing machine or even a plow. However, along the spectrum of low-cost innovation, where a simple DIY hack may lie on one extreme, frugal science sits on the other. Thus, while using a pen knife as a screwdriver may solve a pressing problem, it does not necessarily utilize (or require) the scientific method or open transformational new avenues of scientific discovery or platform innovation (see the discussion on Feynman challenge analogy below).
Figure 1.
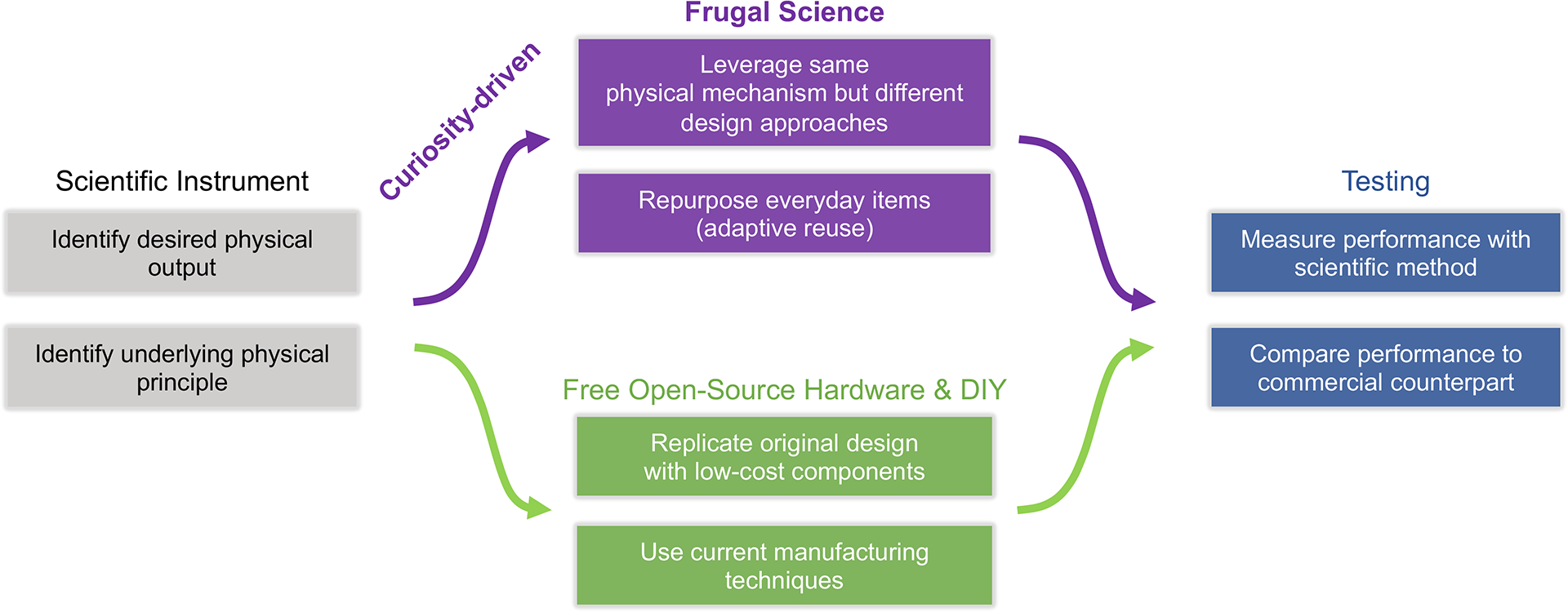
Design steps in frugal science and innovation. Process chart outlining the overall process of designing a “frugal science” device and its contrast from free open-source hardware/do-it-yourself (FOSH/DIY) approaches.
Frugal science thus involves (1) defining the underlying physical mechanism powering a certain scientific instrument, as well as the corresponding intended outcome, (2) identifying and repurposing common objects leveraging the same physical mechanisms but different design approaches to achieve the desired final functionality and performance metrics, and (3) transforming that object into the desired scientific tool while scrutinizing the development through the scientific method. By leveraging a different design approach meeting a core set of principles and performance metrics, the cost threshold is lowered significantly by orders of magnitude, which would not be possible if restricted to the same design approach (see Table S1 in the Supporting Information). In addition, utilization of the scientific method ensures that the new frugal tool does not compromise on quality and meets the same gold performance standards set by commercial instruments or regulatory bodies.
This approach can be applied in multiple different ways. One could begin with a medical tool and apply frugal science to develop a new ultralow-cost version. It could also be flipped around where an individual may identify the underlying physical mechanism exhibited by an everyday tool and then brainstorm the potential applications of this object. Regardless of the way it is used, frugal science relies on creativity, curiosity, and scientific rigor to develop alternatives to current tools focusing on affordability, accessibility, and scalability. In this Review, we highlight a few key examples of frugal science developed by groups across the world (including ours) and demonstrate how they leverage this novel approach to engineer frugal devices. We conclude by establishing an argument for curiosity-driven research through frugal science to enable broader access in bioengineering research and drive innovation.
DISCUSSION
Case Study 1: From BBQ Lighters to ePatch for Vaccine Delivery.
A common practice in bioengineering and synthetic biology is the manipulation of cells for a particular purpose such as protein production, DNA cloning, or even vaccination, such as with mRNA vaccines. These methods (transformation for bacteria and transfection for mammalian cells) are employed by utilizing either chemical or electrical means to modify cell membranes and allow DNA/RNA to enter a cell. The latter, known as electroporation, uses short, high-voltage pulses to create temporary pores in the cell membrane and allow nucleic acid uptake.14 To do this, machines called “electroporators” are used which rely on complex electronic circuits to discharge pulses. These machines are expensive (costing thousands of dollars), are bulky and not portable, require constant access to electricity, and are difficult to manufacture.15 Given their wide applicability and potential in bioengineering, improving the affordability and accessibility of electroporators could be valuable.
Using the frugal science method, let us first begin by considering the basic physical mechanism by which an electroporator works. Electroporation is dependent on short, high-voltage electric pulses with specific waveforms, magnitudes, and lengths to successfully permeabilize cells. In the case of bacteria, this is generally an exponentially decaying waveform with a pulse magnitude on the order of kilovolts and time constant on the order of milliseconds.16 The optimization of these parameters is key to enabling successful transformations, and deviation from them reduces transformation efficiency. We have now defined the physical mechanism by which electroporation works.
Now, let us consider the design approach through which electroporation can be enabled. Currently, electroporators use capacitors to store electrical energy and then discharge it as an electrical pulse with a specific waveform, magnitude, and length.17 This reliance on circuits contributes to the drawbacks of electroporators today. To engineer a frugal electroporator, this usage of circuits must be eliminated. Given these constraints, consider everyday objects that generate sparks or electric pulses that could be applied to electroporation. A common place where this is found is within a barbeque lighter. The same household item used to ignite gas and produce a flame generates short, high-voltage pulses using piezoelectricity (converts mechanical to electrical energy). What if a lighter could be used as an electroporator?
To test this hypothesis, in the Bhamla Laboratory, we measured pulse outputs from a BBQ lighter to determine the waveform, peak voltage, and time constants.15 Testing of different lighters identified one that produces the necessary parameters for bacterial electroporation. Next, the mechanism being used to generate piezoelectricity was then studied and manipulated to determine theoretical outputs and design principles for the system. The final system, coined ElectroPen, was benchmarked against a commercial electroporator delivering Green Fluorescence Protein-encoding DNA plasmids into bacteria and the corresponding transformation efficiency was determined. Through this series of experimentation, optimization, and scientific rigor, a lighter was demonstrated to successfully electroporate bacterial cells, at a fraction of the cost (23 cents) of its commercial counterpart (Figure 2). However, there are certain limitations to this approach including lack of tunability to specific applications and need for validation through external hardware (oscilloscopes).
Figure 2.
