Abstract
To assess the relative contributions of microbial groups (bacteria, protozoa, and fungi) in rumen fluids to the overall process of plant cell wall digestion in the rumen, representatives of these groups were selected by physical and chemical treatments of whole rumen fluid and used to construct an artificial rumen ecosystem. Physical treatments involved homogenization, centrifugation, filtration, and heat sterilization. Chemical treatments involved the addition of antibiotics and various chemicals to rumen fluid. To evaluate the potential activity and relative contribution to degradation of cell walls by specific microbial groups, the following fractions were prepared: a positive system (whole ruminal fluid), a bacterial (B) system, a protozoal (P) system, a fungal (F) system, and a negative system (cell-free rumen fluid). To assess the interactions between specific microbial fractions, mixed cultures (B+P, B+F, and P+F systems) were also assigned. Patterns of degradation due to the various treatments resulted in three distinct groups of data based on the degradation rate of cell wall material and on cell wall-degrading enzyme activities. The order of degradation was as follows: positive and F systems > B system > negative and P systems. Therefore, fungal activity was responsible for most of the cell wall degradation. Cell wall degradation by the anaerobic bacterial fraction was significantly less than by the fungal fraction, and the protozoal fraction failed to grow under the conditions used. In general, in the mixed culture systems the coculture systems demonstrated a decrease in cellulolysis compared with that of the monoculture systems. When one microbial fraction was associated with another microbial fraction, two types of results were obtained. The protozoal fraction inhibited cellulolysis of cell wall material by both the bacterial and the fungal fractions, while in the coculture between the bacterial fraction and the fungal fraction a synergistic interaction was detected.
Bacteria, protozoa, and fungi have been shown to be the microorganisms involved in plant cell wall digestion in the rumen. However, due to the difficulty of separating each microbial group in the rumen, to difficulties in measuring fungal biomass, and to the complex nature of the rumen ecosystem, the precise role and overall contribution of each microbial group to the degradation and fermentation of plant cell wall material is not understood. In spite of complicated interrelationships among the microorganisms (e.g., bacteria, protozoa, and fungi) in the rumen ecosystem, bacteria are believed to play a major role because of their numerical predominance and metabolic diversity (7). However, protozoa have been shown to digest from 25 to 30% of total fiber. The extent of the involvement of fungi, however, has not yet been estimated. Interaction effects between microorganisms can range from synergism to antagonism and depend on the microbial groups and species involved and the type of substrate used. In vitro examinations to estimate the roles that bacteria, protozoa, and fungi play in plant cell wall digestion in the rumen microbial ecosystem have been attempted. Nevertheless, many methodological problems remain, such as how to prepare the in vitro microbial suspensions and how to simulate the natural environment. Many kinds of artificial rumen ecosystems have been constructed. The objective of our experiments was to estimate the relative roles of bacteria, protozoa, and fungi in plant cell wall digestion under artificial circumstances using physical and chemical treatments to inhibit the growth of or to select for specific microbial groups.
MATERIALS AND METHODS
Preparations of cell wall fractions.
The substrate used in these experiments was cell wall fractions of Orchard grass hay. It was ground and passed through a 1-mm-pore-size screen prior to being used to make cell wall component preparations (cell walls consisting largely of cellulose and hemicellulose). The ground material was boiled for 1 h in a 1% sodium dodecyl sulfate solution, and the insoluble residue (cell wall components) was extensively washed to remove other cellular components and detergent before it was dried.
Collection of rumen contents.
Rumen contents used to fractionate the microbial group were collected from the rumen of a lactating Jersey cow (450 kg [live weight]). The animals were prepared with a permanent cannula into the rumen and were housed untethered in pens. Diets were fed in two equal meals at 06:00 and 16:00 h, and the ration consisted of 60% rolled barley, 22% dried alfalfa pellet, 16% canola meal (<1% salt plus dicalcium), and 2% corn steep powder. Water was available ad libitum, and animals received proprietary mineral and vitamin supplements in the form of licks. All samples isolated from the rumen were withdrawn 4 h after the morning ration had been consumed. Collected rumen contents were strained through four layers of cheesecloth and brought immediately to the laboratory.
Separation of microbial fractions.
