Abstract
Background
In this pre-clinical study, we designed a candidate vaccine based on severe acute respiratory syndrome-related -coronavirus 2 (SARS-CoV-2) antigens and evaluated its safety and immunogenicity.
Methods
SARS-CoV-2 recombinant protein antigens, including truncated spike protein (SS1, lacking the N-terminal domain of S1), receptor-binding domain (RBD), and nucleoprotein (N) were used. Immunization program was performed via injection of RBD, SS1 +RBD, and SS1 +N along with different adjuvants, Alum, AS03, and Montanide at doses of 0, 40, 80, and 120 μg at three-time points in mice, rabbits, and primates. The humoral and cellular immunity were analyzed by ELISA, VNT, splenocyte cytokine assay, and flow cytometry.
Results
The candidate vaccine produced strong IgG antibody titers at doses of 80 and 120 μg on days 35 and 42. Even though AS03 and Montanide produced high-titer antibodies compared to Alum adjuvant, these sera did not neutralize the virus. Strong virus neutralization was recorded during immunization with SS1 +RBD and RBD with Alum. AS03 and Montanide showed a strong humoral and cellular immunity; however, Alum showed mild to moderate cellular responses. Ultimately, no cytotoxicity and pathologic change were observed.
Conclusion
These findings strongly suggest that RBD with Alum adjuvant is highly immunogenic as a potential vaccine.
Keywords: COVID-19, SARS-CoV-2, Immunogenicity, Subunit vaccine, Receptor binding domain
1. Introduction
Coronavirus disease 2019 (COVID-19) is a new viral respiratory illness caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) which has been identified as a pandemic on March 11, 2020 (Dikid et al., 2020). COVID-19 is identified as a main public health emergency that has infected more than 240 million people and caused about 4.89 million deaths until October 17, 2021 (Coronavirus, 2019, Koutsakos and Kedzierska, 2020). So, public vaccination with a universal safe vaccine against most SARS-CoV-2 mutants is considered an emergency (Folegatti et al., 2020). SARS-CoV-2 is an enveloped, positive-sense, single-stranded RNA virus (Walls et al., 2020, Sternberg and Naujokat, 2020); containing a receptor-binding domain (RBD) as a fragment of the spike glycoprotein which mediates binding to angiotensin-converting enzyme 2 (ACE2); resulting in virus entry into host cells (Liu et al., 2020, Yang et al., 2020a, Dong et al., 2020). the nucleocapsid (N) proteins of numerous coronaviruses are extremely immunogenic (Cong et al., 2020) and high concentrations of IgG antibodies against N proteins have been identified in serum from patients with SARS (Leung et al., 2004). Remarkably, these proteins are considered as representative antigens for triggering the proliferation of T-cell and cytotoxic activity in a vaccine setting (Gao et al., 2003, Okada et al., 2005). All vaccine platforms including mRNA-based, DNA-based, recombinant proteins-based, adenovirus vector-based, and even killed vaccines are based on evoking the host immune system against spike or RBD protein to prevent the virus from entry to the host cells. For example, Sputink V is a recombinant adenovirus types 26 and 5 (rAd26 and rAd5 which used separately in two doses) vector-based SARS-CoV-2 vaccine which displays a full-length spike protein (Logunov et al., 2020a). Oxford–AstraZeneca ChAdOx1 nCoV-19 vaccine; a genetically changed adenovirus that presents spike protein, is another attempt in this regard (Knoll and Wonodi, 2020). In BioNTech/Moderna and BioNTech/Pfizer nucleoside-modified mRNAs have been used (mRNA-1273 & BNT162b2, respectively); demonstrating high neutralizing antibody titers as well as strong antigen-specific CD4+ and CD8+ T-cell responses against SARS-CoV-2. Studies have shown 50% serum geometric mean neutralizing antibody titers surpassing the geometric mean neutralizing antibody titers reported in convalescent human sera (Sahin et al., 2020a, Walsh et al., 2020, Jackson et al., 2020). However, older individuals showed a lower neutralizing response compared to younger ones (Walsh et al., 2020, Polack et al., 2020). Protein subunit vaccines including RBD-dimer vaccine (Anhui Zhifei Longcom) (Yang et al., 2021a) and NVX-CoV2373 recombinant nanoparticulated vaccine (Novavax) have shown to induce remarkable immune responses (Keech et al., 2020). A study conducted in Iran demonstrated appropriate immunity against the disease achieved by an inactivated whole virus SARS-CoV-2 vaccine (COVIran Barekat) (Abdoli et al., 2021).
Due to the need for a safer and more potent vaccine for new variant SARS-CoV-2, many researchers and pharmaceutical companies are trying to introduce new vaccine platforms. Accordingly, in this study, we tried to design a new recombinant vaccine based on the immunogenic parts of the virus. Since the nucleocapsid (N) and spike (especially RBD of the spike) proteins of coronaviruses could induce high IgG antibody titers and proliferation of cytotoxic T cells in COVID-19 patients (Cong et al., 2020; Leung et al., 2004, Gao et al., 2003, Okada et al., 2005), we selected virus protein subunits N, SS1 (a fragment of spike protein without the N-terminal domain), and RBD separately and in combination forms, to determine the potential immunogenicity and protection against SARS-CoV-2. Regarding the nature of viral infections and the critical role of cell-mediated immunity, it was necessary to use adjuvants that could induce cellular immunity besides humoral immunity. Literature review showed that AS03 (GSK, UK) and Montanide (SEPPIC, France) are promising adjuvants that could potentially stimulate cellular immunity through activation and proliferation of T helper 1 cells (Prompetchara et al., 2020, Wang et al., 2020a, Yang et al., 2020b, Arunachalam et al., 2021). Accordingly, we aimed to assess the safety and immunogenicity of SARS-CoV-2 proteins N, RBD, and SS1 in a vaccine setting in animal models including mice, rabbits, and primates.
2. Material and methods
2.1. Designing and construction of SARS-CoV-2 recombinant proteins
Amino acid sequences of SARS-CoV-2 subunits including RBD, SS1 (Gen Bank accession: NC_045512; Gene ID: 43740568), and N (Gen Bank accession: NC_045512; Gene ID: 43740575) were obtained from the NCBI database (http://www.ncbi.nlm.nih.gov). The full-length sequence of the n gene, and the gene sequence encoding amino acids 319–543 for the RBD, and the gene sequence encoding amino acids 150–836 for SS1 were selected. The amino acid arrangement and physicochemical characterizations of the proteins including estimated half-life, net charge at pH7, molecular weight, theoretical isoelectric point, instability index, solubility, and aliphatic index were determined through Protparam (http://web.expasy.org/cgi-bin/protparam/protparam). Moreover, the features of the messenger RNA including secondary structure, folding, and thermodynamic properties of the native and optimized mRNAs were evaluated by the mfold tools. The optimum Gene TM Algorithm software was used for the optimization of the gene to obtain high expression of the protein in E. coli BL21 DE3. The restriction sites for EcoRI, HindIII, BamHI, XhoI, NdeI, and HindIII enzymes were embedded. The restriction sites were considered at 5 ´ and 3 ´ ends of rbd, ss1, and n genes. The sequences were synthesized by the GenScript Company and subcloned in pET SUMO expression vectors. The constructs were confirmed through digestion with restriction enzymes and sequencing.
2.2. Protein expression and purification in E. coli BL21 DE3
To express the suggested proteins in E. coli BL21 DE3, we prepared the competent cells applying the calcium chloride method and the plasmids containing genes were transferred to the host, cultured in Luria-Bertani (LB), and complemented with 1 mM isopropyl ß-D-1-thiogalactopyranoside (IPTG). The bacterial pellets were resuspended in lysis buffer (NaH2PO4 100 mM, Tris-HCl 10 mM, urea 8 M) and sonicated. The supernatant was purified by SUMO-tagged proteins from E. coli BL21 DE3 under denaturing conditions (Qiagen). The column-bound protein was eluted; using the elution buffer (100 mM NaH2PO4, 8 M Urea, 10 mM Tris-Cl, pH:4.5). The denaturant agent (8 M urea) was removed from the purified proteins by stepwise dialysis and the SUMO-tag was cleaved by SUMO protease. Finally, the recombinant proteins were confirmed by 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and western blotting.
