Abstract
Interleukin-10 (IL-10) is an immunomodulatory cytokine that plays important roles in terminating inflammatory responses and preventing tissue damage resulting from autoimmunity. Although these anti-inflammatory actions have led to considerable clinical interest, efforts to exploit IL-10 therapeutically have been hindered by the highly pleiotropic nature of IL-10 and its ability to elicit pro-inflammatory effects in vivo. In this structural snapshot, we review the recent cryo-EM structure of the IL-10 receptor signaling complex, highlighting its unique structural features, insights into the mechanism of receptor sharing by the IL-10 cytokine family, and the implications for manipulating IL-10 signaling therapeutically.
Keywords: cytokine, IL-10, receptor, signaling, JAK, STAT, immunology, inflammation, protein engineering
Introduction
Cytokines are secreted signaling proteins that coordinate nearly all aspects of innate and adaptive immunity (1). The cytokine interleukin-10 (IL-10) serves a unique immunoregulatory role and is required for the termination of inflammatory responses and the prevention of autoimmunity (2–4). Consistent with this, mice or humans lacking IL-10 or the IL-10 receptor (IL-10R) exhibit in severe and persistent autoinflammatory disease (5, 6). The anti-inflammatory effects of IL-10 are primarily due to its ability to potently inhibit inflammatory cytokine production and antigen presentation by activated myeloid cells, including monocytes, macrophages, and dendritic cells (3, 4, 7). Although these features have led to clinical interest in using recombinant IL-10 as a treatment for autoimmune disease, IL-10 is also highly pleiotropic and can elicit significant pro-inflammatory side effects when administered systemically, limiting its therapeutic utility (3, 4). In particular, IL-10 can stimulate inflammatory activities of both CD4+ and CD8+ T cells, potentiating the production of the pro-inflammatory molecules such as interferon-γ (IFN-γ) and granzyme B (8–10).
IL-10 is secreted as a non-covalent homodimer and signals on immune cells by engaging two copies of a dimeric cell surface receptor complex comprising IL-10Rα and IL-10Rβ (3, 4). Whereas IL-10Rα is a private receptor subunit that engages IL-10 with high affinity, IL-10Rβ is a shared receptor subunit that engages IL-10 with extremely low affinity (11, 12). The IL-10 mediated dimerization of IL-10Rα and IL-10Rβ in turn brings together the intracellular receptor-associated kinases JAK1 and TYK2, resulting in the phosphorylation and activation of the transcription factor signal transducer and activator of transcription (STAT) 3 (3, 4).
Assembly and cryo-EM analysis of the IL-10 receptor complex
In order to stabilize the IL-10 receptor complex for structural studies we employed yeast-display based directed evolution to engineer an IL-10 variant with enhanced affinity for IL-10Rβ (13). We designed a mutagenesis library targeting solvent exposed residues in helices α1 and α3 of IL-10, and displayed this library of IL-10 variants on the surface of yeast for selection against the fluorescently labelled extracellular domain (ECD) of IL-10Rβ (13). After five rounds of selection we obtained an IL-10 variant, called “super-10,” which contained four-point mutations IL-10 (N18Y, N92Q, T100D, R104W) and exhibited an over 10,000-fold increase in binding affinity for IL-10Rβ (13). Unlike wild-type (WT) IL-10, super-10 formed a stable hexameric complex with IL-10Rα and IL-10Rβ, enabling structure determination by single particle cryo-electron microscopy (cryo-EM) (13).
Initial cryo-EM analysis of the IL-10 receptor complex was hampered by the presence of both preferred particle orientation as well as apparent flexibility at the central linker of IL-10, which bridges the two trimeric subcomplexes. However, generating 3D reconstructions without applying C2 symmetry to the complex, together with the incorporation of the fluorinated octyl maltoside in sample preparation, mitigated these issues and enabled the 3D reconstruction of the full complex to 3.5Å resolution (Fig. 1A) (13). Although the break in symmetry resulted in substantial differences in map quality between the two ternary subcomplexes, the map for one subcomplex was of sufficient quality to observe side chain densities at the ligand receptor interface, enabling the molecular characterization of the IL-10–IL-10Rα–IL-10Rβ interaction (Fig. 1, A and B) (13).
