Abstract
Causal analysis of observational studies requires data that comprise of a set of covariates, a treatment assignment indicator, and the observed outcomes. However, data confidentiality restrictions or the nature of data collection may distribute these variables across two or more datasets. In the absence of unique identifiers to link records across files, probabilistic record linkage algorithms can be leveraged to merge the datasets. Current applications of record linkage are concerned with estimation of associations between variables that are exclusive to one file and not causal relationships. We propose a Bayesian framework for record linkage and causal inference where one file comprises all the covariate and observed outcome information, and the second file consists of a list of all individuals who receive the active treatment. Under certain ignorability assumptions, the procedure properly propagates the error in the record linkage process, resulting in valid statistical inferences. To estimate the causal effects, we devise a two-stage procedure. The first stage of the procedure performs Bayesian record linkage to multiply impute the treatment assignment for all individuals in the first file, while adjustments for covariates’ imbalance and imputation of missing potential outcomes are performed in the second stage. This procedure is used to evaluate the effect of Meals on Wheels services on mortality and healthcare utilization among homebound older adults in Rhode Island. In addition, an interpretable sensitivity analysis is developed to assess potential violations of the ignorability assumptions.
Keywords: Record Linkage, Missing Data, Causal Inference, Multiple Imputation, Bayesian Data Analysis
1. Introduction.
Meals on Wheels (MOW) programs are community-based social service organizations that provide home-delivered meals to homebound older adults in order to reduce hunger and food insecurity, promote socialization, and promote community independence. Providing home-delivered meals to these populations is associated with beneficial nutritional outcomes, decreased risk of falls, and improved mental health (Thomas and Mor (2013), Campbell et al. (2015), Thomas, Akobundu and Dosa (2015), Thomas et al. (2018a), Thomas et al. (2018b), Thomas et al. (2018c)). However, healthcare payers, providers, and policy makers are also interested in the effects of community based organizations, like MOW, on pre-mature mortality and healthcare utilization, such as hospitalizations, emergency department visits, and nursing home placement.
One major challenge to performing such research is that MOW, by the nature of the services provided, do not submit medical claims or maintain clients’ health records. Medicare enrollment records and claims data contain comprehensive information on patients’ demographics, diagnoses, healthcare utilization, and long-term health outcomes, but exclude information about receipt of social services, such as MOW. In order to estimate the effects of MOW services on the healthcare utilization of its clients, linkage of Medicare claims data to MOW client data is required. However, data confidentiality restrictions prevent unique identifiers to be available for linking.
Many studies seek to draw causal conclusions about the impact of an intervention. Randomized experiments are the gold standard for inferring causality; however, they are sometimes infeasible because of logistical, ethical, or financial considerations. In these instances, researchers are limited to use non-randomized observational studies to estimate the causal effects. Causal analysis of observational studies requires data that comprise a set of covariates, a treatment assignment indicator, and the observed outcome (Rubin (1973a), Rubin (1973b), Rubin (1978), Rubin (1979)). In some studies, because of confidentiality restrictions or the nature of the data collection, the covariates, treatment assignment, and outcome information are distributed across two or more data sources without unique identifiers to link records that belong to the same entity. One way to overcome this limitation is to incorporate an initial record linkage step in the design phase of the study.
Record linkage is a set of statistical procedures that identifies individuals or entities that are shared across datasets (Jaro (1989), Winkler (1993)). Record linkage techniques can be classified into two broad classes: deterministic and probabilistic. Deterministic record linkage methods identify records that represent the same entity based on deterministic agreement functions between data elements common to both records. Probabilistic methods calculate probabilities or weights that a pair of records represents the same entity. The Fellegi-Sunter procedure estimates the probabilities of observed agreement patterns of data elements between a pair of records if these records were true links or false links (Fellegi and Sunter (1969)). These probabilities are commonly used in an iterative algorithm that classifies the pair of records with the highest probability as a link, and then removes these records from the pool of possible links. This process continues until a certain threshold is reached (Larsen and Rubin (2001)). The remaining records are either sent for clerical review or are declared non-links. Deterministic methods are used widely in practice and are reported to yield a higher proportion of true links than probabilistic methods (Gomatam et al. (2002), Campbell, Deck and Krupski (2008)). However, when the data elements are subject to variations in spelling, data entry inaccuracies, or incompleteness, deterministic methods may miss more true links than probabilistic linking methods (Gomatam et al. (2002), Campbell, Deck and Krupski (2008)). In applications involving large public health datasets, these missed links may limit the practicality of deterministic linking methods (Newman et al. (2009)). Probabilistic linkage methods are less sensitive to errors among the identifying fields, and some of these methods such as hit-miss models can account for measurement error or missingness within records (Copas and Hilton (1990)).
Both probabilistic and deterministic methods may suffer from incorrectly linked entities. Neter, Maynes and Ramanathan (1965) noted that in finite population sampling, a small amount of incorrectly linked records can lead to biased regression estimates and inflated variances. Multiple methods have been developed to reduce bias and propagate linkage errors using linear and generalized linear models (Scheuren and Winkler (1993), Scheuren and Winkler (1997), Lahiri and Larsen (2005), Chambers et al. (2009), Hof and Zwinderman (2012)). However, these methods are model specific and assume that the linkage probabilities are known or estimable. Further, all of these methods rely on the non-informative linkage assumption, which states that the outcome of interest is conditionally independent from the linkage process given the variables that appear in both files.
Bayesian file linkage procedures are probabilistic record linkage techniques that were proposed as possible solutions to overcome some of these limitations (Fortini, Liseo and Nuccitelli (2001), Larsen (2005), Tancredi and Liseo (2011), Gutman, Afendulis and Zaslavsky (2013), Steorts (2015), Steorts, Hall and Fienberg (2016), Sadinle (2017)). These methods introduce a latent structure indicating the pairing of records from one file with records from another file. By generating multiple samples from the posterior distribution of the latent linking structure, Bayesian record linkage procedures account for the uncertainty in the linkage process.
The objective of the previously mentioned Bayesian and Frequentist methods is to estimate marginal and conditional associations and not causal effects. Wortman and Reiter (2018) proposed a method for estimating causal effects using linked observational data sources and described the possible effects of errors in linkage when estimating causal effects. The proposed method incorporates record pair selection strategies that can improve treatment effect estimation when using propensity score subclassification. However, this method does not adjust for error in the linkage process, is limited to the application of propensity score subclassification, and only applies to settings in which the treatment assignment and covariates are in one file and the outcomes are in another.
Based on the potential outcome framework originally proposed by Neyman (1923) in the context of randomization-based inference in randomized experiments and generalized to other settings in Rubin (1978), we propose a joint Bayesian framework for record linkage and causal inference where one file comprises all of the covariates and observed outcome information, and the second file consists of a list of all individuals who receive the active treatment. To estimate the causal effects, we propose a computationally efficient two-stage procedure that accounts for the uncertainty in the linkage process and the unobserved potential outcomes under certain ignorability assumptions. Bayesian record linkage is performed in the first stage to inform the treatment assignment for all units in the first file. Adjustments for covariates’ imbalance and imputation of the unobserved potential outcomes are performed in the second stage. In addition, we develop a procedure to examine the sensitivity of our results to the ignorability assumptions. We apply this procedure to estimate the effect of MOW services on mortality and healthcare utilization among homebound older adults in Rhode Island, and find that MOW receipt does not have a significant effect on 30 day hospitalization rates or nursing home stays.
2. Framework.
2.1. Notation.
The potential outcomes framework posits that for a population of size , where can be infinite, the effect of a binary treatment W on outcome Y for unit i is the comparison of two “potential” outcomes, Yi(0) and Yi(1), which correspond to the two possible levels of W: Wi = 1 indicates the receipt of the active level of the treatment, and Wi = 0 indicates the receipt of the control level. We assume the stable unit treatment value assumption (SUTVA) (Rubin (1980), Rubin (1990)), so that this notation is functionally well defined. For each unit i, there is also a vector of P covariates that are unaffected by Wi, Xi = (Xi1,…, XiP).