2.3. Characterization of antigens by ELISA and Western blot analysis
For enzyme-linked immunosorbent assay (ELISA), purified RBD, SS1, and N antigens as well as combinations of SS1/RBD, SS1/N, and N/RBD antigens were coated (0.3 µg/100 µl/well) and incubated with SARS-CoV-2 convalescent human sera (1:100). Then, anti-human IgG-HRP was added and the absorbance was measured by ELISA reader (BioRad USA). For western blot, purified RBD, SS1, and N antigens were transferred into a membrane, treated with a 1:100 dilution of SARS-CoV-2 convalescent human serum; followed by adding anti-human IgG-HRP. Then, the chromogenic reaction was performed; using 3, 3'-diaminobenzidine.
2.4. Antigen formulation and animal immunization
Three hundred and sixty female BALB/c mice (Razi Vaccine and Serum Research Institute, Iran) were allocated into three groups of 120 mice for three distinct adjuvants including Alum (Alhydrogel® adjuvant 2% InvivoGen, USA), AS03 (GSK, UK), and Montanide/ISA720 (SEPPIC, France). To assess immunization, each adjuvant group (Alum, AS03, and Montanide), was sub-divided into four antigen groups (SS1, RBD, SS1 + N, and SS1 + RBD) with three different doses (40, 80, and 120 μg) ( Table 1). The control groups (30 mice) received phosphate-buffered saline (PBS) and the desired adjuvant. The animals received different concentrations for three time-points on days 0, 21, and 35. Blood samples were obtained after each immunization. Furthermore, 6 New Zealand White rabbits (Razi Vaccine and Serum Research Institute, Iran) were grouped for receiving 120 μg of each antigen (RBD and SS1 +RBD) along with Alum at the same administration intervals and compared to the control group ( Table 2).
Table 1.
Different formulation of recombinant proteins and adjuvants for mice immunization.
| Adjutants | Groups | Dose finding | Number of mice | Number of injections |
|---|---|---|---|---|
| Alum | RBD | 40 | 10 10 10 |
3 |
| 80 | ||||
| 120 | ||||
| SS1 | 40 | 10 10 10 |
||
| 80 | ||||
| 120 | ||||
| SS1 +RBD | 40 | 10 10 10 |
||
| 80 | ||||
| 120 | ||||
| SS1 +N | 40 | 10 10 10 |
||
| 80 | ||||
| 120 | ||||
| AS03 | RBD | 40 | 10 10 10 |
3 |
| 80 | ||||
| 120 | ||||
| SS1 | 40 | 10 10 10 |
||
| 80 | ||||
| 120 | ||||
| SS1 +RBD | 40 | 10 10 10 |
||
| 80 | ||||
| 120 | ||||
| SS1 +N | 40 | 10 10 10 |
||
| 80 | ||||
| 120 | ||||
| Montanide | RBD | 40 | 10 10 10 |
3 |
| 80 | ||||
| 120 | ||||
| SS1 | 40 | 10 10 10 |
||
| 80 | ||||
| 120 | ||||
| SS1 +RBD | 40 | 10 10 10 |
||
| 80 | ||||
| 120 | ||||
| SS1 +N | 40 | 10 10 10 |
||
| 80 | ||||
| 120 | ||||
| Control | Alum | – | 10 | 3 |
| AS03 | – | 10 | ||
| Montanide | – | 10 |
Table 2.
Different formulation of recombinant proteins and adjuvants for rabbit immunization.
| Adjutants | Groups | Dose of injection | Number of primates |
|---|---|---|---|
| Alum | SS1 +RBD | 120 | 2 |
| RBD | 120 | 2 | |
| Control | – | 2 |
A similar procedure was performed for 6 non-human primates (Rhesus macaque). All monkeys were checked for weight, nutritional regiment, and their health condition such as hematology, biochemistry, clotting time, and inflammation parameters before and during the study. Previous infections by SARS-CoV-2, hepatitis B and C, and HIV were studied as well (Supplementary T1 to T9). These animals were also immunized with 120 μg of each antigen (RBD and SS1 +RBD) along with Alum at the same administration intervals ( Table 3).
Table 3.
Different formulation of recombinant proteins and adjuvants for primate immunization.
| Adjutants | Groups | Dose of injection | Number of primates |
|---|---|---|---|
| Alum | SS1 +RBD | 120 | 2 |
| RBD | 120 | 2 | |
| Control | – | 2 |
2.5. Evaluation of antibody titer by ELISA
ELISA was performed to detect specific total IgG and IgG subclasses (IgG1 and IgG2a) in sera samples. Briefly, the plates were coated (0.3 µg/100 µl/well) with SS1, RBD, or N and blocked. Then, diluted sera samples (1:100–1:204800) were added to antigen-coated plates and detected; using anti-mouse-IgG-HRP, anti-rabbit-IgG-HRP, anti-monkey-IgG-HRP, or IgG1 and IgG2a-HRP (1:1000) (Abcam, UK).
2.6. Cytokine and flow cytometry assay
ELISA was performed to assess the titration of interleukin (IL)-4, IL-12, and interferon-gamma (IFN-γ) of splenocytes according to the manufacturer's instructions (Murine: Diaclone, France; Monkey: Biosciences, Netherlands). In brief, cells were isolated from the spleens of immunized and control animals. Cell suspensions (105/well) were separately incubated with 10 μg/ml of SS1, RBD, and N proteins for 72 h. IFN-γ and ILs were detected in the supernatant culture according to the supplier’s protocol. Moreover, the CD3 + , CD4 + , and CD8 + T cells (Abcam, UK) were counted; using the fluorescence-activated cell sorting (FACS) calibur flow cytometer (Becton Dickinson, San Jose, USA). Flowjo7 software was used to analyze the results.
2.7. Virus neutralization assay
To analyze the neutralization capacity of immunized sera, we treated Vero cells with the serial dilutions of both the virus and pseudo-virus, and the IC50 dilution was determined. The convalescent human sera as well as control and immunized animal sera (1:10 and 1:100 serum dilutions) were incubated 45 min with 100 TCID50 and exposed to Vero cell culture. In addition, the neutralizing antibody was measured by Abnova ELISA kit (Abnova, USA) according to the supplier’s protocol.
2.8. Safety assessment and challenge test
Limulus amoebocyte lysate (LAL) test was used to detect leukocytic pyrogen contamination. To assess the protective efficacy of the candidate vaccine, the researchers challenged all primates with 106 virus particles two months after vaccination and then used real-time polymerase chain reaction (PCR) to quantify replicating viruses. The monkeys were anesthetized by a veterinarian and 106 virus induced in the nasopharyngeal. RT-PCR was done with referral virology laboratory through nasopharyngeal sampling. A novel coronavirus (2019-nCoV) Nucleic Acid Diagnostic Kit (PCR fluorescence probing, Sansure Biotech) was used for gene detection. For this purpose, 5 µl of purified RNA was added to the 20 µl PCR mix and the qRT-PCR thermocycling program (50 C for 20 min, followed by one cycle, 95 C for 60 s, followed by one cycle, 95 C for 15 s, and 60 C for 60 s, followed by 45 cycles) was done on a Corbett Instrument. Three months after the last immunization of mice and primates, lung, heart, liver, kidney, skin, and brain tissues were harvested and any inflammation or injury was assessed with Hematoxylin and Eosin staining.
2.9. Statistical analyses
All experiments were performed three times and the resulting data were evaluated by the one-way ANOVA, repeated measure ANOVA, and paired T test. In case of non-parametric variable, Kruskal-Wallis test and Wilcoxon test were used. A p-value < 0.05 was considered statistically significant.