Figure 1: Cryo-EM structure of the IL-10 receptor complex.
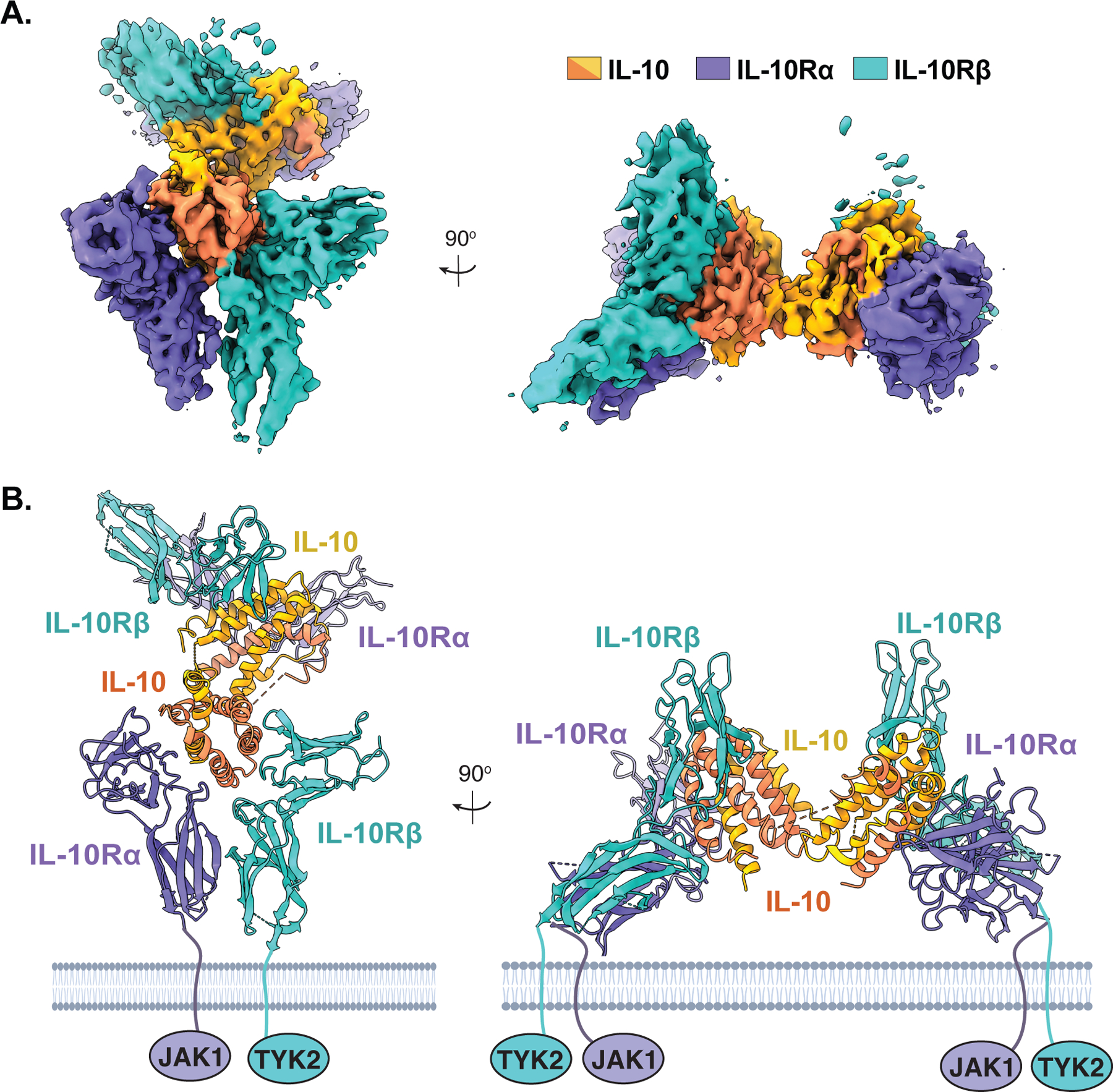
(A) 3.5 Å resolution segmented cryo-EM maps of the hexameric IL-10 receptor complex with density corresponding to IL-10 in yellow/orange, IL-10Rα in purple, and IL-10Rβ in cyan (EMDB-22098). (B) Molecular model of the IL-10 receptor complex with views corresponding to those in (A) (PDB ID: 6X93). Figures were rendered using ChimeraX.
