Abstract
N2 fixation by diazotrophic bacteria associated with the roots of the smooth cordgrass, Spartina alterniflora, is an important source of new nitrogen in many salt marsh ecosystems. However, the diversity and phylogenetic affiliations of these rhizosphere diazotrophs are unknown. Denaturing gradient gel electrophoresis (DGGE) of PCR-amplified nifH sequence segments was used in previous studies to examine the stability and dynamics of the Spartina rhizosphere diazotroph assemblages in the North Inlet salt marsh, near Georgetown, S.C. In this study, plugs were taken from gel bands from representative DGGE gels, the nifH amplimers were recovered and cloned, and their sequences were determined. A total of 59 sequences were recovered, and the amino acid sequences predicted from them were aligned with sequences from known and unknown diazotrophs in order to determine the types of organisms present in the Spartina rhizosphere. We recovered numerous sequences from diazotrophs in the γ subdivision of the division Proteobacteria (γ-Proteobacteria) and from various anaerobic diazotrophs. Diazotrophs in the α-Proteobacteria were poorly represented. None of the Spartina rhizosphere DGGE band sequences were identical to any known or previously recovered environmental nifH sequences. The Spartina rhizosphere diazotroph assemblage is very diverse and apparently consists mainly of unknown organisms.
Low elevations of salt marsh ecosystems along the Atlantic and northern Gulf coasts of temperate North America are characterized by extensive, typically monoculture stands of the smooth cordgrass, Spartina alterniflora (Spartina hereafter) (65). Spartina marshes support high rates of macrophyte primary production and microbially mediated nutrient cycling, contributing to global carbon (15, 44) and nitrogen (12) budgets. The consensus of numerous studies is that primary production (43, 67) and decomposition (40, 46, 68) in Spartina marshes are nitrogen limited. In these systems, diazotrophy (N2 fixation) is a key source of new nitrogen (27, 52, 71).
The importance of diazotrophy to Spartina marsh productivity has led to numerous studies of in situ rates of this process (19, 27, 52). Environmental variables that can influence diazotrophy, including host primary production and root exudation (7, 36, 55, 70) and edaphic physicochemical parameters (51, 54, 71) have also been intensively studied, as have the diazotrophic organisms themselves. Many different physiological types of diazotrophs have been isolated from the Spartina rhizoplane and rhizosphere (4, 20, 41, 52), but the true extent of the diversity of these organisms has not been determined. It is reasonable to assume that, as is typical of most types of natural samples (9, 60), only a small fraction of Spartina rhizosphere bacteria can be readily isolated into pure culture. It is also likely that many of the organisms that have been isolated, while able to grow rapidly on laboratory culture media, may be relatively unimportant in the natural environment. However, it is clear that the Spartina rhizosphere diazotroph assemblage is quite diverse, highly active under most conditions, and that diazotrophy by these organisms can be responsive to both host primary production and several key edaphic environmental variables. It is also clear that the diversity of this assemblage is poorly characterized at present, as is the case for most microbial groups (37).
The application of molecular biological methods has greatly facilitated the study of natural bacterial communities and the identification of functionally significant organisms within them (29, 64, 69, 74). Numerous researchers have employed various PCR primers specific for segments of nifH, the structural gene encoding the nitrogenase iron protein, to amplify partial nifH sequences from diazotrophic pure cultures (4, 5, 28, 33, 49, 76) and from various environmental samples, including marine plankton (8, 78), termite hindguts (34, 48), microbial mats and aggregates (50, 77), terrestrial soils (57, 72), and the rhizoplanes of rice (Oryza sativa) (66) and of shoal grass (Halodule wrightii) (33). These studies have yielded a diverse array of nifH sequences representing many, mostly unknown, lineages of diazotrophic Bacteria and Archaea. PCR amplification of nifH sequences, followed by their separation through denaturing gradient gel electrophoresis (DGGE), has recently been used to examine the complexity and stability of the diazotroph assemblage found in the Spartina rhizosphere (54–56). The Spartina rhizosphere diazotroph assemblage was shown to have a consistent species composition over substantial spatial scales, to be composed of a quite diverse array of organisms, and to be stable in composition over a seasonal cycle of host ontogeny and edaphic variability (56). Furthermore, the composition of this assemblage did not change dramatically in response to short-term manipulations of inorganic nutrient levels (54) or host root exudate levels (55). The extent to which the Spartina rhizosphere diazotroph assemblage has already been characterized by both classical pure culture methods (4) and molecular biological analyses (3, 4, 54–56; C. E. Bagwell and C. R. Lovell, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. N-215, p. 490, 1999) provides a strong foundation for the determination of the function-specific diversity of these organisms, i.e., the numbers of diazotrophic species and the physiological and phylogenetic groups of these species detectable in the rhizosphere.
In this study, we have determined the diversity of diazotrophic bacteria in the rhizosphere of Spartina as defined by recoverable partial nifH sequences resolved by DGGE. We also compared the sequences recovered from Spartina rhizosphere to those from several other sources in order to identify sequences broadly distributed among plant-associated and/or marine habitats.
MATERIALS AND METHODS
Reference cultures.
Acetobacterium woodii (ATCC 29683) was provided by Lars Ljungdahl, University of Georgia. Azomonas agilis (ATCC 7494) was purchased from the American Type Culture Collection (Rockville, Md.). Azospirillum lipoferum Sp 59b was provided by Peter van Berkum, United States Department of Agriculture. Four pure cultures previously isolated from the rhizoplanes of tall and short form Spartina alterniflora (TS210, SC16, SG21) and the black needle-rush, Juncus roemerianus (JC110) (4), were also used. These rhizoplane isolates are all gram negative and rod shaped, but differ substantially in their substrate utilization patterns and other physiological features (4). SC16 is a facultative anaerobe, SG21 is a microaerophile, and TS210 and JC110 are aerobes. These organisms have not been definitively identified to date, and physiological testing does not establish their placement in any known genus of free-living nitrogen-fixing bacteria. Cultivation conditions and DNA extraction procedures for these organisms have been described previously (4, 13, 38).
nifH-specific PCR primer design and specificity.
PCR primer design was based on analysis of nifH sequences from the NCBI GenBank database (6) by using the Wisconsin Genetics Computer Group software (18). In order to maximize the specificity of the primers for amplification of free-living diazotroph and rhizobium-like nifH sequences and to limit primer degeneracy as much as possible, nifH sequences from cyanobacteria, Frankia, and methanogens were excluded from our analysis (56). Most of the representatives of these groups have nifH sequences so divergent from those of other diazotrophs, that their inclusion results in excessive primer degeneracy. Also, Frankia and diazotrophic cyanobacteria, although very important in other environments, would not be expected to be prevalent in the rhizosphere of Spartina alterniflora. To further reduce degeneracy, the primers were synthesized with the artificial nucleotides P {6-(β-d-ribofuranosyl)-3,4-dihydro-8H-pyrimido[4,5-c-]-[1,2]oxazin-7-one} (35) and K [2-amino-9-(2-deoxy-β-ribofuranosyl)-6-methoxyaminopurine] (Glen Research, Sterling, Va.) (10). P pairs with either purine, and K pairs with either pyrimidine. Duplexes formed with primers containing these bases are more stable than would be the case for comparable primers containing a weakly pairing nucleotide, such as inosine (10, 35). The forward primer [5′-TACGG(P/K)AAKGG(P/G)GG(P/K)ATPGG-3′; primer 1] corresponds to Klebsiella pneumoniae (GenBank accession no. X13303) nifH position 25 to 44. The reverse primer (5′-CGCCCGCCGCGCCCCGCGCCCGTCCCGCCGCCCCCGCCCG(G/C)ACGATGTAGATPTCCTG-3′; primer 2) sequence (underlined; the balance is the GC clamp) corresponds to K. pneumoniae position 436 to 453. Primer degeneracy was eightfold for the forward primer and twofold for the reverse primer. Primers were previously tested against DNA from known diazotrophs and nondiazotrophs (56) to establish their effectiveness and specificity.
nifH sequences from natural assemblages of Spartina alterniflora diazotrophs.