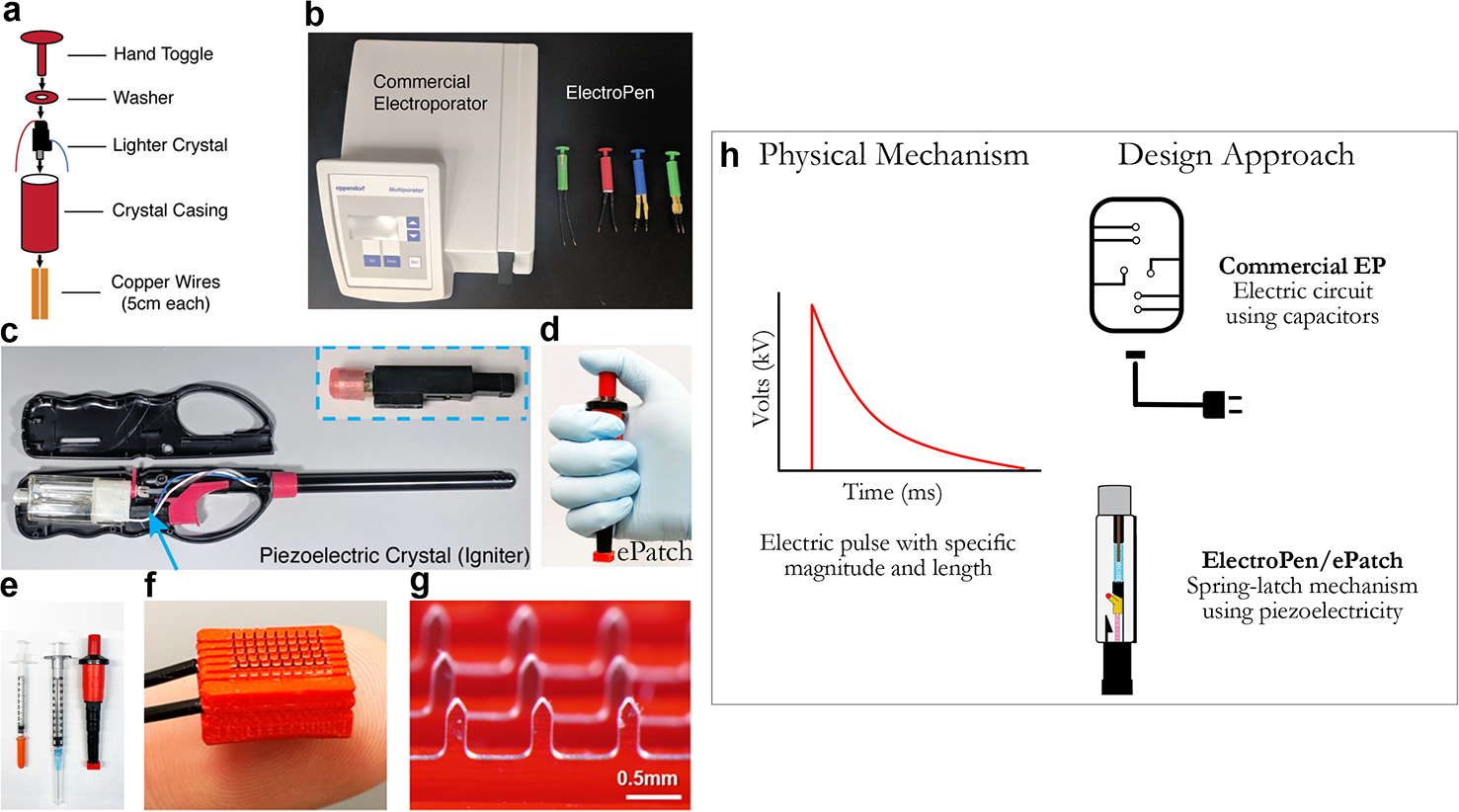
(a) Depiction of the ElectroPen device including its parts, (b) comparison against a commercial electroporator, (c) source of the piezoelectric crystal in a lighter, (d) an image of the ePatch vaccine delivery platform,18 (e) image of ePatch compared to traditional syringe needles for scale, and (f/g) images of the microneedle electrode array. (h) While both approaches rely on electric pulses, the difference lies in the use of piezoelectricity enabling frugal hardware, compared to traditional capacitance discharge. [Panels (a–c) were reproduced (or adapted) with permission from ref 15. Copyright 2020, PLOS Biology. Panels (d–g) were reproduced (or adapted) with permission from ref 18. Copyright 2021, PNAS.]
Building on our ElectroPen work, we decided to expand to other applications of electroporation in the drug/vaccine delivery space, where a breadth of prior work exists. One area of interest where electroporation is being explored in clinical trials is for DNA vaccines, the nucleic acid counterpart to mRNA vaccines. Similar to mRNA vaccines, DNA vaccines encode the antigen of choice but need to reach the nucleus to be expressed. Therefore, without a delivery enhancer, they do not produce potent immune responses. Electroporation is the preferred method of delivery for these vaccines; however, similar challenges persist in the form of expensive and bulky hardware, complex manufacturing, need for highly trained personnel, and pain at delivery site. By leveraging similar principles to ElectroPen and combining it with another attractive vaccine delivery technology called microneedles, we developed a new platform called ePatch: an ultralow-cost, portable microneedle electroporator to deliver DNA vaccines.18 Through benchmarking against state-of-the-art electroporation technologies, we have shown comparable efficacy and better safety delivering GFP-encoding plasmids. In a mice vaccination study with SARS-CoV-2, we have shown robust immune responses with at least 10-fold dose sparing, relative to intradermal and intramuscular injection alone, as well as protection against a SARS-CoV-2 pseudovirus elevated from 20% to 90%. Building on our success with ePatch for DNA vaccines, we plan to expand its applications to broader fields within nucleic acid delivery, including other DNA therapeutics and mRNA medicines. Ultimately, ePatch serves as a prime example of frugal science project evolving through continuous iteration and innovation into a global health tool with high potential for improving the affordability and accessibility of cutting-edge medicines.
Thus, by using a tool (BBQ lighter) that is already produced at scale, inexpensive to purchase, widely accessible, and does not require access to electricity, a new frugal electroporator (costing less than $1) was engineered to address challenges in synthetic biology research, as well as DNA vaccine delivery for pandemic response.
Case Study 2: From Whirligigs to Paper Centrifuges.
Centrifuges are ubiquitous devices in biological laboratories, often used in diagnostics for separating plasma from blood, disease diagnostics, etc.19 Using centrifugal forces, these devices can separate samples based on their density, hence their wide-ranging applicability in molecular biology and medical diagnostics. However, use of centrifugation is currently limited in resource-poor and field settings, because of high costs, the need for electricity, and poor portability. Given their usage, a frugal centrifuge could enable wider access to medical diagnostics.
First, consider the basic physical mechanism by which a centrifuge works. Centrifuges rely on high-speed rotations in a circular motion to produce a centrifugal force. This continued rotation results in sedimentation via gravitational force (g-force), thereby separating particles based on their density, often through pelleting at the bottom of a tube and supernatant on top. Successful centrifugation is dependent on the application and often requires different parameters. This generally involves controlling the relative centrifugal force (rcf), g-force, revolutions per minute (rpm), and diameter of the rotor. We have now defined the key mechanism of centrifugation.
Next, consider the design approach through which this can be enabled. Currently, centrifuges rely on circuits to power a rotor and rotate it at a specified speed. The samples are placed in tubes within slots inside the rotor so multiple samples can be spun at the same time. The speed of rotation can be manipulated as needed to adjust for a given application. This reliance on circuits and power contributes to their poor affordability and accessibility; therefore, a non-circuit-based centrifuge would be ideal. Given these constraints, what everyday items generate high-speed rotations that could be used for centrifugation.? One thought would be using a salad spinner or an eggbeater.20,21 However, these still rely on rigid mechanical gears to convert linear hand motion to rotational motion in an inefficient manner. Thus, the resulting rotational speeds are slower than what is normally used for centrifugation, which would lead to a long usage time and be impractical in the field. Consider another toy that is existed for thousands of years and uses a supercoiling string to convert linear motion to ultrafast rotations: a whirligig or button-on-a-string. This ancient 5000-year-old toy rotates incredibly fast in the blink of an eye.22
To test this hypothesis, Bhamla et al. measured the rotational speed of the whirligig using high-speed video and mathematical analysis.23 A new device, called a “paperfuge”, was designed by using a similar design approach but with a capillary tube attached to a paper wheel with strings passing through the wheel and wooden handles at the ends (Figure 3). After discovering that these objects were able to rotate at very high speeds (>125 000 rpm), the mechanics behind the system were uncovered. Through detailed experimentation and physical modeling, theoretical limits for the paperfuge were predicted (up to 1 000 000 rpm) and variations of different materials and applications were analyzed. Finally, the paperfuge was demonstrated to successfully separate blood and enable malaria diagnosis, and it was benchmarked against commercial centrifuges. However, there are certain limitations with this approach that should be noted, including limitation to small volume samples (10 μL), lack of precise control of rpm/rcf over a rotated sample, and lower throughput.
