Abstract
A field-collected colony of the diamondback moth, Plutella xylostella, had 31-fold resistance to Cry1C protoxin of Bacillus thuringiensis. After 24 generations of selection with Cry1C protoxin and transgenic broccoli expressing a Cry1C protein, the resistance that developed was high enough that neonates of the resistant strain could complete their entire life cycle on transgenic broccoli expressing high levels of Cry1C. After 26 generations of selection, the resistance ratios of this strain to Cry1C protoxin were 12,400- and 63,100-fold, respectively, for the neonates and second instars by a leaf dip assay. The resistance remained stable until generation 38 (G38) under continuous selection but decreased to 235-fold at G38 when selection ceased at G28. The Cry1C resistance in this strain was seen to be inherited as an autosomal and incompletely recessive factor or factors when evaluated using a leaf dip assay and recessive when evaluated using Cry1C transgenic broccoli. Saturable binding of 125I-Cry1C was found with brush border membrane vesicles (BBMV) from both susceptible and Cry1C-resistant strains. Significant differences in Cry1C binding to BBMV from the two strains were detected. BBMV from the resistant strain had about sevenfold-lower affinity for Cry1C and threefold-higher binding site concentration than BBMV from the susceptible strain. The overall Cry1C binding affinity was just 2.5-fold higher for BBMV from the susceptible strain than it was for BBMV from the resistant strain. These results suggest that reduced binding is not the major mechanism of resistance to Cry1C.
Microbial insecticides based on Bacillus thuringiensis can provide a good combination of safety and effectiveness for pest control. In 1992, over 2 million acres of U.S. crops were treated with B. thuringiensis sprays (16). Transgenic insecticidal crops expressing B. thuringiensis toxins entered the U.S. market in 1996, resulting in several important economic and environmental advantages. For example, since the commercialization of B. thuringiensis-transgenic cotton in 1996, insecticide sprays on cotton have been reduced by approximately 3.8 million liters of formulated product per year in the United States, and this has led to a significant reduction in the use of more hazardous organophosphate and pyrethroid insecticides (according to the U.S. Environmental Protection Agency and U.S. Department of Agriculture position paper on insect resistance management in B. thuringiensis crops [http://www.epa .gov/oppbppd1/biopesticides/otherdocs/bt_position_paper _618.htm]). However, there is concern that widespread use of B. thuringiensis-transgenic crops may increase the risk of resistance to both transgenic crops and B. thuringiensis microbial spray formulations (16).
There have been no cases of insects developing resistance on B. thuringiensis-transgenic plants in the field, but the diamondback moth, Plutella xylostella (L.), developed resistance to B. thuringiensis toxins in foliar sprays under field conditions (27, 31). Laboratory populations of Cry1A-resistant diamondback moth can also survive on transgenic crucifers expressing a high level of Cry1Ac (17, 22, 38).
Although intensive research has been conducted on resistance of the diamondback moth to B. thuringiensis, most studies have emphasized resistance to Cry1A or B. thuringiensis formulations. Tabashnik et al. proposed a model 1 of B. thuringiensis resistance, which is characterized by more than 500-fold resistance to at least one Cry1A toxin, recessive inheritance, little or no cross-resistance to Cry1C, and reduced binding of at least one Cry1A toxin (33). It was suggested that the gene(s) conferring resistance to Cry1C segregates independently of the gene conferring resistance to Cry1Ab in the diamondback moth (11).
To date, only a few cases of Cry1C resistance have been reported. A high level of Cry1C resistance (>500-fold) developed in Spodoptera exigua and Spodoptera littoralis after selection in laboratories (18, 19). Binding experiments in S. exigua indicated that, though binding of Cry1C was slightly reduced in the Cry1C-resistant insects compared to susceptible insects, reduced binding was not a major mechanism of resistance to Cry1C in the resistant strain (18). In the diamondback moth, only low to moderate levels of resistance to Cry1C developed either in the field (23-fold) or the laboratory (62-fold) (11, 12). Again, Cry1C-resistant insects did not differ from susceptible insects in terms of Cry1C binding (14).
The objective of our study was to obtain a strain of diamondback moth with a level of resistance to Cry1C high enough to allow the insects to complete their entire life cycle on transgenic broccoli expressing high levels of Cry1C. Once we succeeded, we examined the pattern of inheritance of this resistance and whether Cry1C binding site modification was the biochemical mechanism involved.
MATERIALS AND METHODS
Insects.
We used two strains of diamondback moth for these studies: our standard susceptible strain (Geneva 88) and a resistant strain (Cry1C-Sel). Geneva 88 insects were collected in 1988 from cabbage at the New York State Agricultural Experiment Station, Robbins Farm, Geneva, New York, and have been maintained on a wheat germ-casein artificial diet (26) for over 200 generations. While on diet, the strain was kept in an environmental chamber at 27 ± 1°C, 50 ± 2% relative humidity, and a photoperiod of 16:8 (light/dark hours). The Cry1C-Sel strain was originally collected in March 1997 from a collard field in Lexington, S.C., where B. thuringiensis subsp. aizawai and B. thuringiensis kurstaki products were reported as failing. Using a leaf dip bioassay (27), we determined it had 31-fold resistance to Cry1C before the selection.
B. thuringiensis toxins.
The Cry1C protoxin used for bioassays and selections during generation 2 (G2) to G14 of the Cry1C-Sel strain was provided by W. Moar, Auburn University, Auburn, Ala. The cry1C gene was from B. thuringiensis subsp. entomocidus and was expressed in an acrystalliferous strain of HD-1 (18). A liquid formulation of Cry1C protoxin expressed in and encapsulated by transgenic Pseudomonas fluorescens (15% active ingredient; lot no. 100021276; Mycogen, San Diego, Calif.) was used thereafter. Binding experiments were performed with activated Cry1C toxin from the recombinant B. thuringiensis strain EG1081 (Ecogen Inc.).
