Abstract
Pelagic marine viruses have been shown to cause significant mortality of heterotrophic bacteria, cyanobacteria, and phytoplankton. It was previously demonstrated, in nearshore California waters, that viruses contributed to up to 50% of bacterial mortality, comparable to protists. However, in less productive waters, rates of virus production and removal and estimates of virus-mediated bacterial mortality have been difficult to determine. We have measured rates of virus production and removal, in nearshore and offshore California waters, by using fluorescently labeled viruses (FLV) as tracers. Our approach is mathematically similar to the isotope dilution technique, employed in the past to simultaneously measure the release and uptake of ammonia and amino acids. The results indicated overall virus removal rates in the dark ranging from 1.8 to 6.2% h−1 and production rates in the dark ranging from 1.9 to 6.1% h−1, corresponding to turnover times of virus populations of 1 to 2 days, even in oligotrophic offshore waters. Virus removal rates determined by the FLV tracer method were compared to rates of virus degradation, determined at the same locations by radiolabeling methods, and were similar even though the current FLV method is suitable for only dark incubations. Our results support previous findings that virus impacts on bacterial populations may be more important in some environments and less so in others. This new method can be used to determine rates of virus degradation, production, and turnover in eutrophic, mesotrophic, and oligotrophic waters and will provide important inputs for future investigations of microbial food webs.
Over a decade ago, pelagic marine viruses were first reported to exist in high numbers in the marine environment, exceeding the typical abundance of bacteria (2, 29). More recently, they have been shown to cause significant mortality of heterotrophic bacteria, cyanobacteria, and phytoplankton (10, 34, 36, 46). Specifically, it has been shown by a variety of researchers that viruses are capable of causing up to 50% of the bacterial mortality in a range of aquatic environments (12, 14, 31, 43). The variability of the impact that viruses have on bacterial assemblages can be high, even over short periods in the same study area (4, 31). With their influence upon bacterial populations, viruses appear to have the potential to affect the flow of energy and matter in marine ecosystems. By infection of bacterial cells and subsequent cell lysis, viral infection leads to a “short circuit” in the microbial loop where recycling fuels bacterial production and respiration and reduces the amount of organic matter available to macroorganisms (8, 39). Previous estimates of virus production and decay rates have provided the confirmation that viruses are active members of the marine community (16, 32). Measurements of virus replication rates are also useful for assessing the contribution of viruses to bacterial mortality and organic matter cycling in the ocean. Accurate measurements of virus productivity and turnover will permit researchers to properly model their dynamics and impact within the microbial food web.
Recent studies have demonstrated the use of epifluorescence microscopy with fluorescent stains like DAPI (4′,6′-diamidino-2-phenylindole), SYBR Green I, and Yo-Pro I for enumeration of bacteria and viruses (17, 26, 41, 45). However, information on rates of virus production and removal has been historically limited by methodological constraints. Fuhrman and Noble (12) used a combination of approaches and demonstrated that marine viruses contributed to up to half of the bacterial mortality in the coastal waters of Santa Monica Bay, California, showing the comparable contributions of viruses and protists to bacterial mortality with enclosure experiments. Later, when similar experiments were done on board ship in more oligotrophic offshore waters, we encountered problems in attempting to determine rates of virus production by the method of Steward et al. (33), which involves measuring the amount of [3H]thymidine that is incorporated into the virus-size fraction. When this method was used to measure virus production in offshore waters, the amount of radiolabel incorporated into the virus-size fraction was never greater than the background signal. Experiments published by Steward et al. (32) also demonstrated rates of virus production that were not significantly different from zero in oligotrophic offshore waters. However, Proctor and Fuhrman (29) demonstrated that the percentage of bacteria infected in offshore waters is not necessarily lower than that in nearshore waters, indicating the possibility that the radiolabel incorporation method was not sensitive enough to detect low levels of virus production.
Here, we present a new method, using fluorescently labeled viruses (FLV) as tracers for determining the rates of production and removal. Throughout the rest of this report, it will be referred to as the FLV tracer method. Rates of virus production and concomitant removal were determined using calculations previously used for the isotope dilution technique to measure rates of release and uptake of amino acids or dissolved ammonium by using radioisotopes or stable isotopes, respectively, as tracers (3, 9, 13). Basically, the FLV are analogous to labeled molecules used as tracers in these earlier studies. When FLV are added into the seawater at tracer levels (<10% of the ambient virus concentration), removal processes decrease the number of FLV and unstained viruses in relative proportion to the total virus abundance. However, virus production produces only unlabeled viruses, thereby diluting the initial pool of FLV. Using the rate of change of both FLV (tracer viruses) and the total virus concentration over time, we calculated rates of virus production and removal. The results gathered using this method indicate that it is suitable for measuring virus production and removal, especially in more oligotrophic offshore waters where other methods have proven unsuccessful (15, 32). It is unique in that it permits simultaneous determination of rates of virus production and removal using epifluorescence microscopy. Utilization of this method should improve our understanding of the impacts of viruses on the structure of the microbial food web and our ability to form conceptual and numerical models of how viruses affect the microbial loop and the turnover of dissolved organic matter, particularly in offshore, oligotrophic environments.
MATERIALS AND METHODS
Total counts of bacteria and viruses using epifluorescence microscopy.
SYBR Green I has a proprietary formula, and its manufacturer (Molecular Probes, Inc., Eugene, Oreg.) does not report its molecular weight. The dye is supplied as a 10,000× concentrate. The SYBR Green I used for this study (lot no. 3142-1), when diluted 1,000-fold in sterile water, had an optical density at 494 nm of 0.42. The method of Noble and Fuhrman (26) was used to determine the total virus abundance in the seawater samples. Briefly, for total virus counts, samples were filtered through a 0.02-μm-pore-size Anodisc filter and stained for 15 min with SYBR Green I. From each filter, 10 to 20 fields were selected randomly, and a total of >200 viruses and >200 bacteria were counted on an Olympus Vanox epifluorescence microscope with a 100× D Plan Apochromat UV objective, under blue excitation. Virus particles were distinctly shaped pinpricks and fluoresced bright green. Bacterial cells could easily be distinguished from viruses because of their relative size and brightness. FLV concentrations were determined by filtering 10 ml of seawater onto a 0.02-μm-pore-size Al2O3 Anodisc 25 membrane filter (Whatman), backed by a 0.8-μm-pore-size cellulose mixed-ester membrane (Millipore type AA) at approximately 20 kPa in a vacuum. Whole fields were counted, and >200 FLV were counted per filter.