We assume that the observed data is distributed across two files: A and B, which comprise nA and nB records, respectively. The P covariates are partitioned into P1 covariates that only appear in file A, XA = (X1,…, XP1), P2 covariates that only appear in file B, XB = (XP1+1,…, XP1+P2), and P3 covariates that appear in both files and can be used as semi-identifying information ZA = ZB = (XP1+P2+1,…, XP), where P1 + P2 + P3 = P. Record l ∈ {1,…,nA} in file A comprise XAl, ZAl, and the observed outcome . Record j ∈ {1,…, nB} in file B comprise XBj and ZBj. We further assume that file B represents a list of records that all receive the active treatment, such that Wj = 1, ∀j = 1 … , nB, and that all records receiving the active treatment in file A also appear in file B, or {l ∈ A : Wl = 1} ∈ B.
We introduce a latent structure C = (C1,…, CnA), which represents the link designations in file B for each record in file A:
| (1) |
An immediate consequence of this definition is that all records in A with Cl > 0 receive the active treatment (Wl = 1), and the remaining records with Cl = 0 receive the control treatment (Wl = 0). The observed potential outcomes are defined as , where . Note that the information in XB for the units with Cl = 0 is missing. We define , where represents the observed information for records in A with Cl > 0 and the records in B with j ∉ C that do not link with any record in A, and represents the unobserved information for records in A with Cl = 0 that do not link with any record in B.
To summarize, XA, ZA, and Yobs are observed in A, and XB, ZB, and W are observed in B. The additional unobserved variables are C and , where . The joint distribution of the observed and unobserved data across both files is
| (2) |
2.2. Causal Estimand.
Causal treatment effects are commonly summarized by estimands, τ = τ(Y(0), Y(1), W) = τ(Yobs, Ymis, W), which are functions of the unit level potential outcomes of all units, on a common subset of units (Rubin (1978)). A Bayesian inference for the effects of an exposure using linked data source considers the observed values XA, , ZB, ZA and Yobs as a realization of random variables, and , Ymis, C, and W to be unobserved random variables. This perspective explicitly confronts the observed and missing random variables by conditioning on the observed variables in A and sampling from the posterior distribution of τ:
| (3) |
Equation (3) shows that obtaining the posterior distribution of τ involves integrating over
| (4) |
2.3. Simplifying Assumptions.
To perform the integration in Equation (3), we make the following simplifying assumptions. Some of these assumptions are made explicitly in many applications and some are made implicitly.
Assumption 1. The covariates that are unique to file B are independent from the potential outcomes given XA and ZA. Formally,
| (5) |
This assumption implies that only the covariates in A are required to predict the potential outcomes, and that XB and ZB may only be informative for the linkage process. This assumption is valid in our application, because ZB represents the same covariates as ZA, and XB is only composed of administrative variables that are not influencing the health outcomes given the clinical and demographic covariates defined in XA. It is possible to incorporate additional covariate information in XB by using values of for linked records in A with Cl > 0 and imputing for records with Cl = 0.
Assumption 2. The treatment assignment mechanism is a deterministic function of the linkage structure.
Because file B comprise of all units that received the active treatment, the treatment assignment for units l ∈ A can be derived as a deterministic function of their linkage status, Wl = g(Cl), where
| (6) |
This implies that f(W∣XA, XB, ZA, ZB, Y(0), Y(1), C) is a degenerate distribution that is completely defined by C, and that τ = τ(Y(0), Y(1), W) = τ(Yobs, Ymis, C).
Assumption 3. The linkage is strongly non-informative.
This assumption states that C is conditionally independent from the potential outcomes and any unobserved data components. Formally,
| (7) |
This is a modified version of the non-informative linkage assumption commonly made when estimating non-causal associations using linked data. The implicit non-informative linkage assumption in the Fellegi-Sunter record linkage model implies that the linkage structure is conditionally independent from Yobs, XA, and XB given ZA and ZB (Harron, Goldstein and Dibben (2015)). Equation (7) also implies that the treatment assignment is unconfounded (Rubin (1990)).
By combining Assumptions 1-3, Equation (2) can be expressed as:
| (8) |
These assumptions simplify Equation (3) to:
| (9) |
2.4. Parametric Models.
Given the linkage structure C, we can assume the distributions of XAl, XBl, ZAl, ZBl, Yl(0), and Yl(1) are row exchangeable. Using de-Finetti’s theorem, Equation (8) can be written as a product of independent random variables given the parameters Θ = (θX, θY·X, θC) where θX = (θXA, θXB, θZA, θZB), θY·X = (θY1·X, θY0·X), and θC = (θCM, θCU). When the parameters θX, θY·X, and θC are a-priori independent with prior distribution p(Θ) = p(θX)p(θY·X)p(θC), Equation (8) for linked and unlinked data can be expressed as:
| (10) |
The information in the second line in Equation (10) represents the data components for linked records in A, the third line reflects the information for unlinked records in A, and the last product represents the unlinked records in B. The integrals over θX pass through the distributions for Yl(0), Yl(1), and C such that the marginal distribution f(XA, XB, ZA, ZB) is irrelevant for θY·X and θC.
We will make the assumption of no contamination of imputation across treatments (Rubin (2007)):
Assumption 4. The conditional distribution of potential outcomes for the exposed and unexposed units are independent given baseline covariates, and the parameters governing their distributions are a-priori independent.
| (11) |
Based on this assumption, the conditional distribution for the potential outcomes is:
| (12a) |
| (12b) |
| (12c) |
| (12d) |
| (12e) |
The first factor in Equation (12d) and the first factor in Equation (12e) are the posterior predictive distributions of the unobserved potential outcomes and the remaining terms in each line are the posterior distributions of θY0·X and θY1·X, respectively.
By combining Equation (9) with Equation (10-12), the causal estimand is:
| (13) |
2.5. Record Linkage Models.
To model f(C, θC∣XA, , ZA, ZB), we will rely on the record linkage framework initially proposed by Fellegi and Sunter (1969). This framework considers the set of all possible nA×nB pairs of records from A and B as the union of two disjoint sets of links M = {(l, j) : l ∈ A, j ∈ B, Cl = j} and non-links U = {(l, j) : l ∈ A, j ∈ B, Cl ≠ j}. Without a loss of generality, assume nA ≥ nB. To ensure that each record in file A is linked to at most one record in file B and vice versa, we introduce the following constraint on , ∀ j = 1,…, nB (Larsen (2005), Sadinle (2017)).
For records l ∈ A and j ∈ B, let Γ(ZAl, ZBj) = (γlj1,…,γljP3) be an agreement vector for the k = 1,…, P3 identifying variables that exist in both files. The agreement of field k between two values can be evaluated on an ordinal scale with rk = 1,…, Rk levels, where 1 represents complete disagreement, and Rk represents complete agreement (Winkler (1990), Sadinle (2017)). Let θCM = {θCMk} and θCU = {θCUk} represent the parameters governing the distributions of the comparison functions for record pairs in M and U, respectively, such that θCMk = {θCMkr}, where θCMkr = Pr(γljk = r∣Cl = j) for k = 1,…,P3 and r = 1,…,Rk, and similarly, θCUk = {θCUkr} and θCUkr = Pr(γljk = r∣Cl ≡ j).
Mixture models have been proposed to estimate θCM, θCU, and C based on Γ(ZAl, ZBj) (Jaro (1989), Larsen and Rubin (2001)):
| (14) |
where represents the number of true links.
Let π represent the expected proportion of records that represent the same entities in both files, such that nm ~ Binomial(nB, π) and π ~ Beta(απ, βπ) a-priori. Sadinle (2017) proposes a Beta-Binomial prior for C and nm that marginalizes over π:
| (15) |
A simplifying assumption that is frequently made in the Fellegi and Sunter record linkage model is that each of the comparison functions are conditionally independent given C (Winkler (1988), Jaro (1989)). Under this assumption, the likelihood for C, θCM, θCU is
| (16) |
Independent conjugate priors θCMk ~ Dirichlet(αMk1,…, αMkRk) and θCUk ~ Dirichlet(αUk1,…, αUkRk) for k = 1,…, K can be specified to complete the Bayesian model.
To sample from , we will use the data augmentation algorithm (Tanner and Wong (1987)). The I-Step involves drawing C[t+1] from and the P-step will update values of from f(θC∣ZA, ZB, C[t+1]) (see Appendix A for a detailed description).
2.6. Causal Treatment Effect Estimation.
The record linkage facilitates the identification of the exposed and unexposed units, but it does not guarantee that units are similar across treatment groups. When the distribution of covariates between the treatment and control groups are different, simple comparison of the two groups may result in biased estimates of the treatment effect (Rubin (1973a), Rubin (1973b)). Several types of procedures have been proposed to address this issue when the treatment effect is unconfounded (Imbens and Rubin (2015), Gutman and Rubin (2017)).