Cry1C preparation for binding experiments.
Production, solubilization, and trypsin activation of Cry1C from the recombinant B. thuringiensis strain EG1081 (Ecogen Inc.) have been described elsewhere (25). For binding analyses, purification is required to remove adsorbed protoxin fragments that block iodination and affect binding parameters (15, 39). Cry1C activated toxin (15 ml) was dialyzed against 4 liters of Tris buffer (20 mM Tris-HCl, pH 8.6) overnight at 4°C. The dialyzed Cry1C solution (about 2 mg/ml) was filter sterilized and loaded onto a MonoQ HR 5/5 anion-exchange column (FPLC system; Pharmacia, Uppsala, Sweden) previously equilibrated with Tris buffer. Cry1C was eluted with a linear NaCl gradient (0.033 mM/min). Analysis of chromatographic fractions by sodium dodecyl sulfate-polyacrylamide gel electrophoresis showed two peaks (at 10 and 18 min) with the same molecular weight as the activated Cry1C. Preliminary experiments showed that only the protein of the peak at 18 min provided specific binding to brush border membrane vesicles (BBMV) after 125I labeling (data not shown). Therefore, fractions of the peak at 18 min were pooled (5-ml total volume) and dialyzed against 200 ml of carbonate buffer (50 mM Na2CO3, pH 8) containing 16% of polyethylene glycol 6000 for 4 h at 4°C. Dithiotreitol was added to the sample to a final concentration of 6.5 mM. After overnight incubation at 4°C, the sample was centrifuged at 16000 × g for 5 min at 4°C. The supernatant was applied to a Superose 12 gel filtration column (Pharmacia FPLC system) and eluted with carbonate buffer containing 1 mM dithiothreitol. Cry1C was finally dialyzed for 5 h against 4 liters of phosphate-buffered saline (8 mM Na2HPO4, 2 mM KH2PO4, 150 mM NaCl, pH 7.5).
Transgenic plants expressing Cry1C toxin.
A synthetic truncated cry1C gene (1.9 kb in length) was introduced into broccoli (Brassica oleracea subsp. italica, Green Comet hybrid) by Agrobacterium sp.-mediated transformation (3). The Cry1C protein in the leaves of transgenic broccoli plants used (lines H12 and H14) was ca. 0.4% of total soluble protein (3).
Selection on Cry1C-Sel strain.
Selection on the Cry1C-Sel diamondback moth strain by Cry1C started in the laboratory at G2 after collection from the field. The Cry1C protoxin in cabbage leaf dip assays (27, 36) was used for the initial 13 generations of selection (G2 to G14). For each selection, >1,000 second instars were infested onto cabbage leaf disks treated with Cry1C and incubated in plastic containers for 3 days at 27 ± 1°C. Survivors from the leaf disks were counted and transferred onto rape plants until adult eclosion. Leaves or plants of transgenic broccoli expressing Cry1C toxin (3) were used for the next 23 generations (G15 to G37). The average mortality from selection to adult eclosion caused by the first 22 selections was 76.8% (Table 1). The mortality in G18 to G22 was estimated based on approximate egg numbers infested on transgenic broccoli.
TABLE 1.
Selection process of the Cry1C-Sel strain in the laboratory
| Generations selecteda | Instar for selection | Selection method | Mean % mortality |
|---|---|---|---|
| 1–13 | 2 | Leaf dip assay with Cry1C protoxin | 81.2 |
| 14–17 | 2 | Detached leaves of Cry1C transgenic broccoli | 68.1 |
| 18–22 | 1 | Cry1C transgenic broccoli plants | 72.3 |
| 23–36 | 1 | Cry1C transgenic broccoli plants | (Not evaluated) |
The first generation for selection was G2.
Leaf dip bioassays.
Cabbage leaf dip bioassays, as previously reported (27, 37), were used for each strain of diamondback moth using second instars for most tests. Larvae of each strain for a bioassay were reared on oilseed rape (Brassica napus, Dwarf Essex variety; L. L. Olds Seed, Madison, Wis.) plants in a greenhouse. Five to six concentrations plus a control and six disks for each concentration were included in each bioassay. Five second instars (0.2 to 0.3 mg/larva) were placed on each of the leaf disks. Bond spreader-sticker (Loveland Industry, Loveland, Colo.) was added at 0.1% to all test concentrations and to the water control. Mortality was determined after 72 h at 27 ± 1°C. Data were analyzed with a probit model using the POLO program (23). Where resistance ratio (RR) values were calculated (RR = LC50 / LC50 of Geneva 88, where LC50 is the 50% lethal concentration), the resistant and susceptible strains were tested concurrently.
Survival of resistant strain on transgenic broccoli.
Neonates of the Cry1C-Sel strain after 24 generations of selection (G26) were infested onto Cry1C transgenic broccoli until pupation. Nontransgenic broccoli (Green Comet hybrid) was used as a control. There were 10 replicates for each treatment and 10 neonates for each replicate. Each surviving pupa was weighed and placed into a 30-ml plastic cup until eclosion. To determine the number of eggs laid per female, 15 single male-female pairs from each treatment group were placed in 473-ml styrofoam containers. A 10% sugar solution was provided for the moths, and egg sheets of cabbage-treated aluminum foil (26) were provided for laying eggs in each container. Thirteen pairs in each treatment group laid eggs. Egg sheets were collected at 2, 4, and 6 days, and eggs were counted. To determine the percent hatch, 50 to 60 eggs laid on day 2 were selected from an egg sheet with 5 replicates for each treatment group. Neonates were counted after 2 and 3 days at 27 ± 1°C. Data were analyzed with analysis of variance using the SAS program (24). Data were transformed using arcsin √P for proportion (P) of mortality and using log(x) for other data (x) before each analysis of variance was performed.