Concentration of viruses and preparation of fluorescently labeled tracer viruses.
To prepare each FLV concentrate, 20 liters of fresh seawater was collected either from four 5-liter Niskin bottles or by triple acid- and sample-rinsed bucket into an acid-rinsed 20-liter low-density polyethylene carboy. All virus concentration steps were performed either on ice or in a centrifuge held at <10°C so as to minimize degradation of virus particles during the concentration steps. The sample was filtered at 5 kPa through a 142-mm-diameter, 0.22-μm-pore-size Durapore filter to remove bacteria and protists. The virus-sized fraction (material between 0.22 μm and 30 kDa) was concentrated to ca. 150 ml using a spiral cartridge concentration system (37) (Amicon, Inc.). This was further concentrated, using Centriprep-30 centrifugal concentration units (Amicon, Inc.), to a final volume of ca. 5 ml. To each of these virus concentrates, the nucleic acid stain SYBR Green I (Molecular Probes, Inc.) was added at a final concentration of 2.5% (vol/vol) and was incubated in the dark for at least 8 h at 4°C. After the staining period, the unbound SYBR Green I was rinsed away by adding an equal volume of 0.02-μm-pore-size-filtered seawater (prepared by filtering fresh seawater from the same location through an acid-rinsed, autoclaved Nalgene filtration unit housing a 47-mm, 0.02-μm-pore-size Anodisc filter) to the concentrate and centrifuging the concentrate in Centriprep-30 ultraconcentration units at 3,000 × g for 15 min. This rinse was done three times. Each time, we reused the same Centriprep-30 concentration unit and resuspended the FLV in a total of 5 ml of 0.02-μm-pore-size-filtered seawater. The final FLV concentrates were resuspended in a total of 5 ml of 0.02-μm-pore-size-filtered seawater. To determine the concentration of FLV, 10 μl of concentrate was diluted to a final volume of 2 ml with 0.02-μm-pore-size-filtered seawater, filtered through a 0.02-μm-pore-size Anodisc filter, and counted by epifluorescence microscopy under blue excitation. At the same time, we determined the total virus abundance in the seawater to be used for each experiment, so as to permit calculation of the proper amount of FLV concentrate to be added that would represent a tracer level (<10% of original total virus concentration).
Bacterial production measurements.
Thymidine and leucine incorporation methods were modified from the work of Fuhrman and Azam (11), Kirchman et al. (22), and Simon and Azam (30). Both thymidine and leucine incorporation methods were used to determine bacterial production rates in experiments performed on board ship. Only thymidine incorporation methods were used to determine rates of bacterial production in experiments performed in the laboratory. At t0 of each experiment, duplicate 42-ml seawater samples and 1% formalin-killed controls were subsampled into well-rinsed sterile 50-ml polypropylene tubes (VWR brand). Samples were inoculated with 5 nM [methyl-3H]thymidine or [3H-3,4,5]leucine (both obtained from Dupont New England Nuclear). Subsamples were incubated in either an on-deck incubator with running seawater under ambient sunlight or a fluorescently lighted (during the daytime; dark at night) incubator at ambient seawater temperature. After 30-min incubations, duplicate 20-ml samples from each tube were filtered through HAWP Millipore filters (mixed cellulose acetate and cellulose nitrate, 0.45-μm nominal pore size, 25-mm diameter) at the base of cold stainless steel filtration funnels on a 10-place manifold (Hoefer Scientific). Filtration valves were closed, and 2 ml of ice-cold 5% trichloroacetic acid (TCA) was added. After 2 min, the TCA was filtered through and the filters and funnels were rinsed three times with 1 ml of cold 5% TCA. The funnels were removed, and the edges of the filters were rinsed three times with 1 ml of 5% TCA. Filters were placed in a glass 20-ml vial and 1 ml of 1 N HCl was added, followed by heating to 90 to 100°C for 1 h (to hydrolyze the nucleic acids and proteins). After the vials cooled, 5 ml of Ecoscint (National Diagnostics) was added and the samples were counted by liquid scintillation with disintegrations-per-minute correction (Packard). Conversion factors used to calculate production from the moles of thymidine or leucine incorporated were the averages reported by Fuhrman and Azam (11) at 2 × 1018 cells mol of thymidine incorporated−1 and by Chin-Leo and Kirchman (6) and Kirchman (20) at 1.5 × 1017 cells produced per mol of leucine incorporated−1. Bacteria were counted with SYBR Green I, and counts were used to calculate the turnover time (days) of the bacterial population by dividing the average number of bacterial cells by the production rate (cells per liter per day) (26). Because the thymidine incorporation method was used for every experiment, and since the thymidine and leucine incorporation methods yielded similar bacterial growth rate results, only bacterial growth rates determined by thymidine incorporation were used to estimate the percent bacterial mortality.
Calculation of virus production and removal rates.
Production and removal rates were calculated from the equations of Glibert (13) and Fuhrman (9). The decay constant, k, is calculated as k = [ln(R0/Rt)/t], where t is the incubation time and R0 and Rt are the ratios of FLV to the total viral abundance at time zero and time t, respectively. The first two time points for each experiment were t0 and t1. For example, R0 is equal to FLV0, divided by the total concentration of virus particles, C0, at time zero. The mean specific activity,  , is then calculated as
, is then calculated as
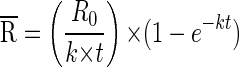 |
The viral decay or removal rate, Dv, is calculated as
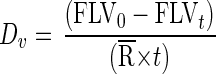 |
where FLV0 and FLVt are the concentrations of FLV at t0 and time t, respectively. The viral production rate, Pv, is calculated as
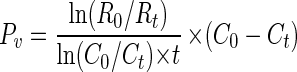 |
where C0 and Ct are the total concentrations of virus particles at t0 and time t, respectively. If the virus abundance does not change over time, then the removal rate is equal to the production rate (and the equation is not used). For each experiment, initial rates (using the first two time points, t0 and t1), and overall rates (using the entire time course) of production and decay were calculated. Initial rates of decay and/or production are closest to in situ rates, as all of the experiments were started at dusk and held under ambient natural conditions. Overall rates represent decay and/or production under natural conditions for approximately 12 h, but samples were held in the dark the following morning and not exposed to natural sunlight. Calculations for total virus production, virus turnover rates, and estimated bacterial mortality used overall virus production rates because initial rate calculations were based upon only two time points.