Matching is a design phase causal estimation technique that reduces bias by identifying units with similar covariate values between the two treatment groups (Stuart (2010)). With a single covariate, it is often easy to identify a match. This task is more complicated with multiple covariates (Stuart (2010)). Matching on the propensity score (Rosenbaum and Rubin (1983)), which is the probability of Wl = 1 given the covariates, was proposed as a possible solution. Formally, the propensity score for unit l is defined as e(XAl, ZAl) ≡ f(Wl∣XAl, ZAl, ϕ), where ϕ are the parameters governing this distribution. Point estimates of τ using matching have been shown to be consistent, but may underestimate its sampling variance when ignoring the variability in the matching procedure (Abadie and Imbens (2011), Gutman and Rubin (2017)). In addition, because matching on e(XAl, ZAl) is not exact, some covariates may still suffer from minor imbalances, which is often addressed using regression adjustments (Imbens and Rubin (2015)).
A different approach to estimate τ is to combine matching with a Bayesian imputation framework (Rubin (2007), Gutman and Rubin (2013), Gutman and Rubin (2015)). This combination reduces the bias resulting from minor covariate imbalances and increases precision by using modeling to impute Ymis. Under the Bayesian causal inference framework, the missing potential outcomes are taken to be unobserved random variables that can be sampled from their posterior predictive distribution. Because sampling from a posterior distribution is complex, we use a multiple imputation procedure as an approximation of the posterior distribution of Ymis (Gutman and Rubin (2013), Gutman and Rubin (2015)).
2.7. Two-Stage Multiple Imputation Estimation Procedure.
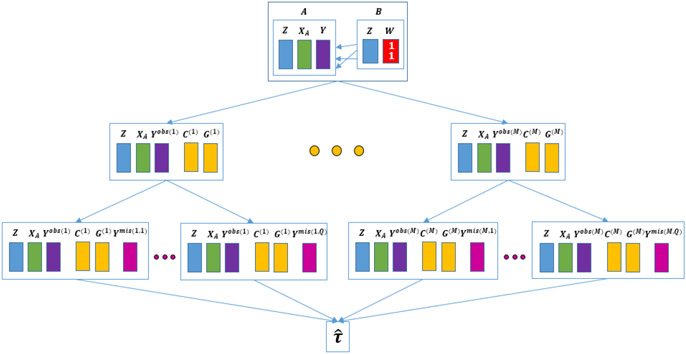
Equation (13) suggests a two-step estimation procedure. In the first step, the record linkage structure is sampled. Using this linkage structure, the potential outcomes are imputed in the second step.
We now explicate and summarize this two stage multiple imputation approach (Shen (2000), Rubin (2003)) to estimate τ
Sample C(m) from f(C, θC∣XA, , ZA, ZB) for m = 1,…,M random draws (Appendix A).
- For C(m), m = 1,…,M,
- Perform nearest neighbor matching using the estimated the propensity score to obtain a sample of exposed and unexposed units with similar covariate distributions. Let G(m) = 1 represent the units in this matched sample.
- For the units with , partition Y(0), Y(1) into Yobs(m) and Ymis(m).
- Sample , q = 1,…, Q, from p(θY0·X∣XA, ZA, Y(0)obs(m), G(m) = 1) and from p(θY1·X∣XA, ZA, Y(1)obs(m), G(m) = 1).
- For each q = 1,…, Q, use and to independently impute the missing potential outcomes for each unit in G(m) = 1 from the posterior predictive distributions and . This will result in Q datasets with imputed Y(0)mis and Y(1)mis for the matched units.
For each of the M×Q imputations, estimate the treatment effect and the within imputation sampling variance, U(m,q).
The point estimate of the treatment effect τ is calculated as . The total variance can be derived according to Shen (2000) as , where , , and . Inference for is based on a t-distribution with .
An illustration of this two-stage multiple imputation procedure is presented in Figure 1.
Fig 1.
Two Stage Multiple Imputation Procedure
3. Application to Meals on Wheels Data.
We applied the proposed procedure to estimate the effects of Meals on Wheels (MOW) programs on mortality and healthcare utilization among Medicare beneficiaries. We compared the difference in mortality rate after 30 days of initiating meal delivery service, and the difference in the frequency of acute inpatient, emergency department (ED), and nursing home (NH) events between MOW recipients and non-recipients who were alive after 30 days of enrollment in both the observed treatment and predicted control arms. Because we do not expect MOW receipt to influence mortality, identification of such an effect may indicate potential violations of the unconfoundedness assumption.
3.1. Data Description.
A comprehensive list of all clients who received home-delivered meals between January 1, 2010 and December 31, 2013 was submitted by Meals on Wheels Rhode Island (MOWRI), which serves the entire state of Rhode Island. This list contained information on each client’s sex, date of birth, start and end date of service, and the 9-digit ZIP code corresponding to the address where their meal was delivered. While MOW serves clients of a variety of ages, we restricted the analysis only to individuals older than 65 at enrollment in MOW, because only those individuals are expected to be enrolled in Medicare. This resulted in a total of nB = 3,916 MOW recipients.
The Medicare Master Beneficiary Summary File (MBSF) is a comprehensive database that contains demographic information on all Medicare enrollees including gender, date of birth, date of death, and the 9-digit ZIP code corresponding to their mailing address. In addition, the MBSF identifies pre-existing chronic medical conditions including congestive heart failure, kidney failure, diabetes, pelvic fracture, stroke, dementia or Alzheimer’s disease, chronic obstructive pulmonary disease, coronary artery disease, and various types of cancer. Using unique identifiers, the MBSF is linked to Medicare inpatient, outpatient, skilled nursing facility, and home health claims as well as the nursing home Minimum Data Set (MDS) between calendar years 2009 and 2014. These claims and assessment data were used to calculate the frequency and cost of inpatient events, emergency department visits, nursing home stays, and home health utilization in the 30, 90, and 180 days prior to enrollment in MOW, as well as the frequency of acute inpatient, ED, and NH events 30 days following MOW enrollment. A total of nA = 179,269 Medicare beneficiaries over the age of 65 resided in the 5-digit ZIP codes serviced by MOWRI.
3.2. Record Linkage of MOW and Medicare Data.
Comparison of all possible record pairs, where one record appears in the MOW file and another record in the Medicare file would result in over 700 million possible record pairs. “Blocking” is a common record linkage to reduce the computational complexity by only considering record pairs that agree on specific blocking fields (Newcombe et al. (1959), Newcombe (1988)). We generated blocks based on 5-digit ZIP codes and gender (Herzog, Scheuren and Winkler (2007)). We assumed that θCM and θCU do not differ across blocks, and we restricted record pairs such that the MOW enrollment date precedes the date of death in the Medicare file. These criteria reduced the total number of possible record pairs to 13,786,172. A sensitivity analysis of our results to this blocking criteria is provided in Appendix D.
A recipient’s date of birth (DOB) and 9-digit ZIP code were used as linking variables to classify record pairs into links and non-links. Ordinal agreement patterns were used for both linking variables, whose levels of agreement are described in Table 1. In addition, we modeled the interaction between agreement on DOB and ZIP code (Winkler (1989)). The resulting record linkage likelihood is
| (17) |
where Blj is an indicator that record pair (l, j) was successfully blocked and met the MOW enrollment date constraint, and θCM = {θCMD, θCMZ} and θCU = {θCUD, θCUZ} are the parameters governing the distribution of agreement functions, such that θCMD = {θCMDrD}, θCMZ = {θCMZrZ∣rD}, θCUD = {θCUDrD}, θCUZ = {θCUZrZ∣rD}. Each θCMDrD = Pr(γljD = rD∣Cl = j, Blj = 1), θCMZrZ∣rD = Pr(γljZ = rZ∣γljD = rD, Cl = j, Blj = 1), θCUDrD = Pr(γljD = rD∣Cl ≠ j, Blj = 1), and θCUZrZ∣rD = Pr(γljZ = rZ∣γljD = rD, Cl ≠ j, Blj = 1) for rD = 1,…,4 and rZ = 1,…,5. A total of M = 100 different linkage structures were imputed from the linkage algorithm.
Table 1.