Figure 3.
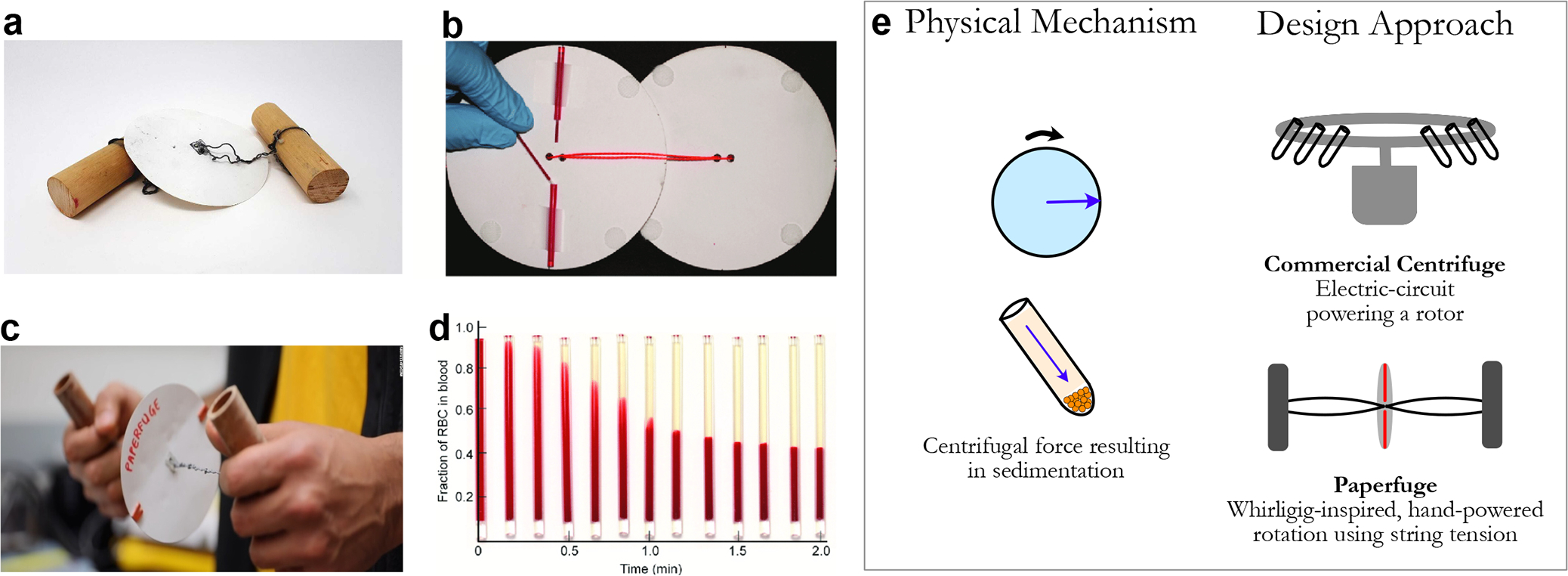
(a) Image of the paperfuge tool, (b) paper disks with loaded capillary tubes, (c) usage of paperfuge, and (d) blood separation over 2 min of usage. (e) While both traditional centrifuges and the paperfuge leverage centrifugal force, the difference lies in the fact that the Paperfuge uses the mechanism of a whirligig to produce the necessary rotational force. [Reproduced (or adapted) with permission from ref 23. Copyright 2017, Springer Nature.]
In later studies, our laboratory also built a 3D-printed version, called the 3D-Fuge, to expand the sample capacity from 10 μL to 2 mL.24 Other groups across the world have also built on these principles for new design approaches and applications.25–29 Through the frugal science approach, an ancient childhood toy was transformed to a powerful tool for synthetic biology and medical diagnostics.
Case Study 3: From Paper and Microfluidics to Diagnostics.
To properly treat diseases, accurate diagnoses must first be obtained. However, diagnostic technologies designed for developed countries often poorly translate to economically challenged and resource-limited settings.30 This is due to the cost of these tests and reliance on medical equipment to perform these tests, bringing additional hurdles through poor portability, and the need for electricity, refrigeration, and trained personnel.31,32 Therefore, for accurate disease monitoring and treatment, affordable and accessible diagnostic tools are needed.
Generally, a diagnostic test works by separating biological samples such as blood, urine, or stool (often via centrifugation). An enzymatic reaction occurs next to produce a readout in the form of a colorimetric output or further analysis is conducted through microscopy or other methods. Thus, diagnostic tests rely on controlled flow of a sample as well as controlled reactions with specific sample sizes, which culminate in final analysis.33
The design approach of these diagnostic tests is dependent on their type, but generally involve separation of the sample, controlled reactions, and final readouts. These often may require multiple different medical devices, which can be time-consuming, expensive, bulky, and require electricity. While this method is acceptable for developed countries, guidelines from the World Health Organization (WHO) indicate their acceptability outside these environments will be minimal. The WHO specifies that diagnostic tests must follow the ASSURED criteria of “affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and deliverable to end-users”.34 Given the parameters initially described, consider what other tools/methods could be leveraged to adhere to these constraints. An immediate thought is microfluidics, where much prior work has been demonstrated including in the diagnostics space.35 However, they are still constrained by high infrastructure costs (clean rooms) and poor scalability. Another approach is paper chromatography tests, which are widely used but require more control and precision.36,37 Now, imagine if these two could be combined so that the simplicity and affordability of paper tests and precision and integration of microfluidics were all in one system.
To test this hypothesis, the Whitesides Laboratory reengineered chromatography paper to create channels (using hydrophilic and hydrophobic barriers) analogous to a microfluidic device and allow controlled flow of samples.38 This was demonstrated to mostly occur because of capillarity and evaporation, thereby restricting fluid movement in microliter amounts. Reagents for diagnostic assays were spotted by hand or inkjet-printed onto paper where the channels directed the samples into test zones containing reagents. These were then dried and ready for use. After modeling and experimentation of the fluid dynamics of this system, assays were performed and benchmarked against currently approved methods. Successful colorimetric assays for glucose, proteins, pH, and alkaline phosphatase were performed on these microfluidic paper-based analytical devices (μPADs) (Figure 4). However, there are certain limitations that should be noted, including a higher threshold for colorimetric output, poor sample retention, effect of paper variations, and shelf life of reagents.39,40 We note that there are equally pioneering efforts in the context of paper-based point-of-care diagnostics and novel synthetic biology-based diagnostics by the Yager Laboratory and Collins Laboratory, respectively.35,41–46
Figure 4.