The Spartina rhizosphere nifH sequences analyzed in this study were obtained from denaturing gradient gels produced for previous studies of Spartina rhizosphere diazotroph assemblages (54–56). Briefly, rhizosphere samples were collected from an intertidal marsh zone on Goat Island in the North Inlet estuary near Georgetown, S.C. (33°20′N: 79°12′W). Sampling transects were established parallel to a small tidal creek within the tall form Spartina growth zone (near the creek bank) and the short form Spartina growth zone (inland from the tall form zone). Cores (2.4 cm in diameter by approximately 5 cm in length) were collected from the Spartina sod along these transects on several sampling dates in 1997. The Spartina rhizosphere includes the soil directly influenced by plant roots and rhizomes (see reference 11) and supports elevated levels of many microbial activities, including diazotrophy, relative to unvegetated soils and sediments (17, 73). It should be noted that these core samples contained high levels of live and dead roots and rhizomes (54, 55), as well as sediment and decaying plant-derived organic matter. Acetylene reduction rates measured in intact rhizosphere cores during the sampling period ranged from 0.35 to 2.63 μmol of ethylene produced per liter of sediment day−1 (56). Experimental plots for manipulations of inorganic nutrients (54) and host exudates (55) were established in the short-form Spartina zone, and cores were collected from these plots as well. DNA was extracted from the cores by using a previously described direct lysis procedure (39, 56).
Specific nifH sequence segments were amplified from bacterial nifH sequences for DGGE with primers 1 and 2. PCR was performed with rTth DNA polymerase XL (Perkin-Elmer, Foster City, Calif.), which has proofreading capability and high tolerance for various types of contaminants that might occur in DNA preparations (1). The reaction conditions and touchdown thermal cycling program have been described previously, as have the reagents and methods for DGGE (56). See references 55 and 56 for images of the gels sampled for this study. Gel plugs were taken from well-resolved bands from several gels by using wide-orifice micropipette tips. The gel plugs were stored in 100 μl of TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA) at −20°C until used in this study. Homoduplex and heteroduplex bands (Fig. 1) were previously identified by reamplification of 1-μl samples of the TE from each band and resolution of the amplimer(s) by DGGE (56). A homoduplex band is composed of two fully complementary strands and will yield a single DGGE band on reamplification. A heteroduplex band is composed of strands that are not completely complementary (i.e., from different parental sequences) and will yield three DGGE bands, one for each of the partially complementary strands, and one for the heteroduplex (23). Prior to adoption of the PCR and DGGE conditions described above, some reactions were performed with the Advantage GC PCR system (Clontech, Palo Alto, Calif.) and resolved with somewhat different denaturant gradients (75 to 95% denaturant). Since the bands from these gels did not correspond in position to bands from gels obtained with the optimized system, sequences from them were designated separately (X1 to X8 and Y1 to Y4). These sequences were included in our analysis to expand the nifH sequence database available for the Spartina rhizosphere.
FIG. 1.
Selected denaturing gradient gel lane showing relative positions of nifH amplimers analyzed in this study. PCR and DGGE conditions are described in Materials and Methods and reference 56.
Amplimer cloning and identification of different cloned amplimer sequences.
Amplimers from gel plugs taken from DGGE gel bands were recovered by reamplification. The forward primer used in these reamplifications was 5′-GGTAT(C/T)GG(C/T)AA(A/G)TG(G/C)AC(G/C)AC-3′ (primer 3), and the reverse primer was 5′-GACGATGTAGAT(C/T)TCCTG-3′ (primer 4). These primers were selected to provide inexpensive alternatives to the P- and K-containing primers 1 and 2. Primer 3 had lower redundancy than primer 1 would have without P and K substitutions and corresponds to K. pneumoniae positions 37 to 56. Primer 4 is colinear with the non-GC clamp portion of primer 2. Reamplification employed the Expand High Fidelity System (error rate of 8.5 × 10−6) by using the reaction mixture recommended by the manufacturer (Boehringer Mannheim, Indianapolis, Ind.) and 1 μl of TE from each stored band plug. The following thermocycling program was used: initial denaturation at 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 47°C for 30 s, and 72°C for 30 s. This was followed by a final extension step at 70°C for 2 min. The same PCR primers and procedures were also used to amplify partial nifH sequences from DNA purified from A. woodii, A. agilis, and A. lipoferum and from four pure culture isolates from the Spartina and Juncus rhizoplanes (4). The amplimers from these reactions were ligated into the pGEM-T vector (Promega, Madison, Wis.), and the ligation reactions were used to transform competent Escherichia coli strain JM109. Recombinant colonies were selected on Luria-Bertani agar plates containing 80 μg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) ml−1, 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and 100 μg ampicillin ml−1. Plates were incubated overnight at 37°C.
Small amounts of growth from selected recombinant colonies were collected with sterile toothpicks and transferred into PCR tubes. PCRs employing primers specific for the T7 and Sp6 RNA polymerase binding sites and AmpliTaq (Perkin-Elmer) were used to amplify the cloned sequences in a reaction volume of 25 μl. These reactions employed the same thermocycling program as that used in the DGGE gel band reamplification. Five microliters of each reaction mixture was run on 1.5% agarose–TBE (89 mM Tris, 89 mM boric acid, 2 mM EDTA) gels to verify amplification of the insert DNA. Restriction digests were set up with 10 μl of PCR product and HaeIII or MspI in separate reaction mixtures having a final volume of 20 μl. Digests were incubated at 37°C overnight, and restriction fragments were resolved by electrophoresis in 4% NuSeive–3:1 agarose (FMC, Rockland, Maine)–0.5× TAE (1× TAE is 40 mM Tris, 20 mM acetic acid, 1 mM EDTA) gels. Amplimers having the same restriction fragment length polymorphism (RFLP) patterns for both digests were considered to be the same sequence. Any difference in either digest pattern was taken to indicate different amplimer sequences. All amplimers yielding unique RFLP patterns were sequenced.
DNA sequencing.
Recombinant plasmids were purified from selected clones by using the Qiagen Plasmid Mini Kit (Santa Clarita, Calif.). Plasmid concentrations were determined fluorometrically. Sequencing reactions used 0.2 pmol of template DNA, fluorescently tagged T7 and Sp6 primers (LiCor, Lincoln, Nebr.), and the Thermosequenase DYEnamic Direct Cycle Sequencing kit with 7-deaza-GTP (Amersham, Cleveland, Ohio). The thermocycling program used was 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 45°C for 30 s, and 72°C for 1 min. After amplification, loading dye was added to the samples, which were then analyzed with a Li-Cor DNA4000LS sequencer. Sequences were determined for both strands of each cloned amplimer.
Sequence analysis.
NCBI GenBank nifH sequences from a variety of diazotrophs were selected for use in phylogenetic analyses and are listed in Table 1. Sequences from two vanadium nitrogenase iron protein genes were also included. Additional sequences from various nif-like genes, including anfH (Fe nitrogenase iron protein) and frxC and bchX (light-independent chlorophyllide reductase) were examined, but had low similarities to all of the nifH sequences analyzed and were dropped from further consideration. Environmental nifH sequences from the NCBI GenBank database that aligned with known cyanobacteria and Archaea were dropped from further analysis (see above), as were sequences belonging to other microbial groups, but having low similarities to nifH sequences from the Spartina rhizosphere. In all cases, primer sequences were removed prior to sequence analysis.
TABLE 1.