Linking Variable Description and Agreement Level
| Agreement Type | Level |
|---|---|
| Disagreement on DOB | rD = 1 |
| Agree on DOB Year only | rD = 2 |
| Agree on DOB Year and Month only | rD = 3 |
| Agree on DOB Year, Month, and Day | rD = 4 |
| Agree on first 5 digits of ZIP code only | rZ = 1 |
| Agree on first 6 digits of ZIP code only | rZ = 2 |
| Agree on first 7 digits of ZIP code only | rZ = 3 |
| Agree on first 8 digits of ZIP code only | rZ = 4 |
| Agree on all 9 digits of ZIP code | rZ = 5 |
3.3. Propensity Score Matching.
Each of the m = 1,…, M linked datasets identifies Medicare beneficiaries who received MOW and those who did not. Prior research suggested that MOW programs target older adults who have higher social and economic needs and are at higher risk for institutional care (Lloyd and Wellman (2015), Lee, Shannon and Brown (2015)). Thus, enrollment in MOW programs may be confounded with pre-existing health conditions or prior healthcare utilization. To reduce covariates’ imbalances, matching on the estimated propensity score was performed.
Prior to matching, all linked individuals who were enrolled in a Medicare Advantage (MA) program in the six months prior to receiving MOW, or were enrolled in MA during the month they began MOW, were removed. This truncation was implemented because MA plans are not required to submit claims, and it was not possible to fully observe the prior history of chronic conditions or healthcare utilization for these individuals. A start date for individuals who are not enrolled in MOW programs is not available. Instead, for individuals who were not linked to MOW records, we calculated the medical history and prior healthcare utilization at the start of each quarter for every year in our study period. This resulted in sixteen sets of pre-treatment covariates calculated at different potential enrollment dates for each unlinked individual.
Matching was implemented by enforcing exact agreement on patients’ gender, race, age categories, and whether the patients had any inpatient, ER, or SNF claim in the 90 days prior to enrollment. The remaining covariates were balanced using propensity score models that included pre-existing medical conditions, prior healthcare utilization frequency, and prior healthcare costs. We selected nearest pair matches without replacement based on the propensity score within each quarter (Stuart (2010)). This process was replicated on each of the M = 100 linked datasets to identify beneficiaries that resemble MOW recipients, but did not enroll in the program.
3.4. Imputation of Unobserved Outcomes.
To assess the impact of MOW on mortality, we examine the average treatment effect on the treated (ATT) among linked MOW clients who are matched to a control individual. Let Dl(1) and Dl(0) represent the potential 30 day mortality for individual l had they received meals or not, respectively. The estimand of interest is τATT = E(D(1) − D(0)∣W = 1, G = 1), which can be estimated within each imputation as
| (18) |
where represents the number of linked MOW clients matched with a control. To predict the unobserved 30 day mortality for MOW clients had they not received meals, we used a Bayesian logistic regression model that included the covariates used in matching, XAl1, … XAlP1, and all of their two-way interactions:
| (19) |
A one-level hierarchical Normal-Gamma shrinkage prior (Griffin and Brown (2017)) was constructed for the coefficients of the main effect and interaction terms such that , Φ1 ~ Gamma(1, 1), , and Φ2 ~ Gamma(1, 2). These prior distributions attenuate interaction terms more aggressively. We also assumed that β0 ~ N(0, 10000).
To examine the impact of MOW on healthcare utilization, we estimate the survivor average treatment effect on the treated (SATT), which compares the effect of MOW receipt among MOWRI clients who would be alive 30 days after their enrollment date irrespective of whether they received services from MOW or not (Frangakis and Rubin (2002), Rubin (2006), Frangakis et al. (2007)). The SATT is defined as τSATT = E(H(1) − H(0)∣W = 1, D(0) = D(1) = 0, G = 1), where H(1) and H(0) denote the potential utilization frequency among MOW clients and controls, respectively. An estimate for the SATT within each imputation is estimated as
| (20) |
where is the number of linked individuals that were matched to a control and who are alive after 30 days following enrollment according to their observed and predicted mortality status.
Bayesian zero-inflated negative binomial models were fitted to impute the frequency of inpatient admissions, emergency department visits, and nursing home stays among MOW clients had they not received meals within each imputed linkage structure. The zero-inflated negative binomial distribution for count response Hl(0) is given by
| (21) |
where α represents the shape parameter,
| (22) |
and
| (23) |
The negative binomial component is modeled in Equation (22) and Equation (23) models the zero-inflation. To complete the Bayesian model, we assumed that , Φ3 ~ Gamma(1, 1), , Φ4 ~ Gamma(1, 2), , Φ5 ~ Gamma(1, 1), , and Φ6 ~ Gamma(1, 2). Lastly, τ ~ U(0, 1000), ζ0 ~ N(0, 10000) and ψ0 ~ N(0, 10000). All models were fit using Rstan version 2.17.2 (Stan Development Team (2018)). All outcomes were imputed for 100 datasets within each of the 100 linked datasets, resulting in M × Q = 10,000 complete datasets.
4. Results.
Of the nB = 3916 MOW clients eligible for linkage to Medicare data, an average of records (95% CI: 3570.91, 3645.14) were linked over M = 100 imputations. Among the Medicare beneficiaries that were linked to MOW clients, an average of 1748.35 (95% CI: 1735.28, 1761.44) individuals had at least 1 month of MA coverage in the 6 months prior to enrollment and were excluded from our analysis. Of the remaining linked individuals, an average of 1859.67 (95% CI: 1835.63, 1883.70) treated units were matched to a control unit that did not receive meals.
Figure 2 displays the median and range of absolute standardized differences for each of the pre-treatment variables between the MOW and control samples before and after matching for the M = 100 linked datasets. The absolute standardized difference exceeded 0.25 in 22 out of the 40 covariates prior to matching. After matching, all absolute standardized differences are less than 0.25 for all covariates, which suggests that the covariates are adequately balanced (Rosenbaum and Rubin (1985), Imbens and Rubin (2015)).
Fig 2.
Covariate balance before and after propensity score matching for M = 100 linked data sets. The points represent the median absolute standardized differences, while the horizontal lines represent the range between the minimum and maximum absolute standardized difference for each covariate.
The observed and imputed potential outcomes for MOW clients, and the estimated treatment effects are provided in Table 1. The estimated ATT on mortality using our two-stage multiple imputation procedure is 0.008 (95% CI: −0.067, 0.083). This indicates that there is no significant difference between the observed and predicted mortality rate among the linked and matched MOW clients. An average of 51.21 individuals died within 30 days of their MOW enrollment or were predicted to die within 30 days without MOW services. This results in an average of 1808.46 individuals who are alive whether they received MOW or not across imputations. Among these individuals, the estimated SATTs are 0.010 (95% CI: −0.174, 0.194) on inpatient acute admissions, −0.013 (95% CI: −0.236, 0.209) on ED visits, and 0.003 (95% CI: −0.268, 0.274) for NH stays. This suggests that among MOW recipients who would be alive 30 days after enrollment irrespective of whether they received MOW or not, no significant differences in the number of acute inpatient admissions, ED visits, or NH stays are detected.
5. Sensitivity Analysis.
5.1. Sensitivity of the Strongly Non-Informative Linkage Assumption.
We examine the sensitivity of our results to the strongly non-informative linkage assumption (Assumption 3). Under Assumption 3, errors in the linkage only depend on comparisons of semi-identifying information that exists in both files. Thus, the probability of record l ∈ A forming a link with record j ∈ B given C(−l) = (Cl′ : l′ ≠ l} is (see Appendix B for additional details)
| (24) |
To examine the impact of potential violations of the strongly non-informative linkage assumption on the estimation of the treatment effect, we assume that the errors in the linkage model depend on 30-day mortality status. Let be an indicator that is equal to 1 if individual l ∈ A died within 30 days of the start date indicated by record j ∈ B. Let λM and λU represent parameters governing the distribution of for links and non-links, respectively. We assume that the distribution of is
The posterior probability of individual l ∈ A linking with individual j ∈ B given C(−l) will then take the form
| (25) |
A violation of the strongly non-informative linkage assumption influences the likelihood of record pairs by a sensitivity factor of . When Δ = 0, Assumption 3 is valid. Values of Δ > 1 suggest that MOWRI was more likely to enroll individuals who may die shortly after enrollment. Values of Δ < 1 represent scenarios where MOWRI may have selectively avoided enrolling individuals who may die shortly after enrollment.