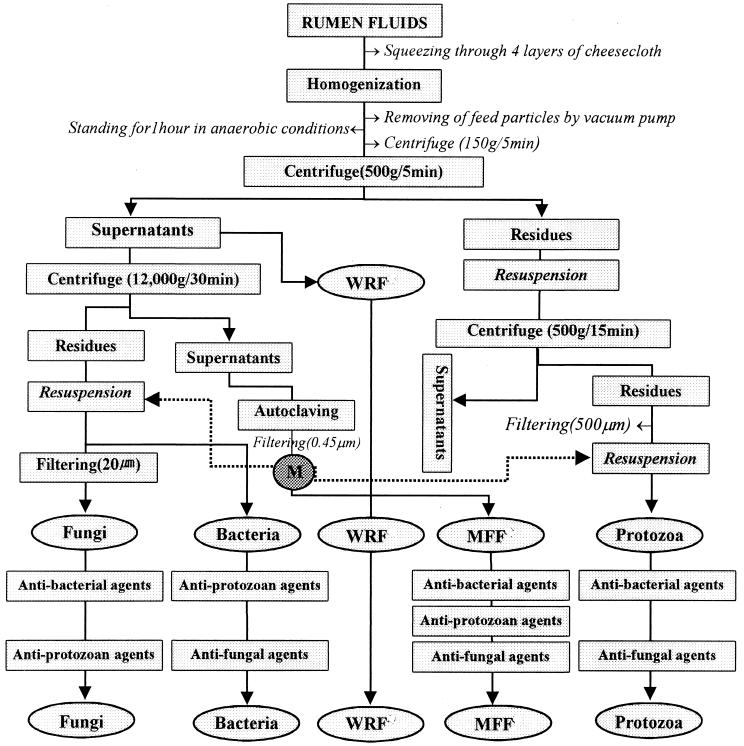
For the separation of microbial fractions from the rumen contents, we used physical and chemical treatments as shown in Fig. 1. All subsequent operations were conducted under anaerobic conditions as described by Bryant (6). Physical treatments involved homogenization, centrifugation, filtration, and heat sterilization. Strained rumen contents were homogenized by an electric mixer (Brinkmann homogenizer, Model-PT 10/35; Brinkmann Instruments Co., Geneva, Switzerland) and poured into a separating funnel that had been gassed with oxygen-free CO2. The sample was incubated under anaerobic conditions at 39°C for up to 60 min to allow small feed particles to buoy up and the microbial fraction to sediment at the bottom.
FIG. 1.
Separation protocols for microbial fractions (bacteria, protozoa, and fungi) from rumen fluids by chemical treatment. M, materials from autoclaved ruminal fluid (MFF); WRF, whole rumen fluid.
Small feed particles that had risen to the surface were removed by using a vacuum tube, and most of the lower liquid portion was then centrifuged at slow speed (150 × g, for 5 min). The remaining residue was resuspended in microbe-free-fraction (MFF) solution (see below) and used to prepare the protozoal fraction. The supernatant was carefully collected to prepare the whole rumen fluid fraction, the bacterial fraction, and the fungal fraction. The resuspended protozoal fraction was washed by centrifugation (500 × g, 15 min) five times in the MFF solution in order to remove bacterial cells and fungal zoospores as completely as possible and finally resuspended in the same solution. A portion of the supernatant centrifuged at slow speed (150 × g, 5 min) was used as the whole rumen fluid (positive system). Bacterial and fungal fractions were recovered from the other aliquots of the supernatant by centrifuging them at 12,000 × g for 30 min. A portion of the supernatant was autoclaved and microfiltered using a sterilization filter (0.45 μm [pore size]; Nalgene Co., Rochester, N.Y.) and is referred to as the MFF; it was used for the negative system or to suspend the collected cell fraction. The bacterial and fungal pellet was resuspended in MFF solution, gassed with oxygen-free CO2, and warmed to 39°C before use. After these various physical treatments, chemical treatments were also performed. The following antibiotics and other chemicals were used: antibacterial agents (streptomycin sulfate, penicillin G, potassium, and chloramphenicol [0.100 mg/ml each]), antiprotozoal agents (copper sulfate [0.15 mg/ml], sodium lauryl sulfate [0.010 mg/ml], and dioctyl sulfosuccinate sodium salts [0.200 mg/ml]), and antifungal agents (cychloheximide [0.05 mg/ml] and nystatin [200 U/ml]).
Culture conditions and treatments.
The anaerobic culture techniques of Hungate as described by Bryant (6) were used for all incubations. The medium used in the experimental cultures was based on the liquid semidefined medium B of Lowe et al. (20), except that antibiotics were omitted and soluble carbon sources were replaced with 75 mg of Orchard grass cell wall material. The following monocultural systems were prepared to evaluate the potential activities and relative contributions to degradation of Orchard grass cell wall by specific microbial fractions: a positive system (whole ruminal fluid without chemical treatment to measure activity of all microbial groups), a bacterial (B) system, a protozoal (P) system, a fungal (F) system, and a negative system (autoclaved ruminal fluid plus the antimicrobial agents listed above). The following cocultural treatments were also prepared to assess the interactions between specific microbial groups: a B+P system (physically fractionated bacterial and protozoal groups plus antifungal agent), a B+F system (physically fractionated bacterial and fungal groups plus antiprotozoal agent), and a P+F system (physically fractionated protozoal and fungal groups plus antibacterial agent). Antimicrobial agents were prepared so that 0.1 ml of solution was added per ml of broth to give the desired concentrations. In the positive system or treatments with only one or two antimicrobial agents, water was added to maintain equivalent volumes. Antimicrobial agents were added to the incubation tubes before inoculating them with microbial fractionates.