Stability of resistance.
A substrain of Cry1C-Sel was established from survivors of the G26 selection (i.e., G27). Larvae of G28 to G38 were reared on oilseed rape plants in the greenhouse without selection. Bioassays were conducted at G28, G29, G30, G32, G34, and G38 of the reversion substrain using second instars to determine levels of resistance to Cry1C.
Inheritance tests.
Insects of Cry1C-Sel from G28 after 26 generations of selection were used as the resistant strain for both the crosses and backcrosses (BCs) to study the inheritance of Cry1C resistance. Crosses and strains used for the inheritance study were as follows: F1 = Geneva 88 (S, G240) × Cry1C-Sel (R, G28); BC = F1 × R (G28). Following Stone's method (28), the degree of dominance (D) for resistance was calculated using the reciprocal F1 crosses and the pooled data. The single-concentration method (12) to determine estimated dominance h was also used for analysis of dominance. Both the neonates and second instars were tested in all bioassays. There were 50 neonates (10 leaf disks and five neonates on each disk) for each concentration, with a procedure similar to that described above. Broccoli leaves (Green Comet hybrid) grown in the greenhouse were used for bioassays of neonates.
The χ2 goodness-of-fit test was used to determine how well the BC mortality data of the second instars observed at each concentration fit mortality as predicted by each model of inheritance. For direct testing of monogenic inheritance, calculations of expected mortality for the BC offspring were based on experimental data (21, 29). For indirect tests of monogenic and polygenic inheritance, we used the methods described by Tabashnik (29) and Tang et al. (37). There were nine concentrations and 30 larvae for each concentration in the bioassay.
The efficacy of Cry1C transgenic broccoli on different instars of F1 progeny was also tested for the dominance of resistance on B. thuringiensis-transgenic broccoli with nontransgenic broccoli as a control. First, second, third, and fourth instars of F1 progeny were used, with five replicates for each treatment and 10 larvae for each replicate. Mortality was determined after 72 h at 27 ± 1°C.
Binding assays.
Last-instar larvae were frozen at −80°C at Cornell University's New York State Agricultural Experiment Station and shipped frozen to the University of Valencia. BBMV were prepared from whole larvae by the differential magnesium precipitation method (4, 41) and then frozen in liquid nitrogen and kept at −80°C until used. Protein concentration in the BBMV was determined with the Bio-Rad reagent (2).
Cry1C toxin was 125I labeled by the IODO-BEADS (Pierce) method (14), a specific activity of 0.4 mCi/mg was obtained. BBMV were incubated with 125I-Cry1C (2.5 nM) in 0.1 ml of binding buffer (PBS–0.1% bovine serum albumin) at room temperature for 90 min. Bound toxin was separated from free toxin by centrifugation (9), and the pellet was washed twice with 500 μl of binding buffer. Radioactivity in the pellet was measured in a Compugamma CS gamma irradiation counter (LKB Pharmacia).
Binding parameters were obtained using the LIGAND computer program (20). Statistical analyses of differences (P < 0.05) of the mean values of binding parameters were performed with a Student's t test using the Prism software computer program (GraphPad Software, San Diego, Calif.).
RESULTS
Resistance development.
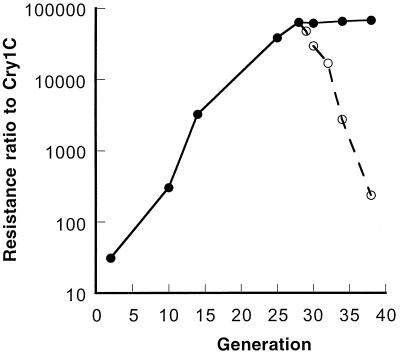
The initial RR of the Cry1C-Sel strain to Cry1C protoxin based on the LC50 was 31-fold (13.7 versus 0.437 mg of the active ingredient [AI]/liter) at the second generation (G2) reared in the laboratory after collection. After 26 generations of selection, the RR of the Cry1C-Sel at G28 increased to 63,100-fold (Fig. 1; also see Table 3). The RR remained stable between G28 and G38 under continuous selection. The RR to Cry1C for the neonates at G28 was 12,400-fold.
FIG. 1.
Development of resistance to Cry1C in the Cry1C-Sel strain (●) of the diamondback moth and stability of resistance after selection ceased at G28 (○).
TABLE 3.