Mechanism of receptor engagement by IL-10 and implications for signal transduction
The hexameric IL-10 receptor complex forms a stelliform structure with the domain swapped, all alpha-helical IL-10 homodimer linking two copies of the IL-10Rα and IL-10Rβ receptor subunits, each of which engage IL-10 via their cytokine-receptor homologous (CRH) domains (Fig. 1B). This assembly results in three unique contact sites, formed between IL-10 and IL-10Rα (site 1), IL-10 and IL-10Rβ (site 2), and “stem contacts” between IL-10Rα and IL-10Rβ (site 3) (Fig. 2A). At site 1, the high affinity private receptor subunit IL-10Rα engages IL-10 helices α1 and α5 via a network of side chain specific polar and electrostatic contacts, as described previously (11). At site 2, loops L2 and L3 of IL-10Rβ clasp helix α3 of IL-10 via both shape complementarity as well as side chain specific contacts, with a total of 15 hydrogen bonds and 9 salt bridges at the interface of super-10 and IL-10Rβ (13). Most notably, the aromatic side chain of Tyr82 in IL-10Rβ inserts between helices α1 and α3 of IL-10 to form key hydrophobic contacts with Asn21 and Met22 in IL-10 (Fig. 2A). Interestingly, the key affinity enhancing mutations in super-10 (N18Y and R104W) both appear to contribute additional hydrophobic surface to accommodate Tyr82, supporting the notion that this residue forms a key interaction hotspot. IL-10Rβ residue Tyr59 similarly inserts between helices α2 and α3 to engage IL-10 residues Leu73, Met77, and Glu81 (Fig. 2A) (13).