Reference nifH sequences from the GenBank database used in the phylogenetic analyses
| Organism or source | GenBank accession no. |
|---|---|
| Known diazotrophic bacteria | |
| Acetobacter diazotrophicus | AF030414 |
| Alcaligenes faecalis | X96609 |
| Anabaena azollae | L34879 |
| Anabaena sp. strain PCC7120 | AF012326 |
| Azoarcus communis | U97116 |
| Azoarcus indigens | U97118 |
| Azoarcus tolulolyticus | U97122 |
| Azospirillum brasilense | M64344 |
| Azotobacter chroococcum | M73020 |
| Azotobacter chroococcum vnfH | X51756 |
| Azotobacter vinelandii | M20568 |
| Azotobacter vinelandii vnfH | M32371 |
| Bradyrhizobium japonicum | E01169, K01620 |
| Chlorobium tepidum | AF065617 |
| Clostridium cellobioparum | U59414 |
| Clostridium pasteurianum | M21537 |
| Desulfobacter curvatus | AF065619 |
| Desulfonema limicola | AF065618 |
| Desulfovibrio gigas | U68183 |
| Frankia alni | L41344 |
| Herbaspirillum seropedicae | Z54207 |
| Klebsiella pneumoniae | J01740 |
| Klebsiella sp. | M63691 |
| Marichromatium purpuratum | AF059648 |
| Methanobacterium thermautotrophicum | AE00916 |
| Methanosarcina barkeri | X56072 |
| Pseudomonas sp. | AF117976 |
| Pseudomonas stutzeri | AF117977, AF117978 |
| Rhizobium leguminosarum biovar phaseoli | M10587 |
| Rhizobium leguminosarum biovar trifolii | K00490 |
| Rhizobium sp. | M16709, M26961 |
| Rhodobacter capsulatus | M15270, X07866 |
| Rhodobacter sphaeroides | AF031817 |
| Rhodospirillum rubrum | M33774 |
| Sinorhizobium meliloti | J01781 |
| Thiobacillus ferrooxidans | M15238 |
| Trichodesmium thiebautii | U23507 |
| Vibrio diazotrophicus | U23650 |
| Unnamed cultivated diazotrophs | |
| Marine microbial mats | AF046827-AF046854, U43438, U43442–U43445 |
| Nonaxenic cyanobacterial cultures | U43436, U43437, U43439 |
| Environmental nifH sequences | |
| Halodule wrightii (shoal grass) rhizoplane | M63688, M63689 |
| Oryza sativa (rice) rhizoplane | D26284-D26306 |
| Douglas Fir forest soil and litter | AF099775–AF099798 |
| Marine microbial mats | U23633–U23646 |
| Marine plankton | AF016592–AF016615, AF016617, AF016618, AF059621–AF059623, AF059629, AF059643–AF059647, U26186 |
| Antarctic diazotrophic microbial consortia | AF049033–AF049040, AF049043 |
Nucleotide sequences were aligned by using ClustalW (63) and translated directly into amino acid sequences by using the “export file” options in MEGA (version 1.0; The Pennsylvania State University, University Park, Pa.). Alignments were hand corrected when necessary. Neighbor-joining phylogenies (58) were constructed in PAUP* (version 4.0b2) by using percent dissimilarity distances and pairwise deletion of gaps and missing data. The use of alternative amino acid distance measures (e.g., Poisson and gamma correction for multiple substitutions) or nucleotide sequences (first and second positions only) had no significant effect on the resulting dendrogram topology (data not shown). A pronounced G+C bias precluded the use of codon third positions in phylogenetic analyses. NifH amino acid sequences from Methanobacterium thermoautotrophicum and Methanosarcina barkeri were used as outgroup taxa. Bootstrapping (22) was used to estimate the reliability of phylogenetic reconstructions (1,000 replicates).
Multiple nifH sequences were recovered from some DGGE bands. Analysis of duplex melting profiles of selected sequences from several bands was performed with WinMelt (version 2.0; Bio-Rad, Hercules, Calif.). This software package calculates duplex melting profiles by using the melting temperature of each base pair in the sequence, effects of neighboring nucleotides on those melting temperatures, positions and effects of major sequence domains, and the effects of secondary structure on the melting behavior of the sequence as a whole.
Nucleotide sequence accession number.
The nifH sequence segments determined in this study are available in the GenBank database under accession no. AF216874 to AF216939.
RESULTS AND DISCUSSION
There were usually 11 prominent, well-resolved bands per lane of typical DGGE gels loaded with nifH amplimers from field Spartina alterniflora rhizosphere samples (Fig. 1) (54–56). This banding pattern was highly reproducible, but the intensity of any given band often varied among replicate sample lanes within a gel. Band intensities also varied among samples from different plant height zones, dates of sample collection, or experimental manipulation. The best-resolved and strongest bands representing those designated in Fig. 1 were sampled from four gels. An additional band, designated band B prime (BP) appeared in some gel lanes from a manipulative experimental study (55), and the best-resolved, strongest example of this band was also sampled. Amplimers from all of these prominent bands were successfully recovered by reamplification and were cloned. It should be noted that additional, typically faint bands were observed sporadically in the DGGE gels, but since these bands were not seen consistently, only two (A2 and A5) were examined in this study.
RFLP analysis of cloned amplimers was employed to differentiate sequences recovered from a given DGGE gel band. In preliminary studies, nifH sequences from the same well-resolved DGGE gel band and having identical RFLP patterns were either identical in nucleotide sequence or differed only slightly. None of these minor differences resulted in different amino acid sequences. Some bands contained a single RFLP pattern, but multiple patterns were recovered from others (Table 2). Note that band F was originally thought to be a doublet (56), but RFLP analysis revealed two distinct groups of sequences (bands F and G).
TABLE 2.
Percent similarities of Spartina rhizosphere NifH amino acid sequences to the most similar sequence(s) from known diazotrophic bacterial speciesa
| Spartina sequence | Organism with most similar sequence(s) | % Similarity |
|---|---|---|
| Presumed γ-Proteobacteria | ||
| A2 | Azotobacter chroococcum | 94.2 |
| B2 | Azoarcus indigens | 97.3 |
| B5 | Azoarcus indigens | 97.3 |
| BP2, HD2-4 | Pseudomonas stutzeri | 97.5 |
| BP3, HD4-1, HD4-2, X1, X3, X5, X8, Y1, Y2 | Pseudomonas stutzeri | 98.3 |
| BP4 | Pseudomonas stutzeri | 96.6 |
| C1 | Azotobacter chroococcum | 96.4 |
| E2, E4, F5, F9 | Azomonas agilis, Azotobacter chroococcum, Klebsiella sp., Marichromatium purpuratum | 93.5 |
| F1 | Azomonas agilis, Azotobacter chroococcum | 87.8 |
| F2 | Azoarcus indigens | 95.6 |
| F3 | Klebsiella sp. | 92.5 |
| F4 | Marichromatium purpuratum | 92.5 |
| F6 | Marichromatium purpuratum | 90.7 |
| F7 | Marichromatium purpuratum | 91.6 |
| F8 | Marichromatium purpuratum | 89.7 |
| HD2-2, X7 | Azotobacter chroococcum, Azotobacter vinelandii | 85.6 |
| HD2-3 | Azoarcus indigens | 92.9 |
| HD3-6 | Marichromatium purpuratum | 89.7 |
| HD3-8 | Pseudomonas stutzeri | 97.5 |
| X2 | Pseudomonas stutzeri | 98.3 |
| X6 | Pseudomonas stutzeri | 95.8 |
| Y3 | Pseudomonas stutzeri | 94.1 |
| Y4 | Azotobacter chroococcum | 95.0 |
| JC110 | Klebsiella sp., Vibrio diazotrophicus | 97.2 |
| SG21 | Azomonas agilis, Azotobacter chroococcum | 95.0 |
| TS210 | Pseudomonas stutzeri | 96.6 |
| Presumed α-Proteobacteria | ||
| A4 | Azoarcus tolulolyticus | 95.5 |
| E1 | Bradyrhizobium japonicum | 87.0 |
| G1 | Azospirillum brasilense, Sinorhizobium meliloti | 90.6 |
| G2 | Azospirillum brasilense | 90.6 |
| G3 | Azospirillum brasilense, Sinorhizobium meliloti | 92.0 |
| G4 | Azospirillum brasilense | 92.8 |
| SC16 | Acetobacter diazotrophicus, Herbaspirillum seropedicae, Rhizobium sp. M16709 | 93.5 |
| Presumed anaerobes | ||
| A1 | Desulfonema limicola | 87.6 |
| A3 | Desulfonema limicola | 88.6 |
| A5 | Desulfonema limicola | 85.7 |
| B1 | Desulfonema limicola | 90.5 |
| B3, HD1-1 | Desulfovibrio gigas | 84.2 |
| B4 | Desulfonema limicola | 87.6 |
| D1, D2, HD3-1, HD3-2 | Desulfovibrio gigas | 83.5 |
| HD2-1 | Desulfonema limicola | 85.7 |
| HD3-3 | Desulfonema limicola | 88.6 |
| HD3-4 | Desulfovibrio gigas | 84.1 |
| HD3-5 | Desulfonema limicola | 86.7 |
| HD3-7 | Desulfobacter curvatus | 88.8 |
| X4 | Desulfonema limicola | 90.5 |
Percent similarities are from the distance matrix constructed in PAUP* (version 4.0b2). Spartina sequences are listed by the DGGE gel bands from which they were recovered (see Materials and Methods) and grouped into the major sequence clusters shown in Fig. 2 to 4. Note that these clusters are named for the predominant types of organisms they contain, but some sequences from other phylogenetic groupings can occur. Pure culture isolates from the Spartina rhizoplane are also included.