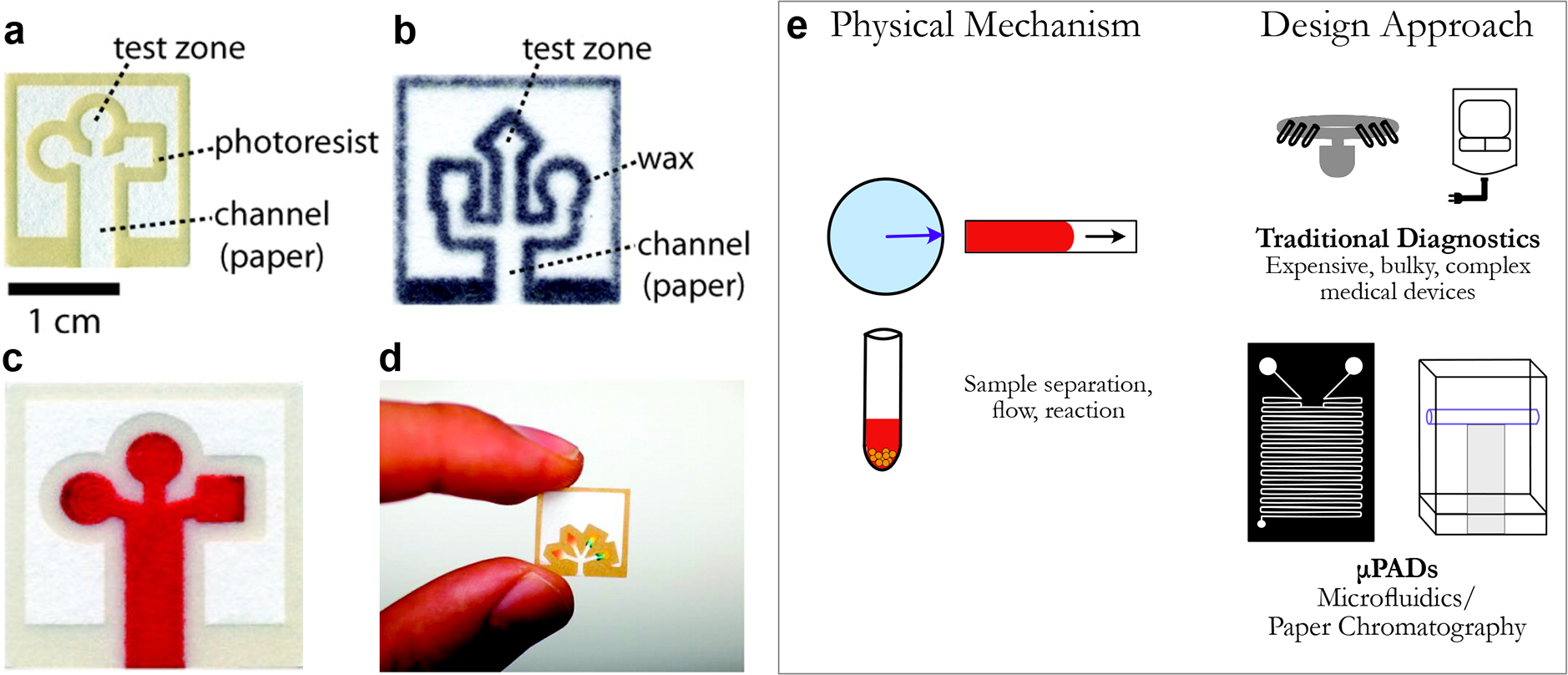
(a, b) Images of paper-based microfluidic-based diagnostic devices in different manners, including (c, d) examples of colorimetric results from the μPADs. [Reproduced (or adapted) from ref 38. Copyright 2010, American Chemical Society, Washington, DC.] (e) While both rely on similar physical mechanisms, μPADs leverage paper chromatography and microfluidics to power low-cost diagnostics.
Here, existing technologies (paper, microfluidics) were successfully repurposed through a novel approach to improve the affordability, scalability, and accessibility of medical diagnostics. By replacing bulky, expensive medical devices and inaccessible kits for diagnostic tests with a simple, inexpensive, and easy-to-use point-of-care solution, μPADs offer another example of frugal science innovation with direct applications for diagnostics in underserved and rural communities across the globe.38,47
Case Study 4: From Paper Origami to an Everyday Microscope.
Microscopes are ubiquitous tools across scientific disciplines; they are used for applications varying from medical diagnostics, microbiology, science education, etc. Since their invention more than 300 years ago, microscopes have both revolutionized and advanced a vast array of sciences, beginning with microbiological discoveries made by Antonie Van Leeuwenhoek and Robert Hooke through the first microscopes.48,49 The field of microscopy has advanced considerably over time with the emergence of new variations, such as electron, scanning probe, fluorescence, and super-resolution microscopy.50 However, for basic light microscopes, current costs and form factors of microscopes make them inaccessible for certain environments such as field research (bulky), and high school science education (cost). Given the wide applicability of microscopes, there is a strong case for frugal microscopes to enable these applications and magnify scientific curiosity across younger generations of scientists.
First, consider the basic physical mechanisms powering a microscope. Despite the large developments in optics over the centuries, the basic principles of light microscopy have remained unchanged. In its simplest form, a light microscope is composed of three main parts: light source, stage, and lens. It relies on manipulating light using a convex lens to amplify the minute details of a microscopic object. The stage is typically moved vertically to improve focus and obtain sharp images of the specimen (fixed to a slide). For tough-to-visualize biological samples, such as those that are almost transparent, different imaging modalities such as darkfield and fluorescence have been developed for enhanced contrast.
Next, consider the current design approach for microscopes. Microscopes currently consist of an eyepiece to view the specimen, an objective to relay the image to the eyepiece, a large lens to magnify the specimen image, a three-axis (x-y-z) movable stage to hold the specimen, and a light source for illumination, all powered by batteries or electrical outlets. The size of these components and the principles through which they are designed contribute to the bulky size of microscopes, their cost, and their poor scalability. What if there was another design approach to miniaturize these components and reduce their costs while retaining the same principles of optical design? Origami has been practiced for over a thousand years, generating complex figures and dynamic shapes through the intricate art of folding paper.51,52 The next step is to evaluate whether this approach could be used to design a new microscope.
To test this idea, the Prakash Laboratory developed a foldable paper-based microscope, or “Foldscope”.53 This device effectively combined movable stages cut from an A4 sheet of paper and assembled via folding, a ball lens, lens-holder, LED, battery, and an electrical switch (Figure 5). Through rigorous testing, the Foldscope was demonstrated to provide “over 2000× magnification with submicron resolution” while enabling bright-field, fluorescence, lens-array, and darkfield microscopy through various designs. The Foldscope is inexpensive, uses minimalistic and scalable origami-based manufacturing, is lightweight and portable (<10 g), and highly durable for harsh environments. Since the publication53 and creation of a company called “Foldscope Instruments”, it has been scaled to over 1 million microscopes that have been distributed across the world and created a new microbiology citizen-science community. Despite being an exciting ultralow-cost alternative to state-of-the-art microscopes, the foldscope comes with inherent performance shortcomings, such as narrow focal range, small visual field (with spherical aberration due to ball lens), and exclusivity to applications in light microscopy.
Figure 5.
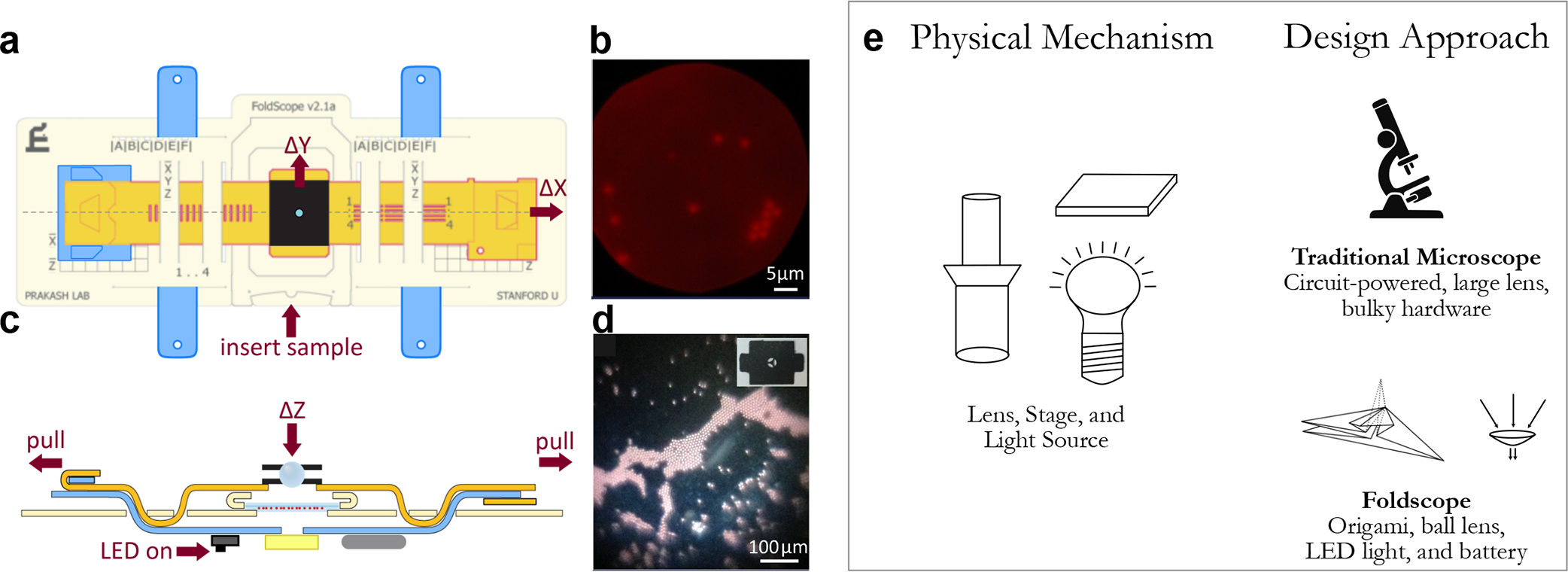
(a) Schematic images of the Foldscope device and (c) how the stage is moved, along with (b, d) sample images of specimen viewed under various types of Foldscopes. (e) While the physical mechanism of both traditional microscopes and Foldscope are the same, Foldscope leverages origami with low-cost components to create a portable, low-cost, and robust basic microscope. [Reproduced (or adapted) with permission from ref 53. Copyright 2014, PLOS One.]