Estimates of virus-induced bacterial mortality were calculated using overall rates of virus production, mean virus abundance, mean bacterial abundance and growth rates, and burst size. In brief, virus production rates were divided by the estimated burst size (we used a range from 20 to 50), to determine the bacterial cells killed per liter per day. We divided the bacterial cells killed per liter per day by the bacterial growth rate in cells per liter per day to determine the portion of the bacterial community killed due to viral lysis. This assumes steady state.
Tracer experiments.
Seawater samples were taken from mesotrophic and oligotrophic marine environments at four locations; Two Harbors, Santa Catalina Island (meso-oligotrophic, latitude 33°27′, longitude 118°30′); mid-San Pedro Channel (meso-oligotrophic, ca. 19 km off the coast of San Pedro, Calif., 33°34′N, 118°24′W); Offshore Station (oligotrophic, near San Juan Seamount, 190 km offshore, 32°51′N, 120°42′W); and Playa del Rey Jetty in Playa del Rey, Calif. (mesotrophic, 34°03′N, 118°29′W).
The first four experiments were performed on board the R/V Point Sur during the week of 20 May to 27 May 1997. The other four experiments were performed in the laboratory after retrieval of the water samples. FLV concentrates were freshly prepared at each new site and for each new experiment. After determination of the concentration of the FLV in the concentrate and the ambient concentration of viruses in the seawater to be used for the experiment, the proper amount of FLV concentrate was added at tracer levels (<10% of original ambient virus concentration) into 400-ml seawater samples treated in various ways (see Table 1). In six of the eight experiments, a formalin-treated (FT) killed control was used, where 0.02-μm-pore-size-filtered formalin was added at a final concentration of 2%. In the experiments at mid-San Pedro Channel (26 May 1997) and at the Offshore Station (San Juan Seamount), we used a heat-treated (HT) killed control. The seawater was boiled for 10 min and then cooled to ambient seawater temperature. The heat treatment denatures active proteins and enzymes and kills most vegetative bacteria (19). Any measurable rate of disappearance of FLV in FT or HT controls was subtracted from that seen in the untreated bottles. Because SYBR Green I stain fades quickly in sunlight, the samples were incubated at ambient seawater temperatures in the dark. However, experiments were started at dusk, and so for approximately the first 12 h, the experiments were done under simulated in situ conditions. At each time point, total virus and FLV tracer virus numbers were determined from duplicate 45-ml subsamples taken into sterile, 50-ml polyethylene tubes. Subsamples were immediately fixed with 2% (final concentration) 0.02-μm-pore-size-filtered formalin. Slides were made immediately, and the remainder of the subsample was stored at 4°C. Dates, depths at which samples were taken, and information on ambient environmental parameters for each of the experiments are outlined in Table 1.
TABLE 1.
Experimental parameters
| Location | Date | Water depth (m) | Depth (m) | Temp (°C) | Treatment | Control treatment | Viral abundance (1010 liter−1) at t0 | Bacterial abundance (109 liter−1) at t0 |
|---|---|---|---|---|---|---|---|---|
| Two Harbors, Santa Catalina Island | 21 May 1997 | 60 | 5 | 17.8 | Untreated | FT | 1.37 | 0.85 |
| Two Harbors, Santa Catalina Island | 21 May 1997 | 60 | 5 | 17.8 | 1.0-μm-pore-size filtered | FT | 1.60 | 0.57 |
| San Pedro Channel | 26 May 1997 | 895 | 5 | 17.9 | Untreated | HT | 0.75 | 0.69 |
| San Pedro Channel | 13 July 1998 | 895 | 5 | 18.6 | Untreated | FT | 0.84 | 1.14 |
| San Pedro Channel | 13 July 1998 | 895 | 20 | 17.1 | Untreated | FT | 0.69 | 0.90 |
| San Pedro Channel | 13 July 1998 | 895 | 60 | 16.5 | Untreated | FT | 0.66 | 0.37 |
| Offshore Station | 26 May 1997 | 3,600 | 5 | 18.0 | Untreated | HT | 1.8 | 1.10 |
| Playa del Rey Jetty | 13 July 1998 | 3 | Surface | 19.8 | Untreated | FT | 4.14 | 4.98 |
Test of SYBR Green I staining and virus decay.
An experiment was done to determine whether viruses stained with SYBR Green I degrade (or lose their ability to be stained) at different rates than those of naturally found, unstained viruses. A 1-liter seawater sample was taken from surface water (5 m) of San Pedro Channel, and a virus concentrate was prepared as described previously. Half of the concentrate was stained, and half was not. Of two replicate seawater samples, stained viruses were added to one, and unstained viruses were added to the other. The incubations (replicate) were 0.02-μm-pore-size-filtered seawater and 0.02-μm-pore-size-filtered seawater with 0.01 nM pronase K and DNase (Sigma Chemical, Inc.). SYBR Green I-stained viruses were counted by filtering 1 ml of seawater through a 0.02-μm-pore-size Anodisc filter and mounting the filter with a coverslip and antifade mounting solution (see below). Counts of unstained viruses were made as described previously for the SYBR I staining method (26). Viruses were counted over a 24-h period.
RESULTS
An experiment was done to determine if stained virus particles degrade (or lose their ability to be stained) faster or slower than unstained viruses due to effects of the SYBR Green I stain. We examined degradation processes by determining the rates of removal of stained and unstained viruses in untreated seawater and in seawater treated with active proteases and nucleases. The results demonstrated that unstained and SYBR Green I-stained viruses in untreated seawater did not degrade at significantly different rates, (1.23 ± 0.3)% and (1.22 ± 0.2)% h−1, respectively (t test, P > 0.01). Also, the SYBR Green I-stained and unstained viruses in the enzyme-treated seawater did not degrade at significantly different rates (t test, P > 0.01); the unstained viruses degraded at a rate of 3.5 ± 0.6% h−1, and the stained viruses degraded at a rate of 3.2 ± 0.4% h−1.
FLV were used as tracers of virus production and removal using calculations based upon the isotope dilution technique. The basis for these calculations is that removal processes include losses of both tracer viruses and unstained virus particles in proportion to their abundance but production yields only unstained virus particles. FLV experiments revealed overall rates of virus production ranging from 1.9 to 6.1% h−1, with the highest rate at San Pedro Channel, 20-m depth, and the lowest rates at a 60-m depth (San Pedro Channel) and at the Offshore Station (Table 2). Virus production ranged from 2.8 × 109 to 2.8 × 1010 virus liter−1 day−1. Estimated turnover times of the virus populations were based upon virus production rates and ranged from 0.68 to 2.2 days (Table 3). Bacterial production rates as determined by both thymidine and leucine incorporation methods were very similar to one another and ranged from 2.36 × 108 to 6.26 × 108 cells liter−1 day−1, respectively. Bacterial production rates corresponded to estimated turnover times of the bacterial population ranging from 1.0 to 4.6 days (Table 3). Using an assumed range of burst sizes, from 20 to 50, viruses were estimated to be responsible for from 59 to 125% and 24 to 50% of the total bacterial mortality, respectively (Table 3). Using values from the work of Lee and Fuhrman for bacterial carbon per cell (20.0 fg per cell), our estimated bacterial mortality values equated a release of 1.12 to 12.4 μg of C liter−1 day−1 (23).