Sequences from DGGE gel bands and laboratory cultures were initially translated and examined for key, highly conserved amino acid residues that are important in nitrogenase iron protein structure and function (16, 53). Within the segments analyzed, 11 amino acids including (Klebsiella pneumoniae numbering) Lys 15 and Ser 16 (within the Mg ATP binding domain), Arg 100 (the ADP ribosylation site), Asp 125 (possibly involved in protein conformation changes), Asp 129 (involved in ATP hydrolysis), Arg 140 and Lys 143 (contribute to salt bridge formation), and four conserved Cys residues (no. 38, 85, 97, and 132, two of which coordinate the Fe4S4 cluster) were used as markers for determining sequence accuracy. Only two sequences (BP1 and E3) had substitutions at more than one of these positions, and in both cases, residues considered essential to protein function (i.e., Cys 132 for BP1 and Arg 100 for E3) were affected. Only two sequences (A2 and G1) had a single substitution each, neither of which would be expected to severely impact protein activity (16). Sequences BP1 and E3 were not included in phylogenetic analyses; sequences A2 and G1 were used.
Due to the pronounced G+C bias in the third positions of codons and the similar outcomes of phylogenetic analyses employing polypeptide sequences or first and second nucleotide position sequences, all phylogenetic analyses reported employed the NifH polypeptide sequences (47, 75). Phylogenetic trees were constructed from Spartina rhizosphere sequences (59 in all), sequences from pure culture isolates from the Spartina rhizoplane (4 sequences), and sequences from reference strains of known diazotrophs (45 sequences). Relatively few sequences from cyanobacteria or from diazotrophic Archaea were included, since the primers used to recover Spartina rhizosphere nifH sequences for DGGE analysis would be unlikely to amplify sequences from these organisms (56). As has been observed in other studies (8, 48, 66, 74, 77, 78), there are several large clusters of NifH sequences representing important major groups of diazotrophs. The neighbor-joining algorithm yielded a topology that contained three major NifH sequence clusters that contained Spartina rhizosphere sequences and were found in greater than 60% of all bootstrap replicates.
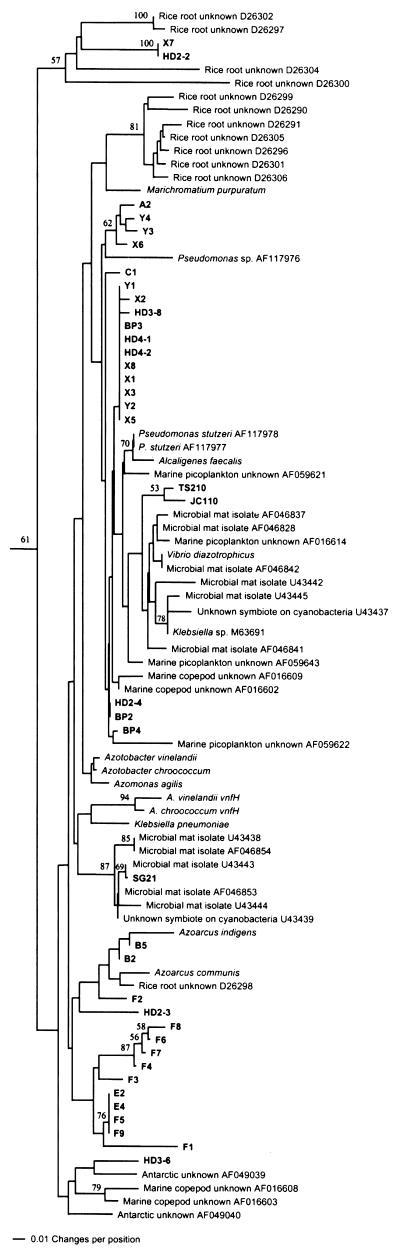
The first major cluster was well supported by bootstrapping (61%) and contained sequences from known members of the γ subdivision of the division Proteobacteria (γ-Proteobacteria [and some β-Proteobacteria]), 36 sequences from Spartina rhizosphere, and three sequences from rhizoplane pure culture isolates (Fig. 2). This cluster was designated the γ-Proteobacteria cluster due to the preponderance of sequences deriving from known organisms in that group. Pairwise similarities among sequences in this cluster averaged 92.1%. Several of the Spartina sequences were highly similar to the NifH sequences from Pseudomonas stutzeri and may represent species in the genus Pseudomonas or closely related genera. However, most sequences segregated into distinct clades to the exclusion of sequences from known species. Among these was a cluster of sequences from bands E and F that had substantial similarity to the sequence from SG21, an unnamed Spartina rhizoplane isolate.
FIG. 2.
Phylogenetic analysis of Spartina rhizosphere NifH amino acid sequences from presumed γ-Proteobacteria, from various known γ-Proteobacteria, and from selected unknown, presumed γ-Proteobacteria from other plant-associated and/or marine environments. The percentage of 1,000 bootstrap samples that supported each branch is shown. Bootstrap values below 50% are not shown.
Numerous NifH sequences from unknown bacteria inhabiting plant-associated and/or marine environments also fell into the γ-Proteobacteria cluster (Fig. 2). Many of these had relatively small similarity scores to sequences from the Spartina rhizosphere and are not shown. However, one sequence from the rice rhizoplane (D26298) was placed in a clade containing two band B sequences, and one sequence from an Antarctic diazotrophic consortium (AF049039) was sister to rhizosphere sequence HD3-6 (93.1% similarity). Several sequences from γ-Proteobacteria isolated from marine microbial mats were also highly similar to the SG21 sequence. These observations may imply that some of the γ- and/or β-proteobacterial diazotrophs represented by our DGGE gel bands and sequences may be amenable to isolation and laboratory cultivation.