Through the frugal science approach, Foldscope has enabled significantly wider access to microscopes by leveraging a unique origami approach for microscopy design. Foldscope also serves as as pioneering example for the positive impact on science education54,55 and disease diagnostics56,57 through the commercialization of frugal hardware.
Case Study 5: From Cellophane Tape to X-rays.
An imaging technique that has quickly become a cornerstone of modern medical diagnostics is X-ray radiography. X-rays were discovered by accident in 1895 by physics professor Wilhelm Roentgen in Bavaria, Germany, which later led to him being awarded the first Nobel Prize in Physics in 1901.58 Clinical X-ray imaging is now an extremely common technique for noninvasively diagnosing bone fractures and oral health. Outside the realm of medicine, X-rays are used by customs at airports to probe inside luggage, to detect pentimento in old paintings and to determine the structure of crystals using crystallography. However, X-ray machines are exorbitantly expensive and complex to manufacture. While their current form factors are suitable for developed countries, their translation to resource-poor environments is challenging. Given their utility in the medical field, there is critical need for the development of a frugal X-ray approach.
First, consider the basic physical mechanisms through which X-rays are generated. X-rays are commonly produced by bombarding a target (typically tungsten) with electrons created with a high voltage (20–150 kV) differential across an X-ray tube. As they interact with the target material, the decelerating electrons emit X-ray photons with energy, reaching values as high as the energy of the incoming electrons. Like visible light, X-rays are electromagnetic waves. However, they have a wavelength on the order of 10−10 m and subsequently carry a much higher energy. Given their small wavelength, they can penetrate soft biological tissues and air but are blocked when they encounter much denser objects, such as tumors or bones. They are typically detected on silver-coated film plates located behind the specimen that showcase a grayscale picture reflecting the density contrast within the intermediate composite material.
Currently, X-rays are produced through special vacuum tubes where a high voltage causes electrons to travel from the cathode to the anode through a potential difference. These are then concentrated and emitted toward a specific specimen/target with a film placed behind it and an image is then captured. The power necessary is produced through high-voltage power sources, which are precisely controlled to the necessary range through complex circuitry. This design approach contributes to their cost and bulkiness. Consider if there are other ways to generate these X-rays without relying on high-voltage power sources and these vacuum tubes. One proposed alternative is using pressure-sensitive adhesive tape, which, when unpeeled, generates visible light through a phenomenon called triboluminescence.
To test this hypothesis, the Putterman Laboratory revealed that unpeeling sticky cellophane tape emits visible light and X-rays due to a phenomenon where closely adhered objects emit light when they are pulled apart (triboluminescence)59,60 (Figure 6). Through measurement via high-speed X-ray detection equipment, they discovered that scintillations contain nanosecond X-ray pulses with the origin of the pulses being found near the vertex of peeling, confirming that peeling released X-rays. They showed that these rays (~15 keV) in a vacuum chamber (10−5 Torr) were strong enough to generate an X-ray image of a finger.
Figure 6.
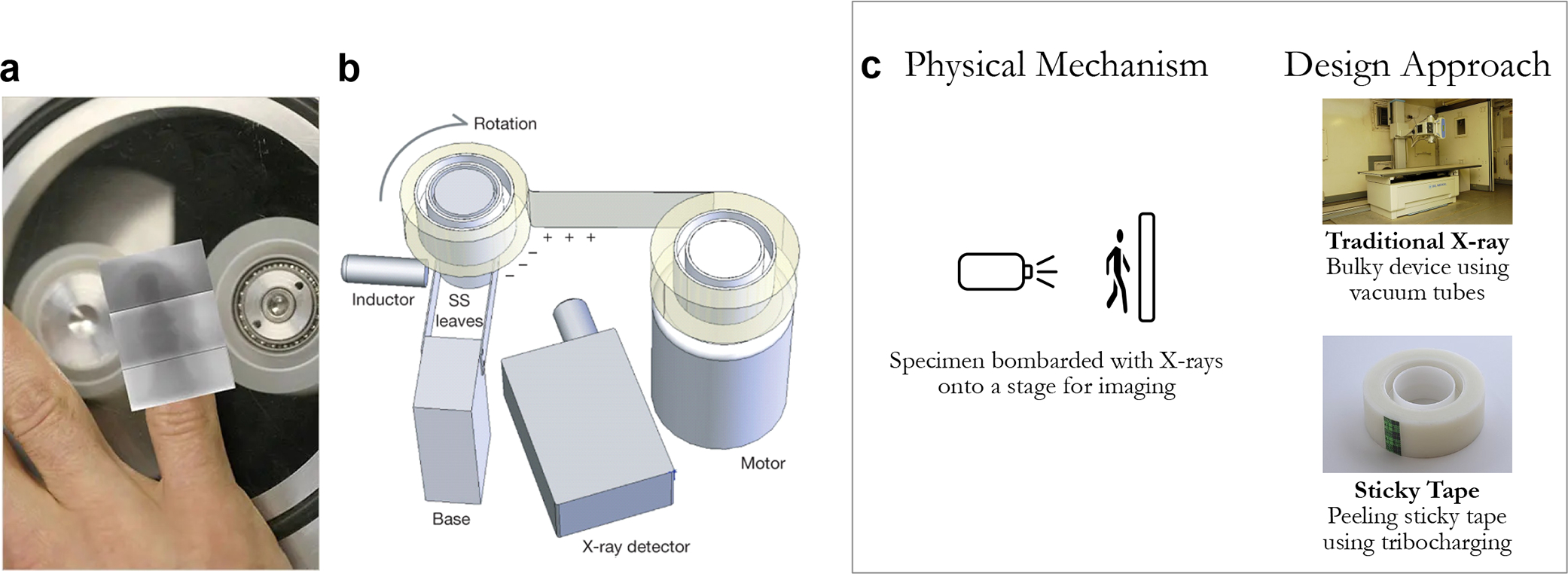
(a) Image of proposed X-ray using sticky tape and (b) image of the setup used to visualize X-rays produced by peeling sticky cellophane tape. (c) The distinction between traditional X-rays and sticky tape is the use of tribocharging and triboluminescence for X-ray production, compared to traditional vacuum tubes with high energy potentials. [Reproduced (or adapted) with permission from ref 59. Copyright 2008, Springer Nature.]
Although triboluminescence-based X-ray technology has not yet been commercialized, this radically different approach holds potential to create an efficient, cost-effective, and energy-efficient X-ray imaging device. Startup companies such as tribogenics seek to exploit this triboluminescence for the development of a battery-powered, portable, and cost-efficient X-ray machine.61 Therefore, this alternative approach of generating X-rays from cellophane tape demonstrates another application of the frugal science method with the potential to transform modern-day X-ray devices.
Case Study 6: From Bubble Wrap to Analytical Assays.
The collection, manipulation, and storage of microliter samples and reagents in controlled and enclosed microenvironments is a common practice in biochemical assays, point-of-care diagnostics, and any experimental analysis of biological specimens. Rigid and sterile containers such as plate wells, sealed vials, and Petri dishes are routinely used in mass during these experiments. Some of these containers may also be equipped with specialized optical and mechanical characteristics, enabling other experimental tasks such as imaging and calorimetric studies. However, these standard containers are often expensive, challenging to sterilize and dispose, and unsustainable, therefore making it challenging to conduct basic and serious scientific experiments in resource-limited settings.
Traditional containers are manufactured using glass or rigid plastics in specific form factors for their respective applications. These are custom-designed for each use case, resulting in higher production costs and thereby higher product costs. Examples of such products include sealed vials, microcentrifuge tubes, cell culture tubes, well plates, cuvettes, etc. Given their ubiquitous usage in biological research, it would be useful to have alternative storage containers that are inexpensive, mass-produced, and easy to dispose of. Consider what everyday items could be used as storage containers that meet these criteria. The Whitesides group tested if bubble wrap could be used as sealed storage containers for biologics, or maybe even further bioanalyses.