TABLE 2.
Mean viral and bacterial abundances and rates of virus and bacterial removal and productiona
| Location | Mean viral abundance (1010/liter) | Mean bacterial abundance (109/liter) | Virus removal (% h−1)
|
Virus production (% h−1)
|
Virus production (1010 virus liter−1 day−1) | Bacterial production (108 cells liter−1 day−1)
|
|||
|---|---|---|---|---|---|---|---|---|---|
| Initial | Overall | Initial | Overall | TdR | Leu | ||||
| Two Harbors | 1.20 | 1.07 | 6.5 | 6.2 | 1.0 | 3.2 | 0.92 | 6.26 | 4.15 |
| Two Harbors (1.0-μm-pore-size filtered) | 1.29 | 0.86 | 3.7 | 3.6 | * | 3.0 | 0.93 | 3.73 | 3.24 |
| San Pedro Channel | 1.86 | 1.02 | 3.9 | 3.2 | 0.6 | 3.2 | 1.43 | 6.24 | 6.57 |
| San Pedro Channel (13 July 1998) | 0.97 | 0.83 | 5.6 | 2.2 | 5.6 | 5.1 | 1.19 | 4.98 | ND |
| San Pedro Channel (13 July 1998, 20 m) | 0.79 | 0.84 | 16.4 | 4.9 | 19.5 | 6.1 | 1.15 | 6.21 | ND |
| San Pedro Channel (13 July 1998, 60 m) | 0.59 | 0.58 | 1.7 | 3.1 | 0.0 | 2.0 | 0.28 | 1.89 | ND |
| Offshore Station | 0.81 | 0.84 | 2.8 | 1.8 | 0.0 | 1.9 | 0.37 | 2.80 | 2.40 |
| Playa del Rey Jetty (surface) | 2.99 | 3.10 | 6.0 | 4.5 | 3.4 | 3.9 | 2.80 | 2.36 | ND |
All experiments were performed with seawater from a 5-m depth, unless otherwise noted. Seawater was unfiltered unless otherwise specified. TdR, thymidine incorporation; Leu, leucine incorporation; ND, not determined. *, the rate of FLV tracer disappearance was higher than the rate of disappearance of unstained virus particles.
TABLE 3.
Estimated virus turnover time, bacterial turnover time, and virus-induced bacterial mortalitya
| Location | Virus turnover time (days) | Bacterial turnover time (days)
|
Virus-induced bacterial mortality (burst size of 20, % of total mortalityb) | Virus-induced bacterial mortality (burst size of 50, % of total mortalityb) | |
|---|---|---|---|---|---|
| TdR | Leu | ||||
| Two Harbors | 1.3 | 1.35 | 2.04 | 74 | 29 |
| Two Harbors (1.0-μm-pore-size-filtered seawater) | 1.4 | 1.58 | 1.80 | 125 | 50 |
| San Pedro Channel (22 May 1997) | 1.3 | 1.04 | 1.02 | 114 | 46 |
| San Pedro Channel (13 July 1998) | 0.82 | 2.28 | ND | 119 | 48 |
| San Pedro Channel (13 July 1998, 20 m) | 0.68 | 1.45 | ND | 93 | 37 |
| San Pedro Channel (13 July 1998, 60 m) | 2.1 | 1.95 | ND | 74 | 30 |
| Offshore Station | 2.2 | 3.95 | 4.56 | 66 | 26 |
| Playa del Rey Jetty (surface) | 1.1 | 2.1 | ND | 59 | 24 |
TdR, thymidine incorporation; Leu, leucine incorporation; ND, not determined.
Assuming steady state, i.e., the total bacterial mortality equals the bacterial production.
Initial rates of virus removal and production, calculated from the first two time points in each experiment, ranged from 1.7 to 16.4% and from 0.0 to 19.5% h−1, respectively (Table 2). Initial rates were generally very similar to the overall rates and represent in situ rates, as incubations were under natural conditions. One exception was at San Pedro Channel at a 20-m depth, where the initial rates of virus removal and production were 16.1 and 19.5% h−1, indicating turnover times of the virus population of 5 h (Table 2). However, the entire experiment was performed in as close to in situ conditions as possible, because the light levels at 20 m (and at 60 m) would have been significantly attenuated during the early morning hours. In two cases, the initial virus production rate was not determinable, as the drop in total virus abundance occurred faster than the drop in FLV abundance. In one of those cases, the drop in the total virus abundance and the drop in the FLV counts were not significantly different from zero.
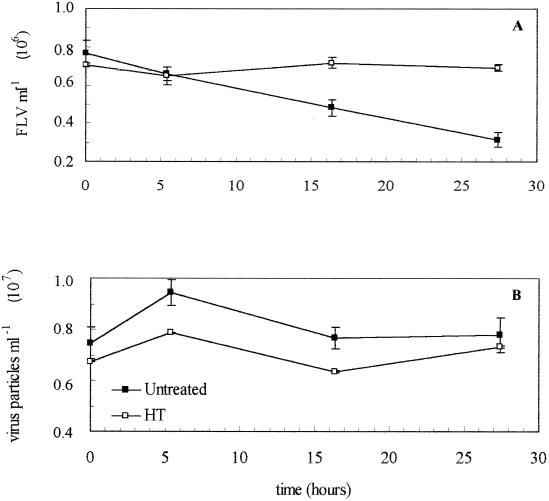
At the Offshore Station, rates of virus production and removal were lower than at any other location, indicating viral turnover times of slightly more than 2 days. FLV counts decreased at a rate of ca. 2% h−1 in the untreated sample but were invariant in the HT sample (Fig. 1A). Total virus abundance increased within the first 5 h in the untreated sample and returned to original levels by 16 h, while the virus counts in the HT control changed only slightly over the course of the experiment (Fig. 1B). At this location, the estimated turnover times of the bacterial population as determined by thymidine and leucine incorporation were 3.95 and 4.56 days, respectively, and were also longer than those seen in nearshore waters (Table 3). Rates of virus production ranged from about 6 × 109 to 9 × 109 virus liter−1 day−1 (Table 2) and are similar to those reported by Steward et al. for waters in the Southern California Bight (SCB) (32). Viruses appeared to be responsible for a smaller fraction of bacterial mortality, between 26 and 66% (Table 3).