Inheritance of Cry1C resistance in the diamondback moth after 26 generations of selection
| Group | Strain | n | Slope (SE) | LC50 (mg of AI/liter) | 95% FL | RR | D |
|---|---|---|---|---|---|---|---|
| Neonate | G88 (S) | 300 | 1.05 (0.15) | 0.339 | 0.166–0.578 | 1.0 | |
| Cry1C-Sel (R) | 300 | 0.89 (0.10) | 4,200 | 2,270–7,760 | 12,400 | ||
| S(f) × R | 300 | 1.10 (0.16) | 12.0 | 7.09–19.0 | 35.4 | −0.24 | |
| R(f) × S | 300 | 1.18 (0.18) | 15.2 | 6.72–29.9 | 44.8 | −0.19 | |
| Pooled F1 | 600 | 1.13 (0.12) | 13.6 | 6.88–23.8 | 40.1 | −0.22 | |
| Second instar | G88 (S) | 150 | 1.90 (0.33) | 0.108 | 0.068–0.156 | 1.0 | |
| Cry1C-Sel (R) | 150 | 2.22 (0.35) | 6,820 | 4,920–8,780 | 63,100 | ||
| S(f) × R | 150 | 1.20 (0.18) | 5.78 | 3.72–9.65 | 53.5 | −0.28 | |
| R(f) × S | 150 | 1.70 (0.30) | 8.20 | 4.69–12.4 | 75.9 | −0.22 | |
| Pooled F1 | 300 | 1.49 (0.22) | 7.08 | 4.69–9.80 | 65.6 | −0.24 |
When neonates of G26, after 24 generations of selection, fed on Cry1C transgenic or on nontransgenic plants, there were no significant differences in mortality until pupation, weight per pupa, eggs per female, or percentage of eggs hatched (Table 2). Neonates of the Cry1C-Sel strain could complete their entire life cycle on transgenic broccoli with a high expression level of Cry1C, although there were evident cumulative disadvantages on B. thuringiensis broccoli. The mortality of Geneva 88 was 100% when the neonates fed on the same Cry1C transgenic broccoli until pupation (unpublished data; 12% on nontransgenic control).
TABLE 2.
Survival and development of neonates of a Cry1C-Sel strain on Cry1C B. thuringiensis-transgenic and a nontransgenic (control) broccolia
| Treatment group | Mean % mortality (SEM)
|
Mean wt (mg)/pupa (SEM) | Mean no. of eggs/female (SEM) | Mean % eggs hatched (SEM) | |
|---|---|---|---|---|---|
| At 3 days | To pupation | ||||
| B. thuringiensis-transgenic broccoli | 12.0 (1.3) A | 23 (4.2) A | 5.26 (0.12) A | 128.2 (19.7) A | 76.7 (5.9) A |
| Control | 6.0 (2.7) B | 15 (4.0) A | 5.55 (0.13) A | 164.4 (16.9) A | 85.2 (4.5) A |
Values in a column followed by same letter are not significantly different (P > 0.05).
Stability of resistance.
After selection ceased at G28 of the Cry1C-Sel strain, the Cry1C resistance decreased from 63,100-fold at G28 to 16,900-, 2,760- and 235-fold, respectively, at G32, G34, and G38 (Fig. 1). The LC50s (95% fiducial limit [FL], milligrams of AI/liter) for the second instars of the Cry1C-Sel strain were 3,020 (2,230 to 3,980) at G32, 663 (359 to 1,080) at G34, and 38.7 (15.5 to 69.3) at G38, respectively.
Inheritance of Cry1C resistance.
Genetic analysis of F1 larvae indicated that the Cry1C resistance in the Cry1C-Sel colony was inherited as an autosomal and incompletely recessive factor or factors at the LC50 (Table 3). The degree of dominance (D) of resistance based on pooled F1 was −0.22 for the neonate and −0.24 for the second instars. The h of resistance using the single-concentration method (12) tended to be more recessive as the concentration increased for both the neonates and second instars. For the neonates, resistance was partially recessive at concentrations of 10 to 100 mg of AI/liter (h = 0.49 to 0.14) and partially dominant at 0.316 to 3.16 mg of AI/liter (h = 0.78 to 0.61). For the second instars, resistance was recessive at concentrations of 100 mg of AI/liter or above (h = 0), partially recessive at 31.6 (h = 0.14) or 10 (h = 0.44) mg of AI/liter, and partially dominant at 0.316 to 3.16 mg of AI/liter (h = 0.94 to 0.73) based on pooled F1.
The mortality of all instars of the F1 progeny was 100% when they were fed Cry1C transgenic broccoli for 72 h (2 to 14% on nontransgenic control), indicating that Cry1C resistance in the Cry1C-Sel strain was recessive.
In the direct test of monogenic inheritance for Cry1C resistance, five of the nine concentrations evaluated resulted in significant deviation between the observed and expected mortality (Table 4). Using the indirect method for testing inheritance, five of the nine concentrations also resulted in significant deviation when the one-locus model was used (Table 5), but deviations were significant at all nine concentrations tested when two- or five-locus models were used.
TABLE 4.
Direct test of monogenic inheritance for Cry1C resistance in Cry1C-Sel strain by comparing expected and observed mortality of second instars of the BC progenya
| Conc (mg of AI/liter) | No. of deaths
|
χ2 (df = 1) | P > χ2 | |
|---|---|---|---|---|
| Observed | Expected | |||
| 3.16 | 6 | 4.1 | 1.02 | 0.31 |
| 10 | 11 | 8.4 | 1.12 | 0.29 |
| 31.6 | 13 | 13.0 | 0 | 1 |
| 100 | 17 | 14.5 | 0.83 | 0.36 |
| 200 | 21 | 15.0 | 4.80 | 0.028b |
| 316 | 22 | 15.5 | 5.64 | 0.018b |
| 1,000 | 26 | 16.3 | 12.6 | 0.0004b |
| 3,160 | 28 | 18.1 | 13.7 | 0.0002b |
| 10,000 | 30 | 25.1 | 5.86 | 0.016b |
For each concentration, 30 larvae were tested.
Observed mortality significantly deviated from the model prediction (P < 0.05).
TABLE 5.