3. Results
3.1. Bioinformatics design of the gene construct encoding SS1, RBD, and N antigens
The complete amino acid sequence of N, partial amino acid sequence (150–836) of SS1, and partial amino acid sequence (319–543) of RBD were selected. Guanine-cytosine (GC) contents of native and optimized sequences of RBD, SS1, and N proteins were 35.61% and 53.61%, 47.29% and 51.2%, and 36.7% and 52.5%, respectively. After optimization (SupplementaryS1 to S6), codon adaptation index of RBD, SS1, and N cassettes were altered from 0.63 to 0.91, 0.61–0.91, and 0.63–0.79, respectively. In addition, the frequency of codon with 91–100 frequency distributions of RBD, SS1, and N cassettes was raised from 42% to 80%, 38–81%, and 44–60%, respectively (Suppl. Fig. 6). The mfold webserver was used to analyze mRNA by the minimum free energy base-pairing (MFE) and probability matrix (BLAST-like) methods. Following optimization, the start codon at the 5′-end had a minimum of pseudoknot and interior loop size at 37 °C, and the ΔG of RBD, SS1, and N mRNA structures were declined to − 245.16, − 623.50, and − 469.11 kcal/mol, respectively. Epitope prediction factors including exposed surfaces, external availability, hydrophilicity, secondary structure, flexibility, and polarity were determined for all proteins. The results of IEDB software for the prediction of final epitopes and their MHC-I and MHC-II binding capacities, and the antigenic, non-toxic, and non-allergenic T-cell/ B-cell epitopes for inducing inflammatory cytokines are shown in supplementary T10 and T11. The physiochemical properties of three antigens including RBD (225 amino acids), SS1 (675 amino acids), and N protein (419 amino acids) were determined by the ProtParam program and, as demonstrated in Table 4.
Table 4.
Physiochemical properties of RBD, SS1, and N antigens.
| RBD | SS1 | N | |
|---|---|---|---|
| Molecular weight (Da) | 25,203.50 | 67,301.00 | 45,625.70 |
| Theoretical pI | 8.79 | 8.00 | 10.07 |
| Number of negatively charged residues (Asp + Glu) | 16 | 52 | 36 |
| Number of positively charged residues (Arg + Lys) | 22 | 55 | 60 |
| Estimated half-life (Escherichia coli,in vivo) | > 10 h | > 10 h | > 10 h |
| Instability index | 21.96 | 31.12 | 55.09 |
3.2. Expression, purification, and characterization of antigens
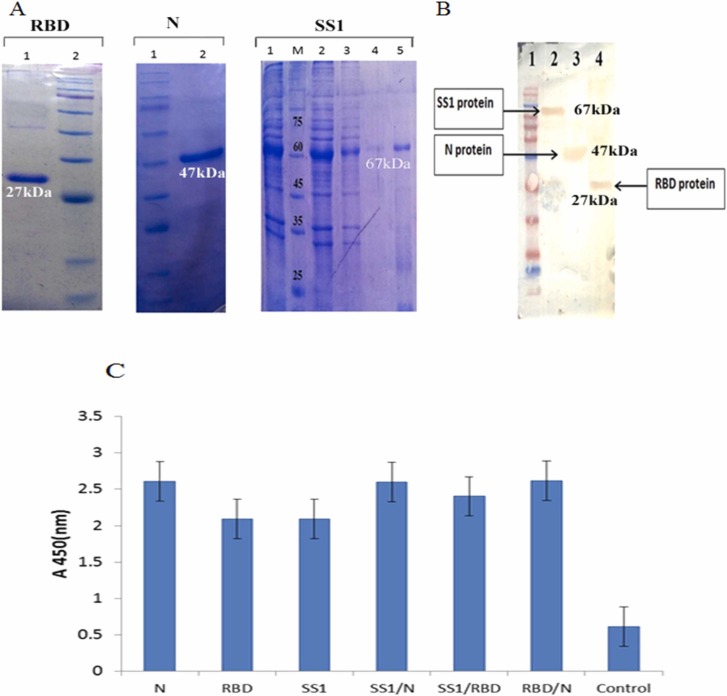
The genes were cloned into a pET SUMO expression vector and expressed in E.coli BL21 DE3 under optimized culture and induction conditions. The proteins were purified and after cleavage, were analyzed; using SDS-PAGE 12%. Accordingly, 27, 47, and 67 kDa protein bands were representative of RBD, N, and SS1, respectively ( Fig. 1A). As indicated in Fig. 1B-C, western blot and ELISA analysis could correctly recognize all three recombinant proteins in convalescent COVID-19 patients’ sera; containing anti-SARS-CoV-2 antibodies.
Fig. 1.
The results of purification and characterization of SS1, receptor-binding domain (RBD), and nucleoprotein (N) antigens A: Purification of the proteins through affinity chromatography; using Ni-NTA resin columns. RBD, Lane 1: purified RBD by elution buffer, Lane 2: molecular weight marker; N, Lane 1: molecular weight marker, Lane 2: purified nucleoprotein by elution buffer; SS1, Lane 1: before column, M: molecular weight marker, Lane 2: flow through, Lanes 3 and 4: washing buffer, Lane 5: purified SS1 by elution buffer, B: SS1, RBD, and N antigens were confirmed by Western blotting using convalescent human sera. Lane 1: protein marker; lane2: SS1 protein; Lane 3: N protein; Lane 4: RBD protein. C: ELISA was used for convalescent patients sera with produced COVID-19 antigens compared to controls.
3.3. Determination of antibody titer in mice sera
The sera collected from mice on days 0, 14, 42, and 65 were evaluated for IgG, IgG1, and IgG2a subclasses against all three SARS-CoV-2 antigens by ELISA. The results showed that the 120 and 80 μg doses produced higher antibody titers than groups with other doses and the control group (p < 0.05). In addition, to produce high IgG antibody titers in mice (approximately 2 folds), the third booster was critical. Regarding antigen groups, the results showed that the RBD group, the SS1 group, and then the SS1 +RBD group could induce higher antibody titers, respectively (p < 0.05). As indicated in Fig. 2, AS03 and Montanide adjuvants could induce remarkably higher antibody responses against SARS-CoV-2 antigens compared to the Alum adjuvant (p < 0.05). In addition, the type of adjuvant could affect the nature of antigenicity of the SARS-CoV-2 proteins. For example, the combination of SS1 +N proteins induced the highest antibody response in favor of the N antigen when injected with Montanide compared to AS03 or Alum.
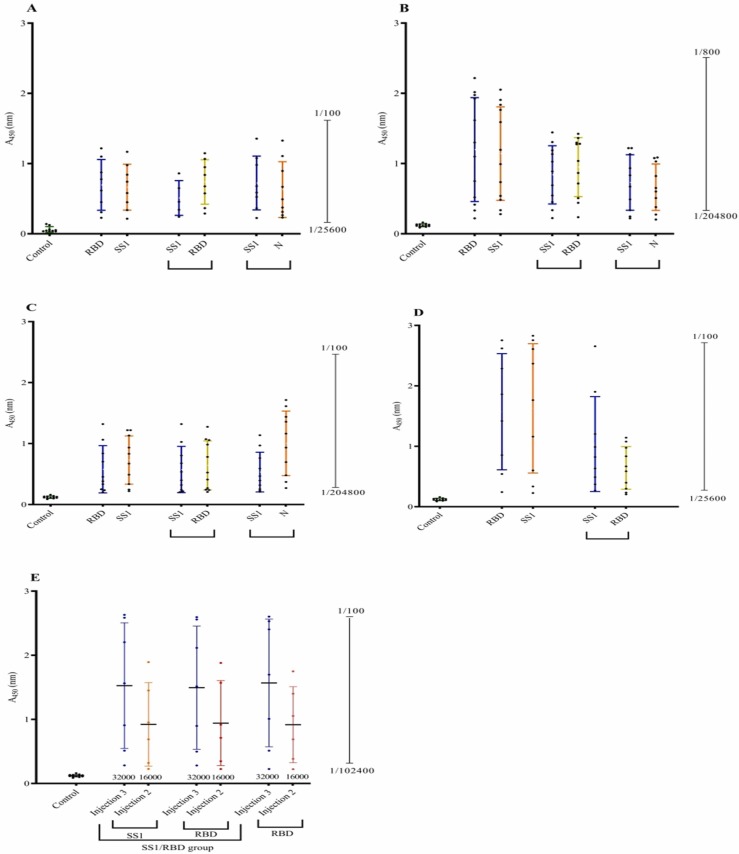
Fig. 2.