FIG. 1.
(A) FLV abundances in seawater from the Offshore Station that was either untreated (■) or HT (□). (B) Total viral abundance in untreated (■) or HT (□) seawater from the Offshore Station.
One of our experiments examined the differences in virus production and removal rates with and without the influence of protist grazing upon the heterotrophic bacterial population. In this experiment (at Two Harbors, Santa Catalina Island), 1.0-μm-pore-size filtration was used to remove the protists. Virus production and removal rates were 3.2 and 6.2% h−1 in the unfiltered sample and 3.0 and 3.6% h−1 in the 1.0-μm-pore-size-filtered sample, respectively (Table 2). In the unfiltered sample, viruses were estimated to be responsible for 29 to 74% of the bacterial mortality, whereas in the 1.0-μm-pore-size-filtered sample viruses were estimated to be responsible for 50 to 125% of the loss of the bacterial population (Table 3). Total viral counts decreased by ca. 3% h−1 in the unfiltered, untreated sample but were nearly invariant in the FT sample (data not shown). Bacterial counts in the untreated, FT, and 1.0-μm-pore-size-filtered and FT samples were invariant, but counts increased by a factor of 2 over the 9-h period in the 1.0-μm-pore-size-filtered seawater, probably from the removal of protist grazers (data not shown).
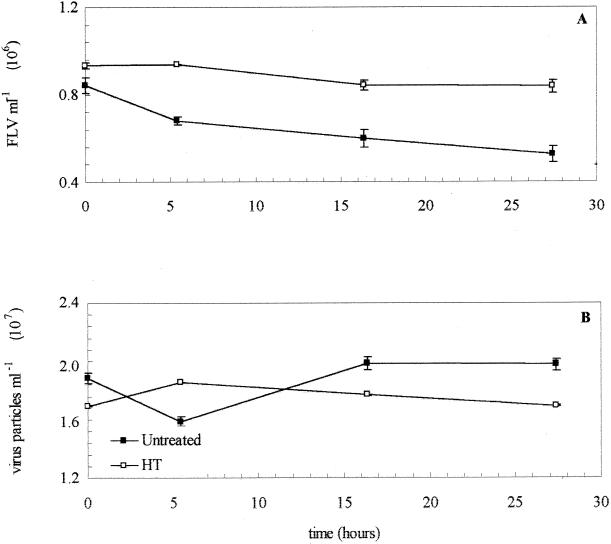
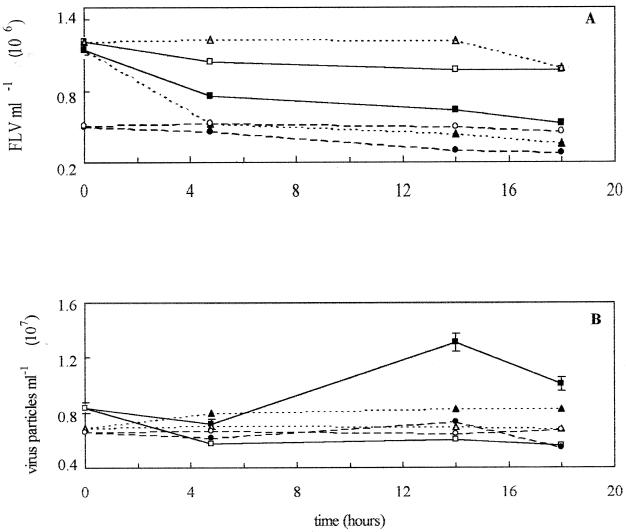
In the mid-San Pedro Channel experiment on 21 May 1997, the rate of virus production of 3.2% h−1 was matched by the rate of virus removal at 3.2% h−1 (Fig. 2; Table 2). There was a slight decrease in the FLV numbers in the HT seawater (Fig. 2A). The total viral abundance decreased dramatically within the first 5 h but then rebounded to the original levels by 15 h. The estimated turnover time of the viral population was 1.3 days (Table 3). Viruses were determined to be responsible for 46 to 114% of the bacterial mortality (Table 3). In the experiment on 13 July 1998 at the same location, samples were taken at three different depths. FLV counts decreased by different rates, with the 20-m sample demonstrating the fastest decrease, and the 60-m depth demonstrating the slowest (Fig. 3A). For example, the FLV count dropped dramatically within the first 4 h in the 20-m-depth seawater sample (resulting in the high initial rate [Fig. 3A; Table 2]). Initial total virus counts were similar at all three depths at time zero (Fig. 3B). The FT samples demonstrated only minimal change over time in FLV or total virus counts at any depth (Fig. 3). The rates of production and removal were higher at the 20-m depth (at 6.1 and 4.9% h−1, respectively) than in surface waters and at the 60-m depth (Table 2; Fig. 3). The estimated virus-mediated bacterial mortality in surface, 20-m-depth, and 60-m-depth seawater ranged from 48 to 119, 37 to 93, and 30 to 74%, respectively (Table 3).
FIG. 2.
(A) FLV abundance from mid-San Pedro Channel seawater that was either untreated (■) or HT (□). (B) Total viral abundance in seawater from mid-San Pedro Channel that was either untreated (■) or HT (□).
FIG. 3.
(A) FLV abundance from mid-San Pedro Channel seawater at 5 m, untreated (▴) or FT (▵); 20 m, untreated (■) or FT (□); and 60 m, untreated (●) or FT (○). (B) Total viral abundance in seawater from 5, 20, and 60 m in mid-San Pedro Channel that was either untreated or FT; the symbols are the same as those in panel A.
DISCUSSION
Using FLV as tracers in an approach mathematically similar to the isotope dilution technique, we have demonstrated virus production rates of 109 to 1010 virus liter−1 day−1 and indicated that the turnover of virus populations in the nearshore and offshore waters of the SCB occurs in ca. 1 to 2 days. These rates of virus production are within the range reported using the method of [3H]thymidine incorporation in a <0.2-μm-size fraction for waters of the SCB (32).
Both initial and overall rates of virus production and removal were determined for each of the experiments. Since all of the experiments were started at dusk, the initial rates are in situ production and removal rates, whereas the overall rates calculated over the entire course of the experiment are not in situ rates because samples were incubated in the dark at the end of the experiments during natural daylight hours. In general, the initial rates of removal and production were very similar to the overall rates. However, in a few cases, initial rates of removal and production were different (by up to a factor of 3) and are reported in Table 2 so as to show in situ rates.