Indirect tests for monogenic and polygenic inheritance of Cry1C resistance in Cry1C-Sel strain using second instars by χ2 analysis (df = 1)
| Conc (mg of AI/liter) | Genetic modela
|
|||||
|---|---|---|---|---|---|---|
| One locus
|
Two loci
|
Five loci
|
||||
| χ2 | P | χ2 | P | χ2 | P | |
| 3.16 | 0.58 | 0.45 | 10.8 | 0.001b | 152 | <0.001b |
| 10 | 0.76 | 0.38 | 10.2 | 0.001b | 188 | <0.001b |
| 31.6 | 0.03 | 0.85 | 7.24 | 0.007b | 153 | <0.001b |
| 100 | 0.94 | 0.33 | 16.3 | <0.001b | 153 | <0.001b |
| 200 | 5.17 | 0.02b | 30.6 | <0.001b | 105 | <0.001b |
| 316 | 6.69 | 0.01b | 30.1 | <0.001b | 57.2 | <0.001b |
| 1000 | 14.8 | <0.001b | 30.0 | <0.001b | 36.5 | <0.001b |
| 3160 | 12.9 | <0.001b | 12.2 | <0.001b | 15.8 | <0.001b |
| 10000 | 6.50 | 0.01b | 4.69 | 0.03b | 8.30 | 0.04b |
Models assume equal and additive effects of loci.
Observed mortality significantly deviated form the model prediction (P < 0.05).
Binding of 125I-Cry1C to BBMV.
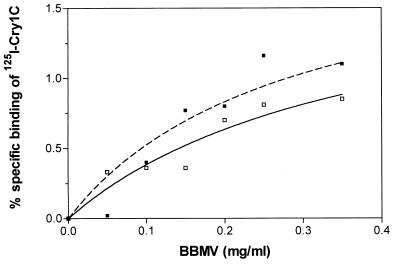
Cry1C-specific binding was tested by incubating 125I-Cry1C with various concentrations of BBMV from the Geneva 88 and the Cry1C-Sel strains (Fig. 2). Saturable binding was found in both strains, with a maximum specific binding level of around 1% of the total radioactivity added. Saturable binding in the Cry1C-Sel strain indicated that the highly resistant insects possessed specific binding sites for Cry1C.
FIG. 2.
Specific binding of 125I-Cry1C as a function of BBMV concentration. Specific binding was calculated by subtracting the nonspecific binding from the total binding. Nonspecific binding was determined in the presence of a 120-fold excess of unlabeled toxin. Each data point represents the mean of duplicate samples. Symbols: □, Geneva 88 strain; ■, Cry1C-Sel strain.
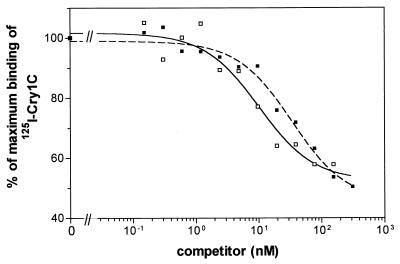
Experiments of competition of 125I-Cry1C with unlabeled Cry1C for binding to BBMV-specific sites (Fig. 3) were performed to obtain quantitative estimates of the equilibrium dissociation constant (Kd) and binding site concentration (Rt). Significantly different values for these binding parameters were obtained for both strains (Table 6). Cry1C showed about a sevenfold lower affinity for BBMV from the Cry1C-Sel strain than for that from the Geneva 88 strain. Moreover, BBMV from Cry1C-Sel had about a threefold higher binding site concentration than BBMV from the Geneva 88 strain. The overall binding affinity of the BBMV for Cry1C, estimated as the Rt/Kd ratio, was also significantly different for both strains (Table 6), being 2.5-fold higher for the Geneva 88 strain.
FIG. 3.
Binding of 125I-Cry1C to diamondback moth BBMV (0.2 mg of vesicle protein/ml) at increasing concentrations of unlabeled Cry1C. Nonspecific binding was not subtracted. Each point represents the mean of two independent experiments. Symbols: □, Geneva 88 strain; ■, Cry1C-Sel strain.
TABLE 6.
Estimates of Kd and Rt for Cry1C toxin in different strains of diamondback motha
| Strain | Kd ± SE (nM) | Rt ± SE (pmol/mg) | Rt/Kd ± SE |
|---|---|---|---|
| Geneva 88 | 7.09 ± 4.12 | 0.75 ± 0.38 | 0.112 ± 0.012 |
| Cry1C-Sel | 50.5 ± 6.4 | 2.20 ± 0.25 | 0.045 ± 0.010 |
Values for each strain were calculated from two experiments.
DISCUSSION
Although several insect species have developed resistance to B. thuringiensis formulations or toxins (30), there are only three reported species for which resistant strains can survive on transgenic insecticidal plants. Survival to maturity has been reported for resistant strains of diamondback moth on B. thuringiensis-transgenic broccoli and B. thuringiensis-transgenic canola (17, 22, 38) and for tobacco budworm (Heliothis virescens) and pink bollworm (Pectinophora gossypiella) on B. thuringiensis-transgenic cotton (6, 13). However, in all these reports the resistant strains did not develop directly from selection on B. thuringiensis-transgenic crops.
After 24 generations of selection with the Cry1C protoxin or transgenic broccoli expressing a Cry1C protein, the Cry1C resistance in our diamondback moth strain was so high that neonates could complete their entire life cycle on transgenic broccoli expressing high levels of Cry1C. This contrasts with the F1 progeny, for which the mortality of all instars was 100% when fed the same Cry1C transgenic broccoli. At an early stage of our laboratory selection with the Cry1C protoxin, the RR of the Cry1C-Sel strain after six generations of selections increased to ca. 100-fold, but the larvae could not survive on Cry1C broccoli (3). B. thuringiensis-transgenic broccoli with a high expression level of Cry1C rapidly accelerated the development of resistance. Considering that there may be 20 generations per year of the diamondback moth in tropical areas (34, 35), B. thuringiensis-transgenic crucifers containing Cry1A or Cry1C toxins could face the potential of rapid development of resistance in the field as they enter commercial application in areas in which B. thuringiensis products containing Cry1A or Cry1C toxins had been used extensively. Although in our trials we gradually increased the frequency of resistance (and perhaps the intensity of resistance of individuals, if such does exist), it is unknown whether populations in the field could eventually adapt to such high expression levels as in our plants. However, our results clearly show the potential for development of a high level of resistance to Cry1C.