Bubble wraps are made of a polymeric film and are readily available across the globe and inexpensive (approximately $0.6/m2), making them an attractive frugal substitute. Through a series of experiments, the Whitesides team showed that bubble wrap can perform remarkably well in storing biological samples and performing various analytical assays.13 The fluid-based samples and compounds are introduced into the inner space of individual bubbles using a needle or pipet with a plastic tip to minimize any possible damage to the thin polymeric film. Afterward, each of the bubbles is sealed with nail hardener to prevent potential spillage and contamination during experiments. The bubble containers exhibit favorable mechanical, chemical, and optical properties. Each bubble is sterile, inert, and gas-permeable, creating an ideal environment for growing microorganisms and culturing bacteria. Because of their transparency and flexibility, they can also be used as a cuvette for spectroscopic measurement of fluorescence and absorbance. Through this, bubble wrap was successfully demonstrated to store reagents in a sterile environment, grow microorganisms through cell culturing, serve as electrochemical cells, and conduct bioanalyses through colorimetric assays (Figure 7). However, bubble wrap comes with its own set of disadvantages, which may compromise its overall practicality, including the fact that bubble wrap is light-sensitive, fragile, and relatively bulky. In addition, it relies on external materials (such as syringe needles) for filling and extraction.
Figure 7.
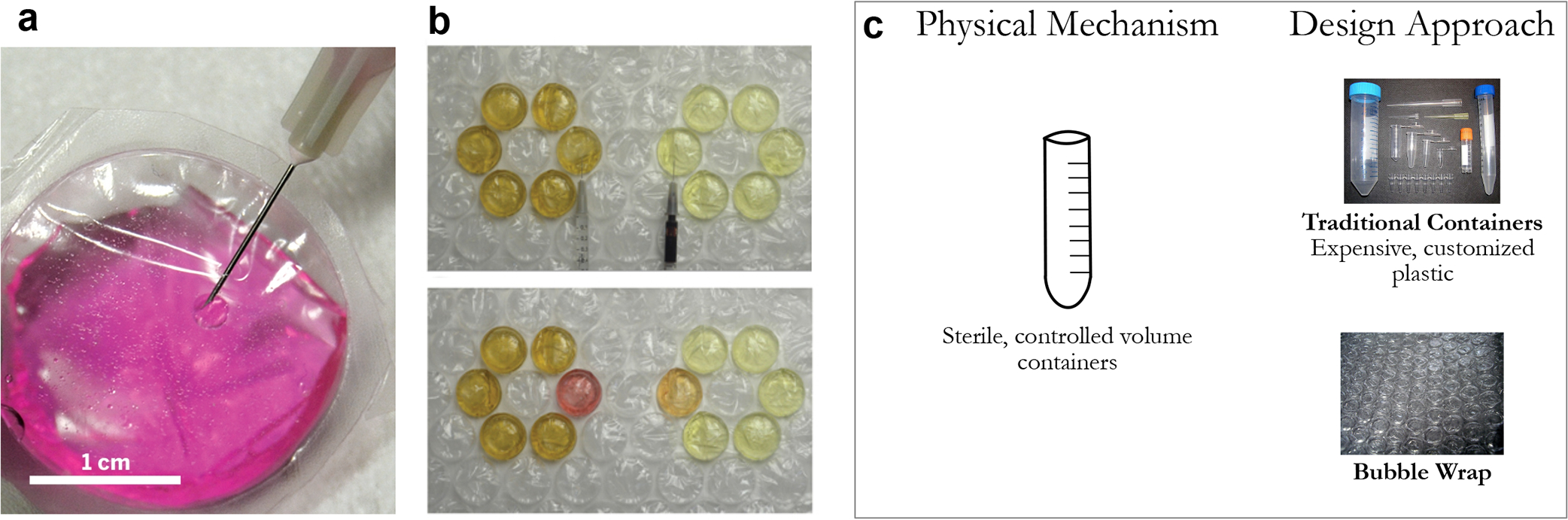
(a) Image of loading bubble wrap container and (b) images of various samples loaded into bubble wrap including bacterial cultures grown in Luria–Bertani broth. [Reproduced (or adapted) from ref 13. Copyright 2016, American Chemical Society, Washington, DC.] (c) While traditional containers and bubble wrap require the same constrains of precise volumes and sterility, bubble wrap is able to provide those in a much more cost-effective and scalable manner, relative to currently manufactured laboratory containers.
The ability of bubble wrap to match the capabilities of their state-of-the-art analogues coupled with its widespread availability and low-cost is a prime example of how a frugal design can result in an inexpensive option without totally jeopardizing the quality of scientific research. In addition, this repurposing of polymeric bubble wraps has far-reaching implications for improving the accessibility of diagnostics and biological research in resource-limited settings.
CONCLUDING REMARKS
Feynman Challenge, FOSH and Frugal Science.
In 1959, Richard Feynman offered a prize of $1000 to anyone (as a high school competition) who could produce a miniature electric motor with sides that did not exceed 1/64th of an inch.62 Feynman knew that, given the state of the art of the technology, his rather audacious dare was extremely difficult to overcome and therefore that his money would be secure for some time.63 However, his true intention was to spark the curiosity of the scientific community and prompt innovative approaches in manufacturing and design to realize the tiny motor. To his surprise, Bill McLellan, a mechanical engineer, and alumnus from Caltech, was able to create the tiny motor with a combination of high level of craftsmanship and using a series of rudimentary crude tools such as toothpicks, fine paintbrush, and tweezers. Feynman conceded and wrote the check, but he was initially disappointed to see that the main points of the challenge were missed.
By analogy, through FOSH or jugaad innovations, although low-cost devices are developed to address the challenges of high cost of scientific hardware, they do not necessarily open new areas of science and technological innovation as the case studies we discuss above. There is no single path to innovate low-cost and functional designs. We classify innovations to create novel and low-cost scientific devices into two main approaches:
Find less expensive but equally effective analogues (to a certain extent) to create a simplified replica of the original device. This method typically relies on existing manufacturing technologies and methodologies and retains the same underlying physical mechanisms. The resulting product would at the most completely reproduce the original design and, at the least, capture some of its essential functionalities. This approach is more common and has grown popular in both the DIY, biohacker culture and FOSH communities that are driven to disseminate information regarding all design aspects of the hardware (e.g., schematics, bills, PCB layouts, source code). This is especially facilitated with the accessibility to relatively low-cost and modular manufacturing techniques such as 3D printing and laser cutting and microcontroller boards such as Arduino and Raspberry Pi. In addition, open-source designers may receive relatively instant feedback on potential improvements via community support websites such as Stack Overflow, social media platforms such as Twitter, and workflow applications such as Slack.
The second approach that we referred to herein as “frugal science” seeks to produce an ultralow-cost device while preserving essential mechanistic functionalities of the original design. Combining these two objectives is typically difficult and adds another level of complexity to the design aspect. This usually involves leveraging seemingly unrelated devices to achieve functionalities that are different from their original intended ones. These frugal devices use the same physical mechanisms as their commercial equivalents but completely different physics, materials, engineering, design, and manufacturing approaches to drive their functionality. As a result, the cost threshold is reduced by orders of magnitude, and generally the tool is scalable, portable, and accessible. Note that both approaches are not entirely mutually exclusive. For example, frugal science tools are also often also open-source and widely disseminated with collaborative communities developing new tools.
There's Plenty of Tools To Reinvent: An Invitation To Join a New Field of Bioengineering Powered by Curiosity.
The past year and a half during the COVID-19 pandemic has witnessed some of the greatest strides in biological innovation, particularly around rapid viral sequencing and the first mRNA vaccine ever granted authorization. The growth in bioengineering and synthetic biology has resulted in numerous advances in medical diagnostics, disease treatments, and even cures for conditions that were previously considered untreatable in the form of cell and gene therapies. Despite these great achievements, a significant portion of the local and global community is excluded, because of economic and infrastructure barriers, where these technologies could have the largest impacts.
With the emerging synthetic biology revolution, engineering biology is easier than ever. Now, with the right tools and knowledge, we are strategically poised as never before to combat global challenges from climate change to pandemics. This is the right time for making scientific and biomolecular engineering tools accessible and affordable for everyone. By radically reducing the cost of scientific tools, we can democratize access to science for everyone and empower them to participate in addressing urgent planetary scale challenges.