After physical and chemical treatments, microbial populations were enumerated using a roll tube and microscopy, and the microbial markers were also detected. Total bacteria and fungal zoospores were enumerated microscopically with a glass slide using a modification of the procedures of Holdman et al. (13). Viable cells were counted by the cell- or thallus-forming unit method for bacteria or fungi, respectively, using a roll tube (14, 38) with five replicates per dilution. Samples were also fixed in methylgreen formalin salt (MFS) solution and TBFS solution (distilled water, 900 ml; 35% formaldehyde solution, 100 ml; trypan blue, 2 g; NaCl, 8 g; dark blue solution) for the enumeration of dead or viable protozoa by the methods of Okimoto and Imai (27). Protozoa fixed in MFS and TBFS solutions were appropriately diluted in the same solution and counted with a plankton counter desk glass by microscopy.
The determination of DAPA (2,6-diaminopimelic acid), AEP (aminoethylphosphonic acid), and chitin as microbial markers for bacteria, protozoa, and fungi was done according to the methods of Olubobokun et al. (28), Julian and Czerkawski (19), and Orpin (31), respectively.
Sampling and analysis.
The degradation rate of Orchard grass cell wall material and enzyme activities were determined in triplicate for each treatment. Cultures were harvested after 12, 24, 36, 48, 72, and 96 h of incubation. Supernatant from each microbial culture was separated from sedimentable material by centrifugation at 3,000 rpm for 20 min. Supernatants from three replicate cultures were analyzed for enzyme activity. One-half milliliter of the supernatant (crude enzyme solution) was mixed with 0.5 ml of 1% carboxymethyl cellulose (CMC) solution in 0.05 M citrate buffer (pH 5.5). The reaction proceeded for 1 h at 55°C without shaking, and the reaction was stopped by boiling for 5 min. Boiled samples were centrifuged at 7,000 rpm for 5 min, and reducing sugar produced in the supernatants was measured colorimetrically by using the dinitrosalicylic acid method of Miller et al. (22). One unit of enzyme activity was defined as the amount of enzyme that produced 1 mmol of glucose equivalent of reducing sugar per min. Xylanase activity was assayed with 1 ml of 2% (wt/vol) oat spelts xylan in 0.5 M potassium phosphate buffer (pH 6.5). Reducing sugar was assayed as described above. After treatment with 1 M NaOH at 100°C to remove adherent microorganisms, three rinses with absolute alcohol at 60°C, and two rinses in running distilled water, the pelleted cell wall material was dried to a constant weight at 78°C for 12 h and used to calculate the degradation rate of Orchard grass cell wall.
Statistical analysis was performed by using Duncan's new multiple range test according to the general linear model procedures of SAS (36).
RESULTS
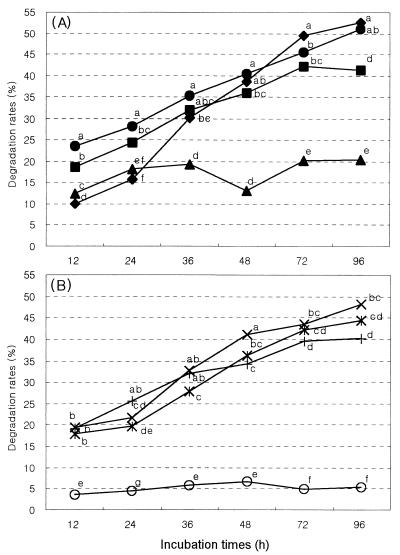
Microbial populations and markers in each microbial fraction obtained by the treatment of physical and chemical are presented in Tables 1 and 2, respectively. Fungal populations existed in the bacterial and protozoal fractions in small amounts, but chitin was not detected in the bacterial and protozoal fractions. This result supported the idea that fractionation methods for separating of bacterial, protozoal, and fungal fractions were enough to proceed with the experiment. Time course analyses of the degradation rates of Orchard grass cell wall by mono- or cocultures of the various microbial fractions are presented in Fig. 2. The various monocultural treatments used to evaluate the potential roles and relative contributions of bacterial, protozoal, and fungal fractions to the degradation of Orchard grass cell wall resulted in three distinct rates as follows: positive and F systems > B system > P and negative systems. The greatest overall degradation rate occurred in the positive and F systems (50.82 and 52.18%, respectively, after 96 h of incubation), indicating that fungal activity was potentially sufficient to account for all of the observed degradation. The bacterial fraction (B system) alone resulted in significantly (P < 0.05) less degradation after prolonged incubation (46% after 96 h incubation). The protozoal fraction (P system) alone did not degrade the cell wall material. Degradation (ca. 4 to 8%) also occurred in the negative system in the absence of microbial activity. Since the cell wall material contained no soluble components, this small amount of apparent degradation may be due to measuring errors.
TABLE 1.