Antibody (IgG) response against various severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) antigens + adjuvants at the dose of 120 μg in different animal models. A: Antibody response against various SARS-CoV-2 antigens + Alum in mice. B: Antibody response against various SARS-CoV-2 antigens + AS03 in mice. C: Antibody response against various SARS-CoV-2 antigens + Montanide in mice. D: Antibody response against various SARS-CoV-2 antigens + Alum in rabbit. E: Antibody response against various SARS-CoV-2 antigens + Alum in primates.
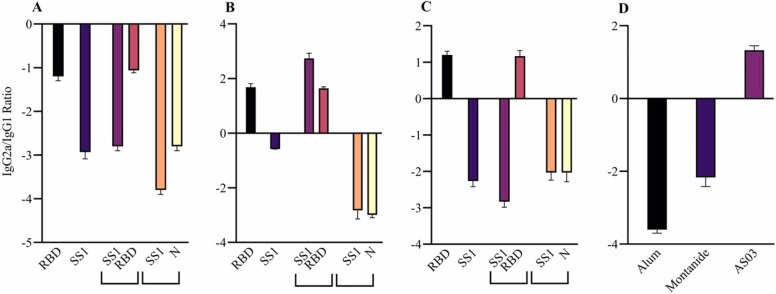
Furthermore, this finding was better observed in IgG2a/IgG1 ratio. In Alum adjuvant, all antigens raised IgG1 antibody as Th2 cell immune response. AS03 induced an IgG2a response against RBD and S antigen, and it seemed that the antibody response was affected by Th1 cells. Montanide adjuvant could raise IgG2a against RBD antigen. Overall, AS03, as expected, induced a predominant Th1 response against both RBD and S1, whereas Montanide could induce only Th1 response against S1 antigen. It seems that the Alum adjuvant exclusively evoked Th2 response due to its salty nature. However, these results should be analyzed regarding the virus neutralization test and the cytokine assay ( Fig. 3).
Fig. 3.
Different IgG2a/IgG1 ratios and humoral responses against severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) antigens in three adjuvants, namely, Alum, AS03, and Montanide at the dose of 120 μg of the candidate vaccine in mice. A: COVID-19 specific IgG2a/IgG1 ratio in Alum at the dose of 120 μg of the candidate vaccine. B: COVID-19 specific IgG2a/IgG1 ratio in AS03at the dose of 120 μg of the candidate vaccine. C: COVID-19 specific IgG2a/IgG1 ratio in Montanide at the dose of 120 μg of the candidate vaccine. D: Different humoral responses between three adjuvants. Receptor-binding domain (RBD) antigen with AS03 adjuvant-induced predominant Th1 response.
3.4. Titration of immunized rabbit sera by ELISA
According to the results of antibody response in mice, 120 μg was selected as the best dose and Alum was selected as the best adjuvant for rabbit and primate assays. Rabbits were divided into two groups, the first group was injected with the SS1 +RBD antigen and the other group received RBD. ELISA assay on blood samples collected one week after each injection, showed that the RBD group could induce higher IgG antibody titrations (Fig. 2D).
3.5. Titration of immunized primates’ sera by ELISA
In three monkeys groups, one group received RBD antigen, one group received SS1 +RBD, and a control group received only the Alum adjuvant. Similar to mice and rabbits, primates also developed higher antibody titers against SS1 and RBD with Alum (Fig. 2E). As indicated in Fig. 2E, the second injection was not sufficient for the elevation of antibody titer and the third injection was necessary.
3.6. Cytokine assay and flow cytometry analysis
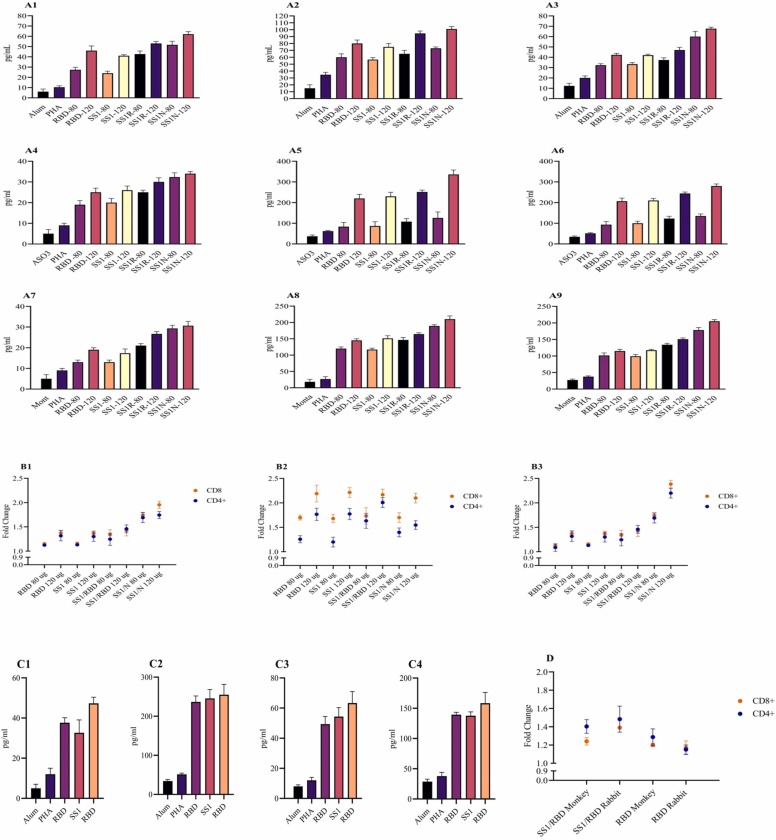
Among different adjuvants and SARS-CoV-2 antigen combinations, AS03, Montanide, and then Alum induced the highest levels of cytokines including IL-12, IFN-γ, and IL-4, respectively. However, Alum adjuvant could induce more IL-4 levels. Regarding antigen combinations, N antigen and then RBD could induce the highest levels of Th1 cytokines including IL-12 and IFN-γ. In general, comparing Montanide and Alum, the results showed that AS03 adjuvant could more significantly shift the immune responses toward Th1 (p < 0.05) ( Fig. 4A and C). The results of Flow cytometry in all animal models confirmed the results of the cytokine assay. AS03 and Montanide adjuvants could prime moderate and high levels of CD4 + and CD8 + T cells, respectively. In Alum adjuvant, mild CD4+ and CD8+ T cells were primed by antigens (Fig. 4B and D). All results of the cytokine assay and Flow cytometry were confirmed by the IgG2a/IgG1 ratio.
Fig. 4.
The results of cytokines assay and flow cytometry analysis in animal models vaccinated against severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) antigens. A: Cytokine production against SARS-CoV-2 antigens + Alum in mice (1: IL-4, 2: IL-12, 3: IFNγ), AS03 (4: IL-4, 5: IL-12, 6: IFNγ), and Montanide (7: IL-4, 8: IL-12, 9: IFNγ). B: The results of flow-cytometry analysis against SARS-CoV-2 antigens in mice (1: Alum, 2: AS03, 3:Montanide). C: Cytokine production against SARS-CoV-2 antigens + Alum in rabbit (1: IL-4, 2: IFNγ), SARS-CoV-2 antigens + Alum in monkey (3: IL-4, 4: IFNγ). D: The results of flow cytometry analysis against SARS-CoV-2 antigens in the monkey model.