Previous experiments have demonstrated that there is a wide range of rates of virus removal, depending upon the type of marine virus and the environmental conditions (for example, see references 27, 38, and 47). Most notably, UV light, heat-labile dissolved organic matter, and levels of particulate matter and enzymes appear to influence viral removal (see the review by Wommack and Colwell [48]). In our FLV tracer experiments, we have used concentrates of natural assemblages of native marine viruses, so as to calculate a decay constant, k, that is representative of a range of virus types in a given sample. During the process of preparing and staining the virus concentrate, degradation of the most unstable viruses probably occurs. We have attempted to minimize the effects of this degradation by limiting the amount of time to concentrate the sample and by performing the concentration steps on ice or at temperatures less than 10°C. The limited degradation that does occur during the preparation steps probably results in an overall underestimate of average rates of virus decay. However, we feel that the use of natural assemblages of viruses provides a snapshot of representative rates of virus removal in seawater. We have demonstrated that the process of staining with SYBR Green I does not appear to cause differences in the rates of degradation of virus particles. However, the SYBR Green I method (26) could have masked the inability of certain viruses to retain their stain or to be stained by SYBR Green I.
Overall rates of virus removal were about half as high as rates of loss of virus infectivity seen in bacteriophage isolated from Santa Monica Bay waters in other experiments (27). This is not surprising, as the ability to initiate infection can be hindered by a variety of mechanisms, many of them involving minuscule changes to the exterior of the virus (e.g., tail fiber damage and disruption of glycoprotein conformation). Rates of virus removal as determined by the FLV tracer method were generally higher, but within a factor of two of the rates of virus degradation determined by radiolabeling methods in the same or nearby geographic locations (25). For example, experiments run at Two Harbors in 1995 revealed rates of degradation of 2.9 and 3.4% h−1, where the rate of virus removal was 6.2% h−1. At the Offshore Station, the rates of degradation and removal were 1.0 and 1.8% h−1, respectively (25) (Table 2). Rates of virus removal as determined by the FLV tracer method were also generally within a factor of two of those reported previously as determined by viral direct counts (mostly by transmission electron microscopy [TEM]) for both specific marine bacteriophage types (49) and whole marine virioplankton (47) (see review by Wommack and Colwell [48]). For example, using TEM counts, Wommack et al. (49) demonstrated removal of two specific bacteriophage types, CB38Φ and CB7Φ, at rates of 2.8% h−1 in estuarine waters incubated in the dark, very similar to the rates reported here. Rates of loss of infectivity reported by Suttle and Chen (38) for experiments performed in the dark were also similar to rates of virus removal in this report. However, other experiments, by Suttle and Chen (38), Suttle et al. (35), Noble and Fuhrman (27), and Wilhelm et al. (47), have demonstrated highly variable rates of loss of infectivity in sunlight. Suttle et al. (35) also demonstrated a stark contrast between rates of loss of virus infectivity in sunlight-incubated samples and rates of virus particle removal (even in waters subjected to only a few percent surface irradiance). Even though virus particle removal appears to be affected by sunlight to a lesser extent than is loss of virus infectivity, rates of removal determined by FLV are likely to be underestimates due to the lack of exposure to sunlight or indirectly due to processes dependent upon sunlight.
Rates of virus removal exceed rates of virus production in half of the experiments reported (Table 2). Because the experiments are performed during dark hours, this may indicate that virus removal rates are lower during the day and higher at night. One possible explanation for this is that, during dark hours, bacteria are likely to be more metabolically active. Aas et al. (1) and Herndl et al. (18) have demonstrated reduced bacterial activity during exposure to sunlight, indicating the likelihood of heightened metabolic activity at night. Therefore, bacteria may be more likely to produce virus-degrading proteases and nucleases during the dark hours, speeding up processes of virus removal.
Burst size is an important component for calculations of bacterial mortality. For the Northern Adriatic Sea, Weinbauer et al. (42) reported a range of burst sizes of from 6 to 140 phage per host cell, with an average of ca. 23 phage per host cell, but the burst size did not vary with season or year. They also found that the burst size was significantly higher in more active waters (eutrophic) than in mesotrophic waters, a trend that we have observed in Southern California waters. An even wider range of burst sizes, from 10 to 300, has been demonstrated in other work (14, 42). For our calculations, we have employed a range of burst sizes, 20 to 50, that reflects the burst sizes that we have observed empirically by TEM in Southern California waters (12).
Using the rate of virus production, the ambient virus concentration, and assumed values for burst size, we can estimate the influence of virus infection by calculating the percentage of the bacterial community lysed by viruses per day, or virus-mediated bacterial mortality. The virus-mediated bacterial mortality ranged from 24 to 125% in the seven experiments, indicating the importance of viruses to bacterial mortality in a variety of types of seawater. A few of these experiments were performed in the same geographic locations as those in the study published by Fuhrman and Noble (12) and are consistent with their findings of roughly half of the bacterial mortality being due to virus-mediated processes.
Our findings of significant bacterial mortality support previous findings, further supporting the importance of viruses to bacterial mortality in a variety of marine environments (12, 29, 32, 34). If viruses are responsible for such bacterial mortality, then they potentially cause the release of a significant amount of organic carbon (our calculations show 1.1 to 12.4 μg of C liter−1 day−1), along with nutrients, cofactors, and trace elements, into the surface and near-surface waters of the ocean. The productivity of heterotrophic bacterioplankton in marine environments is thought to be limited by the availability of organic carbon and, in certain environments, nitrogen and phosphorus (7, 21, 40). Even though viral lysis may be responsible for only a small fraction of the total carbon in the system, the material released could be important to nutrient regeneration. Cell components released by viral lysis are rich in organic carbon, nitrogen, and phosphorus; can be highly labile; and are usable for growth by other noninfected bacterioplankton (24, 25, 28).