Microbial populations of bacterial, protozoan, and fungal fractions separated from rumen fluids by physical and chemical treatments
| Microbial population | Mean microbial fraction ± SEa of:
|
||||
|---|---|---|---|---|---|
| WRF | Bacteria | Protozoa | Fungi | MFF | |
| Bacteria | |||||
| Total cell count (1011) | 36.27 ± 11.14b | 69.54 ± 13.49a | 3.75 ± 0.08c | 0.02 ± 0.01d | ND |
| CFU (108) | 17.32 ± 2.18a | 6.98 ± 3.87b | ND | ND | ND |
| Protozoa | |||||
| Live cells (104) | 11.26 ± 0.49a | ND | 3.87 ± 1.95b | ND | ND |
| Dead cells (105) | 35.26 ± 9.17a | ND | 2.42 ± 1.21b | ND | ND |
| Fungi | |||||
| Total cells (105) | 4.26 ± 1.13b | 0.17 ± 0.02c | 3.75 ± 0.08b | 13.38 ± 0.15a | ND |
| Zoospore TFUb (104) | 9.28 ± 3.66 | ND | ND | 6.32 ± 1.16 | ND |
WRF, whole rumen fluid; ND, not detected. Values in the same row with different lowercase roman superscripts differ significantly (P < 0.05).
TFU, thallus-forming unit(s).
TABLE 2.
Concentrations of microbial markers (DAPA, AEP, and chitin) in bacterial, protozoan, and fungal fractions separated from rumen fluids after physical and chemical treatments
| Microbial markers | Mean microbial fractions (μg/ml) ± SEa of:
|
||||
|---|---|---|---|---|---|
| WRF | Bacteria | Protozoa | Fungi | MFF | |
| DAPA | 414.56 ± 45.47 | 519.13 ± 33.25 | – | – | – |
| AEP | 298.14 ± 63.56 | – | 119.26 ± 21.08 | – | – |
| Chitin | 813.47 ± 39.68 | – | – | 643.93 ± 44.69 | – |
WRF, whole rumen fluid; –, not detected or trace amounts.
FIG. 2.
Degradation rates of cell wall extracted from Orchard grass by the monoculture system to assess the relative contributions of digestion by bacterial (■), protozoan (▴), and fungal (⧫) systems with the positive system (●) as a control (A) and a mixed culture system to assess their interactions: B+P (+), B+F (×), and P+F (✠) systems with negative system (○) as a control (B). Various microbial fractions were separated from bovine rumen fluids by physical and chemical treatments as shown in Fig. 1. The lowercase letters above the spots indicate statistical significance; mean values with different letters are significantly different (P < 0.05).
The coculture systems (B+P, B+F, and P+F) were used to assess the interactions of component microbial groups. In general, coculture systems showed a decrease in cellulolysis compared to the monoculture systems. The protozoal fraction seemed to inhibit the degradation rate of cell wall material by both the bacterial and the fungal fractions. In contrast, cocultures between the bacterial fraction and the fungal fraction seemed to display a synergistic interaction.
Within all of the treatments, cell wall degradation was accompanied by a decrease in supernatant pH. The initial pH of the culture media was 6.67 ± 0.01. After fermentation, the pH of the culture fluid from the positive, B, P, F, and negative systems were 6.23, 6.38, 6.55, 6.21, and 6.65, respectively. The pH values for the coculture systems of B+P, B+F, and P+F were 6.41, 6.38, and 6.53, respectively (Fig. 2). The pH values of the culture fluids from the P and negative systems were highest and did not change significantly throughout the incubation periods. These results indicated that the protozoal fraction cannot ferment Orchard grass cell wall extracts. The amount of reducing sugar in the culture medium increased throughout the incubation period, except for the protozoan monoculture. The high correlation coefficient (92.37%) between cell wall digestion and reducing sugar content in the culture supernatant suggests that reducing sugar was released from the cell wall material by microbial degradation (data not shown).
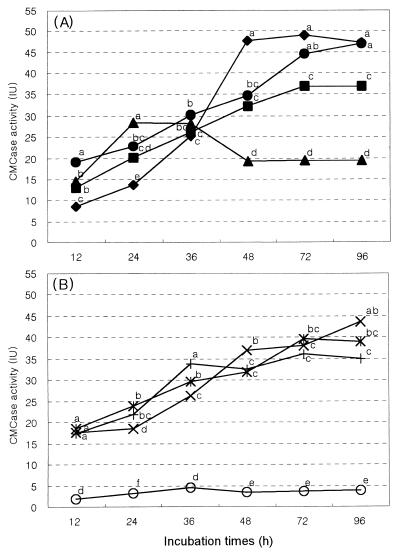
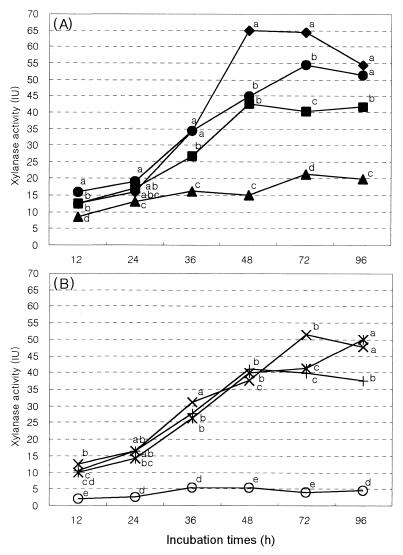
Measurement of endoglucanase (β-1,4-glucan glucanohydrolase, EC3.2.1.4), activity was made with CMC as the assay substrate. The carboxymethyl cellulase (CMCase) activities of the culture supernatants for the positive and F systems were higher than that for the other monoculture systems, similar to the trend observed with cell wall degradation rates (Fig. 3). The results also show that the amount of CMCase activity released from the bacterial fraction is not much greater than that released by the fungal fraction. CMCase activity was lowest in the protozoan fraction, except for the negative system. There was little or no activity (usually <5 μmol ml−1 min−1) in the negative system. There was little or no activity (usually <5 μmol ml−1 min−1) in the negative system. Xylanase production in the F system developed more rapidly and was higher than that in the B system (Fig. 4). After 48 h of incubation, xylanase activity was 1.3 times higher than that of the B system. There was little xylanase activity (usually <20 μmol ml−1 min−1) in the P system.