3.7. Virus neutralization test and challenge
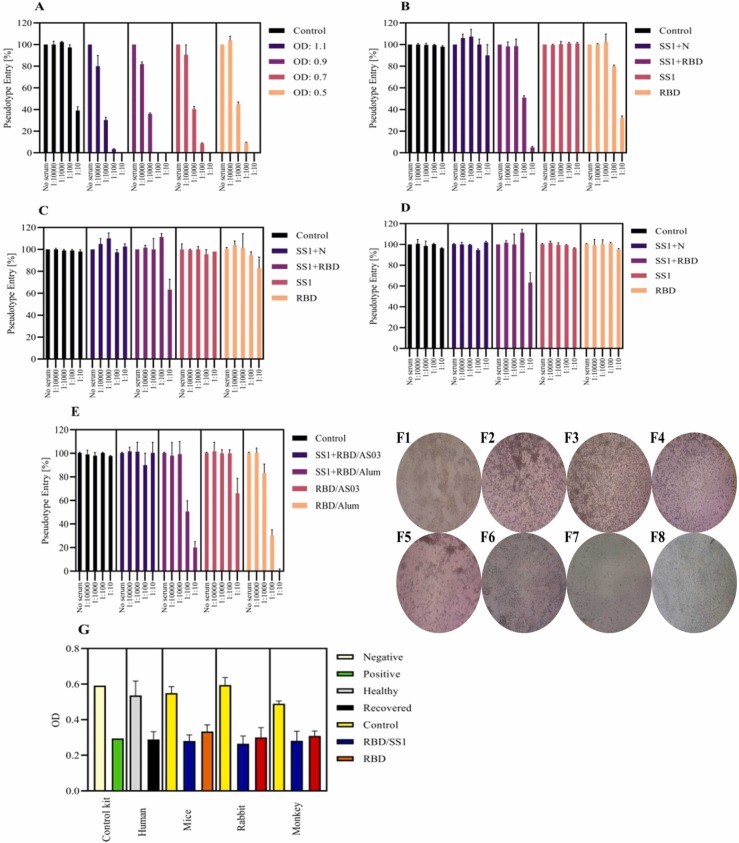
According to the virus neutralization test, only the Alum group could significantly raise the neutralizing antibodies after the third injection in all animal models (p < 0.05). Regarding the antigen groups, although both RBD and SS1 +RBD groups could effectively neutralize the pseudo-virus and SARS-CoV-2 (p < 0.05), protection in the RBD and SS1 +RBD groups has been different in independent dilutions (100% in 1/10 serum dilution and 80% in 1/100 serum dilution) ( Fig. 5A-F). These data were in agreement with the data obtained from Abnova ELISA neutralizing assay (Fig. 5G). The results of the viral load after the challenge with SARS-CoV-2 on days 2, 7, and 14 are summarized in Table 5. In immunized monkeys, the viral load was significantly reduced from day 2 (CT:15) to day 7 (CT: 28). In contrary, non-immunized monkeys could not prevent the localization of the virus.
Fig. 5.
Effect of incubated Vero cells with neutralizing titers of the immunized sera with various severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) antigens. A: Sera from positive humans was collected after immunization and analyzed. 1: Control, 2: OD: 1.1, 3: OD: 0.9, 4: OD: 0.7, 5: OD: 0.5. B: Neutralizing antibodies sera from the immunized mice with various SARS-CoV-2 antigens + Alum adjuvant. 1: Control, 2: SS1 +N, 3: SS1 +RBD, 4: SS1, 5: RBD. C: Neutralizing antibodies sera from the immunized mice with various SARS-CoV-2 antigens + AS03 adjuvant. 1: Control, 2: SS1 +N, 3: SS1 +RBD, 4: SS1, 5: RBD. D: Neutralizing antibodies sera from the immunized mice with various SARS-CoV-2 antigens + Montanide adjuvant. 1: Control, 2: SS1 +N, 3: SS1 +RBD, 4: SS1, 5: RBD. E: Next, according to the results of mice, primates were treated as following and neutralizing antibodies sera from the immunized primates were collected and analyzed. 1: Control, 2: SS1 +RBD with AS03 adjuvant, 3: SS1 +RBD with Alum adjuvant, 4: RBD with AS03 adjuvant, 5: RBD with Alum adjuvant. F: Morphological changes in Vero cells F1: Negative control; F2:Positive control after treatment with 100 TCID50; F3: after treated with SS1 +RBD and Alum mice; F4: after treated with SS1 +RBD and Alum rabbit; F5: after treated with SS1 +RBD and Alum monkey; F6: after treated with RBD and Alum mice; F7: after treated with RBD and Alum rabbit; F8: after treated with RBD and Alum monkey. Images were taken by a phase-contrast microscope (40 ×). G: The results of neutralization titers measured by ELISA. Human: human convalescent plasma and control: serum healthy person; in mice rabbit and monkey: negative: control (PBS+adjuvants); serum test 1: SS1 +RBD; serum test 2: RBD.
Table 5.
The results of challenge with SARS-CoV-2.
| Specimen | Nasopharyngeal and throat swab | ||
|---|---|---|---|
| Days | 2 | 7 | 14 |
| Control | Negative | Negative | Negative |
| Non-Immunized monkey (CT) | 15 | 18 | 27 |
| Immunized monkey (CT) | 15 | 28 | 36 |
3.8. Safety outcomes
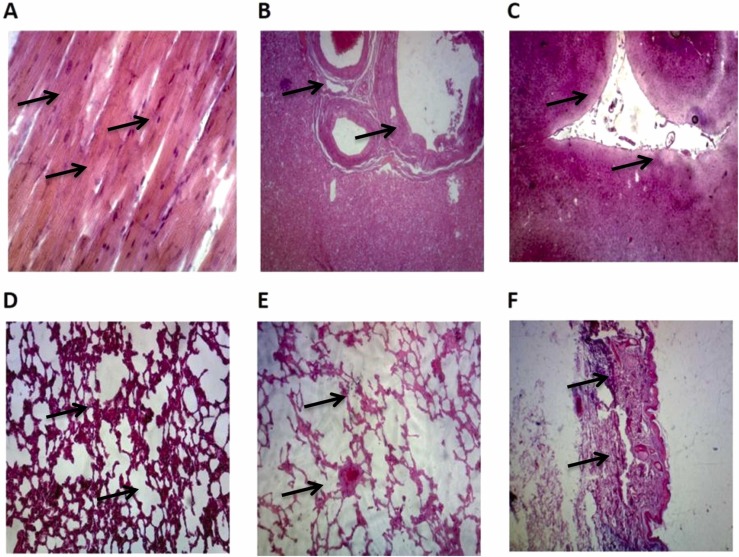
In the LAL test, endotoxin was estimated to be less than < 0.125 EU/µg ( Table 6). Safety experiments showed no serious adverse consequence or specific pathologic change in the heart, liver, kidney, skin, and brain samples of mice and primates ( Fig. 6).
Table 6.
The results of LAL test.
| Protein | Result | Unit | Standard method | Reference |
|---|---|---|---|---|
| SS1 | Negative | EU/ml | Gel clot lonza kit sensitivity | USP |
| RBD | Negative | EU/ml | Gel clot lonza kit sensitivity | USP |
| N | Negative | EU/ml | Gel clot lonza kit sensitivity | USP |
Fig. 6.
The microscopic findings of immunized monkeys collected tissues (heart, liver, brain, lung, kidney, and skin). A: Dense connective tissue with elastic fibers was present in the cardiac/fibrous skeleton. Cardiomyocytes were normal and branched, contained intercalated disks, and were mononucleated. B: Fat vacuoles were seen in the hepatocytes' cytoplasm. Some sinusoids and central veins were congested but portal tracts were seen in normal condition. There was no increase in Kupffer cells and inflammation or dilation of sinusoids. C: No inflammation in meninges and neutrophils was observed. Molecular, granular, and pyramidal layers were intact and no neuron degeneration was noticed. D: No lesion in peribronchial and interalveolar spaces were seen. E: There was no evidence regarding inflammatory cells infiltration and no other lesion was noticed. F: Stratified squamous epithelium was intact with a thin layer of stratum corneum. Irregular dense connective tissue in the dermis layer with hair follicles and no inflammation, congestion, hyperemia, swelling, or other disorders were noticed.
4. Discussion
Despite the cutting-edge progress in the field of SARS-CoV-2 vaccine development and many clinical trials on viral components including mRNA, S1, S2, RBD, and N subunits, a global effort is necessary to achieve public health goals through vaccination (Thevarajan et al., 2020). At the beginning of the SARS-CoV-2 pandemic, WHO officially stated that no worldwide vaccination is expected to enter the market until the next two years (Wang et al., 2020b). However, the WHO has now developed several vaccines. In the present study, we tried to develop a potential vaccine; using different SARS-CoV-2 antigens (SS1, RBD, “SS1/RBD”, and “SS1/N”) with three different adjuvants including Alum, AS03, and Montanide at three doses (40, 80, and 120 μg) in mice, rabbits, and primates.