We champion a new approach to develop affordable, accessible, and scalable medical tools for people across the world (in rich and poor countries) called “frugal science”. Simply put, frugal science is a novel approach to leverage the complex physics of everyday items for a new functional purpose, driven primarily by curiosity. We live in a world where there are mysteries in items lying around us every day, and it takes the curiosity and enthusiasm of an individual to “think outside the box” on what their possibilities are. Throughout this Review, we have highlighted encouraging examples of frugal science from various groups on how seemingly mundane, ubiquitous items can be transformed to marvelous, powerful tools for bioengineering; however, this is just the beginning. We hope these serve as inspiration for young scientists, engineers, educators, inventors, and even high school students to leverage this approach and reinvent new frugal tools to tackle emerging challenges in global health.
We ask you: look closely at the pen on your table, the lighter in your kitchen, and the paper on your desk and think, how can you make a difference in the world?
Supplementary Material
Funding
M.S.B. acknowledges funding support from NSF Grant Nos. CAREER 1941933 and 1817334 and NIH Award No. R35GM142588.
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.iecr.1c02868.
Table showing cost comparisons between the frugal tools outlined in this Review, relative to their commercial counterparts (PDF)
Contributor Information
Gaurav Byagathvalli, School of Chemical and Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia 30332, United States.
Elio J. Challita, Mechanical Engineering, Georgia Institute of Technology, Atlanta, Georgia 30311, United States.
M. Saad Bhamla, School of Chemical and Biomolecular Engineering, Georgia Institute of Technology, Atlanta, Georgia 30332, United States.
REFERENCES
- (1).This Month in Physics History; available via the Internet at: https://www.aps.org/publications/apsnews/200606/history.cfm (accessed July 13, 2021).
- (2).Dolgin E How to Start a Lab When Funds Are Tight. Nature 2018, 559 (7713), 291–293. [DOI] [PubMed] [Google Scholar]
- (3).Marder E The Haves and the Have Nots. eLife 2013, 2, e01515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Sheikh AB; Pal S; Javed N; Shekhar R COVID-19 Vaccination in Developing Nations: Challenges and Opportunities for Innovation. Infect. Dis. Rep. 2021, 13 (2), 429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Maia Chagas A; Molloy JC; Prieto-Godino LL; Baden T Leveraging Open Hardware to Alleviate the Burden of COVID-19 on Global Health Systems. PLoS Biol. 2020, 18 (4), e3000730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Maia Chagas A Haves and Have Nots Must Find a Better Way: The Case for Open Scientific Hardware. PLoS Biol. 2018, 16 (9), e3000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Ravindran S How DIY Technologies Are Democratizing Science. Nature 2020, 587 (7834), 509–511. [DOI] [PubMed] [Google Scholar]
- (8).Ledford H Garage Biotech: Life Hackers. Nature 2010, 467 (7316), 650–652. [DOI] [PubMed] [Google Scholar]
- (9).Pearce JM Laboratory Equipment: Cut Costs with Open-Source Hardware. Nature 2014, 505 (7485), 618. [DOI] [PubMed] [Google Scholar]
- (10).Pearce JM Building Research Equipment with Free, Open-Source Hardware. Science 2012, 337, 1303. [DOI] [PubMed] [Google Scholar]
- (11).Brennan MD; Bokhari FF; Eddington DT Open Design 3D-Printable Adjustable Micropipette That Meets the ISO Standard for Accuracy. Micromachines (Basel) 2018, 9 (4), 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Radjou N; Prabhu J; Ahuja S Jugaad Innovation: Think Frugal, Be Flexible, Generate Breakthrough Growth; John Wiley & Sons, 2012. [Google Scholar]
- (13).Bwambok DK; Christodouleas DC; Morin SA; Lange H; Phillips ST; Whitesides GM Adaptive Use of Bubble Wrap for Storing Liquid Samples and Performing Analytical Assays. Anal. Chem. 2014, 86 (15), 7478–7485. [DOI] [PubMed] [Google Scholar]
- (14).Prasanna GL; Panda T Electroporation: Basic Principles, Practical Considerations and Applications in Molecular Biology. Bioprocess Eng. 1997, 16 (5), 261–264. [Google Scholar]
- (15).Byagathvalli G; Sinha S; Zhang Y; Styczynski MP; Standeven J; Bhamla MS ElectroPen: An Ultra-Low-Cost, Electricity-Free, Portable Electroporator. PLoS Biol. 2020, 18 (1), e3000589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Miller JF; Dower WJ; Tompkins LS High-Voltage Electroporation of Bacteria: Genetic Transformation of Campylobacter Jejuni with Plasmid DNA. Proc. Natl. Acad. Sci. U. S. A. 1988, 85 (3), 856–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).McCormick CA; Toll MO; Marshall WH A Low Cost Microprocessor-Controlled Electrofusion and Electroporation System. J. Chem. Technol Biotechnol. 1992, 54 (2), 159–169. [DOI] [PubMed] [Google Scholar]
- (18).Xia S; Jin R; Byagathvalli G; Yu H; Ye L; Lu C-Y; Bhamla MS; Yang C; Prausnitz MR An Ultralow-Cost Electroporator with Microneedle Electrodes (EPatch) for SARS-CoV-2 Vaccination. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (45), No. e2110817118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Mabey D; Peeling RW; Ustianowski A; Perkins MD Diagnostics for the Developing World. Nat. Rev. Microbiol. 2004, 2 (3), 231–240. [DOI] [PubMed] [Google Scholar]
- (20).Brown J; Theis L; Kerr L; Zakhidova N; O’Connor K; Uthman M; Oden ZM; Richards-Kortum R A Hand-Powered, Portable, Low-Cost Centrifuge for Diagnosing Anemia in Low-Resource Settings. Am. J. Trop. Med. Hyg. 2011, 85 (2), 327–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Wong AP; Gupta M; Shevkoplyas SS; Whitesides GM Egg Beater as Centrifuge: Isolating Human Blood Plasma from Whole Blood in Resource-Poor Settings. Lab Chip 2008, 8 (12), 2032–2037. [DOI] [PubMed] [Google Scholar]
- (22).van Beek GW The Buzz: A Simple Toy from Antiquity. Bull. Am. Schools Orient. Res. 1989, 275, 53–58. [Google Scholar]
- (23).Bhamla MS; Benson B; Chai C; Katsikis G; Johri A; Prakash M Hand-Powered Ultralow-Cost Paper Centrifuge. Nat. Biomed. Eng. 2017, 1, 0009. [Google Scholar]
- (24).Byagathvalli G; Pomerantz A; Sinha S; Standeven J; Bhamla MSA 3D-Printed Hand-Powered Centrifuge for Molecular Biology. PLoS Biol. 2019, 17 (5), e3000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Ponce-Rojas JC; Costello MS; Proctor DA; Kosik KS; Wilson MZ; Arias C; Acosta-Alvear D A Fast and Accessible Method for the Isolation of RNA, DNA, and Protein to Facilitate the Detection of SARS-CoV-2. J. Clin. Microbiol. 2021, 59 (4), DOI: 10.1128/JCM.02403-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Gu Y; Xu D; Zou K; Zhou T; Zhu G; Yang G; Qu L-L Combined Paper Centrifugal Chromatographic Separation and SERS Detection for Multicomponent Substances. Anal. Chem. 2021, 93 (25), 8693–8697. [DOI] [PubMed] [Google Scholar]
- (27).Liu C-H; Chen C-A; Chen S-J; Tsai T-T; Chu C-C; Chang C-C; Chen C-F Blood Plasma Separation Using a FidgetSpinner. Anal. Chem. 2019, 91 (2), 1247–1253. [DOI] [PubMed] [Google Scholar]
- (28).Michael I; Kim D; Gulenko O; Kumar S; Kumar S; Clara J; Ki DY; Park J; Jeong HY; Kim TS; Kwon S; Cho Y-K A Fidget Spinner for the Point-of-Care Diagnosis of Urinary Tract Infection. Nat. Biomed Eng. 2020, 4 (6), 591–600. [DOI] [PubMed] [Google Scholar]
- (29).Li B; Qi J; Fu L; Han J; Choo J; deMello AJ; Lin B; Chen L Integrated Hand-Powered Centrifugation and Paper-Based Diagnosis with Blood-in/Answer-out Capabilities. Biosens. Bioelectron. 2020, 165, 112282. [DOI] [PubMed] [Google Scholar]