FIG. 3.
CMCase activity (IU, μmol of glucose min−1 ml−1) in the culture supernatants of a monoculture system to assess the relative contributions of digestion by bacterial (■), protozoan (▴), and fungal (⧫) systems with a positive system (●) as a control (A) and a mixed culture system to assess their interactions: B+P (+), B+F (×), and P+F (✠) systems with a negative system (○) as a control grown with Orchard grass cell wall as a substrate (B). Various microbial fractions were separated from bovine rumen fluids by physical and chemical treatments as shown in Fig. 1. The lowercase letters above the spots indicate statistical significance; mean values with different letters are significantly different (P < 0.05).
FIG. 4.
Xylanase activity (IU, μmol of glucose min−1 ml−1) in the culture supernatants of a monoculture system to assess the relative contributions of digestion by bacterial (■), protozoan (▴), and fungal (⧫) systems with a positive system (●) as a control (A) and a mixed culture system to assess their interactions: B+P (+), B+F (×), and P+F (✠) systems with a negative system (○) as a control grown with Orchard grass cell wall as a substrates (B). Various microbial fractions were separated from bovine rumen fluids by physical and chemical treatments as shown in Fig. 1. The lowercase letters above the spots indicate statistical significance; mean values with different letters are significantly different (P < 0.05).
CMCase activity was higher in the B+F coculture system than in the other cocultures (i.e., the B+P and P+F systems). Coculture between the bacterial fraction and the fungal fraction (B+F system) also increased xylanase activity to a level higher than that in the cultures of the bacterial or fungal fractions alone. Thus, increased cell wall-degrading enzyme (CMCase and xylanase) activity parallels the increase in cell wall digestion by microbial fractions.
DISCUSSION
Although the interactions that occur among the rumen microbes (bacteria, protozoa, and fungi) have been reviewed by Wolin and Miller (42) and interactions involved in fiber degradation have also been reviewed by Jouany (18), the relative contributions of bacteria, protozoa, and fungi to cell wall degradation are still poorly understood. Ours is the first study conducted to assess the relative contribution to the overall process of cell wall digestion by microbial fractions in rumen fluids.
With the monocultures (i.e., the bacterial, protozoal, or fungal fraction alone), cellulolysis of Orchard grass cell wall by the bacterial fraction was significantly highest (P < 0.05) during the early stages of incubation, but cellulolysis by the fungal fraction was highest during the late stages of incubation. The protozoal fraction alone did not degrade the cell wall material. These results suggest that rumen bacteria quickly die and lyse after prolonged incubation and that anaerobic rumen fungi show a marked lag in their in vitro ability to degrade cell wall materials. The relative contributions of microbial fractions to the overall process of cell wall digestion are thus in the following order: fungal fraction > bacterial fraction > protozoal fraction. Although the rumen bacteria are believed to be responsible for most of the feed digestion in the rumen because of their numerical predominance and metabolic diversity (7), the results obtained in our study suggested that the contribution of the fungal fraction to cell wall degradation may greatly exceed that of the bacteria. The ability of the anaerobic fungi to penetrate deeply into plant tissues that are not normally accessible to bacteria (2) suggests that they have a special role in fiber digestion. In the present study, the ability of a fungal fraction to utilize cell wall components of plant material has been demonstrated. Fungal activity could potentially be sufficient to account for all of the observed degradation.
Onodera et al. (29) showed that mixed rumen protozoa participate in cellulose digestion in the rumen ecosystem with an endogenous 1,4-β-glucanase. Coleman (8, 9, 10), Newbold et al. (26), and Williams and Withers (41) suggested that as much as 62% of the cellulolytic activity associated with plant material in the rumen may be protozoal in origin. However, in our experiments, the protozoal fraction alone did not progressively degrade cell wall material. Protozoa are able to digest bacterial and fungal cells, nutrients from the culture medium, microbial fermentation products, and other protozoa. Small feed particles are also readily ingested by protozoa (11). However, our results indicate that the protozoal fraction failed to uptake the insoluble large feed particles prepared for our experiments. We therefore may not have assessed the direct quantitative contribution of the protozoal fraction to cell wall degradation. Bacteria adsorb nutrients onto the cell wall and hydrolysis occurs at this site (40). Hydrolysis of nutrients by the rumen protozoal fraction can occur intracellularly, and the factors affecting engulfment are more important. In future experiments, differences in the mechanisms by which different microorganisms access feed particles should be taken into account when assessing the relative contributions of each organism group to nutrient digestion. The relative contributions to cell wall digestion from the bacterial and protozoal fractions may be underestimated in our results since the substrate we used was composed of relatively large particles.