Previous experiments have demonstrated that viruses and protists were each responsible for about half of the bacterial mortality in coastal waters (12). However, few experiments have directly compared the influence of each in the same samples. We ran one experiment to examine the relationship between viruses and protists which used filtration to remove protists. In this experiment, the rate of virus production increased with the removal of protists, thereby indicating increased bacterial mortality due to virus infection. Virus infection, as opposed to protist grazing, is thought to be highly specific, with viruses infecting only bacterial species or families (5). Therefore, the mechanisms that control the relationships between protist grazing and virus infection are likely to be quite different. It has been speculated previously that the role that viruses play in bacterial mortality increases when grazing pressure is reduced (14, 44). Although it appears that virus-mediated bacterial mortality is not directly related to trophic status, as was previously suggested for virus degradation and removal rates (25), it does appear that virus infection may be a less important factor at the Offshore Station, where our estimates of bacterial mortality due to viruses ranged from 24 to 59%. In these waters, two factors may negatively influence the likelihood of successful virus infection, dramatically lower rates of encounter between viruses and bacteria and increased grazing pressure. Our experiments indicate a potential increased impact of viruses on bacteria in the absence of protists in nearshore, meso-oligotrophic waters and a possible decrease in the impact of viruses in offshore waters, but more experiments are needed to further examine the relationships between viruses and protist grazers and their control of bacterioplankton.
The FLV tracer method provides a sensitive way to determine rates of virus production, making it particularly suitable in areas of very low productivity (oligotrophic waters). Incorporation of radiolabel into the virus-sized fraction is difficult to determine effectively at low radioisotope incorporation rates as in the [3H]thymidine method outlined by Steward et al. (33). This method involves a large conversion factor to convert the moles of [3H]thymidine incorporated into the number of viruses produced (6.17 × 1020 virus produced per mol of [3H]thymidine for waters in Southern California). There are uncertainties inherent in this empirically derived conversion factor, and it is likely that a new conversion factor should be empirically derived for each new set of experiments, a difficult task (32).
The inability to use the FLV method in sunlit samples currently limits its application under true simulated in situ conditions to nighttime incubations. During daylight hours, our dark incubations could potentially lead to underestimates of virus loss (e.g., due to sun exposure) or possibly underestimates of production (due to reduced microbial food web activity). On the other hand, bacterial activity could be temporarily enhanced in the dark due to a reduction of solar damage to bacterial cells, such as in work by Aas et al. (1) which demonstrated diminished bacterial activity in seawater due to intense sunlight.
The use of FLV as tracers of viral processes offers multiple advantages to studies of virus-mediated processes. This method is valuable because it is sensitive and relatively inexpensive and can be performed on board ship and in the field without the use of radioisotopes. In addition, the method has the potential to be adapted for any aquatic system. The results presented here support previous work indicating the importance of viral processes, and the measurable turnover of virus populations, even in water with slowly growing bacterial assemblages. Continued measurements of viral and bacterial abundances, with the use of this new method to determine rates of virus production and removal in oligotrophic waters, and along depth gradients, will help to elucidate the significance of virus-mediated processes in the oceans.
ACKNOWLEDGMENTS
We thank C. C. Ouverney, A. A. Davis, J. F. Griffith, X. Hernandez, and the crew of the R/V Point Sur for assistance with sample collection.
R.T.N. was supported by NSF grants OCE-9634028 and OCE-9906989, a USC sea grant, and by an ARCS Fellowship during the course of this work.
REFERENCES
- 1.Aas P, Lyons M, Pledger R, Mitchell D L, Jeffrey W H. Inhibition of bacterial activities by solar radiation in nearshore waters and the Gulf of Mexico. Aquat Microb Ecol. 1996;11:229–238. [Google Scholar]
- 2.Bergh O, Børsheim K Y, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340:467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn T H. Method for measuring rates of NH4+ turnover in anoxic marine sediments, using a 15N-NH4+ dilution technique. Appl Environ Microbiol. 1979;37:760–765. doi: 10.1128/aem.37.4.760-765.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bratbak G, Heldal M, Thingstad T F, Tuomi P. Dynamics of virus abundance in coastal seawater. FEMS Microb Ecol. 1996;19:263–269. [Google Scholar]
- 5.Calendar R. The bacteriophages. New York, N.Y: Plenum Press; 1988. [Google Scholar]
- 6.Chin-Leo G, Kirchman D L. Estimating bacterial production in marine waters from the simultaneous incorporation of thymidine and leucine. Appl Environ Microbiol. 1988;54:1934–1939. doi: 10.1128/aem.54.8.1934-1939.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ducklow H W, Carlson C A. Oceanic bacterial production. Adv Microb Ecol. 1992;12:113–181. [Google Scholar]
- 8.Fuhrman J A. Bacterioplankton roles in cycling of organic matter: the microbial food web. In: Falkowski P G, Woodhead A D, editors. Primary productivity and biogeochemical cycles in the sea. New York, N.Y: Plenum Press; 1992. pp. 361–383. [Google Scholar]
- 9.Fuhrman J A. Close coupling between release and uptake of dissolved free amino acids in seawater studied by an isotope dilution approach. Mar Ecol Prog Ser. 1987;37:45–52. [Google Scholar]
- 10.Fuhrman J A. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–548. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 11.Fuhrman J A, Azam F. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results. Mar Biol. 1982;66:109–120. [Google Scholar]
- 12.Fuhrman J A, Noble R T. Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol Oceanogr. 1995;40:1236–1242. [Google Scholar]
- 13.Glibert P M. Regional studies of daily, seasonal, and size fractionation variability in ammonium regeneration. Mar Biol. 1982;70:209–222. [Google Scholar]
- 14.Guixa-Boixareu N, Calderon-Paz J I, Heldal M, Bratbak G, Pedros-Alio C. Viral lysis and bacterivory as prokaryotic loss factors along a salinity gradient. Aquat Microb Ecol. 1996;11:215–227. [Google Scholar]