- (30).Nkengasong JN; Yao K; Onyebujoh P Laboratory Medicine in Low-Income and Middle-Income Countries: Progress and Challenges. Lancet 2018, 391 (10133), 1873–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Schito M; Peter TF; Cavanaugh S; Piatek AS; Young GJ; Alexander H; Coggin W; Domingo GJ; Ellenberger D; Ermantraut E; Jani IV; Katamba A; Palamountain KM; Essajee S; Dowdy DW Opportunities and Challenges for Cost-Efficient Implementation of New Point-of-Care Diagnostics for HIV and Tuberculosis. J. Infect. Dis. 2012, 205, S169–S180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).McNerney R Diagnostics for Developing Countries. Diagnostics 2015, 5 (2), 200–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Valones MAA; Guimaraes RL; Brandao LAC; de Souza PRE; de Albuquerque Tavares Carvalho A; Crovela S Principles and Applications of Polymerase Chain Reaction in Medical Diagnostic Fields: A Review. Braz. J. Microbiol. 2009, 40 (1), 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Peeling RW; Holmes KK; Mabey D; Ronald A Rapid tests for sexually transmitted infections (STIs): the way forward. Sex. Transm. Infect. 2006, 82, v1–v6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Yager P; Edwards T; Fu E; Helton K; Nelson K; Tam MR; Weigl BH Microfluidic Diagnostic Technologies for Global Public Health. Nature 2006, 442 (7101), 412–418. [DOI] [PubMed] [Google Scholar]
- (36).Poonia NS Paper Chromatography and Spot Tests: A Useful Combination of Techniques. J. Chem. Educ. 1966, 43 (8), 423. [Google Scholar]
- (37).Mills JS; Werner AEA Paper Chromatography of Natural Resins. Nature 1952, 169 (4312), 1064. [DOI] [PubMed] [Google Scholar]
- (38).Martinez AW; Phillips ST; Whitesides GM; Carrilho E Diagnostics for the Developing World: Microfluidic Paper-Based Analytical Devices. Anal. Chem. 2010, 82 (1), 3–10. [DOI] [PubMed] [Google Scholar]
- (39).Li X; Ballerini DR; Shen W A Perspective on Paper-Based Microfluidics: Current Status and Future Trends. Biomicrofluidics 2012, 6 (1), 011301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Nishat S; Jafry AT; Martinez AW; Awan FR Paper-Based Microfluidics: Simplified Fabrication and Assay Methods. Sens. Actuators, B 2021, 336, 129681. [Google Scholar]
- (41).Pardee K; Green AA; Takahashi MK; Braff D; Lambert G; Lee JW; Ferrante T; Ma D; Donghia N; Fan M; Daringer NM; Bosch I; Dudley DM; O’Connor DH; Gehrke L; Collins JJ Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components. Cell 2016, 165 (5), 1255–1266. [DOI] [PubMed] [Google Scholar]
- (42).Pardee K; Green AA; Ferrante T; Cameron DE; DaleyKeyser A; Yin P; Collins JJ Paper-Based Synthetic Gene Networks. Cell 2014, 159 (4), 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Takahashi MK; Tan X; Dy AJ; Braff D; Akana RT; Furuta Y; Donghia N; Ananthakrishnan A; Collins JJ A Low-Cost Paper-Based Synthetic Biology Platform for Analyzing Gut Microbiota and Host Biomarkers. Nat. Commun. 2018, 9 (1), 3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Yager P; Domingo GJ; Gerdes J Point-of-Care Diagnostics for Global Health. Annu. Rev. Biomed. Eng. 2008, 10 (1), 107–144. [DOI] [PubMed] [Google Scholar]
- (45).Huang A; Nguyen PQ; Stark JC; Takahashi MK; Donghia N; Ferrante T; Dy AJ; Hsu KJ; Dubner RS; Pardee K; Jewett MC; Collins JJ BioBits Explorer: A Modular Synthetic Biology Education Kit. Sci. Adv. 2018, 4 (8), eaat5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Lafleur LK; Bishop JD; Heiniger EK; Gallagher RP; Wheeler MD; Kauffman P; Zhang X; Kline EC; Buser JR; Kumar S; Byrnes SA; Vermeulen NMJ; Scarr NK; Belousov Y; Mahoney W; Toley BJ; Ladd PD; Lutz BR; Yager P A Rapid, Instrument-Free, Sample-to-Result Nucleic Acid Amplification Test. Lab Chip 2016, 16 (19), 3777–3787. [DOI] [PubMed] [Google Scholar]
- (47).Reboud J; Xu G; Garrett A; Adriko M; Yang Z; Tukahebwa EM; Rowell C; Cooper JM Paper-Ba Microfluidics for DNA Diagnostics of Malaria in Low Resource Underserved Rural Communities. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (11), 4834–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).van Leewenhoeck A Observations, Communicated to the Publisher by Mr. Antony van Leewenhoeck, in a Dutch Letter of the 9th of Octob. 1676. Here English’d: Concerning Little Animals by Him Observed in Rain-Well-Sea. and Snow Water; as Also in Water Wherein Pepper Had Lain Infused. Philos. Trans. (1665–1678) 1677, 12, 821–831. [Google Scholar]
- (49).Hooke R Micrographia: Or Some Physiological Descriptions of Minute Bodies Made by Magnifying Glasses, with Observations and Inquiries Thereupon; Courier Corporation, 2003. [Google Scholar]
- (50).Spector DL; Goldman RD Basic Methods in Microscopy: Protocols and Concepts from Cells: A Laboratory Manual; CSHL Press, 2006. [Google Scholar]
- (51).McArthur M; Lang RJ; Harrison MR; Sheppard C Folding Paper: The Infinite Possibilities of Origami; Tuttle Publishing: Rutland, VT, USA, 2013. [Google Scholar]
- (52).Meloni M; Cai J; Zhang Q; Sang-Hoon Lee D; Li M; Ma R; Parashkevov TE; Feng J Engineering Origami: A Comprehensive Review of Recent Applications, Design Methods, and Tools. Adv. Sci. 2021, 8 (13), 2000636. [Google Scholar]
- (53).Cybulski JS; Clements J; Prakash M Foldscope: Origami-Based Paper Microscope. PLoS One 2014, 9 (6), e98781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Yesudhason BV; Selvan Christyraj JRS; Ganesan M; Subbiahanadar Chelladurai K; Venkatachalam S; Ramalingam A; Benedict J; Paulraj VD; Selvan Christyraj JD Developmental Stages of Zebrafish (Danio Rerio) Embryos and Toxicological Studies Using Foldscope Microscope. Cell Biol. Int. 2020, 44 (10), 1968–1980. [DOI] [PubMed] [Google Scholar]
- (55).Kaur T; Dahiya S; Satija SH; Nawal SJ; Kshetrimayum N; Ningthoujam J; Chahal AK; Rao A Foldscope as a Primary Diagnostic Tool for Oral and Urinary Tract Infections and Its Effectiveness in Oral Health Education. J. Microsc. 2020, 279 (1), 39–51. [DOI] [PubMed] [Google Scholar]
- (56).Ephraim RKD; Duah E; Cybulski JS; Prakash M; D’Ambrosio MV; Fletcher DA; Keiser J; Andrews JR; Bogoch II Diagnosis of Schistosoma Haematobium Infection with a Mobile Phone-Mounted Foldscope and a Reversed-Lens CellScope in Ghana. Am. J. Trop. Med. Hyg. 2015, 92 (6), 1253–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Prusty JS; Kumar A Innovative Screening and Drug Susceptibility Analysis on Candida Albicans Using Foldscope Microscopy. Rend. Lincei Sci. Fis. Nat. 2021, 32, 163. [Google Scholar]
- (58).Röntgen WC On a New Kind of Rays. Science 1896, 3 (59), 227–231. [DOI] [PubMed] [Google Scholar]
- (59).Camara CG; Escobar JV; Hird JR; Putterman SJ Correlation between Nanosecond X-Ray Flashes and Stick-Slip Friction in Peeling Tape. Nature 2008, 455 (7216), 1089–1092. [Google Scholar]
- (60).Hird JR; Camara CG; Putterman SJ A Triboelectric X-Ray Source. Appl. Phys. Lett. 2011, 98 (13), 133501. [Google Scholar]
- (61).Tribogenics; available via the Internet at: https://www.boombang.com/tribogenics (accessed Sept. 29, 2021).
- (62).Feynman RP There’s Plenty of Room at the Bottom: An Invitation to Enter a New Field of Physics. In Handbook of Nanoscience, Engineering, and Technology; CRC Press, 2018; pp 26–35, DOI: 10.1201/9781315217178-6. [DOI] [Google Scholar]
- (63).Ball P Miniature Motors. Nat. Mater. 2004, 3, 428–428. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