Figure 2: Structural basis for receptor sharing by IL-10 family cytokines.
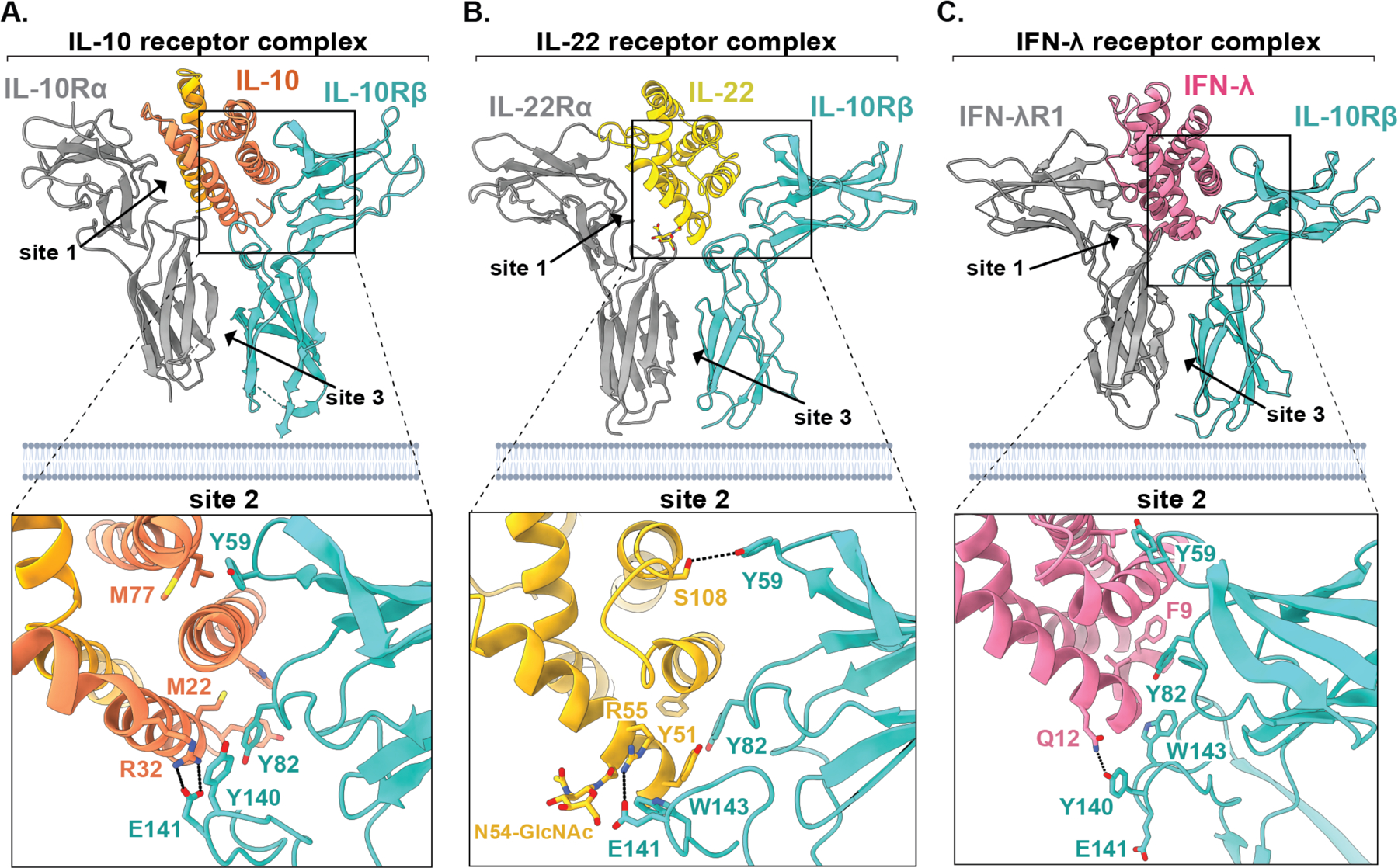
(A-C) Models of the ternary cytokine-receptor complexes for IL-10 (A, PDB ID: 6X93), IL-22 (B, PDB ID: 6WEO), and IFN-λ (C, PDB ID: 5T5W), with closeup views for the site 2 contact between the cytokine and IL-10Rβ (bottom) showing conserved ligand-receptor contact sites between IL-10 family cytokines. Figures were rendered using ChimeraX.
Adjacent to these anchoring aromatic contacts the side chains of Asp25 and Glu96 in IL-10 form electrostatic contacts with the side chains of Lys65 and Lys81 in IL-10Rβ, respectively. Consistent with the importance of these contacts observed in our structure, mutation of IL-10 residues Asp25 or Glu96 to alanine or lysine substantially reduced IL-10 signaling activity, as measured by STAT3 activation across multiple cell types (13).
The highly disparate affinities of IL-10 for the private receptor IL-10Rα (KD ~1 nM) compared to the shared receptor IL-10Rβ (KD = undetectable) suggests a two-step model whereby IL-10 first engages two copies of IL-10Rα to form a 2:2 partial complex on the cell surface. Once captured on the 2-dimensional plasma membrane, the IL-10–IL-10Rα partial complex can then recruit IL-10Rβ subunits, triggering intracellular signal transduction. Thus, whereas IL-10Rα primarily serves to recruit IL-10 to the cell surface, it is the extent of engagement of IL-10Rβ that dictates the subsequent extent of intracellular STAT3 activation (13).