Previously, the S subunit was reported as a target for antibody responses and several studies on SARS-CoV-2 or Middle East respiratory syndrome-related coronavirus (MERS-CoV) have observed both CD4+ and CD8+ T cell epitopes in this subunit (Liu et al., 2017, Grifoni et al., 2020). In this regard, currently, three COVID-19 vaccines have been listed by WHO for emergency use or in clinical trials with the capability to express the full-length S subunit. Among them, Russia worked on recombinant rAd26 and rAd5 vector-based SARS-CoV-2 vaccines (Logunov et al., 2020a). The other candidate is from the UK; using recombinant chimpanzee ChAdOx1 nCoV-19 vaccine with lower efficacy results. This vaccine was able to prevent COVID-19-associated pneumonia in primates and significantly decreased the viral load in the respiratory tract (Knoll and Wonodi, 2020). Ultimately, the last one has been studied in China based on Ad5 (Zhu et al., 2020a, Zhu et al., 2020b). It should be noted that the phase II clinical trials of these studies indicated a potential neutralizing antibody and T-cell responses in human subjects (Folegatti et al., 2020, Zhu et al., 2020a, Logunov et al., 2020b). The fact that using S subunit-vaccines with N terminal domain exhibit lower neutralizing potency; especially when compared to RBD-based vaccines, is of great importance (Chi et al., 2020). Here, we designed and used this subunit without the N-terminal domain as SS1 to strengthen its immunogenic effect.
On the other hand, the highest proportion of neutralizing antibodies specific for SARS-CoV-2 was reported to target the RBD part of the virus and block the interaction between RBD and ACE2 (Dai and Gao, 2021). According to previous studies related to MERS-CoV and SARS-CoV-2, this part of the virus has a variety of epitopes for T cells and has demonstrated a high-quality antibody response and lower antibody-dependent enhancement (ADE) (Yang et al., 2020a, Liu et al., 2017, Zhang et al., 2020, Zhou et al., 2006). Considering these data, here, we used SS1 along with RBD to increase the antibody response of our candidate vaccine. Accordingly, we observed excellent results regarding the production of neutralizing antibodies. Recently, many studies have started to develop RBD-based vaccines for SARS-CoV-2. It was reported that using RBD in vaccines has resulted in the production of a high amount of neutralizing antibodies in mice, rabbits, and primates. Moreover, they indicated that the immune sera showed a protective effect in mice (Yang et al., 2020a). Another study on primates applied DNA producing RBD and observed that neutralizing antibodies levels are associated with immunity against SARS-CoV-2 (Yu et al., 2020). As mentioned before, the phase I/II studies of BioNTech/Pfizer used a nucleoside-modified RNA-based vaccine named BNT162b2 to express RBD and showed high neutralizing antibodies as well as strong T-cell responses against SARS-CoV-2 (Mulligan et al., 2020, Sahin et al., 2020b).
Nevertheless, it should be considered that RBD might also show lower immunogenicity due to its small size and plausible mixed forms of peptides as monomer, dimer, or trimer (Dai and Gao, 2021). Therefore, this could lead to another challenge in designing a practical vaccine. Hence, we applied the SS1, RBD, and N antigens to augment the safety and immunogenicity of our candidate vaccine against SARS-CoV-2 and remarkable results were observed (Dai and Gao, 2021, Bachmann and Zinkernagel, 1997). It is worth bearing in mind that the structure of RBD in E.coli is notably similar to what is expressed in the mammalian cells and both can cause immunity response and protective immunity against SARS-CoV challenge without Fc tag (Du et al., 2009). However, the expression of RBD in E.coli is much more economical, simpler, and faster than in eukaryotes. Remarkably, RBD does not show any post-translational changes in E.coli (Prahlad et al., 2021). E.coli; as a superior system for the mass-production of recombinant proteins, is considered as an appropriate model for producing functional RBD as a vaccine (Boosani and Sudhakar, 2006, Farinha-Arcieri et al., 2008, Marblestone et al., 2006, Saitoh et al., 2009), and RBD-based vaccines could lead to a notable antibody response with long-term protective immunity in different models (Du et al., 2007). Indeed, the N subunit is the most highly expressed and immunogenic viral part of SARS-CoV-2 (Long et al., 2020). Consequently, it is another likely target for inducing the neutralizing antibodies and T cell responses. There are several studies conducted in this area of research with inconsistent results. Among them, one study demonstrated that N subunit-based vaccine resulted in CD4+ and CD8+ T cell responses in mice model (Liu et al., 2006). Additionally, another study indicated that vaccination with Venezuelan equine encephalitis replicon expressing a SARS-CoV CD4+ T cell epitope results in specific protection against the disease by IFN-γ production in mice (Zhao et al., 2016). Nevertheless, earlier studies have questioned the possibility of N subunit as a practical vaccine against SARS-CoV-2 by suggesting that not only N subunit-based vaccines are not able to induce protection against the virus, but also increase pneumonia and enhance respiratory diseases (ERD); followed by infection through enhancing Th2 cell-based response and pulmonary eosinophil infiltration (Deming et al., 2006, Yasui et al., 2008). Moreover, a recent study on mice also showed that anti-N immune sera did not show protection against SARS-CoV-2 (Sun et al., 2020). In this manner, using N subunit in vaccine setting is a debatable issue which should be fully investigated by further investigations. To date, as far as the authors are concerned, no vaccine is completely progressed based on N subunit for SARS-CoV-2 (Dai and Gao, 2021). Based on previous studies which used a high dose of recombinant vaccine (such as NVX-CoV2373 and Zhifei), (Yang et al., 2021b, Tian et al., 2021), we used a higher dose in our animal experiment to investigate the effect of our candidate recombinant vaccine on the immunity of animals. However, the obtained results suggested the potential of these subunits along with other adjuvants to be a candidate vaccine against SARS-CoV-2. According to the results, the 80 and 120 μg concentration groups could induce superior antibody responses compared to other doses. Although the best immune response was induced through the AS03 adjuvant, unexpectedly, no significant neutralizing antibodies were evaluated in the VNT assay. Instead, despite lower humoral and cellular immune responses were detected in the Alum adjuvant, neutralizing antibody was produced against the RBD antigen. Furthermore protection has been just seen in RBD and combination of RBD with SS1. In this regard, the role of the adjuvant is critical as high stimulation of immune cells such as B cells to induce more neutralizing antibodies with a higher affinity. Adjuvants could shape and modulate the epitope recognition profile of antibodies (Wang et al., 2016, Maeda et al., 2017, Ghalavand et al., 2021, Rezaie et al., 2019).
In addition, according to the VNT results, induced neutralizing antibodies could recognize native viral RBD protein domain completely similar to the recombinant RBD antigen. As expected, due to the composition of the Alum adjuvant (aluminum hydroxide), a cell-mediated immune response including CD4/CD8 T cells ratio, IgG2a/IgG1, IL-12, and IFNγ was lower for Alum compared to AS03 and Montanide; demonstrating switching the immune responses toward the Th2 T cell. As pathologic findings indicated, no inflammation and injuries were observed in the brain, lung, heart, liver, kidney, and skin of experimental animal models. In addition, during the whole process of animal vaccinations, no alterations were found in the biochemical parameters (especially creatine phosphokinase), hematological parameters (especially platelets), clotting parameters (including prothrombin time, partial thromboplastin time, and international normalized ratio), and inflammatory factors (including CRP, ESR, and ANA).
As mentioned earlier, the S subunit contains two parts, S1 and S2, both of which have been investigated for developing a potential vaccine. Recently, it was illustrated that SS1 +Alum caused a stronger antibody response against SARS‐CoV‐2 in comparison with RBD. The reason might be due to the presence of neutralization epitopes in SS1 but outside the RBD (Wang et al., 2021).