To estimate the effect of the sensitivity parameter on the outcomes, we implement the algorithm in Section 2.7 after replacing f(C, θC∣XA, , ZA, ZB) in Step 1 with Equation (25).
5.2. Sensitivity Analysis Results.
The average number of records linked for different values of Δ is presented in Table 3. Because a small proportion of the linked individuals die within 30 days after enrollment when Δ = 1, decreasing the sensitivity parameter does not yield significantly lower amounts of linked records. However, increasing the sensitivity parameter significantly increases the number of linked records, specifically record pairs that indicate death within 30 days of enrollment. At Δ = 100, the narrow confidence interval indicates the linkage algorithm links almost all of the MOW records to the Medicare enrollment file.
Table 3.
Sensitivity Analysis Linkage Results
| Δ | 95% CI | |
|---|---|---|
| 1/100 | 3566.74 | (3521.15, 3612.33) |
| 1/50 | 3571.50 | (3529.74, 3613.26) |
| 1/10 | 3580.75 | (3537.22, 3624.29) |
| 1/5 | 3589.74 | (3549.78, 3629.70) |
| 1/2 | 3607.69 | (3563.14, 3652.24) |
| 1 | 3608.02 | (3570.91, 3645.14) |
| 2 | 3610.76 | (3575.98, 3645.54) |
| 5 | 3662.55 | (3621.70, 3703.40) |
| 10 | 3714.30 | (3677.96, 3750.64) |
| 50 | 3844.21 | (3821.23, 3867.19) |
| 100 | 3874.40 | (3860.50, 3888.31) |
The difference in 30-day mortality rate and inpatient acute utilization rate for the various sensitivity levels are presented in Table 4. The estimated mortality difference is similar for values of Δ ≤ 1. While the difference in mortality between MOW clients and controls is negative when Δ = 1/100, this effect is small and insignificant. When Δ ≥ 50, the mortality difference is significant and MOW is estimated to increase mortality among their clients. This results from clients who die within 30 days being added to the linked sample, and subsequently being removed from the potential control cohort to be matched. When Δ = 100, the sensitivity parameter trumps the linkage likelihood such that many MOW clients are linked to Medicare records indicating 30 day mortality despite major disagreements between the linking information in ZA and ZB. Assuming that MOW beneficiaries are 50-100 times more likely to die within 30 days of enrollment is highly implausible. The SATT of MOW on inpatient acute admission frequency does not significantly differ across the sensitivity scenarios. These results imply that our analysis is robust to the strongly non-informative linkage assumption with regards to mortality. Similar results were observed for inpatient acute hospitalization (data not shown).
Table 4.
Sensitivity Analysis Causal Treatment Effect Estimates
| 30 Day Mortality | 30 Day Inpatient Acute | |||
|---|---|---|---|---|
| Δ | 95% CI | 95% CI | ||
| 1/100 | −0.006 | (−0.055, 0.044) | 0.006 | (−0.125, 0.137) |
| 1/50 | 0.004 | (−0.070, 0.078) | 0.009 | (−0.178, 0.197) |
| 1/10 | 0.005 | (−0.070, 0.081) | 0.009 | (−0.178, 0.195) |
| 1/5 | 0.006 | (−0.068, 0.081) | 0.009 | (−0.177, 0.196) |
| 1/2 | 0.007 | (−0.067, 0.082) | 0.010 | (−0.178, 0.197) |
| 1 | 0.008 | (−0.067, 0.083) | 0.010 | (−0.176, 0.196) |
| 2 | 0.010 | (−0.066, 0.086) | 0.010 | (−0.174, 0.194) |
| 5 | 0.016 | (−0.059, 0.092) | 0.010 | (−0.173, 0.193) |
| 10 | 0.025 | (−0.052, 0.102) | 0.010 | (−0.175, 0.194) |
| 50 | 0.086 | (0.002, 0.170) | 0.014 | (−0.170, 0.198) |
| 100 | 0.549 | (0.412, 0.686) | 0.079 | (−0.120, 0.277) |
6. Discussion.
We have proposed a novel Bayesian framework to estimate causal treatment effects using linked data sources. We examine a linkage scenario that combines covariate information and outcome information from one file with the treatment assignment defined by a second file. Under a series of conditional independence and ignorability assumptions, we provide a two-stage multiple imputation procedure to obtain statistically valid treatment effect point and interval estimates. This procedure accounts for both the errors in the linkage and the unobserved outcomes. The first stage of the procedure imputes the linkage structure, and the missing potential outcomes are imputed in the second stage. Because the strong non-informative linkage assumption cannot be examined using observed data, we developed a sensitivity analysis to assess its violations.
In the linkage setting that we considered, all of the records in file B receive the active treatment. This allows us to derive the treatment assignment as a deterministic function of the linkage status for records in file A. More research and possibly stronger assumptions are required to estimate treatment effects in different linkage scenarios, such as when one file contains the treatment assignment and some of the covariates, while the other file includes the rest of the covariates and the observed outcomes. In addition, the proposed linkage algorithm is based on the Fellegi-Sunter framework, which does not account for relationships between variables that are exclusive to one file. A possible extension would be to incorporate such relationships as described in Gutman, Afendulis and Zaslavsky (2013). The modularity of our procedure allows for adjustment of the record linkage algorithm without the need to adjust the causal inference component.
We applied our framework to estimate the effect of receiving services from a MOW program in Rhode Island on mortality and healthcare utilization for its clients. Our analysis suggested that MOW does not have a significant impact on reducing 30 day mortality among its clients. Furthermore, among clients who would be alive after 30 days irrespective of MOW services, no significant differences in the frequency of acute inpatient admissions, ED visits, or NH stays are observed. A sensitivity analysis that examined the strong non-informative linkage assumption showed that our analysis is robust against potential violations of this assumption. However, this assumption may not be valid in different applications, and designing procedures that relax this assumption is an area for future research.
A major limitation of Bayesian record linkage procedures is that they are computationally intensive, and commonly require specialized programming that most non-technical researchers cannot implement. We have addressed these issues by utilizing two-stage multiple imputation and blocking. Both of these techniques reduce the computational complexity and enable nontechnical researchers to perform the causal inference analyses while scaling down the record linkage complexity. Multiple imputation has been widely used in other missing data applications, and was shown to provide valid inferences (Rubin (1996)). We have examined the performance of our proposed two-stage imputation in simulation analyses (Appendix C). The results show that the procedure provides valid statistical inference, but that the coverage is higher than nominal. This implies that the proposed procedure may overestimate the sampling variance. Increased efficiency of such procedures is a future area of research.
Using blocking increases the computational efficiency and scalability of the record linkage procedure. However, it may exclude true links and influence subsequent inferences (Murray (2015)). We have examined the performance of our two-stage procedure with stricter and looser blocking criteria (Appendix D). Using a single CPU, the current runtime of our linkage algorithm was approximately 18 days. Loosening the blocking criteria such that the number of record pairs was more than 4 times larger resulted in a runtime of approximately 30 days. The point estimates were relatively similar for the strict and loosened blocking criteria, but the stricter criteria had larger sampling variance because less record pairs were linked. This shows that our algorithm is relatively efficient and that inferences were robust to blocking. Use of parallel computing, more efficient programming languages, and improvement of the MCMC sampling procedure based on ideas proposed by Zanella (2019) may improve the scaling of the proposed algorithm to even larger settings.
In conclusion, this manuscript describes a statistical framework to estimate causal effects using linked datasets where one file contains the covariates and the observed outcome and the second file contains the treatment assignment. Under the strongly non-informative linkage assumption we develop a two stage multiple imputation procedure that provides statistically valid treatment effect estimates, and we describe a sensitivity analysis for this assumption.
Supplementary Material
Table 2.
Estimated Causal Treatment Effects on MOW Recipients.
| 30 Day Outcome | 95% CI | ||||
|---|---|---|---|---|---|
| Mortality | 1859.67 | 0.023(0.001) | 0.015(0.004) | 0.008 | (−0.067, 0.083) |
| 30 Day Outcome | 95% CI | ||||
| Inpatient Acute | 1808.46 | 0.085(0.002) | 0.075(0.010) | 0.010 | (−0.174, 0.194) |
| Emergency Care | 1808.46 | 0.073(0.002) | 0.088(0.012) | −0.013 | (−0.236, 0.209) |
| Nursing Home | 1808.46 | 0.078(0.003) | 0.075(0.015) | 0.003 | (−0.268, 0.274) |
is the average sample size of MOW individuals linked and matched to a control individual across M = 100 imputations. is the average number of linked and matched MOW individuals who are observed and predicted to be alive irrespective of their treatment, across M × Q = 10,000 imputations. and represent the mortality rates for linked and matched MOW individuals if they received MOW or not, respectively. and are the estimated healthcare utilization rates for linked and matched individuals who are alive for 30 days irrespective of whether they received MOW or not, respectively. is the estimated average treatment effect on the treated and is the survivor average treatment effect on the treated
Acknowledgments
Supported in part by a grant from the Gary and Mary West Foundation and a grant from the Patient-Centered Outcomes Research Institute (PCORI/ME-2017C3-9241).