One prediction of this model is that modulating the affinity of IL-10 for IL-10Rβ would specifically alter the Emax of IL-10 signaling, with minimal influence on the EC50, which would instead be controlled by the IL-10Rα-mediated capture of IL-10 on the cell surface. Consistent with this, we found that structure-guided mutation of key IL-10 residues at the IL-10Rβ-binding interface resulted in partial agonism, with these variants eliciting sub-maximal STAT3 activation even at saturating ligand concentrations, with relatively minor differences in EC50 (13). Moreover, super-10, which has a substantially higher affinity for IL-10Rβ compared to WT IL-10 (KD ~100 nM), displayed enhanced STAT3 activation on several cell lines, again with only slight differences in EC50 (13). Importantly, however, the extent of both partial agonism and superagonism elicited by our engineered IL-10 variants was reduced on cells with high IL-10Rβ expression, indicating that signal strength is a function of both receptor affinity and expression level (13).
Together, these observations have important therapeutic implications for the selective targeting of IL-10 to particular cell types. This is exemplified by the finding that our engineered partial agonist IL-10 variants are able to potently suppress inflammatory monocyte and macrophage activation without stimulating pro-inflammatory IFN-γ or granzyme B production by CD8 T cells (13).
Structural basis for receptor sharing by IL-10 family cytokines
IL-10Rβ is a shared cytokine receptor that, in addition to IL-10, engages several other cytokines such as IFN-λ and IL-22 in the context of distinct private receptor subunits (4, 14). Comparison of the cryo-EM structure of the IL-10 receptor complex discussed here to the previously reported crystal structure of the IFN-λ complex (15), as well as to our recently reported crystal structure of the ternary IL-22 receptor complex (16), provides an opportunity to directly compare how IL-10 engages each of these distinct ligands.
Comparison of the site 2 contact between IL-10Rβ and the cytokine ligand across these three structures surprisingly reveals that the cytokine-binding interface in the IL-10 complex is notably larger, burying a total surface area of approximately 2,000 Å2, compared to just 1,500 Å2 and 1,400 Å2 for IFN-λ and IL-22, respectively (Fig 2 A–C). By contrast, the site 3 “stem” contact between IL-10Rβ and the private receptor IL-10Rα (~512 Å2) is substantially smaller than that formed between IL-10Rβ and IFN-λR1 (1,556 Å2) or IL-22 (876 Å) (Fig. 2A–C). Thus, in the context of IL-10, IL-10Rβ relies more heavily on direct cytokine engagement rather than association with the high affinity private receptor subunit, suggesting that receptor binding by IL-10 may be less cooperative than for the other members of the IL-10 family. This is consistent with the observation that pre-association of IL-10 with IL-10Rα does not substantially increase the affinity of IL-10 for IL-10Rβ in vitro, in contrast to what has been observed with IFN-λ and many other cytokine receptor complexes (13, 15, 17).
Analysis of the interaction between IL-10Rβ and these three cytokine ligands also reveals several similarities. In particular, in all three complexes IL-10Rβ uses conserved aromatic residues in loops L2 and L3 to engage helix α3 of the cytokine ligand (Fog. 2 A-C). In particular, the positions of Tyr59 loop L2 and Tyr82 in loop L3 are highly similar across these three structures and make critical contacts with the bound cytokine. The aromatic residues Tyr140 and/or Trp143 in loop L5 of IL-10Rβ also make important contacts in all three complexes, although the relative position of loop L5 varies considerably. When bound to IFN-λ, loop L5 makes a minor contact with the bottom of helix α1 (Fig. 2C), whereas in both the IL-10 and IL-22-bound structures it forms an extensive contact with the front face of the cytokine, utilizing not only Tyr140 but also the adjacent acidic side chain of E141 (Fig. 2, A and B). Notably, unlike IL-10 and IFN-λ, IL-22 contains an N-linked glycosylation at residue N54 in helix α1, which forms an essential contact with E141 in loop L5 of IL-10Rβ (Fig. 2B) (16). In IL-10, this contact is replaced by an apparent salt bridge between E141 and R32 of IL-10 (Fig. 2A), whereas this contact is absent altogether in the context of the IFN-λ structure (Fig. 2C).