5. Conclusions
In the present study, we evaluated the immunogenicity of different SARS-CoV-2 antigens including SS1, RBD, and N along with various adjuvants, namely Alum, AS03, and Montanide in a vaccine setting in mice, rabbits, and primates. It was observed that our candidate vaccines elicited superior antibody responses at the concentration of 80 and 120 μg compared to other doses, especially when used in combination with AS03. However, a high level of neutralizing antibodies against the SARS-CoV-2 virus was observed in the sera of the animal models immunized with Alum adjuvant. The obtained results were confirmed by observing moderate levels of Th1/Th2 cytokines. Also, pathology, cytotoxicity, and other laboratory findings indicated the safety of our candidate vaccine. Ultimately, our statistical findings suggested that RBD or RBD+SS1 in combination with alum as the adjuvant could lead to remarkable antibody production, T-cell response, and significant immunity in various animal models.
Funding
This research was supported by Baqiyatallah University of Medical Sciences, Tehran, Iran with Grant No: 5121.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Ethics approval statement for human and/or animal studies
This research was approved by the National Research Ethics Committee with the ethics code: Pre-Clinical:IR.BMSU.REC.1399.471, Clinical Phase I: IR.NREC.1400.004, Clinical Phase II: IR.NREC.1400.009, Clinical Phase III: IR.NREC.1400.016. The ethics of research involving animals also was followed based on the protocol of National Research Ethics Committee.
Patient consent statement
The human sera obtained after signing the informed consent form.
Permission to reproduce material from other sources
Not available.
Clinical trial registration
Not available.
CRediT authorship contribution statement
Shahram Nazarian: Conceptualization of the project, Data analysis, Writing – original draft. Gholamreza Olad: Data curation, To collect tissues and clinical data. Raziyeh Abdolhamidi: Laboratory procedures. Mohammad Javad Motamedi: Laboratory procedures. Rouhollah Kazemi: Laboratory procedures. Emad Kordbacheh: Laboratory procedures. Alireza Felagari: Laboratory procedures. Hanieh Olad: Laboratory procedures. Ali Ahmadi: Writing – original draft, Writing – review & editing. Alireza Bahiraee: Data curation, Writing – original draft. Parisa Farahani: Laboratory procedures. Leila Haghighi: Laboratory procedures, To collect tissues and clinical data. Faezeh Hassani: Laboratory procedures, To collect tissues and clinical data. Vahideh Hajhassan: Laboratory procedures. Mona Nadi: Data curation. Abdolkarim Sheikhi: To collect tissues and clinical data. Jafar Salimian: Conceptualization of the project, Data curation, Data analysis, Interpretation of data, Writing – original draft, Writing – review & editing. Jafar Amani: Conceptualization of the project, Data curation, Data analysis, Interpretation of data, Writing – original draft, Writing – review & editing. All authors read and approved the final manuscript.
Acknowledgments
We thank the Flow cytometry section, pathology lab, molecular biology, and medical diagnostic laboratory of Baqiyatallah Hospital, Neuroscience Research Center, and Animal Room for their cooperation. All authors have read the journal's authorship agreement and that the manuscript has been reviewed and approved.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.molimm.2022.06.007.
Appendix A. Supplementary material
Supplementary material
References
- Abdoli A., Aalizadeh R., Aminianfar H., Kianmehr Z., Azimi E., Emamipour N. Safety and potency of COVIran barekat inactivated vaccine candidate for SARS-CoV-2: a preclinical study. bioRxiv. 2021 doi: 10.1002/rmv.2305. 2021.06.10.447951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arunachalam P.S., Walls A.C., Golden N., Atyeo C., Fischinger S., Li C. Adjuvanting a subunit COVID-19 vaccine to induce protective immunity. Nature. 2021;594(7862):253–258. doi: 10.1038/s41586-021-03530-2. [DOI] [PubMed] [Google Scholar]
- Bachmann M.F., Zinkernagel R.M. Neutralizing antiviral B cell responses. Annu. Rev. Immunol. 1997;15(1):235–270. doi: 10.1146/annurev.immunol.15.1.235. [DOI] [PubMed] [Google Scholar]
- Boosani C.S., Sudhakar A. Cloning, purification, and characterization of a non-collagenous anti-angiogenic protein domain from human α1 type IV collagen expressed in Sf9 cells. Protein Expr. Purif. 2006;49(2):211–218. doi: 10.1016/j.pep.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369(6504):650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y., Ulasli M., Schepers H., Mauthe M., V’kovski P., Kriegenburg F. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J. Virol. 2020;94:4. doi: 10.1128/JVI.01925-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronavirus, N., 2019. Situation Report, 22. World Health Organization. 〈https://wwwwhoint/docs/default-source/coronaviruse/situation-reports/20200211-sitrep-22-ncovpdf〉.
- Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021;21(2):73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming D., Sheahan T., Heise M., Yount B., Davis N., Sims A. Vaccine efficacy in senescent mice challenged with recombinant SARS-CoV bearing epidemic and zoonotic spike variants. PLoS Med. 2006;3(12) doi: 10.1371/journal.pmed.0030525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikid T., Chaudhary S., Goel K., Padda P., Sahu R., Kumar T., et al. Responding to COVID-19 pandemic: Why a strong health system is required. Indian J. Med. Res. 2020;151(2):140. doi: 10.4103/ijmr.IJMR_761_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct. Target. Ther. 2020;5(1):1–14. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., He Y., Guo Y., Zheng B.-J., Jiang S. Receptor-binding domain of SARS-CoV spike protein induces long-term protective immunity in an animal model. Vaccine. 2007;25(15):2832–2838. doi: 10.1016/j.vaccine.2006.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L., Zhao G., Chan C.C., Sun S., Chen M., Liu Z., et al. Recombinant receptor-binding domain of SARS-CoV spike protein expressed in mammalian, insect and E. coli cells elicits potent neutralizing antibody and protective immunity. Virology. 2009;393(1):144–150. doi: 10.1016/j.virol.2009.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinha-Arcieri L.E., Porchia B.M., Carromeu C., Simabuco F.M., Tamura R.E., Ferreira L.C. Expression and purification of a recombinant adenovirus fiber knob in a baculovirus system. Intervirology. 2008;51(3):189–195. doi: 10.1159/000151532. [DOI] [PubMed] [Google Scholar]
- Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396(10249):467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao W., Tamin A., Soloff A., D’Aiuto L., Nwanegbo E., Robbins P.D., et al. Effects of a SARS-associated coronavirus vaccine in monkeys. Lancet. 2003;362(9399):1895–1896. doi: 10.1016/S0140-6736(03)14962-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghalavand M., Saadati M., Salimian J., Abbasi E., Hosseinzadeh G., Ghaleh H.E.G., et al. Biological properties the novel application of N-trimethyl chitosan nanospheres as a stabilizer and preservative in tetanus vaccine. Clin. Exp. Vaccin. Res. 2021;10(1):24. doi: 10.7774/cevr.2021.10.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181(7):1489–1501. doi: 10.1016/j.cell.2020.05.015. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N. Engl. J. Med. 2020;383(20):1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keech C., Albert G., Cho I., Robertson A., Reed P., Neal S., et al. Phase 1-2 trial of a SARS-CoV-2 recombinant spike protein nanoparticle vaccine. N. Engl. J. Med. 2020;383(24):2320–2332. doi: 10.1056/NEJMoa2026920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll M.D., Wonodi C. Oxford-AstraZeneca COVID-19 vaccine efficacy. Lancet. 2020 doi: 10.1016/S0140-6736(20)32623-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koutsakos M., Kedzierska K. A Race to Determine What Drives COVID-19 Severity. Nature Publishing Group; 2020. [DOI] [PubMed] [Google Scholar]