APPENDIX A: RECORD LINKAGE GIBBS SAMPLING ALGORITHM
A Gibbs sampling algorithm proposed by Sadinle (2017) can be used to iterate between sampling the posterior distributions of the linking parameters and the linking configuration given the observed data. Starting with initial values for C, we sample from f(C, θC∣XA, , ZA, ZB) using the following procedure:
- Sample new values of from
and from(26) (27) - Sample a new state of C[t+1] by iterating through each entry and proposing updates one entry at a time. Define as the collection of link designations without the lth entry, and let be the number of designated links at iteration [t + 1] excluding the lth entry. The posterior distribution of Cl given C(−l) is a multinomial distribution where the labels are {j : j ∉ C, 0}. The probability for l ∈ A to pair with the unlinked record j ∈ B and increase the total number of links by 1 is
Similarly, the probability for l ∈ A not pairing with any record from B and is(28) (29)
APPENDIX B: DERIVATION OF POSTERIOR LINKAGE PROBABILITIES
The posterior distribution of Cl and nm given the remaining link designations C(−l) is
| (30) |
where p(C, nm) takes the form of Equation (15).
The marginal posterior probability for record l ∈ A to form a true link with j ∈ B given C(−l) is
| (31) |
and the marginal posterior probability for record l ∈ A to remain unlinked given C(−l) is
| (32) |
Dividing the numerator and denominator of Equation (31) by
| (33) |
results in Equation (28). Similarly, dividing the numerator and denominator of Equation (32) by Equation (34) results in Equation (29). Therefore, we see that
and
APPENDIX C: SIMULATION STUDY FOR TWO-STAGE MULTIPLE IMPUTATION PROCEDURE
We examine the operating characteristics of a t-distribution approximation for inference of the treatment effect in our two-stage multiple imputation procedure described in Section 2.7 using a simulation study. We consider a simulation setting with nA = 20,000 records in file A, nB = 2,000 records in file B, and nm = 1,600 true links between both files. The treatment effect is simulated for continuous, count, and binary outcomes. Appendix Table 5 depicts the linking variables, covariates, and response surfaces used to generate the simulations. Linking variables for true-links were simulated according to f(ZA). No errors were simulated among the linking variables, such that ZA and ZB took the same values among true-links in both files. When the intervention has no effect, we assumed that f(Y(0)∣XA, ZA) = f(Y(1)∣XA, ZA). When the treatment effect exists, we assumed that it is constant on the linear, logarithm, or logistic scale for continuous, binary, and count outcomes. The response surfaces were calibrated such that E(Y(0)) is equal to 10 for the continuous outcome, 2 for the count outcome, and 0.3 for the binary outcome. A total of 100 simulated sets of data were generated from this simulation configuration.
Table 5.
Simulated Linking Variables, Covariates, and Outcomes
| Linking Variables | f(ZA) | f(ZB) | |
|---|---|---|---|
| Age | N(50,52) | N(45, 102) | |
| Gender | Bernoulli(0.5) | Bernoulli(0.5) | |
| ZIP Code | 1st Digit ~Discrete Uniform(6) | 1st Digit ~Discrete Uniform(6) | |
| 2nd Digit ~Discrete Uniform(7) | 2nd Digit ~Discrete Uniform(7) | ||
| 3rd Digit ~Discrete Uniform(7) | 3rd Digit ~Discrete Uniform(7) | ||
| 4th Digit ~Discrete Uniform(7) | 4th Digit ~Discrete Uniform(7) | ||
| f(XA) | l : Cl > 0 | l : Cl = 0 | |
| Prior Hospitalization | Poisson(1) | Poisson(0.75) | |
| Prior Log-Healthcare Cost | N(10,32) | N(6,42) | |
| Outcome Type | τ ATT | f(Y(1)∣XA, ZA) | |
| Continuous | 0 | N(7.95-0.7*Gender+0.4*PriorHosp+.2*PriorCost, 0.1) | |
| Continuous | 0.05 | N(8.00-0.7*Gender+0.4*PriorHosp+.2*PriorCost, 0.1) | |
| Continuous | 0.10 | N(8.05-0.7*Gender+0.4*PriorHosp+.2*PriorCost, 0.1) | |
| Count | 0 | Poisson(exp(0.3431-0.7*Gender+0.2*PriorHosp+0.05*PriorCost)) | |
| Count | 0.15 | Poisson(exp(0.4155-0.7*Gender+0.2*PriorHosp+0.05*PriorCost)) | |
| Count | 0.20 | Poisson(exp(0.4384-0.7*Gender+0.2*PriorHosp+0.05*PriorCost)) | |
| Binary | 0 | Bernoulli(expit(−1.1973-0.7*Gender+0.2*PriorHosp+0.05*PriorCost)) | |
| Binary | 0.04 | Bernoulli(expit(−1.0132-0.7*Gender+0.2*PriorHosp+0.05*PriorCost)) | |
| Binary | 0.08 | Bernoulli(expit(−0.8395-0.7*Gender+0.2*PriorHosp+0.05*PriorCost)) | |
To conduct record linkage of the pairs of simulated files, an individual’s continuous age values were converted to a date of birth (DOB) with a year, month, and day value. Four levels of similarity are used to compare the elements of DOB: no agreement on DOB year, agreement on DOB year only, agreement on DOB year and month only, and agreement on all elements of DOB. Five levels of similarity are used to compare ZIP codes: disagreement on the first ZIP digit, agreement on the first ZIP digit only, agreement on the first and second ZIP digits only, agreement on the first through third ZIP digits only, and agreement on all ZIP digits. Exact agreement is used to compare values of gender. Conditional independence is assumed between agreement on the three linking variables, and Equation (16) is used as the record linkage likelihood. Independent Dirichlet(1, …, 1) prior distributions are used for each θCM and θCU. M = 100 imputations of the linkage structure was taken for each simulated dataset.
Propensity score models were fitted on linked and unlinked records in A using patients’ age, gender, prior hospitalization, and prior log-healthcare cost. Nearest neighbor matching without replacement was performed based on the propensity score to identify a set of controls with similar covariate distributions as the linked records. We calculated the average treatment effect on the treated for the continuous, count, and binary outcomes by fitting Bayesian linear, Poisson, and logistic regression models using the set of matched records. Each Bayesian model contained the covariates used in matching and all of their two-way interactions similar to the model specified in Equation (19). This results in all of the imputation models being misspecified, but there is no unmeasured confounding. Non-informative prior distributions were placed on all of the parameters in each model.
Appendix Table 6 displays the , , , and Coverage over the 100 simulated datasets for the different types of outcomes and treatment effect sizes. In settings where and are approximately 1,600, our proposed two-stage procedure can accurately estimate linear, count, and binary treatment effects with minimal bias. Interval estimates according to a t-distribution approximation provides nominal type 1 error and valid statistical inference for true treatment effect for all three types of outcomes and across different treatment effect sizes. However, these intervals seem to be too wide, because the coverage probabilities are close to 1 and are not around the expected nominal coverage of 0.95.
Table 6.