The Cry1C resistance in this strain was seen to be inherited as an incompletely recessive factor (or factors) when evaluated using a leaf dip assay and as recessive when using Cry1C transgenic broccoli. Our results differ from another study in which Cry1C resistance in the diamondback moth (NO-95C) was much lower (62-fold) and inherited in an incompletely dominant fashion (12). The h using the single-concentration method resulted in similar trends; i.e., in both studies the resistance tended to be more recessive as the concentration increased. The original level of Cry1C resistance in NO-95 collected from Hawaii was 22-fold, and there were nine generations of selection using Cry1C protoxin by a method similar to the bioassay (11, 12). The different results on the dominance of Cry1C resistance between the two studies may have resulted from the large difference in levels of resistance and/or from different resistance genes in the two strains.
We could not reach a clear conclusion on monogenic or polygenic inheritance of the Cry1C resistance based on the direct and indirect tests. In the direct tests of monogenic inheritance, there were significant deviations at and near the LC50 of the BC, which suggested nonadditive polygenic inheritance or experimental error (29). Our binding studies also suggested that there should be other important mechanisms responsible for the Cry1C resistance apart from reduced binding. But in the indirect tests, the monogenic model provided a better fit than either the two-locus or five-locus models. Alternative approaches, including repeated BC tests and molecular techniques, will be helpful for determining the mode of inheritance for Cry1C resistance (29).
It is important that the mode of inheritance be known since the “high dose-refuge” strategy for managing the development of insect resistance to B. thuringiensis transgenic crops (7; U.S. Environmental Protection Agency website) was proposed on the basis of recessive inheritance. Our results using Cry1C transgenic broccoli may provide the first experimental evidence for the recessive inheritance of resistance which was selected primarily on B. thuringiensis-transgenic plants.
A previous report indicated that Cry1C resistance in S. littoralis decreased from >500-fold to 11-fold eight generations after selection ceased (19). A similar decline of Cry1C resistance occurred in S. exigua eight generations after selection ceased (18). Our results also suggest the instability of Cry1C resistance in the diamondback moth after release from the selection. One possible reason for the instability might be the fitness cost of the resistance. Significant reduction was observed in the weight per pupa and eggs per female in the Cry1C-Sel strain compared with Geneva 88 and F1 progeny when fed on the same nontransgenic broccoli leaves (Zhao et al., unpublished data), hinting at some fitness costs for Cry1C resistance. However, a precise evaluation for the fitness cost of Cry1C resistance in the Cry1C-Sel strain is difficult, as the genetic background between each strain of the diamondback moth is different in characteristics besides the Cry1C resistance.
Binding site modification is thought to be the major mechanism of resistance to Cry1A toxins in Pectinophora interpunctella (40), H. virescens (10), and the diamondback moth (5, 25, 32, 36, 42), and has also been proposed to be responsible for Cry1F resistance in the diamondback moth (1, 8). However, this seems not to be the case for Cry1C resistance. In S. exigua, insects highly resistant to Cry1C (100-fold to >500-fold resistant) showed no change in Cry1C binding site concentration and a fivefold decrease in Cry1C binding affinity (18). Insects from a diamondback moth population 300-fold resistant to B. thuringiensis subsp. aizawai (which contains Cry1C, besides other toxins) did not show any change in Kd or Rt for Cry1C binding sites (42). No significant difference in Cry1C binding was found between two diamondback moth strains from Hawaii, one susceptible and another selected for Cry1C resistance in the laboratory (resistance ratios at the time of the experiments were 48-fold resistance to Cry1C protoxin and 19-fold resistance to Cry1C toxin) (14). We have found significant differences between our resistant strain and susceptible strain in both Kd and Rt for Cry1C binding sites. Previous estimates of Cry1C binding parameters in the diamondback moth gave Kd values from 4.8 to 9.4 nM (mean value of 7.9 nM) and Rt values from 2.8 to 10.8 pmol/mg (mean value of 5.4 pmol/mg) (see Table 1 in reference 14). The Cry1C-Sel strain has an Rt close to the above interval, but the Kd is considerably higher (50.5 nM), which supports the argument for some kind of modification in the binding site. However, taking together the decrease of affinity and the increase in binding site concentration, the results suggest that reduced binding is not the major mechanism of resistance to Cry1C in the Cry1C-Sel strain. The differences in the primary sequences of the Cry1C protein from different sources of gene (B. thuringiensis subsp. aizawai or entomocidus) may affect the outcome of binding results, and more binding assays using Cry1C toxin with different sources would be helpful for a better understanding of reduced binding mechanism. More tests on other potential mechanisms, especially on altered gut protease patterns, are needed for a better understanding of the Cry1C resistance.
ACKNOWLEDGMENTS
This research was supported by USDA-NRI grant 990-2697.
We thank Y. X. Li for technical assistance, P. Smith for collecting the insects, W. J. Moar for providing Cry1C protoxin for initial selections, Mycogen for providing the M-C formulation for tests, and Ecogen Inc. for providing the bacterial strain used to prepare the Cry1C toxin for binding assays. We also thank B. E. Tabashnik and an anonymous reviewer for comments.