- Leung D.T.M., Chi Hang T.F., Chun Hung M., Sheung Chan P.K., Cheung J.L.K., Niu H., et al. Antibody response of patients with severe acute respiratory syndrome (SARS) targets the viral nucleocapsid. J. Infect. Dis. 2004;190(2):379–386. doi: 10.1086/422040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.-J., Leng C.-H., Lien S.-p, Chi H.-Y., Huang C.-Y., Lin C.-L. Immunological characterizations of the nucleocapsid protein based SARS vaccine candidates. Vaccine. 2006;24(16):3100–3108. doi: 10.1016/j.vaccine.2006.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W.J., Zhao M., Liu K., Xu K., Wong G., Tan W., et al. T-cell immunity of SARS-CoV: implications for vaccine development against MERS-CoV. Antivir. Res. 2017;137:82–92. doi: 10.1016/j.antiviral.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Xu W., Xia S., Gu C., Wang X., Wang Q., et al. RBD-Fc-based COVID-19 vaccine candidate induces highly potent SARS-CoV-2 neutralizing antibody response. Signal Transduct. Target. Ther. 2020;5(1):1–10. doi: 10.1038/s41392-020-00402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logunov D.Y., Dolzhikova I.V., Tukhvatullin A.I., Shcheblyakov D.V. Safety and efficacy of the Russian COVID-19 vaccine: more information needed - authors’ reply. Lancet. 2020;396(10256):e54–e55. doi: 10.1016/S0140-6736(20)31970-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logunov D.Y., Dolzhikova I.V., Zubkova O.V., Tukhvatullin A.I., Shcheblyakov D.V., Dzharullaeva A.S., et al. Safety and immunogenicity of an rAd26 and rAd5 vector-based heterologous prime-boost COVID-19 vaccine in two formulations: two open, non-randomised phase 1/2 studies from Russia. Lancet. 2020;396(10255):887–897. doi: 10.1016/S0140-6736(20)31866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.-X., Liu B.-Z., Deng H.-J., Wu G.-C., Deng K., Chen Y.-K. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Maeda D.L.N.F., Batista M.T., Pereira L.R., de Jesus Cintra M., Amorim J.H., Mathias-Santos C., et al. Adjuvant-mediated epitope specificity and enhanced neutralizing activity of antibodies targeting dengue virus envelope protein. Front. Immunol. 2017;8:1175. doi: 10.3389/fimmu.2017.01175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marblestone J.G., Edavettal S.C., Lim Y., Lim P., Zuo X., Butt T.R. Comparison of SUMO fusion technology with traditional gene fusion systems: enhanced expression and solubility with SUMO. Protein Sci. 2006;15(1):182–189. doi: 10.1110/ps.051812706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M.J., Lyke K.E., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586(7830):589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- Okada M., Takemoto Y., Okuno Y., Hashimoto S., Yoshida S., Fukunaga Y., et al. The development of vaccines against SARS corona virus in mice and SCID-PBL/hu mice. Vaccine. 2005;23(17–18):2269–2272. doi: 10.1016/j.vaccine.2005.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahlad J., Struble L., Lutz W.E., Wallin S.A., Khurana S., Schnaubelt A., et al. Bacterial expression and purification of functional recombinant SARS-CoV-2 spike receptor binding domain. bioRxiv. 2021 doi: 10.1002/pro.4152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: Lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38(1):1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- Rezaie E., Nekoie H., Miri A., Oulad G., Ahmadi A., Saadati M. Different frequencies of memory B-cells induced by tetanus, botulinum, and heat-labile toxin binding domains. Microb. Pathog. 2019;127:225–232. doi: 10.1016/j.micpath.2018.12.003. [DOI] [PubMed] [Google Scholar]
- Sahin U., Muik A., Vogler I., Derhovanessian E., Kranz L.M., Vormehr M., et al. BNT162b2 induces SARS-CoV-2-neutralising antibodies and T cells in humans. medRxiv. 2020 [Google Scholar]
- Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M. COVID-19 vaccine BNT162b1 elicits human antibody and T(H)1 T cell responses. Nature. 2020;586(7830):594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- Saitoh H., Uwada J., Azusa K. Strategies for the expression of SUMO-modified target proteins in Escherichia coli. SUMO Protoc. 2009:211–221. doi: 10.1007/978-1-59745-566-4_14. [DOI] [PubMed] [Google Scholar]
- Sternberg A., Naujokat C. Structural features of coronavirus SARS-CoV-2 spike protein: targets for vaccination. Life Sci. 2020 doi: 10.1016/j.lfs.2020.118056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Zhuang Z., Zheng J., Li K., Wong R.L.-Y., Liu D., et al. Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell. 2020;182(3):734–743. doi: 10.1016/j.cell.2020.06.010. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevarajan I., Nguyen T.H., Koutsakos M., Druce J., Caly L., van de Sandt C.E., et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat. Med. 2020;26(4):453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J.H., Patel N., Haupt R., Zhou H., Weston S. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat. Commun. 2021;12(1):372. doi: 10.1038/s41467-020-20653-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020 doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Frenck Jr.R.W., Falsey A.R., Kitchin N., Absalon J., Gurtman A. Safety and immunogenicity of two RNA-based Covid-19 vaccine candidates. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yu R., Fang T., Yu T., Chi X., Zhang X. Tetanus neurotoxin neutralizing antibodies screened from a human immune scFv antibody phage display library. Toxins. 2016;8(9):266. doi: 10.3390/toxins8090266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Peng Y., Xu H., Cui Z., Williams R.O. The COVID-19 vaccine race: challenges and opportunities in vaccine formulation. AAPS PharmSciTech. 2020;21(6):1–12. doi: 10.1208/s12249-020-01744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Jiang W., He Q., Wang C., Liu B., Zhou P., et al. Early, low-dose and short-term application of corticosteroid treatment in patients with severe COVID-19 pneumonia: single-center experience from Wuhan, China. medRxiv. 2020 2020.03.06.20032342. [Google Scholar]
- Wang Y., Wang L., Cao H., Liu C. SARS-CoV-2 S1 is superior to the RBD as a COVID-19 subunit vaccine antigen. J. Med. Virol. 2021;93(2):892–898. doi: 10.1002/jmv.26320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Wang W., Chen Z., Lu S., Yang F., Bi Z., et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586(7830):572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- Yang L., Tian D., Liu W. Strategies for vaccine development of COVID-19. Sheng wu gong cheng xue bao= Chin. J. Biotechnol. 2020;36(4):593. doi: 10.13345/j.cjb.200094. [DOI] [PubMed] [Google Scholar]
- Yang S., Li Y., Dai L., Wang J., He P., Li C., et al. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect. Dis. 2021 doi: 10.1016/S1473-3099(21)00127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Li Y., Dai L., Wang J., He P., Li C., et al. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect. Dis. 2021;21(8):1107–1119. doi: 10.1016/S1473-3099(21)00127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui F., Kai C., Kitabatake M., Inoue S., Yoneda M., Yokochi S., et al. Prior immunization with severe acute respiratory syndrome (SARS)-associated coronavirus (SARS-CoV) nucleocapsid protein causes severe pneumonia in mice infected with SARS-CoV. J. Immunol. 2008;181(9):6337–6348. doi: 10.4049/jimmunol.181.9.6337. [DOI] [PubMed] [Google Scholar]
- Yu J., Tostanoski L.H., Peter L., Mercado N.B., McMahan K., Mahrokhian S.H., et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369(6505):806–811. doi: 10.1126/science.abc6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N.-N., Li X.-F., Deng Y.-Q., Zhao H., Huang Y.-J., Yang G. A thermostable mRNA vaccine against COVID-19. Cell. 2020;182(5):1271–1283. doi: 10.1016/j.cell.2020.07.024. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., et al. Airway memory CD4+ T cells mediate protective immunity against emerging respiratory coronaviruses. Immunity. 2016;44(6):1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M., Xu D., Li X., Li H., Shan M., Tang J., et al. Screening and identification of severe acute respiratory syndrome-associated coronavirus-specific CTL epitopes. J. Immunol. 2006;177(4):2138–2145. doi: 10.4049/jimmunol.177.4.2138. [DOI] [PubMed] [Google Scholar]
- Zhu F.-C., Guan X.-H., Li Y.-H., Huang J.-Y., Jiang T., Hou L.-H., et al. Immunogenicity and safety of a recombinant adenovirus type-5-vectored COVID-19 vaccine in healthy adults aged 18 years or older: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet. 2020;396(10249):479–488. doi: 10.1016/S0140-6736(20)31605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.-C., Li Y.-H., Guan X.-H., Hou L.-H., Wang W.-J., Li J.-X., et al. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395(10240):1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material