Average bias, SE, coverage, and type I error or power for different outcome distributions and treatment effect sizes.
| Outcome Type | τ ATT | Coverage | |||
|---|---|---|---|---|---|
| Linear | 0 | 0.0006 | 0.0041 | 0.0006 | 1 |
| 0.05 | 0.0695 | 0.0102 | 0.0195 | 1 | |
| 0.10 | 0.1187 | 0.0102 | 0.0187 | 1 | |
| Count | 0 | 0.0296 | 0.0608 | 0.0296 | 1 |
| 0.15 | 0.1688 | 0.0606 | 0.0188 | 1 | |
| 0.20 | 0.2608 | 0.0611 | 0.0608 | 1 | |
| Binary | 0 | 0.0005 | 0.0182 | 0.0005 | 1 |
| 0.04 | 0.0410 | 0.0182 | 0.0010 | 1 | |
| 0.08 | 0.0784 | 0.0182 | −0.0016 | 1 |
APPENDIX D: SENSITIVITY ANALYSIS OF BLOCKING CRITERIA
In our linkage of MOW clients to Medicare enrollment records, blocks were generated based on clients’ gender and the first 5 digits of ZIP code to reduce the number of possible record pairs and increase the efficiency of the linkage procedure. Sadinle and Fienberg (2013) demonstrates that blocking can significantly increase the accuracy of the linkage and subsequent inference even when record linkage is computationally feasible without blocking, especially when the linking variables are limited or prone to error. However, Murray (2015) notes that blocking on variables that may be recorded with error can exclude true matches and influence the subsequent inference on the linked data. We examine the sensitivity of our results to different blocking criteria based on ZIP code digits.
Let nZ represent the number of ZIP code digits used in the blocking criteria, where nZ = 5 in our application. To examine the potential impact of different blocking restraints on the estimation of the causal treatment effect, we consider different values of nZ = (4, 6, 7). Altering the blocking criteria shifts the number of ZIP code digits available as linking variables. The record linkage likelihood in Equation (17) can be re-expressed in terms of nZ as
| (34) |
Treatment effects were estimated according to the algorithm in Section 2.7 after replacing the record linkage likelihood under each blocking scenario with Equation (34).
D.1. Sensitivity Analysis Results.
A comparison of the computational complexity of the linkage for different blocking scenarios and the linkage results are shown in Table 7. While increasing the number of ZIP digits used for blocking significantly reduces the computational complexity, there is also a significant decrease in the number of records that are linked. This is likely due to potential errors or discrepancies in how the 6-9 ZIP code digits are recorded across both files. When blocking on these error-prone ZIP code digits, many true links are classified as non-links. Increasing the blocking constraints also reduces the number of available linking variables, which increases the efficiency of the computation as well. Loosening the blocking criteria to the first 4 ZIP code digits results in more than a 4-fold increase in the number of possible record pairs, as well as an increase in the number of records linked.
The estimates of the causal treatment effects for mortality and acute inpatient admissions for different blocking criteria are presented in Appendix Table 8. Overall, we see that the use of blocking on the first 5 ZIP code digits in our application provides similar results compared to less strict blocking criteria. Gender and the first 5 digits are ZIP code are well defined and unlikely to be reported with errors, which would not result in biased point estimates or suboptimal interval estimates if used as blocking criteria. Using stricter blocking criteria generally results in fewer false links but possibly more true links missed. The point estimates for the stricter criteria were practically the same, but the interval estimates were larger because of the smaller number of true links that were identified.
Table 7. Blocking Sensitivity Analysis Linkage Complexity and Results.
nA represents the number of unique records in the Medicare data, nAnB represents the total number of possible record pairs that are partitioned into a gender and ZIP code block, nm represents the average number of linked records over M = 100 imputations, and run time reflects the approximate time in days our Bayesian record linkage algorithm required to complete 500 iterations using a single CPU core on a Linux system.
| Blocking Criteria | nA | nAnB | 95% CI | Run time | |
|---|---|---|---|---|---|
| 4 digit ZIP | 251,285 | 56,706,359 | 3829.67 | (3814.40, 3844.94) | 30 days |
| 5 digit ZIP | 247,724 | 13,786,172 | 3608.02 | (3570.91, 3645.14) | 18 days |
| 6 digit ZIP | 234,331 | 2,666,450 | 2807.21 | (2728.63, 2885.79) | 2 days |
| 7 digit ZIP | 144,230 | 436,049 | 2767.80 | (2543.67, 2991.93) | < 1 day |
Table 8.
Blocking sensitivity analysis causal treatment effect estimates
| 30 Day Mortality | 30 Day Inpatient Acute | |||
|---|---|---|---|---|
| Blocking Criteria | 95% CI | 95% CI | ||
| First 4 digits | 0.005 | (−0.067, 0.077) | 0.007 | (−0.168, 0.182) |
| First 5 digits | 0.008 | (−0.067, 0.083) | 0.010 | (−0.174, 0.194) |
| First 6 digits | 0.010 | (−0.074, 0.094) | 0.016 | (−0.180, 0.213) |
| First 7 digits | 0.007 | (−0.083, 0.096) | 0.009 | (−0.203, 0.222) |
Footnotes
SUPPLEMENTARY MATERIAL
Supplemental Code: A Multiple Imputation Procedure for Record Linkage and Causal Inference to Estimate the Effects of Home-delivered Meals
(). We provide supplemental code to demonstrate the implementation of the Bayesian Record Linkage algorithm, propensity score matching, imputation of the missing potential outcomes, and calculation of the two-stage multiple imputation treatment effects as proposed in this manuscript.
References.
- Abadie A and Imbens GW (2011). Bias-Corrected Matching Estimators for Average Treatment Effects. Journal of Business & Economic Statistics 29 1–11. [Google Scholar]
- Campbell KM, Deck D and Krupski A (2008). Record linkage software in the public domain: a comparison of Link Plus, The Link King, and a ‘basic’ deterministic algorithm. Health, Informatics Journal 14 5–15. [DOI] [PubMed] [Google Scholar]
- Campbell AD, Godfryd A, Buys DR and Locher JL (2015). Does Participation in Home-Delivered Meals Programs Improve Outcomes for Older Adults? Results of a Systematic Review. Journal of Nutrition in Gerontology and Geriatrics 34 124–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers R, Chipperfield J, Davis W and Kovacevic M (2009). Inference Based on Estimating Equations and Probability-Linked Data. Centre for Statistical and Survey Methodology, University of Wollongong Working Paper Series 18–09. [Google Scholar]
- Copas JB and Hilton FJ (1990). Record Linkage: Statistical Models for Matching Computer Records. Journal of the Royal Statistical Society. Series A (Statistics in Society) 153 287–320. [PubMed] [Google Scholar]
- Fellegi IP and Sunter AB (1969). A Theory for Record Linkage. Journal of the American Statistical Association 64 1183–1210. [Google Scholar]
- Fortini M, Liseo B and Nuccitelli a. (2001). On Bayesian Record Linkage. 4 185–198. [Google Scholar]
- Frangakis CE and Rubin DB (2002). Principal Stratification in Causal Inference. Biometrics 58 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frangakis CE, Rubin DB, An M-W and MacKenzie E (2007). Principal Stratification Designs to Estimate Input Data Missing Due to Death. Biometrics 63 641–649. [DOI] [PubMed] [Google Scholar]
- Gomatam S, Carter R, Ariet M and Mitchell G (2002). An empirical comparison of record linkage procedures. Statistics in Medicine 21 1485–1496. [DOI] [PubMed] [Google Scholar]
- Griffin J and Brown P (2017). Hierarchical Shrinkage Priors for Regression Models. Bayesian Anal. 12 135–159. [Google Scholar]
- Gutman R, Afendulis CC and Zaslavsky AM (2013). A Bayesian Procedure for File Linking to Analyze End-of-Life Medical Costs. Journal of the American Statistical Association 108 501 34–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman R and Rubin DB (2013). Robust estimation of causal effects of binary treatments in unconfounded studies with dichotomous outcomes. Statistics in Medicine 32 1795–1814. [DOI] [PubMed] [Google Scholar]
- Gutman R and Rubin DB (2015). Estimation of causal effects of binary treatments in unconfounded studies. Statistics in Medicine 34 3381–3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman R and Rubin D (2017). Estimation of causal effects of binary treatments in unconfounded studies with one continuous covariate. Statistical Methods in Medical Research 26 1199–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harron K, Goldstein H and Dibben C (2015). Methodological Developments in Data Linkage. Wiley Series in Probability and Statistics. Wiley. [Google Scholar]