REFERENCES
- 1.Ballester V, Granero F, Tabashnik B E, Malvar T, Ferré J. Integrative model for binding of Bacillus thuringiensis toxins in susceptible and resistant larvae of the diamondback moth (Plutella xylostella) Appl Environ Microbiol. 1999;65:1413–1419. doi: 10.1128/aem.65.4.1413-1419.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Cao J, Tang J D, Strizhov N, Shelton A M, Earle E D. Transgenic broccoli with high level of Bacillus thuringiensis protein control diamondback moth larvae resistant to Cry1A or Cry1C. Mol Breeding. 1999;5:131–141. [Google Scholar]
- 4.Escriche B, Silva F J, Ferré J. Testing suitability of brush border membrane vesicles prepared from whole larvae from small insects for binding studies with Bacillus thuringiensis CryIA(b) crystal protein. J Invertebr Pathol. 1995;65:318–320. [Google Scholar]
- 5.Ferré J, Real M D, Van Rie J, Jansens S, Peferoen M. Resistance to the Bacillus thuringiensis bioinsecticide in a field population of Plutella xylostella (L.) is due to change in the midgut receptor. Proc Natl Acad Sci USA. 1991;88:5119–5123. doi: 10.1073/pnas.88.12.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gould F, Anderson A, Jones A, Sumerford D, Heckel D G, Lopez J, Micinski S, Leonard R, Laster M. Initial frequency of alleles for resistance to Bacillus thuringiensis toxins in field populations of Heliothis virescens. Proc Natl Acad Sci USA. 1997;94:3519–3523. doi: 10.1073/pnas.94.8.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gould F. Sustainability of transgenic insecticidal cultivars: integrating pest genetics and ecology. Annu Rev Entomol. 1998;43:701–726. doi: 10.1146/annurev.ento.43.1.701. [DOI] [PubMed] [Google Scholar]
- 8.Granero F, Ballester V, Ferré J. Bacillus thuringiensis crystal proteins Cry1Ab and Cry1Fa share a high affinity binding site in Plutella xylostella (L.) Biochem Biophys Res Commun. 1996;224:779–783. doi: 10.1006/bbrc.1996.1099. [DOI] [PubMed] [Google Scholar]
- 9.Lee M K, Milne R E, Ge A Z, Dean D H. Location of a Bombyx mori receptor binding region on a Bacillus thuringiensis delta-endotoxin. J Biol Chem. 1992;267:3115–3121. [PubMed] [Google Scholar]
- 10.Lee M K, Rajamohan F, Gould F, Dean D H. Resistance to Bacillus thuringiensis CryIA δ-endotoxins in a laboratory-selected Heliothis virescens strain is related to receptor alteration. Appl Environ Microbiol. 1995;61:3836–3842. doi: 10.1128/aem.61.11.3836-3842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y-B, Tabashnik B E, Pusztai-Carey M. Field-evolved resistance to Bacillus thuringiensis toxin Cry1C in diamondback moth (Lepidoptera: Plutellidae) J Econ Entomol. 1996;89:798–804. [Google Scholar]
- 12.Liu Y-B, Tabashnik B E. Inheritance of resistance to Bacillus thuringiensis toxin Cry1C in the diamondback moth. Appl Environ Microbiol. 1997;63:2218–2223. doi: 10.1128/aem.63.6.2218-2223.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y-B, Tabashnik B E, Dennehy T J, Patin A J, Bartlett A C. Development time and resistance to B. thuringiensis crops. Nature. 1999;400:519. doi: 10.1038/22919. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y-B, Tabashnik B E, Masson L, Escriche B, Ferré J. Binding and toxicity of Bacillus thuringiensis protein Cry1C to susceptible and resistant diamondback moth (Lepidoptera: Plutellidae) J Econ Entomol. 2000;93:1–6. doi: 10.1603/0022-0493-93.1.1. [DOI] [PubMed] [Google Scholar]
- 15.Luo K, Adang M J. Removal of adsorbed toxin fragments that modify Bacillus thuringiensis Cry1C delta-endotoxin iodination and binding by sodium dodecyl sulfate treatment and renaturation. Appl Environ Microbiol. 1994;60:2905–2910. doi: 10.1128/aem.60.8.2905-2910.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mellon M, Rissler J, editors. Now or never: serious new plans to save a natural pest control. Cambridge, Mass: Union of Concerned Scientists; 1998. [Google Scholar]
- 17.Metz T D, Roush R T, Tang J D, Shelton A M, Earle E D. Transgenic broccoli expressing a Bacillus thuringiensis insecticidal crystal protein: implications for pest resistance management strategies. Mol Breeding. 1995;1:309–317. [Google Scholar]
- 18.Moar J W, Pusztai-Carey M, Faassen H V, Bosch D, Frutos R, Rang C, Luo K, Adang M J. Development of Bacillus thuringiensis CryIC resistance by Spodoptera exigua (Hubner) (Lepidoptera:Noctuidae) Appl Environ Microbiol. 1995;61:2086–2092. doi: 10.1128/aem.61.6.2086-2092.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller-Cohn J, Chaufaux J, Buisson C, Gilois N, Sanchis V, Lereclus D. Spodoptera littoralis (Lepidoptera:Noctuidae) resistance to CryIC and cross-resistance to other Bacillus thuringiensis crystal toxins. J Econ Entomol. 1996;89:791–797. [Google Scholar]
- 20.Munson P J, Rodbard D. LIGAND: a versatile computerized approach for characterization of ligand-binding systems. Anal Biochem. 1980;107:220–239. doi: 10.1016/0003-2697(80)90515-1. [DOI] [PubMed] [Google Scholar]
- 21.Preisler H K, Hoy M A, Robertson J L. Statistical analysis of modes of inheritance for pesticide resistance. J Econ Entomol. 1990;83:1649–1655. [Google Scholar]