In the coculture systems (B+P, B+F, and P+F) there was a decrease in cellulolysis compared to the monoculture systems. The protozoal fraction inhibited cellulolysis of cell wall material by both the bacterial and the fungal fractions. In the cocultures between bacteria and fungi, a synergistic interaction was detected.
The protozoal fraction alone did not progressively degrade the cell wall material in our experiments, and in coculture with either the bacterial fraction (B+P system) or the fungal fraction (P+F system) degradation was inhibited compared to the bacterial or fungal monoculture. In general, in the early stages (1 to 2 days) of incubation, differences in degradation rates were not marked, but as the incubation time increased the differences between the monocultures and cocultures became more pronounced. When the fungal fraction was incubated with the protozoal fraction, a steady decline in the degradation rate was observed, accounting for a 15.58% reduction at the end of the incubation period. These results differ from earlier studies. Yoder et al. (43), for example, reported that the addition of washed rumen protozoa to a washed suspension of rumen bacteria substantially increased cellulose digestion and acid production. Onodera et al. (30) also observed that the addition of protozoa to bacteria increased cellulose digestion. Moreover, Orpin (32) reported that anaerobic fungi and rumen protozoa may be complementary rather than competitive in a nature system.
The negative effects observed in the B+P and P+F systems may be a consequence of the culture conditions used in our experiments. Another possible explanation is that fungal sporangium can be degraded by protozoal chitinolytic enzymes (23), although these were not observed in the present study. Our results also indicated that controlling the population size in rumen protozoal fractions offers an opportunity for altering rumen fermentation and the productivity of ruminant animals. Anaerobic fungal numbers have been shown to increase in defaunated animals. Romulo et al. (34, 35) showed two- to fourfold increases in zoospores and zoosporangia of anaerobic fungi in defaunated sheep. Soetanto et al. (37) and Ushida et al. (39) found increased fungal populations in defaunated animals and observed increased digestion of the high-fiber diet fed to these animals. In contrast, Newbold and Hillman (25) observed only small increases in fungal zoospores in defaunated ruminants.
The rumen is a highly complex ecosystem that contains many different microbial species and has a great potential for intermicrobial associations. In interactions in the B+P system, we observed a synergistic interaction by detecting higher enzyme activities in the B+P system than in the fungal monoculture. Many relationships are known to exist among microorganisms in the rumen. Various workers have shown that anaerobic fungi interact with hydrogen-utilizing bacteria (3, 4, 12, 15, 16, 17, 21, 24, 33). In the presence of hydrogen-utilizing bacteria such as methanogens, anaerobic fungi are more effective at degrading cellulose. However, in a more recent study on the interactions between anaerobic fungi and rumen cellulolytic bacteria, the bacteria were observed to inhibit the ability of fungi to hydrolyze cellulose (4, 5). The inhibition of fungal activity is caused by an extracellular protein released by cellulolytic bacteria (5).
It is well known that the enzymatic activities of fungi, combined with the particular penetrating growth of the rhizoidal system, leads to weakening and particle size reduction of plant cell walls (1, 3, 32). Perhaps these activities contribute to the high rate of cell wall degradation observed in our study.
ACKNOWLEDGMENTS
This research was partially supported by High-Technology Development Project of the Ministry of Agriculture and Forestry in Korea, the Brain Korea 21 Project, and KOSEF (Korea Science and Engineering Foundation, Taejon, Korea).
We thank K. Jacober, Research Centre, Agriculture and Agri-Food Canada, Lethbridge, Alberta, for helping with manuscript preparation.