It is interesting in this regard that no Spartina rhizosphere NifH sequences were highly similar to NifH sequences from the Azotobacteriaceae. Based on pure-culture isolation methods, Azotobacter-like organisms have been reported as abundant diazotrophs in vegetated salt marsh sediments (20). Organisms belonging to the Azotobacteriaceae can be readily isolated from Spartina and Juncus roemerianus rhizoplanes (4), and nifH sequences from them are efficiently amplified with the primers employed in this study (data not shown). It seems likely from these results that members of the family Azotobacteriaceae, while efficiently recovered from vegetated salt marsh sediments and easily cultivated in the laboratory, may not be numerically important diazotrophs in the Spartina rhizosphere. Similarly, Klebsiella sp. and their relatives, while readily isolated from rhizoplanes of wetlands plants (4, 14), were not represented by any of the nifH sequences we recovered. It is also possible that PCR biases prevented recovery of these sequences from nifH sequence mixtures, but quantitative determination of the abundance of Azotobacter- and Klebsiella-like organisms (Bagwell and Lovell, Abstr. 99th Gen. Meet. Am. Soc. Microbiol.) will be required to resolve this issue.
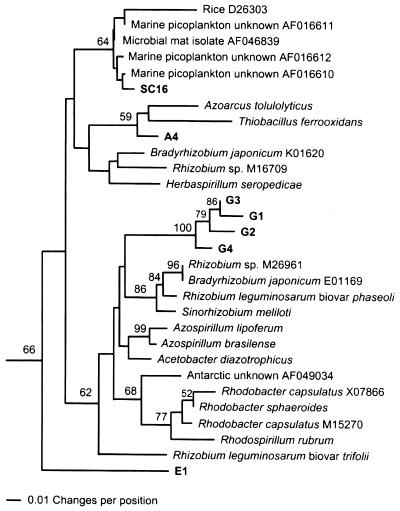
The second major cluster was found in 66% of 1,000 bootstrap replicates and contained sequences from many familiar diazotrophs, including species of the Rhizobiaceae and the purple non-sulfur bacteria (Fig. 3). In addition to known α-Proteobacteria, some β-Proteobacteria (Azoarcus tolulolyticus and Herbaspirillum seropedicae) and the γ-proteobacterium Thiobacillus ferrooxidans are also represented in this cluster, as well as six sequences from the Spartina rhizosphere, and a single sequence from a rhizoplane isolate. Lateral transfer of nifH is considered probable among diazotrophs belonging to several subgroups of the α- and β-Proteobacteria (21, 28, 31) and may explain the appearance of sequences from β- and γ-Proteobacteria in this cluster. This cluster was designated the α-Proteobacteria cluster due to the preponderance of sequences derived from known organisms in that group. Overall similarity among sequences in the α-Proteobacteria cluster was 89.8%.
FIG. 3.
Phylogenetic analysis of Spartina rhizosphere NifH amino acid sequences from presumed α-Proteobacteria, from various known α-Proteobacteria, and from selected unknown, presumed α-Proteobacteria from other plant-associated and/or marine environments. The percentage of 1,000 bootstrap samples that supported each branch is shown. Bootstrap values below 50% are not shown.
Inclusion of unknown environmental NifH sequences from other sample types substantially expanded the α-Proteobacteria cluster (Fig. 3). For example, almost all NifH sequences recovered from Douglas Fir forest soil and litter (72) and one sequence from the rice rhizoplane fell into this well-supported cluster. Numerous sequences in this cluster have also been recovered from various planktonic marine environments (78), but few from rhizoplanes (33, 66) or rhizospheres (this study) of wetland plants to date. Most Spartina rhizosphere NifH sequences formed a monophyletic group to the exclusion of sequences from unknown, presumed α-Proteobacteria. In contrast, the NifH sequence from the rhizoplane isolate SC16 was identical over the length it shared with a sequence from an unknown diazotrophic bacterium in marine picoplankton (AF016612) and from a pure culture isolate from a marine microbial mat (AF046839). It should be noted that the algorithms used to construct dendrograms produce an adjusted “grand average” representation of the various pairwise distance values from the sequence distance matrix. In our analyses, gaps were treated as missing data and the sequences listed as identical (for the sequence stretches they have in common) were of unequal lengths. The software scoring all of the sequences against each other can score otherwise identical sequences differently based on differences in the “missing data” among the sequences that include those data. This can result in horizontal distances in the dendrogram between sequences having 0% dissimilarity in the distance matrix. SC16 is physiologically similar to species of the Rhizobiaceae (4), but has not been definitively identified as yet.
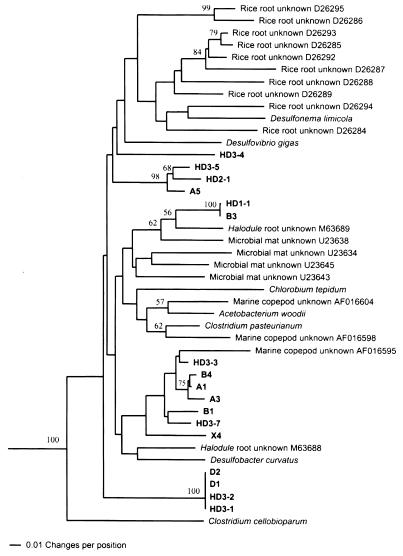
The third cluster contained NifH sequences from known obligate anaerobes and 17 sequences from unknown bacteria from the Spartina rhizosphere (Fig. 4). Although completely supported by bootstrap analysis (100%), the anaerobe cluster contained many highly divergent lineages with an average similarity of only 85.5%. Most of the Spartina rhizosphere sequences formed well-supported monophyletic groups, and none were closely related to any sequence from a known diazotrophic anaerobe. The largest similarity score between any Spartina NifH sequence and any sequence from a known anaerobic diazotroph was 90.5%, and was between two DGGE band sequences (B1 and X4) and Desulfonema limicola. An additional 10 sequences from rice rhizoplane (66) and 2 sequences from the rhizoplane of shoal grass (Halodule wrightii) (33) were included in this cluster. The largest similarity score between any Spartina sequence and any NifH sequence from any other type of environmental sample was 93.1% and involved DGGE band sequence HD3-3 and a sequence recovered from an unknown bacterium associated with a marine copepod (AF016595). We view this relationship with caution, however, since the branch representing the copepod-associated unknown sequence is quite long.
FIG. 4.
Phylogenetic analysis of Spartina rhizosphere NifH amino acid sequences from unknown, presumed anaerobic bacterial sequences, from various known anaerobic bacteria, and from selected unknown, presumed anaerobic bacteria from other plant-associated and/or marine environments. The percentage of 1,000 bootstrap samples that supported each branch is shown. Bootstrap values below 50% are not shown.
The finding of numerous NifH sequences from anaerobes is consistent with the known characteristics of the Spartina rhizosphere. While the rhizoplane of Spartina and sediments in close proximity to roots receive substantial, transient oxygen input through the aerenchyma system (2, 62), rhizosphere sediments not closely associated with live plant roots are likely to be anoxic (30), and our samples included live and dead roots and rhizomes along with surrounding sediment and organic matter. Even the Spartina rhizoplane would be expected to rapidly become suboxic after the onset of darkness. Inhibitor studies of nitrogen fixation associated with Spartina roots and rhizosphere sediments have revealed a potentially significant role of diazotrophic sulfate-reducing bacteria (26), and this seems to be supported by our results. While almost all of the Spartina anaerobe NifH sequences were less than 90% similar to those from any known anaerobe, the highest similarities were all between Spartina anaerobe sequences and sulfate reducer sequences (Table 2). Low representation of organisms very similar to the known clostridia, as reported by Dicker and Smith (20) for surface sediments from various vegetated salt marsh zones, may also be indicated. We have recovered gram-positive bacterial 16S ribosomal DNA sequences from DNA purified from other types of marine sediment samples by using the same direct lysis method used here (G. Matsui and C. R. Lovell, unpublished observations). We have also successfully amplified nifH sequences from gram-positive fermentative anaerobes (including Acetobacterium woodii, a non-spore-forming gram-positive organism related to Clostridium [61]). We surmise that the clostridia were either numerically insignificant in the Spartina rhizosphere or that nifH sequences from these organisms were recovered too inefficiently to permit their detection on our DGGE gels. Ueda et al. (66), using quite different DNA extraction procedures and PCR primers, also recovered few nifH sequences from rice rhizoplane that had substantial similarity to those from the clostridia, while sequences clustering with those from sulfate reducers were common. Anaerobe NifH sequences are highly divergent, even within some defined, monophyletic clusters (e.g., the δ-Proteobacteria, the low G+C Firmicutes). It is also possible that the membership of some Spartina anaerobe sequences in these groups may be obscured by such divergence.