- 22.Ramachandran S, Buntin G D, All J N, Tabashnik B E, Raymer P L, Adang M J, Pulliam D A, Stewart C N., Jr Survival, development, and oviposition of resistant diamondback moth on transgenic canola producing a Bacillus thuringiensis toxin. J Econ Entomol. 1998;91:1239–1244. [Google Scholar]
- 23.Russell R M, Robertson J L, Savin N E. POLO: a new computer program for probit analysis. Bull Entomol Soc Am. 1977;23:209–213. [Google Scholar]
- 24.SAS Institute. SAS user's guide: statistics. 5th ed. Cary, N.C: SAS Institute Inc; 1985. [Google Scholar]
- 25.Sayyed A H, Howard R, Herrero S, Ferré J, Wright D J. Genetic and biochemical approach for characterisation of resistance to Bacillus thuringiensis toxin Cry1Ac in a field population of the diamondback moth. Appl Environ Microbiol. 2000;66:1509–1516. doi: 10.1128/aem.66.4.1509-1516.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shelton A M, Cooley R J, Kroening M K, Wilsey W T, Eigenbrode S D. Comparative analysis of two rearing procedures for diamondback moth. J Entomol Sci. 1991;26:17–26. [Google Scholar]
- 27.Shelton A M, Robertson J L, Tang J D, Perez C, Eigenbrode S D, Preisler H K, Wilsey W T, Cooley R J. Resistance of diamondback moth to Bacillus thuringiensis subspecies in the field. J Econ Entomol. 1993;86:697–705. [Google Scholar]
- 28.Stone B F. A formula for determining degree of dominance in cases of monofactorial inheritance of resistance to chemicals. Bull W H O. 1968;38:325–326. [PMC free article] [PubMed] [Google Scholar]
- 29.Tabashnik B E. Determining the mode of inheritance of pesticide resistance with backcross experiments. J Econ Entomol. 1991;84:703–712. doi: 10.1093/jee/84.3.703. [DOI] [PubMed] [Google Scholar]
- 30.Tabashnik B E. Evolution of resistance to Bacillus thuringiensis. Annu Rev Entomol. 1994;39:47–79. doi: 10.1146/annurev.ento.54.110807.090518. [DOI] [PubMed] [Google Scholar]
- 31.Tabashnik B E, Cushing N L, Finson N, Johnson M W. Field development of resistance to Bacillus thuringiensis in diamondback moth (Lepidoptera: Plutellidae) J Econ Entomol. 1990;83:1671–1676. [Google Scholar]
- 32.Tabashnik B E, Liu Y-B, Malvar T, Heckel D G, Masson L, Ballester V, Granero F, Ménsua J L, Ferré F. Global variation in the genetic and biochemical basis of diamondback moth resistance to Bacillus thuringiensis. Proc Natl Acad Sci USA. 1997;94:12780–12785. doi: 10.1073/pnas.94.24.12780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tabashnik B E, Liu Y B, Malvar T, Heckel D G, Masson L, Ferré J. Insect resistance to Bacillus thuringiensis: uniform or diverse? Phil Trans R Soc Lond B. 1998;353:1751–1756. [Google Scholar]
- 34.Talekar N S, Griggs T D, editors. Diamondback moth management: proceedings of the 1st international workshop. Shanhua, Taiwan: Asian Vegetable Research and Development Center; 1986. [Google Scholar]
- 35.Talekar N S, Shelton A M. Biology, ecology, and management of the diamondback moth. Annu Rev Entomol. 1993;38:275–301. [Google Scholar]
- 36.Tang J D, Shelton A M, Van Rie J, de Roeck S, Moar W J, Roush R T, Peferoen M. Toxicity of Bacillus thuringiensis spore and crystal protein to resistant diamondback moth (Plutella xylostella) Appl Environ Microbiol. 1996;62:564–569. doi: 10.1128/aem.62.2.564-569.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang J D, Gilboa S, Roush R T, Shelton A M. Inheritance, stability and lack of fitness costs of field-selected resistance to Bacillus thuringiensis in diamondback moth from Florida. J Econ Entomol. 1997;90:732–741. [Google Scholar]
- 38.Tang J D, Collins H L, Roush R T, Metz T D, Earle E D, Shelton A M. Survival, weight gain, and oviposition of resistant and susceptible Plutella xylostella (L.) on broccoli expressing Cry1Ac toxin of Bacillus thuringiensis. J Econ Entomol. 1999;92:47–55. [Google Scholar]
- 39.Van Rie J, Jansens S, Höfte H, Degheele D, Van Mellaert H. Receptors on the brush border membrane of the insect midgut as determinants of the specificity of Bacillus thuringiensis delta-endotoxins. Appl Environ Microbiol. 1990;56:1378–1385. doi: 10.1128/aem.56.5.1378-1385.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Rie J, McGaughey W H, Johnson D E, Barnett B D, Van Mellaert H. Mechanism of insect resistance to the microbial insecticide Bacillus thuringiensis. Science. 1990;247:72–74. doi: 10.1126/science.2294593. [DOI] [PubMed] [Google Scholar]
- 41.Wolfersberger M G, Luthy P, Maurer A, Parenti P, Sacchi V F, Giordana B, Hanozet G M. Preparation and partial characterization of amino acid transporting brush border membrane vesicles from the larval midgut of the cabbage butterfly (Pieris brassicae) Comp Biochem Physiol Ser A. 1987;86:301–308. [Google Scholar]
- 42.Wright D J, Iqbal M, Granero F, Ferré J. A change in a single midgut receptor in the diamondback moth (Plutella xylostella) is only in part responsible for the field resistance to Bacillus thuringiensis subsp. kurstaki and B. thuringiensis subsp. aizawai. Appl Environ Microbiol. 1997;63:1814–1819. doi: 10.1128/aem.63.5.1814-1819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]