REFERENCES
- 1.Akin D E, Barton II F E, Coleman S W. Structural factors affecting leaf degradation of old world blue stem and weeping love grass. J Anim Sci. 1983;56:1434–1446. [Google Scholar]
- 2.Bauchop T. The anaerobic fungi in rumen fibre digestion. Agric Environ. 1981;6:339–348. [Google Scholar]
- 3.Bauchop T, Mountfort D O. Cellulose fermentation by a rumen anaerobic fungus in both the absence and the presence of rumen methanogens. Appl Environ Microbiol. 1981;42:1103–1110. doi: 10.1128/aem.42.6.1103-1110.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernalier A, Fonty G, Bonnemoy F, Gouet P. Degradation and fermentation of cellulose by the rumen anaerobic fungi in axenic cultures or in association with cellulolytic bacteria. Curr Microbiol. 1992;25:143–148. [Google Scholar]
- 5.Bernalier A, Fonty G, Bonnemoy F, Gouet P. Inhibition of the cellulolytic activity of Neocallimastix frontalis by Ruminococcus flavefaciens. J Gen Microbiol. 1993;139:873–880. doi: 10.1099/00221287-139-4-873. [DOI] [PubMed] [Google Scholar]
- 6.Bryant M P. Nutritional features and ecology of predominant anaerobic bacteria of the intestinal tract. Am J Clin Nutr. 1973;27:1313–1319. doi: 10.1093/ajcn/27.11.1313. [DOI] [PubMed] [Google Scholar]
- 7.Cheng K-J, Forsberg C W, Minato H, Costerton J W. Microbial ecology and physology of feed degradation within the rumen. In: Tsuda T, Sasaki Y, Kawashima R, editors. Physiological aspects of digestion and metabolism in ruminants. Toronto, Ontario, Canada: Academic Press; 1991. pp. 595–624. [Google Scholar]
- 8.Coleman G S. The cellulase content of 15 species of entodiniomorphid protozoa, mixed bacteria and plant debris isolated from the ovine rumen. J Agric Sci Camb. 1985;104:349–360. [Google Scholar]
- 9.Coleman G S. Protozoal-bacterial interaction in the rumen. In: Nolan J V, Leng R A, Demeyer D I, editors. The roles of protozoa and fungi in ruminant digestion (OECD/UNE International Seminar). Armidale, Australia: Penambul Books; 1989. pp. 13–26. [Google Scholar]
- 10.Coleman G S. The distribution of carboxymethylcellulase between fractions from the rumen of sheep containing no protozoa or one of five different protozoal populations. J Agric Sci Camb. 1986;106:121–127. [Google Scholar]
- 11.Coleman G S. The rate of uptake and metabolism of starch grains and cellulose particles by Entodinium species, Eudiplodinium maggii, some other entodiniomorphid protozoa and natural protozoal populations taken from the ovine rumen. J Appl Bacteriol. 1992;73:507–513. doi: 10.1111/j.1365-2672.1992.tb05013.x. [DOI] [PubMed] [Google Scholar]
- 12.Fonty G, Gouet P, Sante V. Influence d'une bactérie méthanogène sur l'activit cellulolytique de duex espèces de champignons du rumen, in vitro. Résultats préliminaires. Reprod Nutr Dev. 1988;28:133–134. [Google Scholar]
- 13.Holdman L V, Gato E P, Moore W E C. Anaerobic laboratory manual. 4th ed. Blacksburg, Virginia: Virginia Polytechnic Institute and State University; 1977. [Google Scholar]
- 14.Hungate R E. The rumen and its microbes. New York, N.Y: Academic Press, Inc.; 1966. [Google Scholar]
- 15.Irvine H L, Stewart C S. Interactions between anaerobic cellulolytic bacteria and fungi in the presence of Methanobrevibacter smithii. Lett Appl Microbiol. 1991;12:62–64. [Google Scholar]
- 16.Joblin K N, Naylor G E, Williams A G. The effect of Methanobrevibacter smithii on the xylanolytic activity of rumen fungi. Appl Environ Microbiol. 1990;56:2287–2295. doi: 10.1128/aem.56.8.2287-2295.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Joblin K N, Williams A G. Effects of cocultivation of ruminal chytrid with Methanobrevibacter smithii on lucerne stem degradation and extracellular fungal enzyme activities. Lett Appl Microbiol. 1991;12:121–124. [Google Scholar]
- 18.Jouany J P. Effects of diet on populations of rumen protozoa in relation to fiber digestion. In: Nolan J V, Leng R A, Demeyer D I, editors. The roles of protozoa and fungi in ruminant digestion (OECD/UNE International Seminar). Armidale, Australia: Penambul Books; 1989. pp. 59–74. [Google Scholar]
- 19.Julian C J, Czerkawski J W. The isolation of rumen anaerobic bacteria and protozoa using a marker. J Anim Sci. 1974;56:1100–1108. [Google Scholar]
- 20.Lowe S E, Theodorou M K, Trinci A P J, Hespell R B. Growth of anaerobic rumen fungi on defined and semidefined media lacking rumen fluid. J Gen Microbiol. 1985;131:2225–2229. [Google Scholar]
- 21.Marvin-Sikkema F D, Richardson A J, Stewart C S, Gottschal J C, Prins R A. Influence of hydrogen consuming bacteria on cellulose degradation by anaerobic fungi. Appl Environ Microbiol. 1990;56:3793–3797. doi: 10.1128/aem.56.12.3793-3797.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J L, Blum R, Glennon W E, Burton A L. Measurement of carboxymethyl cellulase activity. Anal Biochem. 1960;1:127–132. [Google Scholar]