NifH sequences are quite conservative, so similarity values for closely related species, such as Azotobacter chroococcum and A. vinelandii (99.3%) or Azospirillum brasilense and A. lipoferum (99.3%), are typically very high. Thus, even the highest similarity scores observed between Spartina rhizosphere NifH sequences and those from known diazotrophic bacteria are most likely too low to reflect species-level relationships (Table 2). In addition, many of the Spartina NifH sequences were found in distinct, well-supported clades that were monophyletic with respect to known NifH sequences. The largest similarity score between any Spartina rhizosphere NifH sequence and any NifH sequence from any known diazotroph was between several sequences (HD4-1, HD4-2, X1 to X3, X5, X8, Y1, and Y2) and Pseudomonas stutzeri and amounted to 98.3%. As expected from the primer design, no clustering of Spartina rhizosphere NifH sequences with any archaeal, cyanobacterial, or Frankia NifH sequences was observed (data not shown).
Numerous NifH sequences from rhizoplanes of other wetlands plants, particularly from rice, also fell into the three major sequence clusters containing the Spartina sequences. Like Spartina, rice also introduces oxygen into the rhizosphere via aerenchyma transport (32), but the existence of reduced microzones in close proximity to the roots is likely. Clearly, the superficial similarities among the rhizoplane and rhizosphere microenvironments of these wetlands plants did not lead to the development very similar diazotroph assemblages. However, it is noteworthy that the assemblages from the only two wetlands plants from whose rhizosphere diazotroph assemblages have been examined in some detail, Spartina (this study) and rice (66), yielded numerous sequences from the γ-Proteobacteria and the anaerobes, but few from the α-Proteobacteria.
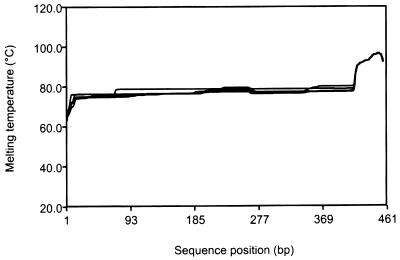
Several bands from the DGGE gels contained heterogeneous sequences. While DGGE can separate sequences differing by as little as a single nucleotide (25), a given band can contain a mixture of sequences (45, 56). This is due at least in part to the fact that the denaturant gradient and gel running conditions are optimized to yield a profile of bands encompassing the total recoverable diversity of sequences from a sample. However, DGGE separates sequences based on their melting profiles (25, 59), and it is possible for nonidentical sequences to have profiles sufficiently similar to allow their near codenaturation in the gel. The five band B sequences had some substantial sequence differences (Table 2), but extremely similar melting profiles (Fig. 5), permitting their effective codenaturation in our DGGE gels. Melting profiles of sequences within more homogeneous bands (bands F and G) were effectively identical (data not shown). The occurrence of DGGE bands containing nonidentical sequences would greatly hinder attempts to interpret changes in organism abundance on the basis of DGGE band intensity. For this reason, Piceno et al. (56) and Piceno and Lovell (54, 55) examined only band numbers and positions in their analyses of natural diazotroph assemblages.
FIG. 5.
Melting profiles of the partial nifH sequences recovered from denaturing gradient gel band B (B1 to B5) (Table 2). The sequences analyzed included both primers and the GC clamp to reflect their behavior in DGGE gels.
The finding of radiations of highly similar nifH sequences in certain DGGE bands also has an interesting implication for the population dynamics and ecological functions of rhizosphere diazotrophs. Bagwell et al. (4) recovered numerous strains of diazotrophs from Spartina and Juncus rhizoplanes, and in several cases, these strains formed distinct clusters of physiologically similar, but distinguishable organisms. Some closely related strains having high levels of genomic DNA homology differed substantially in key physiological characteristics (3). The occurrence of groups of strains or very closely related species (i.e., ecotypes) (23, 24), all able to inhabit similar niches (such as different locations on the rhizoplane), may result in a spectrum of organism physiological features that could support diazotrophy at different stages of host plant ontogeny or under different edaphic conditions. This microdiversity (3, 42) could provide an important foundation for functional redundancy in a highly dynamic microenvironment, such as the Spartina rhizosphere, where conditions can vary over relatively short time frames.
The micro- and macrodiversity of diazotrophs occurring in the Spartina rhizosphere and the dissimilarity of assemblages from presumably comparable habitats underscore the largely undescribed diversity of plant-associated diazotrophic bacteria. The diversity of diazotrophs inhabiting the rhizospheres of wetlands plants clearly reflects a plethora of functioning microniches, and these microenvironments may be much more dynamic than is generally appreciated. This complex array of unknown species and undefined ecotypes is certainly capable of maintaining high levels of nitrogen fixation, the unifying ecological function of the group, across a broad range of host-driven and abiotic environmental variability.
ACKNOWLEDGMENTS
We acknowledge George Matsui and Hongyue Dang for assistance with DNA sequencing.
This research was supported by NSF awards DEB-9407596 and DEB-9903623 to C.R.L.