Conclusions
The cryo-EM structure of the IL-10 receptor complex described here provides important insights into the mechanisms of receptor binding and the initiation of signaling by the cytokine IL-10, while also offering insights into the nature of degenerate ligand binding by the shared receptor IL-10Rβ. Importantly, the information provided by this structure also enabled the design partial agonist variants of IL-10 with altered cell type selectivity and enhanced therapeutic potential. This use of a cryo-EM structure as a molecular blueprint for the design of novel protein therapeutics represents an important step in development of cryo-EM based drug discovery, with implications for the targeting of other receptor signaling complexes.
Acknowledgements
K.C.G. is an investigator of the Howard Hughes Medical Institute (HHMI). R.A.S. is an HHMI Fellow of the Helen Hay Whitney Foundation. This work was supported by NIH grant NIH R37-AI51321. The cryo-EM map for the IL-10 receptor complex has been deposited in the Electron Microscopy Data Bank (EMDB) under accession code EMD-22098, and the model coordinates have been deposited in the Protein Data Bank (PDB) under accession code 6X93.
Abbreviations:
- Cryo-EM
cryogenic electron microscopy
- IL-10
interleukin-10
- IL-22
interleukin-22
- IFN
interferon
- JAK1
Janus Kinase 1
- TYK2
tyrosine kinase 2
- IL-10R
IL-10 receptor
- K D
dissociation constant
- Emax
maximal effect
- EC50
half-maximal effective concentration
Footnotes
Conflicts of interest: R.A.S. and K.C.G. are inventors on a patent application covering therapeutic IL-10 variants described here. K.C.G. is a founder of Synthekine therapeutics.
References
- 1.Morris R, Kershaw NJ, Babon JJ, The molecular details of cytokine signaling via the JAK/STAT pathway. Protein science : a publication of the Protein Society 27, 1984–2009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A, Interleukin-10 and the interleukin-10 receptor. Annual review of immunology 19, 683–765 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Saraiva M, Vieira P, O’Garra A, Biology and therapeutic potential of interleukin-10. The Journal of experimental medicine 217, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ouyang W, Rutz S, Crellin NK, Valdez PA, Hymowitz SG, Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annual review of immunology 29, 71–109 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W, Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75, 263–274 (1993). [DOI] [PubMed] [Google Scholar]
- 6.Zigmond E et al. , Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity 40, 720–733 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O’Garra A, IL-10 inhibits cytokine production by activated macrophages. Journal of immunology (Baltimore, Md. : 1950) 147, 3815–3822 (1991). [PubMed] [Google Scholar]
- 8.Mumm JB et al. , IL-10 elicits IFNgamma-dependent tumor immune surveillance. Cancer cell 20, 781–796 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Tilg H et al. , Treatment of Crohn’s disease with recombinant human interleukin 10 induces the proinflammatory cytokine interferon gamma. Gut 50, 191–195 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan IH et al. , The Potentiation of IFN-gamma and Induction of Cytotoxic Proteins by Pegylated IL-10 in Human CD8 T Cells. Journal of interferon & cytokine research : the official journal of the International Society for Interferon and Cytokine Research 35, 948–955 (2015). [DOI] [PubMed] [Google Scholar]
- 11.Josephson K, Logsdon NJ, Walter MR, Crystal structure of the IL-10/IL-10R1 complex reveals a shared receptor binding site. Immunity 15, 35–46 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Yoon SI et al. , Structure and mechanism of receptor sharing by the IL-10R2 common chain. Structure (London, England : 1993) 18, 638–648 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saxton RA et al. , Structure-based decoupling of the pro- and anti-inflammatory functions of interleukin-10. Science 371, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Lupardus P, Laporte SL, Garcia KC, Structural biology of shared cytokine receptors. Annual review of immunology 27, 29–60 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendoza JL et al. , The IFN-lambda-IFN-lambdaR1-IL-10Rbeta Complex Reveals Structural Features Underlying Type III IFN Functional Plasticity. Immunity 46, 379–392 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saxton RA et al. , The tissue protective functions of interleukin-22 can be decoupled from pro-inflammatory actions through structure-based design. Immunity 54, 660–672.e669 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spangler JB, Moraga I, Mendoza JL, Garcia KC, Insights into cytokine-receptor interactions from cytokine engineering. Annual review of immunology 33, 139–167 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]