- 23.Morgavi D P, Sakurada M, Tomita Y, Onodera R. Presence in rumen bacterial and protozoal populations of enzymes capable of degrading fungal cell walls. Microbiology. 1994;140:631–636. doi: 10.1099/00221287-140-3-631. [DOI] [PubMed] [Google Scholar]
- 24.Mountfort D O, Asher R A, Bauchop T. Fermentation of cellulose to methane and carbon dioxide by a rumen anaerobic fungus in a triculture with Methanobrevibacter sp. strain RA1 and Methanosarcina barkeri. Appl Environ Microbiol. 1982;44:128–134. doi: 10.1128/aem.44.1.128-134.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Newbold C J, Hillman K. The effect of ciliate protozoa on the turnover of bacterial and fungal protein in the rumen of sheep. Lett Appl Microbiol. 1990;11:100–102. [Google Scholar]
- 26.Newbold C J, Griffin P W, Wallace R J. Interactions between rumen bacteria and ciliate protozoa in their attachment to barley straw. Lett Appl Microbiol. 1989;8:63–66. [Google Scholar]
- 27.Ogimoto K, Imai S. Rumen protozoa. In: Ogimoto K, Imai S, editors. Atlas of rumen microbiology. Tokyo, Japan: Japan Scientific Societies Press; 1981. pp. 9–67. [Google Scholar]
- 28.Olubobokun J A, Craig W M, Nipper W A. Characteristics of protozoal and bacterial fractions from microorganisms associated with ruminal fluid or particles. J Anim Sci. 1988;66:2701–2710. doi: 10.2527/1990.68103360x. [DOI] [PubMed] [Google Scholar]
- 29.Onodera R, Yamasaki N, Murakami K. Effect of inhibition by ciliate protozoa on the digestion of fibrous materials in vivo in the rumen of goats and in an in vitro rumen microbial ecosystem. Agric Biol Chem. 1988;52:2635–2637. [Google Scholar]
- 30.Onodera R, Murakami K, Ogawa K. Cellulose-degrading enzyme activities of mixed rumen ciliate protozoa from goats. Agric Biol Chem. 1988;52:2639–2640. [Google Scholar]
- 31.Orpin C G. The occurrence of chitin in the cell-walls of the rumen organisms Neocallimastix frontalis, Piromonas communis and Sphaeromonas communis. J Gen Microbiol. 1977;99:215–218. doi: 10.1099/00221287-99-1-215. [DOI] [PubMed] [Google Scholar]
- 32.Orpin C G. The role of ciliate protozoa and fungi in the rumen digestion of plant cell walls. Anim Feed Sci Technol. 1984;10:121–143. [Google Scholar]
- 33.Roger V, Bdrnalier A, Grenet E, Fonty G, Jamot J, Gouet P. Degradation of wheat straw and maize stem by a monocentric and a polycentric rumen fungi, alone or in association with rumen cellulolytic bacteria. Anim Feed Sci Technol. 1993;42:69–82. [Google Scholar]
- 34.Romulo B H, Bird S H, Leng R A. The effects of defaunation on digestibility and rumen fungi counts in sheep fed high-fibre diets. Proc Aust Soc Anim Prod. 1986;16:327–330. [Google Scholar]
- 35.Romulo B H, Bird S H, Leng R A. Combined effects of defaunation and protein supplementation on intake, digestibility, N retention and fungi counts in sheep fed straw based diet. In: Nolan J V, Leng R A, Demeyer D I, editors. The roles of protozoa and fungi in ruminant digestion (OECD/UNE International Seminar). Armidale, Australia: Penambul Books; 1989. pp. 285–288. [Google Scholar]
- 36.SAS Institute. User's guide: statistics, version 6 editions. Cary, N.C: SAS Institute, Inc.; 1996. [Google Scholar]
- 37.Soetanto H, Gordon G L R, Hume I D, Leng R A. The role of protozoa and fungi in fibre digestion in the rumen of sheep. Proc. 3rd Cong. Asian Aust Anim Prod Soc. 1985;2:805–807. [Google Scholar]
- 38.Theodorou M K, Gill M, King-Spooner C, Beever D E. Enumeration of anaerobic chytridiomycetes as thallus forming units: a novel method for the quantification of fibrolytic fungal populations from the digestive tract ecosystem. Appl Environ Microbiol. 1990;56:1073–1078. doi: 10.1128/aem.56.4.1073-1078.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ushida K, Tanuka H, Kojima Y. A simple in situ method for estimating fungal population size in the rumen. Lett Appl Microbiol. 1989;9:109–111. [Google Scholar]
- 40.Wallace R J. Adsorption of soluble proteins to rumen bacteria and the role of adsorption in proteolysis. Br J Nutr. 1985;53:399–407. doi: 10.1079/bjn19850047. [DOI] [PubMed] [Google Scholar]
- 41.Williams A G, Withers S E. Effect of ciliate protozoa on the activity of polysaccharide-degrading enzymes and fibre breakdown in the rumen ecosystem. J Appl Microbiol. 1991;70:144–155. doi: 10.1111/j.1365-2672.1991.tb04440.x. [DOI] [PubMed] [Google Scholar]
- 42.Wolin M J, Miller T L. Microbe-microbe interactions. In: Hobson P N, editor. The rumen microbial ecosystem. Amsterdam, The Netherlands: Elsevier; 1988. pp. 361–386. [Google Scholar]
- 43.Yoder R D, Trenkle A, Burroughs W. Influence of rumen protozoa and bacteria upon cellulose digestion in vitro. J Anim Sci. 1966;25:609–612. doi: 10.2527/jas1966.253609x. [DOI] [PubMed] [Google Scholar]