REFERENCES
- 1.Al-Soud W A, Rådström P. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl Environ Microbiol. 1998;64:3748–3753. doi: 10.1128/aem.64.10.3748-3753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armstrong W. Root aeration in the wetland condition. In: Hook D D, Crawford R M, editors. Plant life in anaerobic environments. Ann Arbor, Mich: Ann Arbor Science Series; 1978. pp. 269–298. [Google Scholar]
- 3.Bagwell C E, Lovell C R. Microdiversity of culturable diazotrophs from the rhizoplanes of the salt marsh grasses Spartina alterniflora and Juncus roemerianus. Microb Ecol. 2000;39:128–136. doi: 10.1007/s002480000017. [DOI] [PubMed] [Google Scholar]
- 4.Bagwell C E, Piceno Y M, Ashburne-Lucas A, Lovell C R. Physiological diversity of the rhizosphere diazotroph assemblages of selected salt marsh grasses. Appl Environ Microbiol. 1998;64:4276–4282. doi: 10.1128/aem.64.11.4276-4282.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben-Porath J, Zehr J P. Detection and characterization of cyanobacterial nifH genes. Appl Environ Microbiol. 1994;60:880–887. doi: 10.1128/aem.60.3.880-887.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson D A, Boguski M S, Lipman D J, Ostell J, Ouellette B F F. GenBank. Nucleic Acids Res. 1998;26:1–7. doi: 10.1093/nar/26.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyle C D, Patriquin D G. Carbon metabolism of Spartina alterniflora Loisel in relation to that of associated nitrogen-fixing bacteria. New Phytol. 1981;89:275–288. [Google Scholar]
- 8.Braun S T, Proctor L M, Zani S, Mellon M T, Zehr J P. Molecular evidence for zooplankton-associated nitrogen-fixing anaerobes based on amplification of the nifH gene. FEMS Microbiol Ecol. 1999;28:273–279. [Google Scholar]
- 9.Brock T D. The study of microorganisms in situ: progress and problems. Symp Soc Gen Microbiol. 1987;41:1–17. [Google Scholar]
- 10.Brown D M, Lin P K T. Synthesis and duplex stability of oligonucleotides containing adenine-guanine analogues. Carbohydr Res. 1991;216:129–139. doi: 10.1016/0008-6215(92)84156-m. [DOI] [PubMed] [Google Scholar]
- 11.Campbell R, Greaves M P. Anatomy and community structure of the rhizosphere. In: Lynch J M, editor. The rhizosphere. New York, N.Y: John Wiley & Sons, Ltd.; 1990. pp. 11–34. [Google Scholar]
- 12.Capone D G, Carpenter E J. Nitrogen fixation in the marine environment. Science. 1982;217:1140–1142. doi: 10.1126/science.217.4565.1140. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y P, Lopez-de-Victoria G, Lovell C R. Utilization of aromatic compounds as carbon and energy sources during growth and N2-fixation by free-living nitrogen fixing bacteria. Arch Microbiol. 1993;159:207–212. [Google Scholar]
- 14.Currin C A, Paerl H W, Suba G K, Alberte R S. Immunofluorescence detection and characterization of N2-fixing microorganisms from aquatic environments. Limnol Oceanogr. 1990;35:59–71. [Google Scholar]
- 15.Dame R F, Kenny P D. Variability of Spartina alterniflora primary production in the euhaline North Inlet estuary. Mar Ecol Prog Ser. 1986;32:71–80. [Google Scholar]
- 16.Dean D R, Jacobson M R. Biochemical genetics of nitrogenase. In: Stacey G, Burris R H, Evans H J, editors. Biological nitrogen fixation. New York, N.Y: Chapman and Hall; 1992. pp. 763–834. [Google Scholar]
- 17.de Souza M P, Yoch D C. Spartina alterniflora dieback recovery correlates with increased acetylene reduction activity in saltmarsh sediments. Estuar Coast Shelf Sci. 1997;45:547–555. [Google Scholar]
- 18.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dicker H J, Smith D W. Acetylene reduction (nitrogen fixation) in a Delaware U.S.A. salt marsh. Mar Biol. 1980;57:241–250. doi: 10.1007/BF02010555. [DOI] [PubMed] [Google Scholar]
- 20.Dicker H J, Smith D W. Enumeration and relative importance of acetylene-reducing (nitrogen-fixing) bacteria in a Delaware salt marsh. Appl Environ Microbiol. 1980;39:1019–1025. doi: 10.1128/aem.39.5.1019-1025.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eardly B D, Young J P W, Selander R K. Phylogenetic position of Rhizobium sp. strain Or 191, a symbiont of both Medicago sativa and Phaseolus vulgaris, based on partial sequences of the 16S rRNA and nifH genes. Appl Environ Microbiol. 1992;58:1809–1815. doi: 10.1128/aem.58.6.1809-1815.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 23.Ferris M J, Ward D M. Seasonal distributions of dominant 16S rRNA-defined populations in a hot spring microbial mat examined by denaturing gradient gel electrophoresis. Appl Environ Microbiol. 1997;63:1375–1381. doi: 10.1128/aem.63.4.1375-1381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferris M J, Nold S C, Revsbech N P, Ward D M. Population structure and physiological changes within a hot spring microbial mat community following disturbance. Appl Environ Microbiol. 1997;63:1367–1374. doi: 10.1128/aem.63.4.1367-1374.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer S G, Lerman L S. DNA fragments differing by single base-pair substitutions are separated in denaturing gradient gels: correspondence with melting theory. Proc Natl Acad Sci USA. 1983;80:1579–1583. doi: 10.1073/pnas.80.6.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gandy E L, Yoch D C. Relationship between nitrogen-fixing sulfate reducers and fermenters in salt marsh sediments and roots of Spartina alterniflora. Appl Environ Microbiol. 1988;54:2031–2036. doi: 10.1128/aem.54.8.2031-2036.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanson R B. Nitrogen fixation activity (acetylene reduction) in the rhizosphere of salt marsh angiosperms, Georgia, U.S.A. Bot Mar. 1983;26:49–59. [Google Scholar]
- 28.Haukka K, Lindström K, Young J P W. Three phylogenetic groups of nodA and nifH genes in Sinorhizobium and Mesorhizobium isolates from leguminous trees growing in Africa and Latin America. Appl Environ Microbiol. 1998;64:419–426. doi: 10.1128/aem.64.2.419-426.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Head I M, Saunders J R, Pickup R W. Microbial evolution, diversity, and ecology: a decade of ribosomal RNA analysis of uncultivated microorganisms. Microb Ecol. 1998;35:1–21. doi: 10.1007/s002489900056. [DOI] [PubMed] [Google Scholar]
- 30.Howes B L, Teal J M. Oxygen loss from Spartina alterniflora and its relationship to salt marsh oxygen balance. Oecologia. 1994;97:431–438. doi: 10.1007/BF00325879. [DOI] [PubMed] [Google Scholar]
- 31.Hurek T, Egener T, Reinhold-Hurek B. Divergence in nitrogenases of Azoarcus spp., Proteobacteria of the β subclass. J Bacteriol. 1997;179:4172–4178. doi: 10.1128/jb.179.13.4172-4178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kirk G J D. Root ventilation, rhizosphere modification, and nutrient uptake by rice. In: Penning de Vries F W T, Teng P S, editors. Systems approaches for agricultural development. Dordrecht, The Netherlands: Kluwer; 1993. pp. 221–232. [Google Scholar]
- 33.Kirshtein J D, Paerl H W, Zehr J. Amplification, cloning, and sequencing of a nifH segment from aquatic microorganisms and natural communities. Appl Environ Microbiol. 1991;57:2645–2650. doi: 10.1128/aem.57.9.2645-2650.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kudo T, Ohkuma M, Moriya S, Noda S, Ohtoko K. Molecular phylogenetic identification of the intestinal anaerobic microbial community in the hindgut of the termite, Reticulitermes speratus, without cultivation. Extremophiles. 1998;2:155–161. doi: 10.1007/s007920050055. [DOI] [PubMed] [Google Scholar]
- 35.Lin P K T, Brown D M. Synthesis and duplex stability of oligonucleotides containing cytosine-thymine analogues. Nucleic Acids Res. 1989;17:10373–10383. doi: 10.1093/nar/17.24.10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livingstone D C, Patriquin D G. Nitrogenase activity in relation to season, carbohydrates and organic acids in a temperate zone root association. Soil Biol Biochem. 1980;12:543–546. [Google Scholar]
- 37.Lovell C R. Diversity, microbial. In: Lederberg J, editor. Encyclopedia of microbiology. 2nd ed. San Diego, Calif: Academic Press; 2000. pp. 55–70. [Google Scholar]