- Herzog T, Scheuren F and Winkler W (2007). Data Quality and Record Linkage Techniques. Springer, New York. [Google Scholar]
- Hof MHP and Zwinderman AH (2012). Methods for analyzing data from probabilistic linkage strategies based on partially identifying variables. Statistics in Medicine 31 4231–4242. [DOI] [PubMed] [Google Scholar]
- Imbens GW and Rubin DB (2015). Causal Inference for Statistics, Social, and Biomedical Sciences: An Introduction. Cambridge University Press. [Google Scholar]
- Jaro MA (1989). Advances in Record-Linkage Methodology as Applied to Matching the 1985 Census of Tampa, Florida. Journal of the American Statistical Association 84 414–420. [Google Scholar]
- Lahiri P and Larsen MD (2005). Regression Analysis with Linked Data. Journal of the American Statistical Association 100 222–230. [Google Scholar]
- Larsen MD (2005). Hierarchical Bayesian Record Linkage Theory. [Google Scholar]
- Larsen MD and Rubin DB (2001). Iterative Automated Record Linkage Using Mixture Models. Journal of the American Statistical Association 96 32–41. [Google Scholar]
- Lee JS, Shannon J and Brown A (2015). Characteristics of Older Georgians Receiving Older Americans Act Nutrition Program Services and Other Home- and Community-Based Services: Findings from the Georgia Aging Information Management System (GA AIMS). Journal of Nutrition in Gerontology and Geriatrics 34 168–188. [DOI] [PubMed] [Google Scholar]
- Lloyd JL and Wellman NS (2015). Older Americans Act Nutrition Programs: A Community-Based Nutrition Program Helping Older Adults Remain at Home. Journal of Nutrition in Gerontology and Geriatrics 34 90–109. [DOI] [PubMed] [Google Scholar]
- Murray JS (2015). Probabilistic Record Linkage and Deduplication after Indexing, Blocking, and Filtering. Journal of Privacy and Confidentiality 7. [Google Scholar]
- Neter J, Maynes ES and Ramanathan R (1965). The Effect of Mismatching on the Measurement of Response Error. Journal of the American Statistical Association 60 1005–1027. [Google Scholar]
- Newcombe HB (1988). Handbook of record linkage: Methods for health and statistical studies, administration, and buisiness. Oxford University Press, Oxford. [Google Scholar]
- Newcombe HB, Kennedy JM, Axford SJ and James AP (1959). Automatic Linkage of Vital Records. Science 130 954–959. [DOI] [PubMed] [Google Scholar]
- Newman LM, Samuel MC, Stenger MR, Gerber TM, Macomber K, Stover JA and Wise W (2009). Practical Considerations for Matching STD and HIV Surveillance Data with Data from other Sources. Public Health, Reports 124 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyman J (1923). Sur les applications de la thar des probabilities aux experiences agaricales: Essay de principle. English translation of excerpts by Dabrowska, D. and Speed, T. (1990). Statistical Science 5 465–472. [Google Scholar]
- Rosenbaum PR and Rubin DB (1983). The Central Role of the Propensity Score in Observational Studies for Causal Effects. Biometrika 70 41–55. [Google Scholar]
- Rosenbaum PR and Rubin DB (1985). Constructing a Control Group Using Multivariate Matched Sampling Methods That Incorporate the Propensity Score. The American Statistician 39 33–38. [Google Scholar]
- Rubin DB (1973a). Matching to Remove Bias in Observational Studies. Biometrics 29 159–183. [Google Scholar]
- Rubin DB (1973b). The Use of Matched Sampling and Regression Adjustment to Remove Bias in Observational Studies. Biometrics 29 185–203. [Google Scholar]
- Rubin DB (1978). Bayesian Inference for Causal Effects: The Role of Randomization. The Annals of Statistics 6 34–58. [Google Scholar]
- Rubin DB (1979). Using Multivariate Matched Sampling and Regression Adjustment to Control Bias in Observational Studies. Journal of the American Statistical Association 74 318–328. [Google Scholar]
- Rubin DB (1980). Randomization Analysis of Experimental Data: The Fisher Randomization Test Comment. Journal of the American Statistical Association 75 591–593. [Google Scholar]
- Rubin DB (1990). Formal mode of statistical inference for causal effects. Journal of Statistical Planning and Inference 25 279 – 292. [Google Scholar]
- Rubin DB (1996). Multiple Imputation After 18+ Years. Journal of the American Statistical Association 91 473–489. [Google Scholar]
- Rubin DB (2003). Nested multiple imputation of NMES via partially incompatible MCMC. Statistica Neerlandica 57 3–18. [Google Scholar]
- Rubin DB (2006). Causal Inference Through Potential Outcomes and Principal Stratification: Application to Studies with “Censoring” Due to Death. Statist. Sci 21 299–309. [Google Scholar]
- Rubin DB (2007). Statistical Inference for Causal Effects, With Emphasis on Applications in Epidemiology and Medical Statistics. In Epidemiology and Medical Statistics, (Rao CR, Miller JP and Rao DC, eds.). Handbook of Statistics 27 28 – 63. Elsevier. [Google Scholar]
- Sadinle M (2017). Bayesian Estimation of Bipartite Matchings for Record Linkage. Journal of the American Statistical Association 112 600–612. [Google Scholar]
- Sadinle M and Fienberg SE (2013). A Generalized Fellegi–Sunter Framework for Multiple Record Linkage With Application to Homicide Record Systems. Journal of the American Statistical Association 108 385–397. [Google Scholar]
- Scheuren F and Winkler W (1993). Regression analysis of data files that are computer matched - Part I. 19. [Google Scholar]
- Scheuren F and Winkler W (1997). Regression analysis of data files that are computer matched - Part II. 23 157–165. [Google Scholar]
- Shen ZJ (2000). Nested Multiple Imputation, PhD thesis, Department of Statistics, Harvard University, Cambridge, MA. [Google Scholar]
- Steorts RC (2015). Entity Resolution with Empirically Motivated Priors. Bayesian Analysis 10 849–875. [Google Scholar]
- Steorts RC, Hall R and Fienberg SE (2016). A Bayesian Approach to Graphical Record Linkage and Deduplication. Journal of the American Statistical Association 111 1660–1672. [Google Scholar]
- Stuart EA (2010). Matching Methods for Causal Inference: A Review and a Look Forward. Statist. Sci 25 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tancredi A and Liseo B (2011). A Hierarchical Bayesian Approach to Record Linkage and Population Size Problems. The Annals of Applied Statistics 5 1553–1585. [Google Scholar]
- Tanner MA and Wong WH (1987). The Calculation of Posterior Distributions by Data Augmentation. Journal of the American Statistical Association 82 528–540. [Google Scholar]
- Stan Development Team (2018). RStan: the R interface to Stan. R package version 2.17.3 [Google Scholar]
- Thomas KS, Akobundu U and Dosa D (2015). More Than A Meal? A Randomized Control Trial Comparing the Effects of Home-Delivered Meals Programs on Participants’ Feelings of Loneliness. The Journals of Gerontology: Series B 71 1049–1058. [DOI] [PubMed] [Google Scholar]
- Thomas KS and Mor V (2013). Providing More Home-Delivered Meals Is One Way To Keep Older Adults With Low Care Needs Out Of Nursing Homes. Health Affairs 32 1796–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KS, Parikh RB, Zullo AR and Dosa D (2018a). Home-Delivered Meals and Risk of Self-Reported Falls: Results From a Randomized Trial. Journal of Applied Gerontology 37 41–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KS, Gadbois EA, Shield RR, Akobundu U, Morris AM and Dosa DM (2018b). “It’s Not Just a Simple Meal. It’s So Much More”: Interactions Between Meals on Wheels Clients and Drivers. Journal of Applied Gerontology 0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas KS, Parikh RB, Zullo AR and Dosa D (2018c). Home-Delivered Meals and Risk of Self-Reported Falls: Results From a Randomized Trial. Journal of Applied Gerontology 37 41–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler WE (1988). Using the EM Algorithm for Weight Computation in the FellegiSunter Model of Record Linkage. Proceedings of the Section on Survey Research Methods 667–671. [Google Scholar]
- Winkler W (1989). Near Automatic Weight Computation in the Fellegi-Sunter Model of Record Linkage. [Google Scholar]
- Winkler WE (1990). String Comparator Metrics and Enhanced Decision Rules in the Fellegi-Sunter Model of Record Linkage. Proceedings of the Section on Survey Research Methods 354–359. [Google Scholar]
- Winkler WE (1993). Improved decision rules in the fellegi-sunter model of record linkage. [Google Scholar]
- Wortman JH and Reiter JP (2018). Simultaneous record linkage and causal inference with propensity score subclassification. Statistics in Medicine 37 3533–3546. [DOI] [PubMed] [Google Scholar]
- Zanella G (2019). Informed Proposals for Local MCMC in Discrete Spaces. Journal of the American Statistical Association 0 1–27. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.