- 15.Guixa-Boixareu N, Vaqué D, Gasol J, Pedrós-Alió C. Distribution of viruses and their potential effect on bacterioplankton in an oligotrophic marine system. Aquat Microb Ecol. 1999;19:205–213. [Google Scholar]
- 16.Heldal M, Bratbak G. Production and decay of viruses in aquatic environments. Mar Ecol Prog Ser. 1991;72:205–212. [Google Scholar]
- 17.Hennes K P, Suttle C A. Direct counts of viruses in natural waters and laboratory cultures by epifluorescence microscopy. Limnol Oceanogr. 1995;40:1050–1055. [Google Scholar]
- 18.Herndl G J, Müller-Niklas G, Frick J. Major role of ultraviolet-B in controlling bacterioplankton growth in the surface layer of the ocean. Nature. 1993;361:717–719. [Google Scholar]
- 19.Karner M, Rassoulzadegan F. Extracellular enzyme activity: indications for high short-term variability in a coastal marine ecosystem. Microb Ecol. 1995;30:143–156. doi: 10.1007/BF00172570. [DOI] [PubMed] [Google Scholar]
- 20.Kirchman D L. Incorporation of thymidine and leucine in the subarctic Pacific: application to estimating bacterial production. Mar Ecol Prog Ser. 1992;82:301–309. [Google Scholar]
- 21.Kirchman D L. The uptake of inorganic nutrients by heterotrophic bacteria. Microb Ecol. 1994;28:255–271. doi: 10.1007/BF00166816. [DOI] [PubMed] [Google Scholar]
- 22.Kirchman D L, K'Nees E, Hodson R E. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl Environ Microbiol. 1985;49:599–607. doi: 10.1128/aem.49.3.599-607.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S, Fuhrman J A. DNA hybridization to compare species compositions of natural bacterioplankton assemblages. Appl Environ Microbiol. 1990;56:739–746. doi: 10.1128/aem.56.3.739-746.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Middelboe M, Jorgensen N O G, Kroer N. Effects of viruses on nutrient turnover and growth efficiency of noninfected marine bacterioplankton. Appl Environ Microbiol. 1996;62:1991–1997. doi: 10.1128/aem.62.6.1991-1997.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noble R T, Fuhrman J A. Breakdown and microbial uptake of marine viruses and other lysis products. Aquat Microb Ecol. 1999;20:1–11. [Google Scholar]
- 26.Noble R T, Fuhrman J A. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol. 1998;14:113–118. [Google Scholar]
- 27.Noble R T, Fuhrman J A. Virus decay and its causes in coastal waters. Appl Environ Microbiol. 1997;63:77–83. doi: 10.1128/aem.63.1.77-83.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Noble R T, Middelboe M, Fuhrman J A. Effects of viral enrichment on the mortality and growth of heterotrophic bacterioplankton. Aquat Microb Ecol. 1999;18:1–13. [Google Scholar]
- 29.Proctor L M, Fuhrman J A. Viral mortality of marine bacteria and cyanobacteria. Nature. 1990;343:60–62. [Google Scholar]
- 30.Simon M, Azam F. Protein content and protein synthesis rates of planktonic marine bacteria. Mar Ecol Prog Ser. 1989;51:201–213. [Google Scholar]
- 31.Steward G F, Smith D C, Azam F. Abundance and production of bacteria and viruses in the Bering and Chukchi Sea. Mar Ecol Prog Ser. 1996;131:287–300. [Google Scholar]
- 32.Steward G F, Wikner J, Cochlan W P, Smith D C, Azam F. Estimation of virus production in the sea: II. Field results. Mar Microb Food Webs. 1992;6:79–90. [Google Scholar]
- 33.Steward G F, Wikner J, Smith D C, Cochlan W P, Azam F. Estimation of virus production in the sea: I. Method development. Mar Microb Food Webs. 1992;6:57–78. [Google Scholar]
- 34.Suttle C A. The significance of viruses to mortality in aquatic microbial communities. Microb Ecol. 1994;28:237–243. doi: 10.1007/BF00166813. [DOI] [PubMed] [Google Scholar]
- 35.Suttle C A, Chan A M, Chen F, Garza R D. Cyanophages and sunlight: a paradox. In: Guerrero R, Pedros Alio C, editors. Trends in microbial ecology. Proceedings of the 6th ISME, Barcelona, Spain, 1993. 1993. pp. 303–307. [Google Scholar]
- 36.Suttle C A, Chan A M, Cottrell M T. Infection of phytoplankton by viruses and reduction of primary productivity. Nature. 1990;387:467–469. [Google Scholar]
- 37.Suttle C A, Chan A M, Cottrell M T. Use of ultrafiltration to isolate viruses from seawater which are pathogens of marine phytoplankton. Appl Environ Microbiol. 1991;57:721–726. doi: 10.1128/aem.57.3.721-726.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suttle C A, Chen F. Mechanisms and rates of decay of marine viruses in seawater. Appl Environ Microbiol. 1992;58:3721–3729. doi: 10.1128/aem.58.11.3721-3729.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thingstad T F, Heldal M, Bratbak G, Dundas I. Are viruses important partners in pelagic food webs? Trends Ecol Evol. 1993;8:209–213. doi: 10.1016/0169-5347(93)90101-T. [DOI] [PubMed] [Google Scholar]
- 40.Thingstad T F, Zweifel U L, Rassoulzadegan F. P limitation of heterotrophic bacteria and phytoplankton in the northwest Mediterranean. Limnol Oceanogr. 1998;43:88–94. [Google Scholar]
- 41.Weinbauer M G, Beckmann C, Höfle M G. Utility of green fluorescent nucleic acid dyes and aluminum oxide membrane filters for rapid epifluorescence enumeration of soil and sediment bacteria. Appl Environ Microbiol. 1998;64:5000–5003. doi: 10.1128/aem.64.12.5000-5003.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weinbauer M G, Fuks D, Puskaric S, Peduzzi P. Diel, seasonal, and depth-related variability of viruses and dissolved DNA in the Northern Adriatic Sea. Microb Ecol. 1995;30:25–41. doi: 10.1007/BF00184511. [DOI] [PubMed] [Google Scholar]
- 43.Weinbauer M G, Höfle M G. Significance of viral lysis and flagellate grazing as factors controlling bacterioplankton production in a eutrophic lake. Appl Environ Microbiol. 1998;64:431–438. doi: 10.1128/aem.64.2.431-438.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinbauer M G, Peduzzi P. Significance of viruses versus heterotrophic nanoflagellates for controlling bacterial abundance in the northern Adriatic Sea. J Plankton Res. 1995;17:1851–1856. [Google Scholar]
- 45.Weinbauer M G, Suttle C A. Comparison of epifluorescence and transmission electron microscopy for counting viruses in natural marine waters. Aquat Microb Ecol. 1997;13:225–232. [Google Scholar]
- 46.Wilhelm S W, Suttle C A. Viruses and nutrient cycles in the sea. BioScience. 1999;49:781–788. [Google Scholar]
- 47.Wilhelm S W, Weinbauer M G, Suttle C A, Jeffrey W H. The role of sunlight in the removal and repair of viruses in the sea. Limnol Oceanogr. 1998;43:586–592. [Google Scholar]
- 48.Wommack K E, Colwell R R. Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev. 2000;64:69–114. doi: 10.1128/mmbr.64.1.69-114.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wommack K E, Hill R T, Muller T A, Colwell R R. Effects of sunlight on bacteriophage viability and structure. Appl Environ Microbiol. 1996;62:1336–1341. doi: 10.1128/aem.62.4.1336-1341.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]