- 38.Lovell C R, Hui Y. Design and testing of a functional group-specific DNA probe for the study of natural populations of acetogenic bacteria. Appl Environ Microbiol. 1991;57:2602–2609. doi: 10.1128/aem.57.9.2602-2609.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lovell C R, Piceno Y M. Purification of DNA from estuarine sediments. J Microbiol Methods. 1994;20:161–174. [Google Scholar]
- 40.Marinucci A C, Hobbie J E, Helfrich J V K. Effect of litter nitrogen on decomposition and microbial biomass in Spartina alterniflora. Microb Ecol. 1983;9:27–40. doi: 10.1007/BF02011578. [DOI] [PubMed] [Google Scholar]
- 41.McClung C R, Patriquin D G, Davis R E. Campylobacter nitrofigilis sp. nov., a nitrogen-fixing bacterium associated with roots of Spartina alterniflora Loisel. Int J Syst Bacteriol. 1983;33:605–612. [Google Scholar]
- 42.Moore L R, Rocap G, Chisholm S W. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature. 1998;393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 43.Morris J T. Effects of nitrogen loading on wetland ecosystems with particular reference to atmospheric deposition. Annu Rev Ecol Syst. 1991;22:257–279. [Google Scholar]
- 44.Morris J T, Haskin B. A 5-year record of aerial primary production and stand characteristics of Spartina alterniflora. Ecology. 1990;71:2209–2217. [Google Scholar]
- 45.Muyzer G, de Waal E C, Uitterlinden A G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Newell S Y, Hopkinson C S, Scott L A. Patterns of nitrogenase activity (acetylene reduction) associated with standing, decaying shoots of Spartina alterniflora. Estuar Coast Shelf Sci. 1992;35:127–140. [Google Scholar]
- 47.Normand P, Bousquet J. Phylogeny of nitrogenase sequences in Frankia and other nitrogen-fixing microorganisms. J Mol Evol. 1989;29:436–447. doi: 10.1007/BF02602914. [DOI] [PubMed] [Google Scholar]
- 48.Ohkuma M, Noda S, Usami R, Horikoshi K, Kudo T. Diversity of nitrogen fixation genes in the symbiotic intestinal microflora of the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:2747–2752. doi: 10.1128/aem.62.8.2747-2752.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olson J B, Litaker R W, Paerl H W. Ubiquity of heterotrophic diazotrophs in marine microbial mats. Aquat Microb Ecol. 1999;19:29–36. [Google Scholar]
- 50.Olson J B, Steppe T F, Litaker R W, Paerl H W. N2-fixing microbial consortia associated with the ice cover of Lake Bonney, Antarctica. Microb Ecol. 1998;36:231–238. doi: 10.1007/s002489900110. [DOI] [PubMed] [Google Scholar]
- 51.Patriquin D G, Keddy C. Nitrogenase activity (acetylene reduction) in a Nova Scotian salt marsh: its association with angiosperms and the influence of some edaphic factors. Aquat Bot. 1978;4:227–244. [Google Scholar]
- 52.Patriquin D G, McClung C R. Nitrogen accretion, and the nature and possible significance of N2 fixation (acetylene reduction) in a Nova Scotian Spartina alterniflora stand. Mar Biol. 1978;47:227–242. [Google Scholar]
- 53.Peters J W, Fisher K, Dean D R. Nitrogenase structure and function: a biochemical-genetic perspective. Annu Rev Microbiol. 1995;49:335–366. doi: 10.1146/annurev.mi.49.100195.002003. [DOI] [PubMed] [Google Scholar]
- 54.Piceno Y M, Lovell C R. Stability in natural bacterial communities. I. Nutrient addition effects on rhizosphere diazotroph assemblage composition. Microb Ecol. 2000;39:32–40. doi: 10.1007/s002489900192. [DOI] [PubMed] [Google Scholar]
- 55.Piceno Y M, Lovell C R. Stability in natural bacterial communities. II. Plant resource allocation effects on rhizosphere diazotroph assemblage composition. Microb Ecol. 2000;39:41–48. doi: 10.1007/s002489900191. [DOI] [PubMed] [Google Scholar]
- 56.Piceno Y M, Noble P A, Lovell C R. Spatial and temporal assessment of diazotroph assemblage composition in vegetated salt marsh sediments using denaturing gradient gel electrophoresis analysis. Microb Ecol. 1999;38:157–167. doi: 10.1007/s002489900164. [DOI] [PubMed] [Google Scholar]
- 57.Rosado A S, Duarte G F, Seldin L, Van Elsas J D. Genetic diversity of nifH gene sequences in Paenibacillus azotofixans strains and soil samples analyzed by denaturing gradient gel electrophoresis of PCR-amplified gene fragments. Appl Environ Microbiol. 1998;64:2770–2779. doi: 10.1128/aem.64.8.2770-2779.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 59.Sheffield V C, Cox D R, Lerman L S, Myers R M. Attachment of a 40-base-pair G + C-rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci USA. 1989;86:232–236. doi: 10.1073/pnas.86.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Staley J T, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 61.Tanner R S, Stackebrandt E, Fox G E, Gupta R, Magrum L J, Woese C R. A phylogenetic analysis of anaerobic eubacteria capable of synthesizing acetate from carbon dioxide. Curr Microbiol. 1982;7:127–132. [Google Scholar]
- 62.Teal J M, Kanwisher J W. Gas transport in the marsh grass, Spartina alterniflora. J Exp Bot. 1966;17:355–361. [Google Scholar]
- 63.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tunlid A. Molecular biology: a linkage between microbial ecology, general ecology and organismal biology. Oikos. 1999;85:177–189. [Google Scholar]
- 65.Turner R E. Geographic variations in salt marsh macrophyte production: a review. Contrib Mar Sci. 1976;20:47–68. [Google Scholar]
- 66.Ueda T, Suga Y, Yahiro N, Matsuguchi T. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J Bacteriol. 1995;177:1414–1417. doi: 10.1128/jb.177.5.1414-1417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valiela I, Teal J M. Nutrient limitation in salt marsh vegetation. In: Reimold R J, Queen W H, editors. Ecology of halophytes. New York, N.Y: Academic Press; 1974. pp. 547–563. [Google Scholar]
- 68.Valiela I, Teal J M, Allen S D, van Etten R, Goehringer D, Volkmann S. Decomposition in salt marsh ecosystems: the phases and major factors affecting disappearance of above-ground organic matter. J Exp Mar Biol Ecol. 1985;89:29–54. [Google Scholar]
- 69.Ward D M, Ferris M J, Nold S C, Bateson M M. A natural view of microbial biodiversity within hot spring cyanobacterial mat communities. Microbiol Mol Biol Rev. 1998;62:1353–1370. doi: 10.1128/mmbr.62.4.1353-1370.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whiting G J, Gandy E L, Yoch D C. Tight coupling of root-associated nitrogen fixation and plant photosynthesis in the salt marsh grass Spartina alterniflora and carbon dioxide enhancement of nitrogenase activity. Appl Environ Microbiol. 1986;52:108–113. doi: 10.1128/aem.52.1.108-113.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Whiting G J, Morris J T. Nitrogen fixation (C2H2 reduction) in a salt marsh: its relationship to temperature and an evaluation of an in situ chamber technique. Soil Biol Biochem. 1986;18:515–521. [Google Scholar]
- 72.Widmer F, Shaffer B T, Porteous L A, Seidler R J. Analysis of nifH gene pool complexity in soil and litter at a Douglas fir forest site in the Oregon Cascade Mountain Range. Appl Environ Microbiol. 1999;65:374–380. doi: 10.1128/aem.65.2.374-380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wolfenden J, Jones K. Seasonal variation of in situ nitrogen fixation (C2H2 reduction) in an expanding marsh of Spartina anglica. J Ecol. 1987;75:1011–1021. [Google Scholar]
- 74.Zehr J P, Capone D G. Problems and promises of assaying the genetic potential for nitrogen fixation in the marine environment. Microb Ecol. 1996;32:263–281. doi: 10.1007/BF00183062. [DOI] [PubMed] [Google Scholar]
- 75.Zehr J P, Mellon M T, Hiorns W D. Phylogeny of cyanobacterial nifH genes: evolutionary implications and potential applications to natural assemblages. Microbiology. 1997;143:1443–1450. doi: 10.1099/00221287-143-4-1443. [DOI] [PubMed] [Google Scholar]
- 76.Zehr J P, McReynolds L A. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol. 1989;55:2522–2526. doi: 10.1128/aem.55.10.2522-2526.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zehr J P, Mellon M, Braun S, Litaker W, Steppe T, Paerl H W. Diversity of heterotrophic nitrogen fixation genes in a marine cyanobacterial mat. Appl Environ Microbiol. 1995;61:2527–2532. doi: 10.1128/aem.61.7.2527-2532.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zehr J P, Mellon M T, Zani S. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl Environ Microbiol. 1998;64:3444–3450. doi: 10.1128/aem.64.9.3444-3450.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]